Abstract
Why we sleep remains one of the greatest mysteries in science. In the past few years, great advances have been made to better understand this phenomenon. Human genetics has contributed significantly to this movement, as many features of sleep have been found to be heritable. Discoveries about these genetic variations that affect human sleep will aid us in understanding the underlying mechanism of sleep. Here we summarize recent discoveries about the genetic variations affecting the timing of sleep, duration of sleep and EEG patterns. To conclude, we also discuss some of the sleep-related neurological disorders such as Autism Spectrum Disorder (ASD) and Alzheimer’s Disease (AD) and the potential challenges and future directions of human genetics in sleep research.
Introduction
The field of human genetics began to gain momentum as a powerful approach for defining the causes of diseases beginning in the 1980s. This approach continues to be fruitful nearly 3 decades later. Over the past thirty years, human genetics has revolutionized the field of biomedical research and medicine in general. The identification of genetic causes for diseases generated a dramatic paradigm shift in the process of studying disease pathophysiology. The great hope is that understanding of genetics and biology of specific diseases will lead to a more rational approach to devising better treatments.
Sleep is known to have a large impact on human health but remains a great mystery today. Studies of human behaviors, including sleep, are more challenging than studies of diseases because behavioral phenotypes are typically more complex and are generally subject to many environmental factors. Nonetheless, an opportunity arose in the late 1990s with identification of the first familial circadian phenotype (familial advanced sleep phase syndrome-FASP) that made it possible to begin genetic mapping and cloning of genes/mutations that have strong effects on human circadian timing. Less than twenty years after the recognition of these families [1], we have made great strides into understanding regulatory mechanisms of human sleep behavior. Growing evidence has accumulated over the last two decades and revealed that a number of sleep traits in humans are heritable, such as timing of sleep, total daily sleep requirement, response to sleep deprivation, and various EEG measurements/patterns. In these Mendelian sleep phenotypes, single mutations of large effect were shown to be causative for different phenotypes. Therefore, mutations identified using genetics in human families have led to new insights into the detailed molecular mechanisms regulating sleep behavior.
Here, we summarize recent discoveries in the field of human genetics implicating genes in sleep regulation. We will focus primarily on natural variations in sleep traits including the timing, duration and the EEG characteristics of sleep (Figure 1).
Fig 1.
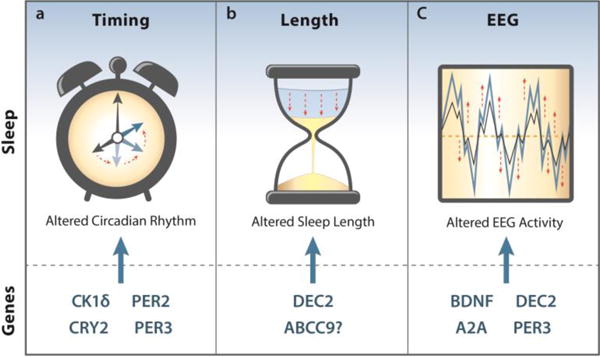
Genes highlighted in this review that affect the timing, duration, or EEG characteristics of sleep.
1a) Mutations in CK1δ, CRY2, PER2, and PER3 have all been shown to shift the timing of sleep forward in their carriers, causing Familial Advanced Sleep Phase (FASP).
1b) DEC2 mutations have been linked to reduced sleep length or sleep deprivation resistant traits. Carriers of one mutation require only around six hours of sleep per night. This trait was named as Natural Short-Sleep (NSS).
1c) Multiple genes have been implicated in changing the EEG characteristics of sleep, primarily through affecting changes in Slow-Wave Amplitude (SWA) or the spectral power of the delta (.25–4 Hz) and theta (6–8 Hz) bands
Timing of sleep
The timing of sleep is determined by the circadian clock, which is entrained to the environment primarily by light. At a molecular level, the periodicity of biological clocks is generated by transcriptional-translational feedback loops [2–5]. A growing list of core clock genes have been discovered that encode proteins participating in this feedback loop. Components of the molecular clock are highly conserved in vertebrates [3,4]. Theoretically, mutations that alter the molecular clock feedback loops may result in altered circadian timing. Indeed, our lab has identified several genetic mutations including casein kinase 1 delta (CK1δ) T44A and H46R, period2 (PER2) S662G, period3 (PER3) P415A/H417R and cryptochrome2 (CRY2) A260T from subjects affected by familial advance sleep phase (FASP) (Table 1) (Fig 2) [6–11]. Sleep onset and offset times are significantly advanced in individuals with FASP. Most of the mutations characterized to date accelerate the clock and shorten the period, which leads to the advanced phase phenomena [7,8,10]. In addition, the importance of clock protein post-translation modifications was elucidated by mutations found in PER2 and CK1δ [7,8]. The fact that control of clock protein turnover and stability is critical for sleep regulation is demonstrated repeatedly by studies of PER2, PER3, and CRY2 mutations [7,9,10]. Each of the mutations found by this approach contributed to a better overall picture of regulatory clock mechanisms.
Table 1.
sleep related genetic variations highlighted in this review
| Genes | Pathology | Experimental Design | Associated SNP, Allele, or Mutation | Notes |
|---|---|---|---|---|
| Csnk1d | FASP & Migraine | Candidate gene sequencing | T44A and H46R | Autosomal-dominant transmission; validation in in vitro and mice models |
| PER2 | FASP | Linkage analysis and Candidate gene sequencing | S662G | Autosomal-dominant transmission; validation in in vitro and mice models |
| PER3 | FASP & SAD (seasonal affective disorder) | Candidate gene sequencing | P415A/H417R | Autosomal-dominant transmission; validation in in vitro and mice models |
| CRY2 | FASP | Candidate gene sequencing | A260T | Autosomal-dominant transmission; validation in in vitro and mice models |
| DEC2 | NSS | Candidate gene sequencing | P384R and Y362H | Only P384R was tested in mice models. |
| ABCC9 | NSS | GWAS | rs11046205 A in intron | Potentially affected the expression levels; not fully testified. |
| PER3 | EEG variations & SAD (seasonal affective disorder) | polymorphism described in previous studies | 54-nucleotide variable number tandem repeat | PER3(5/5) carriers showed increaseddelta activities in NREM sleep and a greater detrimental impact of sleep deprivation. |
| ADA | EEG variations &Multiple Sclerosis & Depression | polymorphism described in previous studies | D8N | Carriers had deeper sleep and were under higher sleep pressure. |
| ADORA2A | EEG variations´ anxiogenic response to caffeine | polymorphism described in previous studies | polymorphisms at 3′ UTR | Power in the high-theta/low-alpha range was invariably enhanced in the carriers in NREM, REM sleep, and wakefulness. Power in the waking EEG was higher in frequency bins between 11.5 – 17.5 Hz. |
| BDNF | EEG variations & autism spectrum disorders | polymorphism described in previous studies | V66M | Carriers showed reduced SWS and decreased spectral power in specific bands at different stages of NREM sleep. |
Fig 2.
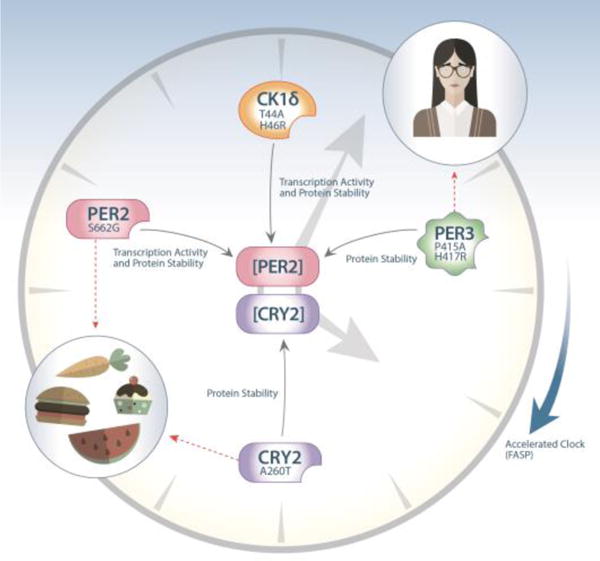
Schematic diagram of the known FASP mutations found in humans. PER2 and CRY2 are core clock components and their protein levels are tightly regulated in a circadian manner to ensure stable clock oscillation. All mutations discovered to date impact the protein levels of these two critical core clock components. The PER2 S662G mutation results in hypophosphorylated of PER2 by casein kinase I (CKI), which in turn causes both increased repressor activity and decreased protein stability. Consistent with this theme, two CK1δ mutations (T44A and H46R) were found to decrease its enzyme activity for substrates including the PER2 S662 site. Thus, these mutations also lead to altered transcription repressor activity and protein stability of PER2. Another FASP mutations, PER3 P415A/H417R, reduces the ability of PER3 to stabilize PER2, resulting in decreased protein stability once again. In addition to this PER2 axis, a more recent finding revealed that the CRY2 A260T mutation increases its accessibility and affinity for ubiquitin E3 ligase, thus promoting its degradation. Both the PER2 and CRY2 mutations have provided insight into the connection between circadian clock and metabolism, while the PER3 mutation offers possible revelation in linking clock and mood regulation. In all the cases mentioned here, the mutations accelerate the endogenous clock, causing mutation carriers to wake up around 4:00 o’clock in the morning. As PERs and CRYs are the major repressive factors in the circadian feedback loop, these genetic findings strongly imply that these repressors play a dominant role in regulating the human sleep wake cycle.
Importantly, some people who have the strong sleep phase advance phenotype do not carry any mutation in known clock genes (unpublished data). Although mutations in (as yet) unidentified (novel) clock genes may be responsible for this phenotype, other possibilities exist. Altered phase in the setting of a normal core clock may result from altering the connection between the environment and the clock system in the body or the coupling of the core clock to physiological outputs. For example, the light entrainment pathway from retina to SCN may be affected by the potential genetic mutation. Consistent with this hypothesis, circadian and phototransduction genes and pathways were enriched in the recent genome-wide association analysis of self-reported morning-ness [12,13]. Therefore, discovery and study of such novel genes with more intensive genetic tools promise to be fruitful.
Duration of sleep
The duration of sleep and response to sleep loss varies among individuals. We are particularly interested in a cohort of individuals known as Natural Short-Sleepers (NSS). These people sleep significantly less than the normal population. Moreover, this phenotype is heritable in some families. Importantly, unlike the patients suffering from sleep disorders, Natural Short-Sleepers are often healthy and free of any apparent detrimental consequence caused by short sleep duration. Previously, we identified a mutation (P384R) in the basic helix-loop-helix family member e41 (BHLHE41 or DEC2) gene that is associated with a human familial natural short sleep phenotype (FNSS) [14]. The habitual self-reported total sleep time (average 6.25 hours) for affected individuals per day was much shorter than the non-carrier controls (average 8.06 hours).Mouse models carrying the exact human mutation largely recapture the short sleep phenotype, further confirming the causative role of the mutation [14]. An independent study from another human cohort found other DEC2missense mutations. One DEC2 mutation (Y362H) occurred in one member of afraternal twin pair who slept one hour less and showed more resistance to sleep deprivation than his mutation-negative twin. The second mutation, identified in three unrelated individuals, resulted in a glutamine (rather than arginine, P384N) substitution at the same codon as our finding, but led to no obvious phenotype [15]. At the molecular level, Y362H and P384R, but not P384N, reduced the ability of DEC2 to suppress CLOCK/BMAL1 transactivation, which suggested a possible underlying mechanism [14,15]. Together, these findings highlight the role of the DEC2 gene in regulation of human sleep duration.
Although several other intriguing candidate genes have been identified, statistical and functional evidence is lacking for many implicated cases [16–19]. Interestingly, ion-channel genes have been shown to regulate duration of sleep or sleep-like behavior in model organisms [20–22], though it is still unclear whether such genes also regulate sleep duration in humans. A plausible case was the ATP-binding cassette sub-family C member 9 (ABCC9) gene which encodes a pore-forming subunit of an ATP-sensitive potassium channel, but the SNPs found by different studies were all intronic [16,17,23].
Recent report from the UK Biobank study suggested two association polymorphisms for sleep duration [13]. One of the polymorphisms is located upstream and the other downstream of the Vaccinia Related Kinase 2 gene. Although intriguing, further validation by both in vitro and in vivo characterizations of these polymorphisms are needed to confirm their roles in sleep duration.
Characteristics of EEG (electroencephalogram) during sleep
The architecture of sleep is defined by EEG based parameters. Although not fully confirmed, it is believed that some characteristics of EEG may reflect the quality or efficiency of sleep. The features of EEG can also be modulated by the genes that regulate the duration and timing of sleep (Table 1). One example is the DEC2 gene mentioned above. DEC2 Y362H mutation carriers showed higher delta power during NREM and less REM sleep compared to the non-carriers [15]. Another example is a polymorphism in PER3 that also influences EEG variability. This polymorphism is a 54-nucleotide variable number tandem repeat (VNTR) in exon 18 of PER3 that encodes 18 amino acids. Approximately 10% of the population is homozygous for the 5-repeat allele PER35/5 [24]. EEG slow wave activity in NREM sleep, theta and alpha activity during wakefulness, and REM sleep were all increased in PER35/5 compared to PER34/4 individuals [25]. In another study of older subjects (55–75 years), PER35/5 carriers also showed increased EEG frontal delta activity and decreased EEG frontal sigma activity during NREM sleep compared with PER34/4 subjects [26]. Furthermore, sleep deprived PER35/5 individuals had elevated sleep homeostatic pressure as measured, physiologically, by EEG slow-wave energy, and showed a greater detrimental impact of sleep deprivation [24,25,27,28]. Thus, different genetic alterations of the PER3 gene give rise to different effects on human sleep. Recent studies also suggested a possible role for PER3 in mood regulation, which provides the first direct molecular genetic evidence for the long suspected connection between sleep and mood [9,29–31]. It will be of interest to probe whether (and how) PER3 serves as a nexus for sleep and mood regulation.
The neuromodulator adenosine is known to contribute to sleep homeostasis [32]. A genetic variant of adenosine deaminase (ADA) D8N, which is associated with the reduced metabolism of adenosine to inosine, specifically enhances deep sleep and SWA during sleep [33,34]. A larger population-based study corroborated some findings that variant carriers have a deeper sleep and are under higher sleep pressure although other details were not identical [35]. In addition, a distinct polymorphism of adenosine A2A receptor (ADORA2A), which occurred in the 3′ UTR region and changed the expression level of this gene, affected EEG during sleep and wakefulness in a non-state-specific manner [33].
Another example is the brain-derived neurotrophic factor (BDNF) gene. A V66M mutation in the encoded protein is linked to impairment of dendritic trafficking and synaptic localization of BDNF and a reduced activity-dependent BDNF secretion [36]. Mutation carriers showed reduced SWS [37] and decreased spectral power in specific bands at different stages of NREM sleep [38]. Thus, BDNF is likely to modulate the electrical activity of the brain, predicting the inter-individual variation of sleep EEG parameters.
Perspectives on Human Genetics in sleep studies
We have focused here on normal variants of human circadian timing and total sleep requirement. These are not diseases per se. Some individuals find it troublesome to wake up in the early morning hours while others feel virtuous for getting up early. Separate from this, there are many primary sleep disorders like restless leg syndrome, obstructive sleep apnea or narcolepsy. In addition, sleep problems also are seen in many disorders that lead to abnormal brain development or degeneration of normal brain. For example, autism spectrum disorder (ASD) is a neurodevelopmental disorder with evidence for strong genetic susceptibility and a high prevalence of insomnia [39].Defects in synaptic pruning during the development of neural circuits disrupt the excitatory/inhibitory balance of synapses, and abnormal synaptic pruning may underlie the atypical neurodevelopment in ASD [40]. Thus, it is intriguing to consider whether sleep problems exacerbate atypical synaptic pruning or if severe neurodevelopmental problem leads to sleep disorders in ASD. Another example is Alzheimer’s disease (AD). Most patients with AD have severe sleep problems but recent evidence suggests that sleep disruption is a major contributing factor to AD. Aβ plaques, the hallmark of AD, are formed by Aβ accumulation. The level of Aβ in brain interstitial space is high while awake and lowest upon awakening from a night of sleep [41,42]. Importantly, sleep disruption abolishes this Aβ reduction, suggesting a neurotoxin clearance function for sleep [43]. These examples highlight the importance of understanding the regulatory mechanisms of sleep and sleep functions. Although much remains to be learned about sleep abnormalities in people with brain disorders, it is likely the case that in general, brain dysfunction leads to alterations in sleep. At the same time, chronic sleep deprivation or desynchrony of the clock from the solar day contributes to development or progression of brain disorders.
One issue for many sleep studies conducted in humans is the use of self-reported phenotypes like sleep duration, timing, etc. Several environmental factors such as drugs, seasonal cycle, modern lifestyle and even lunar cycle may affect human sleep [44–46]. Such factors are not easily controlled and can confound phenotyping. Usually, genetic association studies only provide suggestions that the disease could result from an interaction of environmental factors on a susceptible genetic background. Thus, introducing the genetic mutations into laboratory-housed animals is a powerful approach to test the contribution of a gene to a phenotype. Moreover, in contrast to many well established model organisms, the human population is genetically more heterogeneous, which adds another layer of complexity. In recent years, we and others have mainly focused on single-gene phenotypes, especially those where mutations have dominant effects. However, mutant alleles that segregate as autosomal dominant traits must have a large enough effect to arise on a heterogeneous genetic background. These families with FASP and FNSS are in the ‘tails’ of the normal distributions in general human populations. We speculate that most of the variation in the middle of the normal distribution for human sleep phenotypes results from a combination of many genetic variants of small effect that, together, contribute to each individual’s unique phenotype. More extensive sequencing efforts for exomes or whole genomes in large populations of individuals representing the spectrum of human phenotypes will be necessary to address this hypothesis.
Conclusion
Here we have summarized published genetic mutations discovered to affect the timing, duration and EEG features of human sleep behaviors. Today, the sleep field has expanded its focus from mammalian model organisms to Drosophila, zebrafish, and even worms [47]. Genetic tools in these systems have allowed researchers to undertake large-scale screens to identify new genes for regulation of sleep-like behavior. Such progress has further provided opportunities to probe sleep circuitry and sleep function on a molecular level. Nonetheless, due to the large variation of sleep characteristics among different species and the unique features of human sleep, human genetics will continue to be an indispensable and invaluable source of insight providing critical information on this mysterious phenomenon–sleep.
Highlights.
Human genetics offers essential insight into the characterization of sleep.
Genetic polymorphisms impact sleep timing, duration, and architecture.
A bi-directional relationship exists between sleep and healthy brain.
Acknowledgments
Research in authors’ laboratories are supported by grants from the NIH (HL059596, GM079180, NS072360) to Y.-H.F. and L.J.P
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones CR, Campbell SS, Zone SE, Cooper F, DeSano A, Murphy PJ, Jones B, Czajkowski L, Ptacek LJ. Familial advanced sleep-phase syndrome: A short-period circadian rhythm variant in humans. Nature medicine. 1999;5(9):1062–1065. doi: 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- 2.Gustafson CL, Partch CL. Emerging models for the molecular basis of mammalian circadian timing. Biochemistry. 2015;54(2):134–149. doi: 10.1021/bi500731f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends in cell biology. 2014;24(2):90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowrey PL, Takahashi JS. Genetics of circadian rhythms in mammalian model organisms. Advances in genetics. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288(5468):1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 6.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. An hper2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291(5506):1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptacek LJ. Modeling of a human circadian mutation yields insights into clock regulation by per2. Cell. 2007;128(1):59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH. Functional consequences of a ckidelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434(7033):640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- (*)9.Zhang L, Hirano A, Hsu PK, Jones CR, Sakai N, Okuro M, McMahon T, Yamazaki M, Xu Y, Saigoh N, Saigoh K, et al. A period3 variant causes a circadian phenotype and is associated with a seasonal mood trait. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(11):E1536–1544. doi: 10.1073/pnas.1600039113. This finding suggested that PER3 may be a nexus for sleep and mood regulation while fine-tuning these processes to adapt to seasonal changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (*)10.Hirano A, Shi G, Jones CR, Lipzen A, Pennacchio LA, Xu Y, Hallows WC, McMahon T, Yamazaki M, Ptacek LJ, Fu YH. A cryptochrome 2 mutation yields advanced sleep phase in human. eLife. 2016;5:e16695. doi: 10.7554/eLife.16695. This is the paper that discribed one single mutation in another clock gene CRY2 leads to FASP phenotype in humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan KC, Bates EA, Shapiro RE, Zyuzin J, Hallows WC, Huang Y, Lee HY, Jones CR, Fu YH, Charles AC, Ptacek LJ. Casein kinase idelta mutations in familial migraine and advanced sleep phase. Science translational medicine. 2013;5(183):183ra156, 181–111. doi: 10.1126/scitranslmed.3005784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Y, Shmygelska A, Tran D, Eriksson N, Tung JY, Hinds DA. Gwas of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nature communications. 2016;7:10448. doi: 10.1038/ncomms10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (*)13.Jones SE, Tyrrell J, Wood AR, Beaumont RN, Ruth KS, Tuke MA, Yaghootkar H, Hu Y, Teder-Laving M, Hayward C, Roenneberg T, et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS genetics. 2016;12(8):e1006125. doi: 10.1371/journal.pgen.1006125. This is the most recent GWAS study that suggested several genetic variants that associated with human FASP and FNSS phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y, Jones CR, Fujiki N, Xu Y, Guo B, Holder JL, Jr, Rossner MJ, Nishino S, Fu YH. The transcriptional repressor dec2 regulates sleep length in mammals. Science. 2009;325(5942):866–870. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (**)15.Pellegrino R, Kavakli IH, Goel N, Cardinale CJ, Dinges DF, Kuna ST, Maislin G, Van Dongen HP, Tufik S, Hogenesch JB, Hakonarson H, et al. A novel bhlhe41 variant is associated with short sleep and resistance to sleep deprivation in humans. Sleep. 2014;37(8):1327–1336. doi: 10.5665/sleep.3924. This is an independent report that confirmed the functions of the gene DEC2 in regulating sleep duration and response to sleep deprivation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheinfeldt LB, Gharani N, Kasper RS, Schmidlen TJ, Gordon ES, Jarvis JP, Delaney S, Kronenthal CJ, Gerry NP, Christman MF. Using the coriell personalized medicine collaborative data to conduct a genome-wide association study of sleep duration. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2015;168(8):697–705. doi: 10.1002/ajmg.b.32362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons MJ, Lester KJ, Barclay NL, Nolan PM, Eley TC, Gregory AM. Replication of genome-wide association studies (gwas) loci for sleep in the british g1219 cohort. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2013;162B(5):431–438. doi: 10.1002/ajmg.b.32106. [DOI] [PubMed] [Google Scholar]
- 18.Gottlieb DJ, Hek K, Chen TH, Watson NF, Eiriksdottir G, Byrne EM, Cornelis M, Warby SC, Bandinelli S, Cherkas L, Evans DS, et al. Novel loci associated with usual sleep duration: The charge consortium genome-wide association study. Molecular psychiatry. 2015;20(10):1232–1239. doi: 10.1038/mp.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ollila HM, Kettunen J, Pietilainen O, Aho V, Silander K, Kronholm E, Perola M, Lahti J, Raikkonen K, Widen E, Palotie A, et al. Genome-wide association study of sleep duration in the finnish population. Journal of sleep research. 2014;23(6):609–618. doi: 10.1111/jsr.12175. [DOI] [PubMed] [Google Scholar]
- (**)20.Tatsuki F, Sunagawa GA, Shi S, Susaki EA, Yukinaga H, Perrin D, Sumiyama K, Ukai-Tadenuma M, Fujishima H, Ohno R, Tone D, et al. Involvement of ca(2+)-dependent hyperpolarization in sleep duration in mammals. Neuron. 2016;90(1):70–85. doi: 10.1016/j.neuron.2016.02.032. This work highlighted a group of ion channels in regulating sleep duration in animal models. [DOI] [PubMed] [Google Scholar]
- 21.Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. Identification of sleepless, a sleep-promoting factor. Science. 2008;321(5887):372–376. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in drosophila shaker mutants. Nature. 2005;434(7037):1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 23.Allebrandt KV, Amin N, Muller-Myhsok B, Esko T, Teder-Laving M, Azevedo RV, Hayward C, van Mill J, Vogelzangs N, Green EW, Melville SA, et al. A k(atp) channel gene effect on sleep duration: From genome-wide association studies to function in drosophila. Molecular psychiatry. 2013;18(1):122–132. doi: 10.1038/mp.2011.142. [DOI] [PubMed] [Google Scholar]
- 24.Dijk DJ, Archer SN. Period3, circadian phenotypes, and sleep homeostasis. Sleep medicine reviews. 2010;14(3):151–160. doi: 10.1016/j.smrv.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Viola AU, Archer SN, James LM, Groeger JA, Lo JC, Skene DJ, von Schantz M, Dijk DJ. Per3 polymorphism predicts sleep structure and waking performance. Current biology : CB. 2007;17(7):613–618. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 26.Viola AU, Chellappa SL, Archer SN, Pugin F, Gotz T, Dijk DJ, Cajochen C. Interindividual differences in circadian rhythmicity and sleep homeostasis in older people: Effect of a per3 polymorphism. Neurobiology of aging. 2012;33(5):1010, e1017–1027. doi: 10.1016/j.neurobiolaging.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 27.Goel N, Banks S, Mignot E, Dinges DF. Per3 polymorphism predicts cumulative sleep homeostatic but not neurobehavioral changes to chronic partial sleep deprivation. PloS one. 2009;4(6):e5874. doi: 10.1371/journal.pone.0005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maire M, Reichert CF, Gabel V, Viola AU, Strobel W, Krebs J, Landolt HP, Bachmann V, Cajochen C, Schmidt C. Sleep ability mediates individual differences in the vulnerability to sleep loss: Evidence from a per3 polymorphism. Cortex; a journal devoted to the study of the nervous system and behavior. 2014;52:47–59. doi: 10.1016/j.cortex.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Dallaspezia S, Locatelli C, Lorenzi C, Pirovano A, Colombo C, Benedetti F. Sleep homeostatic pressure and per3 vntr gene polymorphism influence antidepressant response to sleep deprivation in bipolar depression. Journal of affective disorders. 2016;192:64–69. doi: 10.1016/j.jad.2015.11.039. [DOI] [PubMed] [Google Scholar]
- 30.Viena TD, Gobin CM, Fins AI, Craddock TJ, Tartar A, Tartar JL. A per3 polymorphism interacts with sleep duration to influence transient mood states in women. Journal of circadian rhythms. 2016;14:3. doi: 10.5334/jcr.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karthikeyan R, Marimuthu G, Ramasubramanian C, Arunachal G, BaHammam AS, Spence DW, Cardinali DP, Brown GM, Pandi-Perumal SR. Association of per3 length polymorphism with bipolar i disorder and schizophrenia. Neuropsychiatric disease and treatment. 2014;10:2325–2330. doi: 10.2147/NDT.S73765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang ZL, Urade Y, Hayaishi O. The role of adenosine in the regulation of sleep. Current topics in medicinal chemistry. 2011;11(8):1047–1057. doi: 10.2174/156802611795347654. [DOI] [PubMed] [Google Scholar]
- 33.Retey JV, Adam M, Honegger E, Khatami R, Luhmann UF, Jung HH, Berger W, Landolt HP. A functional genetic variation of adenosine deaminase affects the duration and intensity of deep sleep in humans. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15676–15681. doi: 10.1073/pnas.0505414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachmann V, Klaus F, Bodenmann S, Schafer N, Brugger P, Huber S, Berger W, Landolt HP. Functional ada polymorphism increases sleep depth and reduces vigilant attention in humans. Cerebral cortex. 2012;22(4):962–970. doi: 10.1093/cercor/bhr173. [DOI] [PubMed] [Google Scholar]
- 35.Mazzotti DR, Guindalini C, de Souza AA, Sato JR, Santos-Silva R, Bittencourt LR, Tufik S. Adenosine deaminase polymorphism affects sleep eeg spectral power in a large epidemiological sample. PloS one. 2012;7(8):e44154. doi: 10.1371/journal.pone.0044154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu B, Nagappan G, Guan X, Nathan PJ, Wren P. Bdnf-based synaptic repair as a disease-modifying strategy for neurodegenerative diseases. Nature reviews Neuroscience. 2013;14(6):401–416. doi: 10.1038/nrn3505. [DOI] [PubMed] [Google Scholar]
- 37.Bachmann V, Klein C, Bodenmann S, Schafer N, Berger W, Brugger P, Landolt HP. The bdnf val66met polymorphism modulates sleep intensity: Eeg frequency-and state-specificity. Sleep. 2012;35(3):335–344. doi: 10.5665/sleep.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guindalini C, Mazzotti DR, Castro LS, D’Aurea CV, Andersen ML, Poyares D, Bittencourt LR, Tufik S. Brain-derived neurotrophic factor gene polymorphism predicts interindividual variation in the sleep electroencephalogram. Journal of neuroscience research. 2014;92(8):1018–1023. doi: 10.1002/jnr.23380. [DOI] [PubMed] [Google Scholar]
- 39.Veatch OJ, Maxwell-Horn AC, Malow BA. Sleep in autism spectrum disorders. Current sleep medicine reports. 2015;1(2):131–140. doi: 10.1007/s40675-015-0012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas MS, Davis R, Karmiloff-Smith A, Knowland VC, Charman T. The over-pruning hypothesis of autism. Developmental science. 2016;19(2):284–305. doi: 10.1111/desc.12303. [DOI] [PubMed] [Google Scholar]
- 41.Roh JH, Huang Y, Bero AW, Kasten T, Stewart FR, Bateman RJ, Holtzman DM. Disruption of the sleep-wake cycle and diurnal fluctuation of beta-amyloid in mice with Alzheimer’s disease pathology. Science translational medicine. 2012;4(150):150ra122. doi: 10.1126/scitranslmed.3004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y, Potter R, Sigurdson W, Santacruz A, Shih S, Ju YE, Kasten T, Morris JC, Mintun M, Duntley S, Bateman RJ. Effects of age and amyloid deposition on abeta dynamics in the human central nervous system. Archives of neurology. 2012;69(1):51–58. doi: 10.1001/archneurol.2011.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothman SM, Herdener N, Frankola KA, Mughal MR, Mattson MP. Chronic mild sleep restriction accentuates contextual memory impairments, and accumulations of cortical abeta and ptau in a mouse model of Alzheimer’s disease. Brain research. 2013;1529:200–208. doi: 10.1016/j.brainres.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cajochen C, Altanay-Ekici S, Munch M, Frey S, Knoblauch V, Wirz-Justice A. Evidence that the lunar cycle influences human sleep. Current biology: CB. 2013;23(15):1485–1488. doi: 10.1016/j.cub.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 45.Yetish G, Kaplan H, Gurven M, Wood B, Pontzer H, Manger PR, Wilson C, McGregor R, Siegel JM. Natural sleep and its seasonal variations in three pre-industrial societies. Current biology: CB. 2015;25(21):2862–2868. doi: 10.1016/j.cub.2015.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allebrandt KV, Teder-Laving M, Kantermann T, Peters A, Campbell H, Rudan I, Wilson JF, Metspalu A, Roenneberg T. Chronotype and sleep duration: The influence of season of assessment. Chronobiology international. 2014;31(5):731–740. doi: 10.3109/07420528.2014.901347. [DOI] [PubMed] [Google Scholar]
- 47.Sehgal A, Mignot E. Genetics of sleep and sleep disorders. Cell. 2011;146(2):194–207. doi: 10.1016/j.cell.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


