Summary
Human pluripotent stem cells (PSCs) provide an unlimited cell source for cell therapies and disease modeling. Despite their enormous power, technical aspects have hampered reproducibility. Here, we describe a modification of PSC workflows that eliminates a major variable for nearly all PSC experiments: the quality and quantity of the PSC starting material. Most labs continually passage PSCs and use small quantities after expansion, but the “just-in-time” nature of these experiments means that quality control rarely happens before use. Lack of quality control could compromise PSC quality, sterility, and genetic integrity, which creates a variable that might affect results. This method, called CryoPause, banks PSCs as single-use, cryopreserved vials that can be thawed and immediately used in experiments. Each CryoPause bank provides a consistent source of PSCs that can be pre-validated before use to reduce the possibility that high levels of spontaneous differentiation, contamination, or genetic integrity will compromise an experiment.
Keywords: directed differentiation, human pluripotent stem cell, cell therapy, disease modeling, cryopreservation, cell banking, quality control
Highlights
-
•
CryoPause is a new way to perform PSC-based experiments that enhances reproducibility
-
•
Large batches of single-use, cryopreserved cells are used directly post thaw
-
•
Directed differentiation and gene editing can be done without expansion
-
•
Repeated experiments can be done from the same PSCs over time and geographical place
PSC work has traditionally been performed by passaging cells over time while retaining a portion of culture to perform work. Such extended culture creates confounding variables that can skew results. Tomishima and colleagues demonstrate that PSCs can be biobanked as “ready-to-use” aliquots so that PSCs can be thawed before immediate use in directed differentiation or gene editing experiments.
Introduction
Human pluripotent stem cells (hPSCs) are revolutionizing disease modeling and cell replacement therapies. PSCs are now routinely made from patient samples, and genetic alterations suspected to cause disease can be changed at will to explore genotype-phenotype relationships. These technologies are bridging genome-wide associations to causation and provide an unprecedented view into disease mechanisms when combined with our increasingly sophisticated ability to direct PSC differentiation, a necessary step in providing a disease-relevant context. PSCs can also be directed to clinically relevant cell types to provide an unlimited resource for cell replacement therapies. While PSCs are powerful, many practical aspects of hPSC maintenance and differentiation could be improved.
One practical complication in disease-modeling experiments is maintaining hPSC lines while simultaneously directing their differentiation. In induced PSC (iPSC) disease-modeling studies, “best practices” require repeatedly expanding and differentiating many iPSC lines in parallel, a process we define here as “continuous passage.” Synchronizing PSCs is challenging since different lines often expand at different rates. Another complication is that each PSC line can spontaneously differentiate during continuous passage, so phenotypes might reflect the disease state or suboptimal PSC culture. Continuous passage increases the risk of contamination with other cell lines or microorganisms and likely selects for genetic alterations that increase cell division or replating efficiency. The workload associated with continuous passage and parallel differentiation of multiple iPSC lines also increases the chance of human error. A method capable of separating PSC expansion from the differentiation would reduce the workload, allow each iPSC line to be passaged and harvested at an optimal time for the line, permit the initiation of differentiations at a convenient time, and provide time to perform proper quality control before differentiation experiments are initiated. Multiple experiments could be initiated from the same banks of PSCs, increasing reproducibility.
Continuous passage could create an even larger problem for cell therapies. PSCs are banked under good manufacturing practices (GMP) before a battery of expensive tests that verify the integrity and sterility of the cell bank. After validation, banked PSCs are thawed and expanded before initiating differentiation to transform the bank into a clinically relevant cell type. Thawing, expansion, and PSC passage create a major variable and increased complexity during manufacturing. This step can change the timing, yield, and quality of PSCs going into the differentiation process.
Here, we report a new workflow called CryoPause (hereafter referred to as CP: Figure 1). In this method, PSCs are dissociated into single cells and banked as ready-to-use aliquots. Viability post thaw was routinely greater than 90% and no significant difference in stem cell markers, the transcriptome, or epigenetic status post thaw was found. Remarkably, CP cells can be thawed and directly differentiated or genetically modified without recovery and expansion. The CP method provides a simple new way for any laboratory to use PSCs that will increase reproducibility, help synchronize different PSC lines before differentiation, and eliminate the possibility of genetic instability and contamination during expansion of PSCs for cell therapy and disease-modeling applications.
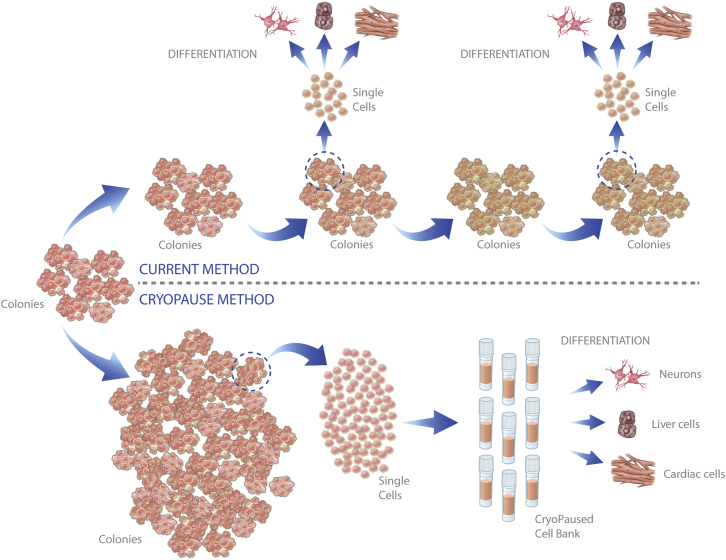
Figure 1.
The CryoPause Method Compared with Conventional PSC Culture
(Top) The conventional (control) workflow recovers colonies from cryopreservation and expands them over long periods of time, periodically using a portion of the culture for specific applications such as directed differentiation into a cell type of interest. Over time, PSCs might acquire genetic changes, contamination, or changes in the amount of spontaneous differentiation, any of which could affect results. (Bottom) CryoPause expands a large pool of PSCs over the least number of passages possible. The large batch is then dissociated into a single-cell suspension before cryopreservation. The freezing process separates the production of PSCs from their use, allowing time to perform proper quality control and characterization of each bank. It also permits the use of identical cells in multiple experiments, and allows shipping anywhere in the world so that other laboratories can initiate independent experiments with the exact same starting population of PSCs.
Results
Developing CryoPause Conditions
Eliminating post-thaw expansion to improve the reproducibility of experiments would require high viability upon thawing to be practical. WA09 cells expanded in Essential 8 medium (E8; Chen et al., 2011) were treated with Accutase to create a single-cell suspension before washing and resuspending in FreSR-S, a commercial medium designed for monodispersed hPSC cryopreservation. Cells were cryopreserved in a controlled-rate freezer using a standard program before long-term storage in liquid nitrogen as “ready-to-use” aliquots (see Experimental Procedures). Pilot experiments using this method demonstrated surprisingly high post-thaw viability. The data indicate that feeder-free culture was the main determinant of the high viability since different PSC lines (Figure S1A), different cryopreservation media (Figure S1B), and conventional slow-rate freezing techniques (Figure S1C) did not appreciably reduce viability. The main factor appeared to be the initial culture conditions since WA09 cells expanded using traditional feeder-based methods had lower viability, although this viability decrease was mitigated in FreSR-S (Figure S1D).
For the balance of the experiments, WA09 cryopreserved in FreSR-S in a controlled-rate freezer was the baseline CP condition unless otherwise noted. This condition routinely yielded post-thaw viability that equaled that of cells before freezing when measured with an automated cell counter using acridine orange/propidium iodide (AOPI) (Figures 2A and 2B). After the creation of multiple banks, it became clear that the viability post thaw was only limited by the input culture's viability before CP (Figure 2B). No loss in viability or pluripotency was observed for up to 1 year of storage in liquid nitrogen, the longest time point tested to date (Figure S1E). Despite exceptional post-thaw viability, a slight decrease in plating efficiency was observed after 24 hr of culture (data not shown), so more CP cells were plated to compensate for this difference in efficiency (400,000 cells/cm2 in CP to 200,000 cells/cm2 in fresh controls, hereafter referred to as control cells).
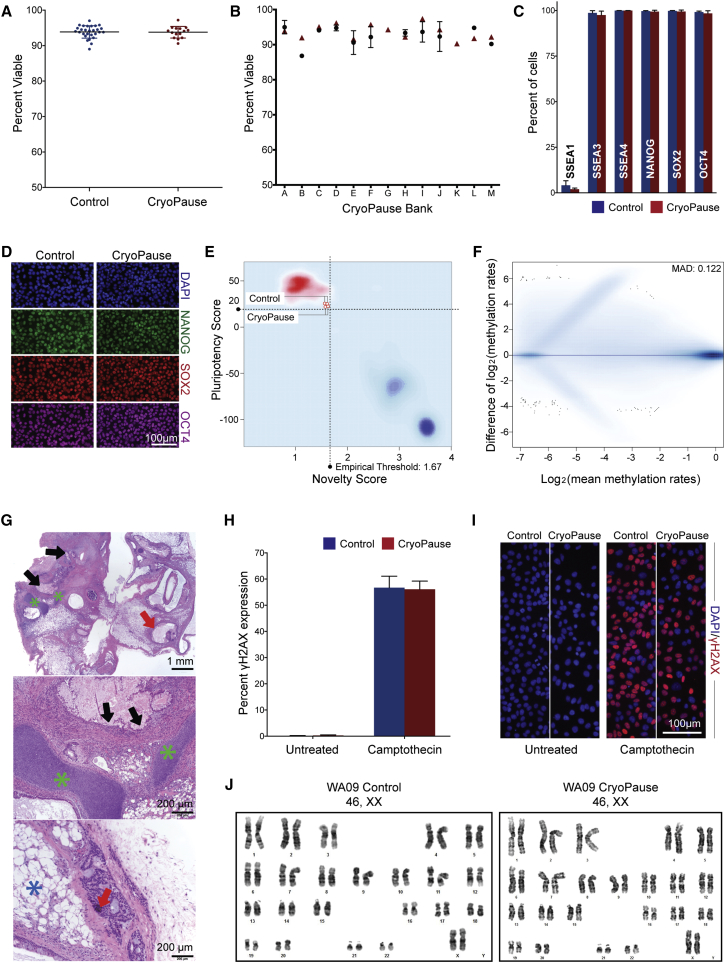
Figure 2.
Validation of CryoPaused PSCs
(A) Post-thaw viability of control and CryoPaused WA09 cells. Viability was measured on an automated cell counter using acridine orange (live) and propidium iodide (dead) fluorescence (n = 15 for CP and n = 28 for control).
(B) Viability before freezing (red triangle) and after thawing (black circles) on 13 independent CryoPaused PSC cell banks.
(C and D) Stem cell marker expression (or spontaneous differentiation, SSEA-1) in control or CryoPaused cells by flow cytometry (C, n = 3; values for independent biological replicates shown as mean ± SD) or immunofluorescence (D).
(E) PluriTest assay to assess pluripotency of control and CryoPaused cells. Control (n = 2) or CryoPaused (n = 2) WA09 cultures were used to obtain RNA before hybridization to a microarray. The array data was processed using the PluriTest algorithm, and each sample is plotted as two related parameters, the Pluripotency score and the Novelty score. All four samples passed PluriTest's assessment of pluripotency. Red cloud is representative of samples that passed the PluriTest while the blue cloud shows samples that failed the PluriTest.
(F) MA plot evaluating the methylation of control and CryoPaused cells. Methylation values for complementary CpG sites (one base apart on opposite strands) were combined to generate CpG-unit methylation. A minimum threshold coverage of ten reads was used to filter CpG-units resulting in 3,714,418 and 3,408,158 CpG units for control and CryoPause samples, respectively. Agreement of methylation levels was evaluated by median absolute deviation (MAD), a robust measure of variability insensitive to outliers that estimates the statistical dispersion in methylation levels of the 3,155,482 common CpG units covered by both samples.
(G) CryoPaused cells are competent to produce teratomas. For all panels, red arrows point to ectoderm, green and blue asterisks denote mesoderm, and black arrows point to endoderm. (Top) Low-magnification image showing an H&E-stained teratoma section derived from CryoPaused WA09 hESCs. Scale bar, 1 mm. (Middle) High-magnification image showing regions containing endoderm (goblet cells, black arrows) and mesoderm (green asterisks). Scale bar, 200 μm. (Bottom) High-magnification image showing ectoderm (melanotic neurectoderm, red arrow) and mesoderm (adipocytes, blue asterisk). Scale bar, 200 μm.
(H) Quantification of DNA damage in control and CryoPaused cells as measured by γH2AX expression on a high content microscope (left) and after treatment with 0.5 μM camptothecin for 1 hr as a positive control for the assay and to test for vulnerability to DNA damage (right, n = 3; values for independent biological replicates shown as mean ± SD).
(I) Representative immunofluorescence of γH2AX expression in control and CryoPaused cells with and without treatment with 0.5 μM camptothecin for 1 hr.
(J) Chromosome analysis was performed on 20 DAPI-banded metaphases, all of which were fully karyotyped. Both control (20/20) and CryoPaused cells (20/20) had a normal 46,XX karyotype.
In (A), (C), and (H), Wilcoxon's signed-rank test was performed with at least three independent experiments, and no statistical difference (p > 0.05) was found between control and CryoPause. See also Figure S1.
Validating CryoPause Cells
To assess the health of CP cells, we examined PSC markers 1 day after plating when directed differentiation would normally be induced. There was no statistical difference in the PSC markers SSEA3, SSEA4, OCT4, SOX2, and NANOG when measured with flow cytometry between control and CP cells (Figure 2C), nor was there an increase in the spontaneous differentiation marker SSEA1 (Figure 2C). Flow results were independently confirmed by immunofluorescence analysis with no discernible difference between the two populations (Figure 2D).
To perform a comprehensive examination of the transcriptome in CP PSCs, we used PluriTest to compare control and CP cultures (Müller et al., 2011, Müller et al., 2012, Williams et al., 2011). PluriTest is based on whole-genome transcriptional profiles and allows for the reliable assessment of pluripotency and spontaneous differentiation in undifferentiated stem cell cultures. In brief, PluriTest analyzes the expression of a large number of pluripotency-associated transcripts with a “Pluripotency score” and tests for the conformity of a tested sample with global transcriptional patterns typically observed in genetically and epigenetically normal hPSCs with a second metric, termed “Novelty score.” Both control and CP WA09 samples pass the empirically defined Pluripotency and Novelty score thresholds and demonstrate fit to the Novelty one-class classifier model, which indicates that both sample groups show gene expression patterns highly similar to those observed in well-characterized human embryonic stem cell (hESC) and induced hPSC cell lines (Figure 2E). The epigenetic landscape of CP cells was also surveyed through reduced representation bisulfide sequencing (RRBS), and strong agreement was found in methylation levels of common CpG units between control and CP cells (Figure 2F; median absolute deviation = 0.122), which is comparable with the concordant methylation levels observed among technical replicates (Kacmarczyk et al., 2016).
Another measure of pluripotency is the creation of teratomas. Control and CP cultures created teratomas with roughly the same size and kinetics (Figure S2). H&E-stained sections of CP teratomas showed derivatives of all three germ layers (Figure 2G: endoderm [black arrows], mesoderm [green and blue asterisks], and ectoderm [red arrows]). Collectively these data demonstrate that there is no difference in the expression of key pluripotency markers by flow cytometry, PluriTest, CpG methylation, and teratoma formation using CP cells directly post thaw.
To address the possibility that CP cultures contain more cells with genetic damage post thaw, we measured DNA damage by performing quantitative immunofluorescence of nuclear γH2AX using a high content microscope and found no statistical difference in the percentage of cells expressing γH2AX (Figures 2H and 2I; for review, see Redon et al., 2002). To validate the assay and determine whether CP cells were more vulnerable to DNA-damaging agents, we added camptothecin, a cytotoxic quinolone alkaloid, to the cells. There was no statistical difference in the percentage of cells expressing γH2AX after camptothecin exposure. Finally, karyotype analysis was performed on CP cells post thaw to test for global genetic abnormalities. Normal karyotypes were found in control (20 of 20 metaphase spreads) and CP cultures (20 of 20 metaphase spreads) (Figure 2J).
Directed Differentiation and Cell Therapies
Because there was little difference between control and CP, the in vitro differentiation capacity of CP cells was assessed. Control and CP WA09 cells were directed to a neural cell fate by dual SMAD inhibition (Chambers et al., 2009). OCT4 and PAX6 were measured over time with immunofluorescence and flow cytometry to assess the efficiency and kinetics of neural differentiation. There was no difference in the timing or the extent of neural induction (Figures 3A–3C). CP cells directed to mesendoderm fates were also indistinguishable in the extent and kinetics of differentiation (Figures 3D–3F). Both of these cell fates are relatively immature and therefore easier to make, so we attempted to make midbrain dopamine neurons from CP cells, a cell type that our group manufactured in clinically compatible conditions as part of Lorenz Studer's NYSTEM consortium group (Barker et al., 2015). WA09 CP cells efficiently created FOXA2/TH double-positive, post-mitotic midbrain dopamine neurons from a clinically compatible “standard operating procedure” and display a gene expression profile similar to that of neurons derived from control cells (Figures 3I and 3J).
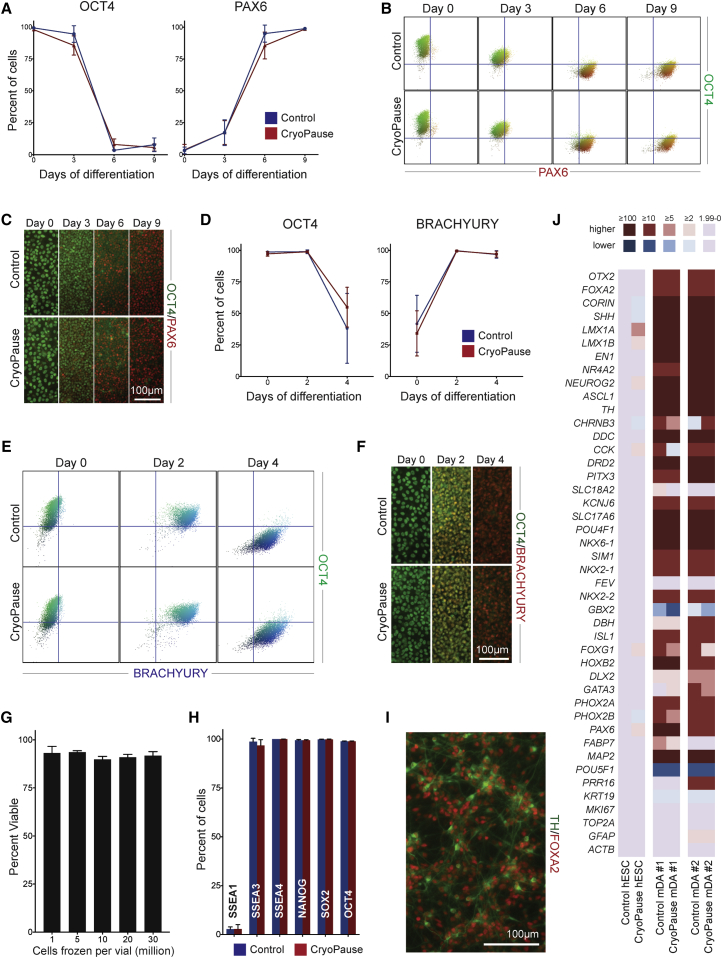
Figure 3.
The Kinetics and Extent of Directed Differentiation of CryoPaused Cells
(A) OCT4 and PAX6 flow-cytometry quantification during neural induction (n = 5; values for independent biological replicates shown as mean ± SD).
(B and C) Representative flow cytometry (B) and immunofluorescence (C) of OCT4 and PAX6 expression at 0, 3, 6, and 9 days after neural induction.
(D) OCT4 and Brachyury expression quantified by flow cytometry during mesendodermal induction (n = 3; values for independent biological replicates shown as mean ± SD).
(E and F) Representative flow cytometry (E) and immunofluorescence (F) at 0, 2, and 4 days after mesendoderm induction.
(G) Viability of CryoPaused cells post thaw when frozen at 1, 5, 10, 20, and 30 million cells/mL. Percentage of viable cells was determined on an automated cell counter with acridine orange (live) and propidium iodide (dead) fluorescence (n = 3; values for independent biological replicates shown as mean ± SD).
(H) Stem cell marker expression (or spontaneous differentiation, SSEA-1) measured by flow cytometry in control and CryoPaused cells expanded in a Cell Factory (n = 3; values for independent biological replicates shown as mean ± SD).
(I) FOXA2 (red) and TH (green) after midbrain dopamine neuron differentiation of CryoPaused cells.
(J) Gene expression of control and CryoPaused WA09 hESC and midbrain dopamine neurons. Samples were normalized to WA09 control hESCs, and the fold change of expression was color coded. Higher levels of expression relative to control hESCs are shown in red, and lower levels are shown in blue.
In (A), (D), (G), and (H), Wilcoxon's signed-rank test was performed with at least three independent experiments, and no statistical difference (p > 0.05) was found between control and CryoPause.
The manufacturer specifies a cell density of 1 million cells/mL when cryopreserving with FreSR-S. This density is adequate for smaller-scale experiments but becomes limiting for applications requiring large cell numbers such as cell therapies, whereby billions of cells are needed per production run. Therefore, the effect of cell density during cryopreservation was assayed. No obvious difference in the viability (Figure 3G) or plating efficiency (data not shown) was found when freezing cells up to a density of 30 million/mL, the highest tested. High-density preparations should provide a reasonable workflow for most therapeutic applications that usually require billions of input PSCs before manufacturing.
The strength of CP is the creation of large, characterized, ready-to-use cell banks. While there are many advantages to our strategy, the disadvantage is that it requires all PSC expansion to be performed at once prior to cryopreservation and quality control. In an effort to scale expansion, “cell factories” were employed: conjoined flasks that allow easy feeding of all layers at one time that can easily be adapted to become a closed system with automation. PSCs were expanded using the 4-layer flask size (2,528 cm2, or around 44 × 6-well plates), and had an average yield of ∼250 million cells per factory (n = 3). A complication of growth in such “factories” is that the morphology is difficult to monitor during expansion since the layered flasks do not permit direct microscopic observation. To verify that CP banks were expanded correctly, we tested PSC markers and found that factory-expanded cells had equivalent marker expression (Figure 3H) and were capable of differentiation (data not shown).
Genetic Modification
Because CP cells behaved normally in directed differentiation, we tested their ability to be genetically manipulated immediately post thaw. The nucleofection efficiency of CP WA09 cells upon thawing was comparable with that of control cells. Twenty-four hours post nucleofection with a GFP plasmid, fluorescent microscopy revealed GFP+ cells in both conditions, and flow cytometry quantitation revealed that >85% expressed GFP (Figures 4A–4C). Directly post thaw CP cells were also successfully transduced by a Sendai viral vector expressing EmGFP, as shown by fluorescence microscopy 24 hr post transduction (Figure 4D). CP-derived, Sendai-transduced subclones could be propagated for at least 15 passages, the longest tested.
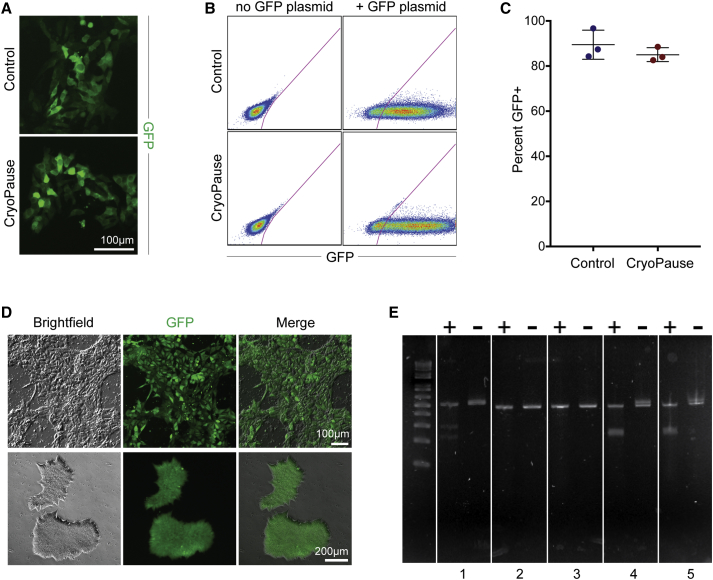
Figure 4.
Genetic Modification of CryoPaused Cells
(A and B) Representative immunofluorescence (A) and flow cytometry (B) of GFP expression in control and CryoPaused cells 24 hr after nucleofection with a GFP plasmid.
(C) GFP expression quantified by flow cytometry in control and CryoPaused cells 24 hr after nucleofection with GFP plasmid (n = 3; values for independent biological replicates shown as mean ± SD).
(D) Representative immunofluorescence of GFP expression in CryoPaused cells after transduction with Sendai virus vector containing EmGFP. Individual subclones from initial transduction could be maintained as GFP+ colonies for at least ten passages.
(E) Agarose gel analysis of cleavage products after using the a genomic cleavage detection kit with HPRT guide RNA in CryoPaused WA01 iCRISPR cells. + or − indicates with or without detection enzyme, respectively. (1) Positive control; (2) iCRISPR cells with Cas9 induced prior to CryoPausing but without guide RNA; (3) iCRISPR cells without Cas9 induction but with HPRT guide RNA; (4) iCRISPR cells with Cas9 induction and HPRT guide RNA; (5) iCRISPR cells (without CryoPausing) with Cas9 induction and HPRT guide RNA.
In (C), Wilcoxon's signed-rank test was performed with three independent experiments, and no statistical difference (p > 0.05) was found between control and CryoPause.
Nucleofection success suggested that CP cells might be able to undergo genome modification upon thawing. To test targeted genome modification, we used PSCs that contain inducible Cas9 engineered into the AAVS1 locus (iCRISPR; González et al., 2014). WA01 (H1) iCRISPR cells were expanded and then treated with doxycycline 24 hr before CP: this created CP cells that pre-expressed Cas9 before cryopreservation. CP-Cas9+ iCRISPR cells were thawed and nucleofected with HPRT guide RNA immediately post thaw. There were no obvious differences in the efficiency of HPRT-targeted mutations between control and CP iCRISPR WA01 (H1) cells (Figure 4E).
Discussion
We describe CryoPause, a new method that eliminates a critical variable for most PSC-based applications: the nature of pluripotent cells before differentiation or genomic modification. It is commonly accepted that cryopreserved hPSCs require recovery, expansion, and passage before use. While this was traditionally required, technical improvements allowed us to challenge this conventional wisdom here. The data show that dissociated hPSCs can be cryopreserved as a single-cell suspension with almost no loss in post-thaw viability and a slight reduction in plating efficiency when compared with parallel “fresh” cells that were not frozen. The main technical driver enabling this paradigm change is the culture system: E8-expanded cells had a higher viability using a number of cryopreservation paradigms (Liu and Chen, 2014; our unpublished data), and others have shared that mTeSR1 also gives very high post-thaw recovery (J. Moore, personal communication). It is likely that many feeder-free culture conditions can support CP. Clusters of PSCs grown under these conditions also give very high recovery rates and likely could also be adapted for many applications (Liu and Chen, 2014; data not shown).
CP provides a number of advantages compared with conventional PSC culture. Disease-modeling studies are best done with multiple iPSC clones derived from numerous healthy and diseased individuals. The conventional parallel culture method is labor intensive and time consuming, since maintenance of multiple lines are necessarily done in parallel with directed differentiations to provide a continuous source of fresh starting material for experiments. iPSC lines that expand at different rates complicate the synchronous initiation of differentiation and parallel passage, usually resulting in a compromise that maximizes the number of cultures that are ready at a point in time: the remainder are often under- or overexpanded. Continuous passage also increases the risk of cross-contamination of cell lines, the accidental introduction of microorganisms during experiments, or the use of cells that acquire a genomic abnormality during extended culture. CP separates the work in PSC expansion from the differentiation experiments. It permits repeated differentiations from an identical pool of PSCs, eliminating variability in the PSC preparation. A full constellation of quality control criteria such as PSC marker status, genetic integrity, sterility, and cell line authentication can validate each bank before use. Most laboratories currently perform “spot checks” during use or perhaps before the serial passage even begins. The variable of “just-in-time” PSC workflows almost certainly reduces the robustness and reproducibility of nearly all PSC applications. It can also be inconvenient, since it complicates when a differentiation can be initiated due to uncertainty in the rate of PSC expansion.
The advantages of CP could be even more profound for manufacturing cell therapies. In a typical cell therapy workflow, hPSCs are expanded and banked in a GMP facility before undergoing expensive and time-consuming tests to validate the cell bank. The conversion of this PSC bank into a therapeutically useful cell type usually requires recovery from the cryopreserved state and a limited number of cell passages before initiating differentiation into the therapeutic cell type. This creates the possibility of initiating the differentiation of a cell bank with PSCs in a suboptimal state, potentially limiting reproducibility and product yield. Manufacturing runs can be exorbitantly expensive in time and money and could potentially cause adverse events in patients. Reproducibility of manufacturing is also one of the key attributes that regulatory authorities examine when assessing a cellular product's safety for human use. Our laboratory recently led the manufacture of a clinically compatible midbrain dopamine neuron product (MSK-DA01) intended for a phase 1 clinical trial after investigational new drug-enabling studies that are currently ongoing (Lorenz Studer's NYSTEM consortium group; Barker et al., 2015). Four at-scale batches of our product were manufactured, and while all met release criteria, the PSC expansion caused challenges in the timing of production and limited scale during particular runs due to the variable yield and timing during expansion. Timing, yield, and quality of PSC expansion can be completely eliminated as variables for cell therapies if CP can be validated for such applications. We show here that CP WA09 cells can be directed to midbrain dopamine neurons using our clinically compatible SOP, but more work is needed before advancing this method to the clinic. If validated, there seems little doubt that manufacturing cell therapies from PSCs will become more robust and reproducible. CP will also be invaluable for “process development,” the systematic exploration of “cleaner” components needed to differentiate cells for clinical use.
One large complication in using PSC derivatives in some cell replacement therapies is matching the human leukocyte antigen (HLA) status of cells to patients. Autologous iPSCs are one strategy to create patient-matched cells and avoid immune mismatches, but there are considerable practical and potential safety complications with this strategy. There are now multiple large-scale efforts to bank large numbers of PSC lines so that they can be carefully quality controlled before clinical use yet still provide a close HLA match to a specific patient for allogeneic transplantation. In this scenario, the use of CP could simplify the procedures used to convert many different PSCs into clinically useful cells. CP could enable a future where ready-to-use, cryopreserved, quality-controlled, HLA-matched PSCs are ordered from a central repository and used directly in local differentiation production runs to derive HLA-matched cells for a patient in need.
PSCs are beginning to transcend our field and are becoming widely used throughout developmental biology, human genetics, and drug discovery. Many laboratories from different disciplines are entering PSC disease modeling, and removing the “art” of PSC culture could help laboratories that lack PSC expertise but are experts in disease pathogenesis. CP could enable biobanks to distribute “ready-to-use” CP aliquots for direct use in differentiations, eliminating the expertise and expense required to expand and cryopreserve local banks.
In summary, we demonstrate a simple change in workflow that eliminates a significant variable to all PSC-based applications. Nearly every laboratory using PSCs could implement this strategy to produce better-controlled, more robust PSC-based experiments, although there are tradeoffs: there is a marginal increase in cost due to reduced plating efficiency, increased workload associated with growing large batches of cells at lower passage, and increased requirement for liquid nitrogen space. Individual laboratories will need to assess the relative merits of increased reproducibility and quality control versus the increase in resources required.
Experimental Procedures
Human Pluripotent Stem Cell Maintenance
The human embryonic stem cell lines WA09 (H9) and WA01 iCRISPR, and the iPSC lines 153.3A and 960.1B were initially maintained in Essential 8 (E8, Thermo Fisher, #A1517001) medium on Geltrex (Thermo Fisher, #A1413202) diluted 1:50 in DMEM/F12 (Thermo Fisher, #11330032), and passaged as clusters every 3–4 days using brief (3 min) 0.05% trypsin-EDTA (Thermo Fisher, #25300054) treatment before scraping (to maintain colony structure). Clusters were washed twice with fresh E8 medium before replating with 10 μM Y-27632 for 24 hr. Cells were used between passages 30 and 55 and no abnormal karyotypes were found. All cell counts and viability were determined using the Nexcelom Cellometer K2 automated cell counter.
Production of CryoPaused Cells
To create CP cell banks, we dissociated PSCs grown in E8 medium with Accutase (Innovative Cell Technologies, #AT-104) for 30 min in a 37°C incubator. Cells were washed with 2 volumes of E8 (relative to the cells/Accutase) and centrifuged to pellet cells (200 × g, room temperature for 5 min). The supernatant was aspirated and the rinse was repeated. Cells were finally resuspended in FreSR-S (Stem Cell Technologies, #05859) at 10 million cells/mL unless otherwise indicated. The FreSR-S/cell mixture was added to pre-chilled cryotubes before freezing in a controlled-rate freezer (see program in Supplemental Experimental Procedures).
Flow Cytometry and Immunofluorescence Analysis for Stem Cell Markers
The Human Pluripotent Stem Cell Sorting and Analysis Kit (BD Biosciences, #560461) and the Human Pluripotent Transcription Factor Analysis Kit (BD Biosciences, #560589) were used as per the manufacturer's protocols to quantify stem cell markers on a BD FACSAria III. For immunofluorescence staining, the following primary antibodies were used: NANOG (1:200, BD Biosciences, #560482); OCT4 (1:200, Santa Cruz Biotechnology, #sc-9081); and SOX2 (1:100, R&D Systems, #AF2018). The appropriate Alexa Fluor-conjugated secondary antibodies were used at 1:400 (Thermo Fisher).
PluriTest Assay
PluriTest is based on whole-genome transcriptome microarray data analysis. In brief, we employed the same procedures as described previously (Müller et al., 2011, Müller et al., 2012, Williams et al., 2011). RNA was isolated from two biological replicates per culture condition (control and CP, 1 × 106 cells per sample) using the Qiagen RNeasy isolation kit following the manufacturer's instructions (Qiagen). Illumina HT12v4 microarrays were hybridized following the manufacturer's instructions (Müller et al., 2011, Müller et al., 2012, Williams et al., 2011). The resulting raw data were processed with the R/Bioconductor lumi-package (Du et al., 2008, Lin et al., 2008, Müller et al., 2011). It has been recognized that due to changes in scanner technology and modifications of the hybridization protocol through Illumina, PluriTest results in recent years tend to show lower Pluripotency and higher Novelty scores (B. Schuldt, personal communication). Even considering this technical bias, both control and CP samples pass the empirical Pluripotency and higher Novelty score thresholds with both biological replicates.
Reduced Representation Bisulfite Sequencing
For sequencing library preparation, 1 μg of high-quality genomic DNA was used with the NEXTFlex Bisulfite-Seq Kit (Bioo, #5119-01), according to the manufacturer’s instructions. Unmethylated λDNA was spiked in at 1% to assess the level of bisulfate conversation. Twelve cycles of PCR were performed. Samples were run on HiSeq 2500 Rapid mode Paired End 125, and an average of 140 million reads was generated per sample.
RRBS DNA Methylation Analysis
FASTQ files were generated by bcl2fastq (V2.17) and filtered for pass filter reads based on Illumina's chastity filter. Sequencing adapters were trimmed by FLEXBAR (V2.4) (Dodt et al., 2012), genomic alignments using Bismark (V0.14.4) (Krueger and Andrews, 2011) and Bowtie2 (V2.2.5) (Langmead and Salzberg, 2012) to reference human genome hg19, and per base CpG methylation metrics were calculated with a custom PERL script (Garrett-Bakelman et al., 2015).
γH2AX Quantitative Immunofluorescence
Single PSCs were plated in E8 medium with 10 μM Y-27632 on Geltrex-coated dishes at 100,000 cells/cm2. After 24 hr, cells were treated with 0.5 μM camptothecin (Sigma, #C9911) in E8 for 1 hr at 37°C. Cells were then stained against phospho-Histone H2A.X (1:300, Millipore, #05-636), and the appropriate Alexa Fluor-conjugated secondary antibodies were used at 1:400. Images were acquired and analyzed on a PerkinElmer Operetta using eHarmony software.
Neural Induction
We used a derivative of Chambers et al. (2009). See Supplemental Experimental Procedures for full protocol details.
Mesendoderm Induction
Single PSCs were plated as above. After 24 hr, the medium was removed and E6 medium with 5 μM CHIR99021 (Stemgent #04-0004-10) was added to create mesendoderm (Lam et al., 2014). E6 with 5 μM CHIR99021 was exchanged every 24 hr for up to 4 days. Efficiency of conversion was quantitated by flow analysis for OCT4 (BD Biosciences, #560186) and Brachyury (R&D Systems, #IC2085A). Cultures were stained using antibodies against OCT4 and Brachyury (1:40, R&D Systems, #AF2085), and the appropriate Alexa Fluor-conjugated secondary antibodies were used at 1:400.
qRT-PCR
RNA was isolated from cell pellets using an RNase-Free DNase set (Qiagen, #79254) and an RNeasy Mini Kit (Qiagen, #74106). Reverse transcription of RNA samples and cDNA synthesis was performed using an RT2 First Strand Kit (Qiagen, #330404) and the Eppendorf Mastercycler PCR machine. cDNA samples were prepared for qPCR analysis using RT2 SYBR Green Mastermix (Qiagen, #330503). Samples were run on a custom RT2 Profiler PCR Array (see Supplemental Experimental Procedures for primer sets.) qPCR was performed using a Bio-Rad c1000 Touch Thermal Cycler CFX96 Real-time System.
Teratomas
NSG mice (Jackson Laboratories) were used for in vivo studies and were cared for in accordance with guidelines approved by MSKCC Institutional Animal Care and Use Committee and Research Animal Resource Center. Eight-week-old female mice were injected subcutaneously with 3 million H9 cells in the flank with Matrigel (BD Biosciences) mixed 1:2 in HEPES-buffered Hank’s balanced salt solution. Mice were observed daily for signs of morbidity/mortality, and body weights were assessed at least twice weekly. Tumors were measured twice weekly using calipers, and volume was calculated using the formula length × width2 × 0.52. At the end of the study, tumors were fixed in 10% formalin, processed, embedded in paraffin, sectioned, and stained with H&E. Microscope slides were reviewed by a pathologist.
Use of Cell Factories
We used Nunc Cell Factory System, four tray layers (Thermo Fisher, #140004) to expand large banks prior to CryoPausing. Cells were fed with 500 mL of E8 medium for the first 2 days after passage and 600 mL on the third day. To create a single-cell suspension we added 140 mL of Accutase for 30 min at 37°C, after which the trays were washed with 100 mL of medium.
Nucleofections/iCRISPR
To nucleofect CP cells, we used the Amaxa Cell Line Nucleofector Kit V (Lonza, #VCA-1003). WA09 CP cells were thawed and washed. Solution V (100 μL) was mixed with 22.2 μL of Supplement 1, and 5 million CP cells were added to 100 μL of this mixture. Tem microliters of the GFP control plasmid was added to the reaction before nucleofecting on program B-016 (Lonza Nucleofector 2b device). Nucleofected cells were added to E8 with 10 μM Y-27632 on Geltrex as above. Live cells were imaged for Figure 4A and the number of fluorescing cells was determined by treating cultures with Accutase, washing, and resuspending in PBS with 0.1% BSA before measuring fluorescence on a BD FACSAria III.
To perform CP iCRISPR gene modification, we expanded WA01 (H1) iCRISPR cells as above before 24 hr of doxycycline treatment to induce Cas9 (González et al., 2014). Cas9-induced cells were harvested and washed as above before CP. CP cells were thawed and washed before nucleofection with 200 ng of HPRT guide RNA purchased from GeneArt (Thermo Fisher). The guide sequence is CAT TTC TCA GTC CTA AAC A.
Sendai Transduction
CryoPaused WA09 cells were thawed, washed, and resuspended in E8 medium supplemented with 10 μM Y-27632 and Sendai viral vectors expressing EmGFP (CytoTune EmGFP, Thermo Fisher Scientific, #A16519) were added at an MOI of 5. Transduced CP cells were replated and expanded in E8. Transduced cells could be expanded for at least 15 passages.
Author Contributions
K.G.W. designed and performed experiments, analyzed data, and helped write the manuscript. S.D.R., K.R., S.A.R., and S.E.M. performed experiments. A.K. and E.D. performed the teratoma analysis. F.J.M. performed the PluriTest analysis. T.J.K., C.Z., and D.B. performed the bisulfide sequencing analysis. M.J.T. funded the project, provided the concept, designed experiments, interpreted results, and wrote the manuscript.
Acknowledgments
New York State's stem cell funding agency (NYSTEM) has been essential for this work and our laboratory. Contract C029153 (M.J.T., PI) has helped fund the basic operation of our facility and allowed us to develop CryoPause. Contract C028503 allowed us to manufacture a hESC-based therapy described in the manuscript (Lorenz Studer, PI). The authors also wish to acknowledge the tremendous contributions of The Starr Foundation for our initial seed funding and for their ongoing support. The authors thank Gouri Nanjangud and staff at the Molecular Cytogenetics Core, and Agnes Viale, Juan Li, Kety Huberman, and their staff at the Integrated Genomics Operation at MSKCC. Both cores provide outstanding service and support, and are supported by the NCI Cancer Center Support grant P30 CA008748. The Integrated Genomics Operation is also supported by Cycle for Survival and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. F.J.M. was supported by grants from the BMBF (13GW0128A and 01GM1513D) and from the Deutsche Forschungsgemeinschaft (DFG MU 3231/3-1). C.Z. and D.B. are supported by Tri-Institutional Research Programs (grant no. 2013-036). We also thank Danwei Huangfu and her entire laboratory (MSKCC) for always sharing reagents and expertise with us, including the iCRISPR WA01 line used here.
Published: June 8, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and two figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.05.010.
Supplemental Information
References
- Barker R.A., Studer L., Cattaneo E., Takahashi J., G-Force PD consortium G-Force PD: a global initiative in coordinating stem cell-based dopamine treatments for Parkinson’s disease. NPJ Parkinsons Dis. 2015;1 doi: 10.1038/npjparkd.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt M., Roehr J.T., Ahmed R., Dieterich C. FLEXBAR-flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel) 2012;1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Garrett-Bakelman F.E., Sheridan C.K., Kacmarczyk T.J., Ishii J., Betel D., Alonso A., Mason C.E., Figueroa M.E., Melnick A.M. Enhanced reduced representation bisulfite sequencing for assessment of DNA methylation at base pair resolution. J. Vis. Exp. 2015:e52246. doi: 10.3791/52246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González F., Zhu Z., Shi Z.D., Lelli K., Verma N., Li Q.V., Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacmarczyk T.J., Fall M.P., Zhang X., Xin Y., Li Y., Alonso A., Betel D. “Same Difference”: comprehensive evaluation of three DNA methylation measurement platforms. bioRxiv. 2016 doi: 10.1186/s13072-018-0190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam A.Q., Freedman B.S., Morizane R., Lerou P.H., Valerius M.T., Bonventre J.V. Rapid and efficient differentiation of human pluripotent stem cells into intermediate mesoderm that forms tubules expressing kidney proximal tubular markers. J. Am. Soc. Nephrol. 2014;25:1211–1225. doi: 10.1681/ASN.2013080831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.M., Du P., Huber W., Kibbe W.A. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Chen G. Cryopreservation of human pluripotent stem cells in defined medium. Curr. Protoc. Stem Cell Biol. 2014;31 doi: 10.1002/9780470151808.sc01c17s31. 1C.17.1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F.-J., Schuldt B.M., Williams R., Mason D., Altun G., Papapetrou E.P., Danner S., Goldmann J.E., Herbst A., Schmidt N.O. A bioinformatic assay for pluripotency in human cells. Nat. Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F.-J., Brändl B., Loring J.F. Harvard Stem Cell Institute; 2012. Assessment of Human Pluripotent Stem Cells with PluriTest. StemBook. [PubMed] [Google Scholar]
- Redon C., Pilch D., Rogakou E., Sedelnikova O., Newrock K., Bonner W. Histone H2A variants H2AX and H2AZ. CurrOpin Genet. Dev. 2002;12:162–169. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- Williams R., Schuldt B., Müller F.-J. A guide to stem cell identification: progress and challenges in system-wide predictive testing with complex biomarkers. Bioessays. 2011;33:880–890. doi: 10.1002/bies.201100073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.