Abstract
Many adult organs grow or shrink to accommodate different physiological demands. Often, as total cell number changes, stem cell number changes proportionally in a phenomenon called “stem cell scaling”. The cellular behaviors that give rise to scaling are unknown. Here we study two complementary theoretical models of the adult Drosophila midgut, a stem cell-based organ with known resizing dynamics. First, we derive a differential equations model of midgut resizing and show that the in vivo kinetics of growth can be recapitulated if the rate of fate commitment depends on the tissue’s stem cell proportion. Second, we develop a 2D simulation of the midgut and find that proportion-dependent commitment rate and stem cell scaling can arise phenomenologically from the stem cells’ exploration of physical tissue space during its lifetime. Together, these models provide a biophysical understanding of how stem cell scaling is maintained during organ growth and shrinkage.
Introduction
Mature organs contain both differentiated cells that execute physiological function and stem cells that generate new differentiated cells. In organ homeostasis, stem cells divide to replace differentiated cells that are lost, and numbers of stem and differentiated cells are constant. Increased functional demand can induce adaptive growth, a transient, nonhomeostatic state in which stem cells divide to generate excess differentiated cells (1, 2, 3, 4). Similarly, decreased demand leads to adaptive shrinkage, in which differentiated cells are reduced in part because stem cells cease to divide (5, 6). Adaptive resizing enables mature organs to maintain physiological fitness in the face of changing environmental conditions (1, 7, 8, 9).
Intriguingly, many organs exhibit altered numbers of stem cells in response to major physiological adaptation or resizing; examples of these include altered numbers of satellite stem cells in muscles after exercise or induced hypertrophy (10, 11, 12), altered numbers of mammary gland stem cells during pregnancy (13, 14), and altered numbers of intestinal stem cells after feeding (15). In particular, O’Brien et al. (15) found that stem cells scale with the size of the organ, that is, stem cells adjust their numbers during resizing to remain a similar proportion of total cells in the organ. Because of scaling, the cellular replacement burden of an individual stem cell stays constant irrespective of organ size. Physiologically, the constant replacement burden may be advantageous because it allows the organ to respond exponentially quickly (at least initially) to environmental changes. This is because the rate of change of the size of the system would typically be proportional to the number of stem cells and therefore proportional to the size of the system, leading to exponential response. In the adult Drosophila midgut, a simple epithelial organ functionally equivalent to the vertebrate small intestine, this scaling behavior is extraordinarily precise; a fourfold increase in differentiated cells, induced by increased dietary load, is matched by a fourfold increase in stem cells (15). Importantly, for stem cell scaling to occur, there must be populationwide coordination between symmetric and asymmetric fate outcomes after cell division (15, 16, 17). Although we know that at the individual cell level, fate outcomes are determined through Delta-Notch signaling, we do not know what mechanisms coordinate stem cell scaling at the population level.
Some prior models of the midgut (16, 18) and of other self-renewing organs (18, 19, 20, 21, 22, 23, 24, 25, 26, 27) have considered homeostasis without adaptive resizing. Some of these as well as other models have considered embryonic development (17, 18, 28, 29) or cancer (19, 24, 27, 30, 31, 32), two growth states that do not exhibit stem cell scaling. To shed light on scaling mechanisms, we develop a set of nonspatial differential equations as well as a 2D simulation of cell dynamics in the Drosophila midgut. Here we find that the physiological kinetics of stem cell scaling during midgut adaptive growth can be recapitulated by a set of ordinary differential equations. The ability of these equations to recapitulate physiological kinetics depends strongly on the inclusion of feedback. Specifically, physiological dynamics of cell populations are captured if the rate at which new cells commit to differentiation depends on the existing proportion of stem cells. Next, we develop a 2D simulation of the midgut and show that this tunable commitment rate can be explained by the concept of a stem cell territory—the physical space that a stem cell explores during its lifetime. We show that territory size is determined by cell-cell adhesion, stochastic motility, and Delta-Notch signaling. We find that stem cell scaling requires a threshold territory size and that systems within this regime fit our differential equations.
Methods
Nonspatial model
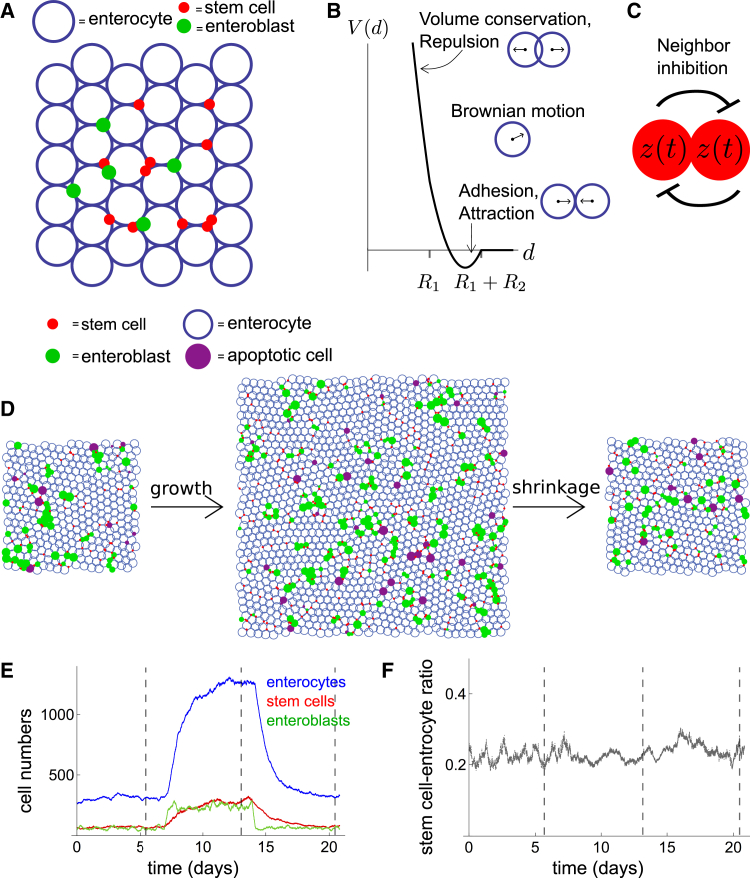
Description of nonspatial model. We propose a mathematical description of the midgut that considers the midgut’s three major cell types: stem cells, enteroblasts, and enterocytes, each of which exhibits a distinct cellular behavior (Fig. 1). Stem cells, s, are the only cells in the midgut that typically divide (33). In our model, all stem cell divisions generate two daughters that are equipotent stem cells (16); this division rate is denoted a. Some stem cells become enteroblasts, u. Enteroblasts are postmitotic and committed to differentiate into enterocytes, but still lack the morphological features of differentiation. The rate that stem cells commit to terminal fate is denoted b. Enterocytes, U, are fully mature epithelial cells that comprise most of the cells in the midgut epithelium. Enteroblasts differentiate into enterocytes at a rate λ. Enterocytes die, or are otherwise lost, at a rate Λ.
Figure 1.
Stem cells in the Drosophila midgut regulate growth and homeostasis. (A) Given here is a schematic of the Drosophila midgut (yellow). (B) Given here is a schematic of midgut epithelium with stem cells (red) and enteroblasts (green) situated among enterocytes (blue). (C) Cellular processes are taken into account by the model in Eq. 1. To see this figure in color, go online.
These cellular relationships lead to a simple mathematical description for numbers of stem cells, enteroblasts, and enterocytes as functions of time s(t), u(t), and U(t), with the following form:
| (1) |
For the system to have a nonzero, finite steady state, either rates of division and commitment must always be equal (b = a), which would imply highly precise regulation of these two processes, or else some rates as given in Fig. 1 and Eq. 1 must depend on cell numbers. We assume the latter case because highly precise regulation is unlikely in a noisy tissue system.
To account for nutrient-driven adaptive growth and shrinkage (15), we propose that cell numbers depend on ingested nutrients, and that the system contains feedback. We denote the total energy of ingested nutrients as Ein and define the energy density, Ed, as follows:
| (2) |
in which A ≡ A1s + A2u + A3U is the total tissue consumption of Ein, and A1, A2, and A3 denote the cell-type specific consumption per cell.
Physiologically, we assume that cells most likely can access only the local value of nutrients, namely the energy density. Therefore, we designate rates of cell division a and cell loss Λ to depend on Ed. In fact, in vivo analysis of the midgut has shown that high levels of nutrients promote divisions and suppress death, whereas low levels of nutrients have the opposite effect, as reflected by levels of insulin signaling (15). Accounting for these observations, denoting am and Λm as maximal rates of division and death, respectively, nondimensionalizing Ed, and choosing Hill functions to represent generic sigmoidal functions, we arrive at nutrient-dependent expressions for division and death rates, as follows:
| (3) |
Note that although we chose second-order Hill functions to represent the sigmoidal dependence of cell division and loss rates on nutrient density, other Hill functions do not significantly alter the results, as detailed in the Supporting Material. Model equations for s, u, and U taking into account nutrient-dependent division and death rates are therefore, as follows:
| (4) |
Importantly, the steady states of Eq. 4 satisfy scaling because leaves the steady-state equations for Eq. 4 invariant. Below, we explore feedback and regulation in this model.
Although it is known that stem cell-to-enteroblast commitment is controlled by the dynamics of Delta-Notch signaling (33, 34, 35, 36), regulation of the differentiation program by which enteroblasts become enterocytes is less understood, and its kinetics had not been measured. Because the source of regulation for differentiation rate λ is unknown, in this article, we take λ to be constant, that is, independent of s, u, or U. An extended model with a regulated differentiation rate is suggested in the Supporting Material.
Given that differentiation rate λ is kept constant, a simplified model with similar dynamics to Eq. 4 could be considered, in which enteroblasts u and enterocytes U are combined into a single population of differentiated cells W = u + U. In this case, the model would be as follows:
| (5) |
where EdW would be analogous to Ed in Eq. 2, as follows:
| (6) |
We explored this model in the Supporting Material, and we found that it gives the same qualitative results as the model in Eq. 4. Given that this simplified model has one fewer equation, one might prefer this model for further analysis. However, to better adhere to biological convention and to provide convenient comparison to experimental data and literature, including our simulation results in the second-half of the article, we proceed for the rest of this article with the model in Eq. 4.
Results
Nonspatial model analysis
Constant versus stem cell proportion-dependent commitment rate: compatibility with empirical measurements
We next sought to compare the solutions of Eq. 4 to the known dynamics of the midgut in vivo. Solving Eq. 4 under steady-state conditions, we find for steady-state stem cell number ratios s0/u0, s0/U0, and s0/(s0 + u0 + U0), as follows:
| (7) |
| (8) |
| (9) |
Full steady-state solutions to Eq. 4 are in the Supporting Material. Note that in this general form, we assume that b may have functional dependence on s0, u0, or U0, and that b cannot surpass am in value.
Important to the discussion of empirical measurements in the midgut is the definition of symmetric and asymmetric fate outcomes. Because, as a first approximation, stem cells are the only cells in the midgut capable of dividing (33), we designate fate outcomes as symmetric-stem if both daughter cells divide before either daughter becomes a committed enteroblast. Using similar logic, we designate fates as symmetric-terminal if both daughters become committed enteroblasts before either divides, and as asymmetric if one daughter becomes a committed enteroblast while the other divides (15).
Because we model division and commitment as Markov processes (uncorrelated) with rates a and b, the probability that a given stem cell undergoes division before commitment is a/(a + b). The frequency of symmetric-stem fate outcomes is therefore as follows:
| (10) |
where a and b can have any functional dependence. Note that, in steady state, similar to the results in Hannezo et al. (18), we have P(sym) = 1/4 because a = b in steady state. The maximum symmetric-stem division rate, often achieved during growth, is Pmax(sym) = (am/(am + b))2.
Constant rate of commitment is incompatible with empirical measurements
Other models of tissue homeostasis have assumed that commitment rate is constant (16, 22). We thus asked whether a constant commitment rate is compatible with growth. Solving Eq. 4 for b = B0 = constant and a 4× increase in food, we find that the key features of midgut growth in vivo are indeed recapitulated: the cell numbers increase, the frequency of symmetric-stem fates increases transiently, and the stem cell number scales (Supporting Material). Here, the value of 4× food increase was chosen to generate a 4× increase in cell numbers so as to best compare to the experimentally measured 4× increase in cell numbers when the gut undergoes feeding (15). As noted, the cell number ratios s0/U0 and s0/u0 and are independent of food input or absolute cell number, so stem cell scaling arises naturally. Thus, in principle, a constant commitment rate is compatible with resizing.
However, the validity of a constant commitment rate model breaks down when we compare the theoretical parameter space with the biologically observed parameter space. Although commitment rates have not been experimentally measured, it is known that in steady state, s0/u0 ≈ 1 and s0/U0 ≈ 0.2 during midgut homeostasis (15, 16); specifically, the numbers of stem cells, enteroblasts and enterocytes are ∼700 ± 200, 500 ± 200, and 2800 ± 800, respectively (15). Using these values in Eqs. 7 and 8, and replacing commitment rate b with the constant B0, we obtain
| (11) |
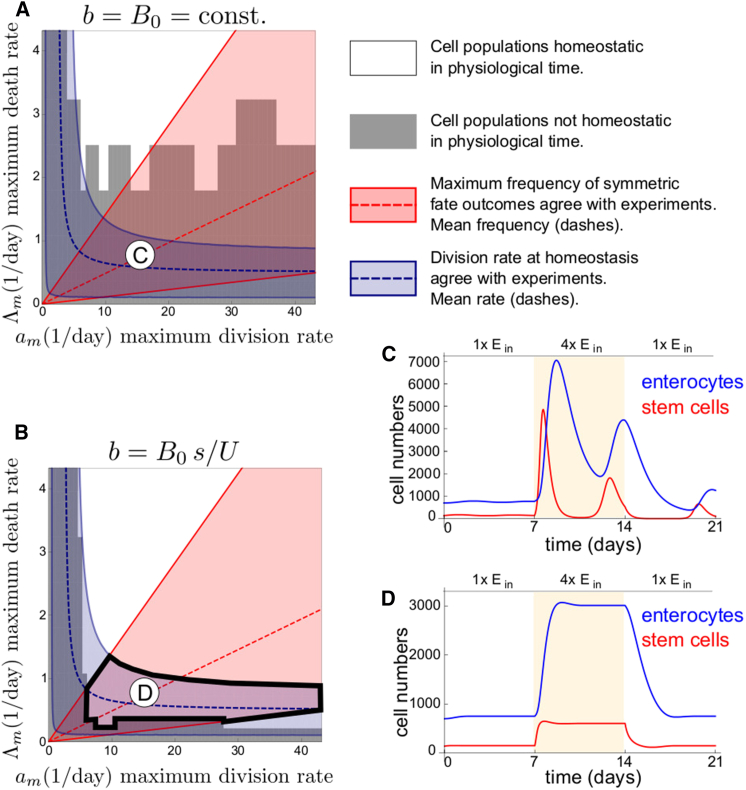
Using these relationships to deduce λ and B0 from am and Λm, we solve Eq. 4 for various values of am and Λm. To focus our parameter range, we apply three criteria. First, homeostatic stem cell division rates a0 vary from 0.5 to 4 times per day (15, 16). Given that division and commitment must be equal at homeostasis (a0 = B0), this biologically relevant range of a0 is obtained from am and Λm using Eq. 11 (Fig. 2 A, blue shading). Second, the maximum reported rate of symmetric-stem fates under physiological conditions is 0.7, which is transiently observed during growth (15). We thus set Pmax(sym) = (am/(am + b))2 ≈ 0.7 (with b = B0) and included a margin of error such that 0.5 ≤ Pmax(sym) ≤ 0.9 (Fig. 2 A, red shading), where the margin of error is estimated from data in O’Brien et al. (15) (see Supporting Material). Third, the time required for the growth phase to reach completion is ∼3.5 days (15), after which homeostasis is reestablished at the organ’s new, larger size. Therefore, only values of am and Λm for which solutions to Eq. 4 approach steady state by t = 3.5 days give rise to steady states with in vivo kinetics (Fig. 2 A, white region). Conditions used to determine whether Eq. 4 has reached approximate equilibrium are in the Supporting Material.
Figure 2.
Stem cell proportion-dependent commitment rate b = B0s/U model satisfies experimental observations whereas constant commitment rate b = B0 model does not. (A and B) For b = B0 (A) and b = B0s/U (B), regions of parameter space (am, Λm) are given that satisfy: homeostatic division rate 0.5/day ≤ a0 ≤ 4.0/day (blue), maximum frequency of symmetric-stem fate outcomes 0.5 < Pmax(sym) < 0.9 (red), and homeostasis within t = 3.5 days (white). Averages a0 = 2.5/day and Pmax(sym) = 0.7 are indicated by dashed lines. Parameter values satisfying all experimental measurements do not exist for the b = B0 model (A), whereas they exist for the b = B0s/U model (B) (solid black outline). Circled letters indicate parameters corresponding to solutions in (C) and (D). (C) Solutions of b = B0 model are highly oscillatory in parameter regimes that satisfy experimental values of division rate and maximum frequency of symmetric-stem fate outcomes. (D) Solutions of b = B0s/U model respond quickly to food changes without large oscillations in parameter regimes that satisfy all experimental measurements. To see this figure in color, go online.
Applying these criteria, we observe that when commitment rate is constant (b = B0), no regions of the theoretical parameter space satisfy all three biological measurements; i.e., there is no region in which blue, red, and white shading all overlap (Fig. 2 A). Specifically, solutions to Eq. 4 with constant b = B0 show strong behaviors of ringing (see Fig. 2 C and Supporting Material), which prevents solutions from approaching steady state within physiological time. This ringing in solutions approaching a new steady state determined by Ein is present for all fold-changes of Ein. Thus, although constant commitment rate is theoretically compatible with midgut adaptive growth, it is incompatible when biologically relevant measurements are applied.
A rate of commitment that depends on stem cell proportion is compatible with scaling and empirical measurements
Because the model with constant commitment rate does not fit experimental measurements, we examine a second possibility, in which commitment rate varies depending on how many stem cells and enterocytes are present in the tissue. Such a relationship can give rise to more realistic dynamics by allowing cell numbers to feed-back into the commitment rate. In the midgut, commitment to enteroblast fate is known to occur through Delta-Notch signaling between stem cells (33, 34, 35, 36). This biological phenomenology suggests that it is reasonable to model the signaling frequency, and consequently commitment rate, such that it depends on the number ratio of stem cells in the tissue s/U. We therefore explore the effects of this additional feedback to our model in Eq. 4.
We find that a commitment rate proportional to stem cell-enterocyte ratio recapitulates midgut resizing within timescales that are biologically relevant. Specifically, we modify three elements: Eq. 4 such that b = B0 s/U, Eq. 11 such that B0 is replaced by B0s0/U0, and the Pmax calculation such that b = B0 s0/U0. Incorporating these modifications, we solve Eq. 4 for various values of am and Λm. The resulting values define a theoretical parameter space that is compatible with the known biological measurements; i.e., the region in which blue, red, and white shading overlap (Fig. 2 B). In addition, stem cell scaling is maintained due to the form of b = B0 s/U. Importantly, the large oscillations and ringing behaviors in the b = B0 model (Fig. 2 C) that delays the approach to steady state are suppressed in the b = B0 s/U model (Fig. 2 D). This result is confirmed by linear stability analysis, detailed in the Supporting Material. Moreover, the parameter region in which oscillations are significant is smaller in the b = B0 s/U compared to the b = B0 model, also detailed in the Supporting Material. We conclude that the known kinetics of midgut resizing can be accounted for by introducing an additional feedback term to Eq. 4, in particular, one that tunes the rate of enteroblast commitment to the existing proportion of stem cells.
Therefore, from these results, we propose that our model equations be modified as
| (12) |
Equation 12 captures stem cell scaling and in vivo resizing dynamics of the midgut. The steady-state solutions to Eq. 12 are presented in the Supporting Material. We note that the feedback through stem cell number ratio s/U does not uniquely allow the resizing model in Eq. 4 to capture realistic dynamics in the midgut; however, proportionality of commitment rate to s/U is one of the simplest forms for feedback that both represents biological phenomenology and captures realistic dynamics. Alternative models for feedback are suggested and explored in the Supporting Material.
2D model
2D model description. Although our nonspatial model implies that a tunable commitment rate b is needed for midgut resizing that fits experimental measurements, it does not provide insight into the cellular mechanisms that underlie this tunability. To explore these potential mechanisms, we develop a 2D model of the midgut that builds upon our nonspatial model. Many mathematical approaches have been used to model 2D epithelia including vertex models (37, 38, 39, 40), cellular automata (41, 42, 43), cellular Potts models (44, 45), and cell-centered models (46, 47, 48). Our 2D model is a cell-centered model based on overlapping spheres (49, 50, 51), where we represent cells as 2D spheres (disks) that are specified by the positions of their centers and that interact physically via forces such as cell-cell adhesion, volume exclusion, and stochastic motion (Fig. 3, A and B). We chose a cell-centered model, among others, because it describes the midgut system at the length scale of cells, which is the length scale of the phenomenology in which we are interested. As shown, in vivo (35, 52), contact-mediated, Delta-Notch signaling between individual stem cells serves to define commitment to terminal fate (Fig. 3 C). Thus, in the 2D model, commitment rate becomes a property that emerges from the fate decisions of individual stem cells, in contrast to the nonspatial model, in which commitment rate is explicitly defined with respect to the tissue-wide ratio of stem cells to enterocytes.
Figure 3.
2D model accounts for spatial tissue dynamics. Midgut cells are represented by physically interacting, soft pseudo-spheres that signal to each other via Delta-Notch interactions. (A) Given here is a schematic: stem cells, enteroblasts, and enterocytes in a 2D domain. (B) Attractive and repulsive forces from neighbor cells and stochastic motility are taken into account. Given here is trapping potential V(d) from neighbor interactions with preferred distance d0 between cells of radii R1 and R2. (C) Given here is a schematic of Delta-Notch lateral inhibition from protein dynamics z(t). (D) Here we have frames from 2D simulation (Movie S1) of the feed-fast cycle: Ein is increased fourfold, then decreased to original value. (E and F) Given here are cell numbers (E) and stem cell-enterocyte ratio (F) as functions of time for the simulation in (D) with timeframes indicated (vertical dotted lines). To see this figure in color, go online.
Physical cell-cell forces. Assuming that the system is highly damped, we set the inertial term to 0 so that external forces are balanced by viscous drag: dx/dt = ηf, where the quantity f is the external force acting on a cell, the quantity x is the position of the cell center, and η is the mobility:
| (13) |
Here, the sum on n∈n.n indicates summing over nearest neighboring cells; if the cell centered at x has radius R1 and another cell has radius R2, the two cells are nearest neighbors if their cell centers are separated by less than R1 + R2. The quantities ρn and γn are magnitudes of repulsion and adhesion forces from neighbor cells, known to be important in epithelial cells, and is a unit vector toward the neighbor cell. The self-generated random force is provided by X(t), which has components sampled from a normal distribution N(0,1); here σ indicates an amplitude factor for X. We refer to σX as an intrinsic stochastic motility. Cell motility has recently been shown to occur in enteroblasts (53), and labeled clones have been shown to split (15, 16). The latter may be due to stochastic motility or random cell shuffling, both of which are captured by the term σX. Forces are indicated in Fig. 3 B. Details are given in the Supporting Material.
Delta-Notch signaling. In the midgut, asymmetric fate outcomes arise through activation of Notch receptor on the surface of one stem cell by Delta ligand on an adjacent stem cell (33, 34, 35, 36, 52, 54). Activation of Notch marks a cell’s commitment to differentiate (52), and is the defining feature of enteroblast identity. Many mathematical models of Delta-Notch interactions separately describe Delta and Notch populations (54, 55, 56, 57), or distinguish between membrane-bound and intracellular Notch (56, 57). To model lateral inhibition signaling, we employed a fictitious protein with values z(t) indicating the “stemness” of a given cell. Delta-Notch interactions between neighboring cells are modeled with the dynamics of z through a delay differential equation. Delta-Notch signaling have been modeled to contain time delays to account for signal transduction (58), and a delayed model of lateral inhibition has been shown to reduce errors in patterning (59).
For each cell (stem cells and enteroblasts) expressing z, we assume that z is produced within the cell in a nonlinear fashion with saturating effects at level p, and z decays exponentially with rate β. To take into account the effects of nearest neighboring cells, we assume that neighbor cells’ protein levels zn decreases z production in a given cell with time delay tn, taking into account protein transport times, see Fig. 3 C. Choosing a sigmoidal Hill function of order m > 1 for the production term, we have for the dynamics of nondimensionalized z:
| (14) |
where gn denotes the switch point for neighbor interactions, and m and k are Hill exponents. Given that m > 1, Eq. 14 gives bistable steady states of z with stable fixed points at z∗ = 0 and z∗ ≠ 0 and an unstable fixed point in between that we denote by zu∗. The nonzero stable fixed point z∗ ≠ 0 is interpreted as the stem cell state, and the trivial stable fixed point z∗ = 0 is interpreted as the enteroblast state. A stem cell (that has z > zu∗) is defined to have permanently committed to terminal fate and become an enteroblast when its z value falls below the value of the unstable fixed point (z becomes z < zu∗). Inhibitory signaling from nearest neighbors cause a stem cell’s z value to be suppressed below zu∗, causing commitment. Therefore, in the 2D model, commitment kinetics and commitment rate are determined by dynamics that drive z below a threshold value. An enteroblast has a small value of z compared to stem cells and therefore signals weakly compared to stem cells; it undergoes growth in size to eventually reach the size of an enterocyte; at this point, the enteroblast is converted into an enterocyte, and its z value is set identically to 0. Note that commitment is still an uncorrelated Markov process, and therefore, the fraction of symmetric fate outcomes in steady state is still P(sym) = 1/4. See Supporting Material for details of implementation of the 2D model.
2D model analysis
Simulations with this 2D model successfully recapitulate essential features of the in vivo fly midgut with respect to growth, shrinking, scaling with food, and the presence of stem cells in every part of the epithelium (Fig. 3, D–F; Movie S1). Additionally, cell populations behave qualitatively similar to the nonspatial b = B0 s/U model in that they are nonoscillatory (similar to Fig. 2 D instead of to Fig. 2 C).
Note that unlike the nonspatial model, the commitment rate b for the 2D model is not defined as a function of cell populations. Rather, for each individual cell, commitment rate arises from the duration and strength of contact-based cell-cell signaling. This dependence implies that global quantities such as stem cell ratios are determined by local dynamics.
Stem cell ratio depends on cells’ physical properties
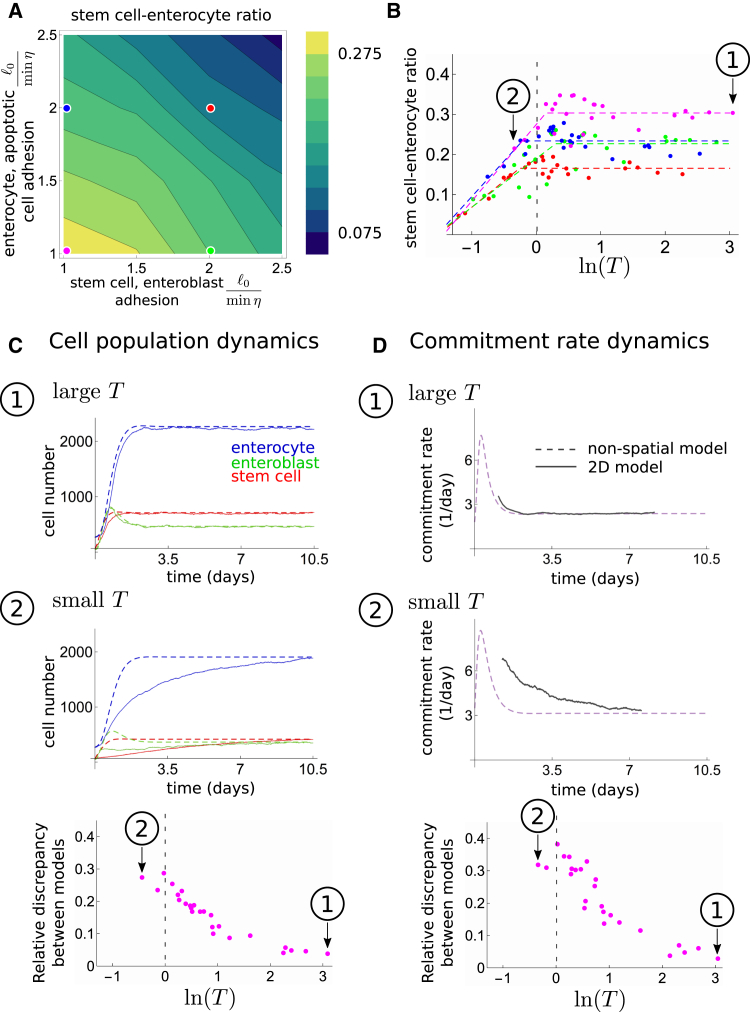
With the 2D model, we can explicitly examine the effects of local dynamics on population-level quantities by changing physical parameters. Specifically, the force magnitudes adhesion γ and motility σ in Eq. 13 determine how fast stem cells move in the tissue. In particular, stochastic motility specifies diffusive motion absent of other interactions. Additionally, these parameters affect the amount of time stem cells are in signaling contact with each other, ultimately affecting the stem cell ratio. Specifically, we found that the stem cell ratio is sensitive to adhesion values (see Fig. 4 A). Here the motilities σsc,eb for stem cells and enteroblasts and σEC for enterocytes were kept constant whereas adhesions were varied. Forces are expressed in units of /(minη), where is the enterocyte diameter without adhesive or repulsive interactions, and η is the inverse viscosity. Note that we use minutes as the unit of time to employ the relevant values of am and Λm that were deduced from nonspatial model (Fig. 2 B) to compare to physiological timescales found in experiments.
Figure 4.
Local physical interactions define a critical territory above which stem cells can sense their density. (A) Given here is a stem cell-enterocyte ratio as a function of adhesion between stem cells and enteroblasts (x axis) and adhesion between enterocytes and apoptotic cells (y axis) for motility σ = 0.2/minη for all cells. Colored dots indicate the adhesion values used in (B). (B) Given here is the stem cell-enterocyte ratio as a function of natural log of stem cell territory ln(T) for various adhesion values. The onset of saturating values for stem cell ratios (dotted line T ≈ 1) corresponds to the lower threshold of T that enables stem cell scaling. (C and D) Given here is a comparison of the 2D model (solid lines, average of six replicate simulations) and the nonspatial b = B0s/U model (dashed lines) using time courses of cell population dynamics (C) and commitment rate dynamics (D). Plots correspond to the magenta parameter set in (B). Circled numbers indicate large versus small T simulations: large T simulations (1) fit well to the b = B0s/U model, whereas small T simulations (2) do not. Calculations of relative discrepancy between models are given in the Supporting Material. To see this figure in color, go online.
Stem cell territory determines whether system scales
In these simulations, we noted that values for adhesion and motility influenced the physical space that a stem cell explores during its lifetime. We denote this space as a “stem cell territory”, T. To gain insight into the relationship between territories and stem cell interactions, we define T as
| (15) |
Here, 1/b is the average lifetime of a stem cell, i.e., the time between the birth of a stem cell and its commitment to terminal fate; the quantity D is a diffusion constant fitted to simulated stem cell tracks (see Supporting Material); and is the average enterocyte diameter. Hence, T is the dimensionless area (number of enterocyte areas) over which a stem cell may influence or respond to other stem cells. Note that Eq. 15 can be generalized for nondiffusive motion.
Investigating resulting stem cell ratios as a function of T (Fig. 4 B), we find a transition from increasing stem cell ratios at lower T toward saturation for larger T. Here, the quantity T was varied via adhesion γ and stochastic motility σ. As expected, larger σ and lower γ lead to larger territories T. Note that this transition occurs around T ≈ 1, i.e., when stem cells start to explore a territory larger than the area of a single enterocyte. For T > 1, different stem cells can come in contact with each other via overlapping territories. The saturation for large T also indicates that additional interactions do not change global behavior. Moreover, the resulting stem cell ratio for large T depends on the intrinsic parameters (adhesion γ); this dependence suggests that the tissue can flexibly regulate its stem cell ratio.
It is insightful to compare the 2D simulations to the nonspatial model (Fig. 4, and see Eq. 12). We find that for large stem cell territories T, the population dynamics of the two models agree (simulation 1) in Fig. 4, C and D. This suggests that when T is large, stem cells can sense their density within the tissue, and hence their commitment rate can contain density feedback through s/U, as in the nonspatial model. When T is small, the two models differ (simulation 2) in Fig. 4, C and D, because the nonspatial model cannot account for the effect, in 2D, that stem cells with small territories do not obtain density knowledge. Additionally, we note that in the latter case, when T is small, the approximate time to reach new homeostatic cell numbers during resizing is longer than observed physiologically (∼7 days instead of 3.5 days (15)); this supports the notion that stem cells in the biological system may display motility to increase their territories.
Finally, we explored how stem cell territory T relates to stem-cell scaling. We performed numerical experiments for different values of food input Ein for various T (Fig. S4). We found that the regime for which stem cell scaling is enabled approximately coincides with the regime for which stem cell ratios saturate: at T ≥ 1 (dotted vertical line in Fig. 4 B). Therefore, when stem cells explore a large enough area to escape the inhibition signaling of its immediate sibling stem cell, stem cell scaling is enabled.
Discussion
Proportional scaling of stem cells to total cells during adaptive growth ensures that the organ has enough stem cells to support its new size after growth is complete. We have shown that the basic features of adaptive organ resizing can be captured by simple mathematical descriptions of the Drosophila midgut in which rates of stem cell division and enterocyte death depend on nutrient density. Importantly, we find that a nonconstant rate of commitment to terminal fate, specifically, a rate tuned to stem cell proportion, reproduces the in vivo kinetics of division and growth.
What biological mechanisms might enable cells to monitor stem cell proportions and tune their commitment rates appropriately? The models presented here show that a mechanism involving stochastic motility is compatible with in vivo measurements. Motility permits stem cells to explore a local tissue area and engage in signaling interactions with other, potentially nonsibling, stem cells. We define this local area as the stem cell’s territory and explore its parameter space by varying cell motility and adhesion.
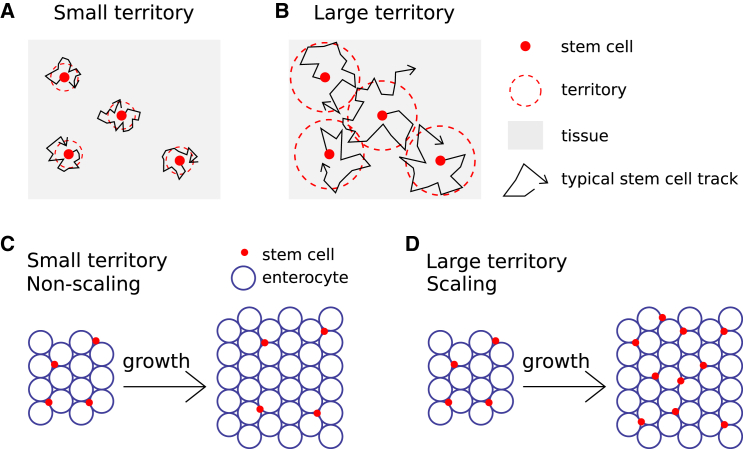
Intriguingly, proportion-dependent commitment and scaling occur only above a threshold territory size. This threshold size can be understood by its impact on commitment rate (i.e., rate of Notch activation), which occurs through cell-cell signaling (Notch-Delta interactions) between pairs of stem cells. Essentially, there are two scenarios: 1) When stem cells are constrained within small territories, two newborn sibling cells contact each other frequently, and their Notch-Delta interactions increase to the point of a near-certain asymmetric outcome. In this case, the total number of stem cells in the tissue remains nearly constant during growth, whereas the number of enterocytes increases, and scaling does not occur (Fig. 5, A and C). 2) When stem cells range in large territories, sibling cells often separate fast enough without Notch-Delta interaction having a differentiating effect, which promotes symmetric fate outcomes, i.e., the total number of stem cells in the tissue can increase. Importantly, the stem cell ratio does not increase beyond a certain value because pairs of nonsibling stem cells come into contact sufficiently frequently for Notch-Delta interactions to induce differentiation. In this case, both stem cells and enterocytes increase in proportion, and scaling does occur (Fig. 5, B and D).
Figure 5.
Scaling occurs when stem cell territories are above a critical threshold. (A and B) Cartoons of small (A) versus large (B) stem cell territory (red dotted line) are as defined by spatial range of physical cell motion (black line). (C) For territories below a threshold size, there is no stem cell scaling. (D) For territories larger than a threshold size, there is stem cell scaling. To see this figure in color, go online.
We make the following suggestions for future experimental tests. The core importance of a tunable commitment rate in these models contrasts with the scant empirical knowledge of commitment rates in vivo. Experimental measurements of commitment rate are currently impractical in many systems. However, the availability of fluorescent Notch reporters (60, 61) may provide a means to measure kinetics of fate commitment in systems such as the midgut in which Notch activation is the committing step. For the midgut in particular, the models here generate specific, testable predictions: 1) If newborn stem cells inherit unequal levels of the Delta ligand or Notch receptor, then because of the delayed nature of Eq. 14, commitment should occur more rapidly in sibling pairs than in pairs of stem cells that come into contact by chance. 2) The stem cell population should undershoot when a tissue undergoes shrinkage (Fig. 2 D). 3) Stem cell scaling should be disrupted by experimental perturbation of stochastic motility, adhesive force, or Delta-Notch signaling (Fig. 4 B). 4) The territory size of individual stem cells should be larger than one enterocyte area. In particular, for the last point (4), methods such as cell tracking in vivo to measure diffusion coefficients of cell motion, or clone-induction methods such as twin-spot mosaic analysis (15) to measure spatial dispersal of cells from a common division, can provide measurements and estimations of the stem cell territory in the midgut.
In summary, we have developed mathematical descriptions of a stem cell-based organ that undergoes adaptive resizing in response to external input. To realistically describe an in vivo system, the Drosophila midgut, we found that the stem cell commitment rate should depend on organwide stem cell proportion. To elucidate this dependence, we suggested local, spatially motivated, cell-level mechanisms such as cell motility, adhesion, and signaling, by which stem cells can detect their density and therefore tune their commitment rate. Importantly, these models naturally give rise to stem cell scaling, and we identify physical regimes in which scaling occurs.
Author Contributions
X.D., L.E.O., and I.R.-K. designed research, performed research, contributed analytic tools, analyzed data, and wrote the manuscript.
Acknowledgments
We thank S. Streichan, A. Lam, J.E. Ferrell, and T. Petty for valuable comments on the manuscript, and members of I.R.-K. and L.E.O. labs for useful discussions. This work was supported by a Stanford Dean’s Fellowship and NIH/NRSA fellowship 1F32GM115065 (to X.D.); NIH grant R01GM116000 and a gift from the G. Harold & Leila Y. Mathers Foundation (to L.E.O.); NIH grant 1R21HD080553, and ACS grant RSG-14-177 (to I.R.-K.).
Editor: Mark Alber.
Footnotes
Supporting Materials and Methods, thirteen figures, three tables, and one movie are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30609-4.
Contributor Information
Lucy Erin O’Brien, Email: lucye@stanford.edu.
Ingmar Hans Riedel-Kruse, Email: ingmar@stanford.edu.
Supporting Material
References
- 1.Piersma T., Lindström A. Rapid reversible changes in organ size as a component of adaptive behaviour. Trends Ecol. Evol. 1997;12:134–138. doi: 10.1016/s0169-5347(97)01003-3. [DOI] [PubMed] [Google Scholar]
- 2.Aldewachi H.S., Wright N.A., Watson A.J. The effect of starvation and refeeding on cell population kinetics in the rat small bowel mucosa. J. Anat. 1975;119:105–121. [PMC free article] [PubMed] [Google Scholar]
- 3.Altmann G.G. Influence of starvation and refeeding on mucosal size and epithelial renewal in the rat small intestine. Am. J. Anat. 1972;133:391–400. doi: 10.1002/aja.1001330403. [DOI] [PubMed] [Google Scholar]
- 4.Koury M.J. Erythropoietin: the story of hypoxia and a finely regulated hematopoietic hormone. Exp. Hematol. 2005;33:1263–1270. doi: 10.1016/j.exphem.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 5.Brown H.O., Levine M.L., Lipkin M. Inhibition of intestinal epithelial cell renewal and migration induced by starvation. Am. J. Physiol. 1963;205:868–872. doi: 10.1152/ajplegacy.1963.205.5.868. [DOI] [PubMed] [Google Scholar]
- 6.Dunel-Erb S., Chevalier C., Le Maho Y. Restoration of the jejunal mucosa in rats refed after prolonged fasting. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2001;129:933–947. doi: 10.1016/s1095-6433(01)00360-9. [DOI] [PubMed] [Google Scholar]
- 7.Meyers L.A., Bull J.J. Fighting change with change: adaptive variation in an uncertain world. Trends Ecol. Evol. 2002;17:551–557. [Google Scholar]
- 8.Carey H.V. Seasonal changes in mucosal structure and function in ground squirrel intestine. Am. J. Physiol. 1990;259:R385–R392. doi: 10.1152/ajpregu.1990.259.2.R385. [DOI] [PubMed] [Google Scholar]
- 9.Secor S.M., Diamond J. A vertebrate model of extreme physiological regulation. Nature. 1998;395:659–662. doi: 10.1038/27131. [DOI] [PubMed] [Google Scholar]
- 10.Dumont N.A., Bentzinger C.F., Rudnicki M.A. Satellite cells and skeletal muscle regeneration. Compr. Physiol. 2015;5:1027–1059. doi: 10.1002/cphy.c140068. [DOI] [PubMed] [Google Scholar]
- 11.Ambrosio F., Kadi F., Huard J. The effect of muscle loading on skeletal muscle regenerative potential: an update of current research findings relating to aging and neuromuscular pathology. Am. J. Phys. Med. Rehabil. 2009;88:145–155. doi: 10.1097/PHM.0b013e3181951fc5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiaffino S., Bormioli S.P., Aloisi M. Cell proliferation in rat skeletal muscle during early stages of compensatory hypertrophy. Virchows Arch. B Cell Pathol. 1972;11:268–273. doi: 10.1007/BF02889406. [DOI] [PubMed] [Google Scholar]
- 13.Visvader J.E. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rios A.C., Fu N.Y., Visvader J.E. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- 15.O’Brien L.E., Soliman S.S., Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147:603–614. doi: 10.1016/j.cell.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Navascués J., Perdigoto C.N., Simons B.D. Drosophila midgut homeostasis involves neutral competition between symmetrically dividing intestinal stem cells. EMBO J. 2012;31:2473–2485. doi: 10.1038/emboj.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itzkovitz S., Blat I.C., van Oudenaarden A. Optimality in the development of intestinal crypts. Cell. 2012;148:608–619. doi: 10.1016/j.cell.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannezo E., Prost J., Joanny J.-F. Growth, homeostatic regulation and stem cell dynamics in tissues. J. R. Soc. Interface. 2014;11:20130895. doi: 10.1098/rsif.2013.0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rulands S., Simons B.D. Tracing cellular dynamics in tissue development, maintenance and disease. Curr. Opin. Cell Biol. 2016;43:38–45. doi: 10.1016/j.ceb.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Krieger T., Simons B.D. Dynamic stem cell heterogeneity. Development. 2015;142:1396–1406. doi: 10.1242/dev.101063. [DOI] [PubMed] [Google Scholar]
- 21.Simons B.D., Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 22.Snippert H.J., van der Flier L.G., Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Klein A.M., Simons B.D. Universal patterns of stem cell fate in cycling adult tissues. Development. 2011;138:3103–3111. doi: 10.1242/dev.060103. [DOI] [PubMed] [Google Scholar]
- 24.Klein A.M., Doupé D.P., Simons B.D. Kinetics of cell division in epidermal maintenance. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007;76:021910. doi: 10.1103/PhysRevE.76.021910. [DOI] [PubMed] [Google Scholar]
- 25.Klein A.M., Doupé D.P., Simons B.D. Mechanism of murine epidermal maintenance: cell division and the voter model. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2008;77:031907. doi: 10.1103/PhysRevE.77.031907. [DOI] [PubMed] [Google Scholar]
- 26.Van Leeuwen I.M., Mirams G.R., Byrne H.M. An integrative computational model for intestinal tissue renewal. Cell Prolif. 2009;42:617–636. doi: 10.1111/j.1365-2184.2009.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston M.D., Edwards C.M., Chapman S.J. Mathematical modeling of cell population dynamics in the colonic crypt and in colorectal cancer. Proc. Natl. Acad. Sci. USA. 2007;104:4008–4013. doi: 10.1073/pnas.0611179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunche S., Yan H., Lander A.D. Feedback, lineages and self-organizing morphogenesis. PLoS Comput. Biol. 2016;12:e1004814. doi: 10.1371/journal.pcbi.1004814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rué P., Kim Y.H., Arias A.M. A framework for the analysis of symmetric and asymmetric divisions in developmental process. BioArxiv. 2015:010835. [Google Scholar]
- 30.Alcolea M.P., Greulich P., Jones P.H. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat. Cell Biol. 2014;16:615–622. doi: 10.1038/ncb2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolahgar G., Suijkerbuijk S.J., Piddini E. Cell competition modifies adult stem cell and tissue population dynamics in a JAK-STAT-dependent manner. Dev. Cell. 2015;34:297–309. doi: 10.1016/j.devcel.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thalheim T., Buske P., Galle J. Stem cell competition in the gut: insights from multi-scale computational modelling. J. R. Soc. Interface. 2016;13:20160218. doi: 10.1098/rsif.2016.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohlstein B., Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- 34.Micchelli C.A., Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 35.Ohlstein B., Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- 36.Bardin A.J., Perdigoto C.N., Schweisguth F. Transcriptional control of stem cell maintenance in the Drosophila intestine. Development. 2010;137:705–714. doi: 10.1242/dev.039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farhadifar R., Röper J.-C., Jülicher F. The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Curr. Biol. 2007;17:2095–2104. doi: 10.1016/j.cub.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 38.Nagai T., Kawasaki K., Nakamura K. Vertex dynamics of two-dimensional cellular patterns. J. Phys. Soc. Jpn. 1988;57:2221–2224. [Google Scholar]
- 39.Honda H., Ogita Y., Kani K. Cell movements in a living mammalian tissue: long-term observation of individual cells in wounded corneal endothelia of cats. J. Morphol. 1982;174:25–39. doi: 10.1002/jmor.1051740104. [DOI] [PubMed] [Google Scholar]
- 40.Kawasaki K., Nagai T., Nakashima K. Vertex models for two-dimensional grain growth. Philos. Mag. B. 1989;60:399–421. [Google Scholar]
- 41.Simpson M.J., Merrifield A., Hughes B.D. Simulating invasion with cellular automata: connecting cell-scale and population-scale properties. Phys. Rev. E Stat. Nonlin. Soft. Matter Phys. 2007;76:021918. doi: 10.1103/PhysRevE.76.021918. [DOI] [PubMed] [Google Scholar]
- 42.Kansal A.R., Torquato S., Deisboeck T.S. Simulated brain tumor growth dynamics using a three-dimensional cellular automaton. J. Theor. Biol. 2000;203:367–382. doi: 10.1006/jtbi.2000.2000. [DOI] [PubMed] [Google Scholar]
- 43.Lee Y., Kouvroukoglou S., Zygourakis K. A cellular automaton model for the proliferation of migrating contact-inhibited cells. Biophys. J. 1995;69:1284–1298. doi: 10.1016/S0006-3495(95)79996-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graner F., Glazier J.A. Simulation of biological cell sorting using a two-dimensional extended Potts model. Phys. Rev. Lett. 1992;69:2013–2016. doi: 10.1103/PhysRevLett.69.2013. [DOI] [PubMed] [Google Scholar]
- 45.Osborne J.M. Multiscale model of colorectal cancer using the cellular Potts framework. Cancer Inform. 2015;14(Suppl 4):83–93. doi: 10.4137/CIN.S19332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buske P., Galle J., Loeffler M. A comprehensive model of the spatio-temporal stem cell and tissue organisation in the intestinal crypt. PLoS Comput. Biol. 2011;7:e1001045. doi: 10.1371/journal.pcbi.1001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osborne J.M., Fletcher A.G., Gavaghan D.J. Comparing individual-based approaches to modelling the self-organization of multicellular tissues. PLOS Comput. Biol. 2017;13:e1005387. doi: 10.1371/journal.pcbi.1005387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basan M., Elgeti J., Levine H. Alignment of cellular motility forces with tissue flow as a mechanism for efficient wound healing. Proc. Natl. Acad. Sci. USA. 2013;110:2452–2459. doi: 10.1073/pnas.1219937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drasdo D., Kree R., McCaskill J.S. Monte Carlo approach to tissue-cell populations. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1995;52:6635–6657. doi: 10.1103/physreve.52.6635. [DOI] [PubMed] [Google Scholar]
- 50.Drasdo D., Höhme S. A single-cell-based model of tumor growth in vitro: monolayers and spheroids. Phys. Biol. 2005;2:133–147. doi: 10.1088/1478-3975/2/3/001. [DOI] [PubMed] [Google Scholar]
- 51.Schötz E.-M., Lanio M., Manning M.L. Glassy dynamics in three-dimensional embryonic tissues. J. R. Soc. Interface. 2013;10:20130726. doi: 10.1098/rsif.2013.0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perdigoto C.N., Schweisguth F., Bardin A.J. Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development. 2011;138:4585–4595. doi: 10.1242/dev.065292. [DOI] [PubMed] [Google Scholar]
- 53.Antonello Z.A., Reiff T., Dominguez M. Robust intestinal homeostasis relies on cellular plasticity in enteroblasts mediated by miR-8-Escargot switch. EMBO J. 2015;34:2025–2041. doi: 10.15252/embj.201591517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guisoni N., Martinez-Corral R., de Navascues J. Diversity of fate outcomes in cell pairs under lateral inhibition. Development. 2017;144:1177–1186. doi: 10.1242/dev.137950. [DOI] [PubMed] [Google Scholar]
- 55.Collier J.R., Monk N.A., Lewis J.H. Pattern formation by lateral inhibition with feedback: a mathematical model of Delta-Notch intercellular signalling. J. Theor. Biol. 1996;183:429–446. doi: 10.1006/jtbi.1996.0233. [DOI] [PubMed] [Google Scholar]
- 56.Sprinzak D., Lakhanpal A., Elowitz M.B. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sprinzak D., Lakhanpal A., Elowitz M.B. Mutual inactivation of Notch receptors and ligands facilitates developmental patterning. PLOS Comput. Biol. 2011;7:e1002069. doi: 10.1371/journal.pcbi.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barad O., Rosin D., Barkai N. Error minimization in lateral inhibition circuits. Sci. Signal. 2010;3:ra51. doi: 10.1126/scisignal.2000857. [DOI] [PubMed] [Google Scholar]
- 59.Glass D.S., Jin X., Riedel-Kruse I.H. Signaling delays preclude defects in lateral inhibition patterning. Phys. Rev. Lett. 2016;116:128102. doi: 10.1103/PhysRevLett.116.128102. [DOI] [PubMed] [Google Scholar]
- 60.Barolo S., Castro B., Posakony J. New Drosophila transgenic reporters: insulated P-element vectors expressing fast-maturing RFP. Biotechniques. 2004;36:436–440. doi: 10.2144/04363ST03. [DOI] [PubMed] [Google Scholar]
- 61.Housden B.E., Millen K., Bray S.J. Drosophila reporter vectors compatible with ΦC31 integrase transgenesis techniques and their use to generate new Notch reporter fly lines. G3. 2012;2:79–82. doi: 10.1534/g3.111.001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.