Abstract
Background
Interferences between pathogenic bacteria and specific commensals are known. We determined the interactions between nasopharyngeal microbial pathogens and commensals during viral upper respiratory tract infection (URI) and acute otitis media (AOM) in infants.
Methods
We analyzed 971 specimens collected monthly and during URI and AOM episodes from 139 infants. The 16S rRNA V4 gene regions were sequenced on the Illumina MiSeq platform.
Results
Among the high abundant genus-level nasopharyngeal microbiota were Moraxella, Haemophilus, and Streptococcus (3 otopathogen genera), Corynebacterium, Dolosigranulum, Staphylococcus, Acinetobacter, Pseudomonas, and Bifidobacterium. Bacterial diversity was lower in culture-positive samples for Streptococcus pneumoniae, and Haemophilus influenzae, compared to cultured-negative samples. URI frequencies were positively associated with increasing trend in otopathogen colonization. AOM frequencies were associated with decreasing trend in Micrococcus colonization. During URI and AOM, there were increases in abundance of otopathogen genera and decreases in Pseudomonas, Myroides, Yersinia, and Sphingomonas. Otopathogen abundance was increased during symptomatic viral infection, but not during asymptomatic infection. The risk for AOM complicating URI was reduced by increased abundance of Staphylococcus and Sphingobium.
Conclusion
Otopathogen genera played the key roles in URI and AOM occurrences. Staphylococcus counteracts otopathogens thus Staphylococcal colonization may be beneficial, rather than harmful. While Sphingobium may play a role in preventing AOM complicating URI, the commonly used probiotic Bifidobacterium did not play a significant role during URI or AOM. The role of less common commensals in counteracting the deleterious effects of otopathogens requires further studies.
Introduction
Otitis media is a common childhood disease; it is the leading cause of doctors’ visits by children, and the most frequent reason children consume antibiotics or undergo surgery [1–3]. Acute otitis media (AOM) occurs as a complication of viral upper respiratory tract infection (URI) [4]. AOM is a polymicrobial disease; its pathogenesis involves complex interactions between bacteria, viruses, and the host inflammatory response [5, 6].
During the first months of life the young infant’s upper respiratory tract gradually acquires complex microbial communities, including pathogens and commensals [7–9]. It is well known that the three otopathogens (Streptococcus pneumoniae, non-typeable Haemophilus influenzae, and Moraxella catarrhalis) colonize the nasopharynx from early infancy but bacterial AOM only occurs after viral URI [10–12]. Less is known about the colonization of commensals and their dynamics during viral URI. Existing data suggest interference between otopathogens and commensals [13–15]. Therefore, enhancement of commensal colonization may interfere with otopathogen colonization leading to disease prevention. Investigators have studied the effect of selected commensal bacteria, e.g. Bifidobacterium and Lactobacillus, the so-called ‘probiotic bacteria’, in preventing URI and recurrent AOM, but the results have been mixed [16–18]. It is possible that effective probiotic component may require combinations of protective commensals. Refinement of probiotics for prevention of URI and AOM depends on better knowledge and understanding of respiratory tract microbiota and pathogen, commensal, and viral interactions.
There have been recent studies of respiratory microbiota in children during URI and AOM [19–21]. These studies were cross-sectional, with no specific viral data, and the children were mostly older than 6 months. Others have performed longitudinal studies of respiratory microbiota but did not focus on viral URI and AOM [7–9, 22]. The purpose of our study was to characterize nasopharyngeal microbiota in infants followed from near birth to the first AOM episode or 12 months of age and elucidate how changing patterns of nasopharyngeal bacterial colonization lead to susceptibility to viral URI and AOM development.
Methods
Study design, subjects and specimens
The subjects were part of a prospective, longitudinal study (2008–2014) of infants in the first year of life to evaluate the prevalence and risks for URI and AOM development [11, 23]. Nasopharyngeal (NP) specimens analyzed in this study included available specimens (average 7 / per subject) from an approximately equal number of subjects with and without AOM. This study was approved by the University of Texas Medical Branch (UTMB) Institutional Review Board and conformed to the human experimentation guidelines of the United States Department of Health and Human Services. Written informed consent was obtained from the parents/guardians of all subjects.
In brief, healthy infants were enrolled from near birth (< 1 month) and completed the study after the first AOM episode was diagnosed, or at age 12 months without AOM; all subjects were followed at least 6 months. Specimens were collected monthly during months 1–6, month 9, and during URI and AOM. Details on data collection, URI and AOM diagnostic criteria, follow-up, and specimen processing are in S1 File.
DNA extraction/ amplification and sequencing
From the nasopharyngeal samples, DNA was extracted using the PowerMag PowerMicrobiome DNA/RNA Isolation kit (MoBio) in the STARlet platform (Hamilton Robotics). The conserved hypervariable 16S rDNA V4 region was amplified by PCR. The amplicons were sequenced using 2 x 250 bp paired end protocol on the MiSeq platform (Illumina), yielding pair-end reads that overlap by ~247 bps. Following sequencing, raw BCL files were retrieved from the MiSeq platform and called into FASTQs by Casava v1.8.3 (Illumina). The read pairs were demultiplexed based on unique molecular barcodes allowing for up to 1 substitution mismatch and reconstituted into two FASTQ files for each. The resulting 250 base long paired end reads were merged together based on the overlapping region. These laboratory procedures were performed at the Alkek Center for Metagenomics and Microbiome Research, Baylor College of Medicine (Joseph Petrosino, Director)[24].
Sequence analysis
To identify the presence of known bacteria and archaea, subsequences were analyzed using CLC Genomics Workbench 8.0.1 Microbial Genomics Module (http://www.clcbio.com). Read containing nucleotides below the quality threshold of 0.05 (using the modified Richard Mott algorithm) and reads with two or more unknown nucleotides or sequencing adapters were filtered. All reads were trimmed to 240 bases for operational taxonomic unit (OTU) classification. Reference based OTU picking was performed using the SILVA SSU v119 97% database [25]. Sequences present in more than one copy but not clustered to the database were then placed into de novo OTUs (97% similarity) and aligned against the reference database with 80% similarity threshold. Chimeras were removed from the results if their absolute crossover cost was 3 using a k-mer size of 6.
Statistics
All analyses were done at the genus level (S2 File). The Shannon diversity index was calculated using the entropy function in the entropy library in R statistical package (cran.r-project.org). To account for the within-subject variability component, a mixed model with a random intercept for subject was used. Finally, abundance was calculated using the mean relative abundance across samples. Significance was declared with P < 0.05. Adjustments for multiple testing were done using the Benjamini-Hochberg adjustment for controlling the false discovery rate (FDR); unadjusted results are also shown to highlight how the similar genera had significant differences across multiple modeling structures. All calculations were done in R (version 3.2.2) and associated libraries (lme4, lmerTest, entropy, p.adjust). Detailed statistical methods are provided in S1 File.
Results
I. Subject characteristics and number of specimens
Characteristics of the subjects and number of specimens are shown in Table 1(Metadata are shown in S3 File). The first specimen in this study was collected in August 2009, and the last, January 2014. Of 139 subjects, 96% had at least 2 healthy (asymptomatic) samples, 77% had (268) URI/ AOM samples, and 60% had at least 1 healthy sample before URI/ AOM samples.
Table 1. Characteristics of subjects and number of specimens.
| Total | Subjects with AOM | Subjects without AOM | |
|---|---|---|---|
| Number of subjects | 139 (100) | 65 (47)a | 74 (53) |
| Male | 83 (60)b | 36 (55) | 47 (64) |
| Female | 56 (41) | 29 (45) | 27 (36) |
| Race | |||
| - White | 119 (86) | 56 (86) | 63 (85) |
| - African American | 18 (13) | 9 (14) | 9 (12) |
| - Asian | 2 (1) | 0 | 2 (3) |
| Ethnicity | |||
| - Hispanic/ Latino | 77 (56) | 35 (54) | 42 (57) |
| - NonHispanic/ Latino | 62 (44) | 30 (46) | 32 (43) |
| Breastfeeding | |||
| - Exclusive breastfeeding for 6 months | 13 (9)b | 5 (8) | 8 (11) |
| - Exclusive breastfeeding for 3 months | 7 (5) | 5 (8) | 2 (3) |
| - Exclusive formula feeding | 62 (45) | 32 (49) | 30 (41) |
| - Mixed feeding | 57 (41) | 23 (35) | 34 (46) |
| One or more siblings at home (% yes) | 55 | 57 | 54 |
| Daycare attendance (% yes) | 33 | 22 | 43 |
| Cigarette smoke exposure (% yes) | 23 | 18 | 27 |
| Number of Specimens | 971c | 432 | 539 |
| Average number of specimens per subject | 7 | 6.6 | 7.3 |
| Age at samples collectiond | |||
| - 1 month | 131 | 57 | 74 |
| - 2 months | 136 | 64 | 72 |
| - 3 months | 148 | 71 | 77 |
| - 4 months | 139 | 65 | 74 |
| - 5 months | 145 | 73 | 72 |
| - 6 months | 137 | 63 | 74 |
| - 7–12 months | 135 | 39 | 96 |
| Specimens collected during URIe | 223 | 119 | 104 |
| Specimens collected during AOM | 45 | 45 | 0 |
| Specimens collected after antibiotic use | |||
| - 7 days | 37 | 30 | 7 |
| - 14 days | 43 | 36 | 7 |
| - 1 month | 71 | 57 | 14 |
| - 2 months | 98 | 72 | 26 |
a- row %
b-column % (% within the same category)
c- all specimens sequenced; 23 (2%) of these had <1000 sequence reads were excluded
d- nearest month, both monthly and URI samples
e- median day of URI at the time of sample collection = day 4, excluded samples collected during AOM diagnosis
II. Nasopharyngeal microbial communities
We obtained a total of 20,976,078 high-quality bacterial sequences comprising 13,982 unique operational taxonomical units (OTUs). Of 971 sequenced samples, 948 (98%) yielded ≥ 1000 sequences (mean = 22,118, median = 19,398, range 1,013–337,402 reads/ sample). Further analyses included only 948 samples with > 1000 sequence reads. Overall, 4 phyla predominated the sequence reads: Proteobacteria (39%), Actinobacteria (26%), Firmicutes (26%), and Bacteroidetes (6%).
Diversity of the microbial communities
Alpha diversity, as measure by Shannon Diversity Index (SDI) by age and type of samples (healthy vs URI/ AOM samples) is shown in Fig 1. There was no association between with age or sample type (p > 0.2 and p > 0.3, respectively).
Fig 1. Shannon diversity index by type of samples.
Healthy (grey circles) or sick visit (URI only or URI with AOM) samples (black circles).
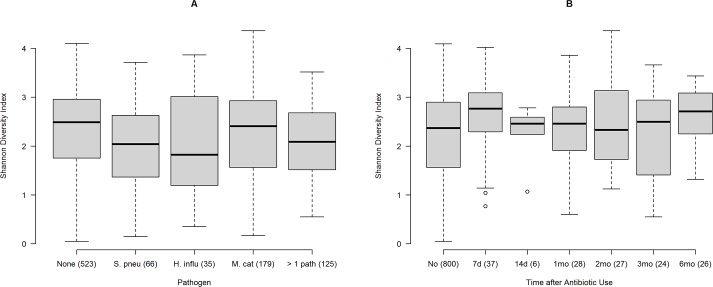
SDI by bacterial culture results are displayed in Fig 2A. The median SDI was 2.49 for samples with no otopathogen, compared to 2.04 in samples positive for S. pneumoniae only (P = 0.022); 1.82 for H. influenzae (P = 0.040); 2.41 for M. catarrhalis (P = 0.620) and 2.09 for 2 or more otopathogens (P = 0.048).
Fig 2.
A-B. Shannon diversity index. Fig 2A. Shannon Diversity Index by positive culture for otopathogens. Number in parentheses are number of samples with positive cultures. None = negative culture; S. pneu = Streptococcus pneumoniae; H. influ = Haemophilus influenzae; M. cat = Moraxella catarrhalis; > 1 path = positive culture for more than one of these pathogens. Fig 2B. Shannon Diversity Index by specific time point after antibiotic use. Samples were grouped based on the time of antibiotic use before sample collection: 7 days, 14 days, 1 month, 2 months, 3 months, and 6 months, comparison was made with samples collected after no history of antibiotic use.
Antibiotic use led to higher diversity overall (p = 0.0029) (Fig 2B); diversity was highest at 7d after antibiotic use (P = 0.011). There was no difference in diversity at 14 days, and at 1, 2, or 3 months, but diversity at 6 months of antibiotic use was higher compared to no antibiotic use (P = 0.016). Diversity was not associated with gender (P = 0.22), race (P = 0.88), breastfeeding (P>0.4), or mode of delivery (P = 0.4).
In the samples from the same subject, diversity at baseline (month 1) was positively associated with diversity at later age (P = 0.02; S1A Fig). Higher diversity at month 1 was associated with increasing frequencies of URI within the first 6 months (P = 0.036; S1B Fig), but baseline diversity was not associated with AOM frequency (P = 0.55; S1C Fig).
Nasopharyngeal microbiota overall
The top 21 most abundant genus-level microbiota (each accounted for ≥ 0.5% abundance) in 948 samples are shown in Table 2. These genera accounted for 70.3% of all identified bacterial genera. Otopathogen genera (Moraxella, Haemophilus, and Streptococcus), Staphylococcus and Pseudomonas were among the most common pathogen genera, while Corynebacterium, Dolosigranulum, and Acinetobacter accounted for the top 3 commensal genera. Samples from infants with AOM in the first year had significantly higher average abundance of Haemophilus, Enterobacter, and Yersinia, and lower abundance of Corynebacterium, and Pseudomonas compared to samples from infants without AOM.
Table 2. Relative abundance of microbiota in samples from subjects without and with AOM in the first year.
| Samples from | Samples from | Samples from | ||||||
|---|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Genus | all subjects a | subjects without AOM | subjects with AOM | P-value (Age-adjusted) |
| (N = 948) | (N = 516) | (N = 432) | ||||||
| Actinobacteria | Actinobacteria | Corynebacteriales | Corynebacteriaceae | Corynebacterium | 17.8% | 19.7% | 15.5% | 0.0071* |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | Moraxella | 9.7% | 9.1% | 10.4% | 0.4309 |
| Firmicutes | Bacilli | Lactobacillales | Carnobacteriaceae | Dolosigranulum | 6.8% | 7.8% | 5.7% | 0.0811 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | 5.7% | 6.0% | 5.4% | 0.8723 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | Acinetobacter | 4.1% | 4.6% | 3.6% | 0.2243 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | Haemophilus | 3.7% | 2.6% | 5.0% | 0.0072* |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | 3.5% | 4.0% | 2.9% | 0.0103* |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | 3.5% | 3.3% | 3.7% | 0.2990 |
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium | 2.9% | 2.1% | 3.9% | 0.0316 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Enterobacter | 2.5% | 1.9% | 3.1% | 0.0019* |
| Actinobacteria | Actinobacteria | Micrococcales | Micrococcaceae | Micrococcus | 1.6% | 1.6% | 1.7% | 0.5602 |
| Proteobacteria | Gammaproteobacteria | Chromatiales | Ectothiorhodospiraceae | Arhodomonas | 1.5% | 1.9% | 1.1% | 0.6486 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 1.3% | 1.0% | 1.6% | 0.1554 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Incertae Sedis | 1.0% | 0.7% | 1.3% | 0.4470 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Burkholderiaceae | Ralstonia | 0.9% | 0.9% | 0.9% | 0.2199 |
| Bacteroidetes | Flavobacteriia | Flavobacteriales | Flavobacteriaceae | Myroides | 0.8% | 1.1% | 0.5% | 0.0202 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Pantoea | 0.6% | 0.5% | 0.8% | 0.8054 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Yersinia | 0.6% | 0.6% | 0.7% | 0.0051* |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae 1 | Clostridium sensu stricto 1 | 0.6% | 0.6% | 0.6% | 0.0688 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | 0.6% | 0.6% | 0.6% | 0.6080 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingobium | 0.6% | 0.6% | 0.6% | 0.1464 |
a-66 subjects had AOM in the first year; 73 subjects did not
Data presented as an average value of relative abundance of the specific genus within each sample
All results which are still significant after adjustment for multiple testing indicated by *.
Effect of antibiotic on the nasopharyngeal microbiota
Of 948 samples, 148 (16%) were collected within 7 days to 6 months of antibiotic use. The usual antibiotic course was 7–10 days; the most commonly used antibiotic was amoxicillin. The abundance of microbiota in relation to the time to prior antibiotic use is shown in S2 Fig. Antibiotics did not significantly affect the otopathogen genera but significantly decreased: 1) Corynebacterium (P = 0.0165) and Dolosigranulum (P = 0.0084) within 7 days; 2) Enterobacter between 7 and 14 days (P = 0.0347); and 3) Staphylococcus at between 14 days and 1 month (P = 0.0161). Antibiotic increased the abundance of Bifidobacterium (P = 0.0146) and Firmicutis Incertae Sedis (P = 0.0140) at the 7day time point.
III Effect of URI/ viral infection on microbiome composition
Microbiota in healthy vs sick visit (URI with and without AOM) samples
Table 3compares microbiome composition in healthy vs sick visit samples. The abundance of all 3 otopathogen genera was significantly higher in sick visit samples, while there was a significant reduction in Pseudomonas, Myroides, Yersinia, and Sphingomonas. Significance remained after adjustment for multiple comparisons for higher abundance of Moraxella and lower abundance of Yersinia in sick visit samples. Comparison between microbiota in specific sample types (healthy vs URI, healthy vs AOM, and URI vs AOM) are shown in Tables A-C in S4 File, respectively. Comparing AOM samples to healthy samples, there was higher abundance of Moraxella and Haemophilus.
Table 3. Relative abundance of microbiota in healthy samples compared to sick visit (URI with or without AOM) samples.
| Genus | Total | Healthy samples | URI / AOM samples | P-value |
|---|---|---|---|---|
| (N = 948) | (N = 685) | (N = 263) | (age-adjusted) | |
| Corynebacterium | 17.8% | 18.3% | 16.4% | 0.6094 |
| Moraxella | 9.7% | 8.4% | 13.1% | 0.0006* |
| Dolosigranulum | 6.8% | 6.8% | 7.0% | 0.4009 |
| Staphylococcus | 5.7% | 6.2% | 4.5% | 0.0755 |
| Acinetobacter | 4.1% | 4.3% | 3.7% | 0.0534 |
| Haemophilus | 3.7% | 2.7% | 6.3% | 0.0314 |
| Pseudomonas | 3.5% | 3.7% | 2.9% | 0.0149 |
| Streptococcus | 3.5% | 3.1% | 4.3% | 0.0278 |
| Bifidobacterium | 2.9% | 2.8% | 3.4% | 0.2694 |
| Enterobacter | 2.5% | 2.5% | 2.4% | 0.4921 |
| Micrococcus | 1.6% | 1.6% | 1.8% | 0.1119 |
| Arhodomonas | 1.5% | 1.6% | 1.3% | 0.6138 |
| Bacteroides | 1.3% | 1.4% | 0.9% | 0.1618 |
| Incertae Sedis | 1.0% | 1.0% | 1.0% | 0.0736 |
| Ralstonia | 0.9% | 1.0% | 0.8% | 0.5836 |
| Myroides | 0.8% | 0.9% | 0.6% | 0.0150 |
| Pantoea | 0.6% | 0.7% | 0.4% | 0.3637 |
| Yersinia | 0.6% | 0.7% | 0.5% | 0.0010* |
| Clostridium sensu stricto 1 | 0.6% | 0.6% | 0.4% | 0.3139 |
| Sphingomonas | 0.6% | 0.6% | 0.5% | 0.0448 |
| Sphingobium | 0.6% | 0.6% | 0.6% | 0.2267 |
* Result still significant at the 0.05 level after adjustment for multiple testing
Effect of virus infections
Virus data were available in 890 of 948 samples (94%); one or more viruses were detected from 42% of samples (500 viruses). Rhinovirus was the most common virus (39%); human coronavirus was detected in 20%; enterovirus, 11%; parainfluenza, 9%; adenovirus, 6%; respiratory syncytial virus, 6%; bocavirus, 4%; metapneumovirus, 4%; and influenza virus, 1%. Table 4compares microbiota in virus-positive vs virus-negative samples. To evaluate the effect of symptomatic virus infection, we compared microbiota in virus-positive URI samples with and virus-positive healthy samples (Table A in S5 File), and with virus-negative healthy samples (Table B in S5 File). Symptomatic virus infection samples had significant increased abundance of Moraxella and Streptococcus. Effect of asymptomatic viral infection is shown in Table C in S5 File. Rhinovirus was the most common virus detected (n = 193) regardless of symptoms; Table D in S5 File compares microbiota in rhinovirus-positive and rhinovirus-negative samples.
Table 4. Relative abundance of microbiota in virus-negative and virus-positive samples.
| Genus | All samples | Virus-negative samples | Virus-positive samples | P-value (age-adjusted)* |
|---|---|---|---|---|
| (N = 872) | (N = 509) | (N = 363) | ||
| Corynebacterium | 17.8% | 18.8% | 16.4% | 0.2444 |
| Moraxella | 10.0% | 8.5% | 12.0% | 0.0143 |
| Dolosigranulum | 6.9% | 7.4% | 6.3% | 0.2987 |
| Staphylococcus | 5.8% | 6.3% | 5.2% | 0.9983 |
| Acinetobacter | 4.1% | 4.3% | 3.9% | 0.5937 |
| Haemophilus | 3.8% | 2.5% | 5.7% | 0.2414 |
| Streptococcus | 3.5% | 2.7% | 4.6% | 0.9299 |
| Pseudomonas | 3.5% | 3.6% | 3.4% | 0.0027 |
| Bifidobacterium | 3.0% | 2.8% | 3.3% | 0.6708 |
| Enterobacter | 2.5% | 2.6% | 2.5% | 0.3380 |
| Micrococcus | 1.6% | 1.7% | 1.5% | 0.7034 |
| Bacteroides | 1.3% | 1.5% | 1.1% | 0.5020 |
| Arhodomonas | 1.2% | 1.5% | 0.9% | 0.2255 |
| Incertae Sedis | 1.0% | 0.9% | 1.2% | 0.9476 |
| Ralstonia | 0.9% | 1.0% | 0.8% | 0.3378 |
| Myroides | 0.9% | 1.1% | 0.7% | 0.0435 |
| Pantoea | 0.7% | 0.7% | 0.6% | 0.1105 |
| Yersinia | 0.6% | 0.6% | 0.6% | 0.6689 |
| Clostridium sensu stricto 1 | 0.6% | 0.7% | 0.5% | 0.9707 |
| Sphingobium | 0.6% | 0.5% | 0.6% | 0.9869 |
| Sphingomonas | 0.6% | 0.6% | 0.5% | 0.1843 |
* All P-values were > 0.05 after adjustment for multiple testing
IV Microbiome composition and frequencies of URI and AOM
We determine the relationship between nasopharyngeal microbiota and URI/ AOM frequencies by comparing the linear trend (from 1–6 months) in microbial presence in samples collected from infants with frequent URIs early in life (e.g. 1, 2, 3, and 4 URI episodes) vs those without URI (control group) (S1 Table). In the first 3 months, 168 infants had ≥ 1 URI; in the first 6 months, 105 infants had 1, 76 had two, 41 had three, and 39 had ≥ 4 URI episodes. Infants with more URIs in the first 3 months had increasing abundance of Streptococcus (Linear Trend P = 0.0112,), while infants with more URI in the first 6 months had increasing abundance of Moraxella and Haemophilus, compared to controls. There was no significant difference in abundance of Staphylococcus or other commensals between groups. A total of 68 infants had ≥ 1 AOM in the first 6 months. Increased number of AOM episodes in the first 6 months was associated with decreasing Micrococcus abundance; there was no significant trend seen with other genera.
V. Changes in microbiome composition during transition from URI to AOM
The availability of longitudinal samples from the subjects allowed us to determine the changes during transition from URI to AOM. We first studied microbiota in samples collected within the first 7 days of URI onset (N = 184); samples were compared based on the follow-up outcome of URI (resolved vs complicated by AOM). Of these, 167 were URI samples from cases that resolved without AOM complication; 17 were from cases that were later complicated by AOM. S2 Table compares microbiota in URI samples from cases that were complicated by AOM vs those that resolved. Although there was higher abundance of Streptococcus in the AOM group, the difference did not reach statistical difference when adjusted for age.
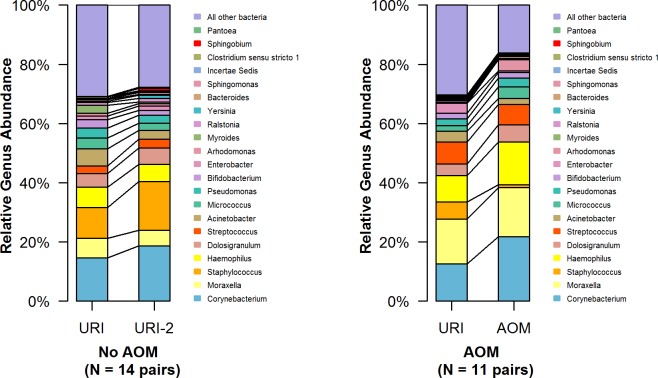
We then studied paired samples from the same subjects collected ≤ 7 days apart; the first sample of the pair was collected as soon as possible and within 7 days of URI onset. Available data included 2 sets of paired samples; the first set of 14 pairs were URI samples collected 1–7 days apart (mean = 3 days) that did not result in AOM; the average age at the sample collection sets was 3.6 (median = 1.9) months. The second set of 11 paired samples, URI was complicated by AOM within 3 days (follow-up samples = AOM samples); the average age was 5.2 (median = 4.9) months. Significant increases in Staphylococcus (P = 0.007) and Sphingobium (P = 0.037) were detected in paired samples of URI that recovered, compared to those that resulted in AOM (Fig 3and S3 Table).
Fig 3. Comparison of the most abundant genera in paired samples collected ≤ 7 days apart.
The left set represents data from 14 pairs from subjects with URI that resolved without AOM complication. The right set represents data from 11 pairs from subjects with URI that were complicated with AOM (all within 3 days of the first URI samples).
Discussion
We studied nasopharyngeal microbiota in nearly one thousand samples, collected longitudinally during health and disease (URI and AOM) from one month of age through the first year. Our results suggested that bacterial otopathogen genera (Haemophilus, Streptococcus, and Moraxella) played the key role in the disease process. The increasing trend in colonization of these otopathogen genera was also correlated positively with frequencies of URI, and their presence was associated with URI symptom expression during viral infection. Interestingly, Staphylococcus, but not the commonly used probiotic bacterium Bifidobacterium, played an important role in counteracting the deleterious effect of otopathogens.
The availability of comprehensive clinical, bacteriologic, and virologic data allowed us to determine the significance of pathogens, commensals, and their interactions with viruses during URI and AOM. Microbiome analyses further expanded the knowledge on commensal bacteria not traditionally cultured or non-culturable. During symptomatic viral infection, there were significant increases in relative abundance of otopathogen genera, while there was no significant change in otopathogen or commensals during asymptomatic viral infection. The finding on symptoms associated with increased otopathogen abundance agrees with a finding from a recent study suggesting that both viruses and bacteria contributed to acute upper and lower respiratory tract symptoms [9]. Microbiome stability found during asymptomatic viral infection helps explain why asymptomatic viral infection did not lead to AOM, which we previously reported from this subject cohort [23].
When comparing microbiota from paired-samples collected during URI episodes that resolved to those complicated by AOM, we found that increased Staphylococcus and Sphingobium abundance prevented the transition from URI to AOM. It must be pointed out that the average age at the time of sample collection was older in the AOM group compared to no AOM group. It is known that infants <6 months are less often diagnosed with AOM compared to older infants. Our data, although from a small number, suggest that one of the factors preventing AOM development in young infants is colonization with Staphylococcus, which helps counteract with the otopathogens. Our previously published report from this cohort [26] has shown the highest S. aureus colonization rate (25%) in samples collected at age 1 month, with declining in rate to 12% by age 6 months, along with increasing rates of colonization of S. pneumoniae, H. influenzae, and M. catarrhalis. Others have also shown the negative association between S. aureus colonization and otopathogen colonization [27–29]. Because S. aureus colonization in young infants is not associated with invasive infections [26–30], Staphylococcus colonization may be beneficial than harmful to these young infants. Not only Staphylococcus protected them from otopathogen colonization but it also helped prevent the transition from URI to AOM.
In other studies, nasopharyngeal bacterial commensals were studied mainly in children > 6 months of age, and the samples were often collected during disease state [19, 20, 31]. Our data were from infants, 86% of whom were 1–6 months. Therefore, commensal genera in our subjects were somewhat different than those reported from older and/ or sick children. In a recent study [7], samples were collected from 102 healthy individuals at 24–36 hours after delivery, 7 and 14 days, and 1, 2, 3 4 and 6 months; the commensal genera shown were more comparable to ours. Not only age has been shown to affect nasopharyngeal microbiota, numerous other factors such as mode of delivery, infant feeding type, antibiotic use, vaccines, etc. have also been found to affect microbiota in healthy children [7, 20, 32, 33]. Nevertheless, Corynebacterium and Dolosigranulum have consistently been found to be the two most common commensal genera in the nasopharynx of children.
During active disease processes when pathogens become predominant, the relative abundance of commensals is reduced, but specific commensals are affected differently. We searched for commensal genera that were affected by the surge in otopathogens; we did not find significant effect on high abundant genera such as Corynebacterium or Dolosigranulum during URI and/or AOM. Aside from Staphylococcus, Proteobacteria such as Acinetobacter, Pseudomonas, Yersinia, and Bacteroides (Myroides) were significantly reduced. Decreasing trend in colonization with Micrococcus was associated with increased AOM frequencies in the first 6 months. The role of these less common commensals in counteracting with otopathogens and preventing AOM deserves further investigations.
This study is limited in that we reported our data to the genus-level, which is not uncommon for this type of study. The otopathogen genera reported may have contained more than the specific otopathogen species. For example, the Streptococcus genus may also contain alpha Streptococci in addition to S. pneumoniae. However, Bosch et al. [7] have shown that S. pneumoniae represented a high proportion of the Streptococcus genus in the samples collected from infants > 1 month. Similarly, Teo et al. [9] showed in a study of infants 2–12 months of age that otopathogen genera were dominated by S. pneumoniae, H. influenzae, and M. catarrhalis. In addition, our bacterial culture data confirmed the relationship between the increased bacterial otopathogen colonization during URI and laboratory-confirmed viral infections.
We studied nasopharyngeal microbiota in a search for important commensal bacteria that may be used as effective intranasal probiotics to prevent AOM. The ideal probiotics should be bacteria that have the most interference (counteracting) with pathogens and cause no harm. Previous studies have used both oral and intranasal probiotics containing alpha streptococci, Lactobacillus rhamnosus GG and Bifidobacterium; results have been mixed [16, 34]. We have not found the Lactobacillus genus to be a major nasopharyngeal commensal and we do not have the data on alpha streptococci. Bifidobacterium was commonly found in our samples, ranked 5th among the commensal genera; we did not find it play a role during URI or AOM process. Furthermore, Bifidobacterium was found more commonly in samples from infants with AOM in the first year of life. Our data suggest that intranasal use of Bifidobacterium may not be helpful as a probiotic. Corynebacterium, the most common commensal was found to be significantly lower in samples from infants with AOM in the first year (Table 2), but we observed no other significant difference during the URI or AOM process. Other bacteria that played a reverse role with otopathogens such as Yersinia and Pseudomonas may not be appropriate probiotics for their possible roles as pathogens.
The mechanisms of AOM pathogenesis are complex; numerous host and environmental factors, as well as interactions between viruses, otopathogenic bacteria and commensals play roles in AOM development. The results of this study emphasize the importance of otopathogens and suggest that prevention of nasopharyngeal otopathogen colonization and viral infection will be the key to preventing AOM.
Supporting information
A. Association between baseline diversity and diversity at later age (month 6), P = 0.02. B. Association between diversity at baseline (month 1) and number of URI in the first 6 months. Higher diversity at month 1 was associated with increasing frequencies of URI within the first 6 months (P = 0.036). C. Association between diversity at baseline (month 1) and number of URI in the first 6 months (P = 0.55).
(TIF)
The time point indicates days or months of antibiotic use prior to nasopharyngeal sample collection. Numbers in parentheses are number of samples collected within specific time of antibiotic use.
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(XLSX)
(XLS)
Table A. Microbiota in healthy samples compared to URI samples. Table B. Microbiota in healthy samples compared to AOM samples. Table C. Microbiota in URI samples compared to AOM samples.
(DOC)
Table A. Microbiota in virus-positive healthy samples and virus-positive URI samples. Table B. Microbiota in virus-negative healthy samples and virus-positive URI samples. Table C. Effect of asymptomatic virus infection: microbiota in healthy virus-negative vs healthy virus-positive samples. Table D. Microbiota in rhinovirus-negative samples and rhinovirus-positive samples.
(DOC)
Acknowledgments
The authors acknowledge and thank Joseph Petrosino, Director of the Alkek Center for Metagenomics and Microbiome Research (CMMR), Baylor College of Medicine for his scientific support; other CMMR experts who contributed to this study are: Nadim Ajami, Daniel P. Smith, Diane S. Hutchison, and Jackie O’Brien. Tonya Bauch provided technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Institutes of Health research grants K18DC13564 and R01DC005841. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- 1.Daly KA, Hoffman HJ, Kvaerner KJ, Kvestad E, Casselbrant ML, Homoe P, et al. Epidemiology, natural history, and risk factors: Panel report from the Ninth International Research Conference on Otitis Media. 2010. Int J Pediatr Otorhinolaryngol; 74: 231–240. doi: 10.1016/j.ijporl.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 2.Grijalva CG, Nuorti JP, and Griffin MR. Antibiotic prescription rates for acute respiratory tract infections in US ambulatory settings. 2009. JAMA; 302:758–766. doi: 10.1001/jama.2009.1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schilder AG, Lok W, Rovers MM. International perspectives on management of acute otitis media: a qualitative review. 2004. Int J Pediatr Otorhinolaryngol; 68(1):29–36. [DOI] [PubMed] [Google Scholar]
- 4.Chonmaitree T, Revai K, Grady JJ, Clos A, Patel JA, Nair S, et al. Viral upper respiratory tract infection and otitis media complication in young children. 2008. Clin Infect Dis; 46:815–823. doi: 10.1086/528685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chonmaitree T. Acute otitis media is not a pure bacterial disease. 2006. Clin Infect Dis; 43: 1423–5. doi: 10.1086/509329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakaletz LO. Immunopathogenesis of polymicrobial otitis media. 2010. J Leukoc Biol; 87:213–222. doi: 10.1189/jlb.0709518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch AA, Levin E, van Houten MA, Hasrat R, Kalkman G, Biesbroek G, et al. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. 2016. EBioMedicine pii: S2352-3964(16)30225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mika M, Mack I, Korten I, Qi W, Aebi S, Frey U, et al. Dynamics of the nasal microbiota in infancy: a prospective cohort study. 2015. J Allergy Clin Immunol; 135(4):905–12.e11. doi: 10.1016/j.jaci.2014.12.1909 [DOI] [PubMed] [Google Scholar]
- 9.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. 2015. Cell Host Microbe;17(5):704–15. doi: 10.1016/j.chom.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faden H, Duffy L, Wasielewski R, Wolf J, Krystofik D. Relationship between nasopharyngeal colonization and the development of otitis media in children. 1997. J Infect Dis; 175:1440–1445. [DOI] [PubMed] [Google Scholar]
- 11.Chonmaitree T, Trujillo R, Jennings K, Alvarez-Fernandez P, Patel JA, Loeffelholz MJ, et al. Acute otitis media and other complications of viral respiratory infection. 2016. Pediatrics; 137 (4) DOI: doi: 10.1542/peds.2015-3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nokso-Koivisto J, Marom T, Chonmaitree T. Importance of viruses in acute otitis media. 2015. Curr Opin Pediatr; 27:110–115. doi: 10.1097/MOP.0000000000000184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimori I, Goto R, Kikushima K, Hisamatsu K, Murakami Y, Yamada T. Investigation of oral alpha-streptococcus showing inhibitory activity against pathogens in children with tonsillitis. 1995. Int J Pediatr Otorhinolaryngol; 33(3):249–55. [DOI] [PubMed] [Google Scholar]
- 14.Tano K, Olofsson C, Grahn-Håkansson E, Holm SE. In vitro inhibition of S. pneumoniae, nontypable H. influenzae and M. catharralis by alpha-hemolytic streptococci from healthy children. 1999. Int J Pediatr Otorhinolaryngol; 47(1):49–56. [DOI] [PubMed] [Google Scholar]
- 15.Tano K, Hellström S. Bacterial adherence to pharyngeal cells: in vitro studies with alpha-haemolytic streptococci and Haemophilus influenzae. 2002. Acta Otolaryngol; 122(7):745–51. [DOI] [PubMed] [Google Scholar]
- 16.Niittynen L, Pitkäranta A, Korpela R. Probiotics and otitis media in children. 2012. Int J Pediatr Otorhinolaryngol;76(4):465–70. doi: 10.1016/j.ijporl.2012.01.011 [DOI] [PubMed] [Google Scholar]
- 17.Nagalingam NA, Cope EK, Lynch SV. Probiotic strategies for treatment of respiratory diseases. 2013. Trends Microbiol; 21(9):485–92. doi: 10.1016/j.tim.2013.04.008 [DOI] [PubMed] [Google Scholar]
- 18.Lehtoranta L, Pitkäranta A, Korpela R. Probiotics in respiratory virus infections. 2014. Eur J Clin Microbiol Infect Dis;33(8):1289–302. doi: 10.1007/s10096-014-2086-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. Microbial communities of the upper respiratory tract and otitis media in children. 2011. MBio;2(1):e00245–10. doi: 10.1128/mBio.00245-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. 2012. Appl Environ Microbiol;78(17):6262–70. doi: 10.1128/AEM.01051-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilty M, Qi W, Brugger SD, Frei L, Agyeman P, Frey PM, et al. Nasopharyngeal microbiota in infants with acute otitis media. 2012. J Infect Dis; 1;205(7):1048–55. doi: 10.1093/infdis/jis024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. 2014. Am J Respir Crit Care Med;190(11):1283–92. doi: 10.1164/rccm.201407-1240OC [DOI] [PubMed] [Google Scholar]
- 23.Chonmaitree T, Alvarez-Fernandez P, Jennings K, Trujillo R, Marom T, Loeffelholz MJ, et al. Symptomatic and asymptomatic viral infections in the first year of life: Association with acute otitis media development. 2015. Clinical Infect Dis; 60(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aagaard K, Petrosino J, Keitel W, Watson M, Katancik J, Garcia N, et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 2013. March;27(3):1012–22. doi: doi: 10.1096/fj.12-220806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. 2013. Nucleic Acids Res; 41 (D1): D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel JA, Alvarez-Fernandez PE, Jennings K, Loeffelholz MJ, McCormick DP, Chonmaitree T. Factors Affecting Staphylococcus aureus Colonization of the Nasopharynx in the First Six Months of Life. 2015. Pediatr Infect Dis J; 34 (8):826–830. doi: 10.1097/INF.0000000000000744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiri T, Nunes MC, Adrian PV, Van Niekerk N, Klugman KP, Madhi SA. Interrelationship of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus colonization within and between pneumococcal-vaccine naïve mother-child dyads. 2013. BMC Infect Dis; 13:483 doi: 10.1186/1471-2334-13-483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodrigues F, Foster D, Nicoli E, Trotter C, Vipond B, Muir P, et al. Relationships between rhinitis symptoms, respiratory viral infections and nasopharyngeal colonization with Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in children attending daycare. 2013. Pediatr Infect Dis J; 32(3):227–32. doi: 10.1097/INF.0b013e31827687fc [DOI] [PubMed] [Google Scholar]
- 29.Lewnard JA, Givon-Lavi N, Huppert A, Pettigrew MM, Regev-Yochay G, Dagan R, et al. Epidemiological Markers for Interactions Among Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus in Upper Respiratory Tract Carriage. J Infect Dis 2016. May 15; 213(10):1596–605. doi: 10.1093/infdis/jiv761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chatzakis E, Scoulica E, Papageorgiou N, Maraki S, Samonis G, Galanakis E. Infant colonization by Staphylococcus aureus: role of maternal carriage. 2011. Eur J Clin Microbiol Infect Dis; 30(9):1111–7. doi: 10.1007/s10096-011-1199-9 [DOI] [PubMed] [Google Scholar]
- 31.Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, et al. Variability and Diversity of Nasopharyngeal Microbiota in Children: A Metagenomic Analysis. 2011. PLoS ONE 6(2): e17035 doi: 10.1371/journal.pone.0017035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biesbroek G, Bosch AA, Wang X, Keijser BJ, Veenhoven RH, Sanders EA, et al. The impact of breastfeeding on nasopharyngeal microbial communities in infants. 2014. Am J Respir Crit Care Med; 190(3):298–308. doi: 10.1164/rccm.201401-0073OC [DOI] [PubMed] [Google Scholar]
- 33.Biesbroek G, Wang X, Keijser BJ, Eijkemans RM, Trzciński K, Rots NY, et al. Seven-valent pneumococcal conjugate vaccine and nasopharyngeal microbiota in healthy children. 2014. Emerg Infect Dis; 20(2):201–10. doi: 10.3201/eid2002.131220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marom T, Marchisio P, Tamir SO, Torretta S, Gavriel H, Esposito S. Complementary and Alternative Medicine Treatment Options for Otitis Media: 2016. A Systematic Review. Medicine (Baltimore); 95(6):e2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Association between baseline diversity and diversity at later age (month 6), P = 0.02. B. Association between diversity at baseline (month 1) and number of URI in the first 6 months. Higher diversity at month 1 was associated with increasing frequencies of URI within the first 6 months (P = 0.036). C. Association between diversity at baseline (month 1) and number of URI in the first 6 months (P = 0.55).
(TIF)
The time point indicates days or months of antibiotic use prior to nasopharyngeal sample collection. Numbers in parentheses are number of samples collected within specific time of antibiotic use.
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(XLSX)
(XLS)
Table A. Microbiota in healthy samples compared to URI samples. Table B. Microbiota in healthy samples compared to AOM samples. Table C. Microbiota in URI samples compared to AOM samples.
(DOC)
Table A. Microbiota in virus-positive healthy samples and virus-positive URI samples. Table B. Microbiota in virus-negative healthy samples and virus-positive URI samples. Table C. Effect of asymptomatic virus infection: microbiota in healthy virus-negative vs healthy virus-positive samples. Table D. Microbiota in rhinovirus-negative samples and rhinovirus-positive samples.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.