Abstract
Mesenchymal stem cells (MSCs) are promising for the regeneration of tendon and ligament tissues. Toward realizing this potential, microenvironment conditions are needed for promoting robust lineage-specific differentiation into tenocytes/ligament fibroblasts. Here, we utilized a statistical design of experiments approach to examine combinations of matrix modulus, composition, and soluble factors in human MSC tenogenic/ligamentogenic differentiation. Specifically, well-defined poly(ethylene glycol)-based hydrogels were synthesized using thiol–ene chemistry providing a bioinert base for probing cell response to extracellular matrix cues. Monomer concentrations were varied to achieve a range of matrix moduli (E ∼ 10 – 90 kPa), and different ratios of integrin-binding peptides were incorporated (GFOGER and RGDS for collagen and fibronectin, respectively), mimicking aspects of developing tendon/ligament tissue. A face-centered central composite response surface design was utilized to understand the contributions of these cues to human MSC differentiation in the presence of soluble factors identified to promote tenogenesis/ligamentogenesis (BMP-13 and ascorbic acid). Increasing modulus and collagen mimetic peptide content increased relevant gene expression and protein production or retention (scleraxis, collagen I, tenascin-C). These findings could inform the design of materials for tendon/ligament regeneration. More broadly, the design of experiments enabled efficient data acquisition and analysis, requiring fewer replicates than if each factor had been varied one at a time. This approach can be combined with other stimuli (e.g., mechanical stimulation) toward a better mechanistic understanding of differentiation down these challenging lineages.
Keywords: Hydrogel, cell microenvironment, mesenchymal stem cell, tendon, design of experiments
Introduction
Tendon and ligament injuries are some of the most common musculoskeletal injuries and comprise approximately 30-50% of all sports injuries.1 To restore function and maintain quality of life, these injuries often necessitate surgery, such as anterior cruciate ligament (ACL) reconstructions (60,000 to 175,000 annually in the United States).2 Unfortunately, high rates of donor site morbidity3 and osteoarthritis4 are associated with reconstruction procedures. These limitations have motivated the search for alternative therapies, such as tissue engineering and regenerative medicine.5,6 In particular, mesenchymal stem cells are a promising cell source for tendon and ligament tissue engineering: they produce relevant extracellular matrix proteins, such as collagens, rapidly proliferate, and survive for weeks in knee joints after surgical implementation.7 Reports of strategies to induce tenogenic/ligamentogenic (T/L) differentiation in vitro and in vivo are growing.8 Methods to induce tenogenic differentiation include mechanical stimulation,9–14 scaffold anisotropy,15–17 co-culture with tendon/ligament fibroblasts,18–21 and growth factor supplementation.22–26 However, the effects of insoluble extracellular cues, such as matrix modulus and the composition of extracellular matrix (ECM) proteins, on human mesenchymal stem cell (hMSC) differentiation into tenocytes or ligament fibroblasts have been the subject of far less investigation. Cues provided by ECM modulus and protein composition, individually and in combination with soluble factors, have been shown to influence more well-studied differentiation lineages, such as osteogenesis,27 chondrogenesis,28 or adipogenesis.29 An improved understanding of the effects of ECM-based cues on tenogenesis/ligamentogenesis would inform the design of relevant tissue engineering scaffolds to regulate hMSC function and fate toward improved clinical outcomes.
Some progress has been made over the past few years in identifying relevant ECM cues for T/L differentiation of hMSCs. Note, tenocytes and ligament fibroblasts are essentially indistinguishable in vitro, and hMSC differentiation down this lineage will be described as tenogenic differentiation for consistency herein. Using collagen-coated polyacrylamide substrates, Sharma and Snedeker established that a modulus on the order of 30 – 50 kPa promoted tenogenic differentiation of hMSCs.30,31 These studies reinforced the importance of matrix modulus for hMSC differentiation and began to define the phase space of insoluble ECM cues for promoting tenogenic differentiation. While informative, naturally-derived collagen may introduce batch-to-batch variability and inherent signals to the cells by presenting a number of biochemical and biophysical cues (e.g., integrin-binding sequences and fibrillar structure).32,33 Synthetic bioinspired materials allow for decoupling the effects of these extracellular cues and could provide additional insight into the differentiation mechanism. Furthermore, soluble factors play a key role in regulating stem cell fate. For example, using a high-throughput screening approach to study more than 1000 different microenvironments, Lutolf and coworkers demonstrated that soluble factors, matrix modulus, and extracellular matrix composition direct embryonic stem cell self-renewal and proliferation; notably, soluble factors accounted for approximately 60% of the variability between conditions, and soluble factors and matrix modulus showed synergistic effects at promoting both self-renewal and proliferation.34 Investigations of the effects of modulus and integrin-binding in the context of soluble factors are needed to elucidate synergies and establish design principles for new materials to deliver and differentiate hMSCs in tendon and ligament regeneration.
Here, we used a bioinert poly(ethylene glycol) (PEG)-based material as a ‘blank-slate’ for the systematic presentation of ECM cues to probe how matrix modulus and integrin-binding peptides influence tenogenesis of hMSCs in the presence of tendon- and ligament-inducing soluble factors. PEG-based materials were selected because they are inherently bioinert and cytocompatible for both two- and three-dimensional cell culture while affording straightforward tuning of mechanical properties and incorporation of bioactive groups.35 The soluble factors selected for this study were ascorbic acid (AA) and the growth factor bone morphogenetic protein-13 (BMP-13; also known as GDF-6). BMP-13 plays an integral role in ligament development, as demonstrated by BMP-13 knockouts that show defective ligament formation,36 and Wei et al. observed that BMP-13 enhances the tenogenic differentiation of rat MSCs.24 Ascorbic acid was added due to its known role in promoting the synthesis of collagen, the predominant ECM protein in tendon and ligament tissues.
A statistical design of experiments (DOE) approach was applied to simultaneously elucidate the effects of individual and combinations of extracellular cues (Figure 1). The response surface design approach allows for concurrent determination of the significance of individual input variables and interactions between those variables while taking into account the possibility of non-linear responses. Here, the input variables are matrix modulus and the concentration of integrin-binding peptides, and the response variables are the expression (i.e., ΔΔCT)37 of the tendon-associated genes (i.e., scleraxis, collagen I, and tenascin-C), markers of hMSC tenogenic differentiation.38
Figure 1.
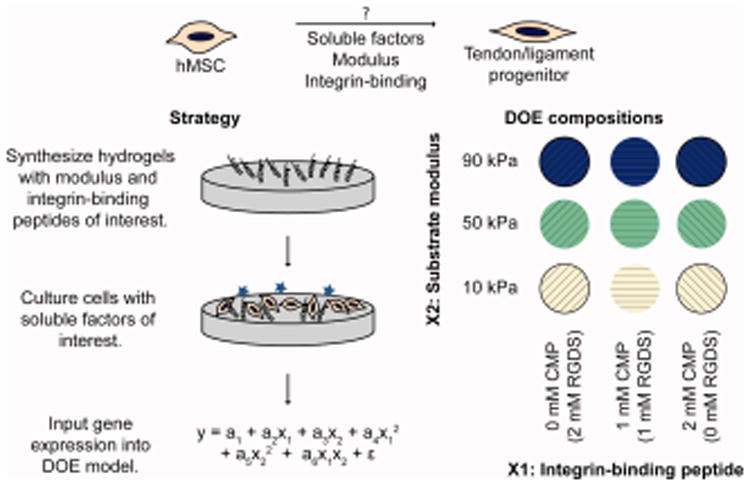
Approach. The purpose of this study was to test the hypothesis that a combination of soluble factors, modulus, and integrin-binding moieties could induce hMSC differentiation into tenocytes/ligament fibroblasts. To study the effects of modulus and integrin-binding peptides on differentiation, well-defined PEG-based hydrogels were synthesized to mimic aspects of developing tendon/ligament, seeded with cells, and cultured in the presence of tendon-inducing soluble factors. A design of experiments approach was applied to simultaneously determine the individual and synergistic effects of the two input factors, a CMP integrin-binding peptide (×1) and substrate modulus (×2). The ΔΔCt values from gene expression measurements were fit as responses in a DOE model, as tenogenic gene expression is generally considered one of the most conclusive markers of tenogenic differentiation. Coefficients a1, a2, a3, a4, a5, a6, and ε were determined in Minitab 17.
For the input variables, we hypothesized that a triple-helical GFOGER sequence, which activates integrins α1β1, α2β1, α10β1, and α11β1 associated with collagen binding,39 would promote hMSC tenogenic differentiation, since both mature and developing tendon are highly collagenous.40 We compared the effect of the GFOGER sequence to a sequence based on the adhesion ligand RGDS found in many ECM proteins, including fibronectin and laminin,41 which are present during ligament development.42 The reported modulus of developing embryonic tendon varies over several orders of magnitude, depending on the method used to take the measurement;43 amongst these, indentation methods may be most relevant for cell sensing, as both the measurement and cell-generated forces are on the nano- to micro-scale. Consequently, we hypothesized that moduli in the kPa range, recently reported for developing tendon by indentation measurements,44 may be appropriate for promoting stem cell tenogenesis. For this study, the modulus range was selected so that the center point was E = 50 kPa, which was previously shown to promote MSC tenogenic differentiation.30 The low modulus was selected to be E = 10 kPa, the modulus of striated muscle,45 as muscles and tendons develop in close spatiotemporal association.46 The high modulus of E = 90 kPa was selected to balance the design of experiments and approach that of mature ligament tissue. This study design contributes to the understanding of microenvironment conditions for hMSC tenogenic differentiation, which has historically been difficult to achieve in vitro. Further, the approach of using a response surface design with tunable synthetic matrices to understand key interactions can be applied in the future in conjunction with other lineage-specific triggers, such as mechanical stimulation or co-culture,8 toward improved strategies for tenogenic differentiation.
Materials and Methods
Materials
Unless otherwise noted, reagents were obtained from the following vendors: Chemical reagents were obtained from Sigma-Aldrich. Cell culture reagents were obtained from Life Technologies. Amino acids were obtained from Chem-Impex International. Solvents were obtained from Fisher Scientific.
MSC Isolation, Culture, and Differentiation
Human MSCs were isolated from bone marrow (Lonza; 26 year old female donor) following a protocol described by Anderson et al.47 Briefly, red blood cells first were lysed using ammonium chloride (STEMCELL Technologies) following the manufacturer's protocol. After red blood cell lysis, hMSCs were grown for two weeks on tissue culture treated polystyrene, where the hMSCs remained as the adherent cell population. The identity of the hMSCs48 was confirmed by i) expression of CD73 and CD105 by flow cytometry, ii) the lack of expression of CD45, CD14, and CD19, and iii) verification of their ability to differentiate down adipogenic, osteogenic, and chondrogenic lineages as confirmed by staining with Oil Red O, Alizarin Red, and Safranin O, respectively. hMSCs were expanded in low glucose Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum, 50 U/mL penicillin, 50 μg/mL streptomycin, 0.2% Fungizone, and 1 ng/mL basic fibroblast growth factor (bFGF; Peprotech). Cells were fed every 2-3 days, passaged at approximately 80% confluency, and used 2 passages after isolation.
For differentiation assays, cells were seeded at 20,000 cells/cm2, allowed to adhere for 24 hours in growth medium (high glucose DMEM with 10% FBS, 50 U/mL penicillin, 50 μg/mL streptomycin, and 0.2% Fungizone), and then serum starved for 24 hours (high glucose DMEM with 1% FBS, 50 U/mL penicillin, 50 μg/mL streptomycin, and 0.2% Fungizone). The medium then was changed to differentiation medium (growth medium supplemented with 50 μg/mL 2-phospho-L-ascorbic acid [Sigma-Aldrich] and 10 ng/mL BMP-13 [Peprotech]). The timeline for the differentiation experiments is summarized in Figure S1.
Synthesis and characterization of monomers and reagents
PEG-tetranorbornene (PEG-4Nb; Mn ∼ 10 kDa) was synthesized based on a modified version of a published protocol.49 PEG-4NH2HCl (10 g, Jenkem USA), 4-dimethylaminopyridine (DMAP; 0.5× molar excess relative to amine groups on PEG), and pyridine (5× molar excess) were dissolved in dichloromethane (DCM; ∼ 40 mL), and the solution was purged with argon for 15 minutes. In a separate flask, N,N'-diisopropylcarbodiimide (5× molar excess) and 5-norbornene-2-carboxylic acid (10× molar excess) were dissolved in DCM (∼ 40 mL) and purged with argon for 15 minutes. After both argon purges were complete, the contents of the two flasks were combined and allowed to react overnight under argon. The next day, the solution was concentrated by rotary evaporation and precipitated in cold diethyl ether (4°C; 9× volume ether relative to DCM). The ether/PEG mixture was stored overnight at -20°C to promote PEG precipitation. The ether/PEG mixture subsequently was centrifuged at 3000 × g for 20 minutes at 4°C. Following centrifugation, the diethyl ether supernatant was discarded, and the PEG precipitate was washed with cold diethyl ether, centrifuged, and dried in a desiccator overnight. Finally, the PEG product was purified by dialysis (MWCO 2000 g/mol, Spectrum Laboratories) against deionized water for 48 hours. Product purity was confirmed by 1H-NMR in DMSO-d6: 400 mHz δ 6.20-5.86 (m, 2H), δ 3.65-3.40 (m, 227H) and disappearance of the peak at δ 3.10-2.90 (t, 2H) (Figure S2).
The peptides CGKGWGKGCG (peptide crosslinker), CGRGDS (RGDS), C(POG)4FOGERG(POG)4G (collagen mimetic peptide [CMP]), and CGRGSD (scrambled peptide [RGSD]) were synthesized using standard Fmoc-chemistry on a peptide synthesizer (Tribute, Protein Technologies, Inc., Tucson, AZ). All amino acids were double-coupled. The CMP was built on H-Rink Amide-Chemmatrix resin (PCAS Biomatrix), and all other peptides were built on Rink Amide MBHA resin (Novabiochem). Peptides were cleaved for 2 hours in 95% trifluoroacetic acid (Acros Organics), 2.5% water, and 2.5% triisopropylsilane (Acros Organics) (all percentages v/v) supplemented with 25 mg/mL phenol and 50 mg/mL dithiothreitol (Research Products International). After cleavage, the peptides were precipitated in cold diethyl ether (9× volume) overnight and purified by reverse-phase high pressure liquid chromatography (HPLC; XBridge BEH C18 OBD 5μm column; Waters, Milford, MA) with a linear water-acetonitrile (ACN) gradient (Water:ACN 95:5 to 45:55; 1.17% change in water per minute). Peptide molecular weight was verified using electrospray ionization mass spectrometry (ESI-MS; LCQ Advantage, Thermo Scientific, Waltham, MA) (Figure S3 – S5). Triple helical confirmation of the CMP peptide at 37°C was verified using circular dichroism (Jasco 810 spectrometer, Jasco, Easton, MD) (Figure S6).
The photoinitiator, lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), was synthesized following a previously published protocol.50 Briefly, stoichiometric amounts of dimethyl phenylphosphonite and 2,4,6-trimethylbenzoyl chloride were mixed, purged with argon, and allowed to react overnight. The next day, lithium bromide (4× molar excess) was dissolved in 2-butanone (∼ 100 mL) and added dropwise by syringe to the reaction mixture; the reaction subsequently was heated to 50°C for 10 minutes, when a solid precipitate formed. The resulting mixture was left at room temperature for four hours and then filtered. The final powder product was dried in a desiccator. Product purity was confirmed by 1H-NMR in D2O: 400 mHz δ 7.59 (m, 2H), 7.44 (m, 1H), 7.36 (m, 2H), 6.78 (s, 2H), 2.12 (s, 3H), and 1.90 (s, 6H) (Figure S7).
Hydrogel synthesis and characterization
Monomer stock solutions were prepared by dissolving each in Dulbecco's phosphate buffered saline (PBS): i) PEG-4Nb (37.5 mM PEG-4Nb for 150 mM Nb functional group), ii) the peptide crosslinker (typically 300 – 400 mM SH), iii) LAP (5 – 10 mM), and iv) RGDS, CMP, and RGSD (each typically 75 – 150 mM). Stock solutions were stored in aliquots at -80°C until use. The free-thiol functionality of the peptides, which varies for each synthesis owing to some amount of disulfide formation during handling steps, was determined for each stock solution using Ellman's assay.
Hydrogel precursor solutions were prepared by diluting stock solutions to final concentrations of 4.5 – 18.5 mM PEG-4Nb (18 – 74 mM Nb functional groups), 8 – 36 mM peptide crosslinker (16 – 72 mM SH functional groups), 0.5 mM LAP, and 2 mM RGDS, CMP, or RGSD in PBS with 50 U/mL penicillin, 50 μg/mL streptomycin, and 0.2% Fungizone. Total thiol:norbornene stoichiometry was maintained at 1:1 throughout all of the cell experiments.
Rheometric measurements were conducted on a rheometer with UV-visible light attachment (AR-G2, TA Instruments) for irradiation with an Omnicure Series 2000 light source (Excilitas, Waltham, MA) with light guide and 365 nm filter (Exfo). Hydrogel crosslinking and gelation was monitored by measuring the shear modulus, a measure of the crosslink density and the degree of polymerization of these hydrogels.51 All of the compositions reached > 95% of their final moduli within 3 minutes of irradiation (5 mW/cm2 at 365 nm). Consequently, for modulus measurements, hydrogels were formed on the rheometer by irradiating precursor solutions for 3 minutes at 5 mW/cm2. All rheometric measurements were taken with 2 mM excess norbornene to mimic the addition of 2 mM of peptide-based biological cues (e.g., RGDS, CMP).
The initial shear modulus (Go) was measured after irradiation at a strain of 1% and a frequency of 2 Hz, which was in the linear viscoelastic regime for these hydrogels. The final equilibrium swollen modulus (Gfinal) was estimated from the measured modulus and equilibrium swelling:52
| (Eq. 1) |
where Tfinal was 310 K (incubator temperature); To was 298 K (room temperature); Qfinal was the equilibrium volumetric swelling ratio, measured using calipers (see Supplemental Materials and Methods); and Qo is the initial volumetric swelling ratio of the hydrogels. The elastic modulus was estimated by rubber elasticity theory:
| (Eq. 2) |
where ν is Poisson's ratio, taken to be 0.5 for these PEG hydrogels.52 The shear modulus, swelling ratio, and elastic modulus for each composition are listed in Table S1.
Hydrogels used for cell culture experiments were photopolymerized (5 mW/cm2 at 365 nm for 3 minutes, Omnicure S2000), washed with PBS, and sterilized with germicidal UV (254 nm) for 20 minutes before use. For qRT-PCR experiments, hydrogels (diameter ∼ 20 mm) were attached to 22 mm cover slips (No. 2 thickness) during gel formation for ease of handling. For these samples, the monomer solution was placed in a cylindrical mold: a 0.01” rubber gasket (McMaster-Carr) between a coverslip and PDMS (Figure S8). Cylindrical PDMS slabs (approximately 3.5 cm in diameter and 1.8 cm in height) were formed using Sylgard 184 (Dow Corning, Midland, MI) following the manufacturer's instructions. Molds containing monomer solution were placed under the light source for polymerization to form a gel. After formation, the gels adhered to the glass cover slips without additional modification and remained attached for at least 15 days during the cell culture experiments. Hydrogels for swelling and imaging experiments were formed in a 1-mL syringe with the tip cut off (25 μL precursor solution per mold; diameter ∼ 5 mm).
Cell adhesion
Cells were seeded on hydrogels at 20,000 cells/cm2, allowed to adhere for 24 hours in growth medium, and then were serum starved for 24 hours. After serum starvation, cell/gel constructs were washed twice with PBS, incubated with 700 nM 4′,6-diamidino-2-phenylindole (DAPI) for 10 minutes to label cell nuclei, and then washed three more times in PBS. Constructs were transferred to fresh 48-well plates, imaged on an Axio Observer Z1 microscope (Zeiss, Germany), and analyzed on ImageJ (National Institutes of Health, Bethesda, MD).
RNA Isolation and qRT-PCR
To examine tenogenesis, cells were seeded and cultured as described for differentiation assays above. RNA was isolated with Trizol (Life Technologies) after 15 days in differentiation medium. For plated cells, RNA was isolated following the manufacturer's protocol. For each hydrogel replicate, two 22 mm gels of the same condition were scraped into 950 μl Trizol using a razor blade and homogenized with a pellet mixer. Subsequently, the RNA was isolated according to the Trizol protocol; the gel separated from the RNA during the first centrifugation step. In both cases, the RNA was further purified using a DNase treatment and phenol-chloroform extractions. Briefly, the RNA pellet was redissolved in water and treated with amplification-grade DNase (Life Technologies) according to the manufacturer's instructions. The RNA was then extracted twice with phenol-chloroform-isoamyl alcohol and then chloroform. RNA in the extracted solution was precipitated in 50% isopropanol with 0.3 M sodium acetate and 15 μg/ml GlycoBlue (Life Technologies) in DNase-free water. The pellet was washed with 75% ethanol, redissolved in water, precipitated again in 50% isopropanol with 0.3 M sodium acetate and 15 μg/ml GlycoBlue, and then washed again with 75% ethanol. Finally, the pellet was redissolved in Tris-EDTA, pH 8.0. RNA content and purity were quantified on a UV-vis spectrophotometer (Nanodrop; Thermo Scientific, Waltham, MA); the A260/A280 value was verified to be equal to or above 1.8 for each sample.
After RNA isolation, cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. Quantitative real-time PCR (qRT-PCR) was run using a SYBR-Green master mix according to the manufacturer's instructions. Primers for each gene are listed in Table S2. The ΔΔCt method37 was used to evaluate gene expression with 18S rRNA as the housekeeping gene. Gene expression was normalized to hMSC gene expression cultured on plates (TCPS) in growth medium. Primer efficiencies were verified to be between 0.9 and 1.1 for each primer pair. PCR product integrity was verified by measuring melt curves of amplified products. cDNA synthesis and qRT-PCR were run on a CFX96 detection system (Bio-Rad, Hercules, CA). These analyses of tenogenic differentiation were performed at the University of Delaware.
To examine expression of an array of genes associated with other differentiation lineages, hMSCs were removed from gel substrates by treatment with 0.05% trypsin-EDTA for 9 minutes at 37°C, centrifuged for 5 minutes at 100 × g, washed with PBS, centrifuged again for 5 minutes at 100 × g, resuspended in RNAlater (Life Technologies), and stored at -80°C until use. RNA was extracted using an RNeasy plus mini kit (Qiagen). RNA quantification was performed using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA). RNA integrity was assessed by Agilent 2200 Tape Station (Agilent, Santa Clara, CA); all samples had RIN values of 9.0 or higher. Reverse transcription of total RNA was performed using an iScript cDNA synthesis kit (Bio-Rad). qRT-PCR was set up in customized Prime PCR plates using SYBR Green qPCR Super Mix (Bio-Rad), and plates were run on a CFX96 Real-Time PCR system (Bio-Rad). Three reference genes (GAPDH, TBP, HPRT1) were used for normalization. This analysis of differentiation to other lineages was performed at the University of California, Davis.
Immunocytochemistry and protein analysis
Protein content was analyzed semi-quantitatively by measuring fluorescence intensities after immunocytochemical staining using a protocol adapted from Tong et al.53 After 15 days of culture in differentiation medium, cell/gel constructs were washed with PBS, fixed by incubation overnight in 4% paraformaldehyde (Alfa Aesar) at 4°C, and stored in PBS at 4°C until use (less than 1 week after fixation). To stain the constructs, samples were permeabilized for 10 minutes in PBS + 0.1% Triton-X (Fisher Scientific), washed with PBS, blocked for 2 hours in PBS + 50 mg/mL bovine serum albumin (Sigma-Aldrich), washed again with PBS, and incubated overnight at 4°C with primary antibody (rabbit polyclonal to collagen I [Abcam ab34710] and mouse monoclonal to tenascin-C [Abcam ab6393] diluted 1:200 in PBS + 15 mg/mL BSA). The following day, samples were washed with PBS + 0.05% Tween-20, incubated for 1 hour in secondary antibody (Alexa Fluor 647 goat anti-rabbit [Life Technologies A21244] and Alexa Fluor 488 goat anti-mouse [Life Technologies A11001] diluted 1:500 in PBS + 15 mg/mL BSA), and stored overnight at 4°C in PBS. Finally, the samples were incubated for 10 minutes in 700 nM DAPI in PBS, washed with PBS, and imaged the same day. All incubations took place with gentle rocking, and all steps took place at room temperature unless otherwise specified.
Samples were imaged using an LSM META 510 confocal microscope (Zeiss, Germany) using small z-stacks (∼ 100 μm thick). The laser power and master gain for the green and deep red fluorescent channels were set to ensure no image saturation and were kept constant for the acquisition of all images. LSM files were saved, stacked using a maximum intensity projection, and analyzed on ImageJ, where the overall fluorescence intensity for each channel was determined using the raw integrated density measurement. Nuclei were counted using the same procedure as in the cell adhesion tests, and the final fluorescence values for each image were reported as the raw integrated density divided by the number of cell nuclei. Each replicate consisted of the average value from 1 - 3 z-stacks taken on a single gel.
Statistical analysis
Results are reported as mean ± standard error. Specific numbers of replicates (n ≥ 3) are noted for each experiment within the Results and Discussion. Statistical design and analysis was conducted using Minitab 17 (Minitab Inc., State College, PA).
A DOE approach was used to assess and model hMSC tenogenic gene expression in response to specific microenvironment conditions:54 modulus (E ∼ 10 – 90 kPa) or integrin-binding peptides (0 – 2 mM CMP). A face-centered response surface design model was applied, as we hypothesized that the relationships between modulus, integrin-binding peptide concentration, and gene expression would be non-linear.54 All statistical analysis of gene expression was performed on ΔΔCt, as ΔΔCt values are often normally distributed.55 Coded coefficients were used so that the effects of each factor could be compared: the high value of each factor was +1; the low value of each factor was -1; and the intermediate value was 0 in the model fit. The postulated model for each response, y, was:
| (Eq. 3) |
where a1 represents the grand average; a2, a3, a4, and a5 are coefficients that are related to the individual effects of each factor (CMP concentration [x1] and modulus [x2]); a6 accounts for the synergistic effects of modulus and CMP concentration; and ε accounts for any variability not captured by the model. For the DOE model, the studentized deleted residual test was applied to detect outliers. A data point was excluded from the model if the studentized deleted residual was greater than the t statistic at α = 0.01;56 however, a potential outlier was retained if its removal would bring the replicate number to n < 3 for any particular condition.
Results
hMSC gene expression in response to tendon-inducing soluble factors
hMSCs first were cultured in the presence of AA and BMP-13 on TCPS to verify that the soluble factors selected for this study promoted hMSC tenogenic gene expression. Expression of the tendon-associated genes scleraxis, collagen I, and tenascin-C were normalized to the expression of the housekeeping gene 18S rRNA and compared to the growth condition (i.e., no AA or BMP-13) (Figure 2). Supplementation with AA and BMP-13 caused an apparent upregulation of scleraxis, a tendon- and ligament-specific transcription factor, although the upregulation was not statistically significant (p = 0.10).57 Supplementation with AA alone or with AA and BMP-13 led to upregulation of collagen I (p < 0.01) and tenascin-C (p < 0.05) gene expression. Collagen I is the predominant ECM protein associated with tendons and ligaments, although it is present in other tissues throughout the body including bone. Tenascin is primarily localized to tendinous junctions during development,58 and, in particular, tenascin-C upregulation is typically considered a marker of tenogenic differentiation.59 Taken together, these results supported the use of AA and BMP-13 to promote modest hMSC tenogenic gene expression.
Figure 2.
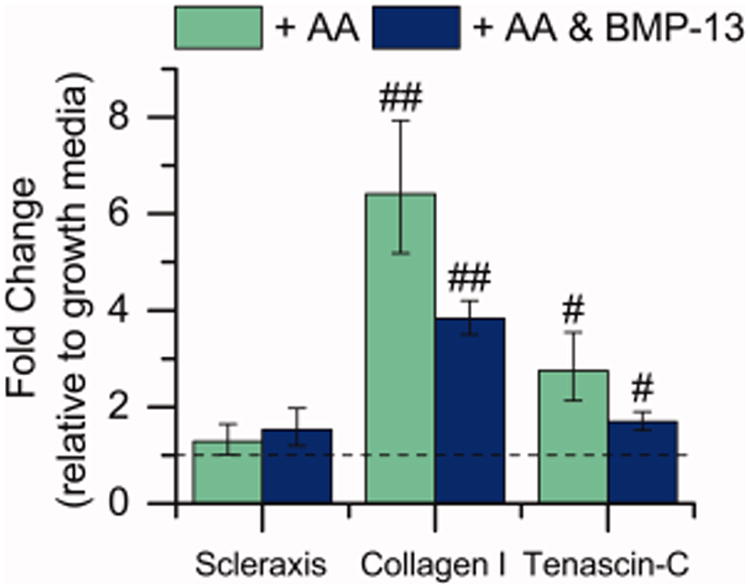
Effect of soluble factors on expression of tendon-associated genes. The gene expression of hMSCs cultured on TCPS cultured with AA (50 μg/mL) and BMP-13 (10 ng/mL) was evaluated, in part to determine a baseline level of expression in the presence of soluble factors for comparison to effects in the presence of matrix mimics. Gene expression was measured after 15 days of culture. Significant upregulation of the tendon-associated genes collagen I and tenascin-C were observed [n = 10 for the growth samples, n = 13 for AA and AA and BMP-13 samples, #p < 0.05, ##p < 0.01 compared with growth medium].
Synthesis and characterization of well-defined synthetic matrix mimics
To characterize the response of hMSCs to insoluble extracellular cues, PEG-peptide hydrogels were synthesized as a blank slate material into which cues of interest (e.g., matrix modulus and biochemical content) could be engineered. Thiol–norbornene chemistry was used to form the hydrogels owing to its cytocompatibility, light-initiated mechanism for rapid and facile gel formation and modification, and translatability for 3-D cell culture and delivery in future studies. PEG-4Nb was reacted with a dithiol peptide crosslinker and monothiol peptide tethers with biological activity of interest (Figure 3A) by radically-mediated step growth polymerization using the photoinitiator LAP.
Figure 3.
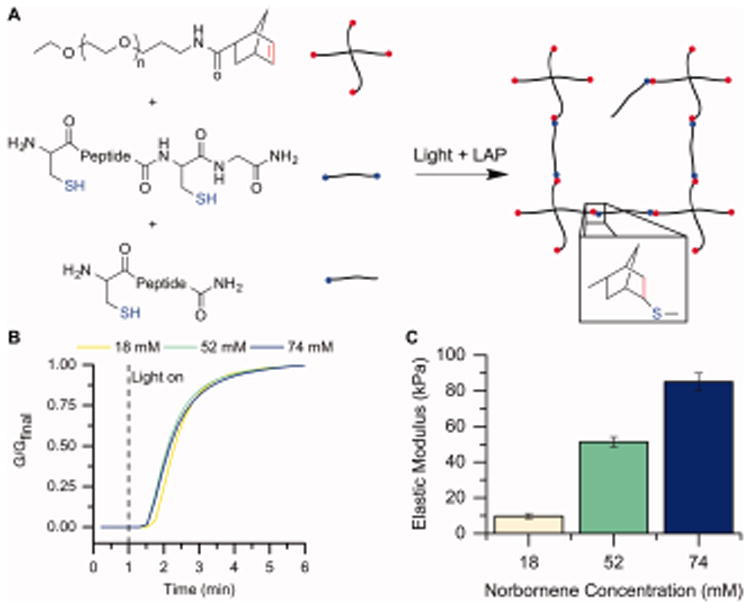
Hydrogel formation and characterization. A) Hydrogels were formed by reaction of PEG-4Nb (chemical structure of one arm shown here), a dithiol crosslinking peptide (CGKGWGKGCG), and monofunctional bioactive or scrambled tethers. Upon addition of a photoinitiator (LAP) and light (365 nm), these macromers react by a thiol–ene reaction to form a water-stable bond and a hydrogel. B) Gelation was measured for the compositions of interest by forming the gels in situ on a rheometer. The change in modulus, as a measure of crosslink density, was monitored over time upon light exposure (5 mW/cm2 at 365 nm) to assess the progress of gelation. Here, the modulus is normalized to the final modulus. Each condition was observed to reach 95% of its final modulus value by 3 min after polymerization commenced (that is, 4 min after the run started), and consequently, 3 min of irradiation was chosen as the gelation time for subsequent experiments. Representative curves are shown for each condition. C) The Young's modulus (E) was determined by rheometry and rubber elasticity theory. As the concentration of PEG-norbornene increases (noted here by the total norbornene concentration), the modulus of the gel also increases. Conditions were identified to achieve moduli of E = 10 kPa, 50 kPa, and 90 kPa, as desired for the design of experiments model [n = 6].
We established an appropriate time for gelation by measuring the change in modulus over time during in situ photopolymerization on a rheometer (Figure 3B). In each case, the precursor solution sample was pipetted onto the rheometer, and solution measurements were collected for 1 minute before irradiation was commenced. All of the compositions reached ∼ 95% of their final modulus within 3 minutes of irradiation. Consequently, 3 minutes was selected as the time for polymerization for all studies.
We then identified compositions leading to the desired modulus range for examining tenogenic differentiation (E = 10 – 90 kPa). The Young's modulus (elastic modulus, E) was calculated using rubber elasticity theory and the measured shear modulus and swelling values (Table S1), as described in the Materials and Methods section. As expected,60 the elastic modulus of these step growth PEG-based hydrogels increased with increasing concentration of monomer in the precursor solution (Figure 3C). Appropriate moduli for the compositions in this study were obtained at 18 mM Nb functional group (4.5 mM PEG; E = 9.69 ± 1.43 kPa; referred to as 10 kPa throughout the text), 52 mM Nb (13 mM PEG; E = 51.38 ± 2.80 kPa; referred to as 50 kPa), and 74 mM Nb (18.5 mM PEG; E = 85.10 ± 4.81 kPa; referred to as 90 kPa).
hMSC seeding density on hydrogels of different compositions
We sought to determine whether changing hydrogel properties would change the ability of hMSCs to attach and adhere to the gels. hMSCs were seeded on PEG hydrogels at 20,000 cells/cm2, allowed to adhere in growth media for 24 hours, and then serum starved for 24 hours, mimicking the beginning of the differentiation protocol. While increasing modulus led to an apparent decrease in initial cell density, the effect was minor and the differences were not statistically significant (p = 0.20; Figure 4A). No differences in adhesion were observed when the peptide tether incorporated into the gel was RGDS or CMP, but the adhesion dropped sharply when the scrambled RGSD sequence was incorporated into the gel (Figure 4B; p < 0.05). Thus, to keep initial cell density on the surface constant, 2 mM total of bioactive peptides (RGDS and CMP in different ratios) were incorporated into the gel. Note, while initial seeding density was not significantly affected by matrix properties, at least in the ranges considered here, some differences in cell density were observed by day 15, potentially due to differentiation altering hMSC proliferation rates or differences in the ability of the cells to adhere and spread to specific synthetic matrices (Figure S9). Further studies could deconvolute these effects and correlate cell density and morphology with differentiation on these synthetic matrices.
Figure 4.
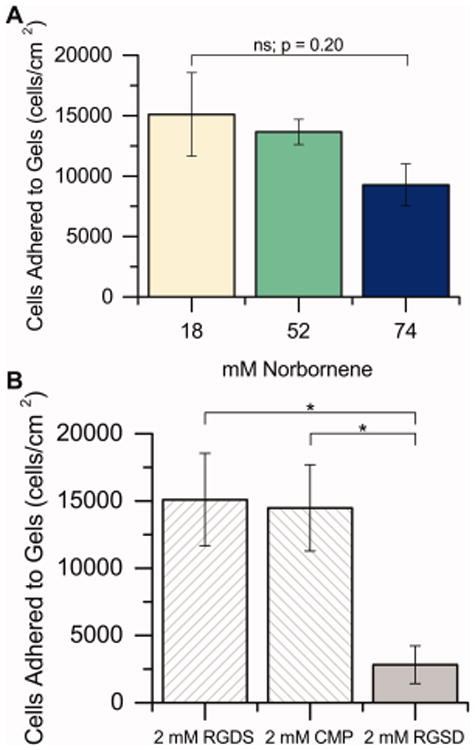
hMSC attachment to substrates. A) hMSCs were seeded on hydrogels with different PEG-norbornene concentrations containing 2 mM RGDS, allowed to adhere for 24 h in growth media (10% FBS), and then starved for 24 h (1% FBS). After starvation, cells were counted to establish the initial seeding density (that is, the number of hMSCs on each hydrogel surface at day 0 of the differentiation study). The initial cell seeding density was statistically equal for all concentrations of PEG-norbornene tested in this study [n = 3 for 18 mM and 52 mM; n = 4 for 74 mM]. B) Similarly, the effect of varying the identity of the peptide tether incorporated into the hydrogels was assessed on 18 mM Nb substrates. No differences in initial cell seeding density were observed between the two bioactive sequences (RGDS and CMP), whereas the scrambled RGSD permitted significantly less hMSC adhesion to the hydrogel at day 0, suggesting some specificity in cell interactions with these synthetic ECMs [n = 3, * p < 0.05 by two-sample t test].
hMSC tenogenic gene expression in response to insoluble microenvironment cues
A DOE approach was applied to investigate hMSC tenogenic gene expression in response to matrix modulus and integrin-binding peptides. Since changing the total concentration of bioactive peptide affected the initial cell density on the surface (Figure 4B), experiments were designed to vary the ratio of CMP to RGDS rather than the total amount of integrin-binding peptide: consequently, the three levels of integrin-binding peptide were i) 2 mM RGDS and 0 mM CMP, ii) 1 mM RGDS and 1 mM CMP, and iii) 0 mM RGDS and 2 mM CMP, referred to by the CMP concentration throughout the article. The coefficients determined by fitting the DOE equation (Eq. 3) to the gene expression measurements, ΔΔCt = ΔCttreated - ΔCtgrowth, are presented in Figure 5. Since the gene expression measurements were fitted as ΔΔCt, rather than fold change, a negative coefficient indicates a positive increase in gene expression; for example, the negative coefficient for scleraxis and modulus indicates that ΔΔCt decreases and fold change increases with increasing modulus. The model was simplified to consider only the statistically significant input parameters (p < 0.05), and the results of the simplifications are also plotted in Figure 5. Fisher's LSD post-hoc test was used to determine differences between the levels after simplification.
Figure 5.
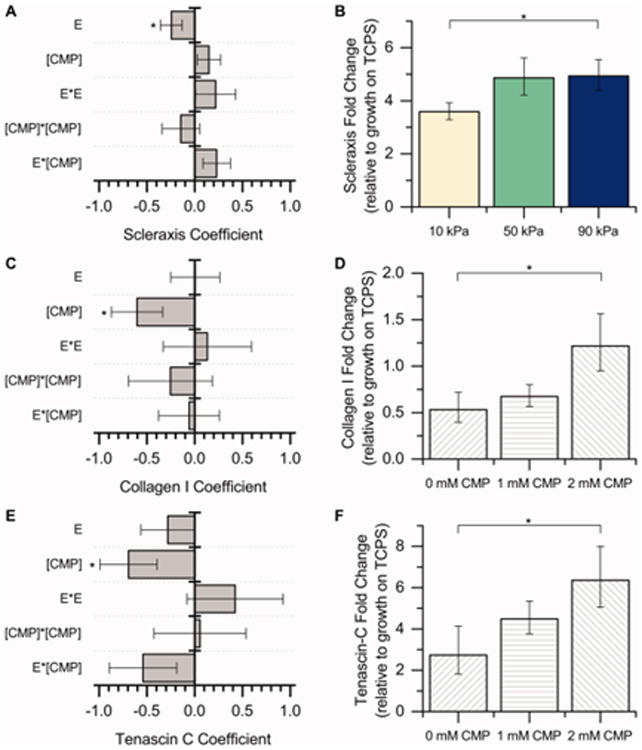
DOE analysis of microenvironment cues on tenogenic differentiation. A response surface methodology was applied to study the effect of modulus (E) and integrin-binding peptides ([CMP]) on hMSC tenogenic gene expression (scleraxis, collagen I, tenascin-C) after culture for 15 days in the presence of soluble factors (BMP-13 and AA). The coefficients for the DOE model [a2–a6 in Eq. (3)] are plotted here (left). The response variable into the model was ΔΔCt, so a negative coefficient value corresponds to an increase in expression. [*p < 0.05 indicates significance of the marked term by ANOVA.] A) Scleraxis gene expression is significantly impacted by hydrogel modulus, while C) collagen I and E) tenascin-C gene expression are significantly impacted by the presence of the CMP integrin-binding peptide. Conditions subsequently were binned to further understand the effects of increasing modulus or CMP alone (right). B) Since the only significant term for the scleraxis coefficient model was modulus, the overall fold change data are presented here for all integrin-binding peptides as a function of modulus. Increasing the hydrogel modulus from 10 kPa to 90 kPa led to significant upregulation of scleraxis in hMSCs. D, F) Since the only significant term for the collagen I and tenascin-C coefficient models was CMP, the overall fold change data are presented here for all moduli as a function of CMP concentration. Increasing the CMP concentration from 0 mM to 2 mM led to significant upregulation of collagen I and tenascin-C in hMSCs. These results suggest that both increasing modulus and increasing CMP concentration increase hMSC tenogenic gene expression, an indicator of tenogenic differentiation [n = 7 for each condition, *p < 0.05 by Fisher's post hoc test].
The negative coefficient for modulus in the model fit for scleraxis indicates that scleraxis expression increases as modulus increases (Figure 5A). This is further demonstrated in Figure 5B: The expression of scleraxis on the 10 kPa substrates is lower than the expression on the 90 kPa substrates (p < 0.05). Thus, a substrate with a modulus of 90 kPa promotes tenogenic differentiation of hMSCs when compared to a substrate with modulus of 10 kPa. Scleraxis expression at 50 kPa is roughly equal to scleraxis expression at 90 kPa (p = 0.94), indicating that substrates with a modulus of 50 kPa and 90 kPa promote tenogenic differentiation of hMSCs to similar levels in the presence of AA and BMP-13.
The negative coefficient for CMP in the model fit for collagen I and tenascin-C indicates that the expression of these tendon-associated ECM proteins increases as CMP concentration increases (and as RGDS concentration decreases) (Figure 5C – 5F). As noted in Figure 4B, a minimum concentration of total bioactive peptide is required for uniform cell attachment, so the concentration of CMP cannot be altered in this system without a compensatory change in RGDS; consequently, we were not able to decouple the effects of increasing CMP concentration and decreasing RGDS concentration in our system. Nevertheless, the presence of 2 mM CMP (0 mM RGDS) led to significant increases in hMSC collagen I and tenascin-C expression when compared to 0 mM CMP (2 mM RGDS; Figure 5D, 5F). This suggests, at least within the composition ranges tested here, that the presence of the CMP sequence, which mimics collagen (the predominant ECM protein in tendon), is beneficial for increasing hMSC tenogenic gene expression.
Semi-quantitative analysis of protein levels
We next sought to verify that increases in hMSC collagen I and tenascin-C gene expression resulted in observable increases in production of these ECM proteins. A subset of samples was selected (10 kPa, 50 kPa, and 90 kPa with 2 mM CMP; 10 kPa, 50 kPa, and 90 kPa with 2 mM RGDS) and analyzed semi-quantitatively for production of collagen I and tenascin-C. This semi-quantitative analysis was performed by: i) immunostaining for collagen I and tenascin-C, ii) imaging samples using confocal microscopy, and iii) quantifying the fluorescence intensity for each protein, normalized to the number of cell nuclei, in ImageJ. The data were analyzed using two-way ANOVA, with modulus and CMP concentration as factors and collagen I and tenascin-C staining as responses. Fisher's LSD post-hoc test was used to identify conditions that had significantly higher or lower fluorescence values.
The results of the ANOVA are summarized in Table 1. The concentration of CMP affects the degree of staining for both collagen I and tenascin-C (p < 0.05 for both), which agrees well with the gene expression measurements presented in Figure 5. Modulus had a significant effect on tenascin-C staining (p < 0.05). The modulus and CMP interaction terms were not significant for either protein, again agreeing with the gene expression measurements in Figure 5.
Table 1.
Two-way ANOVA was run to determine which factors had an effect on the total raw integrated density per cell in the immunostaining images. The presence of the collagen mimetic peptide had a significant effect on the staining intensity (divided by the number of nuclei) for both collagen I (Col I) and tenascin-C (Tn-C). The modulus (E) of the substrate had a significant effect on the staining intensity (divided by the number of nuclei) for tenascin-C but not for collagen I.
| Term | Col I p-value | Tn-C p-value |
|---|---|---|
| Modulus | 0.11 | 0.01 |
| [CMP] | 0.01 | 0.03 |
| E * [CMP] | 0.26 | 0.46 |
Since the interaction between substrate modulus and CMP concentration was not significant for either protein, the factors were analyzed individually, and the results are plotted in Figure 6. The 2 mM CMP condition (0 mM RGDS) had significantly greater degrees of staining for both collagen I (Figure 6A) and tenascin-C (Figure 6B) than the 0 mM CMP condition (2 mM RGDS). Interestingly, the 50 kPa and 90 kPa conditions had significantly greater degrees of staining for tenascin-C than the 10 kPa condition (Figure 6C), despite the gene-level analysis showing no significant impact of modulus on tenascin-C expression (Figure 5F). While gene expression and protein production are generally correlated,61 tenascin-c production also is highly regulated at the post-translational level.62 Here, it is possible that modulus is impacting the rate of tenascin-C translation or post-translational modification or the degree of tenascin-C retention on these synthetic hydrogels.
Figure 6.
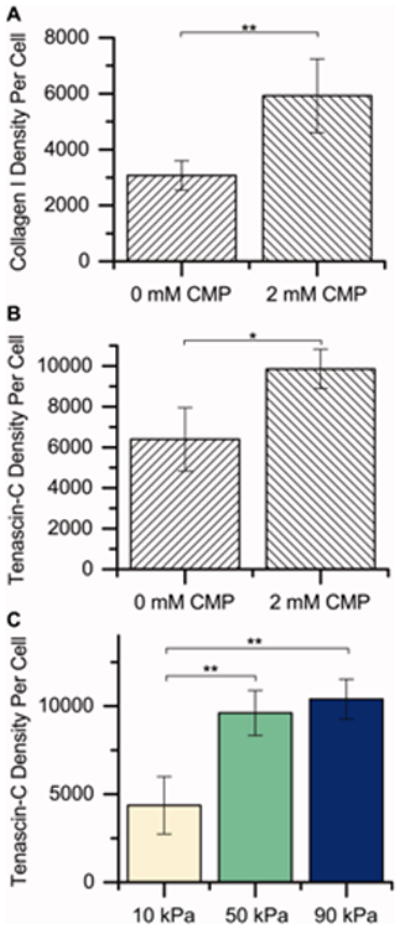
Semiquantitative analysis of protein content by immunocytochemistry. To determine if differences in gene expression resulted in observable differences in protein production, we performed a semiquantitative analysis of protein expression for a subset of samples (each modulus with 0 or 2 mM CMP [2 or 0 mM RGDS, respectively]). Here, we immunostained for collagen I and tenascin-C, imaged the samples, and summed the fluorescence intensities (raw integrated density in ImageJ) in each image, normalizing to the number of nuclei. The factors (CMP and modulus) were analyzed individually to determine their effects on protein expression since the interaction terms (CMP × modulus) by ANOVA were not statistically significant (see Table I). Increasing the CMP concentration from 0 mM CMP (2 mM RGDS) to 2 mM CMP (0 mM RGDS) significantly increased the staining intensity for A) collagen I and B) tenascin-C, indicating increased protein expression, consistent with gene expression observations. C) Increasing the modulus of the substrate from 10 kPa to 50 kPa or 90 kPa also significantly increased the staining intensity for tenascin-C [n = 3 for each condition, *p < 0.05 by Fisher's post hoc test, **p < 0.01 by Fisher's post hoc test].
Characterization of hMSC differentiation to other lineages
Lastly, we wanted to investigate how the most influential factors identified in this study (higher modulus and CMP concentration) affect hMSC differentiation to non-tenogenic lineages. Specifically, we cultured cells for 15 days on 90 kPa substrates with 2 mM CMP in the presence of the tenogenic soluble factors AA and BMP-13 and compared their gene expression with the gene expression of hMSCs cultured for 15 days in growth media on TCPS. The results obtained using a qRT-PCR pathway array are shown in Figure 7 for the adipogenic gene CEBPA (CCAAT/enhancer-binding protein, alpha); the osteogenic genes ALP (alkaline phosphatase), RUNX2 (runt-related transcription factor 2), and BGLAP (osteocalcin); and the chondrogenic gene ACAN (aggrecan). Significant downregulation of the adipogenic and chondrogenic genes were observed, as well as downregulation of the early markers of osteogenesis (ALP and RUNX2). Significant upregulation of BGLAP, a more mature marker of osteogenesis, was observed, suggesting potential osteogenesis occurring on the high-modulus, high-CMP substrates. To further investigate this result, expression of another osteogenic gene, osteopontin, was measured using RNA isolated from the DOE experiments. While the effect of the integrin-binding peptides on osteopontin expression was not significant (Figure S11), the 90 kPa gels promoted substantially more osteopontin expression than the 10 kPa and 50 kPa gels (Figure S12), suggesting a greater degree of osteogenic gene expression on the higher-modulus substrate.
Figure 7.
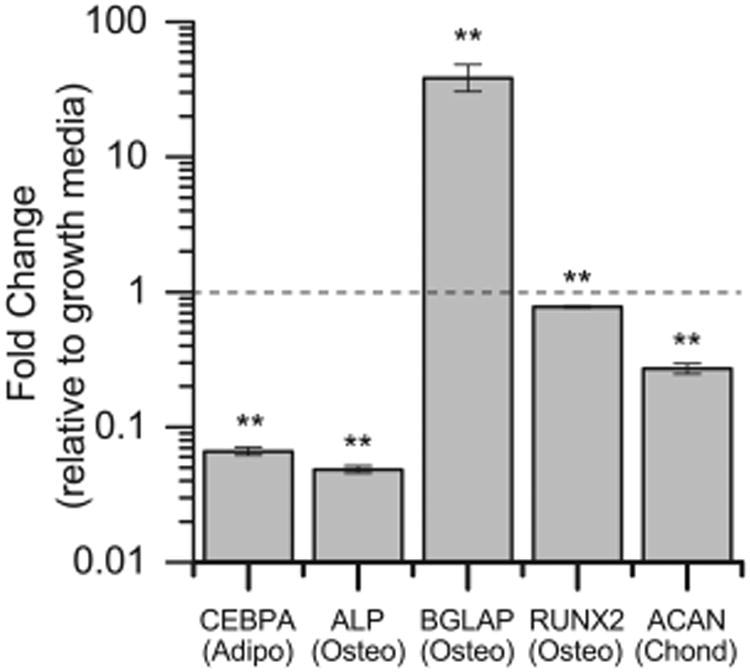
Lineage specificity in controlled microenvironment. hMSCs were cultured on 90 kPa and 2 mM CMP gels with BMP-13 and AA (the condition observed to most significantly affect tenogenic gene expression in the DOE), or in growth media on TCPS (control), for 15 days; expression of genes associated with nontenogenic lineages was measured with a PCR plate. Significant downregulation of the adipogenic gene CEBPA, the osteogenic genes ALP and RUNX2 (early markers), and the chondrogenic gene ACAN were observed. Significant upregulation of the osteogenic gene BGLAP was observed. Thus, the combinations of factors presented here did not lead to significant degrees of adipogenesis or chondrogenesis, but a high modulus and high CMP substrate, in combination with the soluble factors AA and BMP-13, led to an increase in the late osteogenic marker BGLAP [n = 3, **p < 0.01 compared with growth media on TCPS].
Discussion
We used a DOE approach to evaluate the effects of extracellular matrix cues on hMSC tenogenic differentiation. Previous studies have demonstrated the utility of the statistical design approach for the design of biomaterials to direct MSC osteogenic differentiation,63,64 but, to the best of our knowledge, this is the first time this approach has been successfully used to study hMSC tenogenic differentiation. We showed that, within the range of compositions tested in this study, increasing modulus and increasing CMP concentration resulted in increases in expression of the tendon/ligament-associated genes, scleraxis, collagen I, and tenascin-C, and increases in the production or retention of the tendon/ligament-associated proteins collagen I and tenascin-C.
Our modulus result agrees well with the result reported by Sharma and Snedeker, where they observed greater tenogenic differentiation of hMSCs at moduli between 30 - 50 kPa.31 Interestingly, on their 70 - 90 kPa substrates, Sharma and Snedeker observed no significant tenogenic differentiation but marked osteogenic differentiation. They attributed this result to paracrine signaling between differentiated osteoblasts and MSCs, suppressing tenogenic differentiation. While we also observed increased expression of the osteogenic genes BGLAP and osteopontin on our stiffer substrates (Figure 7 and Figure S12), in agreement with results that show osteogenesis with increasing substrate modulus,45 we did not observe a decrease in scleraxis expression at the higher modulus (∼ 90 kPa) when compared to the mid-range modulus (∼ 50 kPa). Upregulation of genes associated with both tenogenesis and osteogenesis on 90 kPa substrates suggests a potential mixed population of tenogenic and osteogenic progenitor cells by day 15. We hypothesize that, while a modulus of ∼ 90 kPa may promote some degree of both osteogenic and tenogenic gene expression, the presence of specific soluble factors in the extracellular environment (here, AA and BMP-13) may be important and pivotal in lineage commitment on these substrates. While further testing is required to test this hypothesis, these findings demonstrate the importance of studying the interplay between soluble factors and matrix modulus for regulating hMSC differentiation.
To the best of our knowledge, no previous study has investigated the effect of CMPs on the tenogenic differentiation of hMSCs. Here, we show that the presence of the CMP increases the expression of genes related to the tenogenic ECM proteins collagen I and tenascin-C. CMPs also have been shown to promote MSC osteogenic65 and chondrogenic differentiation;66 this finding suggests the potential general importance of CMPs in hMSC differentiation down lineages associated with collagenous tissues, where the context in which CMPs are presented may determine fate. RGDS was used to promote cell adhesion on these synthetic matrices for comparison to and in conjunction with CMP, owing to the presence of RGDS in many ECM proteins,67 including fibronectin and laminin that are found in developing tendon/ligament. While RGDS also has been shown to affect hMSC differentiation down chondrogenic28 and osteogenic lineages,68 the demonstration that CMP alone promotes tenogenic gene expression more than the RGDS alone highlights the potential utility of CMPs for regulating hMSC tenogenic differentiation. More broadly, this finding also provides support for the hypothesis that mimicking biologically-relevant ECM compositions regulates hMSC differentiation.
The CMP used in the current work could be interacting with the hMSCs in at least two ways: i) CMPs have been shown to increase collagen production of encapsulated cells and increase collagen retention in PEG-based networks, likely through association with collagen by strand invasion,69 and ii) the CMP used here contains an integrin-binding GFOGER sequence known to regulate ECM protein production by cells.70 In either case, increased activation of collagen-related integrins may play a role; however, the structure of the underlying matrix also may be important if secreted collagen is retained. These findings motivate further studies to understand how CMPs influence hMSC tenogenesis, both in two and three dimensions, which may provide additional insight into the mechanism by which CMPs promote tenogenesis toward the design of scaffolds for hMSC tendon/ligament tissue engineering.
The utility of the DOE approach lies in being able to extract information rapidly from a system about how factors influence response variables using fewer total replicates than the one-variable-at-a-time approach. Here, the DOE approach allowed us to test our hypothesis that modulus and integrin-binding peptides would influence tenogenic gene expression in hMSCs in the presence of tenogenic soluble factors. This statistical analysis was critical in establishing effects of variables within a system that is inherently ‘noisy’ owing to the challenge of achieving hMSC tenogenesis in vitro and heterogeneity within the differentiating cell population. While, to date, relatively few studies have utilized the DOE approach to optimize or understand cell microenvironments,63,64,71,72 this method may prove useful in future studies within the field with the growing focus on studying synergies between cues for directing stem cell differentiation. The data presented in this paper demonstrate the benefits of using a DOE approach to study specific biological responses, as well as the importance of studying combinations of extracellular cues to better understand the hMSC differentiation process.
Conclusion
In sum, this study used a DOE approach to establish individual and synergistic effects of matrix modulus and composition on tenogenesis of hMSCs. Soluble factors BMP-13 and AA were found to upregulate tendon-associated genes, and this upregulation was enhanced by cell culture with physiologically-relevant insoluble matrix cues. Specifically, well-defined hydrogels that provide control of matrix modulus and biochemical content were created that mimicked aspects of developing tendon tissue. With this approach, increasing matrix modulus and concentration of CMP were observed to promote relevant gene expression and ECM protein content in the presence of tenogenic soluble factors. However, one tradeoff with increasing modulus may be some promotion of osteogenesis, where upregulation of osteocalcin was observed along with scleraxis. Taken together, these results motivate the design of materials that mimic developing to mature tendon and a collagen-rich matrix to promote tenogenic differentiation of hMSCs. Further, a framework was established for studying combinatorial effects of multiple microenvironment cues that can be broadly applied toward understanding stem cell differentiation down difficult lineages.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge financial support from the Burroughs Wellcome Fund, the National Institutes of Health (NIH) Chemistry-Biology Interface (CBI) program at the University of Delaware (NIH T32GM008550), and Delaware COBRE programs supported by grants from the National Institute of General Medical Sciences – NIGMS (P30 GM110758, P20 RR016458, P20 GM103541, and P20 GM104316; each at different stages of the presented work) from the NIH. Work conducted at UC Davis was supported by the California Institute for Regenerative Medicine (RN3-06460). The authors would like to thank Prof. Wilfred Chen, Prof. Millicent Sullivan, and the Bio-Imaging Center at the Delaware Biotechnology Institute for use of their instruments and training. The authors would like to thank Alina Marusina and Carolin Hartwig for assistance with initial experiments, Megan Smithmyer for feedback on early versions of this manuscript, and Prathamesh Kharkar and Lisa Sawicki for input in schematic preparation.
References
- 1.September AV, Schwellnus MP, Collins M. Tendon and ligament injuries: the genetic component. Br J Sports Med. 2007;41:241–6. doi: 10.1136/bjsm.2006.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chahal J, Lee A, Heard W, Bach BR. A Retrospective Review of Anterior Cruciate Ligament Reconstruction Using Patellar Tendon: 25 Years of Experience. Orthop J Sport Med. 2013;1:2325967113501789. doi: 10.1177/2325967113501789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kartus J, Movin T, Karlsson J. Donor-site morbidity and anterior knee problems after anterior cruciate ligament reconstruction using autografts. Arthroscopy. 2001;17:971–80. doi: 10.1053/jars.2001.28979. [DOI] [PubMed] [Google Scholar]
- 4.Fleming BC, Hulstyn MJ, Oksendahl HL, Fadale PD. Ligament Injury, Reconstruction and Osteoarthritis. Curr Opin Orthop. 2005;16:354–62. doi: 10.1097/01.bco.0000176423.07865.d2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodrigues MT, Reis RL, Gomes ME. Engineering tendon and ligament tissues: present developments towards successful clinical products. J Tissue Eng Regen Med. 2013;7:673–86. doi: 10.1002/term.1459. [DOI] [PubMed] [Google Scholar]
- 6.Ajibade DA, Vance DD, Hare JM, Kaplan LD, Lesniak BP. Emerging Applications of Stem Cell and Regenerative Medicine to Sports Injuries. Orthop J Sport Med. 2014;2:2325967113519935. doi: 10.1177/2325967113519935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge Z, Goh JCH, Lee EH. Selection of Cell Source for Ligament Tissue Engineering. Cell Transplant. 2005;14:573–83. doi: 10.3727/000000005783982819. [DOI] [PubMed] [Google Scholar]
- 8.Chen JL, Zhang W, Heng BC, Ouyang HW, Dai XS. Physical regulation of stem cells differentiation into teno-lineage: current strategies and future direction. Cell Tissue Res. 2015;360:195–207. doi: 10.1007/s00441-014-2077-4. [DOI] [PubMed] [Google Scholar]
- 9.Altman G, Horan R, Martin I, Farhadi J, Stark P, Volloch V, Vunjak-Novakovic G, Richmond J, Kaplan DL. Cell differentiation by mechanical stress. FASEB J. 2002;16:270–2. doi: 10.1096/fj.01-0656fje. [DOI] [PubMed] [Google Scholar]
- 10.Song G, Luo Q, Xu B, Ju Y. Mechanical Stretch-Induced Changes in Cell Morphology and mRNA Expression of Tendon/Ligament-Associated Genes in Rat Bone-Marrow Mesenchymal Stem Cells. Mol Cell Biomech. 2010;7:165–74. [PubMed] [Google Scholar]
- 11.Kuo CK, Tuan RS. Mechanoactive tenogenic differentiation of human mesenchymal stem cells. Tissue Eng Part A. 2008;14:1615–27. doi: 10.1089/ten.tea.2006.0415. [DOI] [PubMed] [Google Scholar]
- 12.Xu B, Song G, Ju Y. Effect of Focal Adhesion Kinase on the Regulation of Realignment and Tenogenic Differentiation of Human Mesenchymal Stem Cells by Mechanical Stretch. Connect Tissue Res. 2011;52:373–9. doi: 10.3109/03008207.2010.541961. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Tran N, Chen HQ, Kahn CJF, Marchal S, Groubatch F, Wang X. Time-related changes in expression of collagen types I and III and of tenascin-C in rat bone mesenchymal stem cells under co-culture with ligament fibroblasts or uniaxial stretching. Cell Tissue Res. 2008;332:101–9. doi: 10.1007/s00441-007-0564-6. [DOI] [PubMed] [Google Scholar]
- 14.Morita Y, Watanabe S, Ju Y, Xu B. Determination of optimal cyclic uniaxial stretches for stem cell-to-tenocyte differentiation under a wide range of mechanical stretch conditions by evaluating gene expression and protein synthesis levels. Acta Bioeng Biomech. 2013;15:71–9. [PubMed] [Google Scholar]
- 15.Bashur CA, Shaffer RD, Dahlgren LA, Guelcher SA, Goldstein AS. Effect of fiber diameter and alignment of electrospun polyurethane meshes on mesenchymal progenitor cells. Tissue Eng Part A. 2009;15:2435–45. doi: 10.1089/ten.tea.2008.0295. [DOI] [PubMed] [Google Scholar]
- 16.Caliari SR, Harley BAC. Structural and biochemical modification of a collagen scaffold to selectively enhance MSC tenogenic, chondrogenic, and osteogenic differentiation. Adv Healthc Mater. 2014;3:1086–96. doi: 10.1002/adhm.201300646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teh TKH, Toh SL, Goh JCH. Aligned hybrid silk scaffold for enhanced differentiation of mesenchymal stem cells into ligament fibroblasts. Tissue Eng Part C Methods. 2011;17:687–703. doi: 10.1089/ten.tec.2010.0513. [DOI] [PubMed] [Google Scholar]
- 18.Luo Q, Song G, Song Y, Xu B, Qin J, Shi Y. Indirect co-culture with tenocytes promotes proliferation and mRNA expression of tendon/ligament related genes in rat bone marrow mesenchymal stem cells. Cytotechnology. 2009;61:1–10. doi: 10.1007/s10616-009-9233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee IC, Wang JH, Lee YT, Young TH. The differentiation of mesenchymal stem cells by mechanical stress or/and co-culture system. Biochem Biophys Res Commun. 2007;352:147–52. doi: 10.1016/j.bbrc.2006.10.170. [DOI] [PubMed] [Google Scholar]
- 20.Schneider PRA, Buhrmann C, Mobasheri A, Matis U, Shakibaei M. Three-dimensional high-density co-culture with primary tenocytes induces tenogenic differentiation in mesenchymal stem cells. J Orthop Res. 2011;29:1351–60. doi: 10.1002/jor.21400. [DOI] [PubMed] [Google Scholar]
- 21.Fan H, Liu H, Toh SL, Goh JCH. Enhanced differentiation of mesenchymal stem cells co-cultured with ligament fibroblasts on gelatin/silk fibroin hybrid scaffold. Biomaterials. 2008;29:1017–27. doi: 10.1016/j.biomaterials.2007.10.048. [DOI] [PubMed] [Google Scholar]
- 22.Lee JY, Zhou Z, Taub PJ, Ramcharan M, Li Y, Akinbiyi T, Maharam ER, Leong DJ, Laudier DM, Ruike T, Torina PJ, Zaidi M, Majeska RJ, Schaffler MB, Flatow EL, Sun HB. BMP-12 treatment of adult mesenchymal stem cells in vitro augments tendon-like tissue formation and defect repair in vivo. PLoS One. 2011;6:e17531. doi: 10.1371/journal.pone.0017531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haddad-Weber M, Prager P, Kunz M, Seefried L, Jakob F, Murray MM, Evans CH, Nöth U, Steinert AF. BMP12 and BMP13 gene transfer induce ligamentogenic differentiation in mesenchymal progenitor and anterior cruciate ligament cells. Cytotherapy. 2010;12:505–13. doi: 10.3109/14653241003709652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei C, Ming N, Yun-Feng R, Kai-Yi Z, Qiang Z, Liang-Liang X, Kai-Ming C, Gang L, Yan W. Effect of growth and differentiation factor 6 on the tenogenic differentiation of bone marrow-derived mesenchymal stem cells. Chin Med J (Engl) 2013;126:1509–16. [PubMed] [Google Scholar]
- 25.Raabe O, Shell K, Fietz D, Freitag C, Ohrndorf A, Christ HJ, Wenisch S, Arnhold S. Tenogenic differentiation of equine adipose-tissue-derived stem cells under the influence of tensile strain, growth differentiation factors and various oxygen tensions. Cell Tissue Res. 2013;352:509–21. doi: 10.1007/s00441-013-1574-1. [DOI] [PubMed] [Google Scholar]
- 26.Farng E, Urdaneta AR, Barba D, Esmende S, McAllister DR. The effects of GDF-5 and uniaxial strain on mesenchymal stem cells in 3-D culture. Clin Orthop Relat Res. 2008;466:1930–7. doi: 10.1007/s11999-008-0300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gandavarapu NR, Alge DL, Anseth KS. Osteogenic differentiation of human mesenchymal stem cells on α5 integrin binding peptide hydrogels is dependent on substrate elasticity. Biomater Sci. 2014;2:352–61. doi: 10.1039/C3BM60149H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salinas CN, Anseth KS. The enhancement of chondrogenic differentiation of human mesenchymal stem cells by enzymatically regulated RGD functionalities. Biomaterials. 2008;29:2370–7. doi: 10.1016/j.biomaterials.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flynn LE. The use of decellularized adipose tissue to provide an inductive microenvironment for the adipogenic differentiation of human adipose-derived stem cells. Biomaterials. 2010;31:4715–24. doi: 10.1016/j.biomaterials.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 30.Sharma RI, Snedeker JG. Biochemical and biomechanical gradients for directed bone marrow stromal cell differentiation toward tendon and bone. Biomaterials. 2010;31:7695–704. doi: 10.1016/j.biomaterials.2010.06.046. [DOI] [PubMed] [Google Scholar]
- 31.Sharma RI, Snedeker JG. Paracrine interactions between mesenchymal stem cells affect substrate driven differentiation toward tendon and bone phenotypes. PLoS One. 2012;7:e31504. doi: 10.1371/journal.pone.0031504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikonen L, Kerkelä E, Metselaar G, Stuart MCA, de Jong MR, Aalto-Setälä K. 2D and 3D self-assembling nanofiber hydrogels for cardiomyocyte culture. Biomed Res Int. 2013;2013:285678. doi: 10.1155/2013/285678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hutmacher DW. Scaffolds in tissue engineering bone and cartilage. Biomaterials. 2000;21:2529–43. doi: 10.1016/s0142-9612(00)00121-6. [DOI] [PubMed] [Google Scholar]
- 34.Ranga A, Gobaa S, Okawa Y, Mosiewicz K, Negro A, Lutolf MP. 3D niche microarrays for systems-level analyses of cell fate. Nat Commun. 2014;5:4324. doi: 10.1038/ncomms5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeForest CA, Anseth KS. Advances in bioactive hydrogels to probe and direct cell fate. Annu Rev Chem Biomol Eng. 2012;3:421–44. doi: 10.1146/annurev-chembioeng-062011-080945. [DOI] [PubMed] [Google Scholar]
- 36.Settle SH, Rountree RB, Sinha A, Thacker A, Higgins K, Kingsley DM. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev Biol. 2003;254:116–30. doi: 10.1016/s0012-1606(02)00022-2. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Doroski DM, Brink KS, Temenoff JS. Techniques for biological characterization of tissue-engineered tendon and ligament. Biomaterials. 2007;28:187–202. doi: 10.1016/j.biomaterials.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 39.Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–80. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang G, Rothrauff BB, Tuan RS. Tendon and ligament regeneration and repair: clinical relevance and developmental paradigm. Birth Defects Res C Embryo Today. 2013;99:203–22. doi: 10.1002/bdrc.21041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 42.Benjamin M, Ralphs JR. The Cell and Developmental Biology of Tendons and Ligaments. Int Rev Cytol. 2000;196:85–130. doi: 10.1016/s0074-7696(00)96003-0. [DOI] [PubMed] [Google Scholar]
- 43.Schiele NR, Marturano JE, Kuo CK. Mechanical factors in embryonic tendon development: potential cues for stem cell tenogenesis. Curr Opin Biotechnol. 2013;24:834–40. doi: 10.1016/j.copbio.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marturano JE, Arena JD, Schiller ZA, Georgakoudi I, Kuo CK. Characterization of mechanical and biochemical properties of developing embryonic tendon. Proc Natl Acad Sci U S A. 2013;110:6370–5. doi: 10.1073/pnas.1300135110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 46.Schweitzer R, Zelzer E, Volk T. Connecting muscles to tendons: tendons and musculoskeletal development in flies and vertebrates. Development. 2010;137:2807–17. doi: 10.1242/dev.047498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson SB, Lin CC, Kuntzler DV, Anseth KS. The performance of human mesenchymal stem cells encapsulated in cell-degradable polymer-peptide hydrogels. Biomaterials. 2011;32:3564–74. doi: 10.1016/j.biomaterials.2011.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–7. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 49.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. A Versatile Synthetic Extracellular Matrix Mimic via Thiol-Norbornene Photopolymerization. Adv Mater. 2009;21:5005–10. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility. Biomaterials. 2009;30:6702–7. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kloxin AM, Kloxin CJ, Bowman CN, Anseth KS. Mechanical properties of cellularly responsive hydrogels and their experimental determination. Adv Mater. 2010;22:3484–94. doi: 10.1002/adma.200904179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gould ST, Darling NJ, Anseth KS. Small Peptide Functionalized Thiol-ene Hydrogels as Culture Substrates for Understanding Valvular Interstitial Cell Activation and De Novo Tissue Deposition. Acta Biomater. 2012;8:3201–9. doi: 10.1016/j.actbio.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong Z, Sant S, Khademhosseini A, Jia X. Controlling the Fibroblastic Differentiation of Mesenchymal Stem Cells Via the Combination of Fibrous Scaffolds and Connective Tissue Growth Factor. Tissue Eng Part A. 2011;17:2773–85. doi: 10.1089/ten.tea.2011.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogunnaike BA. Design of Experiments. Random Phenom. 2010 [Google Scholar]
- 55.Vandesompele J, Kubista M, Pfaffl MW. Reference Gene Validation Software for Improved Normalization. In: Logan J, Edwards K, Saunders N, editors. Real-time PCR Curr Technol Appl. 2009. [Google Scholar]
- 56.Jennings K. Statistics 512: Applied Linear Models. Internet. [Google Scholar]
- 57.Schweitzer R, Chyung JH, Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, Tabin CJ. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128:3855–66. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- 58.Kardon G. Muscle and tendon morphogenesis in the avian hind limb. Development. 1998;125:4019–32. doi: 10.1242/dev.125.20.4019. [DOI] [PubMed] [Google Scholar]
- 59.Lui PPY, Rui YF, Ni M, Chan KM. Tenogenic differentiation of stem cells for tendon repair —what is the current evidence? J Tissue Eng Regen Med. 2011;5:e144–63. doi: 10.1002/term.424. [DOI] [PubMed] [Google Scholar]
- 60.Toepke MW, Impellitteri NA, Theisen JM, Murphy WL. Characterization of Thiol-Ene Crosslinked PEG Hydrogels. Macromol Mater Eng. 2013;298:699–703. doi: 10.1002/mame.201200119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 62.Giblin SP, Midwood KS. Tenascin-C: Form versus function. Cell Adh Migr. 2015;9:48–82. doi: 10.4161/19336918.2014.987587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen WLK, Likhitpanichkul M, Ho A, Simmons CA. Integration of statistical modeling and high-content microscopy to systematically investigate cell-substrate interactions. Biomaterials. 2010;31:2489–97. doi: 10.1016/j.biomaterials.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 64.Decaris ML, Leach JK. Design of experiments approach to engineer cell-secreted matrices for directing osteogenic differentiation. Ann Biomed Eng. 2011;39:1174–85. doi: 10.1007/s10439-010-0217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Connelly JT, Petrie TA, García AJ, Levenston ME. Fibronectin- and collagen-mimetic ligands regulate bone marrow stromal cell chondrogenesis in three-dimensional hydrogels. Eur Cells Mater. 2011;22:168–77. doi: 10.22203/ecm.v022a13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee HJ, Yu C, Chansakul T, Hwang NS, Varghese S, Yu SM, Elisseeff JH. Enhanced Chondrogenesis of Mesenchymal Stem Cells in Collagen Mimetic Peptide-Mediated Microenvironment. 2008;11:1843–51. doi: 10.1089/ten.tea.2007.0204. [DOI] [PubMed] [Google Scholar]
- 67.Ruoslahti E, Pierschbacher M. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–7. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 68.Yang F, Williams CG, Wang DA, Lee H, Manson PN, Elisseeff J. The effect of incorporating RGD adhesive peptide in polyethylene glycol diacrylate hydrogel on osteogenesis of bone marrow stromal cells. Biomaterials. 2005;26:5991–8. doi: 10.1016/j.biomaterials.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 69.Lee HJ, Lee JS, Chansakul T, Yu C, Elisseeff JH, Yu SM. Collagen mimetic peptide-conjugated photopolymerizable PEG hydrogel. Biomaterials. 2006;27:5268–76. doi: 10.1016/j.biomaterials.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 70.Zutter MM, Santoro SA. Function of Alpha2Beta1 Integrin. In: Gullberg D, editor. I Domains Integrins. 2003. [Google Scholar]
- 71.Lam J, Carmichael ST, Lowry WE, Segura T. Hydrogel design of experiments methodology to optimize hydrogel for iPSC-NPC culture. Adv Healthc Mater. 2015;4:534–9. doi: 10.1002/adhm.201400410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jung JP, Moyano JV, Collier JH. Multifactorial optimization of endothelial cell growth using modular synthetic extracellular matrices. Integr Biol (Camb) 2011;3:185–96. doi: 10.1039/c0ib00112k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


