Abstract
False negative result remains an ongoing problem in direct gene sequencing of cancers. It is important to utilize the appropriate mutation detection method most appropriate to each circumstance and the available tissue. Here, we report a patient with melanoma of unknown primary with metastases to spleen and bone marrow, who was tested negative for Cobas BRAF V600E mutation, whose cancer progressed on anti-programmed death 1 (PD1) receptor monoclonal antibody therapy. Subsequent VE1 immunohistochemistry was positive for BRAF V600E mutation, and the tumor responded dramatically to BRAF/MEK inhibitor combination therapy. This demonstrates how alternative BRAF testing methodology could produce results that can influence treatment choice and the outcome.
Case Presentation
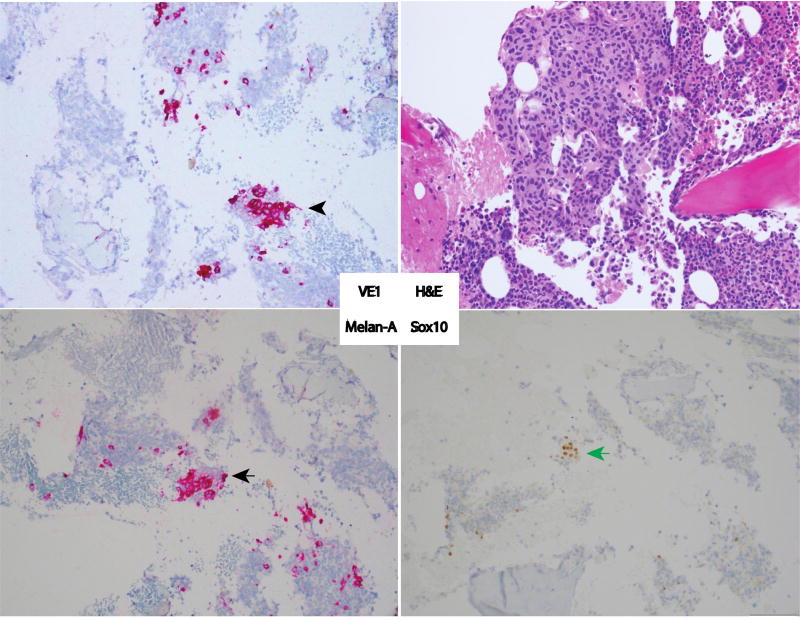
A 67 year-old Caucasian female presented with dyspnea and recent weight loss. Initial evaluation demonstrated splenomegaly. Abdominal CT revealed a 17cm spleen with heterogeneous attenuating lesions, the largest being 6.9 cm in diameter. A bone marrow biopsy showed 10% of marrow tissue infiltrated by non-hematopoietic cells with immunohistochemistry (IHC) positive for S100, Melan-A, and HMB-45, consistent with metastatic melanoma. Further evaluation including imaging showed no evidence of primary melanoma or other metastatic foci. A standard Cobas assay on her bone marrow biopsy was reported as BRAF wild type. (A standard mineral decalcification with formic acid was performed on the bone marrow specimen). She received nivolumab monotherapy at 2mg/kg every 2 weeks as part of a clinical trial protocol. Treatment was stopped after 12 weeks because of progressive disease. Due to the lack of effective treatment options, her bone marrow biopsy specimen was sent for additional immunohistochemical staining including with the VE1 (anti-BRAFV600E) antibody. The specimen stained positive for BRAFV600E with the VE1 antibody (Figure.1).
Figure 1.
A hematoxylin and eosin stained section (Upper Right) of the bone marrow core biopsy shows an infiltrate of large epithelioid cells with ovoid to spindled nuclei, condensed chromatin and abundant cytoplasm arranged in loosely cohesive clusters and present in a background of hematopoietic precursor cells and bony trabeculae. The malignant cells were immunoreactive for VE1 (anti-BRAFV600E) (Upper Left; black arrowhead). Similar cell aggregates were positive for Sox10 (Lower Right; black arrow) and Melan-A (Lower Left; green arrow). The cells are negative for CD45, AE1/AE3, MAK6, CK7 and CK20. Interpretation of the findings is limited by the fact that only rare cells are positive; however, the constellation of findings is in keeping with metastatic melanoma harboring BRAFV600E.
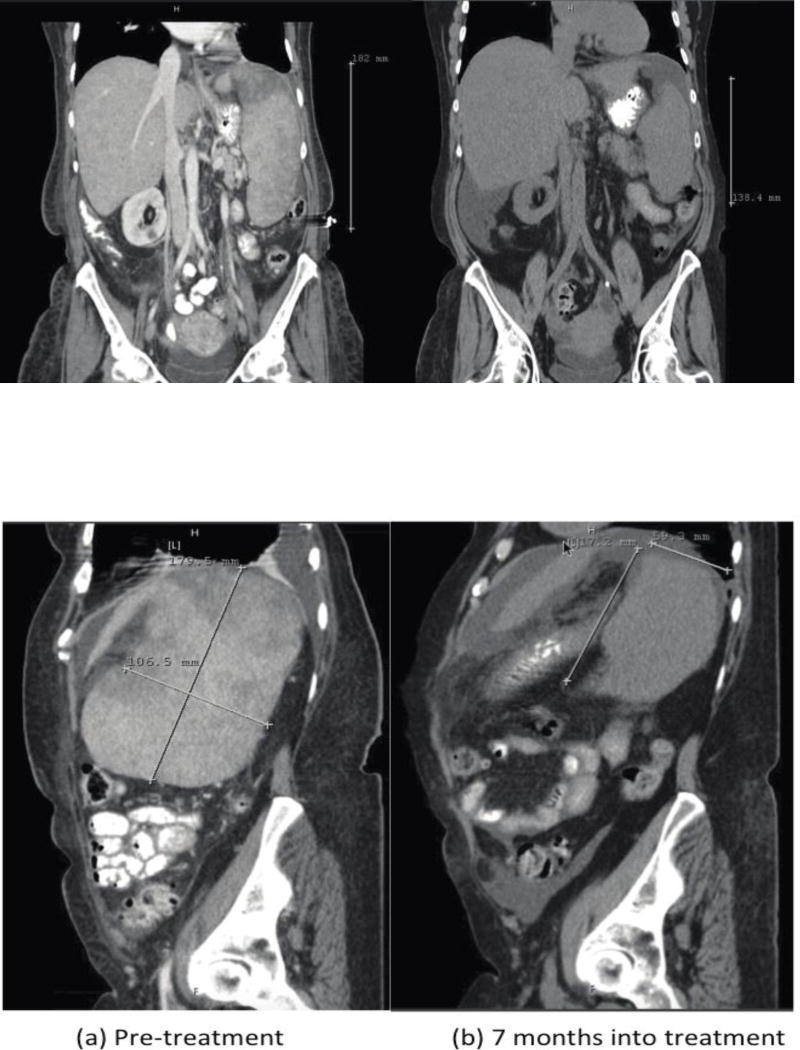
She received a combination of dabrafenib, 150mg twice daily, with trametinib, 2mg daily. Abdominal CT at 32 weeks of treatment showed significant shrinkage splenic metastases and overall spleen size (Figure 2), which was associated with symptomatic improvement.
Figure 2.
Abdominal computed tomography before (a) and after 28 weeks (b) of treatment with dabrafenib and trametinib demonstrates shrinkage in splenic metastases (arrows) with 43% reduction in spleen size and intrasplenic metastases.
Discussion
BRAF inhibitors either alone or in combination with a MEK inhibitor have improved survival of patients with BRAFV600 mutant melanoma (1–3). Treatment response is often quick (median time-to-response of 1.4 months)(4, 5); therefore, treatment is invaluable in patients who require rapid tumor shrinkage for symptomatic relief. Direct gene sequencing for BRAF mutation detection can be performed by multiple different techniques. The lower limit of mutation detection varies for each testing platform - Sanger sequencing 15–20%, Pyrosequencing 10%, TaqMan 2.5%, and Cobas test at 5–10% mutation fraction(6). In general, 100% consensus was observed among testing platforms when BRAF V600E mutation was present in more than 10% of the tissue.
Conventional mutation analyses have several limitations: (i) random tissue sampling leads to normal stromal tissue contamination during genomic DNA extraction steps diluting the BRAF mutant somatic DNA (ii) poor tissue quality due to tumor necrosis or sample preparation (e.g. bone decalcification with inorganic acids leads to degradation of nucleic acids) can produce indeterminate results (7), (iii) lower limit of detection by qPCR with a minimum of 5–10% mutant allele for each base pair, underscoring the need for alternative methods of mutation detection.
A monoclonal BRAF V600E mutation-specific antibody could facilitate the diagnosis of BRAF mutant melanoma in situations with small number of tumor cells in the midst of stromal tissue or nucleic acids damages during tissue processing. Limitations of VE1 antibody IHC assay can include: (i) non-specific background or focal nuclear staining, (ii) loss of mutant antigen expression following tissue ischemia or necrosis (8), (iii) variations between manual staining methods, or even autostainer protocols (e.g. fixation time) and (iv) melanin background affecting the interpretation of bright field IHC. However, this approach has been shown to be sensitive and specific in comparison to direct sequencing techniques(9) and allows direct assessment of the BRAF status of melanoma cells under the microscope. Alternative approaches to detecting BRAFV600E mutations when VE1 is non-diagnostic are to assess for BRAFV600E mutation from available tumor tissue using droplet digital PCR (ddPCR) (10) or Beads, Emulsion Amplification and Magnetic (BEAMing) technology (11–13). Both assays were demonstrated to have a lower limit of detection at or less than 1:10,000 (0.01%), each lower than Cobas BRAF V600E testing. These remain available only on a research basis.
This patient represents an example of the clinical utility of VE1 monoclonal antibody. In her case the standard BRAF sequencing method failed to detect the BRAF mutation in her tumor likely due to the dilutional effect of normal bone marrow cells in the biopsy material or DNA degradation from bone marrow decalcification. The VE1 IHC stain provided an alternative to assess the BRAF mutational status of her tumor cells necessary to safely provide her BRAF inhibitor therapy, which produced clinically significant improvement.
VE1 antibody IHC could also apply to patients with microscopic involvement of sentinel lymph nodes and biopsy inaccessible metastatic disease and those with FNA biopsy where the amount of tumor is insufficient for routine mutational analysis. In addition, the assay could be of value in critically ill patients, where a rapid assessment of BRAF mutation status would facilitate the treatment decision and patients whose tumors are BRAF wild-type by DNA sequencing methods who have exhausted all standard treatment options. Finally, it could have utility in evaluating resistant disease to determine if resistance may involve selection of a subpopulation of cells lacking a BRAF mutation.
Footnotes
Conflicts of Interest/Sources of Funding: none declared by all author
References
- 1.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364(26):2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. The New England journal of medicine. 2012;367(2):107–14. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 3.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. The New England journal of medicine. 2015;372(1):30–9. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 4.McArthur GA, Chapman PB, Robert C, Larkin J, Haanen JB, Dummer R, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. The Lancet Oncology. 2014;15(3):323–32. doi: 10.1016/S1470-2045(14)70012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380(9839):358–65. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 6.Lade-Keller J, Romer KM, Guldberg P, Riber-Hansen R, Hansen LL, Steiniche T, et al. Evaluation of BRAF mutation testing methodologies in formalin-fixed, paraffin-embedded cutaneous melanomas. The Journal of molecular diagnostics : JMD. 2013;15(1):70–80. doi: 10.1016/j.jmoldx.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Wickham CL, Sarsfield P, Joyner MV, Jones DB, Ellard S, Wilkins B. Formic acid decalcification of bone marrow trephines degrades DNA: alternative use of EDTA allows the amplification and sequencing of relatively long PCR products. Mol Pathol. 2000;53(6):336. doi: 10.1136/mp.53.6.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capper D, Berghoff AS, Magerle M, Ilhan A, Wohrer A, Hackl M, et al. Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta neuropathologica. 2012;123(2):223–33. doi: 10.1007/s00401-011-0887-y. [DOI] [PubMed] [Google Scholar]
- 9.Colomba E, Helias-Rodzewicz Z, Von Deimling A, Marin C, Terrones N, Pechaud D, et al. Detection of BRAF p.V600E mutations in melanomas: comparison of four methods argues for sequential use of immunohistochemistry and pyrosequencing. The Journal of molecular diagnostics : JMD. 2013;15(1):94–100. doi: 10.1016/j.jmoldx.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Sanmamed MF, Fernandez-Landazuri S, Rodriguez C, Zarate R, Lozano MD, Zubiri L, et al. Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clin Chem. 2015;61(1):297–304. doi: 10.1373/clinchem.2014.230235. [DOI] [PubMed] [Google Scholar]
- 11.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102(45):16368–73. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci U S A. 2003;100(15):8817–22. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]