Abstract
This introductory article should be viewed as a prologue to the Free Radical Biology & Medicine Special Issue devoted to the important topic of Oxidatively Damaged DNA and its Repair. This special issue is dedicated to Professor Tomas Lindahl, co-winner of the 2015 Nobel Prize in Chemistry for his seminal discoveries in the area repair of oxidatively damaged DNA. In the past several years it has become abundantly clear that DNA oxidation is a major consequence of life in an oxygen-rich environment. Concomitantly, survival in the presence of oxygen, with the constant threat of deleterious DNA mutations and deletions, has largely been made possible through the evolution of a vast array of DNA repair enzymes. The articles in this Oxidatively Damaged DNA & Repair special issue detail the reactions by which intracellular DNA is oxidatively damaged, and the enzymatic reactions and pathways by which living organisms survive such assaults by repair processes.
Keywords: DNA Oxidation, DNA Damage, DNA Repair, Thomas Lindahl, Nobel Prize, Oxidative Stress
Introduction
Tomas Lindahl, Paul Modrich and Aziz Sancar are the 2015 Nobel Prize Laureates in Chemistry [1–4] who were honored for their pioneering and seminal contributions to the delineation of biochemical mechanisms of several DNA repair pathways including base excision repair (BER) [5–10], mismatch repair (MMR) [11–13], nucleotide excision repair (NER) and enzymatic photoreversal [14–16], 21 years after “DNA repair enzyme” was recognized by the Science magazine as the molecule of the year [17].
Susceptibility to Oxidation
Life in an oxygen-rich environment is a constant struggle to minimize and then repair the results of oxidatively induced damage. This is partly because oxygen itself (a bi-radical) is reactive with various metal ions and biological molecules, almost always causing oxidation. Nevertheless, multicellular eukaryotic life forms evolved largely because of the ability to extract some 18 times more energy from food sources via oxygen-dependent complete oxidation in mitochondria than is possible from glycolysis. The very process of mitochondrial respiration, however, actually generates more reactive forms of oxygen, including the superoxide anion radical (O2·−), hydrogen peroxide (H2O2), and even the highly oxidizing hydroxyl radical (·OH). Thus, oxygen represents something of a double-edged sword for aerobic organisms; it is an absolute requirement for life, yet it threatens the very life it supports: This has been referred to as the Oxygen Paradox (18–21).
All organic molecules, very much including DNA, are susceptible to oxidative damage from a wide variety of oxygen-, or nitroxygen-based reactive species. Cells, organs, and organisms utilize a wide variety of antioxidant compounds (largely from and fruit foods) such as vitamins E and C, and a plethora of polyphenols; antioxidant enzymes such as superoxide dismutases, glutathione peroxidases, peroxiredoxins, and glutaredoxins; and reductive enzyme cofactors such as glutathione and thioredoxin to try to minimize the amount of oxidatively generated damage that occurs to DNA, proteins, and lipids; nevertheless, substantial damage does occur on a daily basis (22–24).
Damage Removal & Repair
To cope with the oxidatively induced damage whose formation has escaped antioxidant compounds and enzymes, an enormous variety of damage removal and repair enzymes has evolved. Thus, we find proteolytic enzymes (such as the Proteasome and the mitochondrial Lon protease) that can recognize and degrade oxidatively damaged polypeptides and remove them from cells before they aggregate and cross-link (25–27) and lipolytic enzymes, such as phospholipase A2 that can selectively recognize and remove oxidized phospholipids from biological membranes (28). In addition, heavily oxidized proteins and lipids (often in cross-linked mixed-form aggregates) that have escaped the proteolytic and lipolytic enzymes can be engulfed by phagocytic cells and degraded by a process called autophagy.
As impressive as these damage removal systems for proteins and lipids may be, however, they simply pale in comparison with the vast array of enzymes that recognize and repair, or replace, oxidatively damaged DNA. The evolution of this immense DNA repair armament was presumably driven by the selective advantage conferred by effective conservation of vital genetic information. Today we know of more than 100 DNA repair enzymes and it is quite probable that even more remain to be discovered (1–17).
Transient Stress Adaptation: Hormesis and Adaptive Homeostasis
In addition to this substantial armament of antioxidant compounds, antioxidant enzymes, damage removal enzymes, and repair enzymes, organisms as diverse as bacteria, fungi, worms, flies, mice, and humans all possess extra layers of protection in the form of inducible antioxidant enzymes, and damage removal and repair systems. Simply put, an exposure to low levels of oxidants can induce the increased expression of protective or repair enzymes in processes catalyzed by discrete signal transduction pathways. In some cases small amounts of oxidatively generated damage may induce transient adaptation in a process that has been called hormesis (29). In other cases, the levels of oxidant(s) experienced are too low to cause any damage, and they instead act as pure signal transducing agents for transient adaptation in a process called adaptive homeostasis (30). Adaptive homeostasis involves transient expansion of the physiological, or homeostatic, range of stress resistance that is typically reversed within hours. Major pathways for adaptive homeostasis include the Nrf2 signal transduction pathway in mammals (31), its CncC orthologue in flies (32), and its SKN-1 orthologue in worms (33).
Diminished baseline function of protein, lipid, and DNA repair systems has been suggested as a significant contributor to the aging phenomenon, and was proposed as an amendment to the Free Radical Theory of Aging (34–36). More recently it has become clear that inducible systems for transient adaptation, such as hormesis and adaptive homeostasis, also decline with age (31–33). Indeed, mounting evidence now suggests that diminished ability to modulate homeostasis in response to varying levels of environmental and metabolic stressors, may be a significant contributor to age-related degeneration.
Repair of Oxidatively Modified DNA is Vital for Genome Stability
The effective function of dedicated repair enzymes is essential for maintaining the integrity of the genome against a large variety of endogenous and exogenous reactive oxygen species and agents while allowing accurate replication and preventing mutations to occur [37]. A major finding in the field of DNA repair was the discovery by Tomas Lindahl in 1974 of uracil DNA N-glycosylase that is able to remove uracil, the hydrolytic deamination product of cytosine [38]. This was followed by another key observation showing that a novel DNA glycosylase isolated from E coli cells triggered the enzymatic release from modified DNA of 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine [39], an alkylated model compound of the major ·OH-mediated degradation product of the guanine moiety in cellular DNA [40]. This explains why the enzyme receives the name of formamidopyrimidine DNA N-glycosylase (Fpg), also called MutM which was found later on to recognize and excise 8-oxo-7,8-dihydroguanine [41].
Further investigations concerning in particular assessment of substrate specificity were facilitated by the overproduction of the repair protein through gene cloning by Boiteux et al [42]. It is also worth mentioning that the short-patch BER pathway of uracil DNA N-glycosylase has been reconstituted in Tomas Lindahl’s laboratory [43]. These ground breaking findings have further stimulated numerous research activities in the field of repair of single base lesions since 16 DNA glycosylases have been identified so far with in addition the discovery by other research groups of nucleobase incision repair [44] and hydrolytic dephosphorylation activity of MuT protein toward 8-oxo-7,8-dihydro-2′-deoxyribonucleoside triphosphate in nucleotide pools [45]. These various aspects together with recent developments and the biological role of DNA glycosylases are further discussed in this special issue.
It may be added that Tomas Lindahl has played a leading role in the discovery of the oxidative repair of methyl substituted bases by DNA dioxygenases including AlkB protein and its homologs [46] which have been shown recently to be implicated in the ten-eleven translocation (TET) iterative oxidation of 5-methylcytosine [47,48]. It may be added to this already long list of striking findings that purine 5′,8-cyclo-2′-deoxyribonucleosides were shown by Tomas Lindahl et al to be substrates for NER and not BER enzymes [49]. This has provided a strong stimulus to the development of studies aimed at further delineating the biological significance of these oxidatively generated lesions even if their formation in cellular DNA remains a matter of debate. Other types of complex DNA lesions (tandem base modifications, inter- and intra-strand cross-links, DNA-protein cross-links and clustered damage) induced by ·OH, one-electron oxidants and ionizing radiation have been characterized in cellular DNA together with single modifications involving ·OH, singlet oxygen, hypochlorite, dioxygenases and reactive aldehydes from lipid peroxide decomposition.
ARTICLES INCLUDED IN THIS DNA DAMAGE & REPAIR SPECIAL ISSUE
Formation and repair of oxidatively generated damage in cellular DNA, by Jean Cadet, Kelvin J. A. Davies, Marisa HG Medeiros, Paolo Di Mascio, and J. Richard Wagner [50]
The identification of oxidatively damage in cellular DNA has been until recently hampered by the lack of accurate and sensitive detection methods. As an exception HPLC coupled with electrochemical detection that was applied almost 30 years ago has allowed to monitor the formation in cellular DNA of 8-oxo-7,8-dihydroguanine (8-oxoGua), an ubiquitous guanine oxidation product. However 15 years have been necessary to extend the measurement of 8-oxoGua to about 25% of the other 80 oxidized nucleobases that were characterized in model studies. This has been made possible by the advent of HPLC-MS/MS a versatile method that provided unambiguous assignment of the damage and quantitative measurement through the use of the isotope dilution technique. Thus 16 single base lesions together with more complex damage such as intra- and inter-strand cross-links have been accurately detected in the DNA of cultured cells exposed to biologically relevant oxidizing agents including hydroxyl radical, singlet oxygen, one-electron oxidants and ten-eleven translocation enzymes. In addition several modifications whose generation is triggered by activation of myeloperoxidases and the release of reactive aldehydes from the breakdown of lipid peroxides have been also measured in cellular DNA.
In contrast the formation of secondary oxidation products of 8-oxoGua that has received a strong attention from chemists and biochemist is at best a very minor process in cells, This remark applies as well to the questionable formation of purine 5′,8-cyclo-2′-deoxyribonucleosides that have been shown to be repaired in model studies by the nucleotide excision repair pathway and not by the base excision repair that operates on single oxidized nucleobases. The recently collected data on cytosine and 5-methylcytosine confirm that the steady-state levels of oxidatively induced base lesions are rather low being comprised in most cases within the range of 1 modification par 106 to 107 nucleobases. It may also be noted that there is still a paucity of information on the kinetics of base damage removal in cellular DNA. However as one of the main exceptions it was shown that 8-oxoGua is removed from DNA in cells and animal organs, typically half of the lesion being repaired in 2h-4h time period. Further works should focus on the repair of other single oxidized bases as well as the identification of tandem modifications whose measurement in cellular DNA remains a challenging analytical issue.
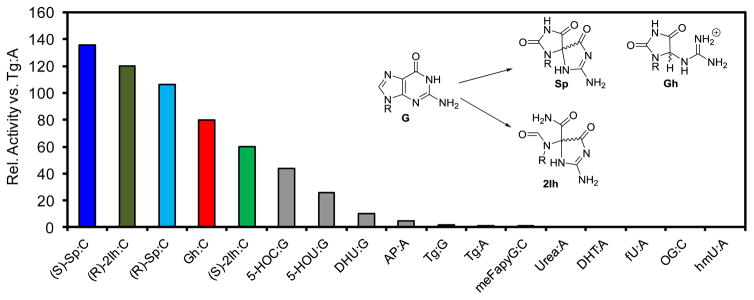
Formation and processing of DNA damage substrates for the hNEIL enzymes, by Aaron M. Fleming and Cynthia J. Burrows [51]
Base excision repair in humans utilizes a DNA glycosylase to initiate lesion removal. Three critical human DNA glycosylases include NEIL1, NEIL2, and NEIL3. The NEILs possess superior kinetic preference for the oxidatively modified guanine (G) lesions spiroiminodihydantoin (Sp), 5-guanidinohydantoin (Gh), and 5-carboxamido-5-formamido-2-iminohydantoin (2Ih); in contrast, the NEILs operate poorly on 8-oxo-7,8-dihydroguanine (OG) and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (Fapy-G; Fig. 1). A unique feature of the NEIL glycosylases resides in their ability to remove hydantoins from single-stranded and double-stranded DNA (ssDNA and dsDNA), as well as G-quadruplex DNA (G4 DNA). More specifically, NEIL1 initiates hydantoin removal best in ssDNA and dsDNA, while NEIL2 removes hydantoins from ssDNA, and NEIL3 liberates hydantoins from ssDNA and G4 DNA, albeit it very slowly. The diverse contexts for which the NEILs function to remove Sp, Gh, or 2Ih supports the hypothesis that NEIL1 is involved in replication-coupled repair, NEIL2 participates in transcription-coupled repair, and the role of NEIL3 remains elusive. Because NEIL3 is a slow glycosylase and its cellular expression is associated with the cellular state it has been proposed to be a regulatory protein, such as a transcription factor. The guanine modifications Sp, Gh, and 2Ih result from DNA oxidation by all of the culprit cellular reactive oxygen species. For instance, G oxidation by hydroxyl radical yields 2Ih in a similar amount as the established product OG. Inflammation-derived carbonate radical oxidizes G to yield Sp in ssDNA or G4DNA and Gh in dsDNA contexts. When G is oxidized by singlet oxygen, OG and Sp are the oxidation products observed. These observations confirm that hydantoins are oxidation products in all contexts in which the NEIL glycosylases have been demonstrated to remove DNA lesions.
Figure 1.
Relative activity for the human NEIL1 DNA glycosylase acting upon a variety of DNA base lesions
Removal of oxidatively generated DNA damage by overlapping repair pathways, by Vladimir Shafirovich and Nicholas E. Geacintov [52]
It is generally believed that the mammalian nucleotide excision repair (NER) pathway removes DNA helix-distorting bulky DNA lesions, while small non-bulky lesions are repaired by base excision repair (BER). Nevertheless, the possibility that there might be some ‘cross-talk’ between BER and NER mechanisms of repair have been raised over the years. This article summarizes recent observations on the excision of oxidatively generated DNA lesions in cell-free HeLa cell extracts that contain active BER and NER proteins.
Interestingly, several oxidatively generated DNA lesions were found to be excellent substrates of not only BER, but also of NER. In cells, the primary target of oxidatively generated damage to DNA is guanine, the most easily oxidizable natural nucleic acid base. The best known products of oxidatively generated guanine transformations is 8-oxo-7,8-dihydroguanine (8-oxoG). Recent work has shown that the oxidativly generated 8-oxoG oxidation products, spiroimininodihydantoin (Sp) and 5-guanidinohydantoin (Gh), which are typical BER substrates, and the intrastrand cross-linked DNA lesion guanine(C8)-thymine(N3) intrastrand crosslinks (G*-T*), are substrates of both BER and NER when incubated in the same repair-active DNA substrates.
Another well known set of NER substrates are the non-bulky 5′,8-cyclopurine lesions that, however, are not incised by BER proteins. Many other BER substrates are not incised by NER mechanisms, and include 5-guanidino-4-nitroimidazole (NIm) that is generated by the oxidation of guanine by the chemical mediator of inflammation peroxynitrite. The latter is structurally similar to Gh, except that the 4-nitro group in NIm is replaced by a keto group in Gh. However, unlike Gh, NIm is an excellent substrate of BER, but not of NER. Both Sp and NIM destabilize DNA duplexes, but the differences in susceptibilities to NER suggest that subtle structural effects play a role in determining whether a given non-bulky lesion is a substrate only of NER (5′,8-cyclopurines), or only of BER (NIm), or of both NER and BER (Sp, Gh, and G*-T*).
The recent results reviewed in this article indicate that the range of oxidatively generated DNA lesions that are substrates of the nucleotide excision repair pathway may be more extensive than previously thought. Understanding the relationships between susceptibility to NER and molecular structure of the lesion is challenging, because the thermodynamic destabilization of the DNA by the lesions is often, but not always correlated with NER activity.
Oxidized C5-methyl cytosine bases in DNA: 5-hydroxymethylcytosine; 5-formylcytosine; and 5-carboxycytosine, by Arne Klungland and Adam B. Robertson [53]
Regulatory DNA modifications types in mammalian cells are relatively sparse. Until recently only one type of DNA modification, 5-methylcytosine (5mC), represented the entirety of known mammalian DNA modifications. More recently stable, enzyme catalyzed oxidation products of 5mC have been described. These oxidized DNA bases are 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxycytosine (5caC). The Ten Eleven Translocation (Tet) enzyme family catalyzes the oxidation of 5mC initially yielding 5hmC; subsequent oxidation of this base using the same enzymes and a similar enzymatic mechanism yields 5fC and 5caC. It is widely accepted that 5mC negatively regulates transcription; however, the function of oxidized 5mC products is less clear. Oxidized 5mC in DNA can repress or enhance transcription, affect stem cell pluripotency and differentiation, and are likely intermediates in active demethylation. The review in this section discusses the enzymes that catalyze the oxidation of 5mC, the molecular effects of oxidized 5mC DNA bases, and finally we discuss the organismal consequences of aberrant 5mC oxidation.
Sensitive detection of oxidative damage to DNA induced by nanomaterials, by Andrew Collins, Naouale El Yamani, and Maria Dusinska [54]
Because of the high ratio of the surface area of nanoparticles (NPs) to their volume, nanomaterials (NMs) tend to be more reactive than the bulk chemicals from which they are derived, and so specific testing is necessary. Toxicity depends on size, shape but also on the surface coating and charge of the NPs. The aim of the EC project NANoREG was to create a regulatory framework for toxicity testing of NMs.
The in vitro comet assay (single cell gel electrophoresis – detecting DNA breaks) is now routinely used to investigate the genotoxicity of NMs. The sensitivity of the assay is increased by incorporating a digestion of DNA with a lesion-specific enzyme, most commonly formamido-pyrimidine DNA glycosylase (Fpg), which converts oxidised purines to strand breaks. This article reviews recent results obtained using this approach; the most common NM studied is TiO2, but factors such as cell type, concentration range, and times of incubation vary too widely among these publications to be able to draw any firm conclusions.
The systematic study of four metallic NMs is described, using a high throughput comet assay to allow testing of a range of concentrations of all four NMs in a single experiment with exposure times of 3 h and 24 h. Cytotoxicity was assessed in parallel with genotoxicity, since secondary DNA-damaging effects can arise in dying cells. Two of the NMs tested, CeO2 and TiO2, showed negligible cytotoxicity, and no effect on colony-forming efficiency, and yet caused significant damage, both DNA breaks and Fpg-sites, which decreased over 24 h. In contrast, ZnO and Ag NMs were cytotoxic and caused cell death at low concentrations. ZnO induced DNA damage at 3 h, mostly Fpg-sites, but by 24 h the damage (except at cytotoxic concentrations) was substantially reduced, even though NPs were still present. Ag NPs also caused DNA damage, with little sign of recovery over 24 h. Such variations in the time course of damage point to differences in cellular mechanisms. Some general advice for the testing of NMs is developed and rationalized as follows:
Thorough characterisation of NMs is crucial – and should be carried out in the dispersion medium, before, during and after incubation with cells.
Fpg is a valuable adjunct to the comet assay, enhancing its ability to detect, specifically, DNA damage arising as a result of oxidative stress induced by the NMs.
Testing for DNA damage at cytotoxic concentrations is likely to lead to false positive results.
Care should be taken to avoid artefacts arising from interference, such as the interaction of photocatalytic NMs with light.
Effects of NMs can vary according to the cell type used to test them.
A wide range of NM concentrations should be tested to establish the concentration-effect relationship.
Short- and long-term incubations should be performed; damage can either increase or decrease over a 24 h period; a decrease could possibly result from an induction of DNA repair enzymes, or a sequestration of the NPs.
Lipid peroxidation in the face of DNA damage, DNA repair and other cellular processes, by Barbara Tudek, Daria Zdżalik-Bielecka, Agnieszka Tudek, Konrad Kosicki, Anna Fabisiewicz, and Elżbieta Speina [55]
Exocyclic DNA adducts are found in the DNA of mammals unexposed and exposed to certain environmental carcinogens, such as vinyl chloride, ethylene oxide or frequent contaminants of ground water polychlorinated biphenyls, which can be converted to vinyl chloride and its metabolite chloroacetaldehyde. Endogenously, exocyclic DNA adducts are formed as a consequence of inflammation and lipid peroxidation (LPO). Complex family of LPO products gives rise to a variety of DNA adducts, which can be grouped in two classes: (i) small etheno-type adducts of strong mutagenic potential, and (ii) bulky, propano-type adducts, with alkyl side chains, which block replication and transcription, and are lethal lesions. In vitro etheno-DNA adducts are removed from the DNA by base excision repair (BER), AlkB oxidative dealkylation enzymes and driven by abasic-sites endonuclease (APE1) nucleotide incision repair (NIR). BER repairs etheno-type adducts with the highest efficiency, while NIR binds to these lesions efficiently, but remove them very slowly, and may not have much importance in cells clearance of these lesions. Substituted propano-type LPO-generated adducts are repaired by both sub-pathways of nucleotide excision repair (NER), global genome repair (GGR) and transcription-coupled repair (TCR), as well as by homologous recombination (HR). Bulky LPO-DNA adducts inhibit polymerization of the DNA and RNA, due to the fact that they both form a steric hindrance for DNA and RNA polymerases, and due to the fact that LPO-derived reactive aldehydes form adducts to various polymerases affecting their activity. LPO products also affect repair proteins performance in cells and influence several pathophysiological states of the organism, such as aging and cancerogenesis. For example, Cockayne syndrome B (CSB) ATPase, as well as Werner syndrome (WRN) helicase and exonuclease activities are abolished by one of the major LPO product, 4-hydroxynonenal (HNE). The balance between consecutive stages of BER is also changed by HNE (increase of APE1 endonuclease but decrease of ligase activities), and this results in sensitization of cells to other genotoxic agents, MMS and H2O2. Such sensitization is accompanied by increase of the level of single strand breaks due to unfinished repair.
Thus, inflammation accompanied lipid peroxidation runs multiple repair pathways, and triggers plejotropic effects on cell functioning. Changes of the level and activity of several enzymes removing exocyclic adducts from the DNA was reported during carcinogenesis. Several other (non-repair) functions of these enzymes were also recently described, which show their participation in regulation of cell proliferation and growth, as well as RNA processing. This review summarizes pathways for exocyclic adducts removal and describes how proteins involved in repair of these adducts can modify pathological states of the organism.
The cyclopurine deoxynucleotides: DNA repair, biological effects, mechanistic insights, and unanswered questions, by Philip J. Brooks [56]
The majority of chemical modifications resulting from the attack of reactive oxygen species on DNA are subject to repair by the base excision repair (BER) pathway. The 8,5-cyclopurine deoxynucleotides (cyPu) are an exception, in that while they result from the reaction of the hydroxyl radical with DNA, they are substrates for nucleotide excision repair (NER) but not BER or any other known DNA repair mechanism. NER is responsible for the removal of DNA lesions resulting from ultraviolet light, such as cyclobutane pyrimidine dimers (CPDs), and is defective in patients with the genetic disease xeroderma pigmentosum (XP). In addition to an increased risk of skin cancer, a subset of XP patients develop a progressive neurodegenerative disease called XP neurologic disease, which is thought to result from the accumulation of endogenous DNA damage that is specifically repaired by NER. As endogenous DNA lesions that are specifically repaired by NER and block transcription, cyPu have emerged as candidate DNA lesions responsible for XP neurologic disease.
This review focuses on the formation, repair and biological effects of these lesions, and highlights the important role of Tomas Lindahl and his laboratory in this research area. Recent insights into the biological effects of these lesions, particularly the mechanistic basis of the effects of cyPu lesions on transcription by RNA polymerase II, and the implications of these findings for understanding the effects of other DNA lesions on transcription are highlighted. The review also includes an updated model of the role for cyPu lesions in XP neurologic disease, and a critical evaluation of the supporting evidence. The final section addresses a key prediction of the hypothesis, which is that the NER defect results in an accumulation of cyPu lesions on the transcribed strand of active genes in neurons, and the conceptual and technologic challenges that must be overcome so that it may be directly tested.
Oxidatively-generated damage to DNA and proteins mediated by UVA photo-sensitization, by Reto Brema, Melisa Guven and Peter Karran [57]
Solar ultraviolet radiation (UV) is a complete carcinogen. The UVB that comprises around 5% of incident UV is absorbed by DNA and causes direct photochemical damage. DNA damage by the remaining UVA radiation is largely indirect and occurs following its interaction with cellular UVA chromophores and the generation of reactive oxygen species. Treatment of cells with exogenous UVA chromophores can amplify the damaging effects of UVA and this provides a means to investigate UVA-induced photodamage. Additionally, these reactions are important because many widely-prescribed medicines, including the thiopurine immunosuppressants and the fluoroquinolone antibiotics are UVA chromophores that are often associated with an increased risk of skin cancer. Introduction of a sulphur atom into canonical purine and pyrimidine bases converts them into potent UVA chromophores. These modified nucleobases can be incorporated in DNA where they act as a DNA-embedded source of reactive oxygen. Various DNA lesions that are produced by UVA activation of DNA thiopurines/thiopyrimidines and by some fluoroquinolones are described. Attention is also drawn to the potentially important contribution of protein damage caused by photosensitized UVA, which leads to the suggestion that this may contribute to the carcinogenic effects of solar radiation.
Consequences of sunlight in cellular DNA: focus on the effects of oxidatively generated DNA damage, by André Passaglia Schuch, Natalia Cestari Moreno, Natielen Jacques Schuch, Carlos Frederico Martins Menck, Camila Carrião, and Machado Garcia [58]
This review presents an overview on the incidence of solar ultraviolet (UV) radiation at the Earth’s surface, and its beneficial and adverse effects on human health. In fact, UV radiation can be absorbed by DNA and non-DNA chromophores present inside the cells, leading to the formation of DNA damage. The direct and indirect mechanisms of DNA damage formation by UVB and UVA wavelengths are presented, with emphasis on the formation of oxidized DNA bases. Once thought to be relatively innocuous, UVA is now considered an important damaging agent for many macromolecules that can also result in harmful consequences, such as carcinogenesis and skin aging. Complex cellular systems prevent the formation of such damage, including antioxidant molecules and enzymes that reduce oxidative processes. If DNA damage are produced, then several DNA repair pathways act on their removal. For unremoved lesions, DNA replication can still bypass the lesions, helping the cells to tolerate sunlight injuries.
The impact of sunlight induced DNA damage is dramatically demonstrated by rare genetic syndromes associated with DNA repair defects, such as xeroderma pigmentosum (XP). XP patients present high levels of skin lesions in the areas exposed to sunlight, including accelerated photoaging and cancer. This syndrome is associated with mutations in the genes of the Nucleotide Excision Repair (NER) pathway, which is a highly versatile and sophisticated DNA repair pathway that removes lesions that distort the DNA double helix, including the pyrimidine dimers induced by UV radiation, or to a translesion synthesis pathway (TLS). Despite the belief that these patients suffer mainly due to UV-induced photoproducts, there is growing evidence indicating that these two pathways can also protect cells from DNA oxidation products. Cells from these patients may be useful to better understand the relative roles of direct photoproducts on DNA or oxidatively generated DNA damage induced by sunlight in photoaging and carcinogenesis processes. This will be helpful not only to understand the impact of UVA and UVB on the skin damage of XP patients, but also on the skin of DNA repair proficient human individuals.
Radiation-induced clustered DNA lesions: repair and mutagenesis, by Evelyne Sage and Naoya Shikazono [59]
This is a review of the formation and repair of so-called clustered DNA lesions, possibly induced by ionizing radiation. Indeed, ionizing radiation is well known to induce oxidatively generated damage to DNA of various types, including single strand breaks (SSB), base lesion, abasic sites, repaired by base excision repair (BER), as well as double strand breaks (DSB), repaired by non-homologous end joining or homologous recombination. All these DNA lesions, when generated sparsely in the genome, are efficiently and rapidly repaired within few hours. However, clusters of ionization may occur at the end of secondary electron tracks, and lead to clustered DNA lesions, whose proportion relative to isolated DNA lesions varies depending on the type of radiation, photons, λ-rays (low Linear Energy transfer, LET, radiation) or alpha or accelerated particles (high LET radiation).
Clustered DNA lesions combine either 2 or more SSB, oxidatively generated base damage and abasic site, distributed within 1 or 2 helix turns (non-DSB clustered lesions) or a DSB surrounded by 1 or more oxidatively generated base lesion or abasic site (complex DSB). The authors explain that a hierarchy is observed in the recognition and excision/incision steps of BER of non-DSB clustered lesions, that repair intermediates such as SSB may prevent the excision of other lesions in the clusters, consequently extending the lifetime of the lesions within the cluster. The ultimate consequences are replication fork collapse and mutations. Alternatively, the processing of bistranded non-DSB clustered lesions by BER proteins may produce DSB with or without base lesions or abasic sites in the close proximity. These complex DSB, as well as those produced directly by the radiation, require ATM signalling and specific proteins of the DSB repair pathways, such as Artemis and Poly ADP ribose polymerase (PARP), and have much longer lifespan than simple DSB, with phosphate and phosphoglycolate termini.
It is proposed that those complex DSB may be repaired by alternative end-joining and lead to deletions. However, most of them are still present at S-phase and are thus repaired by homologous recombination, at the price of large deletions or sister chromatid exchanges. Some may also pass mitosis and are likely responsible for chromosomal aberrations and gross-rearrangements. The high significance of the clustered DNA lesions is also stressed, and the highly predominant role of the clustered DNA lesions in the biological effects and genome instability caused by ionizing radiation is emphasized.
Radiation-induced DNA-protein cross-links: mechanisms and biological significance, by Toshiaki Nakano, Xu Xu, Amir M. H. Salem, Mahmoud I. Shoulkamy, and Hiroshi Ide [60]
DNA is associated with various proteins involved in the folding and transaction of DNA. The reversible association of DNA with proteins ensures faithful expression and propagation of genetic information through transcription, replication, and repair. However, proteins are often covalently trapped in DNA to form DNA–protein cross-links (DPCs) when cells are exposed to DNA-damaging agents such as ionizing radiation, ultraviolet light, aldehydes, bifunctional alkylating or platinum agents. DPCs are also produced by abortive enzymatic reactions. Topoisomerases and DNA methyltransferases are trapped in a covalent complex with DNA in the presence of inhibitors. Accordingly, DPCs are ubiquitous and biologically important DNA lesions like base damage, DNA single-strand (SSBs), and double-strand (DSBs) breaks.
Enzymatic DPCs involving topoisomerases have well-defined DNA structures and have attracted considerable attention for their immediate connection to cancer chemotherapy. In contrast, DPCs produced by DNA-damaging agents have received relatively minor attention. Thus, much remains to learn about the mechanism by which cells mitigate the cytotoxic and genotoxic effects of DPCs produced by DNA-damaging agents and the biological consequences of unrepaired DPCs. In this article, we focus on radiation-induced DPCs and review the current understanding of their induction, properties, repair, and biological consequences.
Radiation-induced DNA and protein radicals react with the proximal constituents of proteins and DNA (i.e., addition of a DNA radical to a protein or vice versa), forming DPCs. While the formation of base damage, SSBs, and DSBs is suppressed under hypoxic conditions, that of DPCs is promoted, suggesting their importance in hypoxic cells, such as those present in tumors. Actin, histones, and other proteins have been identified as cross-linked proteins. In addition, covalent linkages between DNA and protein constituents such as thymine–lysine and guanine–lysine have been identified.
Radiation-induced DPCs are repaired in a biphasic manner, consisting of fast and slow components. The half-time for the fast component is 20 min–2 h and that for the slow component is 2–12 h. Notably, DPCs are repaired much slower than DSBs, suggesting that DPCs persisting in the genome impede DNA transactions and have deleterious effects on cells in conjunction with DSBs. Recently, a novel mechanism underlying DPC repair was proposed that involves the proteolytic degradation of cross-linked proteins by DPC proteases (yeast Wss1 and human Spartan) and the subsequent translesion synthesis of the resulting DNA–peptide cross-links. Cells deficient in Wss1 or Spartan share the sensitivity to formaldehyde. However, it remains to be established whether Wss1 and Spartan play a leading or supporting role in the repair of radiation-induced DPCs.
Risky repair: DNA-protein crosslinks formed by mitochondrial base excision DNA repair enzymes acting on free radical lesions, by Rachel Audrey Caston and Bruce Demple [61]
Mitochondrial DNA is exposed to oxidative damage by its close proximity to the electron transport chain and its lack of a nuclear-style chromatin structure. Mitochondrial DNA damage can cause faults in the electron transport chain, which potentiates further damage. Thus, repairing mitochondrial DNA is essential for maintaining cell function. Although they lack the array of DNA repair pathways found in the nucleus, mitochondria do have base excision DNA repair (BER). BER handles small, non-distorting but frequent lesions in the DNA, such as most of those generated by oxygen radicals. BER is initiated when a DNA glycosylase removes a damaged base to generate an abasic site, followed by incision of the DNA by Ape1 protein and recruitment of the repair DNA polymerase. The simplest pathway replaces a single nucleotide, with the polymerase using a separate lyase activity to remove the 5′-deoxyribose-5-phosphate residue, and finally ligation. “Long-patch” repair instead replaces multiple nucleotides, displacing an oligonucleotide flap that must be excised to permit ligation. In mitochondria, potential flap-excising nucleases are Fen1, DNA2, ExoG, and MGME1.
BER can handle most damage with the simple, single-nucleotide process, but some lesions require long-patch BER. A significant lesion processed in this way is 2-deoxyribonolactone, an oxidized abasic site that otherwise generates a dead-end DNA-protein crosslink with DNA polymerase β in short-patch BER during attempted removal of the 5′-dRP. Other lesions can trap some other repair proteins, such as DNA glycosylases and lysases, so the problem of preventing and handling the crosslinks is a broader one.
Accumulation of these repair-mechanism-driven DNA-protein crosslinks is cytotoxic. In the nucleus, they are tagged with ubiquitin for degradation by the proteasome. Mitochondria do not have a proteasome, but they do possess proteases used for quality control of proteins, notably the AAA proteases. It is also possible that heavily damaged mitochondrial DNA genomes could be eliminated and replaced via the replication of other mitochondrial DNA molecules in the shared space of the mitochondrion.
MTH1 as a nucleotide pool sanitizing enzyme: friend or foe? By Yusaku Nakabeppu, Eiko Ohta and Nona Abolhassani [62]
Among all the nucleobases, guanine is the most susceptible to oxidation by reactive oxygen species, such as ·OH or 1O2. Exposure of guanine in DNA or free 2′-deoxyguanosine 5′-triphosphate (dGTP) to reactive oxygen species adds oxygen to the C-8 carbon to generate 8-oxo-7,8-dihydroguanine (GO) or 8-oxo-dGTP, respectively. Accumulation of GO in nuclear or mitochondrial genomes may cause mutagenesis or programmed cell death. To avoid such deleterious effects of GO, accumulation of GO in cellular genomes is effectively minimized by the actions of MutT homolog-1 (MTH1) with 8-oxo-dGTPase, OGG1 with GO DNA glycosylase and MutY homolog (MUTYH) with adenine DNA glycosylase. Mth1/Ogg1/Mutyh-triple knockout mice are highly mutagenic and developed many tumors in various tissues demonstrating that the defense systems efficiently minimize GO accumulation in cellular genomes. Mth1/Ogg1-double knockout mice also increased GO accumulation in the genome, however, unexpectedly little susceptibility to spontaneous tumorigenesis was observed, and thus revealing that accumulation of GO in cellular genomes induces programmed cell death which is induced by MUTYH-initiated base excision repair.
Cancer tissues are likely to be under increased oxidative conditions, and thus cancer cells accumulate a high level of 8-oxo-dGTP in their nucleotide pools. Cancer cells are highly proliferative, therefore higher levels of GO may be easily incorporated into their genomes. Cancer cells express increased levels of MTH1, and thus eliminate 8-oxo-dGTP from the nucleotide pools to minimize GO accumulation in the genomes. The increased expression of MTH1 protects cancer cells from the deleterious effects of GO, however, it may be detrimental for cancer patients. On the other hand, Mth1/Ogg1-double knockout mice are highly vulnerable to neurodegeneration under oxidative conditions, with increased accumulation of GO in both mitochondrial genomes of neurons and nuclear genome of microglia. Expression of human MTH1 transgene efficiently prevents neurodegeneration by avoiding GO accumulation in the brain, indicating that increased expression of MTH1 is beneficial for neuronal tissues. GO accumulated in nuclear genome of microglia results in microglial activation, which depends on MUTYH-initiated base excision repair of adenine inserted opposite GO during DNA replication, thus exacerbating neurodegeneration.
Chromatin associated mechanisms in base excision repair – nucleosome remodeling and DNA transcription, two key players, by Hervé Menoni, Paolo Di Mascio, Jean Cadet, Stefan Dimitrov, and Dimitar Angelov [63]
DNA within all living cells is constantly damaged by exogenous and endogenous factors such as radiation, alkylating agents and reactive oxygen species. Base excision repair (BER), discovered by Tomas Lindahl, is the main repair pathway of the DNA non-helix disturbing lesions such as the oxidatively generated 8-oxoG, thymine glycols etc. BER enzymes have been extensively characterized and the entire BER pathway reconstituted in vitro with naked DNA substrates However, much less is known about how BER enzymes function in the context of the chromatin that packages DNA in eukaryotes.
The basic subunit of chromatin is the nucleosome core particle that includes approximately 146 bp of DNA wrapped almost twice around a core histone octamer which comprises basically of a pair of each histone H2A, H2B, H3 and H4. The nucleosome core particles are connected by a 20 to 90 bp long linker DNAs. In the presence of the linker histone H1 that binds at the entry and exit site of the DNA the nucleosomal array condense into the so-called 30 nm fiber. The chromatin and in particular the nucleosomes present themselves as obstacles for DNA repair and this repression arises mainly due to the stearic hindrance of the histone octamer around the wrapped DNA. The malleability of the DNA in the chromatin is highly constrained and hence any protein or enzyme that needs to bind or carry out catalysis requires severe alteration of the DNA structure.
BER initiation and completion of a large proportion of DNA lesions or repair-intermediates remain refractory within the nucleosome (chromatin). To override these restrictions and repressions modulation of the chromatin structure takes place mainly by either one of the following epigenetic strategies, namely, the incorporation of histone variants, post translational modifications of core histones and ATP-dependent nucleosome remodeling.
The review of Menoni et al. is dedicated on the state of the art high-lightening of the in vitro and in cellulo chromatin-associated mechanisms of the eukaryote BER. An emphasis on the role of the quasi-stochastically generated partially accessible metastable nucleosomal species “remosomes” by the SWI/SNF family of ATP-dependent chromatin remodeling is presented. Namely, arguments are provided in a favor of hypothesis of quasi-stochastic remodeler-driven access to BER initiation glycosylases through transient chromatin “fluidification” by remosome generation. Based on recent findings of the recruitment of the histone chaperone complex FACT and the remodeler CSB at oxidatively generated DNA damage it is suggested that another mechanism to overcoming chromatin barrier to BER initiation is the pervasive (intergenic) transcription that could play a role as both DNA damage sensor and/or chromatin “permeabilizer”. Briefly, the review of Menoni et al suggests the idea that the cheapest way for the cell to proceed with the removal of a stochastically generated DNA damage is to use stochastically induced repair initiators such as nucleosome remodelers and RNA polymerases. This stochastic chromatin dynamicity is essential to carry out the surveillance of the DNA by making it accessible to repair.
Hide and seek: How do DNA glycosylases locate oxidatively damaged DNA bases amidst a sea of undamaged bases? By Andrea J. Lee and Susan S. Wallace [64]
When Tomas Lindahl published his seminal paper on the discovery of Escherichia coli uracil DNA glycosylase in 1974, those in the “DNA repair field” had no idea that this enzyme, and its soon to be discovered allies, would be the first enzyme in the Base Excision Repair pathway conserved from bacteria to man, and responsible for repairing some 40,000 endogenous DNA damages per human cell per day. E. coli glycosylases that remove DNA lesions produced by free radicals were identified shortly after, Nth by our group, Fpg by Tomas, and much later, Nei by our group. Since those early days, a substantial literature has developed describing the biochemical properties and biological functions of these glycosylases as well as their eukaryotic orthologs and functional homologs. It was obvious at the outset that DNA glycosylases do not require ATP to remove lesions and they apparently located their substrates in DNA using only thermal energy. Since the majority of the damages recognized by DNA glycosylases do not majorly distort the DNA helix, to locate these lesions in the sea of undamaged bases appeared to be a formidable task. It is only very recently, with the advent of high resolution single molecule microscopy, that major inroads have been made into observing the glycosylase search for damage in real time.
This Review describes how the three bacterial DNA glycosylases that recognize free radical-damaged bases, Nth, Fpg and Nei, are able to diffusively scan the DNA molecule, randomly, bidirectionally and rotationally, in an attempt to locate their substrates. These glycosylases periodically insert an amino acid wedge residue into the DNA helix to probe for minor distortions, as they redundantly search tracks 450–600 base pairs in length. Given the number of each of these glycosylases in an E. coli cell, this process is sufficiently efficient to allow the entire E. coli chromosome to be queried for DNA damage once every ten minutes. We now have a much better understanding of how specialized interactions between DNA glycosylases and the DNA helix facilitate efficient repair of lesions.
Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: properties and biological roles of the PFG and Ogg1 8-oxoguanine DNA N-glycosylases. Serge Boiteux, Franck Coste, and Bertrand Castaing [65]
Tomas Lindahl and his coworkers critically advanced our understanding of DNA repair by identifing and characterizing a great number of DNA repair proteins involved in different DNA repair processes. Among these processes, the base excision repair (BER) pathway figures prominently. In 1977, the purification of the Uracil DNA N-glycosylase from E. coli was a seminal study in the field of BER. At that time, one of us (SB) joined the laboratory headed by Dr. Jacques Laval, which was the leader in the field of BER in France. The link between our present review and Tomas Lindahl is obvious, since the 2,6-diamino-4-hydroxy-5N-methylformamidopyrimidine (N7-MeFapyG)-DNA N-glycosylase of E. coli (today known as Fpg) was first described in Lindahl’s laboratory (1979). This work definitely inspired our research in the following years. Finally, in 1987, we cloned the fpg gene from E. coli, coding for the Fpg protein, which is one of the two main topics of our review. It should be noted that at that time, Fpg was associated with the repair of alkylation DNA damage, not oxidative DNA damage. The role of Fpg in the repair of oxidatively damaged DNA emerged later on, when Fpg was shown to release 8-oxo-7,8-dihydroguanine (8-oxoG) and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) from λ irradiated DNA (1992). The transition from DNA alkylation to oxidation field was conducted in collaboration with eminent colleagues, Drs. Jean Cadet, Miral Dizdaroglu, Bernd Epe and Peter Nehls. The transition from bacterial Fpg to human OGG1 was achieved through genetic and biochemical studies using the yeast Saccharomyces cerevisiae as a model (1996–97) and summarized in this review.
The impact of 8-oxoG on genetic stability was revealed by the strong spontaneous mutator phenotype of bacterial and yeast mutants affected in the repair of this lesion in DNA and nucleotide pools. The 8-oxoG damaged base is primarily repaired by BER initiated by a DNA N-glycosylase, Fpg and OGG1 in prokaryotic and eukaryotic cells, respectively. In addition, Fpg and OGG1 were shown to cooperate with other functions, yielding complex networks, which are detailed in this review. Furthermore, mice deficient in 8-oxoG repair functions (OGG1 and MUTYH) develop cancer in various organs at adult age, which points to the critical impact of 8-oxoG repair on genetic stability in mammals. This review focuses on Fpg and OGG1, their biochemical and structural properties as well as their biological roles.
Repair of 8-oxoG:A mismatches by the MUTYH glycosylase: mechanisms, metals & medicine, by Douglas M. Banda, Nicole N. Nunez, Michael A. Burnside, Katie M. Bradshaw, and Sheila S. David [66]
This article summarizes new work on the functional and structural properties of bacterial MutY glycosylases and their human homolog MUTYH. MutY was originally implicated in mismatch repair as activity that led to the restoration of G:A mismatches to G:C base-pairs. Importantly, the discovery of Uracil-DNA glycosylase and Base Excision Repair (BER) by Tomas Lindahl suggested an alternative role for MutY in this apparent mismatch repair activity. Indeed, MutY was found to be a G:A mismatch specific adenine glycosylase that initiates the BER pathway. Later work established that MutY enzymes place a key role in preventing mutations associated with 8-oxoG by removing adenine from pro-mutagenic 8-oxoG:A mismatches. Knowledge of these fundamental features of MutY, BER and 8-oxoG were crucial in the discovery of a colorectal cancer (CRC) predisposition syndrome involving variants of MUTYH, referred to an MUTYH-associated polyposis (MAP).
Surprisingly, despite MutY’s discovery almost 30 years ago, new insight into the structural and functional properties of MutY, MUTYH and MAP variants continue to un-fold. For example, recent investigations of MAP variants and detailed sequence alignments led to the discovery of Zn2+ linchpin binding motif in the linker domain of mammalian homologs of MutY that is required for efficient repair. Studies on MutY and MAP variants have also further established the importance of the [4Fe-4S]2+ cluster cofactor in locating DNA damage. Recent structural and mechanistic insights on MutY enzymes have also prompted revisions to the accepted mechanism for MutY. These studies have shown that MutY is a retaining N-glycosylase, and suggest that by analogy to O-glycosidases that the catalytic mechanism involves formation of a covalent MutY-DNA intermediate. MUTYH has also been implicated to play roles in other diseases beyond MAP, including intriguing links between MUTYH and neurological disorders that are highlighted in this review. These various aspects of MUTYH collectively accentuate how the intricacies of damage recognition and chemistry of base excision of MutY and MUTYH are intimately tied to their biological roles in preserving the genome and preventing disease.
DNA damage related crosstalk between the nucleus and mitochondria, by Mohammad Saki and Aishwarya Prakash [67]
The ~16.5 kb human mitochondrial genome is distinct from the nuclear genome and encodes for 13 polypeptides, 22 tRNAs, and two rRNAs that partake in oxidative phosphorylation. Akin to its nuclear counterpart, mitochondrial DNA (mtDNA) is also subjected to DNA damage caused by damaging agents such as reactive oxygen species that are generated by both endogenous processes and exogenous factors. If left unrepaired, accumulated damaged mtDNA may eventually lead to mitochondrial dysfunction and diseases such as aging related neurodegenerative disorders and some cancers. While multiple repair pathways exist in the nucleus to maintain genomic integrity, the base excision repair (BER) pathway is the primary pathway involved in the repair of damaged mtDNA. The majority of the mitochondrial proteome comprises proteins, replication factors, and repair enzymes that are encoded for by nuclear genes and translocated to the mitochondria for maintenance of the mitochondrial genome.
Since cells respond to a variety of stressors that include but are not limited to DNA damaging agents, nutrient deprivation, and changes in ATP levels, crosstalk between the nucleus and the mitochondria becomes necessary and inevitable. While many strides have been made in furthering our understanding of the dynamic nature of the mitochondrial proteome and stress responses in event of mitochondrial dysfunction, much still remains to be elucidated. This review summarizes current understanding of the DNA repair pathways that occur in mitochondria and highlight the repair enzymes in each pathway that have been observed in this organelle thus far. The bidirectional mitochondrial-nuclear signaling pathways are also examined by focusing on the molecules that mediate crosstalk by acting as messengers between the two organelles.
Coordination of DNA single strand break repair, by Rachel Abbotts and David M. Wilson III [68]
This review by Abbotts and Wilson considers the repair of DNA single strand breaks (SSBs) primarily in the context of two proteins central to pathway coordination, poly(ADP-ribose) polymerase 1 (PARP1) and X-ray cross-complementing protein 1 (XRCC1). Single strand breaks (SSBs) are estimated to occur at a frequency of ~10,000 per cell per day, of which the majority are endogenous in origin. Reactive oxygen species, particularly hydroxyl radicals, are a common source. Modification of the sugar phosphate backbone by reactive oxygen species may result in disintegration of the oxidized sugar, generating a strand break. Indirect SSB formation may occur during the repair of damaged nucleotides by the base excision repair (BER) pathway, which generates a strand nick as an obligate intermediate. Similarly, SSBs may be generated by failed religation of DNA incised by TOP1 or RNase H2 activity. Unrepaired, SSBs can stall replication machinery, which may activate the error-prone damage tolerance mechanism of translesion synthesis, or may lead to fork collapse into a potentially cytotoxic double strand break.
The repair of SSBs is performed by SSB repair (SSBR), often considered a subpathway of BER. The major steps of SSBR are 1) strand break detection; 2) removal of 5′- or 3′-terminal blocking groups; 3) gap-filling repair synthesis; and 4) nick-sealing by a DNA ligase. Recognition and binding of SSBs by PARP1 is a critical step in the initiation of SSBR, resulting in a conformational rearrangement that activates PARP1 catalytic activity. PARP1 catalyzes the transfer of an ADP-ribose subunit from NAD+ to amino acid side chains of acceptor proteins, forming branched chains of poly(ADP-ribose) (PAR) molecules. Although several acceptor proteins have been recognized, the primary target of PARylation is PARP1 itself. Automodification has the primary role of stimulating the recruitment of XRCC1, a non-enzymatic scaffold protein which, in concert with PARP1, recruits and stimulates the proteins involved in SSBR, including DNA polymerase β, ligase 3, and enzymes involved in processing of termini blocking groups. This review summarizes the literature regarding the discovery, structure, function, and protein-protein interactions of the PARP1-XRCC1 ensemble in the context of SSBR, including the role of SSBR in neurological disease.
Mechanistic and biological considerations of oxidatively damaged DNA for helicase-dependent pathways of nucleic acid metabolism, by Jack D. Crouch, Robert M. Brosh, Jr. [69]
Helicases are a class of enzymes responsible for remodeling structured nucleic acids. As such, they regularly encounter a spectrum of oxidatively damaged DNA modifications ranging from small non-helix distorting lesions to bulkier adducts that cause significant structural changes to the DNA double helix. Cells are only able to tolerate a limited extent of oxidatively damaged DNA, relying on DNA damage response and repair pathways to maintain chromosomal stability and cellular homeostasis. While DNA helicases by their very nature share certain fundamental catalytic properties, they vary in terms of their substrate specificity, cellular functional roles, and participation in signaling pathways to help cells cope with oxidatively damaged DNA. For example, redox active Fe-S cluster helicases may be recruited to an oxidative lesion which disrupts normal charge transport through the DNA double helix. Certain specialized DNA helicases are known to deal with guanine (G)-rich sequences known to form an alternative DNA structure designated a G-quadruplex that is highly susceptible to oxidation and potentially pathological. These G-rich sequences are found with high density in promoters, ribosomal DNA repeats, and telomere repeats of nuclear DNA, as well as in the mitochondrial genome. Oxygen radicals produced via oxidative phosphorylation pose a significant threat to macromolecules within the mitochondria, including the organelle’s nucleic acid. Multiple helicases localize to mitochondria to preserve the integrity of the mitochondrial genome, but the precise roles of these helicases have yet to be elucidated. In summary, helicases play prominent roles in disease and cancer, and assist in repairing oxidatively damaged DNA to maintain genomic stability.
Oxidatively generated base modifications in DNA: not only carcinogenic risk factor but also regulatory mark? By Marco Seifermann and Bernd Epe [70]
This review deals with recent unexpected findings which indicate that the generation of 8-oxo-7,8-dihydroguanine (8-oxoG) in the nuclear DNA is not always an accidental and potentially harmful event that causes mutations and initiates carcinogenesis. Rather, it can fulfill important regulatory functions in gene transcription and signal transduction. Evidence for two quite different scenarios have been obtained. Both of them can help to explain that mice deficient in OGG1, the major repair glycosylase for 8-oxoG, not only accumulate 8-oxoG in their genomes and have elevated spontaneous mutations rates, but also show a reduced immune response in several experimental settings.
In the first mechanism, a localized formation of 8-oxoG in regulatory regions of certain genes takes place, which at least in some cases is caused by a specific oxidase, the lysine-specific histone demethylase LSD1, which produces H2O2 as a stoichiometric by-product of its enzymatic activity. The recognition of the locally induced DNA modifications by OGG1 subsequently activates or enhances the transcription of the affected genes, as originally shown for estrogen-dependent genes and later also for genes dependent on the major immune regulatory transcription factor NF-second mechanism is based on the finding that OGG1 forms a tight complex with the excised free base 8-oxoGua and that this complex can act as a guanine nucleotide exchange factor (GEF) for small GTPases such as RAS, RAC and RHO, thus activating the signalling of these proteins. In accordance with this mechanism, low doses of the free base 8-oxoGua can substitute for expression changes of immune response genes observed under oxidative stress.
Several details of the suggested mechanisms are still puzzling. Thus, a stoichiometric production of H2O2 by LSD1 is expected to be a rather inefficient source of 8-oxoG generation in DNA, while a more distant (extranuclear) source of reactive oxygen species would inevitably cause high collateral DNA damage. Also, the advantages of a dual function of OGG1 as repair glycosylase in the nucleus and as regulatory GEF in the cytosol and of an apparently DNA damage dependent regulation of an immune response are not clear. Thus, the convincing indications for a role of 8-oxoG and OGG1 in gene regulation still give rise to challenging questions for future research.
Aberrant base excision repair pathway of oxidatively damaged DNA: implications for degenerative diseases, by Ibtissam Talhaoui, Bakhyt T. Matkarimov, Thierry Tchenio, Dmitry O. Zharkov, and Murat K. Saparbaev [71]
DNA repair systems have evolved to discriminate between regular and modified nucleobases. For example, DNA glycosylases can recognize and excise damaged bases among a vast majority of regular bases in the base excision repair pathway. However, mispairs consisting of two regular bases, which occur due to DNA polymerase errors in replication and spontaneous conversion of 5-methylcytosine to thymine, present a challenge to DNA repair. To counteract these mutagenic threats to genome stability, cells possess specific DNA repair pathways that can target a non-damaged DNA strand to remove mismatched regular DNA bases.
This review describes numerous observations showing that under certain circumstances these repair pathways can go awry and aberrantly remove a non-substrate base (often undamaged). In the ensuing round of repair, some of these aberrant events lead to mutagenic repair; alternatively, the same nucleotide may be incorporated initiating multiple rounds of futile repair. Oxidative stress and environmental insults induce a variety of DNA base lesions, which are gradually accumulated during aging. Indeed, non-dividing cells might amass more oxidative damage to their genome as compared to proliferating ones because DNA is no more replicated. The genotoxic effects of this unrepaired DNA damage can be worsened by the aberrant excision of the undamaged base, which either initiates futile cycle of DNA repair or generates mutations in the non-damaged DNA strand. Based on the analysis of existing data, it is suggested that the removal of spontaneous and induced DNA damage in DNA repair-deficient and non-dividing mammalian cells could proceed in an aberrant manner leading to persistent DNA damage response and the production and accumulation of mutant proteins in the absence of DNA replication. This may be a factor contributing to age-related diseases such as cancer and degenerative conditions and also to natural aging.
Single nucleotide polymorphisms in DNA glycosylases: from function to disease, by Mariarosaria D’Errico, Eleonora Parlanti, Barbara Pascucci, Paola Fortini, Sara Baccarini, Valeria Simonelli, and Eugenia Dogliotti [72]
Numerous single nucleotide polymorphisms (SNPs) have been identified in DNA repair genes and a plethora of studies have attempted to establish the association of SNP-mediated individual susceptibility with disease. However, only a few SNPs have been validated by functional analysis and the investigation of their “causality” is hampered by lack of statistical power in most studies to investigate gene-gene and gene-environment interactions. In this extensive review of studies addressing the role of SNPs of DNA glycosylases, notwithstanding these limitations, a convincing picture emerged that these genetic variants may be susceptibility factors for diseases which all present as major risk factor oxidative stress. The overall disease spectrum includes cancer (mostly of lung, breast and gastrointestinal tract), ocular and cochlear disorders, myocardial infarction, neurodegenerative disorders and obesity which can be all grouped under the umbrella of oxidative-stress-related pathologies. The phenotype of knock-out mice and the clinical features of the few human diseases with full inactivation of these DNA glycosylases support this conclusion. Future research should address the role of genomic variations in DNA repair pathways in a broader spectrum of disease than cancer.
Role of the oxidized form of XRCC1 in protection against extreme oxidative stress, by Julie K. Horton, Hannah J. Seddon, Ming-Lang Zhao, Natalie R. Gassman, Agnes K. Janoshazi, Donna F. Stefanick, and Samuel H. Wilson [73]
The mammalian base excision repair factor XRCC1 is a redox sensitive protein. XRCC1 undergoes a large redox-mediated conformational change in its N-terminal domain as revealed by crystallographic and NMR analyses. The oxidized form is stabilized by a sulfhydryl covalent bond between Cys12 and Cys20 and the oxidized form of XRCC1 binds to its DNA repair partner DNA polymerase β much tighter (50-fold) than the reduced form of XRCC1. However, both forms of XRCC1 are present in cultured mouse fibroblasts in approximately equal amounts.
The interaction between XRCC1 and DNA polymerase β is important for recruitment of DNA polymerase β to sites of laser-induced DNA damage and for successful base excision repair, but differences in the roles of the two forms of XRCC1, as this relates to the oxidative stress response, is not known. In a previous study, evidence was obtained for a deficiency in base excision repair in mouse fibroblasts by removing the ability of XRCC1 to take its oxidized form. This was accomplished by comparing XRCC1 null cells expressing the C12A XRCC1 mutant protein locked in its reduced conformation with cells expressing wild-type XRCC1 protein. Expression levels of the two forms of XRCC1 were similar in the two cells lines.
In the present work, these observations were extended and revealed that the reduced form of XRCC1 (C12A) was successfully recruited to sites of micro-irradiation DNA damage, but provided significantly slower recruitment of polymerase β than wild-type XRCC1. Expression of reduced XRCC1 did not change resistance to MMS or H2O2. Extreme oxidative stress imposed by glutathione depletion resulted in enhanced sensitivity to H2O2 in C12A cells compared with wild-type XRCC1-expressing cells. In addition, elevated cellular PAR levels were found in reduced XRCC1-expressing cells following H2O2 exposure. This suggested a base excision repair deficiency of H2O2-induced DNA damage in the C12A-expressing cells. These results indicate the importance of the redox status of XRCC1 in the cellular response to oxidative stress.
Acknowledgments
KJAD was supported by grant # ES 003598 from the National Institute of Environmental Health Sciences of the US National Institutes of Health, and by grant # AG 052374 from the National Institute on Aging of the US National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cressey D. DNA repair sleuths win chemistry Nobel. Nature. 2015;526:307–308. doi: 10.1038/nature.2015.18515. [DOI] [PubMed] [Google Scholar]
- 2.Stokstad E. Nobel Prizes. DNA’s repair tricks win chemistry’s top prize. Science. 2015;350:266. doi: 10.1126/science.350.6258.266. [DOI] [PubMed] [Google Scholar]
- 3.Carell T. DNA Repair. Angew Chem Int Ed Engl. 2015;54:15330–15333. doi: 10.1002/anie.201509770. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T, Modrich P, Sancar A. The 2015 Nobel prize in chemistry. The discovery of essential mechanisms that repair DNA damage. J Assoc Genet Technol. 2016;42:37–41. [PubMed] [Google Scholar]
- 5.Lindahl T. My journey to DNA repair. Genomics Proteomics Bioinformatics. 2013;11:2–7. doi: 10.1016/j.gpb.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer NC, Corbett AH, Doetsch PW. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 2015;43:10083–10101. doi: 10.1093/nar/gkv1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller JH, Goodman MF. Tomas Lindahl: 2015 Nobel laureate. DNA Repair (Amst) 2016;37:A29–A34. doi: 10.1016/j.dnarep.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Lindahl T. The intrinsic fragility of DNA (Nobel Lecture) Angew Chem Int Ed Engl. 2016;55:8528–8534. doi: 10.1002/anie.201602159. [DOI] [PubMed] [Google Scholar]
- 9.Friedberg EC. A history of the DNA repair and mutagenesis field: The discovery of base excision repair. DNA Repair (Amst) 2016;37:A35–A39. doi: 10.1016/j.dnarep.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Mi S, Klungland A, Yang YG. Base-excision repair and beyond --A short summary attributed to scientific achievements of Tomas Lindahl, Nobel prize laureate in chemistry 2015. Sci China Life Sci. 2016;59:89–92. doi: 10.1007/s11427-015-4983-4. [DOI] [PubMed] [Google Scholar]
- 11.Modrich P. Mechanisms in E. coli and human mismatch repair (Nobel Lecture) Angew Chem Int Ed Engl. 2016;55:8490–8501. doi: 10.1002/anie.201601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radman M. Mismatch repair earns Nobel prize in chemistry 2015 to Paul Modrich for a biochemical tour de force. DNA Repair (Amst) 2016;37:A22–A28. doi: 10.1016/j.dnarep.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Li GM. A personal tribute to 2015 Nobel laureate Paul Modrich. DNA Repair (Amst) 2016;37:A14–21. doi: 10.1016/j.dnarep.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Orren DK. The Nobel prize in chemistry 2015: Exciting discoveries in DNA repair by Aziz Sancar. Sci China Life Sci. 2016;59:97–102. doi: 10.1007/s11427-015-4994-1. [DOI] [PubMed] [Google Scholar]
- 15.Sancar A. Mechanisms of DNA Repair by Photolyase and Excision Nuclease (Nobel Lecture) Angew Chem Int Ed Engl. 2016 Jul 18;55(30):8502–27. doi: 10.1002/anie.201601524. [DOI] [PubMed] [Google Scholar]
- 16.Van Houten B. A tale of two cities: A tribute to Aziz Sancar’s Nobel Prize in Chemistry for his molecular characterization of NER. DNA Repair (Amst) 2016;37:A3–A13. doi: 10.1016/j.dnarep.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koshland DE., Jr Molecule of the year: the DNA repair enzyme. Science. 1994;265:1925. doi: 10.1126/science.7801114. [DOI] [PubMed] [Google Scholar]
- 18.Davies KJA. (1995) Oxidative Stress: the paradox of aerobic life. Biochem Soc Symp. 61:1–31. doi: 10.1042/bss0610001. [DOI] [PubMed] [Google Scholar]
- 19.Davies KJA, Davies JMS, Cadenas E, Packer L, Sevanian A, Forman HJ, Chan TM, Landolph JR, Ursini F. The Oxygen Paradox. In: Davies KJA, Ursini F, editors. The Oxygen Paradox. CLEUP University Press; Padova (Italy): 1995. pp. 1–9. [Google Scholar]
- 20.Davies KJA. (2000) An overview of oxidative stress. IUBMB Life. 2000;50:1–4. doi: 10.1080/713803723. [DOI] [PubMed] [Google Scholar]
- 21.Pryor WA, Houk K, Foote CS, Fucuto J, Ignarro L, Squadrito G, Davies KJA. Free radical biology and medicine: it’s a gas! Am J Physiol Regul Integr Comp Physiol. 2006;291:R491–R511. doi: 10.1152/ajpregu.00614.2005. [DOI] [PubMed] [Google Scholar]
- 22.Davies KJA. The broad spectrum of responses to oxidants in proliferating cells: A new paradigm for oxidative stress. IUBMB Life. 1999;48:41–47. doi: 10.1080/713803463. [DOI] [PubMed] [Google Scholar]
- 23.Davies KJA. Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life. 2000;50:279–289. doi: 10.1080/713803728. [DOI] [PubMed] [Google Scholar]
- 24.Davies KJA. The broad spectrum of responses to oxidative stress in proliferating cells. Handbook of Environmental Chemistry. 2005;2:63–75. [Google Scholar]
- 25.Davies KJA. Protein modification by oxidants and the role of proteolytic enzymes. Biochem Soc Trans. 1993;21:346–353. doi: 10.1042/bst0210346. [DOI] [PubMed] [Google Scholar]
- 26.Davies KJA. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 27.Bota D, Davies KJA. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nature Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 28.Van Kuijk FJGM, Sevanian A, Handelman GJ, Dratz EA. A new role for phospholipase A2: Protection of membranes from lipid peroxidation damage. Trends Biochem Sci. 1987;12:31–34. [Google Scholar]
- 29.Calabrese EJ, Baldwin LA. Toxicology rethinks its central belief – hormesis demands a reappraisal of the way risks are assessed. Nature. 2003;421:691–692. doi: 10.1038/421691a. [DOI] [PubMed] [Google Scholar]
- 30.Davies KJA. Adaptive homeostasis. Mol Aspects Med. 2016;49:1–7. doi: 10.1016/j.mam.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pickering AM, Staab TA, Tower J, Sieburth D, Davies KJA. A conserved role for the 20S proteasome and Nrf2 transcription factor in oxidative-stress adaptation in mammals, C. elegans and D. melanogaster. J Exptl Biol. 2013;216:543–553. doi: 10.1242/jeb.074757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pomatto LC-D, Davies KJA, Tower JG. The mitochondrial lon protease regulates age-specific and sex-specific adaptation to oxidative stress. Curr Biol. 2016 doi: 10.1016/j.cub.2016.10.044. (published on-line ahead of print) http://dx.doi.org/10.1016/j.cub.2016.10.044. [DOI] [PMC free article] [PubMed]
- 33.Raynes R, Juarez C, Pomatto LC-D, Sieburth D, Davies KJA. Aging and SKN-1-dependent loss of 20S proteasome adaptation in C. elegans. J Gerontol: Biol Sci. 2016 doi: 10.1093/gerona/glw093. (published on-line ahead of print June 2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harman D. Aging: A theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 35.Pacifici RE, Davies KJA. Protein, lipid, and DNA repair systems in oxidative stress: the free radical theory of aging revisited. Gerontology. 1991;37:166–180. doi: 10.1159/000213257. [DOI] [PubMed] [Google Scholar]
- 36.Davies KJA. The oxygen paradox, oxidative stress, and ageing. Arch Biochem Biophys. 2015;595:28–32. doi: 10.1016/j.abb.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanawalt PC, Wilson SH. Cutting-edge perspectives in genomic maintenance III: preface. DNA Repair (Amst) 2016;44:1–3. doi: 10.1016/j.dnarep.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 38.Lindahl T. An N-glycosylase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci USA. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chetsanga CJ, Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979;6:3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cadet J, Douki T, Ravanat JL. Oxidatively generated base damage to cellular DNA. Free Radic Biol Med. 2010;49:9–21. doi: 10.1016/j.freeradbiomed.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 41.Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollman AP, Nishimura S. 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci USA. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boiteux S, O’Connor TR, Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987;6:3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dianov G, Lindahl T. Reconstitution of the DNA base excision-repair pathway. Curr Biol. 1994;4:1069–1076. doi: 10.1016/s0960-9822(00)00245-1. [DOI] [PubMed] [Google Scholar]
- 44.Ischenko AA, Saparbaev MK. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature. 2002;415:183–187. doi: 10.1038/415183a. [DOI] [PubMed] [Google Scholar]
- 45.Mo JY, Maki H, Sekiguchi M. Hydrolytic elimination of a mutagenic nucleotide, 8-oxodGTP, by human 18-kilodalton protein: sanitization of nucleotide pool. Proc Natl Acad Sci USA. 89:11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 47.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl Acad Sci USA. 97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Formation and repair of oxidatively generated damage in cellular DNA. Cadet Jean, Davies Kelvin JA, Medeiros Marisa HG, DiMascio Paolo, Richard Wagner J. doi: 10.1016/j.freeradbiomed.2016.12.049. PUBLISHED IN THIS ISSUE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Formation and processing of DNA damage substrates for the hNEIL enzymes. Fleming Aaron M, Burrows Cynthia J. doi: 10.1016/j.freeradbiomed.2016.11.030. PUBLISHED IN THIS ISSUE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Removal of oxidatively generated DNA damage by overlapping repair pathways. Shafirovich Vladimir, Geacintov Nicholas E. doi: 10.1016/j.freeradbiomed.2016.10.507. PUBLISHED IN THIS ISSUE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oxidized C5-methyl cytosine bases in DNA: 5-hydroxymethylcytosine; 5-formylcytosine; and 5-carboxycytosine. Klungland Arne, Robertson Adam B. doi: 10.1016/j.freeradbiomed.2016.11.038. PUBLISHED IN THIS ISSUE. [DOI] [PubMed] [Google Scholar]
- 54.Sensitive detection of oxidative damage to DNA induced by nanomaterials. Collins Andrew, El Yamani Naouale, Dusinska Maria. doi: 10.1016/j.freeradbiomed.2017.02.001. PUBLISHED IN THIS ISSUE. [DOI] [PubMed] [Google Scholar]
- 55.Lipid peroxidation in the face of DNA damage, DNA repair and other cellular processes. Tudek Barbara, Zdżalik-Bielecka Daria, Tudek Agnieszka, Kosicki Konrad, Fabisiewicz Anna, Speina Elżbieta. doi: 10.1016/j.freeradbiomed.2016.11.043. PUBLISHED IN THIS ISSUE. [DOI] [PubMed] [Google Scholar]
- 56.The cyclopurine deoxynucleotides: DNA repair, biological effects, mechanistic insights, and unanswered questions. Brooks Philip J. doi: 10.1016/j.freeradbiomed.2016.12.028. PUBLISHED IN THIS ISSUE. [DOI] [PubMed] [Google Scholar]
- 57.Oxidatively-generated damage to DNA and proteins mediated by UVA photo-sensitization. Brema Reto, Guven Melisa, Karran Peter. doi: 10.1016/j.freeradbiomed.2016.10.488. PUBLISHED IN THIS ISSUE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Consequences of sunlight in cellular DNA: focus on the effects of oxidatively generated DNA damage. Schuch André Passaglia, Moreno Natalia Cestari, Schuch Natielen Jacques, Menck Carlos Frederico Martins, Carrião Camila, Garcia Machado. PUBLISHED IN THIS ISSUE. [Google Scholar]
- 59.Radiation-induced clustered DNA lesions: repair and mutagenesis. Sage Evelyne, Shikazono Naoya. doi: 10.1016/j.freeradbiomed.2016.12.008. PUBLISHED IN THIS ISSUE. [DOI] [PubMed] [Google Scholar]
- 60.Radiation-induced DNA-protein cross-links: mechanisms and biological significance. Nakano Toshiaki, Xu Xu, Salem Amir MH, Shoulkamy Mahmoud I, de Hiroshi I. doi: 10.1016/j.freeradbiomed.2016.11.041. PUBLISHED IN THIS ISSUE. [DOI] [PubMed] [Google Scholar]
- 61.Risky repair: DNA-protein crosslinks formed by mitochondrial base excision DNA repair enzymes acting on free radical lesions. Caston Rachel Audrey, Demple Bruce. doi: 10.1016/j.freeradbiomed.2016.11.025. PUBLISHED IN THIS ISSUE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MTH1 as a nucleotide pool sanitizing enzyme: friend or foe? Nakabeppu Yusaku, Ohta Eiko, Abolhassani Nona. doi: 10.1016/j.freeradbiomed.2016.11.002. PUBLISHED IN THIS ISSUE. [DOI] [PubMed] [Google Scholar]
- 63.Chromatin associated mechanisms in base excision repair – nucleosome remodeling and DNA transcription, two key players. Menoni Hervé, Di Mascio Paolo, Cadet Jean, Dimitrov Stefan, Angelov Dimitar. doi: 10.1016/j.freeradbiomed.2016.12.026. PUBLISHED IN THIS ISSUE. [DOI] [PubMed] [Google Scholar]
- 64.Hide and seek: How do DNA glycosylases locate oxidatively damaged DNA bases amidst a sea of undamaged bases? Lee Andrea J, Wallace Susan S. doi: 10.1016/j.freeradbiomed.2016.11.024. PUBLISHED IN THIS ISSUE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: properties and biological roles of the PFG and Ogg1 8-oxoguanine DNA N-glycosylases. Boiteux Serge, Coste Franck, Castaing Bertrand. doi: 10.1016/j.freeradbiomed.2016.11.042. PUBLISHED IN THIS ISSUE. [DOI] [PubMed] [Google Scholar]
- 66.Repair of 8-oxoG:A mismatches by the MUTYH glycosylase: mechanisms, metals & medicine. Banda Douglas M, Nunez Nicole N, Burnside Michael A, Bradshaw Katie M, David Sheila S. doi: 10.1016/j.freeradbiomed.2017.01.008. PUBLISHED IN THIS ISSUE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DNA damage related crosstalk between the nucleus and mitochondria. Saki Mohammad, Prakash Aishwarya. doi: 10.1016/j.freeradbiomed.2016.11.050. PUBLISHED IN THIS ISSUE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coordination of DNA single strand break repair. Abbotts Rachel, Wilson David M., III doi: 10.1016/j.freeradbiomed.2016.11.039. PUBLISHED IN THIS ISSUE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mechanistic and biological considerations of oxidatively damaged DNA for helicase-dependent pathways of nucleic acid metabolism. Crouch Jack D, Brosh Robert M., Jr doi: 10.1016/j.freeradbiomed.2016.11.022. PUBLISHED IN THIS ISSUE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oxidatively generated base modifications in DNA: not only carcinogenic risk factor but also regulatory mark? Seifermann Marco, Epe Bernd. doi: 10.1016/j.freeradbiomed.2016.11.018. PUBLISHED IN THIS ISSUE. [DOI] [PubMed] [Google Scholar]
- 71.Aberrant base excision repair pathway of oxidatively damaged DNA: implications for degenerative diseases. Talhaoui Ibtissam, Matkarimov Bakhyt T, Tchenio Thierry, Zharkov Dmitry O, Saparbaev Murat K. doi: 10.1016/j.freeradbiomed.2016.11.040. PUBLISHED IN THIS ISSUE. [DOI] [PubMed] [Google Scholar]
- 72.Single nucleotide polymorphisms in DNA glycosylases: from function to disease. D’Errico Mariarosaria, Parlanti Eleonora, Pascucci Barbara, Fortini Paola, Baccarini Sara, Simonelli Valeria, Dogliotti Eugenia. doi: 10.1016/j.freeradbiomed.2016.12.002. PUBLISHED IN THIS ISSUE. [DOI] [PubMed] [Google Scholar]
- 73.Role of the oxidized form of XRCC1 in protection against extreme oxidative stress. Horton Julie K, Seddon Hannah J, Zhao Ming-Lang, Gassman Natalie R, Janoshazi Agnes K, Stefanick Donna F, Wilson Samuel H. doi: 10.1016/j.freeradbiomed.2017.02.005. PUBLISHED IN THIS ISSUE. [DOI] [PMC free article] [PubMed] [Google Scholar]