Abstract
In eukaryotic cells, transport of molecules between the nucleus and the cytoplasm is facilitated by highly selective and efficient biomachines known as nuclear pore complexes (NPCs). The structural details of NPCs vary across species, with many of their constituent proteins exhibiting relatively low sequence conservation; yet the NPC as a whole retains its general architecture and mechanism of action in all eukaryotes from yeast to humans. This functional conservation in the absence of precise molecular conservation suggests that many aspects of the NPC transport mechanism may be understood based on general biophysical considerations. Accordingly, some aspects of NPC function have been recapitulated in artificial nanochannel mimics, even though they lack certain molecular elements of the endogenous NPC. Herein, we review biophysical aspects of NPC architecture and function and cover recent progress in the field. We also review recent advances in man-made molecular filters inspired by NPCs, and their applications in nanotechnology. We conclude the review with an outlook on outstanding questions in the field and biomedical aspects of NPC transport.
Main Text
What is a nuclear pore complex?
In eukaryotic cells, genetic material is compartmentalized inside the nucleus, which is separated from the cytoplasm by the double membrane nuclear envelope (NE) (1). The two compartments can communicate through gateways embedded in the NE, known as nuclear pore complexes (NPCs), which form passageways through the NE to connect the nucleus and the cytoplasm (1). These highly selective and efficient conduits facilitate the bidirectional nucleocytoplasmic transport of a wide range of cargo molecules and regulate a diverse array of fundamental cellular processes. Below we discuss the advances in understanding NPC organization and function—from a brief historical introduction to current research. For further details and original historical articles, we refer the reader to the following reviews (2, 3, 4, 5).
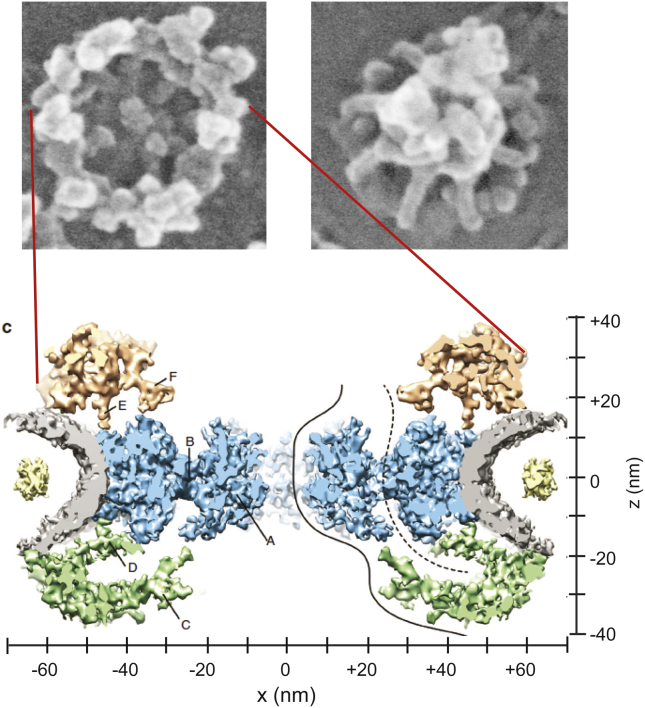
In the 1950s, electron microscopy (EM) was combined with advanced preparation techniques and applied to amphibian cells, providing the first glimpses of both the structure of the NE and the existence of NPCs (6). In the next two decades, extensive EM studies in eukaryotic cells determined the overall shape and dimensions of the NPC, and showed that it has an eightfold radial symmetry (Fig. 1). Several critical tools—including the purification of NE from rat liver, production of monoclonal antibodies against NPC components, and advanced proteomics approaches—enabled identification of the molecular constituents of the NPC in multiple species, and their approximate locations within the complex (7, 8).
Figure 1.
Shown here is the electron microscopy structure of the NPC. (Upper panel) Given here is the electron microscopy image of the cytoplasmic (left) and the nucleocytoplasmic (right) openings of the NPC imaged in Xenopus oocyte; adapted from Goldberg et al. (15) (Springer-Verlag with permission). (Lower panel) Given here is a cryo-TEM reconstruction of the NPC vertical cross-section in Xenopus oocyte (nuclear basket is not shown) (16). The nuclear envelope is shown in gray. Structural proteins forming the central ring are shown in blue, whereas those forming the distal rings are shown in beige and green. To see this figure in color, go online.
Currently, the NPC has been established as the cell’s largest macromolecular complex, with a molecular mass ranging from ∼50 MDa in yeast to ∼112 MDa in vertebrates (5, 9, 10, 11). It is composed of multiple copies of ∼30 different proteins known as “nucleoporins” (3, 4). Transmembrane nucleoporins anchor the NPC in the NE, whereas structural nucleoporins form a scaffold that shapes an hourglass passageway with a diameter ranging from 35 nm in yeast to 50 nm in vertebrates (5, 9, 10, 11). The lumen of this passageway is filled with an assembly of intrinsically disordered nucleoporins harboring repeats of hydrophobic phenylalanine (F) and glycine (G) motifs—hence known as FG nucleoporins or FG nups. FG nup assembly is a critical component of the NPC transport mechanism (3, 4). Recently, localizations of different types of nucleoporins within the NPC have been refined using a combination of biochemistry/molecular biology techniques, powerful biophysical approaches (e.g., (cryo)EM, x-ray crystallography, and superresolution microscopy), and integrative computational biology methods (9, 10, 12, 13, 14, 15, 16). These approaches have provided structural maps of the NPC. However, details about FG nup localizations and morphologies are still limited because the FG nups are disordered in nature, and few experimental methods can probe their conformations at relevant length and timescales.
In parallel with investigating NPC organization and structure, significant effort has been made to understand its function as a nucleocytoplasmic transporter. Research in the 1970s using Xenopus oocytes established that NPCs allow small molecules (up to ∼40 kDa) to pass through without discrimination but translocate large molecules with high selectivity. Subsequent studies in the 1980s and 1990s identified specific signal sequences within cargo molecules allowing them to enter or exit the nucleus—known as nuclear localization sequences (NLS) and nuclear export sequences (NES) (2, 4). Around this time, the newly developed digitonin-permeabilized cell assay in vertebrate cells (17) was used to identify transport proteins—also known as Karyopherins, Importins, Exportins, and Transportins—that bind cargo molecules via NLS/NES and ferry them through the NPC (reviewed in (4); see also (18)). It was established that this directional transport depends on GTP hydrolysis and the nucleocytoplasmic gradient of the small GTPase Ran (4, 19). The RanGTP/GDP gradient is maintained by the guanine nucleotide exchange factor RanGEF (localized in the nucleus), GTPase-activating protein RanGAP1 (localized in the cytoplasm), and a soluble transport protein Nuclear Transport Factor 2 (NTF2) (3, 4).
Strikingly, both the general mechanism of NPC function and the blueprint of its overall architecture are mainly conserved among species, from yeast to humans (8). In particular, NPCs in investigated species contain passageways lined with FG nups. However, molecular details exhibit considerable interspecies variation. This includes the yeast NPC being significantly smaller than the human NPC (5, 9, 10, 11), and the relatively low sequence conservation of FG nups (8). Nevertheless, the degree of functional conservation is so high that: 1) transport proteins (NTF2) from one species (humans) are operational in other species (yeast) (20), and 2) chimeras containing yeast FG nups can restore selective barrier function in Xenopus FG nup deletion mutants (21). This suggests that many aspects of NPC function may be understood based on general biophysical considerations. The following discussion reviews such biophysical aspects of NPC function with an emphasis on protein import, which has served as a test bed for uncovering the biophysical transport mechanisms.
Simple biophysics of the NPC
What drives directional transport through the NPC?
Macroscopically, NPCs can efficiently import and export macromolecular cargoes into and out of the nucleus against the cargoes’ apparent concentration gradients. From the thermodynamic standpoint, this requires energy input, which is provided by GTP hydrolysis during the transport cycle (4, 22). In many other types of transporters (such as ion exchangers or proton pumps), GTP/ATP hydrolysis is directly coupled to directional substrate transport via conformational changes of the transporter (1, 23). By contrast, NPCs do not appear to possess gates transitioning from open to closed states (Fig. 2). Translocation of individual transport protein-cargo complexes is not directly coupled to GTP hydrolysis (4) and occurs by thermal diffusion that is modulated by the interactions with the FG nups (24, 25, 26, 27). Accordingly, transport protein-cargo complexes can move in both directions through the NPC, which often results in abortive translocations, as observed by single-molecule fluorescence microscopy and bulk flux measurements (28, 29, 30). Although the main function of the transport proteins is to ferry cargo, they can also translocate through the NPC without cargo. Multiple copies of cargo-bound and cargo-free transport proteins have been found in the NPC; they may play a role in shaping the NPC structure (4, 28, 31, 32, 33, 34, 35).
Figure 2.
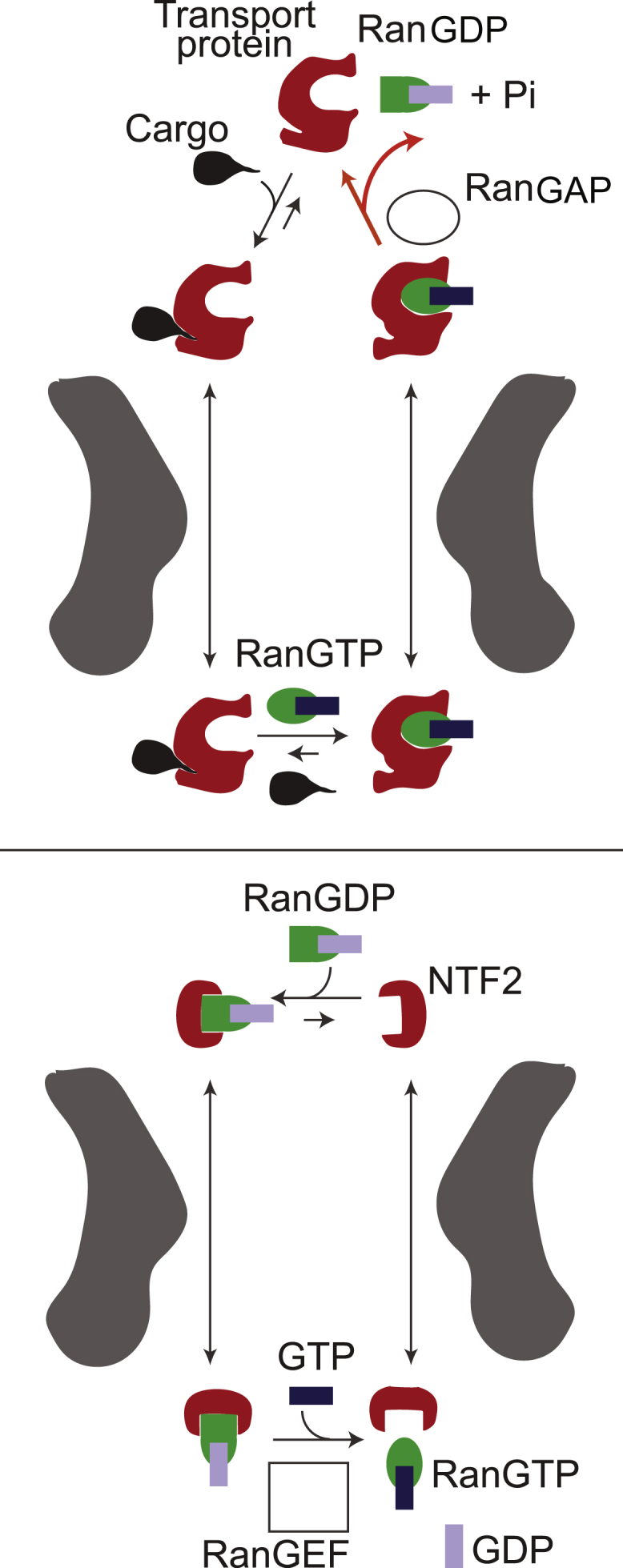
Given here are simplified schematics of the NPC operating cycle. The NPC import process comprises two interlinked cycles. The first, shown in the upper panel, uses the energy released by the hydrolysis of one GTP molecule (catalyzed by RanGAP) to import one cargo molecule into the nucleus and relies on the higher concentration of RanGTP in the nucleus relative to the cytoplasm. The red line is the only nonequilibrium step of the import cycle that depends on the metabolic energy release in the form of GTP hydrolysis. The nucleocytoplasmic RanGTP/RanGDP gradient is maintained by the second cycle, shown in the lower panel, which relies on the high concentration of RanGEF in the nucleus due to its association with chromatin. To see this figure in color, go online.
In the canonical import pathway (Fig. 2), transport proteins bind their cargoes in the cytoplasm and carry them through the NPC passageway. Once the transport protein-cargo complex reaches the nucleus, nuclear factor RanGTP binds the transport protein; as a result, the cargo is released and sequestered in the nucleus. The resulting RanGTP-transport protein complex is capable of translocating through the NPC back into the cytoplasm, where GTP is hydrolyzed by the cytoplasmic factor RanGAP1. Energy released by the conversion of RanGTP to RanGDP leads to a conformational change that sets RanGDP free from the transport protein. The cycle is completed through the import of RanGDP back into the nucleus by a specialized transport protein NTF2. In the nucleus, RanGDP is converted to RanGTP by a chromatin-associated guanine exchange factor RanGEF. Overall, directional transport is driven by the energy released from irreversible GTP hydrolysis in the cytoplasm (22) and maintained by the localization gradient of RanGTP; RanGTP concentration is higher in the nucleus relative to the cytoplasm. In turn, this gradient relies on the constitutive localizations of RanGEF in the nucleus, RanGAP1 in the cytoplasm, and the NTF2-mediated transport of RanGDP (4). The canonical NPC protein export cycle operates in a similar fashion and is reviewed in Cautain et al. (4); see Grünwald et al. (36) for the review of mRNA export.
Why is NPC selective?
Small molecules, such as ATP and ions, freely diffuse through the NPC. In contrast, the ability of macromolecules (or similarly sized nanoparticles) to cross the NPC unaided decreases as their size increases: those >∼40 kDa can efficiently translocate though the NPC only when they are bound to a transport protein or can interact directly with the FG nups (4, 37, 38). The NPC is able to recognize these cognate transport proteins and selectively translocate them despite the vast numbers of other macromolecules present in the cell that they could potentially interact with the NPC non-specifically. There are two main tiers of recognition: 1) the first tier is based on binding of the cargoes (directly or via adaptor proteins) to the transport proteins through a recognition sequence—NLS for import or NES for export; and 2) the second tier relies on the interactions between the transport proteins and the FG nups (3, 4). The NPC is not unique in this regard—molecular recognition commonly relies on the interactions between the transported molecules and the transporter (1, 23). However, unlike the specificity mechanisms of many other transporters, which are commonly conferred by strong lock-and-key interactions, the selectivity of the NPC is based on multiple weak interactions between the transport proteins and the intrinsically disordered and dynamic FG nups, whose conformations likely undergo significant thermal fluctuations (24, 25, 26, 27). The binding of the transport proteins to the FG nups relies mostly on the hydrophobic interactions between the FG motifs and the hydrophobic pockets on the transport proteins. However, electrostatic and possibly other interactions are likely involved as well (39, 40, 41). Although the exact number of binding sites on the transport proteins is not completely established, experimental and computational estimates range from 2–6 for NTF2 to 4–10 for Importins (42, 43).
Many ideas have been discussed in the last two decades regarding the mechanisms of NPC transport and selectivity (for recent reviews, see (3, 4)). Although the proposed mechanisms often differ on the conformational dynamics of FG nups, most of them agree on the fundamental physical picture of transport. The FG nups set up a permeability barrier for the inert molecules and serve as the template for the transient binding of transport proteins. Translocation of the transport protein-cargo complexes relies on Brownian diffusion through the NPC passageway, modulated by the interactions with the FG nups. This fundamental physical picture can be encapsulated in the “Honorary Enzyme” model, which was first coined in the context of cell membrane channels and transporters (23). According to this picture (Fig. 3), the binding of the transport proteins to the FG nups lowers the free energy permeability barrier, thus enabling their partitioning into and translocation through the NPC (21, 39, 44, 45, 46).
Figure 3.
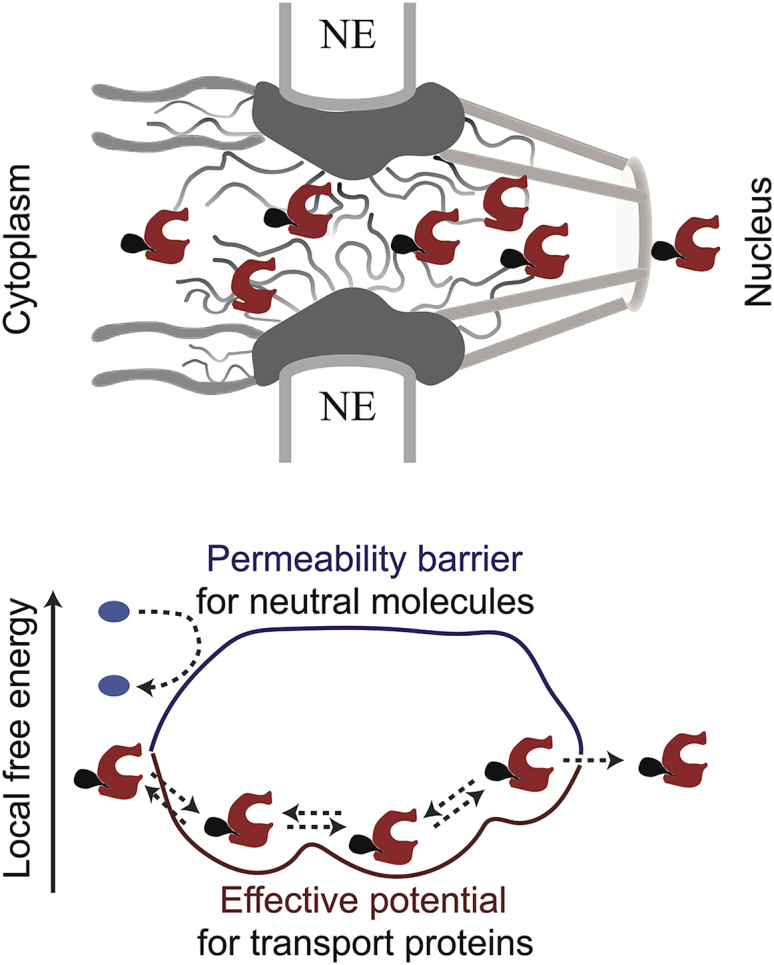
Shown here are schematics of NPC structure and function. (Upper panel) Schematic rendering of the NPC structure in the vertical cross-section is given. The wavy lines denote FG nups whereas the transport proteins and their cargo are shown in red and black, respectively. (Lower panel) Given here is the “Honorary Enzyme” picture of the NPC. The inert molecules (blue) encounter a high entry barrier provided by the FG nups (dark blue) whereas the binding of the transport proteins (red) to the FG nups lowers this barrier, so that they experience an attractive effective potential (burgundy), catalyzing their translocation through the NPC. The dashed lines indicate transport protein-cargo complex entry and diffusion through, and exit from, the NPC. To see this figure in color, go online.
These ideas can be mathematically quantified by formulating the translocation of transport proteins within the NPC as 1D diffusion in an effective potential arising from the interactions with the FG nups (44, 47) (Fig. 3). Similarly, by formulating the translocation as hopping between discrete sites within the NPC, one can account for molecular crowding inside the NPC (44). Mathematical modeling suggests that the binding of the transport proteins to the FG nups can enhance transport flux because binding increases the probability of a transport protein molecule to permeate and translocate through the NPC. However, this happens at the expense of increased individual translocation times, and proteins that bind the NPC very tightly can block the passageway. Consequently, only the transport proteins that bind the FG nups in a particular range of affinities are transported efficiently, providing a natural mechanism of specificity (44, 47, 48). Application of these ideas to single-molecule tracking experiments enables estimation of effective diffusivities of transport proteins and the local free energies within the NPC, arising from different FG nup distributions (49).
These simple ideas are sufficient to explain how the NPC retains selectivity despite the nonspecific competition that is always present in the cell. Through a nonequilibrium kinetic mechanism, strongly bound transport proteins filter out nonspecific competitors—even if competitors are present in orders-of-magnitude excess (48). These ideas have been tested and verified in artificial NPC mimics (33). They highlight NPC transport as a collective process, whereby the crowding of multiple copies of the transport proteins inside the NPC plays a crucial role in its transport mechanism (discussed in more detail below) (31, 33, 34, 35, 48, 50).
Biophysics of the NPC building blocks
Many features of NPC transport have been successfully explained by the simple models of facilitated diffusion described above. Yet comprehensive understanding requires a better picture of the spatial architecture and dynamics of assemblies formed by FG nups and transport proteins. A number of ideas regarding the conformational dynamics of these assemblies during transport have been put forward (3, 35, 47, 51). However, even using cutting-edge experimental techniques (27, 31, 32, 49, 52), it is challenging to directly probe the collective dynamics of the FG nups inside intact NPCs on relevant time (milliseconds) and length (several nanometers) scales. Thus, much of our biophysical knowledge about FG nup and transport protein assemblies has been derived from in vitro experiments augmented by computational and theoretical modeling.
In vitro, individual disordered FG nup chains behave in many ways as conventional polymers. They exhibit a persistence length of a few Ångstroms, a radius of gyration and hydrodynamic radius of several nanometers, and wormlike chain entropic elasticity (24, 51, 53). As shown by atomic force microscopy (AFM) experiments, grafted assemblies of multiple FG nups have nanomechanical properties resembling those of a polymer brush: a layer of grafted polymers whose spatial conformations are stabilized by entropic repulsion between the chains, which arise from their thermal fluctuations (35). Whereas these layers are impenetrable for inert molecules, binding of transport proteins allows them to penetrate FG nup assemblies (35). Penetration of the transport proteins modulates the layer morphology and, depending on the conditions, can either swell the layer or cause its compaction (35, 54, 58). These results underscored the importance of entropic effects in determining the conformations of FG nup assemblies and suggest that the permeability barrier is at least in part entropic, encapsulated in the brush model (3, 35, 55).
The intra- and interchain cohesiveness of the FG nups has also been implicated in shaping their spatial morphology and selective permeability of their assemblies (47, 51, 56). The cohesiveness predominantly arises from hydrophobic interactions between the FG motifs and can be modulated by electrostatic interactions between the charged amino acids and other interactions between the chains (3, 4, 39, 41, 51, 57). The magnitude of the cohesiveness depends on the amino acid sequence of the specific FG nups (51, 58). For some FG nups, notably the centrally located Nup100/116 in yeast and its functional homolog Nup98 in vertebrates, the cohesiveness is strong enough to promote formation of large aggregates in bulk solutions, observed by microscopy and light scattering (21, 25, 47, 56). Similar to the surface-grafted assemblies mentioned above, 3D assemblies of FG nups also possess rudimentary selectivity properties. Although these assemblies are impenetrable for inert molecules, transport proteins are able to penetrate them (56). These results gave rise to the selective phase model known as the “gel” model. The model postulates that disruption of cross-linked FG motifs is energetically unfavorable for inert molecules, and their passage is thus prevented, resulting in the permeability barrier. In contrast, transport proteins that bind to FG motifs can open the cross-links, which allows their passage through the NPC (21, 47).
Although gel-versus-brush models have generated considerable controversy in the past, it is becoming clear that effects from both models, as well as the spatial heterogeneities of the FG nup assembly, play a role in the NPC organization and transport mechanisms. It has also become apparent that the transport proteins play a considerable role in shaping NPC architecture (31, 33, 34, 35, 54, 58). Despite the diversity of observed morphologies of FG nup and transport protein assemblies and their nanomechanical and permeability properties, their main features can be understood based on the statistical thermodynamics of the fundamental interplay between enthalpic and entropic effects (51, 54, 58, 59). Cohesive interactions between the transport proteins and the FG nups favor more compact and less dynamic structures whereas the configurational entropy of the chains favors more diffuse and plastic morphologies (58). Interplay between these factors, combined with different cohesiveness and lengths of individual FG nups and the intrachain sequence heterogeneity, might allow formation of regions of different physical and nanomechanical properties that enable robust selectivity for a wide range of cargoes (32, 39, 49, 51, 57, 58, 67, 68, 69). Overall, good agreement with in vitro results has been obtained with physical models of FG nup and transport factor assemblies that rely on the statistical physics of polymers using coarse-grained FG nup descriptions (39, 54, 57, 58, 59). Such models provide a theoretical underpinning for further understanding of FG nup assemblies.
Crucially, FG nups remain highly dynamic even within dense assemblies. Both the cohesive interactions between the FG nups and the interactions between the FG nups and the transport proteins result from transient contacts that break and reform on very short time scales (24, 25, 26). This is consistent with the agile diffusion of transport proteins observed in the gel-like FG nup aggregates and the short (millisecond) transport times observed in the NPC by single-molecule fluorescence (21, 29, 30). These findings emphasize the fact that both FG-FG and FG-transport protein binding likely rely on multiple weak interactions. On the other hand, other in vitro (35) and in vivo (29, 31) results indicate that some transport proteins are relatively immobile. Thus, the picture of transport protein mobility within FG nup assemblies still remains incomplete.
Spatial architecture of the NPC passageway
In vitro experiments have provided significant insight into the spatial morphologies of FG nup-transport protein assemblies. However, in vitro findings do not directly translate in vivo, where NPCs contain multiple copies of diverse FG nups at different locations within the NPC passageway. Cutting-edge experimental methods, such as superresolution microscopy and high-speed AFM, have started to provide insight into the internal organization and dynamics of intact NPCs (27, 31, 32). These methods can 1) quantify the number of transport proteins in the pore (31), 2) determine regions where they preferentially accumulate (32, 52), and 3) visualize individual FG nup chain dynamics (27). The interpretation of these experiments is substantially assisted by computational modeling, which has become a major tool for describing the NPC spatial architecture, with parameterizations inferred from in vitro measurements. Most models use coarse-grained representations of the flexible FG nups as chains of monomers with appropriate physical attributes, such as charge, hydrophobicity, and degree of interaction with other monomers; the models are typically calibrated by comparison with in vitro data. These models vary in some of their assumptions: parameterization of molecular interactions, implementation of electrostatic and water-mediated interactions, and the degree of coarse-graining (26, 39, 40, 50, 54, 57, 58, 59, 60, 61). Although they differ in some of their predictions regarding FG nup distribution in the pore, these studies have generally produced a set of robust predictions about the expected FG nup organization in the pore. In the absence of transport proteins, FG nups typically appear to form a toroidal cloud of relatively high density near the pore walls; a region of lower FG nup density lies along the pore axis (Fig. 4). Transport proteins that either translocate through or reside within the NPC can partition into the FG nup cloud and significantly change the density distribution within the pore. Some predictions of the coarse-grained models agree with the experimentally observed size dependence of inert particle transport (37, 38), but a consensus has not been reached regarding the spatial distribution of different types of FG nups and transport proteins, and their dynamics within the NPC.
Figure 4.
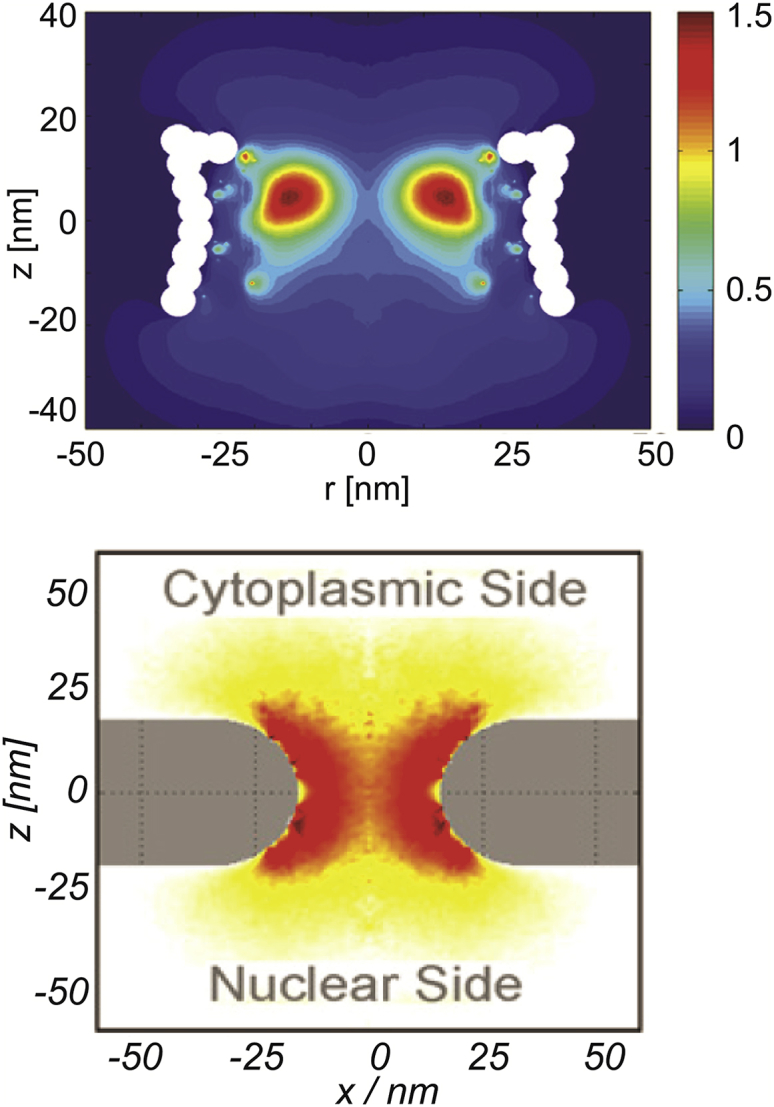
Given here are computational models of FG nup distribution within the NPC passageway. Upper and lower panels show the results of two different computational models of the FG nup distribution within the NPC passageway, reproduced from Ghavami et al. (57) and Tagliazucchi et al. (39), respectively. Each panel shows the vertical cross-section of the FG nup density distribution within the NPC passageway; red indicates higher local density. The results in the upper panel are obtained using Brownian dynamics simulations; the results in the lower panel are based on the density functional theory. Although using different parameterizations and computational techniques, the models agree in the qualitative predictions of the FG nup density distribution, which also agrees with the more coarse-grained models (50, 60). To see this figure in color, go online.
Selective artificial nanochannels and NPC mimics
Many aspects of NPC transport can be recapitulated in vitro using artificial mimics. Because these mimics can be customized with NPC components, they allow us to test aspects of different models in a controlled environment. For example, they can be used to study the impact of pore geometry and environment as well as protein density and composition. NPC mimics are not only excellent tools for understanding basic physical principles of NPC organization and function, but also have potential to advance the design of artificial biomolecular sorters.
The forerunners to the NPC mimics were functionalized nanopores designed to separate molecules based on charge, affinity, hydrophobicity, and size (62, 63). In parallel, nuclei isolated from Xenopus oocytes were attached to nanoporous membranes; this setup was used to investigate NPC transport by optical single-transporter recording method (64). Building upon these approaches, several devices were engineered with NPC components to mimic transport selectivity in the last decade.
Nucleocytoplasmic transport is robustly selective, which is conferred by three essential features: 1) flexible FG nups line the NPC channel and create a permeability barrier; 2) transport proteins transiently bind to the FG nups; and 3) translocating molecules are confined within the transport channel, creating crowding. Artificial nanochannel-based devices with these basic elements reproduce many transport properties of native NPCs. In one example (33), nanopores in polycarbonate membranes were functionalized with two different yeast FG nups, Nsp1 or Nup100, which are involved in forming the selectivity barrier in native NPCs. These FG nup-functionalized pores sustained the efficient passage of transport proteins and transport protein-cargo complexes, whereas the transport of control nonspecific molecules (such as bovine serum albumine) was significantly inhibited. Binding of transport proteins to FG nups was critical for transport selectivity: control pores functionalized with the neutral polymer PEG were not selective, in accord with theoretical understanding (48).
Further insights into transport at the single-molecule level were provided by another mimic, which was developed by functionalizing single SiN solid-state nanopores with vertebrate FG nups (Nup98 or Nup153) (65). Through ionic current measurements with submillisecond temporal resolution, transport proteins were shown to efficiently cross the pore, whereas nonspecific molecules of similar size (bovine serum albumine) were significantly blocked. Furthermore, the data largely supported a model wherein FG nups are densely concentrated around the pore walls, with a relatively open channel along the pore axis. The size of the opening could be changed by varying the pore radius. This agrees with the current theoretical models of the spatial distribution of the polymers in the pore (39, 50, 57, 60).
NPC-like selectivity can be also recapitulated in entirely synthetic, nonbiological nanochannels. In one example, polycarbonate nanoporous membranes were functionalized with polyisopropyl-acrylamide (PNIPAM), whose intra- and interchain cohesiveness can be controlled by temperature (66). In this system, receptor-mediated selective diffusion was modeled through weak interactions between PNIPAM chains lining the nanochannel and soluble PNIPAM segments, which served as the carrier molecules for single-stranded DNA cargo molecules.
Current questions and outlook
Despite the rapid progress of recent years in understanding NPC structure and function, we still do not fully understand the heterogeneity of its spatial organization. Likewise, the dynamics of FG nup and transport protein assemblies is not well understood on the nanoscale level. Notably, the NPC maintains fast and selective transport in both directions whereas its passageway appears to be densely packed with intercalated FG nups and transport proteins. According to recent studies, separating different types of import and export traffic may rely on spatial segregation of different regions within the FG meshwork in the pore (32, 67, 68, 69). To investigate this aspect further, it will be important to integrate in vivo imaging tools and advanced labeling methods with the insights arising from artificial in vitro mimics and computational studies. Higher resolution imaging and microscopy techniques, such as superresolution microscopy, FRET, ultrafast AFM, and fluorescence anisotropy, will be crucial for mapping the locations, mobility, and interactions of individual FG nups and their cooperative dynamics during transport. On the computational side, interpretation of such data will require models with enhanced parameterization and improved descriptions of molecular interactions. This will allow us to further clarify the fundamental concepts and pinpoint the critical molecular factors contributing to the import and export traffic into and out of the nucleus.
As a part of this effort, it will be important to advance our understanding of the biophysical mechanisms of the transport of large cargoes through the NPC. One example is the efficient translocation of very large mRNA particles through the dense environment of the NPC and coordination of their transport with other NPC transport pathways (36, 70). Understanding nuclear translocation of viruses, another class of large cargoes, is an emerging topic with important biomedical applications (71). Of paramount importance in their own right, these studies will provide additional insights into NPC organization and dynamics.
Although most of the studies of NPC selectivity have been focused on FG nups, it has been proposed that conformational changes and dynamics of the scaffold might contribute to the transport mechanism (72). Integrating structural studies using cryo-EM (5, 9, 16), crystallography (11, 12, 13), superresolution imaging (13), and integrative biology (10, 14) with computational models of scaffold dynamics (73) may yield further insights into NPC transport selectivity and answer the ultimate questions about its transport dynamics.
In a broader biomedical context, defects in nucleocytoplasmic transport have been implicated in a number of diseases. In particular, expression levels of certain nucleoporins and transport proteins are commonly altered in cancer and other diseases (74, 75). This makes nuclear trafficking of therapeutic interest; targeting NPC translocation may open new avenues toward inhibiting disease processes. Detailed understanding of the molecular underpinnings of the NPC transport can lead to novel molecular targets for rational drug development.
Given the high effectiveness of the NPC in molecular separation, new insights into the NPC transport mechanism may accelerate nanotechnological advances. We expect that future NPC mimics will incorporate multiple FG nups, include active transport through a RanGTP gradient, support bidirectional transport, and lead to new nanodevices with active molecular separation.
Author Contributions
Both authors conceived the idea and wrote the manuscript.
Acknowledgments
We thank Dr. I. Talisman for manuscript editing. The authors are indebted to colleagues in the field for numerous discussions, and apologize to the authors whose work could not be cited in full.
T.J.-T. acknowledges support from the Beckman Research Institute of the City of Hope and the STOP Cancer Foundation; A.Z. acknowledges support from the National Science and Engineering Research Council of Canada (NSERC).
Editor: Brian Salzberg.
Contributor Information
Tijana Jovanovic-Talisman, Email: ttalisman@coh.org.
Anton Zilman, Email: zilmana@physics.utoronto.ca.
References
- 1.Alberts B., Johnson A., Walter P. Garland Publishing; New York: 1994. Molecular Biology of the Cell. [Google Scholar]
- 2.Dingwall C., Laskey R.A. Protein import into the cell nucleus. Annu. Rev. Cell Biol. 1986;2:367–390. doi: 10.1146/annurev.cb.02.110186.002055. [DOI] [PubMed] [Google Scholar]
- 3.Lim R.Y., Aebi U., Fahrenkrog B. Towards reconciling structure and function in the nuclear pore complex. Histochem. Cell Biol. 2008;129:105–116. doi: 10.1007/s00418-007-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cautain B., Hill R., Link W. Components and regulation of nuclear transport processes. FEBS J. 2015;282:445–462. doi: 10.1111/febs.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck M., Hurt E. The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017;18:73–89. doi: 10.1038/nrm.2016.147. [DOI] [PubMed] [Google Scholar]
- 6.Callan H.G., Tomlin S.G. Experimental studies on amphibian oocyte nuclei. I. Investigation of the structure of the nuclear membrane by means of the electron microscope. Proc. R. Soc. Lond. B Biol. Sci. 1950;137:367–378. doi: 10.1098/rspb.1950.0047. [DOI] [PubMed] [Google Scholar]
- 7.Cronshaw J.M., Krutchinsky A.N., Matunis M.J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeGrasse J.A., DuBois K.N., Chait B.T. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics. 2009;8:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Appen A., Kosinski J., Beck M. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526:140–143. doi: 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alber F., Dokudovskaya S., Rout M.P. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 11.Knockenhauer K.E., Schwartz T.U. The nuclear pore complex as a flexible and dynamic gate. Cell. 2016;164:1162–1171. doi: 10.1016/j.cell.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brohawn S.G., Leksa N.C., Schwartz T.U. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szymborska A., de Marco A., Ellenberg J. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science. 2013;341:655–658. doi: 10.1126/science.1240672. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Martinez J., Kim S.J., Rout M.P. Structure and function of the nuclear pore complex cytoplasmic mRNA export platform. Cell. 2016;167:1215–1228.e25. doi: 10.1016/j.cell.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldberg M.W., Cronshaw J.M., Allen T.D. Nuclear-pore-complex dynamics and transport in higher eukaryotes. Protoplasma. 1999;209:144–156. [Google Scholar]
- 16.Eibauer M., Pellanda M., Medalia O. Structure and gating of the nuclear pore complex. Nat. Commun. 2015;6:7532. doi: 10.1038/ncomms8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adam S.A., Marr R.S., Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blobel G. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore M.S., Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 20.Clarkson W.D., Corbett A.H., Stewart M. Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran. J. Mol. Biol. 1997;272:716–730. doi: 10.1006/jmbi.1997.1255. [DOI] [PubMed] [Google Scholar]
- 21.Hülsmann B.B., Labokha A.A., Görlich D. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell. 2012;150:738–751. doi: 10.1016/j.cell.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 22.Wang C.-H., Mehta P., Elbaum M. Thermodynamic paradigm for solution demixing inspired by nuclear transport in living cells. Phys. Rev. Lett. 2017;118:158101. doi: 10.1103/PhysRevLett.118.158101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein W.D. Academic Press; New York: 1990. Channels, Carriers, and Pumps: An Introduction to Membrane Transport. [Google Scholar]
- 24.Milles S., Mercadante D., Lemke E.A. Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell. 2015;163:734–745. doi: 10.1016/j.cell.2015.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hough L.E., Dutta K., Cowburn D. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. eLife. 2015;4:e10027. doi: 10.7554/eLife.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moussavi-Baygi R., Mofrad M.R. Rapid Brownian motion primes ultrafast reconstruction of intrinsically disordered Phe-Gly repeats inside the nuclear pore complex. Sci. Rep. 2016;6:29991. doi: 10.1038/srep29991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakiyama Y., Mazur A., Lim R.Y. Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high-speed atomic force microscopy. Nat. Nanotechnol. 2016;11:719–723. doi: 10.1038/nnano.2016.62. [DOI] [PubMed] [Google Scholar]
- 28.Kopito R.B., Elbaum M. Reversibility in nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA. 2007;104:12743–12748. doi: 10.1073/pnas.0702690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W., Gelles J., Musser S.M. Imaging of single-molecule translocation through nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 2004;101:12887–12892. doi: 10.1073/pnas.0403675101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dange T., Grünwald D., Kubitscheck U. Autonomy and robustness of translocation through the nuclear pore complex: a single-molecule study. J. Cell Biol. 2008;183:77–86. doi: 10.1083/jcb.200806173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe A.R., Tang J.H., Liphardt J.T. Importin-β modulates the permeability of the nuclear pore complex in a Ran-dependent manner. Elife. 2015;4:e04052. doi: 10.7554/eLife.04052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma J., Goryaynov A., Yang W. Super-resolution 3D tomography of interactions and competition in the nuclear pore complex. Nat. Struct. Mol. Biol. 2016;23:239–247. doi: 10.1038/nsmb.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jovanovic-Talisman T., Tetenbaum-Novatt J., Chait B.T. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature. 2009;457:1023–1027. doi: 10.1038/nature07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters R. Translocation through the nuclear pore: kaps pave the way. Bioessays. 2009;31:466–477. doi: 10.1002/bies.200800159. [DOI] [PubMed] [Google Scholar]
- 35.Kapinos L.E., Schoch R.L., Lim R.Y.H. Karyopherin-centric control of nuclear pores based on molecular occupancy and kinetic analysis of multivalent binding with FG nucleoporins. Biophys. J. 2014;106:1751–1762. doi: 10.1016/j.bpj.2014.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grünwald D., Singer R.H., Rout M. Nuclear export dynamics of RNA-protein complexes. Nature. 2011;475:333–341. doi: 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timney B.L., Raveh B., Rout M.P. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 2016;215:57–76. doi: 10.1083/jcb.201601004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popken P., Ghavami A., Veenhoff L.M. Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol. Biol. Cell. 2015;26:1386–1394. doi: 10.1091/mbc.E14-07-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tagliazucchi M., Peleg O., Szleifer I. Effect of charge, hydrophobicity, and sequence of nucleoporins on the translocation of model particles through the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 2013;110:3363–3368. doi: 10.1073/pnas.1212909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gamini R., Han W., Schulten K. Assembly of Nsp1 nucleoporins provides insight into Nuclear Pore Complex gating. PLoS Comp. Biol. 2014;10:e1003488. doi: 10.1371/journal.pcbi.1003488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colwell L.J., Brenner M.P., Ribbeck K. Charge as a selection criterion for translocation through the nuclear pore complex. PLOS Comput. Biol. 2010;6:e1000747. doi: 10.1371/journal.pcbi.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayliss R., Ribbeck K., Stewart M. Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J. Mol. Biol. 1999;293:579–593. doi: 10.1006/jmbi.1999.3166. [DOI] [PubMed] [Google Scholar]
- 43.Isgro T.A., Schulten K. Binding dynamics of isolated nucleoporin repeat regions to importin-beta. Structure. 2005;13:1869–1879. doi: 10.1016/j.str.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Zilman A., Di Talia S., Magnasco M.O. Efficiency, selectivity, and robustness of nucleocytoplasmic transport. PLoS Comp. Biol. 2007;3:e125. doi: 10.1371/journal.pcbi.0030125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rout M.P., Aitchison J.D., Chait B.T. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13:622–628. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Ghavami A., van der Giessen E., Onck P.R. Energetics of transport through the nuclear pore complex. PLoS One. 2016;11:e0148876. doi: 10.1371/journal.pone.0148876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frey S., Görlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 48.Zilman A., Di Talia S., Magnasco M.O. Enhancement of transport selectivity through nano-channels by non-specific competition. PLOS Comput. Biol. 2010;6:e1000804. doi: 10.1371/journal.pcbi.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tu L.C., Fu G., Musser S.M. Large cargo transport by nuclear pores: implications for the spatial organization of FG-nucleoporins. EMBO J. 2013;32:3220–3230. doi: 10.1038/emboj.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osmanović D., Ford I.J., Hoogenboom B.W. Model inspired by nuclear pore complex suggests possible roles for nuclear transport receptors in determining its structure. Biophys. J. 2013;105:2781–2789. doi: 10.1016/j.bpj.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamada J., Phillips J.L., Rexach M.F. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol. Cell. Proteomics. 2010;9:2205–2224. doi: 10.1074/mcp.M000035-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bestembayeva A., Kramer A., Hoogenboom B.W. Nanoscale stiffness topography reveals structure and mechanics of the transport barrier in intact nuclear pore complexes. Nat. Nanotechnol. 2015;10:60–64. doi: 10.1038/nnano.2014.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim R.Y.H., Huang N.-P.P., Aebi U. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA. 2006;103:9512–9517. doi: 10.1073/pnas.0603521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zahn R., Osmanović D., Richter R.P. A physical model describing the interaction of nuclear transport receptors with FG nucleoporin domain assemblies. Elife. 2016;5:e14119. doi: 10.7554/eLife.14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner R.S., Kapinos L.E., Lim R.Y. Promiscuous binding of Karyopherinβ1 modulates FG nucleoporin barrier function and expedites NTF2 transport kinetics. Biophys. J. 2015;108:918–927. doi: 10.1016/j.bpj.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt H.B., Görlich D. Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. Elife. 2015;4:e04251. doi: 10.7554/eLife.04251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghavami A., Veenhoff L.M., Onck P.R. Probing the disordered domain of the nuclear pore complex through coarse-grained molecular dynamics simulations. Biophys. J. 2014;107:1393–1402. doi: 10.1016/j.bpj.2014.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vovk A., Gu C., Zilman A. Simple biophysics underpins collective conformations of the intrinsically disordered proteins of the nuclear pore complex. Elife. 2016;5:e10785. doi: 10.7554/eLife.10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ando D., Zandi R., Gopinathan A. Nuclear pore complex protein sequences determine overall copolymer brush structure and function. Biophys. J. 2014;106:1997–2007. doi: 10.1016/j.bpj.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nasrabad A.E., Jasnow D., Coalson R.D. Precise control of polymer coated nanopores by nanoparticle additives: insights from computational modeling. J. Chem. Phys. 2016;145:064901. [Google Scholar]
- 61.Mincer J.S., Simon S.M. Simulations of nuclear pore transport yield mechanistic insights and quantitative predictions. Proc. Natl. Acad. Sci. USA. 2011;108:E351–E358. doi: 10.1073/pnas.1104521108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee S.B., Mitchell D.T., Martin C.R. Antibody-based bio-nanotube membranes for enantiomeric drug separations. Science. 2002;296:2198–2200. doi: 10.1126/science.1071396. [DOI] [PubMed] [Google Scholar]
- 63.Kohli P., Harrell C.C., Martin C.R. DNA-functionalized nanotube membranes with single-base mismatch selectivity. Science. 2004;305:984–986. doi: 10.1126/science.1100024. [DOI] [PubMed] [Google Scholar]
- 64.Keminer O., Peters R. Permeability of single nuclear pores. Biophys. J. 1999;77:217–228. doi: 10.1016/S0006-3495(99)76883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kowalczyk S.W., Kapinos L., Dekker C. Single-molecule transport across an individual biomimetic nuclear pore complex. Nat. Nanotechnol. 2011;6:433–438. doi: 10.1038/nnano.2011.88. [DOI] [PubMed] [Google Scholar]
- 66.Caspi Y., Zbaida D., Elbaum M. Synthetic mimic of selective transport through the nuclear pore complex. Nano Lett. 2008;8:3728–3734. doi: 10.1021/nl801975q. [DOI] [PubMed] [Google Scholar]
- 67.Naim B., Brumfeld V., Reich Z. Passive and facilitated transport in nuclear pore complexes is largely uncoupled. J. Biol. Chem. 2007;282:3881–3888. doi: 10.1074/jbc.M608329200. [DOI] [PubMed] [Google Scholar]
- 68.Fiserova J., Richards S.A., Goldberg M.W. Facilitated transport and diffusion take distinct spatial routes through the nuclear pore complex. J. Cell Sci. 2010;123:2773–2780. doi: 10.1242/jcs.070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J., Izadyar A., Amemiya S. Nanoscale mechanism of molecular transport through the nuclear pore complex as studied by scanning electrochemical microscopy. J. Am. Chem. Soc. 2013;135:2321–2329. doi: 10.1021/ja311080j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valkov E., Dean J.C., Stewart M. Structural basis for the assembly and disassembly of mRNA nuclear export complexes. Biochim. Biophys. Acta. 2012;1819:578–592. doi: 10.1016/j.bbagrm.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 71.Fay N., Panté N. Nuclear entry of DNA viruses. Front. Microbiol. 2015;6:467. doi: 10.3389/fmicb.2015.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Solmaz S.R., Blobel G., Melcák I. Ring cycle for dilating and constricting the nuclear pore. Proc. Natl. Acad. Sci. USA. 2013;110:5858–5863. doi: 10.1073/pnas.1302655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolf C., Mofrad M.R. On the octagonal structure of the nuclear pore complex: insights from coarse-grained models. Biophys. J. 2008;95:2073–2085. doi: 10.1529/biophysj.108.130336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ori A., Banterle N., Beck M. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 2013;9:648. doi: 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gould V.E., Orucevic A., Alonso A. Nup88 (karyoporin) in human malignant neoplasms and dysplasias: correlations of immunostaining of tissue sections, cytologic smears, and immunoblot analysis. Hum. Pathol. 2002;33:536–544. doi: 10.1053/hupa.2002.124785. [DOI] [PubMed] [Google Scholar]