Summary
THAP1 (THAP [Thanatos-associated protein] domain-containing, apoptosis-associated protein 1) is a ubiquitously expressed member of a family of transcription factors with highly conserved DNA-binding and protein-interacting regions. Mutations in THAP1 cause dystonia, DYT6, a neurologic movement disorder. THAP1 downstream targets and the mechanism via which it causes dystonia are largely unknown. Here, we show that wild-type THAP1 regulates embryonic stem cell (ESC) potential, survival, and proliferation. Our findings identify THAP1 as an essential factor underlying mouse ESC survival and to some extent, differentiation, particularly neuroectodermal. Loss of THAP1 or replacement with a disease-causing mutation results in an enhanced rate of cell death, prolongs Nanog, Prdm14, and/or Rex1 expression upon differentiation, and results in failure to upregulate ectodermal genes. ChIP-Seq reveals that these activities are likely due in part to indirect regulation of gene expression.
Keywords: Thanatos-associated protein domain-containing apoptosis-associated protein 1, THAP1, dystonia, differentiation, survival, embryonic stem cells, transcriptomics, neuroectodermal differentiation, apoptosis, zinc finger transcription factor
Highlights
-
•
Wild-type THAP1 regulates ESC potential, survival, and proliferation
-
•
THAP1 is essential for ESC differentiation, particularly neuroectodermal
-
•
Thap1C54Y or ΔExon2 ESCs prolong expression of pluripotent genes upon differentiation
-
•
Thap1C54Y or ΔExon2 EBs show increased cell death and abnormal differentiation
Ehrlich and colleagues identified THAP1, mutations of which cause dystonia (DYT6), as an essential regulator of ESC survival, proliferation, and differentiation. As THAP1 mutations result in a neurologic disease, this work examined the effects of a causative mutation, C54Y, and an essentially null allele, ΔExon2, on neuronal differentiation.
Introduction
THAP1 (THAP [Thanatos-associated protein] domain-containing, apoptosis-associated protein 1) is a member of a large family of proteins which are primarily transcription factors (Gervais et al., 2013, Roussigne et al., 2003). The THAP domain, an atypical zinc finger (CysX2-4CysX35-53CysX2His), is highly conserved and is part of the DNA-binding domain (DBD) with homology to P-transposable elements (Majumdar and Rio, 2015). THAP1 mutations cause DYT6 dystonia (Fuchs et al., 2009), and mutations are located throughout the protein, with about 50% in the DNA-binding domain. Importantly, recessive mutations have been identified (Houlden et al., 2010, Schneider et al., 2011, Xiromerisiou et al., 2012). THAP1 functions, targets, and the mechanisms by which its mutations lead to dystonia are largely unknown, including the effects of mutations on DNA binding (Campagne et al., 2012).
Functional studies of THAP1 in human umbilical vein endothelial cells (HUVECs) show a role in the S phase of mitosis via modulation of pRb-E2F cell-cycle target genes, including RRM1 (Clouaire et al., 2005). In vitro, a coiled-coil domain is required for dimerization (Sengel et al., 2011). Other interactors include prostate apoptosis response-4 protein (Par-4), an effector of cell death linked to prostate cancer and neurodegenerative diseases (Roussigne et al., 2003); HCF-1, a transcriptional co-activator involved in cell-cycle regulation; and O-GlcNAc transferase (OGT), which catalyzes the addition of O-GlcNAc and thereby also participates in epigenetic regulation of gene expression with an essential function in dividing cells (Mazars et al., 2010).
In mouse models of DYT6 which harbor either a disease-causing C54Y mutation in the DBD or a null allele (ΔExon2) (Ruiz et al., 2015), rare homozygous embryos survived to day 14. They were small with defects in peripheral organs and brain, which showed deficits in the number and morphology of neurons. To study the impact of the mutant alleles on stem cell maintenance and differentiation, we generated mouse embryonic stem cells (mESCs) homozygous for either the C54Y (Thap1C54Y) or ΔExon2 (Thap1ΔExon2) alleles. Herein, we characterize both ESCs, which are viable with intact stem cell characteristics, but with abnormalities in cell death, cell cycle, and proliferation rate, that are more severe in the ΔExon2 than in the C54Y homozygote. Furthermore, we show that during differentiation of embryoid bodies (EBs), wild-type THAP1 is required for repression of a cohort of core pluripotency-associated genes and survival in ΔExon2 cells, and apparently for terminal neuronal differentiation in C54Y homozygote surviving cells.
Results
Generation of Thap1-Recombinant Embryonic Stem Cells
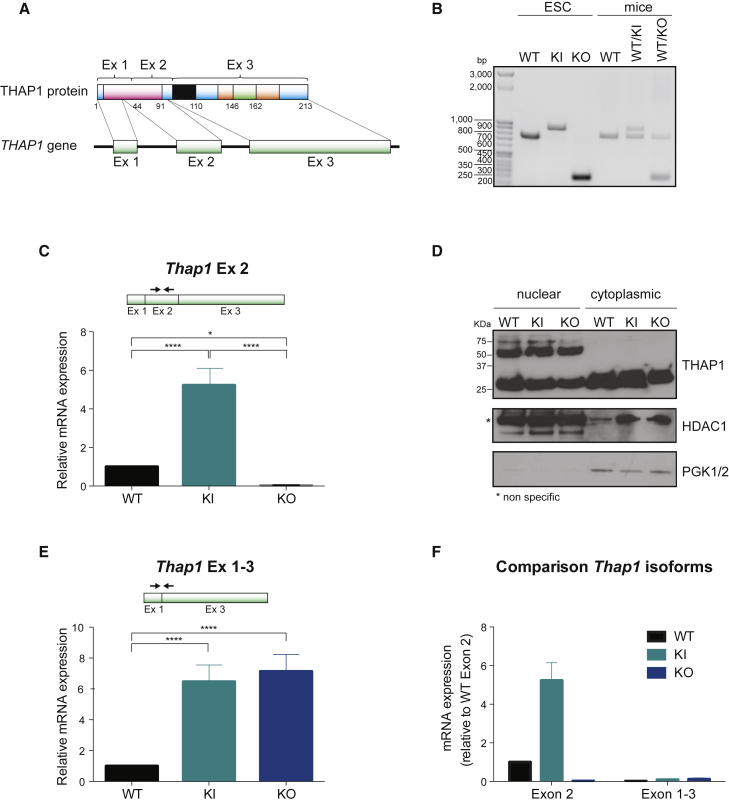
To explore the function of THAP1 in ESCs, we derived ESCs from two mouse alleles, (1) Thap1C54Y, a constitutive knockin (KI) of the C54Y causative mutation in the DBD of THAP1, and (2) Thap1ΔExon2, a constitutive knockout (KO), allele lacking exon 2 (Ruiz et al., 2015; Figure 1A). Genotypes of wild-type (WT), Thap1C54Y/C54Y, and Thap1 ΔExon2/ΔExon2 ESCs were analyzed by PCR and compared with WT, Thap1C54Y/+, and Thap1 ΔExon2/+ heterozygote mice (Figure 1B). Consistent with THAP1 autorepression (Erogullari et al., 2014) and failure of THAP1C54Y to bind at the Tor1a promoter (Gavarini et al., 2010), Thap1C54Y cells exhibited higher levels of Thap1 mRNA than WT ESCs, whereas full-length Thap1 mRNA was undetectable in Thap1ΔExon2 ESCs (Figure 1C). THAP1 antibodies recognize several THAP1-like immunoreactive (THAP1-LIR) species (Ortiz-Virumbrales et al., 2014). Subcellular fractionation followed by western blot analysis revealed the presence of three distinct THAP1-LIR species in the nuclear fraction at ∼29 kDa, ∼50 kDa, and ∼75 kDa (Figure 1D, upper panel). Only the ∼75-kDa species was induced or drastically reduced in Thap1C54Y or Thap1ΔExon2 ESCs, respectively, following the same pattern of the corresponding mRNA as assessed by qRT-PCR (Figure 1C). Thus, the ∼29- and ∼50-kDa species appear to be largely composed of cross-reacting proteins in ESCs. In murine brain, the ∼29-kDa species was also non-specific, whereas the 50- and 75-kDa THAP1-LIR species were nuclear and neuron specific, and virtually undetectable in ΔExon2 embryos (Ortiz-Virumbrales et al., 2014). Primers spanning exon 1 and exon 3 of Thap1 mRNA amplified a de-repressed transcript, i.e., its expression was induced, in Thap1C54Y/C54Y and Thap1ΔExon2 ESCs (Figure 1E). This naturally occurring THAP1ΔExon2 represents less than 1% relative to the major isoform (containing exon 2) (Figure 1F) and in vivo does not substitute for the loss of the full-length isoform.
Figure 1.
Generation of Thap1-Recombinant ESCs
(A) Structure of Thap1 gene and its encoded protein.
(B) Genotyping of wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs by PCR and comparison with the pattern of WT, Thap1C54Y/+ (WT/KI), and Thap1ΔExon2/+ (WT/KO) heterozygote mice.
(C) Thap1 exon 2 (Thap1 Ex 2) transcript level measured by qRT-PCR in wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs. An ANOVA was performed which revealed a significant difference among the genotypes (F(2,23) = 91.69, p < 0.001). The Holm-Sidak multiple comparisons test was performed post hoc, revealing significant differences between the genotypes. Data are presented as mean ± SEM of three independent experiments. ∗p < 0.05; ∗∗∗∗p < 0.001.
(D) Distribution of THAP1 protein in nuclear and cytoplasmic fraction of wild-type (WT), Thap1C54Y/C54Y (KI) and Thap1ΔExon2 ESCs (KO). Histone deacetylase 1 (HDAC1) and phosphoglycerate kinase 1/2 (PGK1/2) were used as a control of nuclear and cytoplasmic extract purity, respectively.
(E) Level of Thap1 transcript spanning exon 1 and exon 3 (Thap1 Ex 1–3) measured by qRT-PCR in wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs. ANOVA revealed a significant difference among the genotypes (F(2,21) = 73.85, p < 0.0001). The Holm-Sidak multiple comparisons test was performed post hoc, revealing significant differences between the genotypes. Data are presented as mean ± SEM of at least three independent experiments. ∗∗∗∗p < 0.001.
(F) Comparison of Thap1 isoforms by qRT-PCR in wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs. Data are presented as mean ± SEM.
Loss of THAP1 Alters ESC Viability
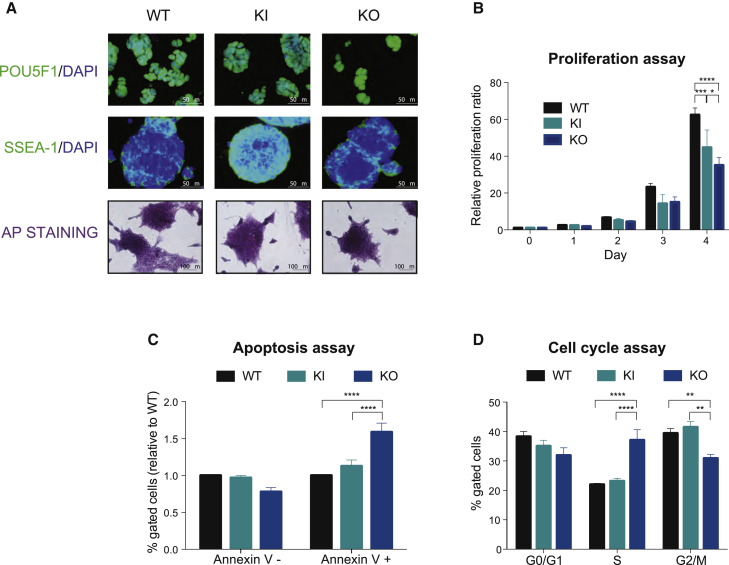
To investigate THAP1 function in self-renewal, we analyzed the expression of stage-specific embryonic antigen 1 (SSEA1) and POU5F1 (Palmqvist et al., 2005) in all three genotypes (Figure 2A, top and middle panels). There were no detectable differences. The level of alkaline phosphatase (AP) staining (Figure 2A, bottom panels), characteristic of undifferentiated ESCs, was also genotype independent. These data suggest that THAP1 is not required to maintain the pluripotency state of ESCs.
Figure 2.
THAP1 Is Not Required for ESC Maintenance
(A) Immunofluorescence analysis of POU5F1 (upper panel) SSEA1 (middle panel), and alkaline phosphatase (AP) staining (lower panel) in wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs.
(B) Proliferation rate of wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs relative to day 0 and measured by counting the cells at the indicated time points. Two-way repeated-measure (RM) ANOVA was performed revealing an interaction effect between days and genotype (F(8,36) = 4.288, p = 0.0011). In addition, significant differences were observed by day (F(4,36) = 139.4, p < 0.0001), while marginal differences were observed by genotype (F(2,9) = 3.685, p = 0.0677). The Holm-Sidak multiple comparisons test was performed post hoc, revealing significant differences between the genotypes. Data are presented as mean ± SEM of four independent experiments. ∗p < 0.05, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001.
(C) Percentage of live (Annexin V−) and dead cells (Annexin V+) in wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs relative to WT. Two-way RM ANOVA was performed revealing an interaction effect (F(2,24) = 21.29, p < 0.0001). In addition, significant differences were observed by apoptotic marker (F(4,36) = 139.4, p < 0.0001) and by genotype (F(2,24) = 4.205, p = 0.0272). The Holm-Sidak multiple comparisons test was performed post hoc, revealing significant differences between the genotypes. Data are presented as mean ± SEM of five independent experiments. ∗∗∗∗p < 0.001.
(D) Cell-cycle distribution of wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs relative examined by DNA content index. Two-way RM ANOVA was performed, revealing an interaction effect (F(4,48) = 14.61, p < 0.0001). In addition, significant differences were observed by cell-cycle phase (F(2,48) = 21.52, p < 0.0001), but not by genotype. The Holm-Sidak multiple comparisons test was performed post hoc, revealing significant differences between the genotypes for S and G2/M cell-cycle phases. Data are presented as mean ± SEM of four independent experiments. ∗∗p < 0.01, ∗∗∗∗p < 0.001.
To assess the proliferation rates of WT, Thap1C54Y, and Thap1ΔExon2 ESCs, we counted the cells at predetermined times after plating. Thap1C54Y ESC proliferation rate was slightly slower than in WT cells, whereas deletion of THAP1 Exon2 severely compromised proliferation (Figure 2B). Viability was not affected in Thap1C54Y ESCs as indicated by annexin V, whereas the number of viable cells was reduced ∼25% in Thap1ΔExon2 ESCs compared with WT (Figure 2C). Propidium iodide (PI) staining followed by DNA flow cytometry revealed a significant reduction in G2/M-phase cell populations and a corresponding increase in the number of cells in S phase (Figure 2D) in the Thap1ΔExon2 ESCs. Again, Thap1C54Y showed no significant differences compared with WT ESCs. Thus, loss of full-length THAP1, but not the C54Y mutation, affects cell viability by increasing the rate of cell death and arresting the cell cycle at the S phase.
Global Expression Profile of Thap1C54Y and Thap1ΔExon2 ESCs
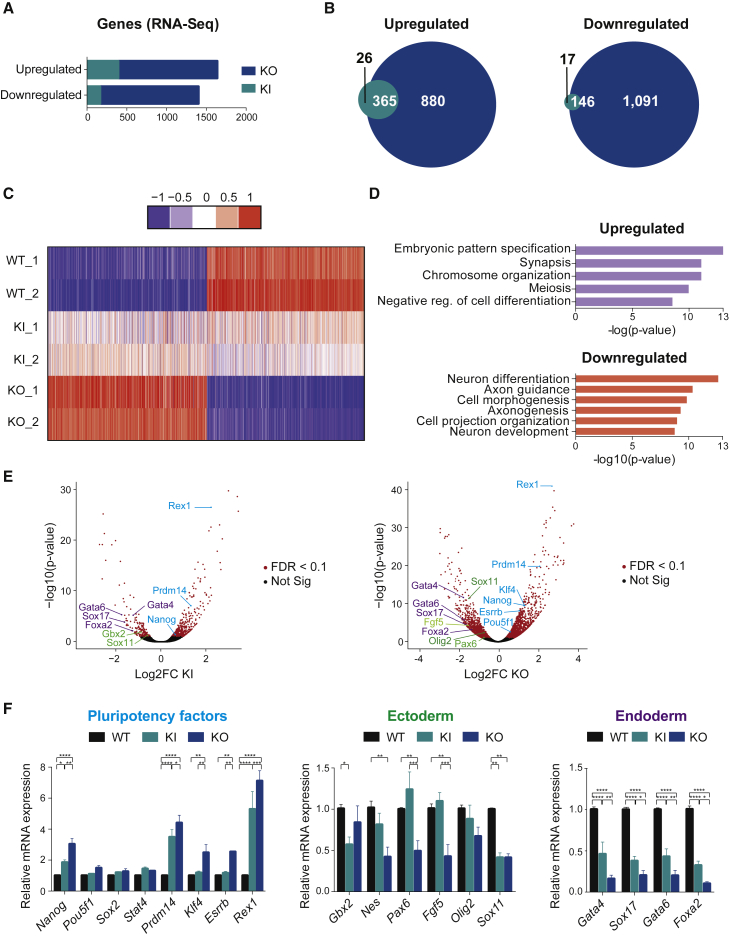
We studied the global transcriptional response in both ESC lines via high-throughput RNA sequencing (RNA-Seq), and identified a larger number of differentially regulated genes in Thap1ΔExon2 than in Thap1C54Y ESCs (Figure 3A and Table S1). Specifically, comparing Thap1C54Y with WT ESCs, there were 391 upregulated and 163 downregulated genes, whereas comparing Thap1ΔExon2 with WT we identified 1,245 upregulated and 1,237 downregulated genes (Figure 3A). Notably, 93% of the upregulated and 89.5% of the downregulated Thap1C54Y genes were common to both genotypes (Figure 3B), but a heatmap illustrated that Thap1C54Y and Thap1ΔExon2 alleles have distinct effects on the transcriptional ESC profile (Figure 3C). These differences were also reflected by gene ontology (GO) analyses of biological processes of uniquely differentially expressed genes (Figure S1), and could not be attributed to metabolic or apoptotic gene expression dysregulation. Interestingly, GO analyses of biological processes of upregulated genes (log2 fold change = +1 of the Thap1ΔExon2 ESC dataset) revealed categories related to embryonic pattern specification, synapsis, chromosome organization, meiosis, and negative regulation of cell differentiation (Figure 3D, upper panel). Analysis of downregulated genes (log2 fold change = −1 of the Thap1ΔExon2 ESC dataset) demonstrated enrichment for processes involved in neuron development, including neuron differentiation, axon guidance, axonogenesis, and cell projection organization (Figure 3D, lower panel). Known pluripotency genes, including Rex1, Prdm14, and Nanog were increased in both Thap1C54Y and Thap1ΔExon2 ESCs, whereas Klf4, Esrrb, and Pou5F1 were increased only in Thap1ΔExon2 ESCs, all compared with WT ESCs (Figure 3E). In contrast, ectodermal markers, e.g., Gbx2 and Sox11, were decreased in Thap1C54Y ESCs, and Pax6, Sox11, Olig2, and Fgf5 were decreased in Thap1ΔExon2 ESCs, all compared with WT. Endoderm specification genes, including Gata4, Gata6, Sox17, and Foxa2, were also significantly decreased in both genotypes compared with WT ESCs (Figure 3E), whereas no differences in mesoderm markers were detected (data not shown). The RNA-Seq results were confirmed by qRT-PCR for selected genes (Figure 3F). Altogether, these results provide initial evidence that pluripotency factors and developmental regulators are differentially, and oppositely, regulated after C54Y mutation of Thap1, and that such changes in the ESC transcriptome are even more apparent after loss of full-length THAP1.
Figure 3.
THAP1 Negatively Regulates the ESC Transcriptome
(A) Bar graph illustrating the numbers of upregulated and downregulated genes in Thap1C54Y/C54Y (KI) and Thap1ΔExon2 (KO) compared with wild-type (WT) ESCs by RNA sequencing (RNA-Seq).
(B) Venn diagram showing the overlap of upregulated (left panel) and downregulated (right panel) genes in Thap1C54Y/C54Y (KI) and Thap1ΔExon2 (KO) ESCs.
(C) Heatmap illustrating the global gene expression performed in duplicates of wild-type 1 and 2 (WT_1 and WT_2), Thap1C54Y/C54Y 1 and 2 (KI_1 and KI_2), and Thap1ΔExon2 1 and 2 (KO_1 and KO_2) ESCs.
(D) Gene ontology (GO) analyses of biological processes of the top (log2 fold change 1) upregulated (upper panel) and downregulated (lower panel) genes of the Thap1ΔExon2 ESCs dataset.
(E) Volcano plots demonstrated differentially expressed genes in Thap1C54Y/C54Y (KI) (left panel) and Thap1ΔExon2 (KO) (right panel) ESCs versus WT ESCs. Significant and not significant hits are shown in red and black, respectively (false discovery rate [FDR] < 0.1). Pluripotency-associated transcripts are shown in blue. Selected ectoderm and mesoderm RNAs are depicted in green and purple, respectively.
(F) qRT-PCR analysis of selected pluripotency factors (left panel), ectoderm (middle panel), and endoderm (right panel) specification markers in WT, Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs. For pluripotency factors, two-way ANOVA was performed, revealing an interaction effect (F(14,170) = 10.92, p < 0.0001). In addition, significant differences were observed by pluripotency gene(s) (F(7,170) = 38.54, p < 0.0001) and by genotype (F(2,170) = 85.22, p < 0.0001). For ectodermal markers, two-way ANOVA was performed, revealing an interaction effect (F(10,107) = 3.178, p = 0.0013). In addition, marginal differences were observed by ectodermal gene(s) (F(5,107) = 2.144, p = 0.0657), and significant differences were observed by genotype (F(2,170) = 22.47, p < 0.0001). For endodermal markers, two-way ANOVA was performed, revealing significant differences by genotype (F(2,81) = 194.2, p < 0.0001); however, no significant differences were observed by endodermal gene(s), and there was no interaction effect. The Holm-Sidak multiple comparisons test was performed post hoc, revealing significant differences between the genotypes for the following genes: Nanog, Prdm14, Klf4, Esrrb, Rex1, Gbx2, Nes, Pax6, Fgf5, Sox11, Gata4, Sox17, and Foxa2. Data are presented as mean ± SEM of four independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001.
THAP1 Genomic Binding Analysis
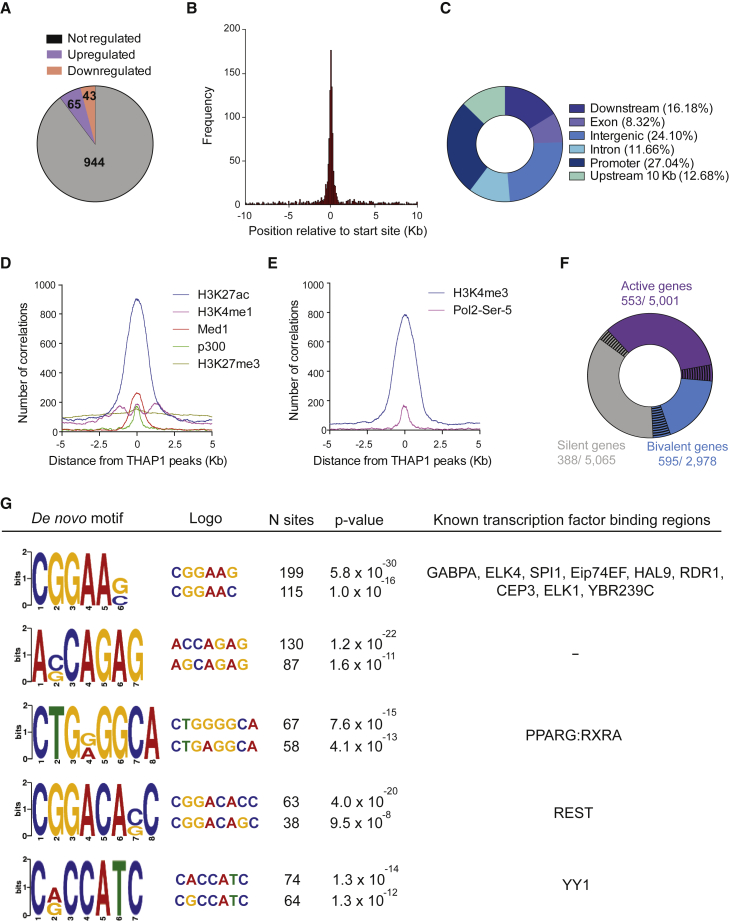
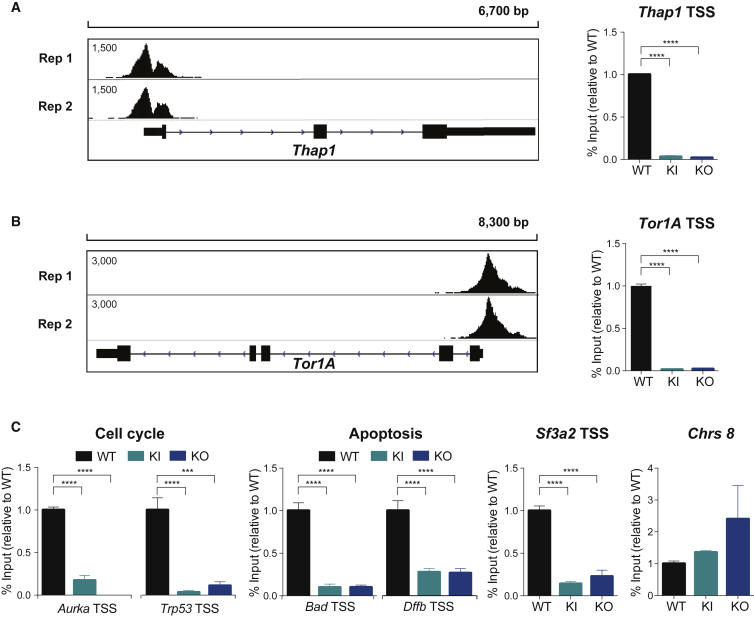
To gain an overview of the global role of THAP1 in ESCs, we performed chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq). We identified 1,731 high-confidence peaks bound by THAP1, corresponding to 1,052 target genes (Table S2), 65 of which were upregulated and 43 were downregulated in Thap1ΔExon2 ESCs (Figure 4A). THAP1 binding was enriched at the transcription start site (TSS), extending over a 2-kb interval (Figure 4B). THAP1 bound only slightly more at promoters (27.04%) than in intergenic regions (24.10%) (Figure 4C). We used public ESC ChIP-Seq datasets for p300, Med1, and monomethylated histone H3 Lys4 (H3K4me1), acetyl histone H3 Lys27 (H3K27ac), and trimethylated histone H3 Lys27 (H3K27me3) (Arnold et al., 2013, Heintzman et al., 2007, Heintzman et al., 2009, Rada-Iglesias et al., 2011, Visel et al., 2009) to determine that THAP1 occupation was detected at activated enhancers but not with the H3K27me3 silent mark (Figure 4D). By clustering the ChIP-Seq enrichment profiles of RNA polymerase II Ser-5 phosphorylated (Pol II Ser-5P) and the histone modification trimethylated histone H3 Lys4 (H3K4me3) with high-confidence THAP1 peaks, we found that THAP1 occupancy occurred at promoter regions of activated genes (Figure 4E) associated with GO categories related to metabolic processes, including nucleobase-containing compound and RNA metabolic, and neurologic system process (Figures 4F and S2A). THAP1 also occupied bivalent or poised genes that are associated with developmental processes, system development, cell communication, and nervous system and ectoderm development, among others (Figures 4F and S2B). Occupancy of THAP1 at silent genes was related to diverse functions, including developmental and immune system process (Figures 4F and S2C). We identified the top DNA motifs enriched at the THAP-domain-binding sequence (THABS) and de novo motifs using the top 1,000 THAP1 peaks and the algorithm MEME (Figure 4G, left panel). Motifs #1 and #3 overlapped with the THABS identified by ENCODE in the human K562 cell line (Kheradpour and Kellis, 2014). Motif #3 contained the core GGCA sequence, essential for recognition by the THAP domain (Clouaire et al., 2005). Secondary motifs overrepresented in THABS overlapped with the binding sequence for RE-silencing transcription factor (REST), which represses the expression of neuronal genes in differentiated non-neuronal cells (Ballas et al., 2005), and Yin Yang 1 (YY1), which plays multiple roles in the development of the central and peripheral nervous systems (He and Casaccia-Bonnefil, 2008; Figure 4G, right panel). Consistent with previous reports, inspection of individual gene tracks showed THAP1 binding at the core promoter of Thap1 and Tor1A (Erogullari et al., 2014, Gavarini et al., 2010, Kaiser et al., 2010), thus validating our approach (Figures 5A and 5B, left panel). ChIP-qPCR using anti-THAP1 antibody confirmed THAP1 occupancy at these sites in WT but not in Thap1C54Y and Thap1ΔExon2 ESCs (Figures 5A and 5B, right panel), and at promoters of the cell-cycle genes Aurora A kinase (Aurka) and tumor protein p53 (Trp53), at the apoptosis genes Bcl-2-associated death promoter (Bad) and DNA fragmentation factor subunit beta (Dffb), and at the splicing factor 3a subunit 2 (Sf3a2). Despite this occupancy, the expression of these genes is not altered in Thap1C54Y and Thap1ΔExon2 ESCs compared with WT (Table S1), suggesting that THAP is present on specific promoters as part of an inactive complex poised to respond to defined developmental stimuli yet to be elucidated. Binding of THAP1 at the non-specific region of chromosome 8 was not detected (Figure 5C).
Figure 4.
THAP1 ChIP-Seq Analysis
(A) Pie chart depicting the number of THAP1 only bound genes, activated and bound, and repressed and bound in Thap1ΔExon2 (KO) ESCs.
(B) Histogram showing the distribution of THAP1-binding peaks relative to the nearest TSS.
(C) Pie chart of the genomic distribution of THAP1-binding peaks including promoters (within 5 kb upstream of TSS), downstream (within 10 kb downstream of the gene), introns, exons, upstream (within 10 kb upstream of the gene), and intergenic regions.
(D) p300, MED1, H3K4me1 (characteristic of predicted enhancer), H3K27ac (active state), and H3K27me3 (repressed state) occupancy around the summit of THAP1 peaks.
(E) Pol II Ser-5P and H3K4me3 (characteristic of promoters) occupancy around the summit of THAP1 peaks.
(F) Pie chart depicting THAP1 binding sites (black lines) at active (purple), bivalent (blue), and silent (gray) genes (defined in Experimental Procedures).
(G) The top 1,000 TF ChIP-Seq peaks were used to identify THAP1 de novo motif (left panel showing the five most significant motifs) and overlapping of THAP1 binding with the indicated transcription factors (right panel). Number (N) of sites and the corresponding p values are indicated.
Figure 5.
ChIP and qRT-PCR Validation of THAP1 Target Genes
(A) ChIP-Seq binding profiles for THAP1 replicates (Rep) 1 and 2 at the Thap1 locus (left panel). ChIP qRT-PCR analysis of THAP1 at Thap1 TSS in WT, Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs (right panel). Data are normalized to percent input and relative to WT. An ANOVA was performed, which revealed a significant difference among the genotypes when examining THAP1 binding (F(2,8) = 1544, p < 0.0001). Holm-Sidak's multiple comparisons test was performed post hoc, revealing significant differences between the genotypes. Data are presented as mean ± SEM of four independent experiments. ∗∗∗∗p < 0.001.
(B) ChIP-Seq binding profiles for THAP1 replicates (Rep) 1 and 2 at the Tor1A locus (left panel). ChIP qRT-PCR analysis of THAP1 at Tor1A TSS in WT, Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs (right panel). Data are normalized to percent input and relative to WT. An ANOVA was performed, which revealed a significant difference among the genotypes when examining THAP1 binding (F(2,0) = 1854, p < 0.0001). The Holm-Sidak multiple comparisons test was performed post hoc, revealing significant differences between the genotypes. Data are presented as mean ± SEM of four independent experiments. ∗∗∗∗p < 0.001.
(C) ChIP-qPCR analysis of THAP1 at cell-cycle genes (Aurka and Trp53), apoptosis (Bad and Dffb), and Sf3a2 TSS in WT, Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs. Chromosome 8 was used as a negative control. Data are normalized to percent input and relative to WT. Significant differences were observed among the genotypes when examining the binding of THAP1 at the cell-cycle genes Aurka (F(2,7) = 118.2, p < 0.0001) and Trp53 (F(2,9) = 37.43, p < 0.0001). Significant differences were observed among the genotypes when examining the THAP1 binding at apoptosis related genes Bad (F(2,9) = 74.64, p < 0.0001) and Dffb (F(2,9) = 39.21, p < 0.0001). Significant differences were observed among the genotypes when examining the THAP1 binding at the RNA splicing related gene, Sf3a2 (F(2,9) = 39.21, p < 0.0001). No differences were observed among the genotypes with ANOVA when examining the THAP1 binding at Chrs8. The Holm-Sidak multiple comparisons test was performed post hoc, revealing significant differences between the genotypes. Data are presented as mean ± SEM of four independent experiments. ∗∗∗p < 0.005, ∗∗∗∗p < 0.001.
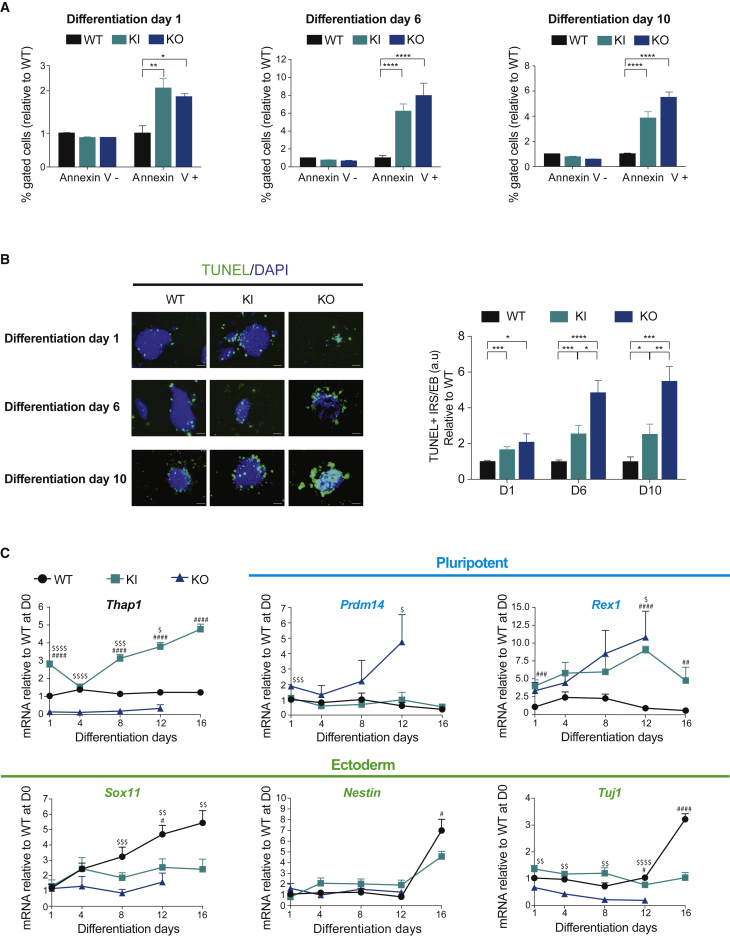
Thap1C54Y and Thap1ΔExon2 EBs Show Increased Cell Death and Fail to Properly Differentiate
To study lineage specification, we assayed EB formation and differentiation (Höpfl et al., 2004). The absence of leukemia inhibitory factor (LIF), replacement of fetal calf serum (FCS) with KnockOut serum replacement, and growth in suspension triggered cell differentiation. WT, Thap1C54Y, and Thap1ΔExon2 ESCs formed EB aggregates (Figure S3A). It was immediately apparent that Thap1ΔExon2 EBs were smaller and all died between days 12 and 16. We assayed mRNA levels of markers that either promote or inhibit cell death (Youle and Strasser, 2008). Compared with WT, the expression of Bax was higher in Thap1C54Y and Thap1ΔExon2 EBs, and Puma also increased in the latter, with a peak at day 8 observed in both genotypes (Figure S3B, upper panel). The expression of the anti-apoptotic markers Xiap and Bcl-2 was also higher in Thap1ΔExon2 EBs compared with Thap1C54Y and WT EBs, with a striking 50- to 75-fold upregulation of Bcl-2 in Thap1ΔExon2 EBs (Figure S3B, lower panel). Cell viability in EBs was actually decreased in both Thap1C54Y and Thap1ΔExon2 ESCs compared with WT ESCs as indicated by annexin V (Figure 6A) and TUNEL staining (Figure 6B), and cleaved caspase-3 staining (Figure S4A), more so in the ΔExon2 EBs. Cell death was not diminished in either genotype when EBs were grown in 40 μM Z-VAD-FMK, a pan-caspase inhibitor (Figure S4B). Thap1ΔExon2 EBs demonstrated the highest percentage of cell death whereas an intermediate level was observed in Thap1C54Y EBs, compared with WT, over the 4-day period (Figure S4B). Failure of Z-VAD-FMK to preserve survival is consistent with literature showing that cellular commitment to apoptosis occurs at the level of BCL-2 family-regulated mitochondrial outer membrane permeabilization (i.e., cytochrome c release), and that subsequent death phenotypes occur whether or not caspases activate (Chipuk and Green, 2005).
Figure 6.
Thap1C54Y/C54Y and Thap1ΔExon2 ESCs Show Abnormal EB Viability and Differentiation
(A) Percentage of live (Annexin V−) and apoptotic cells (Annexin V+) were measured over EB differentiation days 1, 6, and 10 in wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) EBs. On each day, data (mean ± SEM) are presented relative to WT = 1. The two-way ANOVA results for the annexin V assay during EB differentiation days 1, 6, and 10 can be found in Table S3. Bonferroni's multiple comparisons test was performed post hoc, revealing significant differences between the genotypes for each differentiation day. Data are presented as mean ± SEM of three independent experiments. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗∗p < 0.001.
(B) Wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) EBs were examined with TUNEL on differentiation days 1, 6, and 10 (left panel; scale bar, 50 μm). Quantification of TUNEL+ EBs (right panel) was performed by scoring TUNEL-immunopositive cells (immunoreactive species [IRS+]) as a function of total EB area, normalized to WT for each differentiation day. The two-way ANOVA results for TUNEL IRS+/EB quantifications during EB differentiation days 1, 6, and 10 can be found in Table S3. Bonferroni's multiple comparisons test was performed post hoc, revealing significant differences between the genotypes for each differentiation day. Data are presented as mean ± SEM; n = 10 per group for each day, with data pooled from three independent experiments. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001.
(C) qRT-PCR analysis of Thap1, pluripotency markers, and ectodermal markers during EB differentiation of Thap1C54Y/C54Y (KI) and Thap1ΔExon2 (KO) ESCs at the indicated time points. Data are normalized to WT, relative to D1. The two-way ANOVA results for Thap1, pluripotency markers (Prdm14 and Rex1), and ectodermal markers (Sox11, Nestin, and Tuj1) for differentiation days 1, 6, and 10 can be found in Table S3. Bonferroni multiple comparisons test was performed post hoc, revealing significant differences between the genotypes. Data are presented as mean ± SEM of three independent experiments, relative to WT. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. # denotes differences between KI versus WT; $ denotes differences between KO versus WT.
We also used qRT-PCR to assay the expression of differentiation markers in EBs, representing the three germ layers, many of which were already dysregulated in the ESCs. Thap1 mRNA levels were constant in WT EBs up to day 16, the last day examined. In Thap1C54Y ESCs, Thap1 mRNA was higher than in WT, and increased steadily up to day 16. Thap1 mRNA was undetectable in Thap1ΔExon2 EBs (Figure 6C, top left panel). The expression of ESC genes Prdm14 and Rex1 increased progressively between day 1 and day 12 in Thap1ΔExon EBs, and the latter also progressively increased in Thap1C54Y EBs (Figure 6C, top right panel), suggesting that failure to silence pluripotency genes during differentiation may contribute to restriction of developmental potential. Thap1C54Y and Thap1ΔExon2 EBs expressed markers of endodermal lineage (Figure S3C, upper panel) following a pattern similar to that of WT EBs. However, the expression of the mesodermal markers T (i.e., Brachyury) and Flk1 was abnormal in the Thap1C54Y and Thap1ΔExon2 EBs (Figure S3C, lower right panel). Consistent with the GO analysis, the expression of the ectodermal markers Sox11, Nestin, and Tuj1 were decreased in Thap1C54Y and Thap1ΔExon2 EBs, whereas the marker of primitive ectoderm Fgf5 was normally expressed in Thap1C54Y/C54Y and Thap1ΔExon2 EBs compared with WT EBs (Figure S3C, lower panel). Failure of Thap1C54Y to increase expression of Tuj1 suggests a stall at early ectodermal differentiation.
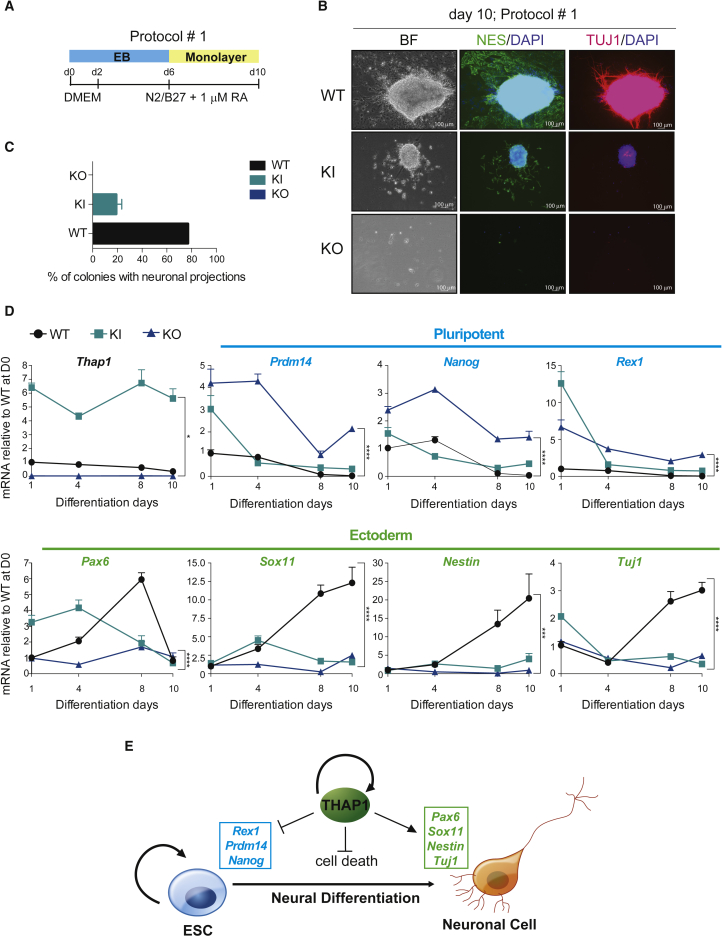
THAP1 Is Essential for Neural Differentiation of Mouse ESCs
To further investigate the role of THAP1 during neural differentiation of mESCs from EBs, we used one protocol based on retinoic acid (RA)-dependent neural differentiation (Figure 7A, protocol #1; Okada et al., 2004), and a second protocol without RA but with the addition of N2 supplemented with epidermal growth factor β and insulin (Figure S5A, protocol #2; Okabe et al., 1996). Survival of mutant cells, particularly ΔExon2, was improved in protocol #2 relative to #1. Using protocol #1, undifferentiated, neural progenitor Nestin-positive cells were detected in Thap1C54Y EBs, but at a lower expression level than in protocol #2, which has a longer culture period. Using protocol #2, Nestin was detected in both WT and Thap1C54Y differentiated cells, but TuJ1-positive neuronal projections, indicative of more mature neurons, were detected almost exclusively in WT ESCs and were abnormally restricted to the internal portion of cell mass in Thap1C54Y ESCs, again consistent with a stall at early ectodermal differentiation (Figure S5B). Thap1ΔExon2 EBs failed to develop Nestin- or TuJ1-immunopositive neuronal projections with either protocol (Figures 7B and S5B). The quantification of neuronal projections in the three genotypes is illustrated in the bar graphs (Figures 7C and S5C). Expression of the ESC genes Prdm14, Nanog, and Rex1 in samples from protocol #1 (Figure 6) was abnormally enhanced in Thap1C54Y and Thap1ΔExon2 EBs, even at the later stages of neuronal differentiation (Figure 7D, upper panel). We also monitored progression of ESC differentiation toward neural fates by analysis of Pax6, Sox11, Nestin, and Tuj1 expression with qRT-PCR. Thap1C54Y and Thap1ΔExon2 failed to activate the expression of these ectodermal markers, suggesting a role for THAP1 in multiple stages of neuronal differentiation (Figure 7D, lower panel). Similar results were obtained by expression analysis in protocol #2 (Figure S5D). Collectively, our results show that THAP1 is required to silence the expression of the pluripotency factors and to activate the expression of the neural factors (Figure 7E).
Figure 7.
Characterization of Neural Differentiation
(A) Schematic representation of cell-culture strategy of neural differentiation using protocol #1.
(B) Immunofluorescence analysis of Nestin (green; middle panel) and TuJ1 (red; right panel) of WT, Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) differentiated cells at day 10 of differentiation. Bright field (left panel) and DAPI stain (blue) are shown. Scale bars, 100 μm.
(C) Bar graph depicting percentage of colonies with neuronal projection.
(D) qRT-PCR analysis of Thap1, pluripotent, and ectoderm marker genes during neural differentiation (protocol #1) of Thap1C54Y/C54Y (KI) and Thap1ΔExon2 (KO) ESCs at the indicated time points. Data are presented relative to WT at day 0. The two-way ANOVA results for Thap1, pluripotent, and ectoderm marker genes during RA-induced differentiation can be found in Table S3. The Holm-Sidak multiple comparisons test was performed post hoc for each gene of interest by day, revealing significant differences between the genotypes. Data are presented as mean ± SEM of at least three independent experiments. ∗p < 0.05, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001.
(E) Schema illustrating that THAP1 is required to silence the expression of the pluripotency factors and to activate the expression of the neural factors, as well as mediate ESC survival.
Discussion
We have identified THAP1 as a major regulator of ESC survival and, to a lesser degree, proliferation and differentiation. As THAP1 mutations result in a neurologic disease, we were particularly interested in the effects of a causative mutation, C54Y, on neuronal differentiation. We had previously posited, based on observations of homozygote mutant embryos, that THAP1 is required in multiple stages and layers during embryogenesis, organogenesis, and maturation of the nervous system, including neurogenesis and neuritogenesis, and that the C54Y variant is frequently unable to substitute for the WT (Ruiz et al., 2015).
In all assays, the homozygous ΔExon2 phenotype was more severe than that of the homozygous C54Y phenotype, confirming that C54Y does not confer a complete loss of function. Both genotypes showed a decreased rate of proliferation, more so in the ΔExon2, consistent with regulation of the cell cycle by THAP1 in HUVECs and lymphoblasts (Cayrol et al., 2007, Vemula et al., 2014). Only the ΔExon2 homozygote showed a decrease in viability in the ESC stage. Thus, a loss of WT THAP1 promotes cell death even though THAP1 is reportedly a pro-apoptotic factor (Roussigne et al., 2003). The lack of a requirement for THAP1 for pluripotency is in marked contrast to RONIN/THAP11, another member of the THAP family to have been characterized in this manner (Dejosez et al., 2008).
We compared this study of the global transcriptome in ESCs harboring a mutation of THAP1 with previous microarray assays of HUVECs with up- or downregulation of THAP1 and of human lymphoblasts harboring a disease-causing intronic variant of THAP1, both of which revealed that the main pathway regulated by THAP1 is the cell cycle. The cell-cycle abnormalities and cell death described herein bear a strong resemblance to these HUVECs and lymphoblasts. Similar to the Thap1ΔExon2 ESCs, the mutant human lymphoblastoid cells had decreased viability and showed a reduced number of cells in the G2 phase (Vemula et al., 2014). In HUVECs, knockdown of THAP1 resulted in an impaired G1/S phase (Cayrol et al., 2007). Despite the presence of cell-cycle abnormalities in the THAP1 null cells, cell-cycle genes were not a highly ranked GO category, but genes pertaining to meiosis and X chromosome inactivation categories were also differentially expressed in Thap1C54Y/C54Y and Thap1ΔExon2 compared with WT ESCs.
We examined the role of THAP1 in ESC differentiation by performing EB formation assays. EB formation and growth were highly compromised in THAP1ΔExon2 cells. Indeed, we showed that cell viability was compromised in both Thap1C54Y and Thap1ΔExon2 EBs, with a greater cell death phenotype after loss of full-length THAP1. The extent of cell death in the ΔExon2 EBs precludes a conclusion regarding an effect on differentiation. Our investigations into the mechanism of cell death implicate the mitochondrial pathway of apoptosis as responsible for cell death commitment and caspase activation. Cell death execution, however, apparently can proceed independently of caspase activation. Delineation of a definitive sequence of events leading to cell death requires further characterization. Although there is increased cell death in the Thap1C54Y EBs, their survival is sufficient to enable some conclusions regarding differentiation. Specifically, mesoderm specification may be delayed, but, more strikingly for a mutation that leads to a neurologic disease, neuronal differentiation appears to stall at an early stage.
The mechanism(s) via which the Thap1 mutations prevent repression of pluripotency markers is not entirely obvious, but there are leads, particularly in the categories of dysregulated genes. An in silico search for THABS motifs in mouse Nanog, Prdm14, Pou5f1, Esrrb, and Klf4 was negative, and ChIP-Seq did not uncover binding sites in these genes. Regulation of the pluripotency network requires cooperation between multiple transcription factors, including the pluripotency factors themselves, signal transduction pathways, and epigenetic modifications. Notably, of the DEGs detected by RNA-Seq, many genes in these categories are altered, e.g., Bmp4, Dnmt3a and Dnmt3b, Satb1, Klf10, Sirt6, and Sox17, with a greater degree of dysregulation in the Thap1ΔExon2 relative to the Thap1C54Y ESCs. Interestingly, Dnmt3a and 3b, Satb1, and Sox17 KO ESCs display phenotypes that are similar in many ways to those of the Thap1 mESCs. Specifically, upon exposure to differentiation factors, they simultaneously fail to downregulate pluripotency genes and upregulate lineage-specific genes, some of which are already decreased prior to differentiation (Bergsland et al., 2011, Chen et al., 2003, Savarese et al., 2009). Thus, the mechanism via which THAP1 regulates these genes requires further examination but may represent a pathway for inverse regulation of pluripotency and differentiation genes.
The low percentage (10%) of overlap between RNA-Seq and ChIP-Seq datasets suggests an alternative role of THAP1 in regulating gene expression other than direct binding at DNA, as exists with other zinc-finger factors (Aguilo et al., 2015). This conclusion may also be supported by the relatively low log2 fold changes observed in the Thap1C54Y ESCs, as much greater changes are seen in Thap1ΔExon2 ESCs, where the full-length transcription factor is eliminated. It is possible that the mutant THAP1 proteins are able to bind to some, but not all, targets of WT THAP1, and in addition, may bind to sequences that are normally not bound by WT THAP1 (Campagne et al., 2012). Also, these mutant proteins may participate in THAP1 protein interactions. For example, THAP1 interacts with HCF-1 via a motif that does not include amino acid 54 (Mazars et al., 2010). Therefore, the C54Y form may still be able to interact with HCF-1, Par-4, and partners as yet to be identified, thereby recruiting such transcription factors to the target DNA. This hypothesis would be consistent with the fact that there are individuals with dystonia with homozygote THAP1 mutations. In the case where THAP1 binding is decreased or eliminated, there may be increased binding of transcription factors with an affinity for similar motifs. As noted, these include YY1 and REST, thereby altering levels of their direct and indirect targets, which include many of the RNA-Seq DEGs. In fact, there is a very strong super-shift with anti-YY1 in electrophoretic mobility shift assays using the Tor1a THABS and embryonic brain nuclear extract (data not shown).
In summary, we have identified THAP1 as an essential regulator of mESC potential, including viability and likely neuronal differentiation, the latter particularly for the naturally occurring C54Y mutation. These activities are likely due in part to direct regulation of gene expression but to a large extent to indirect regulation, the mechanisms of which are important to identify. Notably, dysregulated pathways in ESCs with mutations of THAP1 overlap with those seen in mouse models of DYT6, and the ESC system may potentially be exploited to discover how following differentiation, mutations of the ubiquitously expressed THAP1 protein can lead to a strictly neurologic disease.
Experimental Procedures
Preparation of nuclear extracts and qRT-PCR gene expression analysis were performed as previously described (Ruiz et al., 2015).
Further experimental details and lists of antibodies, public datasets, and primer sets utilized in this study may be found in Supplemental Experimental Procedures and other Supplemental Information.
Cell Culture and EB Formation
Wild-type (WT), Thap1C54Y, and Thap1ΔExon2 ESCs were maintained on 0.1% gelatin-coated tissue culture plates on irradiated mouse embryonic fibroblasts (MEFs) in DMEM containing 15% FCS, 0.1 mM 2-mercaptoethanol, L-glutamine, non-essential amino acids, and LIF (1,000 units/mL) and penicillin-streptomycin (Thermo Fisher). Cells were passaged once onto gelatin-coated dishes and then aggregated to form EBs in 10-cm Petri dishes at 5 × 106/plate in the absence of LIF. Cells were harvested for extraction of total RNA at the indicated time points. All cell cultures were maintained at 37°C with 5% CO2.
Differentiation Assays
Induction of neural differentiation was performed based on generation of EBs, using two different approaches (Okabe et al., 1996, Okada et al., 2004), with and without addition of RA as indicated in Supplemental Experimental Procedures.
Cellular Proliferation, Apoptosis Assay, and Cell-Cycle Analyses
Cellular proliferation, apoptosis assay, and cell-cycle analysis were performed using a Muse Cell Analyzer (Millipore) following the manufacturer's instructions.
Alkaline Phosphatase Activity
AP activity was measured using the Stemgent Alkaline Phosphatase Staining kit (Stemgent) following the manufacturer's recommendations.
Chromatin Immunoprecipitation with High-Throughput Sequencing
THAP1 ChIP-Seq was performed as described in Supplemental Experimental Procedures.
RNA-Seq and Analysis
RNA-Seq library preparation was performed at the Weill-Cornell Medical College Genomic Core Facility using the TrueSeq RNA sample preparation kit (Illumina RS-122-2001) and sequenced by the Illumina HiSeq 2,500 platform as 100-bp pair-ended reads.
Gene Ontology Analysis
For functional profiling of THAP1 binding regions identified by ChIP-Seq, GO analysis was performed using Panther software (http://pantherdb.org) (Mi et al., 2016). GO analysis from RNA-Seq datasets was performed using the web tool The Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) (Huang et al., 2009a, Huang et al., 2009b).
Statistical Analysis
All values are expressed as mean ± SD. Statistical analysis was performed by two-way ANOVA followed by Bonferroni's multiple testing corrections using GraphPad Prism 5 Software. A probability value of p < 0.05 was considered statistically significant.
Author Contributions
Conceptualization, F.A., Z.Z., M.J.W., L.J.O., P.G.-A., T.P.Z., and M.E.E.; Methodology, K.K.; Formal analysis, F.A., Z.Z., R.S., C.W, and W.Z.; Investigation, F.A., Z.Z., K.N., R.W., M.H., and Y.S.; Writing – Original Draft, F.A. and M.E.E.; Writing – Review & Editing, Z.Z., P.G.-A., L.J.O., M.J.W., T.P.Z., and M.E.E.; Funding Acquisition, M.E.E.; Supervision and Project Administration, F.A. and M.E.E.
Acknowledgments
We thank Dr. A. Alonso of the Weill-Cornell College of Medicine's Epigenomic Sequencing Core for expert advice for sequencing and library preparation. This work was funded by NIH R01NS081282 (to M.E.E. and L.J.O.) New York State Stem Cell Science (NYSTEM C029553 to M.E.E.), and the Huffington Foundation (T.P.Z.).
Published: June 1, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.04.032.
Contributor Information
Francesca Aguilo, Email: francesca.aguilo@umu.se.
Michelle E. Ehrlich, Email: michelle.ehrlich@mssm.edu.
Accession Numbers
All next-generation sequencing data are deposited in NCBI GEO database under accession numbers GEO: GSE86947 and GSE86911.
Supplemental Information
References
- Aguilo F., Zhang F., Sancho A., Fidalgo M., Di Cecilia S., Vashisht A., Lee D.-F., Chen C.-H., Rengasamy M., Andino B. Coordination of m(6)A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell. 2015;17:689–704. doi: 10.1016/j.stem.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold C.D., Gerlach D., Stelzer C., Boryń Ł.M., Rath M., Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bergsland M., Ramsköld D., Zaouter C., Klum S., Sandberg R., Muhr J. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25:2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagne S., Muller I., Milon A., Gervais V. Towards the classification of DYT6 dystonia mutants in the DNA-binding domain of THAP1. Nucleic Acids Res. 2012;40:9927–9940. doi: 10.1093/nar/gks703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrol C., Lacroix C., Mathe C., Ecochard V., Ceribelli M., Loreau E., Lazar V., Dessen P., Mantovani R., Aguilar L. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood. 2007;109:584–594. doi: 10.1182/blood-2006-03-012013. [DOI] [PubMed] [Google Scholar]
- Chen Y., He Z.X., Liu A., Wang K., Mao W.W., Chu J.X., Lu Y., Fang Z.F., Shi Y.T., Yang Q.Z. Embryonic stem cells generated by nuclear transfer of human somatic nuclei into rabbit oocytes. Cell Res. 2003;13:251–263. doi: 10.1038/sj.cr.7290170. [DOI] [PubMed] [Google Scholar]
- Chipuk J.E., Green D.R. Do inducers of apoptosis trigger caspase-independent cell death? Nat. Rev. Mol. Cell Biol. 2005;6:268–275. doi: 10.1038/nrm1573. [DOI] [PubMed] [Google Scholar]
- Clouaire T., Roussigne M., Ecochard V., Mathe C., Amalric F., Girard J.-P. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc. Natl. Acad. Sci. USA. 2005;102:6907–6912. doi: 10.1073/pnas.0406882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M., Krumenacker J.S., Zitur L.J., Passeri M., Chu L.-F., Songyang Z., Thomson J.A., Zwaka T.P. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erogullari A., Hollstein R., Seibler P., Braunholz D., Koschmidder E., Depping R., Eckhold J., Lohnau T., Gillessen-Kaesbach G., Grünewald A. THAP1, the gene mutated in DYT6 dystonia, autoregulates its own expression. Biochim. Biophys. Acta. 2014;1839:1196–1204. doi: 10.1016/j.bbagrm.2014.07.019. [DOI] [PubMed] [Google Scholar]
- Fuchs T., Gavarini S., Saunders-Pullman R., Raymond D., Ehrlich M.E., Bressman S.B., Ozelius L.J. Mutations in the THAP1 gene are responsible for DYT6 primary torsion dystonia. Nat. Genet. 2009;41:286–288. doi: 10.1038/ng.304. [DOI] [PubMed] [Google Scholar]
- Gavarini S., Cayrol C., Fuchs T., Lyons N., Ehrlich M.E., Girard J.-P., Ozelius L.J. Direct interaction between causative genes of DYT1 and DYT6 primary dystonia. Ann. Neurol. 2010;68:549–553. doi: 10.1002/ana.22138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervais V., Campagne S., Durand J., Muller I., Milon A. NMR studies of a new family of DNA binding proteins: the THAP proteins. J. Biomol. NMR. 2013;56:3–15. doi: 10.1007/s10858-012-9699-1. [DOI] [PubMed] [Google Scholar]
- He Y., Casaccia-Bonnefil P. The Yin and Yang of YY1 in the nervous system. J. Neurochem. 2008;106:1493–1502. doi: 10.1111/j.1471-4159.2008.05486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höpfl G., Gassmann M., Desbaillets I. Differentiating embryonic stem cells into embryoid bodies. Methods Mol. Biol. 2004;254:79–98. doi: 10.1385/1-59259-741-6:079. [DOI] [PubMed] [Google Scholar]
- Houlden H., Schneider S.A., Paudel R., Melchers A., Schwingenschuh P., Edwards M., Hardy J., Bhatia K.P. THAP1 mutations (DYT6) are an additional cause of early-onset dystonia. Neurology. 2010;74:846–850. doi: 10.1212/WNL.0b013e3181d5276d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser F.J., Osmanoric A., Rakovic A., Erogullari A., Uflacker N., Braunholz D., Lohnau T., Orolicki S., Albrecht M., Gillessen-Kaesbach G. The dystonia gene DYT1 is repressed by the transcription factor THAP1 (DYT6) Ann. Neurol. 2010;68:554–559. doi: 10.1002/ana.22157. [DOI] [PubMed] [Google Scholar]
- Kheradpour P., Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S., Rio D.C. P transposable elements in Drosophila and other eukaryotic organisms. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0004-2014. MDNA3-0004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazars R., Gonzalez-de-Peredo A., Cayrol C., Lavigne A.-C., Vogel J.L., Ortega N., Lacroix C., Gautier V., Huet G., Ray A. The THAP-zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase: a link between DYT6 and DYT3 dystonias. J. Biol. Chem. 2010;285:13364–13371. doi: 10.1074/jbc.M109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Poudel S., Muruganujan A., Casagrande J.T., Thomas P.D. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S., Forsberg-Nilsson K., Spiro A.C., Segal M., McKay R.D. Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech. Dev. 1996;59:89–102. doi: 10.1016/0925-4773(96)00572-2. [DOI] [PubMed] [Google Scholar]
- Okada Y., Shimazaki T., Sobue G., Okano H. Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev. Biol. 2004;275:124–142. doi: 10.1016/j.ydbio.2004.07.038. [DOI] [PubMed] [Google Scholar]
- Ortiz-Virumbrales M., Ruiz M., Hone E., Dolios G., Wang R., Morant A., Kottwitz J., Ozelius L.J., Gandy S., Ehrlich M.E. Dystonia type 6 gene product Thap1: identification of a 50 kDa DNA-binding species in neuronal nuclear fractions. Acta Neuropathol. Commun. 2014;2:139. doi: 10.1186/s40478-014-0139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist L., Glover C.H., Hsu L., Lu M., Bossen B., Piret J.M., Humphries R.K., Helgason C.D. Correlation of murine embryonic stem cell gene expression profiles with functional measures of pluripotency. Stem Cells. 2005;23:663–680. doi: 10.1634/stemcells.2004-0157. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussigne M., Cayrol C., Clouaire T., Amalric F., Girard J.-P. THAP1 is a nuclear proapoptotic factor that links prostate-apoptosis-response-4 (Par-4) to PML nuclear bodies. Oncogene. 2003;22:2432–2442. doi: 10.1038/sj.onc.1206271. [DOI] [PubMed] [Google Scholar]
- Ruiz M., Perez-Garcia G., Ortiz-Virumbrales M., Méneret A., Morant A., Kottwitz J., Fuchs T., Bonet J., Gonzalez-Alegre P., Hof P.R. Abnormalities of motor function, transcription and cerebellar structure in mouse models of THAP1 dystonia. Hum. Mol. Genet. 2015;24:7159–7170. doi: 10.1093/hmg/ddv384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese F., Dávila A., Nechanitzky R., De La Rosa-Velazquez I., Pereira C.F., Engelke R., Takahashi K., Jenuwein T., Kohwi-Shigematsu T., Fisher A.G. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 2009;23:2625–2638. doi: 10.1101/gad.1815709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S.A., Ramirez A., Shafiee K., Kaiser F.J., Erogullari A., Brüggemann N., Winkler S., Bahman I., Osmanovic A., Shafa M.A. Homozygous THAP1 mutations as cause of early-onset generalized dystonia. Mov. Disord. 2011;26:858–861. doi: 10.1002/mds.23561. [DOI] [PubMed] [Google Scholar]
- Sengel C., Gavarini S., Sharma N., Ozelius L.J., Bragg D.C. Dimerization of the DYT6 dystonia protein, THAP1, requires residues within the coiled-coil domain. J. Neurochem. 2011;118:1087–1100. doi: 10.1111/j.1471-4159.2011.07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemula S.R., Xiao J., Zhao Y., Bastian R.W., Perlmutter J.S., Racette B.A., Paniello R.C., Wszolek Z.K., Uitti R.J., Van Gerpen J.A. A rare sequence variant in intron 1 of THAP1 is associated with primary dystonia. Mol. Genet. Genomic Med. 2014;2:261–272. doi: 10.1002/mgg3.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Blow M.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiromerisiou G., Houlden H., Scarmeas N., Stamelou M., Kara E., Hardy J., Lees A.J., Korlipara P., Limousin P., Paudel R. THAP1 mutations and dystonia phenotypes: genotype phenotype correlations. Mov. Disord. 2012;27:1290–1294. doi: 10.1002/mds.25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R.J., Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.