Summary
Primordial germ cells (PGCs) are the earliest embryonic progenitors in the germline. Correct formation of PGCs is critical to reproductive health as an adult. Recent work has shown that primate PGCs can be differentiated from pluripotent stem cells; however, a bioassay that supports their identity as transplantable germ cells has not been reported. Here, we adopted a xenotransplantation assay by transplanting single-cell suspensions of human and nonhuman primate embryonic Macaca mulatta (rhesus macaque) testes containing PGCs into the seminiferous tubules of adult busulfan-treated nude mice. We discovered that both human and nonhuman primate embryonic testis are xenotransplantable, generating colonies while not generating tumors. Taken together, this work provides two critical references (molecular and functional) for defining transplantable primate PGCs. These results provide a blueprint for differentiating pluripotent stem cells to transplantable PGC-like cells in a species that is amenable to transplantation and fertility studies.
Keywords: primordial-germ-cell, PGC, transplant, xenotransplant
Graphical Abstract
Highlights
-
•
Rhesus PGCs in Carnegie stage 23 correspond to human PGCs at 8–10 weeks
-
•
Carnegie stage 23 PGCs co-express cKIT, OCT4, TFAP2C, BLIMP1, SOX17, and VASA
-
•
Carnegie stage 23 PGCs generate colonies following xenotransplantation
-
•
Carnegie stage 23 PGCs do not yield tumors following xenotransplantation
In this article, Clark and colleagues examined the molecular identity and xenotransplantability of primate PGCs in the seminiferous tubules of the mouse testis. This work revealed that following transplant, human and nonhuman primate PGCs generated colonies while not generating tumors. This work establishes a new in vivo test for confirming putative PGC-like cell differentiation from pluripotent stem cells.
Introduction
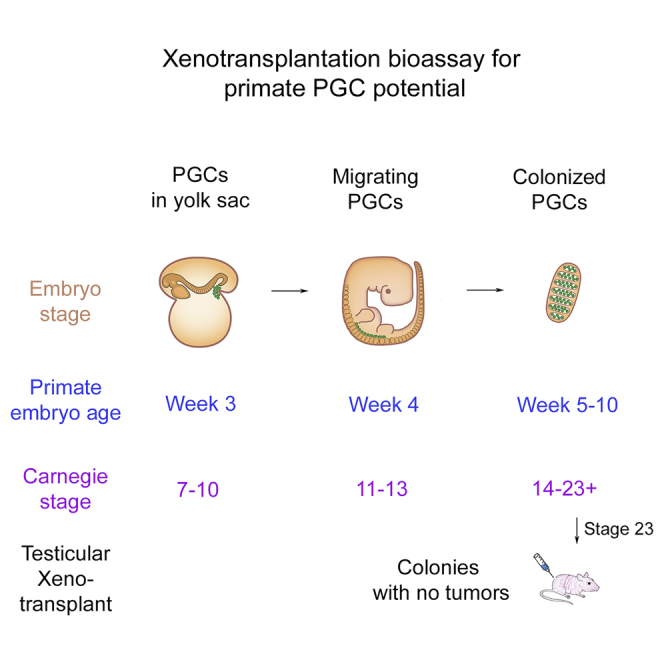
Primordial germ cells (PGCs) are the pioneering cells of the germline. Correct formation of PGCs is necessary for the differentiation of high-quality haploid gametes and ultimately reproductive success as adults. In the mouse, PGC precursors first develop from the epiblast at the end of gastrulation at embryonic day 6.25 (E6.25) (Kurimoto et al., 2008). Definitive PGCs are identified 24 hr later at E7.25 in an extra-embryonic structure called an allantois. At this stage PGCs express transcription factors required for pluripotency as well as germline development (Kurimoto et al., 2008). From E7.5 to E8.5, the PGCs leave their extra-embryonic location, enter the embryo, and migrate toward the genital ridges. The PGCs approach and colonize the genital ridges beginning at E10.5. By E11.5, the PGCs have finished colonizing the ridge, and the gonad is now referred to as an indifferent gonad. During the migration and colonization stages, the nascent PGCs maintain a latent pluripotency program (Hargan-Calvopina et al., 2016, Jameson et al., 2012). At E12.5, male gonads can be distinguished from female gonads by the formation of immature testis cords (Combes et al., 2009). Between E12.5 and E14.5 the male PGCs enter G0 arrest, downregulating most of the transcribed pluripotency-associated genes to become pro-spermatogonia (Western et al., 2008). During the pro-spermatogonia stage, male germ cells undergo epigenetic remodeling in preparation for differentiation into spermatogonia after birth. Given that PGCs are the founding germline cells in human reproduction, and abnormal PGC formation can lead germline loss, understanding the basic cell and molecular biology of PGCs, and creating bioassays that discriminate PGCs functionally are critical areas of investigation.
Capitalizing on their knowledge of mouse PGC development (Ohinata et al., 2009, Saitou et al., 2002), Hayashi and colleagues devised methods to differentiate mouse embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) into mouse PGC-like cells (PGCLCs). Remarkably, the resulting PGCLCs could be transplanted into ovaries or testes, giving rise to fertilization competent eggs, sperm, and viable offspring (Hayashi et al., 2011, Hayashi et al., 2012). The differentiation of human PSCs (hPSCs) into PGCLCs has emerged as a major new model for uncovering the cell and molecular events in human PGC specification (Irie et al., 2015, Sasaki et al., 2015, Sugawa et al., 2015). In the long term, this approach may have implications for treating infertility. However, unlike the mouse model, an approach to prove that human PGCLCs are functional following transplantation is not currently forthcoming. Methods for PGCLC differentiation in humans are based upon similar signaling principles to the mouse, namely the use of bone morphogenetic protein 4 (BMP4) to induce PGCLC fate. However, the transcription factor network downstream of BMP4 may be different in humans (Irie et al., 2015, Sasaki et al., 2015, Sugawa et al., 2015). This suggests either divergence of the transcriptional factor network that drives PGC formation, or an artifact of in vitro differentiation. To discriminate between the two possibilities, it is necessary to investigate the molecular and functional characteristics of bona fide PGCs in higher primates so that appropriate standards can be established for the generation of PGCLCs in primate models.
Human PGCs are specified in the embryo between the second and third week of life after fertilization. Similar to mice, human PGCs are first localized outside of the embryo, then they migrate into the embryo during week 3, ultimately entering the genital ridges and indifferent gonads between the fourth and fifth week of life (Chiquoine, 1954, Witschi, 1948). Gonadal sex determination occurs between week 5 and 6, and male PGCs advance in differentiation toward pro-spermatogonia after week 10 post-fertilization (Gkountela et al., 2013, Guo et al., 2015). During the PGC stage of embryo development, both mouse and human PGCs express the tyrosine kinase receptor cKIT on their surface, with repression of cKIT occurring as PGCs differentiate into pro-spermatogonia (Gkountela et al., 2013, Høyer et al., 2005). In adults, cKIT is again expressed on a subset of differentiating spermatogonia just prior to entering meiosis (Unni et al., 2009). Recent work in the cynomologous (cy) macaque (Macaca fascicularis) suggests that cyPGCs are specified prior to primitive streak formation, at 11 days post-fertilization. At this time, the cyPGCs express cKIT as well as proteins encoding the pluripotency and PGC transcription factors OCT4, SOX17, TFAP2C, and PRDM1 (Sasaki et al., 2016). These transcription factors remain expressed in PGCs until after embryonic day 50. From embryonic day 50 to 70, expression of the pro-spermatogonial marker PLZF (ZBTB16) is initiated in cyPGCs, while genes associated with pluripotency including OCT4 and NANOG are repressed (Sasaki et al., 2016).
Transplanting mouse germ cells into the seminiferous tubules of adult mice was used to determine that spermatogenic potential is initiated as PGCs become pro-spermatogonia between E12.5 and E14.5 (Ohta et al., 2003). In contrast, using neonates as recipients, epiblast cells at E5.5, and embryonic tissues containing PGCs at E8.5, were shown to have both spermatogenic and teratoma-forming potential (Chuma et al., 2005), most likely on account of the latent pluripotency program of nascent PGCs (Matsui et al., 1992, Resnick et al., 1992). Curiously, in the neonatal recipients transplanted with E10.5 tissues, spermatogenesis occurred without teratoma formation. This suggests that neither differentiation into pro-spermatogonia, or repression of the latent pluripotency program per se are required for spermatogenic potential following PGC transplantation. In primates, the degree of latent pluripotency in PGCs is unclear given that neither cyPGCs, nor human PGCs, express SOX2 (Perrett et al., 2008, Sasaki et al., 2016). Testing this hypothesis by transplanting human PGCs into human testicles is not conceivable.
In the monkey model, transplantable testicular stem cells can be identified and quantified using primate-to-nude mouse xenotransplantation, a method that was first described by Nagano and colleagues using monkey and human donor cells over 15 years ago (Nagano et al., 2001, Nagano et al., 2002). Primate-to-nude mouse xenotransplantation is a quantitative bioassay that demonstrates the functional capacity of primate cells to engraft the basement membrane of mouse seminiferous tubules, proliferate to produce characteristic chains and networks of spermatogonia, and persist long term. Primate cells do not produce complete spermatogenesis in mouse tubules, probably due to species differences, but recapitulate many of the unique biological functions of spermatogonial stem cells (SSCs) that are not recapitulated by any other cell type. Based on these criteria, xenotransplantation to mouse seminiferous tubules has emerged as a routine bioassay for nonhuman primate and human SSCs (Dovey et al., 2013, Hermann et al., 2007, Hermann et al., 2009, Izadyar et al., 2011, Maki et al., 2009, Sadri-Ardekani et al., 2009, Sadri-Ardekani et al., 2011, Wu et al., 2009, Zohni et al., 2012).
Xenotransplantation was recently extended to human fetal testis at 22 weeks of gestation (Durruthy-Durruthy et al., 2014), an age where fetal testes are enriched in pro-spermatogonia. In that study, 22 week fetal testicular cells produced colonies of primate cells in the mouse seminiferous tubules that were similar in morphology to those produced by adult human spermatogonia. At the other extreme, transplanting undifferentiated human iPSCs (hiPSCs) or human ESCs (hESCs) directly into the seminiferous tubules of busulfan-treated nude mice resulted in putative germ cell colonies accompanied by proliferating cell masses that correspond to embryonal carcinoma and yolk sac-like tumors (Durruthy-Durruthy et al., 2014, Ramathal et al., 2014), and occasionally teratomas (Durruthy-Durruthy et al., 2014). It is unclear whether xenotransplanting embryonic testes containing PGCs will yield colonies, tumors, or both.
Unlike humans, the nonhuman primate is amenable to autologous and allogeneic transplantation to test the complete spermatogenic potential of donor stem cells (Hermann et al., 2012). Autologous/allogeneic transplantation is not feasible as a routine bioassay due to cost and biological variability among individual outbred animals; xenotransplantation is the preferred method for initial studies. Therefore, in the current study, we used hormone-guided time-mated breeding to obtain accurately staged rhesus macaque embryos in Carnegie stage 23, the end of the PGC period. We chose Carnegie stage 23 because this time point represents E13.5 of mouse embryo development, a known transplantable stage in the mouse.
Results
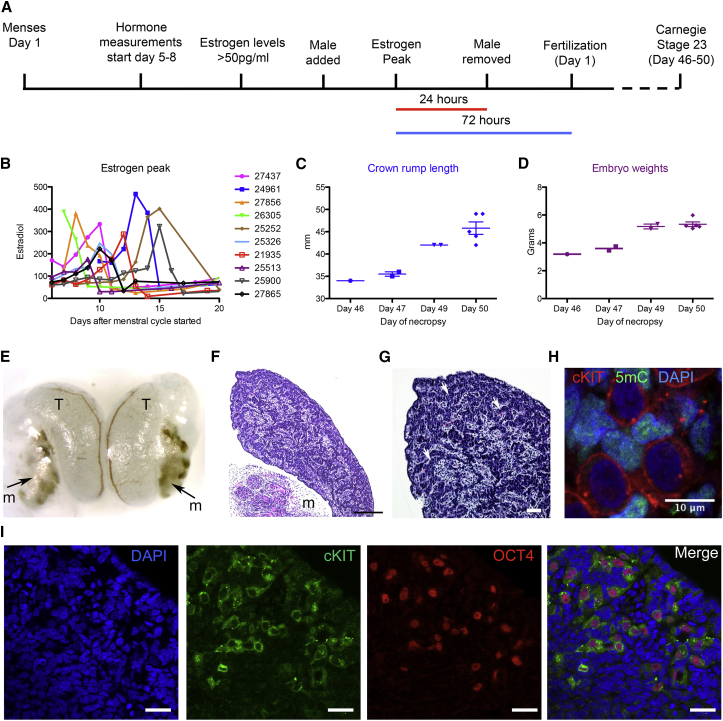
To study germline development we used the time-mated breeding program at the Oregon National Primate Research Center. A total of n = 10 rhesus macaque adult females of reproductive age were used for this project. To time embryonic development, serum estradiol and progesterone were measured daily in naturally cycling females starting between 5 and 8 days after the onset of menses (Figure 1A). Once estradiol levels had risen to >50 pg/mL, indicating the selection and development of the single preovulatory follicle, an adult fertile male was paired with the female until the estrogen peak was detected. The male was removed 24 hr after the estrogen peak. The length of time males and females were paired together ranged from 3 to 7 days total. Based on previous studies, fertilization (day 1 of embryo development) was estimated to occur 72 hr after the estrogen peak (Weick et al., 1973). Pregnancy was confirmed by ultrasound around 3–4 weeks later.
Figure 1.
Time-Mated Breeding to Acquire Rhesus Macaque Embryos at Carnegie Stage 23
(A) Strategy for rhesus macaque time-mated breeding using estrogen to monitor ovulation. Day 1 of embryo development (fertilization) was estimated to occur 72 hr after the estrogen peak.
(B) Estradiol measurements (pg/mL) in the serum of females (animal identification number is shown, n = 10 animals).
(C) Embryo crown rump length (mm) of Carnegie stage 23 embryos (n = 10 embryos).
(D) Embryo weight (g) of Carnegie stage 23 embryos (n = 10 embryos).
(E) A pair of embryonic testicles (T) isolated from a day 47 embryo, and associated mesonephros (m), indicated by the arrow.
(F) H&E stain of a testis at embryonic day 49 and associated mesonephros (m) with glomeruli. Scale bar, 200 μm.
(G) H&E stain of testis cords at embryonic day 49 (arrow points to cords). Scale bar, 50 μm.
(H) Immunofluorescence of embryonic testis at day 49 showing that cKIT-positive PGCs are depleted in 5mC (n = 1 embryonic testis, n = 2 technical duplicates). Scale bar, 10 μm.
(I) Immunofluorescence of embryonic testis at day 49 showing cKIT/OCT4 double-positive PGCs (n = 1 embryonic testis, n = 2 technical duplicate). Scale bar, 20 μm.
Estrogen peaks in individual females varied widely in naturally cycling females ranging from before day 10, up to 15 days after the first day of menses (Figure 1B). This confirms that measuring the estrogen peak, and not date of last menstrual cycle, is a more accurate measure for fertilization. Embryos were collected by C-section at embryonic days 46–50 (days 48–52 post-estrogen peak), corresponding to Carnegie stage 23 of embryo development. Crown rump length (Figure 1C), and embryo weights (Figure 1D) were performed within minutes of C-section. Crown rump length is a classic indicator of embryo age, and combined with embryo weights we show that, based on hormone-guided embryonic staging, the older embryos were larger (Figure 1C) and heavier (Figure 1D) than younger embryos. All pregnancies were singletons, and a total of n = 9 males and n = 1 female embryos were used for this study.
At the time of necropsy, embryos were photographed, and Carnegie staging of external characteristics was performed according to previous comparisons of rhesus macaque embryonic development (Hendrickx and Sawyer, 1975). All males had distinct external genitalia, and upon necropsy the gonads of both male and female embryos were located in the abdominal cavity. Male and female gonads were closely associated with a primitive embryonic kidney called the mesonephros (Figures 1E and 1F).
Histological sectioning of the male gonad at embryonic day 49 revealed cord-like structures in the cortex (Figures 1F and 1G), and all PGCs at this age were positive for the surface receptor cKIT with low genomic levels of 5mC relative to somatic cells of the gonad (Figure 1H). We also determined that the majority of cKIT-positive PGCs were also positive for the pluripotent transcription factor OCT4 (Figure 1I), with only rare examples of cKIT-positive, OCT4-negative PGCs (data not shown). Taken together, hormone-guided time-mated breeding leads to accurate staging of embryo development in the rhesus macaque, where small differences in crown rump length and embryo weights positively correlate with the predicted day of embryo development. Similar to humans (Gkountela et al., 2013), the embryonic gonads at this stage are closely associated with the mesonephros.
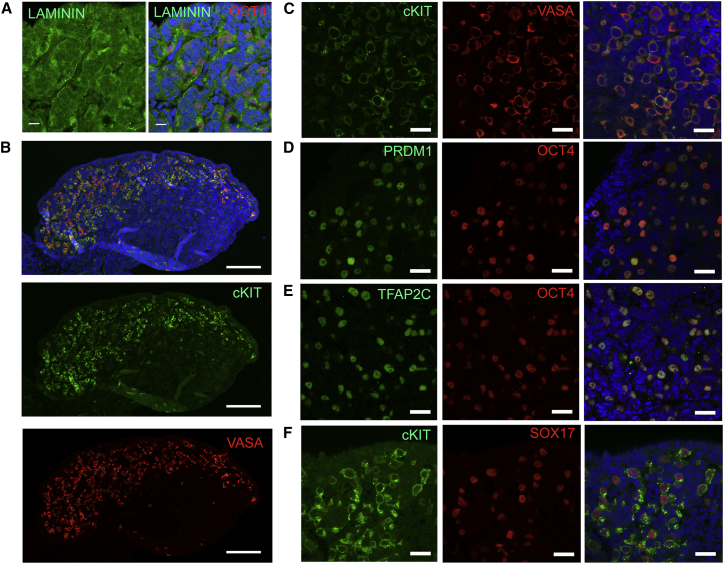
To better characterize the Carnegie stage 23 primate PGCs, we performed immunofluorescence for the basement membrane protein Laminin and the PGC marker OCT4 (Figure 2A). All OCT4-positive PGCs were located within the Laminin-positive cords in the testicular cortex at this age. We also discovered that the cKIT-positive PGCs were all positive for VASA (Figure 2B and higher power Figure 2C). Furthermore, almost every OCT4-positive PGC expressed PRDM1 (BLIMP1) and TFAP2C, and every cKIT-positive PGC expressed SOX17 (Figures 2D–2F). Taken together, the pluripotency and PGC transcription factor expression of Carnegie stage 23 rhesus PGCs is consistent with male PGCs that have not yet progressed to the pro-spermatogonia stage.
Figure 2.
Rhesus PGCs Express PGC Transcription Factors and Are Localized in Testis Cords
(A) Immunofluorescence of embryonic testes at day 49 showing OCT4-positive PGCs within LAMININ-positive cords. Scale bar, 10 μm.
(B) Immunofluorescence for cKIT and VASA showing all cKIT-positive cells are VASA positive and localized in the cortex of the embryonic testes. Scale bar, 100 μm.
(C) Immunofluorescence for cKIT and VASA showing co-localization.
(D) Immunofluorescence for PRDM1 and OCT4 showing co-localization.
(E) Immunofluorescence for TFAP2C and OCT4 showing co-localization.
(F) Immunofluorescence for cKIT and SOX17 showing co-localization.
All experiments were performed on n = 1 embryonic testis at day 49 in technical duplicate. Scale bars, 20 μm unless otherwise stated.
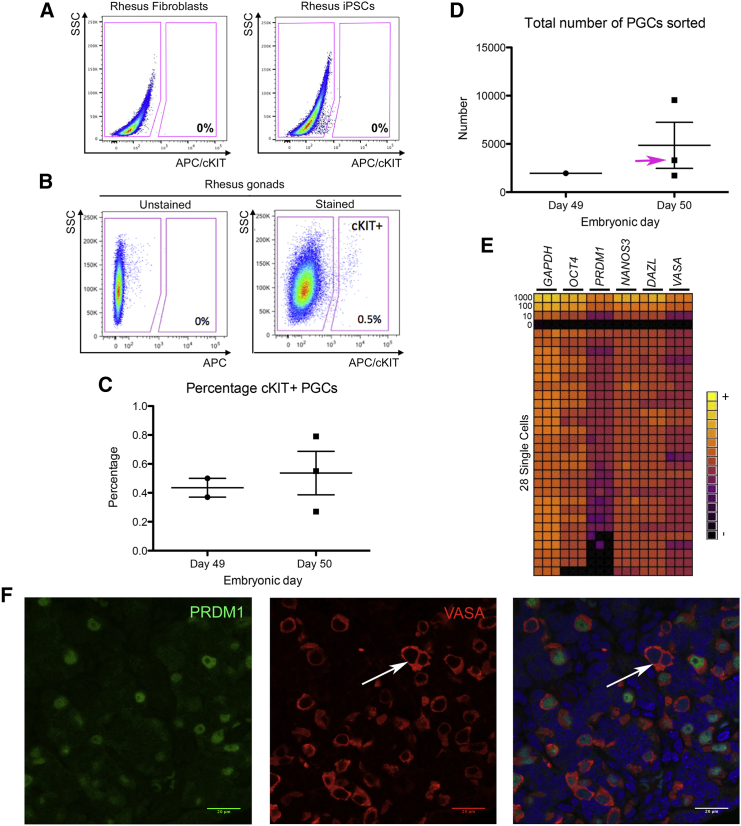
To create a comprehensive resource of PGC identity at Carnegie stage 23, we chose to perform RNA sequencing (RNA-seq) of cKIT-positive PGCs sorted by fluorescence-activated cell sorting (FACS). As a negative control we used rhesus embryonic fibroblasts, which do not express cKIT, and riPSCs which express cKIT at background levels (Figure 3A). When staining embryonic gonads in Carnegie stage 23 (n = 3 pairs of testes, n = 1 single testis and n = 1 pair of ovaries with cKIT-APC-conjugated antibodies), we found cKIT-positive cells in less than 1% of the gonad at day 49–50 (Figures 3B and 3C). The total number of PGCs sorted from a pair of gonads ranged from 1,721 to 9,549 cells (Figure 3D).
Figure 3.
Sorting Rhesus PGCs from Carnegie Stage 23 Gonads using cKIT
(A) FACS plot showing APC-conjugated cKIT antibody binding in rhesus fibroblasts (negative) and rhesus iPSCs (negative).
(B) Representative FACS plot showing APC-conjugated cKIT antibody binding in rhesus embryonic gonads at Carnegie stage 23.
(C) The percentage cKIT-positive rhesus PGCs in embryonic gonads. Each shape corresponds to cKIT-positive cells from the gonads of one embryo (n = 5 embryos total).
(D) Total number of cKIT-positive rhesus PGCs sorted from a pair of testes (n = 3 embryos) and a pair or ovaries (n = 1 embryo, pink arrow) at Carnegie stage 23.
(E) Single-cell analysis of 28 cKIT-APC sorted rhesus PGCs analyzed in technical triplicate for pluripotency and germline genes including OCT4, PRDM1, NANOS3, DAZL, and VASA. A standard curve was established using 1,000, 100, 10, and 0 cells. Yellow, high expression levels; black, background expression levels.
(F) Immunofluorescence for PRDM1 and VASA showing that the majority of VASA-positive cells are PRDM1-positive; however, a minor number of VASA-positive PRDM1-negative cells can be identified (arrow) (n = 1 day 49 embryonic testis, n = 2 technical duplicates). Scale bar, 20 μm.
The purity of the PGC population was confirmed by sorting 28 individual cKIT-positive cells from a day 49 embryonic testis into single wells of a 96-well plate, followed by single-cell, real-time PCR using a panel of diagnostic germ cell genes (Figure 3E). The negative control for this assay involved RT-PCR on a well with no cells (0). Positive controls were performed on wells that contained 10, 100, and 1,000 cKIT-positive sorted PGCs. Single-cell analysis revealed that all PGCs co-expressed DAZL, VASA, and NANOS3, indicating that the sorting strategy yielded a population of highly pure PGCs. Consistent with the immunofluorescence, we discovered that rare cKIT-positive PGCs had repressed OCT4 expression (one cell) and five cKIT-positive PGCs had repressed PRDM1 (Figure 3E). To determine whether some PGCs are also negative for PRDM1 protein, we performed immunofluorescence for VASA together with PRDM1, and show that while the majority of PGCs were VASA/PRDM1 double-positive, we can detect rare examples where PRDM1 is no longer expressed in VASA-positive PGCs (Figure 3F).
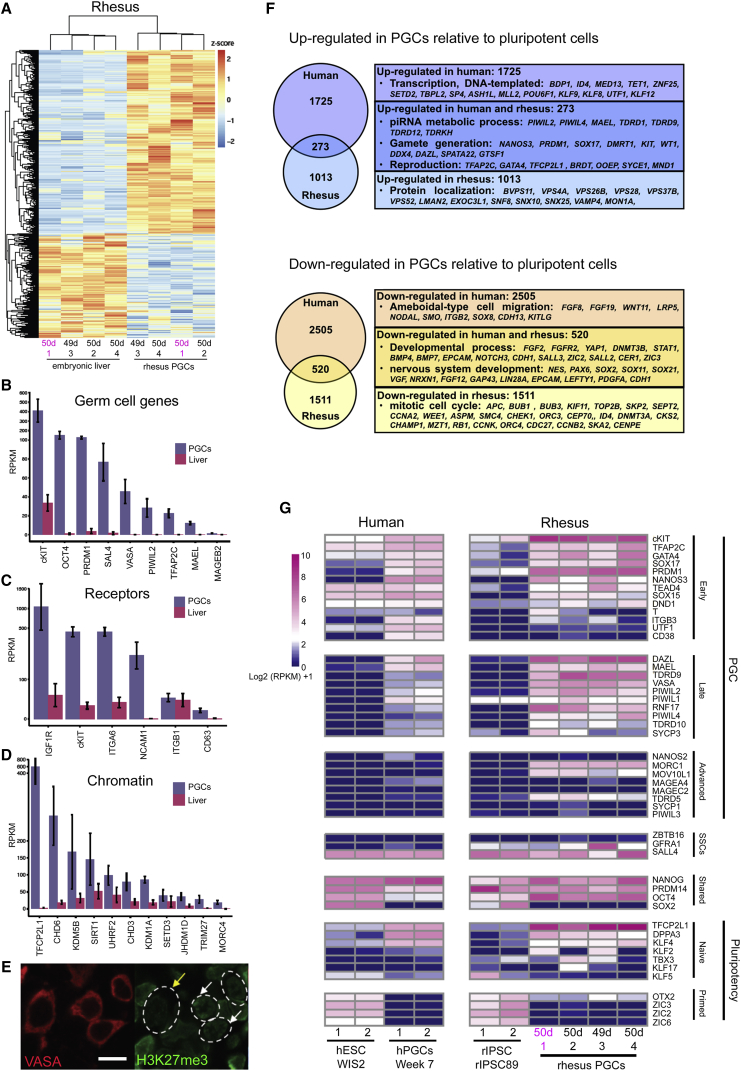
To identify PGC-specific genes that can be used as a reference for PGC identity relative to somatic cells, we performed RNA-seq of cKIT-sorted PGCs and embryonic livers from the same embryos (n = 4 embryos). We identified more than 2,000 differentially expressed genes (DEGs) between PGCs and embryonic liver (≥2-fold) with genes upregulated in PGCs including cKIT, OCT4 (POU5F1), PRDM1, TFAP2C, VASA, (DDX4), as well as Spalt-like transcription factor 4 (SAL4), Piwi-like 2 (PIWIL2), Maelstrom (MAEL), and melanoma-associated antigen B2 (MAGEB2) (Figures 4A and 4B). We also identified genes for receptors that were enriched in PGCs and not somatic cells, notably insulin-like growth factor 1R (IGF1R), integrin α6 (ITGA6), as well as neural cell adhesion molecule (NCAM1) (Figure 4C). Analysis of chromatin modifiers enriched in PGCs included the lysine demethylases (KDM3B, KDM5B, and KDM1A) (Figure 4D). These enzymes are responsible for removing methyl groups from the amino acid lysine, most notably from histone tails. In the mouse, one of the characteristic features of PGCs soon after specification is the depletion of histone H3 lysine 9 dimethylation (H3K9me2) from chromatin (Seki et al., 2007). Staining the rhesus fetal male testis at day 49 of embryo development for H3K9me2 together with VASA revealed that the majority of VASA-positive PGCs had low to undetectable levels of H3K9me2 in the genome (Figure 4E).
Figure 4.
Rhesus PGCs in Carnegie Stage 23 Are Equivalent to Human PGCs at 8–10 Weeks
(A) RNA-seq heatmap of cKIT-sorted PGCs from the gonads of n = 4 embryos. This includes the cKIT-positive population sorted from pairs of testes (n = 3 male embryos), and pairs of ovaries (n = 1 female embryo) with embryonic liver from the same embryos as a somatic cell control (n = 4 embryos). The age of the embryo is shown in days (d). The age shown in pink refers to the female samples. The ages shown in black are male samples. The numbers refer to the sample number. See also Chart S1.
(B) RPKM (reads per kilobase of transcript per million mapped read) of selected genes enriched in rhesus PGCs relative to rhesus embryonic liver cells. Data are shown as average of n=4 embryos with mean and SD.
(C) RPKM of selected genes encoding receptors. Data are shown as average of n=4 embryos with mean and SD.
(D) RPKM values of selected genes enriched in PGCs that are also involved in chromatin regulation. Data are shown as average of n=4 embryos with mean and SD.
(E) Immunofluorescence of histone H3 lysine 9 dimethylation (H3K9me2) in VASA-positive PGCs (circled in white). Most PGCs are dim to negative for H3K9me2 (white arrow) (n = 1 embryonic testis at day 49, n = 2 technical replicates). Scale bar, 10 μm.
(F) Identification of overlapping and distinct genes enriched or depleted >3-fold in rhesus or human PGCs relative to undifferentiated hESCs or riPSCs. See also Charts S2 and S3.
(G) Heatmap of diagnostic genes that discriminate PGCs as being early, late, or advanced, as well as genes that are found in primate spermatogonial stem cells (SSCs) or PSCs. See text for additional details.
To determine the similarity and differences between rhesus PGCs in Carnegie stage 23 (around 7 weeks of rhesus embryo development) and human PGCs at 7 weeks, we first compared the RNA-seq dataset of human PGCs to conventional (primed) hESCs, and identified 1,998 DEGs at ≥3-fold (Irie et al., 2015). Next, we performed this same analysis by comparing the cKIT-positive rhesus PGCs to primed rhesus iPSCs (Sosa et al., 2016), and identified 1,276 DEGs at ≥3-fold (Figure 4F). By comparing the DEG lists from the two species, we discovered 273 upregulated genes in common between human and rhesus PGCs relative to undifferentiated PSCs, including genes involved in reproduction, gamete generation, and piRNA metabolic process. Similarly, we identified 520 genes that were commonly downregulated in human and rhesus PGCs relative to undifferentiated PSCs, including genes involved in developmental processes and specifically neural development (Figure 4F). Species- or stage-specific differences were also revealed in this analysis, with human PGCs more highly enriched in genes associated with transcription, and rhesus PGCs repressed genes involved in mitotic cell cycle relative to their respective PSCs.
To stage rhesus and human PGCs, we created a heatmap of diagnostic genes that represent early PGCs (prior to DAZL and VASA expression), late PGCs (DAZL and VASA positive but prior to the emergence of major sex-specific transcriptional differences), advanced PGCs (enrichment of the male-specific gene NANOS2 and genome-defense pathways), as well as genes expressed by pro-spermatogonia and SSCs, and genes either unique to or shared between primed and naive PSCs (Gkountela et al., 2013, Sasaki et al., 2015; Figure 4G). This heatmap illustrates that rhesus PGCs in Carnegie stage 23 are older than human PGCs at 7 weeks, expressing late stage PGC genes including PIWIL4, as well as some advanced PGC genes including microrchidia 1 (MORC1), MOV10L1, and TDRD5, which in humans are enriched in PGCs after 10 weeks (Gkountela et al., 2015). Notably, the rhesus PGCs have not advanced to pro-spermatogonia or spermatogonia as they do not express NANOS2 or PLZF (ZBTB16) (Sasaki et al., 2016). Therefore, we stage the molecular age of rhesus PGCs in Carnegie stage 23 as being older than 7 weeks and most likely equivalent to human PGCs between 8 and 10 weeks of human development.
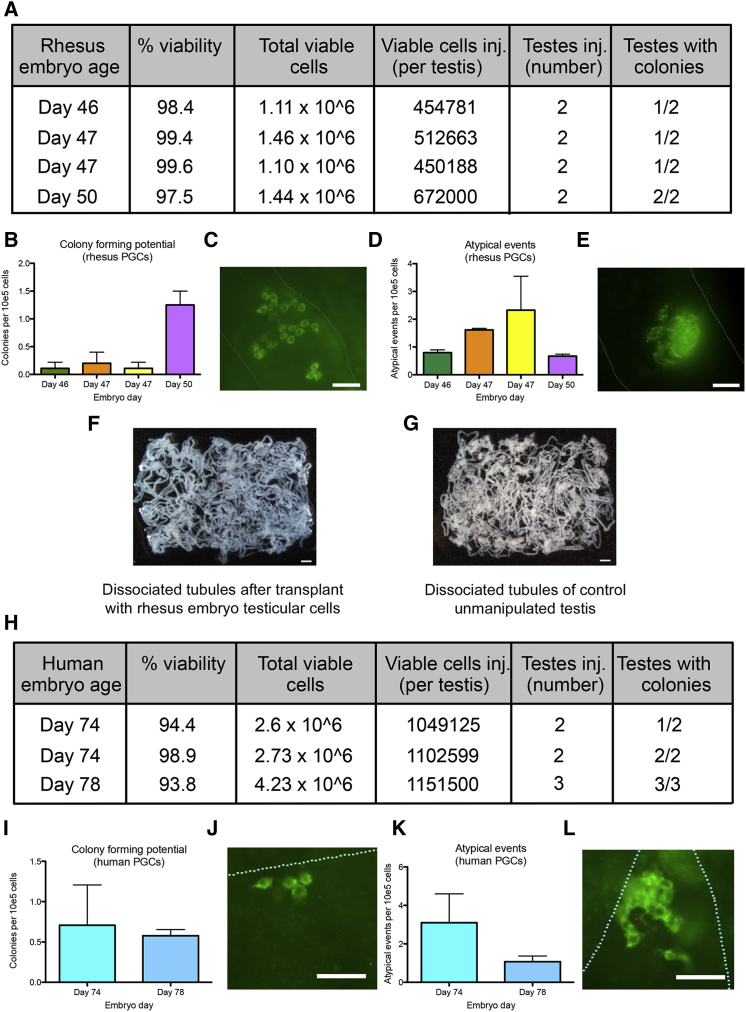
Finally, we characterized colony-forming potential of Carnegie stage 23 testicular cells containing PGCs using the established primate-to-nude mouse xenotransplantation assay. For this experiment n = 4 pairs of embryonic testes at day 46–50 were digested to single cells, and the single-cell suspensions were injected into the rete testis of busulfan-treated nude mice. Primate-derived colonies were identified by whole-mount immunofluorescence using the rabbit antibody that was raised against nonhuman primate (Nhp) testis cells. The average total number of cells in a pair of embryonic testes at Carnegie stage 23 was 1.3 million, and total cellular viability of this population prior to injection was >97%. For each single-cell suspension of embryonic donor testis pairs, n = 2 busulfan-treated nude mice were injected (n = 8 mice total). For embryonic testes at day 46–47, only one of the two recipient testes had evidence of primate donor-derived cells. For the day 50 embryonic testes, both recipient testes exhibited evidence of primate donor-derived cells (Figure 5A). We discovered two types of colonies in the recipient mouse testes after xenotransplant. The first were typical chains or networks of Nhp-positive cells, consistent with colonies produced by adult primate SSCs (Figures 5B and 5C). The second were atypical events within the tubules (Figures 5D and 5E). These atypical events corresponded to cells that stained positive with the anti-NhP antibody, but did not exhibit typical spermatogonia-like colony morphology (Figure 5E). To determine whether transplanting the embryonic testes caused tumors, we inspected the mouse testis at necropsy, followed by dissociating the testis and inspecting the entire tubule system under the light microscope (Figures 5F and 5G). We found no difference in tubule transparency or evidence of tumor formation in the mouse testis transplanted with rhesus cells compared with the unmanipulated mouse testis.
Figure 5.
Rhesus and Human PGCs Are Transplantable
(A) Xenotransplantation of Carnegie stage 23 rhesus testis cells leads to colonies in the seminiferous tubules (n = 4 pairs of embryonic testis).
(B) Quantification of typical colonies following rhesus embryonic testis xenotransplantation. Error bars show the average of technical replicates and SEM.
(C) Morphology of typical rhesus colonies stained with the anti-NhP antibody, Scale bar, 50 μm.
(D) Quantification of atypical events following xenotransplantation of rhesus embryonic testes. Error bars show the average of technical replicates and SEM.
(E) Morphology of an atypical rhesus event stained with the anti-NhP antibody. Scale bar, 50 μm.
(F) Dissociated tubules from a busulfan-treated nude mouse testis that was xenotransplanted with donor embryonic testes. Scale bar, 1 mm.
(G) Dissociated tubules from a busulfan-treated nude mouse testis that was unmanipulated. Scale bar,= 1 mm.
(H) Xenotransplantation of human embryonic testes results in the identification of colonies (n = 3 pairs of embryonic human testes).
(I) Quantification of typical human colonies stained with the anti-NhP antibody. Error bars show average of technical replicates for n = 2 (day 74) or n = 1 (day 78) and SEM.
(J) Morphology of a typical human colony stained with the anti-NhP antibody. Scale bar, 50 μm.
(K) Quantification of atypical events stained with the ani-NHP antibody. Error bars show average of technical replicates for n = 2 (day 74) or n = 1 (day 78) and SEM.
(L) Morphology of an atypical human event stained with the anti-NhP antibody. Scale bar, 50 μm. Error bars show average of technical replicates and SEM.
To evaluate colony-forming potential of human PGCs at a similar molecular age we transplanted n = 3 pairs of human embryonic testes at 74–78 days of development post-fertilization (Figure 5H) and show that human testes containing PGCs have colony-forming potential in this assay (Figures 5I–5J). Similar to xenotransplantation with rhesus macaque, atypical events were also identified following transplantation with human embryonic testes, and these were similarly reduced in number with increasing age (Figures 5K and 5L). Dissected mouse tubules transplanted with human cells also had no evidence of tumors (data not shown), similar to the results in Figures 5F and 5G. Therefore, our results indicate that human and nonhuman primate PGCs are transplantable in busulfan-treated adult nude mice, and are not tumorigenic. This assay could be used in future to determine identity of PGCLCs differentiated in vitro from PSCs.
Discussion
This work describes the use of hormone-guided time-mated breeding to examine the molecular and functional characteristics of rhesus macaque PGCs in Carnegie stage 23 of embryo development using RNA-seq and xenotransplantation into busulfan-treated adult nude mice. This resource should serve as an important reference for the differentiation of primate PGCLCs in vitro from PSCs, and also the behavior of single-cell suspensions of tissues containing bona fide PGCs in the busulfan-treated xenotransplantation model. Furthermore, future studies aimed at characterizing the germline identity of these colonies, and determining whether the colonies represent PGCs, pro-spermatogonia, or spermatogonia are now warranted.
Xenotransplantation of human and nonhuman primate adult testicular cells into mouse testis is one of the major bioassays for germline potential. Using this assay together with FACS to isolate putative SSCs reveals that colony-forming potential is enriched in germ cells with an SSEA4, ITGA6, Thy1dim, EpCAMdim, or CD9 cell surface phenotype (Izadyar et al., 2011, Valli et al., 2014, Zohni et al., 2012). Specifically, xenotransplantation of unsorted adult testicular cells into busulfan-treated nude mice leads to ∼2.9–4.8 colonies per 100,000 cells transplanted, whereas sorting cells for ITGA6, Thy1, and EpCAM results in an average 9.6, 7.3, and 11.9 colonies per 100,000 cells transplanted (Valli et al., 2014). In the current study, the colony-forming potential from unsorted Carnegie stage 23 rhesus embryonic testes was 0.42 colonies per 100,000 cells transplanted (when averaging the results of all four replicates in Carnegie stage 23). However, at day 50 of embryo development, the average colony-forming potential was 1.25 colonies per 100,000 cells transplanted.
In allogeneic mouse transplants, Chuma et al. (2005) reported spermatogenesis and teratoma-forming potential following transplantation of epiblast cells as well as PGCs at E8.5 into the seminiferous tubules of 5–8-day-old W (cKIT mutant) mouse pups. Just 2 days later, at E10.5, teratoma potential was lost when donor tissues containing PGCs were used (Chuma et al., 2005). Using the adult testis as a recipient, our data reveal that primate embryonic testes containing OCT4- and NANOG-positive PGCs do not yield teratomas after xenotransplantation into the adult testis. Therefore, repression of the latent pluripotency program in PGCs is not a requirement for xenotransplantation and colony-forming potential. We did identify atypical microscopic events that were relatively rare. It is not known whether these events are of germ cell origin, or whether they may correspond to somatic cells of the embryonic testis that were able to survive in the seminiferous tubule epithelium. Notably, these events were observed when either human or rhesus macaque embryonic testis were used as donor cells, and in both species the number of atypical events decreased with increasing embryonic age. This was opposite to typical colony-forming potential which appeared to increase with age. One possibility for this is PGC number. Our FACS experiments indicate that PGC number ranges from around 1,700 to ∼10,000 PGCs in Carnegie stage 23. Therefore, it is conceivable that the best transplants originated from testes with the highest number of PGCs. Alternatively, it is also conceivable that the microenvironment of the laminin-positive cords changes abruptly at the end of Carnegie stage 23, leading to a change in PGC colonizing ability. One way to distinguish these possibilities would be to sort PGCs at a range of developmental ages, and normalize PGC number prior to xenotransplant.
Recently, transplanting undifferentiated hiPSCs into the seminiferous tubules of busulfan-treated adult nude mice yielded germ cell colonies as well as nonseminoma-type germ cell tumors including embryonal carcinoma, yolk sac tumors, and teratomas (Durruthy-Durruthy et al., 2014, Ramathal et al., 2014). From this work it was unclear whether these different outcomes were due to the seminiferous tubule microenvironment instructing hiPSCs to differentiate into the tumors, or whether the seminiferous tubule microenvironment first induced the differentiation of PGCLCs that subsequently differentiated into the nonseminoma-type germ cell tumors (Durruthy-Durruthy et al., 2014, Ramathal et al., 2014). In our study, we discovered that embryonic testes containing PGCs do not generate tumors. This argues that the tumors generated from hiPSCs/hESCs originate from the transplanted pluripotent cells and not a PGCLC intermediate.
In summary, the xenotransplant method is a valuable tool for observing and quantifying the colonizing activity of experimental cell populations. Although we did not determine whether the colonies emerging after xenotransplantation are spermatogonia. Future studies are now justified to address this. Importantly, the xenotransplantation method is robust and accessible to most investigators with the expertise to perform transplantation in mice, but is limited because it does not recapitulate complete spermatogenesis. A major strength of the nonhuman primate system is that once methods for producing putative PGCLCs are established and validated using the xenotransplant assay, the full spermatogenic potential of those cells can be tested by autologous or allogeneic transplantation into the testes of recipient monkeys. If successful, the fertilization potential of monkey PGCLC-derived sperm can be tested by fertilization of primate eggs, and transfer of the resulting embryos, with the potential to produce live offspring. These experiments that provide the “gold standard” evidence of spermatogenic potential are not possible in the human system, but should provide a valuable template for development of human PGCLC differentiation protocols.
Experimental Procedures
Time-Mated Breeding of Rhesus Macaque
Time-mated breeding of rhesus macaque males and females was performed by measuring estradiol daily in the female starting from day 5 to 8 after menses began. A known fertile adult male was paired with the female once her estradiol levels had risen above baseline (>50 pg/mL). Twenty-four hours after ovulation, as measured by the estradiol peak (Weick et al., 1973), the male was removed. Day 1 of embryo development was estimated to occur 72 hr after the estradiol peak. Pregnancy was confirmed by measuring progesterone as well as by ultrasound. At the time of necropsy Carnegie staging was performed according to (Hendrickx and Sawyer, 1975). A total of n = 10 rhesus macaque embryos were used in this study. All rhesus macaque time-mated breeding experiments were conducted following Institutional Animal Care and Use Committee Approval.
Staging of Human Embryos for Testis Xenotransplantation
Human embryonic testes were acquired following elected termination and pathological evaluation only after UCLA-IRB review which deemed the project exempt under 45 CRF 46.102(f). The samples were acquired by the University of Washington Birth Defects Research Laboratory (BDRL), under the regulatory oversight of the University of Washington IRB approved Human Subjects protocol combined with a Certificate of Confidentiality from the Federal Government. All consented testes provided to us by BDRL were anonymous and carried no personal identifiers. Developmental age was estimated by prenatal intakes, foot length, Streeter's stage, and crown rump length. Any conceptus with a documented birth defect or chromosomal abnormality was excluded from the study. The embryonic testes were shipped overnight for immediate processing for xenotransplant in Pittsburgh. Three pairs of embryonic testes were used in this study, n = 2 at E74 and n = 1 at E 78.
FACS
Rhesus cells were dissociated with 0.25% Trypsin-EDTA for 5 min at 37°C. Dissociated cells were incubated in 1% BSA in PBS on ice for 20 min containing the APC-conjugated primary immunoglobulin G antibody raised against cKIT (1:100; BioLegend, 313205). Cells were then washed and incubated with 1% BSA in PBS for 5 min on ice. Cells were passed through a 40 μM filter (BD Biosciences) before flow analysis or FACS. DAPI (10 μg/mL; Sigma) was used as viability dye at 1:1,000 dilution and all DAPI-positive cells were excluded. Analysis was performed using LSR II (Becton Dickinson) and FlowJo software (Tree Star).
Single-Cell Real-Time PCR
Single cells were sorted with a BD FACSAria cell sorter equipped for biosafety level 2 sorting, and single-cell analysis was performed as described using rhesus macaque specific assays from TaqMan (Gkountela et al., 2013, Vincent et al., 2013).
RNA-Seq
cKIT-sorted rhesus PGCs, rhesus liver cells, and rhesus iPS cells were lysed in 350 μL RLT buffer (QIAGEN) and RNA was extracted using the RNeasy Micro Kit (QIAGEN) according to the manufacturer's instructions. RNA was amplified and converted to cDNA using the Ovation RNA-Seq System V2 (NuGEN). cDNA was sonicated to DNA fragments within the 200 bp range using a Covaris S2 sonicator according to the manufacturer’s instructions (duty cycle 10%, intensity 5, cycles per burst 200, and time 180 s). Subsequently libraries were generated using the Encore Rapid Library System according to the manufacturer’s instructions. Library concentration was estimated using a Kapa Library Quantification Kit (catalog no. 4827) according to the manufacturer’s instructions. Rhesus macaque PGC and embryonic liver libraries were run using 100 bp paired-end reads on the HiSeq 2000 system (Illumina). The rhesus iPSC libraries were run using a 50 bp single end read. ERCC RNA Spike-In Mix (catalog no. 4456740, Invitrogen) was used according to the manufacturer's instructions in the all samples except rhesus PGCs. ERCC (External RNA Controls Consortium) standards were calculated to have an observed and expected correlation of R > 0.96.
RNA-Seq Analysis
Raw reads in qseq format obtained from sequencer were first converted to fastq format with customized perl script. Reads quality was controlled with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). High-quality reads were then aligned to rhesus macaque genome (MacaM) (Zimin et al., 2014) or Homo sapiens genome (hg19) reference genome using Tophat (Trapnell et al., 2009) (v. 2.0.13) by using a “-no-coverage-search” option, allowing up to two mismatches and only keeping reads that mapped to one location. Reads were first mapped to MacaM or hg19 gene annotation with known splice junction. When reads did not map to the annotated genes, the reads were mapped to entire MacaM or hg19 genome. The number of reads mapping to genes was calculated by HTseq (Anders et al., 2015) (v. 0.5.4) with default parameters. Expression levels were determined by RPKM (reads per kilobase of exons per million aligned reads) in R using customized scripts. For RNA-seq of human PGC and ESC, published datasets GSM1466233, GSM1466234, GSM1574595, and GSM1574596 (Irie et al., 2015) were obtained from GEO and then processed exactly the same as described above.
Differential Gene Expression Calling
Figure 4A
R DESeq package was used to normalize counts per RefSeq transcripts to evaluate differential expression. For comparison between rhesus PGC and rhesus embryonic liver cells, differentiated expressed genes with mean fold-change greater than 4 and adjusted p value less than 0.05 were selected and plotted as heatmaps.
Figure 4F
R DESeq package was used to normalize counts per RefSeq transcripts to evaluate differential expression. For comparison between hESC versus hPGC as well as rhesus PGC versus rhesus iPSC, differentiated expressed genes with mean fold-change greater than 3 and adjusted p value less than 0.05 were selected and plotted as heatmaps. For comparison within different species (rhesus versus human), only homologous genes shared by both species were used for analysis.
Heatmap of Selected Genes
Figure 4G
Diagnostic germ cell, somatic cell, and pluripotency genes were obtained from previously published literature. Log2(RPKM+1) values were plotted.
Immunofluorescence
Immunofluorescence of was performed as described previously (Gkountela et al., 2013, Gkountela et al., 2015). Dilutions and catalog numbers of primary antibodies were: mouse anti-5mC (1:100, AMM99021; AVIVA Biosciences) mouse anti-H3K9me2 (1:100, ab1220; Abcam), goat-anti-VASA (R&D Systems, AF2030, 1:100), rabbit-anti-cKIT (Dako, A4502, 1:100), goat-anti-OCT4 (Santa Cruz Biotechnology, sc-8628 [concentrated], 1:100), rabbit-anti-PRDM1 (Cell Signaling Technology, 9115, 1:100), rabbit-anti-TFAP2C (Santa Cruz Biotechnology, sc-8977), goat-anti-SOX17 (Neuromics, GT15094, 1:100), and rabbit-anti-LAMININ (Abcam, ab11575, 1:100). All samples were incubated with primary antibodies overnight at 4°C. Sections were washed, incubated with FITC/TRITC-conjugated secondary antibodies (Jackson ImmunoResearch) for 30 min and mounted in ProLong Antifade Reagent with DAPI (Invitrogen). Samples were imaged on a Zeiss Axio Imager using AxioVision 4.8 software (Zeiss).
Xenotransplantation
Pairs of testes from a single embryo were shipped overnight on cold packs to Magee Women's Health Research Institute in cold Hank’s balanced salt solution. The testes were trypsinized to single cells and 6–7 μL was injected into the rete testis as described previously (Valli et al., 2014). At 6–8 weeks the animals were euthanized and the testes removed, visually inspected, documented as being soft or hard, and weighed. Testes were then stained using whole-mount immunofluorescence using the rabbit-anti-NhP antibody as described previously (Valli et al., 2014). Colonies were counted according to (Valli et al., 2014). All mouse transplantation studies were conducted following Institutional Animal Care and Use Committee Approval.
Author Contributions
A.T.C. conceived the experiments, performed experiments, maintained Institutional Biosafety Approval for rhesus macaque work, obtained funding and wrote the manuscript; S.G. performed experiments on sorting PGCs; D.C. performed immunofluorescence experiments and bioinformatics; W.L. performed bioinformatics; E.S. performed immunofluorescence experiments; M.S. performed xenotransplantation experiments; J.D.H. performed experiments, obtained funding, maintained IACUC approval, and oversight of rhesus macaque work; K.E.O. planned the experiments, obtained funding, maintained IACUC approval, and oversight of mouse work.
Acknowledgments
The authors would like to thank Felecia Codrea and Jessica Scholes of the UCLA BSCRC FACS core, Suhua Feng of the UCLA BSCRC Sequencing core, and Tiasha Shafiq for immunostaining. This project was funded by NIH grant P01HD075795. Time-mated breeding was supported by NIH OD P51OD011092 (J.D.H.). The Laboratory of Developmental Biology, University of Washington, Seattle is supported by NIH Award Number 5R24HD000836 from the NICHD. W.L. is supported by a Philip J. Whitcome fellowship from the Molecular Biology Institute of University of California, Los Angeles, and a scholarship from the Chinese Scholarship Council. DC is supported by a Postdoctoral Fellowship from the UCLA BSCRC and the Shaffer Family Foundation. Human conceptus tissue requests can be made to: bdrl@u.washington.edu.
Published: June 1, 2017
Footnotes
Supplemental Information includes three charts and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.05.002.
Accession Numbers
The RNA-seq data have been deposited in the GEO under the accession number GEO: GSE95736.
Supplemental Information
References
- Anders S., Pyl P.T., Huber W. HTSeq – a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquoine A. The identification, origin and migration of the primordial germ cells in the mouse embryo. Anat. Rec. 1954;118:135–146. doi: 10.1002/ar.1091180202. [DOI] [PubMed] [Google Scholar]
- Chuma S., Kanatsu-Shinohara M., Inoue K., Ogonuki N., Miki H., Toyokuni S., Hosokawa M., Nakatsuji N., Ogura A., Shinohara T. Spermatogenesis from epiblast and primordial germ cells following transplantation into postnatal mouse testis. Development. 2005;132:117–122. doi: 10.1242/dev.01555. [DOI] [PubMed] [Google Scholar]
- Combes A.N., Lesieur E., Harley V.R., Sinclair A.H., Little M.H., Wilhelm D., Koopman P. Three-dimensional visualization of testis cord morphogenesis, a novel tubulogenic mechanism in development. Dev. Dyn. 2009;238:1033–1041. doi: 10.1002/dvdy.21925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey S.L., Valli H., Hermann B.P., Sukhwani M., Donohue J., Castro C.A., Chu T., Sanfilippo J.S., Orwig K.E. Eliminating malignant contamination from therapeutic human spermatogonial stem cells. J. Clin. Invest. 2013;123:1833–1843. doi: 10.1172/JCI65822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durruthy-Durruthy J., Ramathal C., Sukhwani M., Fang F., Cui J., Orwig K., Reijo-Pera R. Fate of induced pluripotent stem cells following transplantation to murine seminiferous tubules. Hum. Mol. Genet. 2014;23:3071–3084. doi: 10.1093/hmg/ddu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S., Li Z., Vincent J., Zhang K., Chen A., Pellegrini M., Clark A. The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat. Cell Biol. 2013;15:113–122. doi: 10.1038/ncb2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S., Zhang K.X., Shafiq T.A., Liao W.W., Hargan-Calvopina J., Chen P.Y., Clark A.T. DNA demethylation dynamics in the human prenatal germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Yan L., Guo H., Li L., Hu B., Zhao Y., Yong J., Hu Y., Wang X., Wei Y. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Hargan-Calvopina J., Taylor S., Cook H., Hu Z., Lee S.A., Yen M.R., Chiang Y.S., Chen P.Y., Clark A.T. Stage-specific demethylation in primordial germ cells safeguards against precocious differentiation. Dev. Cell. 2016;39:75–86. doi: 10.1016/j.devcel.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ogushi S., Kurimoto K., Shimamoto S., Ohta H., Saitou M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science. 2012;338:971–975. doi: 10.1126/science.1226889. [DOI] [PubMed] [Google Scholar]
- Hendrickx A.G., Sawyer R.H. Vol II. Academic Press; 1975. (Embryology of the Rhesus Monkey). [Google Scholar]
- Hermann B.P., Sukhwani M., Lin C.C., Sheng Y., Tomko J., Rodriguez M., Shuttleworth J.J., McFarland D., Hobbs R.M., Pandolfi P.P. Characterization, cryopreservation, and ablation of spermatogonial stem cells in adult rhesus macaques. Stem Cells. 2007;25:2330–2338. doi: 10.1634/stemcells.2007-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B.P., Sukhwani M., Simorangkir D.R., Chu T., Plant T.M., Orwig K.E. Molecular dissection of the male germ cell lineage identifies putative spermatogonial stem cells in rhesus macaques. Hum. Reprod. 2009;24:1704–1716. doi: 10.1093/humrep/dep073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B.P., Sukhwani M., Winkler F., Pascarella J.N., Peters K.A., Sheng Y., Valli H., Rodriguez M., Ezzelarab M., Dargo G. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell. 2012;11:715–726. doi: 10.1016/j.stem.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyer P., Byskov A., Møllgård K. Stem cell factor and c-Kit in human primordial germ cells and fetal ovaries. Mol. Cell Endocrinol. 2005;234:1–10. doi: 10.1016/j.mce.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Irie N., Weinberger L., Tang W., Kobayashi T., Viukov S., Manor Y.S., Dietmann S., Hanna J., Surani M. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izadyar F., Wong J., Maki C., Pacchiarotti J., Ramos T., Howerton K., Yuen C., Greilach S., Zhao H.H., Chow M. Identification and characterization of repopulating spermatogonial stem cells from the adult human testis. Hum. Reprod. 2011;26:1296–1306. doi: 10.1093/humrep/der026. [DOI] [PubMed] [Google Scholar]
- Jameson S., Natarajan A., Cool J., DeFalco T., Maatouk D.M., Mork L., Munger S., Capel B. Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet. 2012;8:e1002575. doi: 10.1371/journal.pgen.1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K., Yabuta Y., Ohinata Y., Shigeta M., Yamanaka K., Saitou M. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008;22:1617–1635. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki C., Pacchiarotti J., Ramos T., Pascual M., Pham J., Kinjo J., Anorve S., Izadyar F. Phenotypic and molecular characterization of spermatogonial stem cells in adult primate testes. Hum. Reprod. 2009;6:1480–1491. doi: 10.1093/humrep/dep033. [DOI] [PubMed] [Google Scholar]
- Matsui Y., Zsebo K., Hogan B. Derivation of pluripotent embryonic stem cells from murine primordial germ cells. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- Nagano M., McCarrey J., Brinster R. Primate spermatogonial stem cells colonize the mouse testis. Biol. Reprod. 2001;64:1409–1416. doi: 10.1095/biolreprod64.5.1409. [DOI] [PubMed] [Google Scholar]
- Nagano M., Patrizio P., Brinster R. Long-term survival of human spermatogonial stem cells in mouse testes. Fertil. Steril. 2002;78:1225–1233. doi: 10.1016/s0015-0282(02)04345-5. [DOI] [PubMed] [Google Scholar]
- Ohinata Y., Ohta H., Shigeta M., Yamanaka K., Wakayama T., Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Ohta H., Tohda A., Nishimune Y. Proliferation and differentiation of spermatogonial stem cells in the w/wv mutant mouse testis. Biol. Reprod. 2003;69:1815–1821. doi: 10.1095/biolreprod.103.019323. [DOI] [PubMed] [Google Scholar]
- Perrett R.M., Turnpenny L., Eckert J.J., O'Shea M., Sonne S.B., Cameron I.T., Wilson D.I., Rajpert-De Meyts E., Hanley N.A. The early human germ cell lineage does not express sox2 during in vivo development or upon in vitro culture. Biol. Reprod. 2008;78:852–858. doi: 10.1095/biolreprod.107.066175. [DOI] [PubMed] [Google Scholar]
- Ramathal C., Durruthy-Durruthy J., Sukhwani M., Arakaki J., Turek P., Orwig K., Reijo-Pera R. Fate of iPSCs derived from azoospermic and fertile men following xenotransplantation to murine seminiferous tubules. Cell Rep. 2014;7:1284–1297. doi: 10.1016/j.celrep.2014.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick J.L., Bixler L.S., Cheng L., Donovan P.J. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359:550–551. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- Sadri-Ardekani H., Mizrak S.C., van Daalen S.K., Korver C.M., Roepers-Gajadien H.L., Koruji M., Hovingh S., de Reijke T.M., de la Rosette J.J., van der Veen F. Propagation of human spermatogonial stem cells in vitro. JAMA. 2009;302:2127–2134. doi: 10.1001/jama.2009.1689. [DOI] [PubMed] [Google Scholar]
- Sadri-Ardekani H., Akhondi M.A., van der Veen F., Repping S., van Pelt A.M. In vitro propagation of human prepubertal spermatogonial stem cells. JAMA. 2011;305:2416–2418. doi: 10.1001/jama.2011.791. [DOI] [PubMed] [Google Scholar]
- Saitou M., Barton S.C., Surani M.A. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Yokobayashi S., Nakamura T., Okamoto I., Yabuta Y., Kurimoto K., Ohta H., Moritoki Y., Iwatani C., Tsuchiya H. Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell. 2015;17:178–194. doi: 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Nakamura T., Okamoto I., Yabuta Y., Iwatani C., Tsuchiya H., Seita Y., Nakamura S., Shiraki N., Takakuwa T. The germ cell fate of cynomolgus monkeys is specified in the nascent amnion. Dev. Cell. 2016;39:169–185. doi: 10.1016/j.devcel.2016.09.007. [DOI] [PubMed] [Google Scholar]
- Seki Y., Yamaji M., Yabuta Y., Sano M., Shigeta M., Matsui Y., Saga Y., Tachibana M., Shinkai Y., Saitou M. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- Sosa E., Kim R., Rojas E.J., Hosohama L., Hennebold J.D., Orwig K.E., Clark A.T. An integration-free, virus-free rhesus macaque induced pluripotent stem cell line (riPSC89) from embryonic fibroblasts. Stem Cell Res. 2016;17:444–447. doi: 10.1016/j.scr.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawa F., Arauzo-Bravo M., Yoon J., Kim K., Aramaki S., Wu G., Stehling M., Psathaki O., Hubner K., Scholer H. Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J. 2015;34:1009–1024. doi: 10.15252/embj.201488049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unni S., Modi D., Pathak S., Dhabalia J., Bhartiya D. Stage-specific localization and expression of c-kit in the adult human testis. J. Histochem. Cytochem. 2009;57:861–869. doi: 10.1369/jhc.2009.953737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli H., Sukhwani M., Dovey S., Peters K., Donohue J., Castro C., Chu T., Marshall G., Orwig K. Fluorescence- and magnetic-activated cell sorting strategies to isolate and enrich human spermatogonial stem cells. Fertil. Steril. 2014;102:566–580. doi: 10.1016/j.fertnstert.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J.J., Huang Y., Chen P.Y., Feng S., Calvopina J.H., Nee K., Lee S.A., Le T., Yoon A.J., Faull K. Stage-specific roles for Tet1 and Tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell. 2013;12:470–478. doi: 10.1016/j.stem.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick R., Dierschke D., Karsch F., Butler W., Hotchkiss J., Knobil E. Periovulatory time courses of curfulating gonadotrophic and ovarian hormones in the rhesus monkey. Endocrinology. 1973;93:1140–1147. doi: 10.1210/endo-93-5-1140. [DOI] [PubMed] [Google Scholar]
- Western P., Miles D., van den Bergen J., Bruton M., Sinclair A. Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells. 2008;26:339–347. doi: 10.1634/stemcells.2007-0622. [DOI] [PubMed] [Google Scholar]
- Witschi E. Migration of the germ cells of human embryos from the yolk sac to the primitive gonadal folds. Carnegie Inst. Contrib. Embryol. 1948;32:67–80. [Google Scholar]
- Wu X., Schmidt J.A., Avarbock M.R., Tobias J.W., Carlson C.A., Kolon T.F., Ginsberg J.P., Brinster R.L. Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. Proc. Natl. Acad. Sci. USA. 2009;106:21672–21677. doi: 10.1073/pnas.0912432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A.V., Cornish A.S., Maudhoo M.D., Gibbs R.M., Zhang X., Pandey S., Meehan D.T., Wipfler K., Bosinger S.E., Johnson Z.P. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol. Direct. 2014;9:20. doi: 10.1186/1745-6150-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohni K., Zhang X., Tan S.L., Chan P., Nagano M. CD9 is expressed on human male germ cells that have a long-term repopulation potential after transplantation into mouse testes. Biol. Reprod. 2012;87:27. doi: 10.1095/biolreprod.112.098913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.