Abstract
Mesenchymal stem cells (MSCs) are effective in treating several pathologies. We and others have demonstrated that hypoxia or hypoxia-inducible factor 1 alpha (HIF-1α) stabilization improves several MSC functions, including cell adhesion, migration, and proliferation, thereby increasing their therapeutic potential. To further explore the mechanisms induced by HIF-1α in MSCs, we studied its relationship with Notch signaling and observed that overexpression of HIF-1α in MSCs increased protein levels of the Notch ligands Jagged 1–2 and Delta-like (Dll)1, Dll3, and Dll4 and potentiated Notch signaling only when this pathway was activated. Crosstalk between HIF and Notch resulted in Notch-dependent migration and spreading of MSCs, which was abolished by γ-secretase inhibition. However, the HIF-1-induced increase in MSC proliferation was independent of Notch signaling. The ubiquitin family member, small ubiquitin-like modifier (SUMO), has important functions in many cellular processes and increased SUMO1 protein levels have been reported in hypoxia. To investigate the potential involvement of SUMOylation in HIF/Notch crosstalk, we measured general SUMOylation levels and observed increased SUMOylation in HIF-1-expressing MSCs. Moreover, proliferation and migration of MSCs were reduced in the presence of a SUMOylation inhibitor, and this effect was particularly robust in HIF-MSCs. Immunoprecipitation studies demonstrated SUMOylation of the intracellular domain of Notch1 (N1ICD) in HIF-1-expressing MSCs, which contributed to Notch pathway activation and resulted in increased levels of N1ICD nuclear translocation as assessed by subcellular fractionation. SUMOylation of N1ICD was also observed in HEK293T cells with stabilized HIF-1α expression, suggesting that this is a common mechanism in eukaryotic cells. In summary, we describe, for the first time, SUMOylation of N1ICD, which is potentiated by HIF signaling. These phenomena could be relevant for the therapeutic effects of MSCs in hypoxia or under conditions of HIF stabilization.
Keywords: : mesenchymal stem cells, hypoxia-inducible factor, Notch, SUMOylation, migration, proliferation
Introduction
Human mesenchymal stem cells (MSCs) from different sources have attracted great interest in several areas of medicine and biomedical research, particularly in the fields of regenerative medicine and cell therapy. Indeed, we previously demonstrated the capacity of MSCs isolated from dental pulp or bone marrow to treat myocardial infarction [1,2] and other studies have shown the potential of MSCs to treat ischemic diseases in preclinical and clinical models [3]. Sustained growth and maintenance of MSCs are key factors for their therapeutic use as they ultimately undergo replicative senescence after extended periods of normal growth [4,5]. To overcome this limitation, we and others have proposed culturing MSCs at reduced oxygen tensions or under conditions of hypoxia-inducible factor (HIF) overexpression/stabilization since these variables help to maintain cells in an undifferentiated state [6,7]. HIF-1α is a pivotal transcription factor regulating the adaptive response to hypoxia [8], and numerous proteins interact directly with HIF-1 to enhance or reduce its function [9–11]. Among them, HIF-1α interacts with the highly conserved Notch signaling pathway [12,13], and the processes of migration, proliferation, differentiation, and angiogenesis in precursor and tumor cells have all been directly correlated with HIF-1α upstream of Notch signaling [12,14–16]. The molecular mechanisms that regulate Notch activity are, however, complex. Four Notch receptors (Notch1–4) are found in mammals, which are activated by five different ligands: Delta-like 1, 3, and 4 (Dll1, Dll3, and Dll4) and Jagged 1 and 2 [17–19]. Among these, Notch1 and Jagged1 are the most widely studied [20]. Ligand binding to the Notch1 receptor causes a conformational change in its intracellular domain that allows its cleavage by γ-secretase, resulting in the release of the Notch intracellular domain (NICD) [21]. NICD subsequently translocates to the nucleus where it activates the transcription of Notch target genes, including Hes1, Hey1, and Hey2 [17,22].
Recently, an interaction between the Notch1 receptor and the SUMOylation cascade has been described in breast cancer [23]. Small ubiquitin-like modifier (SUMO), a small (11 kDa) ubiquitin-like protein, can covalently attach to proteins as a post-translational modification and influence their stability, localization, or activity [24]. Notch1 signaling can deplete the unconjugated SUMO1 pool, which selectively impairs the growth of Notch1-activated cells. However, whether crosstalk between HIF-1, Notch1, and SUMO signaling pathways is involved in the control of MSC behavior is unknown. The aim of the present study was to examine the regulation of proliferation, migration, and spreading in MSCs through crosstalk between HIF-1α, Notch, and SUMO signaling pathways.
Materials and Methods
All experiments were carried out in accordance with the approved guidelines and approved by the Instituto de Salud Carlos III and institutional ethics and animal care committees.
Cell lines and lentiviral labeling
Human MSCs obtained from dental pulp were purchased from Inbiomed (Inbiobank, San Sebastian, Guipuzcoa, Spain). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM)-low glucose (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Lonza, Basel, Switzerland) and 1% penicillin/streptomycin (Gibco) in a humidified atmosphere of 95% air and 5% CO2 at 37°C. Lentiviral overexpression of HIF-1α was performed as described [25]. Briefly, cells were transduced with pWPI-green fluorescent protein (GFP) or pWPI-HIF-1α-GFP expression vectors (http://addgene.org cat. #12254) daily for 3 days. Transduction efficiency was evaluated by flow cytometry and the percentage of infection was routinely higher than 90%. Cells were expanded and cryopreserved until use.
Transient transfection of HEK293T cells
HEK293T cells (6 × 106) were cultured in 10-cm2 dishes with 10 mL of DMEM-high glucose supplemented with 10% FBS and 1% penicillin/streptomycin. Twenty-four hours after seeding and 1 h before transfection, the medium was replenished and supplemented with 25 μM chloroquine (Sigma-Aldrich, Madrid, Spain). Cells were transduced with a mixture of 20 μg of EF.hICN1.Ubc.GFP, 62 μL of 2 M CaCl2, and 418 μL of H2O added gently dropwise to 500 μL 2 × HBS (10 mM glucose, 40 mM HEPES, 10 mM KCl, 270 mM NaCl, 1.5 mM Na2HPO4, pH 7.1). After incubation for 6–8 h, the medium was replenished without chloroquine, and 2–3 days later, cells were analyzed by flow cytometry and western blotting [anticleaved Notch1 (A-8,1:1,000); Santa Cruz Biotechnology, Santa Cruz, CA) and real time quantitative PCR (RT-qPCR) (Hes1, Hey1, and Hey2].
Induction of hypoxia in HEK293T cells
Cells were cultured under hypoxia (1% O2) in an Invivo2 400 workstation (Ruskinn Technology Ltd., Bridgend, United Kingdom) for 4 h.
Activation/inhibition of Notch signaling pathway
To activate the Notch pathway, 60-cm2 culture plates were coated with anti-human IgG-Fc (stock at 1.3 mg/mL, 7.4 μL/1.5 mL phosphate-buffered saline [PBS]; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) for 30 min at 37°C, washed with PBS twice, and blocked with DMEM plus 10% FBS for 1 h at 37°C. Plates were washed again twice with PBS and coated with Jag1-Fc for 2 h at 37°C (stock at 200 μg/mL, 8.1 μL from stock/1.5 mL PBS; R&D Systems, Abingdon, United Kingdom). Then, control or HIF-MSCs were seeded onto the coated plates for 24 h. In some experiments, the gamma-secretase inhibitor RO4929097 (Xcess Biosciences, Inc., San Diego, CA) was used to inhibit the Notch pathway (10 μM for 48 h; 24 h before seeding cells onto Jagged1-coated plates and repeated after seeding).
Inhibition of SUMOylation
To inhibit protein SUMOylation, cells were treated with 50 μM anacardic acid (AA; cat. #172050 Calbiochem, Merck Millipore, Darmstadt, Germany) for 4 h. AA is a cell-permeable ginkgolic acid analog that inhibits protein SUMO modification by selectively targeting SUMO-activating enzyme E1 and interfering with E1-SUMO intermediate formation. With the inhibition of this first step, subsequent reactions of conjugation and ligation of SUMO to the target proteins are abolished [26].
Migration assay
Cells were seeded in basal medium (DMEM plus 0.5% FBS) at 10,000 cells/cm2 in the top chamber of an 8-μm-pore Transwell chamber (BD Falcon, Bedford, MA). After overnight culture, 25 ng/mL of IL-1β was added to the bottom chamber for 8 h. DMEM plus 0.5% and 10% FBS were used as negative and positive controls, respectively. Nonmigrated cells were removed from the upper side of the membrane with a cotton bud and migrated cells were fixed with 70% cold ethanol for 10 min. The membrane was cut and placed in a glass slide with the bottom side upward, stained with DAPI, and migrated cells were counted.
Proliferation assay
Proliferation of MSCs was assessed by incorporation of 5′-bromo-2′-deoxyuridine (BrdU; Life Technologies, Madrid, Spain). Cells were cultured at 50–60% confluence on 11-mm diameter cover slides coated with Jagged 1 and BrdU was added at 10 μg/mL for 8 h. After this time, cells were washed with PBS and fixed with 70% cold ethanol for 10 min at 4°C. Slides were then washed three times with PBS and 1 N HCl was added for 30 min at 37°C. Subsequently, the acid was neutralized with borate buffer for 10 min at room temperature and, after five washes in PBS, slides were treated with blocking buffer (PBS, 10% normal goat serum, 0.01% Triton X-100) for 1 h at 37°C, followed by addition of an anti-BrdU antibody and incubation overnight at 4°C (1:100 rat anti-BrdU; Abcam, Cambridge, United Kingdom). After five washes, an Alexa Fluor 555 (Invitrogen, Carlsbad, CA) secondary antibody was added for 2 h at room temperature and positive nuclei were then counted.
Scratch-wound assay
To analyze spreading and invasiveness, cells were seeded in 6-well culture plates and, once confluent, a scratch was made on the monolayer with a p10 pipette tip. Debris was removed with 1 mL of growth medium. Invasiveness of the scratch was tracked using time-lapse microscopy and image analysis was performed with ImageJ software (NIH).
Real time quantitative PCR
RNA was extracted using TRIzol reagent (Invitrogen) and purified with the RNeasy Plus Mini Kit (Qiagen, Dusseldorf, Germany). RNA was quantified spectrophotometrically using a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE). cDNA was obtained by reverse transcription using M-MLV reverse transcriptase (Invitrogen). RT-qPCR was performed with the following human-specific sense and antisense primers: Hes1 5′-AAGAAAGATAGCTCGCGGCA-3′ (forward) and 5′-TACTTCCCCAGCACACTTGG-3′ (reverse); Hey1 5′-AGGTAATGGAGCAAGGATCTGC-3′ (forward) and 5′-CCCGAAATCCCAAACTCCGA-3′ (reverse); Hey2 5′-GGATTATAGAGAAAAGGCGTC-3′ (forward) and 5′-GTTTTTCAAAAGCAGTTGGC-3′ (reverse); and ACTB 5′-AGAGCCTCGCCTTTGCCGATCC-3′ (forward) and 5′-CATGCCGGAGCCGTTGTCGAC-3′ (reverse) and SYBR Green I 1 × LightCycler 480 SYBR Green I Master Mix (Roche Molecular Biochemical, Mannheim, Germany). Multiwell plates of 96 wells were run on a LightCycler 480 Instrument (Roche Diagnostics, Mannheim, Germany).
Subcellular fractionation
Cells were seeded onto Jagged1-coated plates to activate Notch signaling for 24 h in DMEM-low glucose supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were treated (+) or not (−) with AA (4 h, μM). Then, cells were washed twice with cold PBS, lysed in 500 μL of subcellular fractionation buffer (250 mM sucrose, 20 mM HEPES pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and protease/phosphatase inhibitor cocktail), and shaken on a tube roller at 30–50 rpm for 30 min at 4°C. The lysate was centrifugated at 720 g for 10 min at 4°C and the pellet was washed twice with the above buffer and centrifuged as before. The supernatant was collected and centrifugated at 10,000 g for 10 min at 4°C. The pellet from the previous step was lysed in nuclear lysis buffer (50 mM Tris-HCl pH8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 10% glycerol, and protease/phosphatase inhibitor cocktail) and shaken on a tube roller at 30–50 rpm for 15 min, 4°C, to obtain the nuclear fraction. Once centrifugated, the supernatant was collected and centrifugated at 100,000 g for 1 h at 4°C. The supernatant from this step corresponded to the cytosolic fraction. Cytosolic and nuclear fractions were quantified by the Pierce® BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL). Equal amounts of protein were resolved by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to PVDF membranes (Thermo Scientific), and blocked with 5% dry milk in Tris-buffered saline. Membranes were incubated overnight at 4°C with the following primary antibodies at 1:1,000 dilution: anti-Notch1 intracellular domain (N1ICD), anti-GAPDH as a loading control for cytosolic fraction, and anti-histone 3 (H3) as a nuclear fraction loading control (all from Cell Signaling Technology, Europe, Schuttersveld, Leiden, The Netherlands). The membranes were then exposed to HRP-conjugated secondary antibodies (1:10,000; Promega, Madison, WI) for 1 h at room temperature. Immunoreactivity was detected with SuperSignal™ WestFemto (Thermo Fisher Scientific, Waltham, MA) and densitometry was performed with ImageJ.
Immunoprecipitation assay
Cells were seeded onto Jagged1-coated plates to activate Notch signaling for 24 h in DMEM-low glucose supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were lysed with 1 mL of ionic buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 0.5% NP40, 0.5% sodium deoxycholate, and 0.05% SDS) supplemented with protease inhibitor cocktail, phosphatase inhibitor, and 10 μM MG132 (all from Roche). Protein A/G Plus-agarose (Santa Cruz Biotechnology, Santa Cruz, CA) and cleaved Notch1 (Val 1744; Cell Signaling Technology) were used for immunoprecipitation. Agarose beads were washed several times with cold Tris buffer containing 1 mM Tris, pH 7.4, and 15 mM NaCl (TBS) and blocked with 3% bovine serum albumin in TBS. Whole lysates were precleared with agarose beads and then incubated overnight (rocking at 4°C) with cleaved Notch1 antibody. Whole cell lysates were added onto new pellets of agarose beads (previously blocked with bovine serum albumin) and incubated for 3 h in a rocking wheel at 4°C. Finally, the supernatant was removed and beads were washed several times with cold 1 × TBS-T (TBS supplemented with 0.05% Tween). Proteins were eluted with 2 × Laemmli buffer supplemented with dithiothreitol and denatured for 5 min at 99°C.
Electrophoresis and western blotting
Cells were washed with PBS and lysed in RIPA buffer (1.5 M NaCl, 10% Nonidet P-40, 500 mM Tris-HCl, 5% deoxycholate, and 1% SDS, pH 8) supplemented with protease inhibitors, PMSF and leupeptin (Roche). Lysis was completed by five freeze–thaw cycles, followed by centrifugation at 12,000 g for 10 min 4°C. Protein concentration determination and electrophoresis were as described above. Membranes were incubated overnight at 4°C with the following primary antibodies (1:1,000 dilution): anti-Jagged1, anti-Jagged2, anti-Dll1, anti-Dll3, and anti-Dll4 (all from Cell Signaling Technology Europe, Schuttersveld, Leiden, The Netherlands), anticleaved Notch1 (A-8; Santa Cruz), and anti-HIF-1α (ab1; Abcam). An antibody to tubulin (T5158; Sigma) was used as a loading control. Exposure to secondary antibodies and immunoreactivity were performed as described above.
Immunocytochemistry and confocal microscopy
Cells in DMEM-low glucose medium were seeded onto 11-mm diameter slides. After 2 days, the slides were washed twice with PBS and cells were fixed with cold 70% ethanol for 10 min at 4°C. After three washes with PBS, the slides were incubated for 1 h at 37°C with blocking buffer consisting of PBS with 10% newborn goat serum (Gibco) and 0.01% Triton X-100 (Pronadisa, Basel, Switzerland). Cells were overlaid with the primary antibodies anti-HIF-1-α (H1alpha67, 1:200; Abcam) or antiactivated Notch1 (1:200; Abcam) diluted in blocking buffer and PBS (1:1) and incubated overnight at 4°C. The slides were then rinsed five times with PBS and overlaid with the secondary antibodies, Alexa Fluor 555 goat anti-rabbit and Alexa Fluor 647 goat anti-mouse (both 1:1,000; Invitrogen). Immunocytochemistry of negative controls was performed to discriminate between autofluorescence and specific signals. Slides were analyzed by vertical microscopy with a × 63 immersion objective (Leica 0.3). Quantification of fluorescence was performed using ImageJ.
InCell Analyzer quantification
N1ICD nuclear translocation was measured by immunolabeling. Cells were cultured in 24-well plates in triplicate in DMEM with 10% FBS and 1% penicillin/streptomycin and then fixed with 70% cold ethanol for 10 min at 4°C. Cells were labeled with antiactivated Notch1 antibody (ab8925; Abcam). The secondary antibody used was Alexa Fluor-555 (Invitrogen) and DAPI was used for nuclear labeling. Images were acquired with an InCell Analyzer 1000 epifluorescence microscope (GE Healthcare, Cardiff, United Kingdom). A 20 × objective was used to collect the fluorescence signals, and a combination of two excitation (EX) and emission (EM) filters was applied to detect DAPI (EX 405 nm/EM 450 nm) and red fluorescence from antiactivated Notch1 (EX 530 nm/EM 620 nm). Fifteen fields were acquired for each well. Analysis was performed with the InCell Analyzer 1000 Workstation software. Cells were first defined using the nuclear segmentation based on DAPI. The red signal was used to define N1ICD localization in the cell.
Statistical analysis
Data are expressed as mean ± standard error of mean or standard deviation as indicated. Comparisons were performed using unpaired Student's t-test. Statistical values were calculated using GraphPad Prism 6 software. Differences were considered statistically significant at P < 0.05 with a 95% confidence interval.
Results
HIF-1α overexpression induces protein and gene expression changes in MSCs
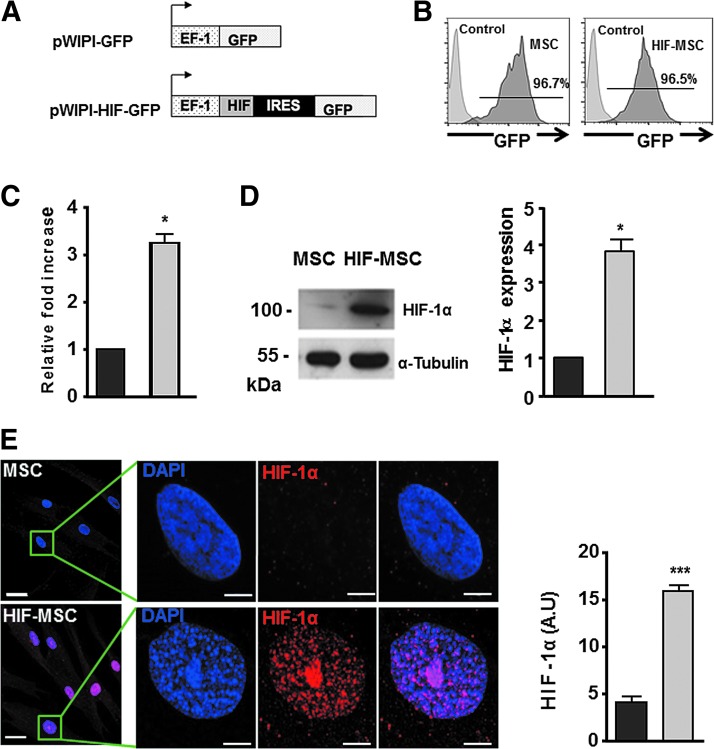
MSCs were transduced with pWPI-GFP (hereafter named MSC) or with pWPI-HIF-1α-GFP (hereafter named HIF-MSC) lentiviral vectors (Fig. 1A) as described [25] and cultured in normoxia. After infection, >95% of cultured cells were GFP positive (Fig. 1B). HIF-1α overexpression was confirmed by qPCR analysis (Fig. 1C) and western blotting (Fig. 1D), and confocal microscopy analysis showed nuclear localization of HIF-1α in HIF-MSCs (Fig. 1E).
FIG. 1.
HIF-1α overexpression in MSCs. (A) pWPI GFP and HIF-GFP lentiviral vectors. (B) Human dental pulp MSC lentiviral labeling with pWIPI-GFP (MSCs) and pWIPI-HIF-GFP (HIF-MSCs) detected by flow cytometric analysis of GFP. (C) HIF-1α gene expression levels detected by RT-qPCR in HIF-MSCs (gray) relative to MSCs (black) and expressed as mean ± SD of fold change. (D) Levels of HIF-1α protein as detected by western blotting with an anti-HIF-1α antibody and quantification by densitometry of exposed films in HIF-MSCs (gray) relative to MSCs (black); α-tubulin was used as a protein loading control. (E) Confocal microscopy (representative images) of MSCs and HIF-MSCs showing HIF-1α nuclear localization. Scale bar: 30 μm in upper and lower left panels and 5 μm in the remaining panels. Data represent mean ± SD of three independent experiments. *P < 0.05, ***P < 0.001. GFP, green fluorescent protein; HIF1α, hypoxia-inducible factor 1 alpha; MSCs, mesenchymal stem cells; RT-qPCR, real time quantitative PCR; SD, standard deviation.
HIF-1α overexpression in MSCs increases Notch signaling activation
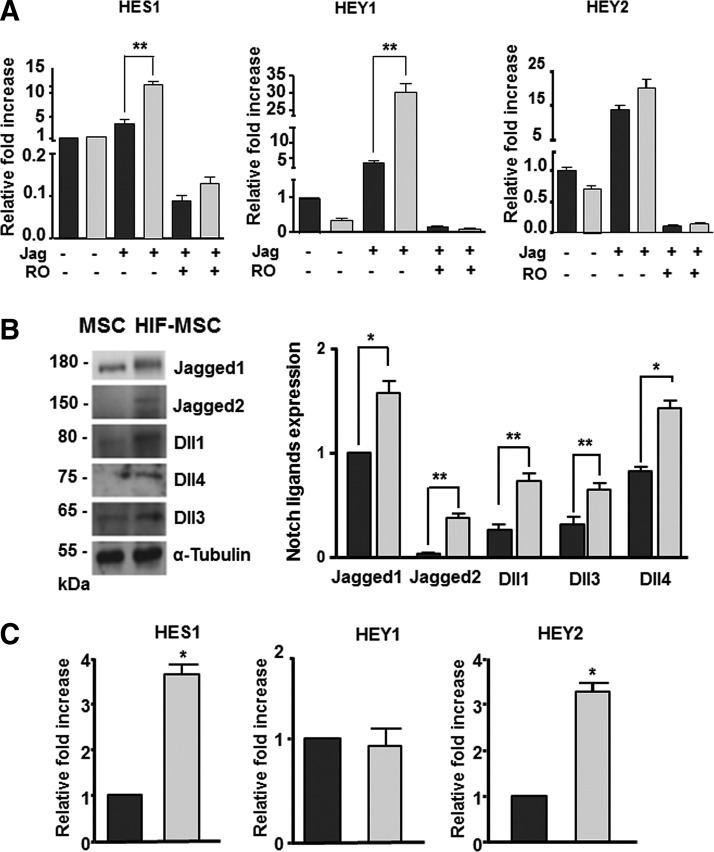
Crosstalk between HIF and Notch signaling has been recently described in some cancer cell lines [16,27], in which Notch relays the hypoxia signal into increased metastatic potential. To address whether a similar system was operating in MSCs, we assessed Notch signaling components in MSC overexpressing HIF-1α and cultured on Jagged1-coated plates at low confluence to prevent Notch activation by cell–cell contact. The mRNA expression levels of the Notch downstream targets, Hes1, Hey1, and Hey2, were significantly increased by Jagged1 stimulation both in MSCs (black bars) and in HIF-MSCs (gray bars), and this increase was higher in HIF-MSCs than in MSCs (Fig. 2A), suggesting a cooperative effect between HIF-1α and Notch signaling. Notch signaling was abolished by treatment with RO4929097, a selective gamma-secretase inhibitor that impairs Notch receptor processing and generation of NICD (Fig. 2A). To evaluate whether MSCs expressed Notch receptor ligands on the cell surface and whether these ligands were functional, we performed western blotting on MSCs and HIF-MSCs. Results showed that the Notch ligands Jagged1, Dll1, Dll3, and Dll4, but not Jagged2, were all expressed in MSCs and all were significantly increased in cells expressing HIF (Fig. 2B). To test if the ligands were functional and could activate Notch, we seeded HIF-MSCs and MSCs at confluence onto plates in the absence of Jagged1 and analyzed the expression of Notch downstream genes. Under these conditions, the expression of Hes1 and Hey2, but not Hey1, was significantly higher in HIF-MSCs than in MSCs (Fig. 2C). These results indicate that Notch signaling is activated in confluent HIF-MSCs and that more than one Notch ligand could be implicated in the process.
FIG. 2.
HIF-1 enhances Notch signaling in MSCs. (A) RT-qPCR analysis of Notch downstream genes Hes1, Hey1, and Hey2 in MSCs (black bars) and HIF-MSCs (gray bars) cultured with (+) or without (−) recombinant Jagged1 (Jag) and RO4929097 (gamma-secretase inhibitor) as negative control. Data represent mean ± SEM of three independent experiments. (B) Representative western blots and quantification of Notch ligands Jagged1, Jagged2, and Delta-like family (Dll1, Dll3, and Dll4) in MSCs and HIF-MSCs. Expression levels of Notch ligands were quantified by densitometry of exposed films (black bars for MSCs and gray bars for HIF-MSCs). Data represent mean ± SEM of three independent experiments. (C) RT-qPCR analysis of basal expression of principal downstream genes of Notch pathway signaling (Hes1, Hey1, and Hey2) in confluent cells after 24-h culture of MSCs (black bars) and HIF-MSCs (gray bars). *P < 0.05; **P < 0.01. SEM, standard error of mean.
Notch mediates migration and spreading in HIF-MSCs
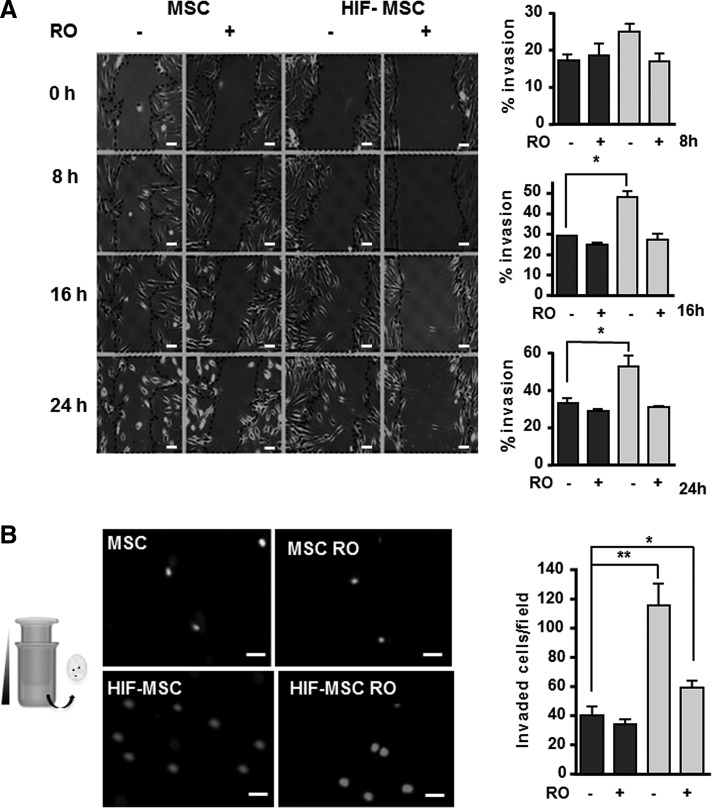
We have previously shown that HIF overexpression in bone marrow-derived MSCs stimulates their migration and invasion [25]. Given the well-documented role of Notch in migration and invasion in homeostasis and cancer [20,28], we investigated whether it participated in HIF-mediated spreading of MSCs. Overexpression of HIF-1α in MSCs significantly increased invasion, as measured by the scratch-wound assay, and promoted the development of lamellipodia and filopodia (Fig. 3A). The addition of RO4929097 to cultures blocked HIF-α-mediated invasion, which was similar to that of control MSCs (Fig. 3A). Likewise, the HIF-mediated increase in migration of HIF-MSCs through a gradient of IL-1β in the Transwell assay was abrogated by treatment with RO4929097, pointing to the participation of NICD in this process (Fig. 3B).
FIG. 3.
Increased migration of HIF-MSCs is dependent on Notch pathway activation. (A) Representative images of scratch-wound assays showing MSC and HIF-MSC migration at different time intervals in the presence or absence of RO4929097. Images were taken at 100 × magnification. Dotted lines define the wound area. Percentage of invasion was calculated at the indicated time intervals (black bars for MSCs and gray bars for HIF-MSCs). Scale bar: 100 μm (B) Migration toward trophic factor IL-1β in the presence or absence of RO4929097. Cells were grown on top of a Transwell membrane and the bottom chamber contained IL-1β at 25 ng/mL. After 8 h, cells that migrated through the membrane were stained with DAPI and counted (black bars for MSCs and gray bars for HIF-MSCs). Images were taken at 40 × magnification. Scale bar: 100 μm Data represent mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01.
HIF-mediated cell proliferation is dependent on SUMO, but not Notch
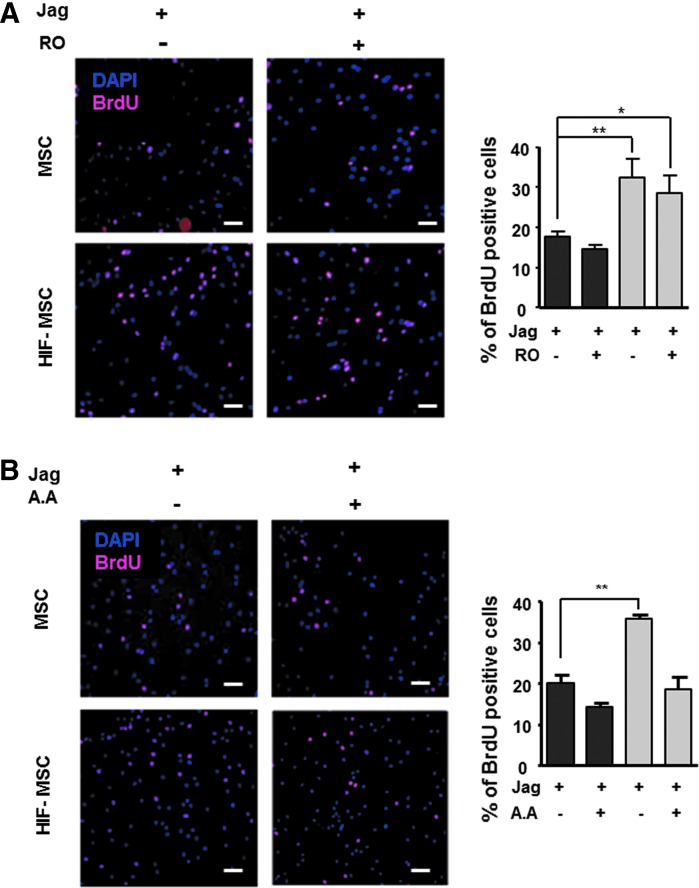
We next sought to evaluate the influence of Notch signaling for MSC proliferation. HIF-MSC and MSC cultures were seeded onto Jagged 1-coated plates to ensure Notch pathway activation and were incubated with BrdU for 8 h in the presence or absence of RO4929097. Subsequently, fixed cells were immunostained with anti-BrdU antibodies. Consistent with our previous results using bone marrow-derived MSCs [25], HIF-1α overexpression increased MSC proliferation as shown by a significant increase in BrdU incorporation; however, this effect was not abolished by RO4929097, indicating that HIF-induced cell proliferation is independent of Notch (Fig. 4A).
FIG. 4.
Increased proliferation of HIF-MSCs is Notch independent, but SUMO1 dependent. MSCs and HIF-MSCs were treated or not with 10 μM RO4929097 (RO) for 48 h (A) or with 50 μM anacardic acid (AA) for 4 h (B), and the proliferation rate was quantified by BrdU incorporation (black bars for MSCs and gray bars for HIF-MSCs). Images were taken at 100 × magnification. Scale bar: 100 μm Data represent mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01. BrdU, 5′-bromo-2′-deoxyuridine; SUMO, small ubiquitin-like modifier.
It has been reported that a fine balance between SUMOylation/deSUMOylation is required for activation of the hypoxia-signaling cascade, possibly through post-translation modifications to HIF-1α [29]. To question the potential role of SUMO in HIF-1α-mediated proliferation in MSCs, we repeated the BrdU incorporation study using 50 μM AA, an inhibitor of SUMO ligase. As expected, cell proliferation was significantly higher in HIF-MSCs than in MSCs; however, the HIF-1α-mediated increase in cell proliferation was completely abolished by AA treatment, demonstrating the relevance of the SUMO pathway for HIF-1α-induced MSC proliferation (Fig. 4B).
HIF and Notch pathways are linked by SUMOylation
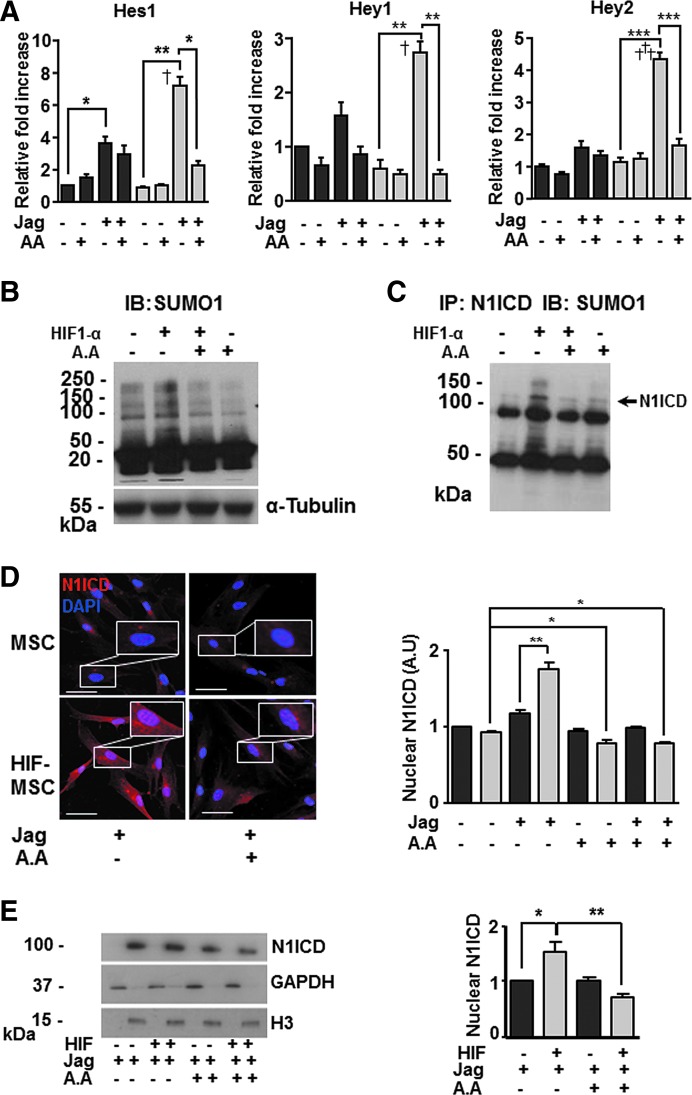
The above results suggested possible interplay between HIF, Notch, and SUMO pathways. To investigate these potential interactions, we first analyzed the influence of SUMO1 for the expression of Notch downstream genes. Treatment of MSCs and HIF-MSCs with AA, under conditions of Notch pathway activation, led to a decrease in the levels of expression of Hes1, Hey1, and Hey2 in both groups, but the decrease was significant only for HIF-MSCs (Fig. 5A). Moreover, global levels of SUMO were higher in HIF-MSCs than in MSCs as measured by western blotting (Fig. 5B), suggesting that HIF increases SUMO protein levels. Consistent with previous studies, these results suggested an association between HIF and SUMO pathways. To explore this functional relationship, we first analyzed the protein sequence of the Notch1 receptor with SUMOplot™ Analysis software, a program that predicts and scores SUMOylation sites in protein sequences. The results of this analysis showed six potential motifs with high probability of representing the SUMO binding site on the N1ICD (not shown). We next immunoprecipitated N1ICD in MSCs and HIF-MSCs with an anti-N1ICD antibody and performed western blotting for SUMO1. We observed that N1ICD was susceptible to SUMOylation and, moreover, levels of SUMOylated N1ICD were higher in HIF-MSCs than in MSCs (Fig. 5C). Additional analysis using an InCell Analyzer to quantify N1ICD nuclear translocation [30] confirmed that levels of N1ICD were significantly higher in the nuclei of Notch-activated HIF-MSCs than in equivalent MSCs and also showed that AA abolished N1ICD nuclear translocation (Fig. 5D). Overall, these results indicate a functional relationship between SUMO and Notch1, whereby cellular SUMO levels can affect N1ICD nuclear translocation. To further corroborate SUMOylation of N1ICD, we performed subcellular fractionation experiments and western blotting using MSCs and HIF-MSCs cultured on Jagged 1-coated plates and treated or not with AA to modulate SUMOylation. Analysis of N1ICD expression in nuclear fractions revealed a reduction in nuclear expression of the protein in the presence of the SUMO inhibitor (Fig. 5E).
FIG. 5.
HIF-1α and SUMO1 increase nuclear translocation of N1ICD. (A) RT-qPCR analysis of Notch downstream genes Hes1, Hey1, and Hey2 in MSCs and HIF-MSCs cultured in the presence (+) or absence (−) of Jagged1 (Jag) and AA. Data represent mean ± SEM of three independent experiments. (B) Analysis of SUMOylation levels in MSCs and HIF-MSCs in the presence (+) or absence (−) of AA. SUMOylation was detected with an anti-SUMO1 antibody. α-tubulin was used as a protein loading control. (C) Immunoprecipitation of N1ICD. MSC and HIF-MSC extracts from cultures seeded on plates coated with Jagged1 to ensure the pathway activation, treated (+) or not (−) with AA, were immunoprecipitated with an anti-N1ICD antibody, resolved by SDS-PAGE, and immunoblotted with anti-SUMO1 antibody. (D) Representative immunofluorescence images of N1ICD (red) in MSCs and HIF-MSCs cultured in plates coated with Jagged1 (24 h) with (+) or without (−) AA (4 h). N1ICD nuclear translocation is increased in HIF-MSCs, which is reversed by AA. Scale bar: 50 μm. Quantification was performed using an InCell Analyzer for measurement of fluorescence units in the nuclei. (E) Representative western blot of MSC and HIF-MSC subcellular fractionation. Cells were coated with Jagged1 to ensure the pathway activation and cells were treated (+) or not (−) with AA. Then, the amount of N1ICD in nuclei under different conditions was quantified by densitometry. GAPDH was used as a loading control of cytosolic proteins and histone 3 (H3) as a loading control of nuclear proteins. Data represent mean ± SEM of three independent experiments. IB denotes immunoblot and IP denotes immunoprecipitation. *P < 0.05; **P < 0.01, ***P < 0.001, †P < 0.05, †††P < 0.001. † denotes significance of HIF-MSCs versus MSCs in the presence of Jagged 1 (Jag+). Black bars for MSCs and gray bars for HIF-MSCs in panels A, D, and E. N1ICD, Notch1 intracellular domain; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
SUMOylation mediates HIF-induced Notch migration and invasiveness
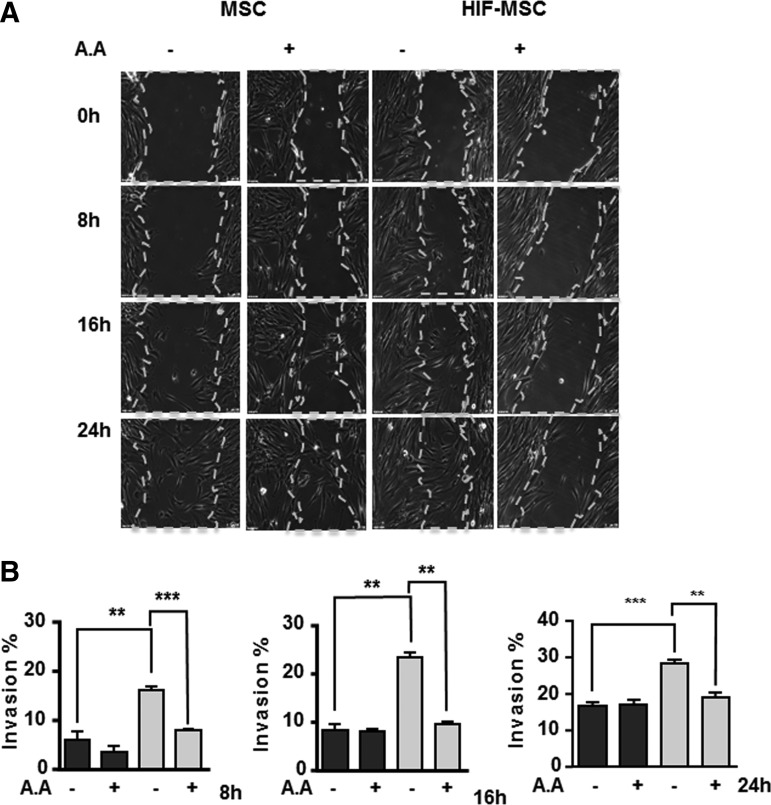
To test the functional effect of SUMOylation for cell invasion, we repeated the scratch-wound assays in HIF-MSC and MSC cultures using AA to inhibit SUMOylation. Results showed that AA treatment completely abolished the increase in migration of HIF-MSC cultures (Fig. 6A, B). These results establish a close relationship between the three pathways.
FIG. 6.
HIF-1α-enhanced cell migration is mediated by SUMO signaling. (A) Representative images of a time-lapse sequence of scratch-wound assays at different time intervals in MSCs and HIF-MSCs with cells treated (+) or not (−) with the SUMO inhibitor AA. Dotted lines define the wound area. (B) Percentage of invasion in MSCs (black bars) and HIF-MSCs (gray bars) with cells treated (+) or not (−) with AA calculated at indicated time intervals. Images were taken at 100 × magnification. Scale bar: 100 μm. Data represent mean ± SEM of four independent experiments. **P < 0.01; ***P < 0.001.
SUMOylation of N1ICD is increased by hypoxia in HEK293T cells
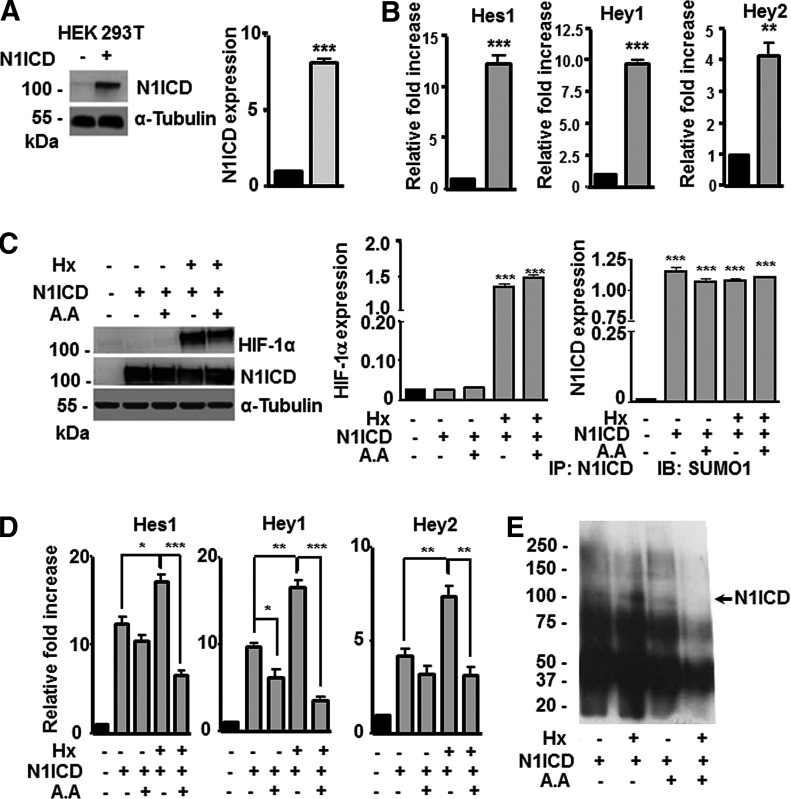
To validate the proposed model of N1ICD SUMOylation in other cell lines, we transiently transfected HEK293T cells with EF.hICN1.Ubc.GFP, a lentiviral vector encoding N1ICD (N1ICD-293T). Flow cytometry analysis showed that 95% of cells were infected with the vector. Subsequently, N1ICD expression was confirmed by western blotting using N1ICD antibodies and quantified by densitometry (Fig. 7A). In addition, genes downstream the Notch signaling pathway (Hes1, Hey1, and Hey2) were analyzed by RT-qPCR (Fig. 7B) and, as expected, were overexpressed in N1ICD-293T cells. Cells were then cultured under hypoxic conditions (1% O2) for 4 h in the presence or absence of AA (same conditions as used for MSCs) and expression of HIF-1α and N1ICD was evaluated by western blotting using anti-HIF-1α and antiactivated Notch1 antibodies and quantified by densitometry. As anticipated, hypoxia increased the steady-state levels of HIF-1α (Fig. 7C). Next, Hes1, Hey1, and Hey2 genes were analyzed by RT-qPCR and an increase in gene expression was observed in N1ICD-293T cells exposed to hypoxia. Moreover, when hypoxic N1ICD-293T cells were treated with AA, Hes1, Hey1, and Hey2 expression decreased significantly in comparison with the levels in N1ICD-293T cells in normoxia (treated or not with AA) (Fig. 7D). These results correlate with previous findings observed in MSCs, suggesting that the intracellular domain of the Notch1 receptor is SUMOylated, which increases its nuclear translocation, and that this SUMOylation is enhanced by hypoxia or by HIF-1α. Finally, to confirm SUMOylation of N1ICD, we performed immunoprecipitation of N1ICD in N1ICD-293T cells under normoxia/hypoxia and treated or not with AA. Protein extracts were immunoprecipitated with an anti-N1ICD antibody and SUMOylated proteins were detected using an anti-SUMO1 antibody. We observed that N1ICD was SUMOylated both in normoxia and hypoxia and that treatment with AA decreased this post-translational modification, especially under hypoxia (Fig. 7E).
FIG. 7.
HIF-1α and SUMO1 increase nuclear translocation of N1ICD in HEK293T cells. (A) Representative western blot and quantification of N1ICD by densitometry in transiently transfected HEK293T cells (black bars for HEK293T and gray bars for N1ICD-293T cells). Data represent mean ± SEM of three independent experiments. (B) RT-qPCR analysis of Notch downstream genes Hes1, Hey1, and Hey2 in HEK293T (black bars) and N1ICD-293T cells (gray bars). Data represent mean ± SEM of three independent experiments. (C) Representative western blot and quantification by densitometry of HIF-1α to assess hypoxia induction and N1ICD in HEK293T and N1ICD-293T cells treated or not with AA. (D) RT-qPCR analysis of Notch downstream genes Hes1, Hey1, and Hey2 in HEK293T and N1ICD-293T cultured in the presence (+) or absence (−) of hypoxia (Hx) and AA. Data represent mean ± SEM of three independent experiments. (E) Immunoprecipitation of N1ICD. HEK293T transduced with an N1ICD lentiviral vector (N1ICD +) cultured under normoxic or hypoxic conditions (Hx +/−) and treated or not with AA (+/−) were lysed and cell extracts were immunoprecipitated with an anti-N1ICD antibody, resolved by SDS-PAGE, and immunoblotted with anti-SUMO1 antibody. Molecular weights of protein markers are indicated at the left side of the panel in kDa. Data represent mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01, ***P < 0.001.
Discussion
MSCs can repair damaged tissues and we have previously demonstrated that infusion of MSC overexpressing HIF-1α promotes myocardial healing in an experimental rat model of myocardial infarction. As described by us and others, signaling pathways related to several paracrine factors and interleukins are upregulated in HIF-MSCs. For example, expression levels of p38 mitogen-activated protein kinase (p38/MAPK) [25,31], JNK/SAP [25], AKT [25], and Notch [32,33], together with fibronectin [34,35], fibroblast growth factor [25,36], angiopoietin-1 (ANGPT1) [37], NGF [38], and insulin growth factor [25,39] signaling pathways, are all induced by HIF-1α. We examined Notch signaling since hypoxia can promote Notch pathway activation and HIF-1α is a key mediator in the adaptation of eukaryotic cells to hypoxia. We first compared Notch downstream target gene expression in HIF-MSCs and MSCs. Activation of Notch signaling in HIF-MSCs was observed when HIF-MSCs were cultured at confluence or seeded at low density on Jagged1-coated plates, indicating that HIF-1α enhanced Notch signaling only after activation of the Notch receptor. This is in accordance with previous studies showing that hypoxia increases activation levels of Notch1 in different tumor cell lines only after coculture with a Jagged1-expressing cell line [27].
We next analyzed the contribution of the Notch pathway to HIF-MSC migration and proliferation since Notch is a major participant in these processes [16,27,40]. We found that migration and invasiveness, but not proliferation, were dependent on Notch signaling. The contribution of HIF-1α to enhanced Notch-dependent migration and invasiveness has been previously described in choriocarcinoma cell lines, suggesting that this is a general mechanism in normal and tumor cells [16]. However, the observation that increased cell proliferation in HIF-MSCs was Notch independent pointed to the participation of other mechanisms and led us to explore alternative pathways. In this regard, a relationship has been described between HIF-1α and the ubiquitin family member, SUMO-1 [41,42]. In contrast to ubiquitination, SUMO-1 modifications enhance the stability of nuclear proteins [43]. Previous studies have shown that HIF-1α undergoes post-translational modification by the three isoforms of SUMO (SUMO-1, -2, and -3) in vitro [44]. Increased SUMO-1 mRNA and protein level have been reported after hypoxic stimulation in various cell lines [45] and in vivo [42]. Moreover, SUMOylation of HIF-1α results in its stabilization and increases its transcriptional activity [41]. To investigate whether SUMOylation was involved in HIF/Notch crosstalk in MSCs, we cultured cells in the presence or absence of AA, a SUMOylation inhibitor. Under these conditions, we observed that both proliferation and migration were abolished and this effect was stronger in HIF-MSCs. Western blotting showed an increase in global SUMO levels in HIF-MSCs. Moreover, immunoprecipitation studies showed higher amounts of SUMOylated N1ICD in HIF-MSCs. Immunofluorescence labeling, followed by InCell quantification, showed decreased nuclear translocation of NICD in HIF-MSCs in the presence of AA, indicating that SUMOylation of N1ICD favors the nuclear translocation of this transcriptional factor and thus confirming that enhanced SUMO pathway by HIF-1α overexpression might contribute to the differential activation of the Notch pathway in MSC-HIF with respect to MSCs.
Several mechanisms may explain the increased SUMOylation of N1ICD in HIF-MSCs. First, it has been described that HIF-1α can increase γ-secretase activity, which would increase N1ICD cleavage leading to higher cytosolic concentrations [46]. Second, the global increase in cytosolic SUMO levels in HIF-MSCs could favor N1ICD SUMOylation. Third, SUMOylated N1ICD could be more easily translocated to the nucleus since it is known that protein SUMOylation promotes nuclear localization [30,47].
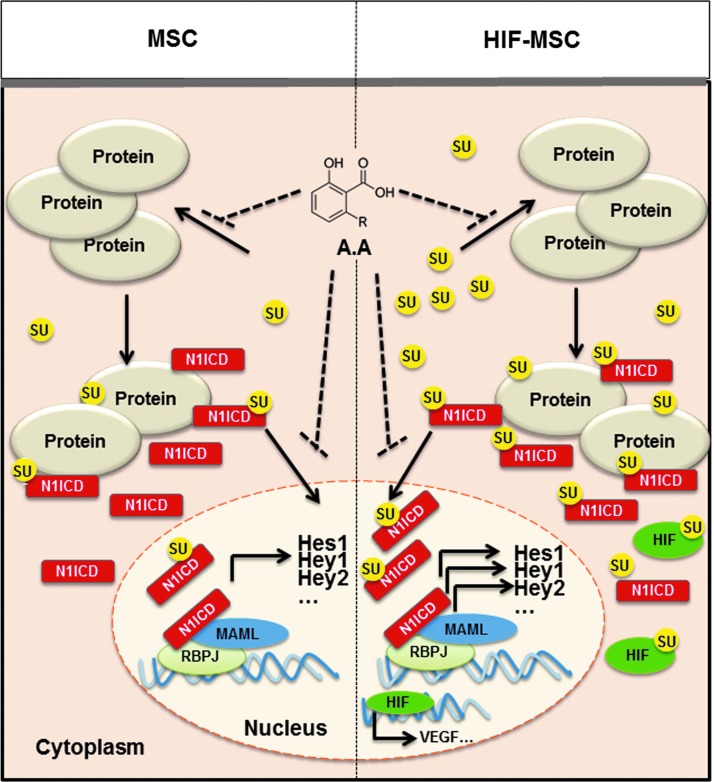
The effect of HIF-1α stabilization on MSCs is summarized in the model proposed in Fig. 8. Overexpression of HIF-1α leads to an increase in N1ICD levels by stabilization of the transcription factor, increasing its half-life, or by enhancing gamma-secretase enzyme activity [46,48,49]. Concurrently, HIF-1α increases the cellular amounts of SUMO1, which is reflected by an increase in the SUMOylation of N1ICD, a phenomenon described for the first time here. This process together with N1ICD nuclear translocation is important for cellular proliferation and migration, as well as for tumoral processes where the independent roles of SUMO, HIF, and Notch are well described. With this model, we are able to explain how HIF-1α acts as a potent enhancer of Notch transcriptional activity, enhancing the therapeutic properties of hMSCs through SUMO1. Moreover, we establish a link between these two highly conserved pathways and the key role of post-translational SUMO modification, which is implicated in tissue renewal, regeneration, proliferation, migration, invasiveness, and tumorigenesis.
FIG. 8.
Proposed model for HIF-1α, Notch, and SUMO crosstalk in hMSCs. Under normoxic conditions, HIF-1α undergoes ubiquitin–proteasomal-mediated degradation, while N1ICD is SUMOylated at a low level. Under hypoxic conditions or after HIF stabilization, levels of SUMO1 are higher and SUMOylation of proteins, including N1ICD, is elevated, which results in increased N1ICD translocation and increased Notch transcriptional activity, leading to enhanced cell migration and proliferation. SUMO1-increased levels by HIF-1α stabilization increased the amount of SUMOylated proteins, including N1ICD. RBPJ, recombining binding protein suppressor of hairless; MAML1: mastermind-like protein-1.
In summary, we demonstrate at the molecular and cellular levels the mechanism underlying HIF-SUMO-Notch crosstalk in hMSCs and how this interaction results in (i) autoactivation of Notch signaling pathway, (ii) increased Notch ligand expression, (iii) increased levels of SUMO and SUMOylation of N1ICD, (iv) increased N1ICD nuclear translocation, and (v) increased migration, invasiveness, and proliferation of HIF-MSCs. The relationship between the Notch pathway and cancer is well recognized [50]. Thus, this novel level of regulation for N1ICD could have potential implications in cancer pathogenesis. Further studies are needed to identify the residues SUMOylated in N1ICD and to elucidate whether higher concentrations of N1ICD by increased γ-secretase activity or higher SUMO-mediated nuclear translocation of N1ICD account for the enhanced Notch activity in HIF-MSCs.
Acknowledgments
This work was supported, in part, by grants from the Instituto de Salud Carlos III PI13/00414, PI16/0107, RETICS RD12/0019/0025 to P.S. and RETICS RD12/0019/0003 (TERCEL) to J.L.D.L.P cofunded by FEDER “una manera de hacer Europa.” It was also supported by the Regenerative Medicine Program of Instituto de Salud Carlos III and Valencian Community to Centro de Investigación Principe Felipe. The authors are grateful to Dr. A. Dorronsoro for critical review of the work and Dr. K McCreath for manuscript editing.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Arminan A, Gandia C, Garcia-Verdugo JM, Lledo E, Trigueros C, Ruiz-Sauri A, Minana MD, Solves P, Paya R, Montero JA. and Sepulveda P. (2010). Mesenchymal stem cells provide better results than hematopoietic precursors for the treatment of myocardial infarction. J Am Coll Cardiol 55:2244–2253 [DOI] [PubMed] [Google Scholar]
- 2.Gandia C, Arminan A, Garcia-Verdugo JM, Lledo E, Ruiz A, Minana MD, Sanchez-Torrijos J, Paya R, Mirabet V, et al. (2008). Human dental pulp stem cells improve left ventricular function, induce angiogenesis, and reduce infarct size in rats with acute myocardial infarction. Stem Cells 26:638–645 [DOI] [PubMed] [Google Scholar]
- 3.Li S, Wang X, Li J, Zhang J, Zhang F, Hu J, Qi Y, Yan B. and Li Q. (2016). Advances in the treatment of ischemic diseases by mesenchymal stem cells. Stem Cells Int 2016:5896061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noh HB, Ahn HJ, Lee WJ, Kwack K. and Do Kwon Y. (2010). The molecular signature of in vitro senescence in human mesenchymal stem cells. Genes Genomics 32:87–93 [Google Scholar]
- 5.Tsai CC, Chen YJ, Yew TL, Chen LL, Wang JY, Chiu CH. and Hung SC. (2011). Hypoxia inhibits senescence and maintains mesenchymal stem cell properties through down-regulation of E2A-p21 by HIF-TWIST. Blood 117:459–469 [DOI] [PubMed] [Google Scholar]
- 6.Basciano L, Nemos C, Foliguet B, de Isla N, de Carvalho M, Tran N. and Dalloul A. (2011). Long term culture of mesenchymal stem cells in hypoxia promotes a genetic program maintaining their undifferentiated and multipotent status. BMC Cell Biol 12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fehrer C, Brunauer R, Laschober G, Unterluggauer H, Reitinger S, Kloss F, Gully C, Gassner R. and Lepperdinger G. (2007). Reduced oxygen tension attenuates differentiation capacity of human mesenchymal stem cells and prolongs their lifespan. Aging Cell 6:745–757 [DOI] [PubMed] [Google Scholar]
- 8.Greijer AE, van der Groep P, Kemming D, Shvarts A, Semenza GL, Meijer GA, van de Wiel MA, Belien JA, van Diest PJ. and van der Wall E. (2005). Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1). J Pathol 206:291–304 [DOI] [PubMed] [Google Scholar]
- 9.Arnesen T, Kong X, Evjenth R, Gromyko D, Varhaug JE, Lin Z, Sang N, Caro J. and Lillehaug JR. (2005). Interaction between HIF-1 alpha (ODD) and hARD1 does not induce acetylation and destabilization of HIF-1 alpha. FEBS Lett 579:6428–6432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ockaili R, Natarajan R, Salloum F, Fisher BJ, Jones D, Fowler AA., 3rd and Kukreja RC. (2005). HIF-1 activation attenuates postischemic myocardial injury: role for heme oxygenase-1 in modulating microvascular chemokine generation. Am J Physiol Heart Circ Physiol 289:H542–H548 [DOI] [PubMed] [Google Scholar]
- 11.Zheng X, Linke S, Dias JM, Zheng X, Gradin K, Wallis TP, Hamilton BR, Gustafsson M, Ruas JL, et al. (2008). Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc Natl Acad Sci U S A 105:3368–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U. and Bondesson M. (2005). Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell 9:617–628 [DOI] [PubMed] [Google Scholar]
- 13.Sainson RC. and Harris AL. (2006). Hypoxia-regulated differentiation: let's step it up a Notch. Trends Mol Med 12:141–143 [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, De Marco MA, Graziani I, Gazdar AF, Strack PR, Miele L. and Bocchetta M. (2007). Oxygen concentration determines the biological effects of NOTCH-1 signaling in adenocarcinoma of the lung. Cancer Res 67:7954–7959 [DOI] [PubMed] [Google Scholar]
- 15.Fang Y, Yu S, Ma Y, Sun P, Ma D, Ji C. and Kong B. (2013). Association of Dll4/notch and HIF-1a-VEGF signaling in the angiogenesis of missed abortion. PLoS One 8:e70667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian Q, Xue Y, Zheng W, Sun R, Ji W, Wang X. and An R. (2015). Overexpression of hypoxia-inducible factor 1alpha induces migration and invasion through Notch signaling. Int J Oncol 47:728–738 [DOI] [PubMed] [Google Scholar]
- 17.Diez H, Fischer A, Winkler A, Hu CJ, Hatzopoulos AK, Breier G. and Gessler M. (2007). Hypoxia-mediated activation of Dll4-Notch-Hey2 signaling in endothelial progenitor cells and adoption of arterial cell fate. Exp Cell Res 313:1–9 [DOI] [PubMed] [Google Scholar]
- 18.Olsauskas-Kuprys R, Zlobin A. and Osipo C. (2013). Gamma secretase inhibitors of Notch signaling. Onco Targets Ther 6:943–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sassoli C, Pini A, Mazzanti B, Quercioli F, Nistri S, Saccardi R, Zecchi-Orlandini S, Bani D. and Formigli L. (2011). Mesenchymal stromal cells affect cardiomyocyte growth through juxtacrine Notch-1/Jagged-1 signaling and paracrine mechanisms: clues for cardiac regeneration. J Mol Cell Cardiol 51:399–408 [DOI] [PubMed] [Google Scholar]
- 20.Bolos V, Grego-Bessa J. and de la Pompa JL. (2007). Notch signaling in development and cancer. Endocr Rev 28:339–363 [DOI] [PubMed] [Google Scholar]
- 21.D'Souza B, Miyamoto A. and Weinmaster G. (2008). The many facets of Notch ligands. Oncogene 27:5148–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier-Stiegen F, Schwanbeck R, Bernoth K, Martini S, Hieronymus T, Ruau D, Zenke M. and Just U. (2010). Activated Notch1 target genes during embryonic cell differentiation depend on the cellular context and include lineage determinants and inhibitors. PLoS One 5:e11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licciardello MP, Mullner MK, Durnberger G, Kerzendorfer C, Boidol B, Trefzer C, Sdelci S, Berg T, Penz T, et al. (2015). NOTCH1 activation in breast cancer confers sensitivity to inhibition of SUMOylation. Oncogene 34:3780–3790 [DOI] [PubMed] [Google Scholar]
- 24.Geiss-Friedlander R. and Melchior F. (2007). Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8:947–956 [DOI] [PubMed] [Google Scholar]
- 25.Cerrada I, Ruiz-Sauri A, Carrero R, Trigueros C, Dorronsoro A, Sanchez-Puelles JM, Diez-Juan A, Montero JA. and Sepulveda P. (2013). Hypoxia-inducible factor 1 alpha contributes to cardiac healing in mesenchymal stem cells-mediated cardiac repair. Stem Cells Dev 22:501–511 [DOI] [PubMed] [Google Scholar]
- 26.Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H, Kimura K, Sodeoka M. and Yoshida M. (2009). Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol 16:133–140 [DOI] [PubMed] [Google Scholar]
- 27.Sahlgren C, Gustafsson MV, Jin S, Poellinger L. and Lendahl U. (2008). Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci U S A 105:6392–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto-Torii K, Torii M, Sarkisian MR, Bartley CM, Shen J, Radtke F, Gridley T, Sestan N. and Rakic P. (2008). Interaction between Reelin and Notch signaling regulates neuronal migration in the cerebral cortex. Neuron 60:273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nunez-O'Mara A. and Berra E. (2013). Deciphering the emerging role of SUMO conjugation in the hypoxia-signaling cascade. Biol Chem 394:459–469 [DOI] [PubMed] [Google Scholar]
- 30.Seeler JS. and Dejean A. (2003). Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol 4:690–699 [DOI] [PubMed] [Google Scholar]
- 31.Sodhi CP, Batlle D. and Sahai A. (2000). Osteopontin mediates hypoxia-induced proliferation of cultured mesangial cells: role of PKC and p38 MAPK. Kidney Int 58:691–700 [DOI] [PubMed] [Google Scholar]
- 32.Cejudo-Martin P. and Johnson RS. (2005). A new notch in the HIF belt: how hypoxia impacts differentiation. Dev Cell 9:575–576 [DOI] [PubMed] [Google Scholar]
- 33.Pear WS. and Simon MC. (2005). Lasting longer without oxygen: the influence of hypoxia on Notch signaling. Cancer Cell 8:435–437 [DOI] [PubMed] [Google Scholar]
- 34.Distler JH, Jungel A, Huber LC, Schulze-Horsel U, Zwerina J, Gay RE, Michel BA, Hauser T, Schett G, Gay S. and Distler O. (2007). Imatinib mesylate reduces production of extracellular matrix and prevents development of experimental dermal fibrosis. Arthritis Rheum 56:311–322 [DOI] [PubMed] [Google Scholar]
- 35.Lee SH, Lee YJ, Song CH, Ahn YK. and Han HJ. (2010). Role of FAK phosphorylation in hypoxia-induced hMSCS migration: involvement of VEGF as well as MAPKS and eNOS pathways. Am J Physiol Cell Physiol 298:C847–C856 [DOI] [PubMed] [Google Scholar]
- 36.Egger M, Schgoer W, Beer AG, Jeschke J, Leierer J, Theurl M, Frauscher S, Tepper OM, Niederwanger A, et al. (2007). Hypoxia up-regulates the angiogenic cytokine secretoneurin via an HIF-1alpha- and basic FGF-dependent pathway in muscle cells. FASEB J 21:2906–2917 [DOI] [PubMed] [Google Scholar]
- 37.Winter SF, Acevedo VD, Gangula RD, Freeman KW, Spencer DM. and Greenberg NM. (2007). Conditional activation of FGFR1 in the prostate epithelium induces angiogenesis with concomitant differential regulation of Ang-1 and Ang-2. Oncogene 26:4897–4907 [DOI] [PubMed] [Google Scholar]
- 38.Nakamura K, Tan F, Li Z. and Thiele CJ. (2011). NGF activation of TrkA induces vascular endothelial growth factor expression via induction of hypoxia-inducible factor-1alpha. Mol Cell Neurosci 46:498–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akeno N, Robins J, Zhang M, Czyzyk-Krzeska MF. and Clemens TL. (2002). Induction of vascular endothelial growth factor by IGF-I in osteoblast-like cells is mediated by the PI3K signaling pathway through the hypoxia-inducible factor-2alpha. Endocrinology 143:420–425 [DOI] [PubMed] [Google Scholar]
- 40.Gaiano N. (2008). Strange bedfellows: Reelin and Notch signaling interact to regulate cell migration in the developing neocortex. Neuron 60:189–191 [DOI] [PubMed] [Google Scholar]
- 41.Bae SH, Jeong JW, Park JA, Kim SH, Bae MK, Choi SJ. and Kim KW. (2004). Sumoylation increases HIF-1alpha stability and its transcriptional activity. Biochem Biophys Res Commun 324:394–400 [DOI] [PubMed] [Google Scholar]
- 42.Shao R, Zhang FP, Tian F, Anders Friberg P, Wang X, Sjoland H. and Billig H. (2004). Increase of SUMO-1 expression in response to hypoxia: direct interaction with HIF-1alpha in adult mouse brain and heart in vivo. FEBS Lett 569:293–300 [DOI] [PubMed] [Google Scholar]
- 43.Desterro JM, Rodriguez MS. and Hay RT. (1998). SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell 2:233–239 [DOI] [PubMed] [Google Scholar]
- 44.Berta MA, Mazure N, Hattab M, Pouyssegur J. and Brahimi-Horn MC. (2007). SUMOylation of hypoxia-inducible factor-1alpha reduces its transcriptional activity. Biochem Biophys Res Commun 360:646–652 [DOI] [PubMed] [Google Scholar]
- 45.Comerford KM, Leonard MO, Karhausen J, Carey R, Colgan SP. and Taylor CT. (2003). Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc Natl Acad Sci U S A 100:986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villa JC, Chiu D, Brandes AH, Escorcia FE, Villa CH, Maguire WF, Hu CJ, de Stanchina E, Simon MC, et al. (2014). Nontranscriptional role of Hif-1alpha in activation of gamma-secretase and notch signaling in breast cancer. Cell Rep 8:1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson VG. and Rangasamy D. (2001). Intracellular targeting of proteins by sumoylation. Exp Cell Res 271:57–65 [DOI] [PubMed] [Google Scholar]
- 48.Cheng YL, Park JS, Manzanero S, Choi Y, Baik SH, Okun E, Gelderblom M, Fann DY, Magnus T, et al. (2014). Evidence that collaboration between HIF-1alpha and Notch-1 promotes neuronal cell death in ischemic stroke. Neurobiol Dis 62:286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pistollato F, Rampazzo E, Persano L, Abbadi S, Frasson C, Denaro L, D'Avella D, Panchision DM, Della Puppa A, Scienza R. and Basso G. (2010). Interaction of hypoxia-inducible factor-1alpha and Notch signaling regulates medulloblastoma precursor proliferation and fate. Stem Cells 28:1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ranganathan P, Weaver KL. and Capobianco AJ. (2011). Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer 11:338–351 [DOI] [PubMed] [Google Scholar]