Summary
The establishment of DNA methylation patterns in oocytes is a highly dynamic process marking gene-regulatory events during fertilization, embryonic development, and adulthood. However, after epigenetic reprogramming in primordial germ cells, how and when DNA methylation is re-established in developing human oocytes remains to be characterized. Here, using single-cell whole-genome bisulfite sequencing, we describe DNA methylation patterns in three different maturation stages of human oocytes. We found that while broad-scale patterns of CpG methylation have been largely established by the immature germinal vesicle stage, localized changes continue into later development. Non-CpG methylation, on the other hand, undergoes a large-scale, generalized remodeling through the final stage of maturation, with the net overall result being the accumulation of methylation as oocytes mature. The role of the genome-wide, non-CpG methylation remodeling in the final stage of oocyte maturation deserves further investigation.
Keywords: single cell, DNA methylome, human oocyte, oocyte maturation, non-CpG methylation, epigenome, in vitro maturation
Highlights
-
•
Broad-scale CpG methylation is largely stable during oocyte maturation
-
•
Non-CpG methylation accumulates as oocytes mature
-
•
Non-CpG methylation changes are genome wide
CpG methylation is established globally in adult immature oocytes and remains stable during maturation. Non-CpG methylation undergoes a genome-wide, generalized remodeling through the final stage of maturation, with the net overall result being accumulation of methylation as oocytes mature.
Introduction
A woman is born with approximately 295,000 oocytes arrested in the prophase of meiosis I, also commonly referred to as the germinal vesicle (GV) stage (Wallace and Kelsey, 2010). Unlike in mice whose oocytes start to grow and mature from the neonatal period, human oocytes stay quiescent in meiosis I until puberty or beyond (Smallwood and Kelsey, 2012). After puberty, 10–15 GV oocytes are recruited for growth and maturation during each menstrual cycle, of which only 1–2 reach the mature metaphase II (MII) stage soon before the ovulation, ready for fertilization, while the rest of the growing oocyte cohort degenerate. The genetic and epigenetic basis for the selection of dominant oocytes destined for ovulation and fertilization from the rest of the growing cohort is poorly understood (Surani, 2015).
DNA methylation is a dynamic process during the growth and development of oocytes. It has been shown in mice that DNA methylation is erased in the primordial germ cells during early embryonic development and re-established after birth as the oocytes start to grow and mature. The wave of DNA methylation erasure and re-establishment is repeated after fertilization in pre-implantation embryos (Smallwood and Kelsey, 2012). These waves of DNA methylation changes in oocytes and embryos are essential for resetting genomic potential, establishing the germline, and marking correct developmental genes (Bogdanovic et al., 2016, Hargan-Calvopina et al., 2016, Hon et al., 2013, von Meyenn and Reik, 2015). In humans, the process of DNA methylome erasure in primordial germ cells and in pre-implantation embryos was recently demonstrated in several genome-wide studies, and this process resembles what was previously described in mice, although some significant differences exist (Gkountela et al., 2015, Guo et al., 2014a, Guo et al., 2015, Smith et al., 2014, Tang et al., 2015, von Meyenn and Reik, 2015). The timing of de novo DNA methylation of oocytes after epigenetic erasure of parental marks in embryos is not only scientifically intriguing, but also presents a clinically important opportunity for guiding the efforts in developing in vitro maturation methods for fertility treatment.
However, the DNA methylome establishment and maintenance during human oocyte growth and maturation beyond the early prenatal stage has not yet been illuminated, partly due to technical limitations of genome-wide studies in cells only available in very small numbers (Yu et al., 2015). Previous studies mapping the DNA methylome in human oocytes (Okae et al., 2014, Smith et al., 2014) had to pool a large number of oocytes in certain stages of development, and therefore were unable to specifically investigate DNA methylome variations among different oocyte maturation stages. Recent advances in single-cell bisulfite sequencing technologies (Farlik et al., 2015, Gravina et al., 2015, Gravina et al., 2016, Schwartzman and Tanay, 2015, Smallwood et al., 2014) now enable DNA methylome analysis in small samples, down to the level of single cells.
Here, we use single-cell whole-genome bisulfite sequencing (SC-WGBS) techniques to explore the patterns of DNA methylation in three different maturation stages of human oocytes, from the least mature GV stage, to intermediate metaphase I (MI) stage, and then to the mature MII stage. We found that while CpG methylation is mostly established during the GV stage, non-CpG methylation continues to accumulate throughout the maturation process. The biological role that may be played by genome-wide non-CpG methylation remodeling in the final stage of oocyte maturation remains to be investigated.
Results
Data Structure and DNA Methylome Survey
We collected a total of 30 human oocytes from 13 individuals during assisted reproductive technologies (ART) cycles. Ten oocytes were included in each maturation stage (GV, MI, and MII) (Table 1). Many individuals contributed multiple oocytes in the same or different maturation stages. A wide range of age, stimulation protocols and response, and fertility diagnoses exist among these individuals (Table 2), reflecting the general ART practices. None of the oocytes collected was ever exposed to sperms or discarded due to quality concerns. Each oocyte was denuded of surrounding cumulus cells and the zona pellucida was removed to ensure a single oocyte was collected as the starting material, before being analyzed using the SC-WGBS method (Figure S1i). Each experiment from cell lysis, bisulfite conversion to library preparation was always performed on three to four oocytes and a negative control (water) simultaneously. In none of these experiments did the negative controls reveal a bioanalyzer peak, ruling out contamination (Figure S1ii). We intentionally used samples from different individuals and different oocyte maturation stages in each experiment to decrease possible bias introduced by experimental batch effects. For the same reason, each of the two Illumina HiSeq runs to sequence these libraries included multiple samples from different individuals and all three maturation stages.
Table 1.
Overview of All Samples
| Patient ID | GV | MI | MII |
|---|---|---|---|
| A | BY20, BY21 | BY13, BY14, BY18 | |
| B | BY06, BY07 | BY05 | |
| C | BY08 | ||
| D | BY02 | BY03 | |
| F | BY16, BY17 | ||
| G | BY19 | ||
| H | BY09 | ||
| I | BY11 | ||
| J | BY23, BY27, BY29 | ||
| K | BY39 | BY40 | |
| L | BY42 | BY41, BY57 | |
| M | BY52, BY55, BY56 | ||
| N | BY50, BY51, BY54 |
See also Figure S1 and Tables S1–S3.
Table 2.
Characteristics of All Individuals
| Individual ID | Clinic ID | Age (years) | Diagnosis | Ovarian Stimulation Protocol | Total No. of Oocytes Retrieved | History of Live Birth |
|---|---|---|---|---|---|---|
| A | 1 | 35 | anovulatory infertility | GnRH antagonist | 20 | yes |
| B | 1 | 36 | severe male factor infertility | GnRH antagonist | 12 | no |
| C | 1 | 42 | age-related infertility | GnRH agonist flare | 9 | no |
| D | 1 | 36 | anovulatory infertility | GnRH antagonist | 21 | yes |
| F | 1 | 38 | anovulatory infertility | GnRH antagonist | 19 | no |
| G | 1 | 37 | tubal factor infertility | luteal Lupron | 14 | no |
| H | 1 | 40 | age and male factor infertility | luteal Lupron | 7 | no |
| I | 1 | 40 | age-related infertility | GnRH antagonist | 8 | yes |
| J | 2 | 38 | fertility preservation | luteal Lupron | 11 | no |
| K | 1 | 41 | age-related infertility | GnRH antagonist | 6 | no |
| L | 1 | 34 | severe male factor infertility | luteal Lupron | 19 | no |
| M | 3 | 28 | oocyte donor | GnRH agonist flare | 26 | yes |
| N | 3 | 29 | oocyte donor | GnRH agonist flare | 15 | yes |
The SC-WGBS method we used was comparable with previously published protocols (Table S1) (Farlik et al., 2015, Smallwood et al., 2014). With a mean mapping efficiency at 29.4% and duplication rate at 83.3%, an average of 1.15 million uniquely mapped reads were obtained for each single oocyte (Table S2). Approximately 1%–5% of the genome was covered in each cell and the coverage was genome wide. Merging the oocytes within each of the three stages increased average genomic coverage to 33.2% for each maturation stage, and the merged analyses were used to compare different maturation stages, which also helped to minimize some of the effects of patient heterogeneity.
Even though multiple measures were taken to eliminate somatic contamination of the oocytes, including removal of zona pellucida which was rarely done in previous genomic studies, we further confirmed the lack of contamination in our oocyte samples using two different approaches. We first correlated the methylation levels of 68 oocyte-specific imprinted regions in each of our oocytes with those of different cell types published in Okae et al. (2014), and found high correlations between the two sets of oocytes and low correlations with all the other cell types (Table S3). Another validation approach was to correlate the average methylation levels of each sliding window in a single oocyte sample with those in the merged sample. Each oocyte showed high concordance with the pooled sample, demonstrating technical reproducibility and the lack of contamination (Table S4).
Some of the individuals contributed multiple oocytes within the same stage, while some other oocytes from two different stages were from the same individual (Table 1). On average, the CpG methylation level was 48.9%, consistent with previous studies in mouse oocytes (Figure S2A). Also similar to previous studies in mouse oocytes (Kocabas et al., 2006, Shirane et al., 2013, Smallwood et al., 2011, Veselovska et al., 2015), gene bodies, especially highly transcribed gene bodies, preferentially acquired DNA methylation compared with the intergenic regions (Figures S3A and S3B).
Previous studies in mouse oocytes, mouse brain tissue, and human pluripotent stem cells (Guo et al., 2014b, Lister et al., 2013, Shirane et al., 2013, Ziller et al., 2011) found non-CpG methylation to be a prominent feature in these cell types. We found the non-CpG methylation in human oocytes to be 5.2% of CHH and CHG sites on average, which is significantly higher than was reported for somatic cells (Ziller et al., 2011) (Figures S2B and S2C). This high fraction of non-CpG methylation is not an artifact of incomplete bisulfite conversion, which was consistently found to be 99% or higher based on spiked-in unmethylated λDNA (Table S2).
Global Methylation Patterns during Oocyte Maturation
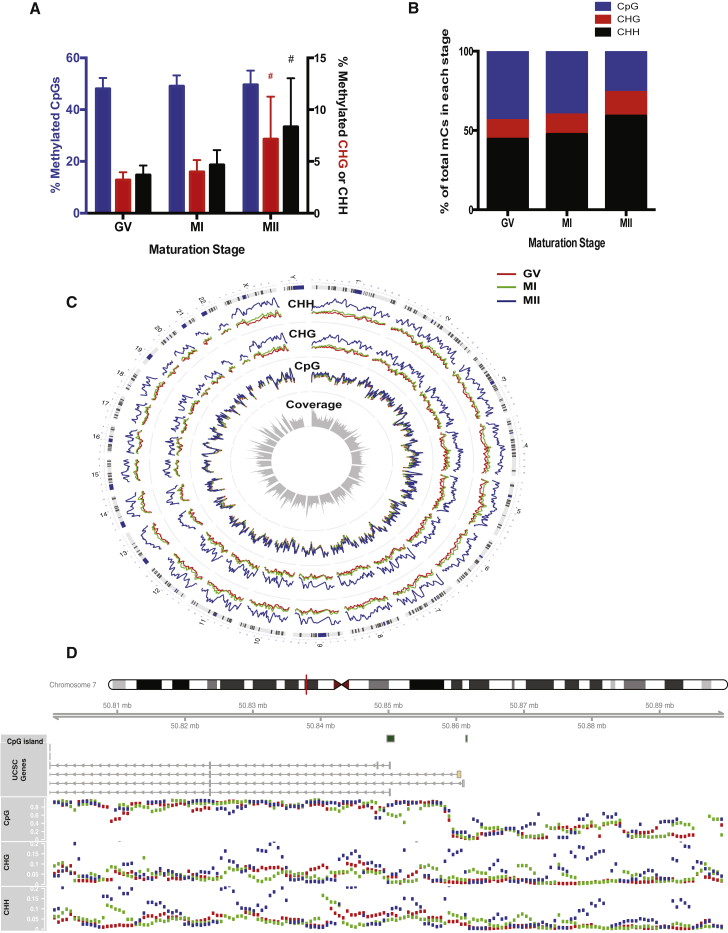
After the initial general survey of the oocyte methylome described above, we next investigated how the CpG and non-CpG methylation levels changed on a genome-wide scale over the course of oocyte maturation. It was previously shown that merging a few individual single-cell methylomes provides an accurate representation of the DNA methylation landscape in the bulk sample, which makes SC-WGBS a powerful tool to investigate the DNA methylome in very rare material (Farlik et al., 2015, Smallwood et al., 2014, Ziller et al., 2011). Hence for the in-depth analyses, we used merged data from each stage to compare the three maturation stages. By merging all CpG or non-CpG sites from all of the oocytes at the same stage of maturation, we find that the global average CpG methylation level stays the same from GV to MII stage (from 48.1% ± 4.2% to 49.6% ± 5.5%, p > 0.5), while the average CHG and CHH methylation levels increase significantly as the oocyte maturity increases (from 3.2% ± 0.8% to 7.2% ± 4.1% for CHG, from 3.7% ± 0.9% to for 8.3% ± 4.7% for CHH, p < 10−4) (Figure 1A). The majority of the methylated cytosines are found in CHH motifs, and the percentages of methylated CHG and CHH continue to increase as the oocytes mature, which is consistent with the increased non-CpG methylation level observed above (Figure 1B).
Figure 1.
Genome-wide CpG and Non-CpG Methylation Patterns
(A) Average methylation levels (mean and SD) of all CpGs, CHGs, or CHHs at each oocyte maturation stage. Hash indicates significant change from GV or MI to MII (Wilcoxon rank sum two-sided test, #p < 0.05). Left y axis: percentage of methylated CpGs (in blue); Right y axis: percentage methylated CHGs (in red) or CHHs (in black).
(B) Composition of methylated cytosines in each maturation stage. From GV or MI to MII stage, significant increases in percentage of CHH and CHG, and significant decreases in percentage of CpG (two-sided t test, p < 0.05).
(C) Circular genomic plot of average DNA methylation level in 1-Mb non-overlapping bins. Circles from outside to the center: cytochromosome, CHH methylation levels (blue, MII; green, MI; red, GV), CHG methylation levels, CpG methylation levels, and average coverage depth per oocyte (scale: 0–1,930 CpG sites). Each peak represents a 1-Mb bin.
(D) An example region (Chr7: 50,800,000–50,900,000) that includes imprinted gene GRB10. Panels from the top down: location of plotted region on Chr7 (red line); CpG islands; GRB10 transcripts on UCSC browser; CpG methylation levels (scale: 0%–100%); CHG methylation levels (scale: 0%–20%); CHH methylation levels (scale: 0%–20%). Blue, MII; green, MI; red, GV.
n = 10 oocytes per maturation stage. See also Figure S2.
When we plotted the average methylation levels of non-overlapping sliding windows of 1 Mb in size along each chromosome, the global changes in non-CpG methylation patterns, as well as the lack of variation in CpG methylation patterns, from immature to mature oocytes can be easily appreciated (Figure 1C). The plot of an example region surrounding a maternally imprinted gene GRB10 is shown in Figure 1D. As evident from the plot, CpG sites were almost fully methylated in all three stages, while non-CpG methylation levels increased in multiple windows from GV or MI to MII stage. These analyses suggest that at least on a global level, CpG methylation is re-established first during oocyte growth, and non-CpG sites are gradually methylated up to the last stage of maturation.
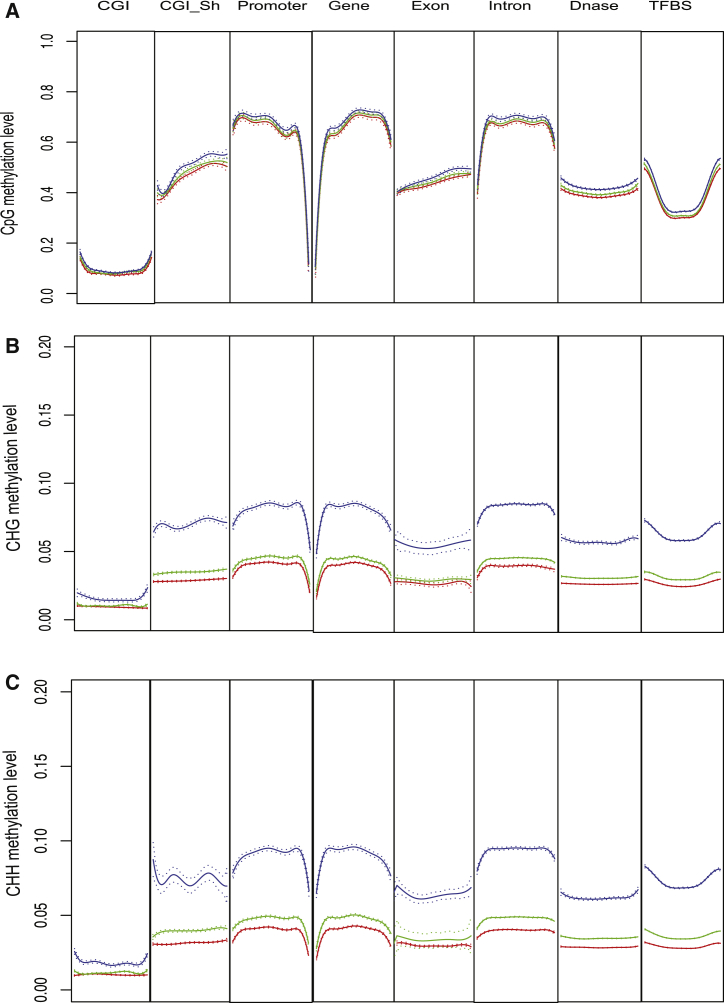
We then investigated whether the difference in average overall methylation levels was due to any particular genomic feature. The mean and 95% confidence interval of the methylation levels of all the CpG or non-CpG sites were plotted for the major genomic features. This analysis revealed that the lack of difference in CpG methylation levels across the three maturation stages was also seen in all genomic features analyzed. Similarly, the increases in CHH or CHG methylation levels from immature to mature oocytes that were detected in the global analysis distribute evenly across various genomic features (Figures 2A–2C). The only exception is in CpG islands, which showed very low CpG and non-CpG methylation across all three maturation stages. Of note, the non-CpG density in CpG islands is similar to that in the rest of the genome. These results confirm that non-CpG methylation re-establishment occurs in a generalized manner, and CpG methylation is essentially completed early during the oocyte maturation process.
Figure 2.
Composite Plot of DNA Methylation Levels across Genomic Features
(A) CpG; (B) CHG; (C) CHH. Blue, MII stage; green, MI; red, GV. Each line consists of mean with 95% confidence interval. CGI (CpG island): a region with at least 200 bp, a GC percentage greater than 50%, and an observed-to-expected CpG ratio greater than 60%, as defined in UCSC genome browser. CGI_Sh (CpG island shore): 2-kb flanking regions of a CpG island. Promoter: 2 kb upstream of transcription start sites. Gene: a gene body from transcription start to end site. Dnase: DNase I hypersensitivity sites, i.e., regions hypersensitive to cleavage by DNase I in various cell types, obtained from ENCODE project. TFBS: transcription factor binding sites, derived from a large collection of chromatin immunoprecipitation sequencing experiments performed by the ENCODE project.
n = 10 oocytes per maturation stage, See also Figure S3.
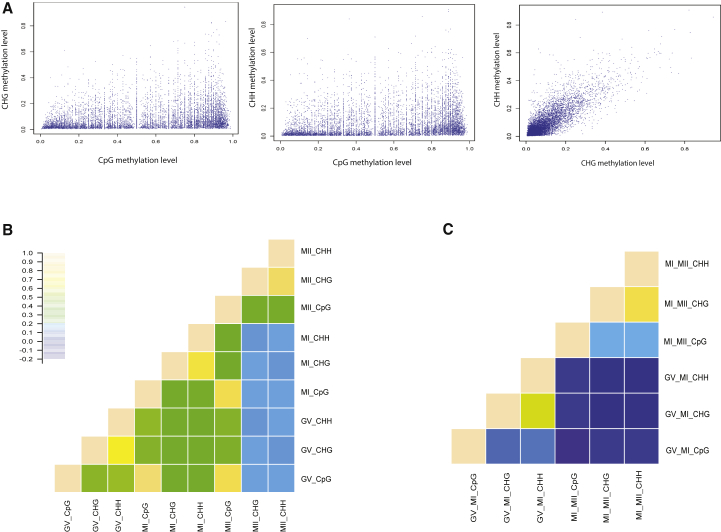
Next, using Pearson correlation coefficients we correlated the methylation levels of non-overlapping sliding windows of 3 kb in size (also called bins hereafter) containing two of the three cytosine categories (CpG, CHG, and CHH), to further examine the question of whether methylation variations associated with oocyte maturation occur independently in CpGs and non-CpGs. As predicted from the global and regional patterns seen above, in each stage of maturation the methylation levels of CHH and CHG of all the bins are closely correlated, but CpG methylation level does not strongly correlate with CHH or CHG methylation level in the same bin in any of the maturation stages (Figures 3A and 3B; Table S5). Similar patterns emerged when we correlated the methylation changes associated with maturation in these overlapping bins. When a bin contains both CHH and CHG, as the oocytes becomes more mature the CHH methylation change is highly correlated with that of CHG. However, the CpG methylation variations across different maturation stages do not correlate with those of non-CpGs in the same bin (Figure 3C and Table S5). These results confirmed the different timing in methylation establishment between CpG and non-CpG methylation. It has been previously suggested that the same enzymes and machineries probably are responsible for methylation establishment of all non-CpG sites, rather than being discriminatory for either CHH or CHG (Guo et al., 2014b), which is consistent with our data.
Figure 3.
Pearson Correlations among CpG, CHG, and CHH Methylation Levels and Stage Differences
(A) Scatterplot of CpG, CHG, or CHH methylation levels in all 3-kb bins that contain the pair of cytosine categories in question. Pearson correlation coefficient in each scenario is calculated and used in (B). Only MII stage is shown here (r = 0.22, 0.21, 0.85, respectively, p < 10−10).
(B) Heatmap of Pearson correlation coefficients in methylation levels. Each coefficient is calculated from pairwise correlation. Scale bar on upper left applies to both (B) and (C).
(C) Heatmap of Pearson correlation coefficients of methylation level differences between two maturation stages.
n = 10 oocytes per maturation stage, See also Table S5 and Figure S4.
Regional Methylation Patterns
Even though the global CpG methylation patterns clearly remain stable from immature GV stage to mature MII stage, regional or subtle differences among the three developmental stages could still exist. For the non-CpG sites, local changes may follow a completely different pattern compared with global changes. Therefore, we used a sliding window (or bin) of 3 kb in size and 600 bp in step to scan the methylome for regional variations among the three maturation stages. Only bins with at least five counts of CpG or non-CpG sites were included in the analysis, and pairwise comparisons (Student's t or Z test based on the site counts of each bin) were performed only at sites that were covered by both maturation stages in question. A statistically significant difference was defined by a p value of <0.0001 and the lower bound of 95% confidence interval of >0.1. These criteria resulted in a false discovery rate (FDR) of ≤2.7%. Bins with only data from a single individual were excluded to decrease individual bias. Using this analytical approach, we identified 4,377 differentially methylated regions (DMRs) between GV and MI stages, and 2,826 between MI and MII stages, for CpG sites (Figure S4A). More DMRs were located in the areas covered by non-CpG sites, with 2,183 and 11,207 for CHG sites, and 71,198 and 205,919 for CHH sites, in the GV versus MI and MI versus MII comparisons, respectively (Figure S4A). More non-CpG DMRs showed increased methylation than decreased methylation from immature to mature stages, which is consistent with the global change of average methylation level.
We then investigated whether these DMRs are preferentially distributed in certain genomic regions. The CpG DMRs with decreased methylation from GV to MI stage were more concentrated in CpG islands (odds ratio 1.71 compared with the genomic mean, 95% confidence interval 1.34–2.15). While the non-CpG DMRs mostly distribute in a generalized pattern across the genome, CHH DMRs are slightly more concentrated in LINE repeat regions (odds ratio 1.20–1.25 compared with the genomic mean, Figure S4B), and this small enrichment in LINE regions is not due to cytosine density.
To further characterize these DMRs, we performed gene ontology (GO) analysis to test whether certain biological pathways were enriched in genes with promoters located in these regions. Non-CpG DMRs were not enriched for any particular pathway, indicating that the differential methylation of non-CpGs associated with oocyte maturation occurs in a generalized manner. However, genes with an increase in CpG methylation in the promoter region from GV to MI stage were enriched in plasma membrane and ion channel-related pathways (Table S6), which may be linked to germinal vesicle breakdown during this transition. Genes with decreased CpG methylation in their promoter regions from MI to MII stage were enriched for extracellular matrix pathways, which could be related to zona pellucida remodeling as the oocyte matures and prepares for fertilization.
The results from all of the above global and regional analyses suggest that while CpG methylation has been largely established by the immature GV stage, there is still some fine-tuning occurring in local genomic regions, especially in regions with high CpG densities, such as CpG islands. Non-CpG, especially CHH, methylation, on the other hand, is undergoing a large-scale remodeling, with the net overall result being accumulation of methylation as oocytes mature. The remodeling of non-CpG methylation occurs across the genome in a generalized manner, rather than selecting for a certain biological pathway or genomic location.
Discussion
We obtained single-cell, single-base-resolution DNA methylomes in human oocytes from three different maturation stages. Global and regional analyses generated one major conclusion: CpG methylation is, for the most part, established before the immature GV stage, although some local fine-tuning still occurs. By contrast, non-CpG methylation in the genome gradually accumulates as the oocyte matures, and this methylation establishment process is generalized, rather than focused on particular genomic regions or functional features.
For the final analyses we merged all the samples from each maturation stage, which not only increased the genomic coverage significantly but also minimized the impact of patient heterogeneity on the final conclusion. However, individual characteristics, such as age or fertility status, and differences in ovarian stimulation from stimulated assisted reproductive technology cycles could still be factors that confound our findings. Nevertheless, our findings of stable CpG methylation during oocyte maturation is reassuringly consistent with an earlier study using unstimulated oocytes and in vitro maturation protocol, which found unchanged CpG methylation of four imprinted genes during in vitro oocyte maturation (Kuhtz et al., 2014). With the rare availability of human oocytes, especially MII oocytes, for research purposes a collective effort from multiple research teams is needed in the future to collect a sample large enough to address all the limitations of sample heterogeneity.
The higher methylation levels of non-CpG sites in human oocytes is consistent with what has been reported for other cell types that possess high potential for maturation or further differentiation, including mammalian brain cell types and human pluripotent stem cells (Lister et al., 2013), as well as mouse oocytes (Tomizawa et al., 2011). The biological roles that non-CpG methylation may play in these specific cell types remain to be investigated. Could non-CpG methylation be marking a poised epigenetic state for future differentiation and continued development in these cell types? This question needs to be answered by future mechanistic studies. Future investigation in single-cell transcriptomic changes associated with human oocyte maturation may help to elucidate the physiological roles played by non-CpG methylation during human oocyte maturation.
The DNA methylome of human oocytes shares many other similar features with mouse oocytes, such as preferentially acquiring DNA methylation within the gene bodies, especially in those genes that are highly transcribed (Kocabas et al., 2006, Shirane et al., 2013, Smallwood et al., 2011, Veselovska et al., 2015). However, to the best of our knowledge, there has not been a study in mouse or other species that directly examines the non-CpG methylation changes during oocyte maturation. Several studies investigated non-CpG methylation in mouse or human oocytes in one of the maturation stages, but none compared oocytes in two or three different stages of maturation.
Specific DNA methyltransferases (DNMTs) involved in genomic non-CpG methylation in mouse oocytes have been examined by Shirane et al. (2013). Using mutant mouse GV oocytes lacking Dnmt1, 3a, 3b, or 3L, it was shown that genomic non-CpG methylation in oocytes depends on the Dnmt3a-Dnmt3L complex, while Dnmt1 and 3b are dispensable. Our study could hold potential clinical significance, because in vitro maturation of oocytes is an important assisted reproductive intervention that has been under intense investigation for many years, and its success will have wide applications in infertility treatments and especially in fertility preservation in childhood cancer patients. By manipulating the non-CpG methylation in immature oocytes such as by upregulating the responsible DNMTs, it may be possible to facilitate or accelerate the maturation process, and subsequently use the resulting mature oocytes for therapeutic purposes.
In conclusion, our study utilized the recently introduced single-cell WGBS technology to investigate the DNA methylome establishment during the human oocyte maturation process. The datasets of these precious human oocyte samples may serve as an important resource for germ cell and stem cell research communities. The results support the notion that genomic CpG methylation is for the most part stable as human oocytes mature, yet non-CpG methylation is dynamic and continues to accumulate during the oocyte maturation process. The biological role that non-CpG methylation may play in human oocyte maturation requires further investigation.
Experimental Procedures
Human Oocyte Collection
All human oocytes used in this study were obtained in embryology laboratories at Saint Barnabas Center for Reproductive Medicine and Sher Institutes for Reproductive Medicine under the regulatory oversight of Institutional Review Board (IRB)-approved Human Subjects protocol at each institution. All consented materials were donated anonymously and carried no personal identifiers. After oocyte retrieval procedures under standard Assisted Reproductive Technology protocols, oocytes that were destined to be discarded were collected under previously obtained written informed consent. To eliminate contaminations from cumulus cells, we removed zona pellucida from each oocyte using either acid Tyrode's solution or mechanical separation techniques. Each oocyte was washed in PBS twice and immediately frozen in 2 μL of PBS in a −80°C freezer until shipment on dry ice. Oocytes were received for further studies at Albert Einstein College of Medicine (AECOM) under the approval of AECOM IRB, which deemed the project exempt under 45 CRF 46.102(f).
None of the oocytes was exposed to sperms or discarded due to fertilization or quality issues. GV and MI oocytes were collected at the time of maturity check. Most of the MII oocytes were collected from oocyte donors who had excess oocytes to dispose, due to rare circumstances that removed them from the donor list. MII oocytes from individual J were donated to research due to a change in her social situation. These oocytes were cryopreserved without any damage before being donated to our research. One MII oocyte (BY03) was MI at the time of maturity check and thus excluded from clinical use, but was found to be MII later at the time of zona pellucida removal for research. We specifically removed the zona pellucida from each oocyte to eliminate the contamination of cumulus cells and other cell types. All oocytes were processed by the same embryologist (T.S.), including the removal of zona pellucida, washing, cryopreserving, and shipping. All WGBS experiments were carried out by the same individual (B.Y.).
SC-WGBS
All experiments were carried out in a clean PCR hood designated for single-cell work. Each experiment from cell lysis, bisulfite conversion to library preparation consisted of three to four single oocytes at different maturation stages, a negative control (water), and a positive control (purified bulk DNA). Each individual oocyte was lysed using 1 μL of proteinase K in M-Digestion buffer (Zymo Research) at 50°C for 30 min. Unmethylated λDNA (Promega) was spiked-in oocyte samples (10 pg/sample) after the cell lysis step. A Pico methylSeq kit (Zymo Research) was used for library preparation. Bisulfite treatment was performed by adding 130 μL of lightening conversion reagent to each sample and incubating at 98°C for 8 min and 54°C for 60 min. This step simultaneously fragmentizes and bisulfite-converts DNA. Complementary strands were then synthesized by using random PreAmp primers, with two cycles of incubation at 98°C for 2 min, 8°C for 5 min, 16°C, 22°C, 28°C, 36°C, and 36.5°C for 1 min each, and 37°C for 8 min. DNA clean-up was carried out on Zymo-Spin IC columns following the manufacturer's protocol. The library amplification step consisted of ten PCR cycles at the settings of 94°C, 45°C, 55°C for 30 s each, and 68°C for 1 min. The library preparation was completed by PCR amplification with Illumina indexed primers. The incubation setting for amplification was 94°C and 58°C for 30 s each, and 68°C for 1 min, for a total of ten cycles.
The indexed libraries were assessed for quality using High-Sensitivity DNA chips on the Agilent Bioanalyzer 2100. Every experiment included in the final analysis had flat bioanalyzer tracing for the negative control. The quantity of each sequencing library was measured with a Qubit fluorometer. Three oocyte libraries were multiplexed and sequenced on one lane of an Illumina HiSeq2500 for 150-bp single-end sequencing. All libraries in this study were sequenced during two HiSeq runs, each of which included multiple oocytes from all three maturation stages and different individuals, in order to decrease technical variations.
DNA Methylation Data Processing
Raw sequence reads were trimmed to remove the adaptor contamination and poor-quality reads using Trim Galore! (v0.3.5, www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Single-cell WGBS libraries are non-directional due to the multiple rounds of random priming. Trimmed sequences were mapped to the human genome (build hg19) using Bismark21 (v0.10.1; parameters: –bowtie2, –non_directional). All trimmed sequences were also mapped to the Enterobacteria phage λ genome (GenBank: J02459.1) to obtain the bisulfite conversion rate of each sample. Duplicate sequences were excluded using Bismark Deduplication. Methylation calls were made on the aligned and de-duplicated sequences using Bismark Methylation Extractor.
Estimation of Regional Methylation Level
We first took a sliding-window approach to estimate the regional methylation level of oocytes (Smallwood et al., 2014). To increase the confidence of estimation, we merged together sequences of individual oocytes from the same maturation stage. The genome was scanned using a 3-kb sliding window with a 600-bp step size. The methylation level of either CpG, CHG, or CHH (calculated separately), within window i from an oocyte in maturation stage j, was calculated based on a binomial distribution with add-one smoothing, as follows:
| (Equation 1) |
where and are methylated and unmethylated cytosine read counts, respectively. The SE was given as
| (Equation 2) |
where ni,j is the sum of and
Partial methylation, or asymmetric allelic methylation, refers to the percentage of single cytosine sites with reads that are not fully methylated or unmethylated.
Identification of Differentially Methylated Regions
To identify DMRs between oocytes from stage a and stage b, we used the following test statistic:
| (Equation 3) |
where sei,ab is the pooled SE of the two samples defined as
| (Equation 4) |
When methylation counts in a window passed all of the four following criteria,
| (c1.1) |
| (c1.2) |
| (c1.3) |
| (c1.4) |
the test statistic si,ab follows a standard normal distribution, and we obtained a two-sided p value for it. Additionally the confidence interval of the methylation level difference was estimated as
| (Equation 5) |
under an α level of 0.05, with z approximated as 1.96.
When a window does not pass all of the criteria (c1.1 to c1.4), the test statistic si,ab follows a Student t distribution, under a degree of freedom
| (Equation 6) |
Additionally the p value and confidence interval were also obtained.
Similar to the method previously reported (Ziller et al., 2013), we applied two cutoffs for DMRs: (1) significance: a p value of <0.0001 indicating significance; and (2) effect size: the absolute value of the upper and lower bounds of 95% confidence interval of the methylation level difference larger than 0.1. To assess FDRs in DMR calling, we applied an R package (fdrtool), and FDRs were estimated from all p values in DMR calling (Strimmer, 2008a, Strimmer, 2008b). The highest estimated FDR was 2.7%.
Of note, when local variations in methylation (<3 kb in size) are present, we may identify false positives in our pairwise comparisons due to the possibility that the cytosine sites covered in the two stages do not overlap. To avoid this situation, we only included the cytosine sites that are shared by both stages when calculating the regional methylation levels for DMR calling.
Author Contributions
B.Y. conceived and designed the study, obtained funding, collected samples from collaborators, conducted all experiments, contributed to data analysis, and wrote the manuscript. X.D. analyzed data and drafted bioinformatics method section of the manuscript. S.G. optimized the SC-WGBS library preparation protocol. Ö.K. contributed to data analysis. T.S., J.C., D.T., and R.Z. provided oocyte samples. R.D.H. edited the manuscript. J.V. supervised the study and edited the manuscript.
Acknowledgments
The authors would like to thank Adam Auton, PhD, for guidance in data analysis and statistics and Moonsook Lee for technical guidance in single-cell WGBS library preparation. This work was supported by grants from the Reproductive Scientist Development Program (NIH 5K12HD000849), American Society for Reproductive Medicine, Howard and Georgeanna Jones Foundation for Reproductive Medicine, Einstein Nathan Shock Center of Excellence in Basic Biology of Aging (to B.Y.); NIH P01AG017242, the Glenn Foundation, and the SENS Foundation (to J.V.).
Published: June 22, 2017
Footnotes
Supplemental Information includes four figures and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.05.026.
Contributor Information
Bo Yu, Email: by26@uw.edu.
Jan Vijg, Email: jan.vijg@einstein.yu.edu.
Accession Numbers
The NCBI GEO accession number for the genome-wide DNA methylation profiles of single oocytes reported in this paper is GEO: GSE71985.
Supplemental Information
References
- Bogdanovic O., Smits A.H., de la Calle Mustienes E., Tena J.J., Ford E., Williams R., Senanayake U., Schultz M.D., Hontelez S., van Kruijsbergen I. Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat. Genet. 2016;48:417–426. doi: 10.1038/ng.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlik M., Sheffield N.C., Nuzzo A., Datlinger P., Schonegger A., Klughammer J., Bock C. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S., Zhang K.X., Shafiq T.A., Liao W.W., Hargan-Calvopina J., Chen P.Y., Clark A.T. DNA demethylation dynamics in the human prenatal germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravina S., Ganapathi S., Vijg J. Single-cell, locus-specific bisulfite sequencing (SLBS) for direct detection of epimutations in DNA methylation patterns. Nucleic Acids Res. 2015;43:e93. doi: 10.1093/nar/gkv366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravina S., Dong X., Yu B., Vijg J. Single-cell genome-wide bisulfite sequencing uncovers extensive heterogeneity in the mouse liver methylome. Genome Biol. 2016;17:150. doi: 10.1186/s13059-016-1011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.U., Su Y., Shin J.H., Shin J., Li H., Xie B., Zhong C., Hu S., Le T., Fan G. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Chung W.Y., Qian M., Pellegrini M., Zhang M.Q. Characterizing the strand-specific distribution of non-CpG methylation in human pluripotent cells. Nucleic Acids Res. 2014;42:3009–3016. doi: 10.1093/nar/gkt1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Yan L., Guo H., Li L., Hu B., Zhao Y., Yong J., Hu Y., Wang X., Wei Y. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Hargan-Calvopina J., Taylor S., Cook H., Hu Z., Lee S.A., Yen M.R., Chiang Y.S., Chen P.Y., Clark A.T. Stage-specific demethylation in primordial germ cells safeguards against precocious differentiation. Dev. Cell. 2016;39:75–86. doi: 10.1016/j.devcel.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon G.C., Rajagopal N., Shen Y., McCleary D.F., Yue F., Dang M.D., Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocabas A.M., Crosby J., Ross P.J., Otu H.H., Beyhan Z., Can H., Tam W.L., Rosa G.J., Halgren R.G., Lim B. The transcriptome of human oocytes. Proc. Natl. Acad. Sci. USA. 2006;103:14027–14032. doi: 10.1073/pnas.0603227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhtz J., Romero S., De Vos M., Smitz J., Haaf T., Anckaert E. Human in vitro oocyte maturation is not associated with increased imprinting error rates at LIT1, SNRPN, PEG3 and GTL2. Hum. Reprod. 2014;29:1995–2005. doi: 10.1093/humrep/deu155. [DOI] [PubMed] [Google Scholar]
- Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okae H., Chiba H., Hiura H., Hamada H., Sato A., Utsunomiya T., Kikuchi H., Yoshida H., Tanaka A., Suyama M. Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet. 2014;10:e1004868. doi: 10.1371/journal.pgen.1004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman O., Tanay A. Single-cell epigenomics: techniques and emerging applications. Nat. Rev. Genet. 2015;16:716–726. doi: 10.1038/nrg3980. [DOI] [PubMed] [Google Scholar]
- Shirane K., Toh H., Kobayashi H., Miura F., Chiba H., Ito T., Kono T., Sasaki H. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet. 2013;9:e1003439. doi: 10.1371/journal.pgen.1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood S.A., Kelsey G. De novo DNA methylation: a germ cell perspective. Trends Genet. 2012;28:33–42. doi: 10.1016/j.tig.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Smallwood S.A., Tomizawa S., Krueger F., Ruf N., Carli N., Segonds-Pichon A., Sato S., Hata K., Andrews S.R., Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 2011;43:811–814. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood S.A., Lee H.J., Angermueller C., Krueger F., Saadeh H., Peat J., Andrews S.R., Stegle O., Reik W., Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z.D., Chan M.M., Humm K.C., Karnik R., Mekhoubad S., Regev A., Eggan K., Meissner A. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- Strimmer K. A unified approach to false discovery rate estimation. BMC Bioinformatics. 2008;9:303. doi: 10.1186/1471-2105-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani M.A. Human germline: a new research frontier. Stem Cell Reports. 2015;4:955–960. doi: 10.1016/j.stemcr.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.W., Dietmann S., Irie N., Leitch H.G., Floros V.I., Bradshaw C.R., Hackett J.A., Chinnery P.F., Surani M.A. A unique gene regulatory network resets the human germline epigenome for development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S., Kobayashi H., Watanabe T., Andrews S., Hata K., Kelsey G., Sasaki H. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138:811–820. doi: 10.1242/dev.061416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselovska L., Smallwood S.A., Saadeh H., Stewart K.R., Krueger F., Maupetit-Mehouas S., Arnaud P., Tomizawa S., Andrews S., Kelsey G. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 2015;16:209. doi: 10.1186/s13059-015-0769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Meyenn F., Reik W. Forget the parents: epigenetic reprogramming in human germ cells. Cell. 2015;161:1248–1251. doi: 10.1016/j.cell.2015.05.039. [DOI] [PubMed] [Google Scholar]
- Wallace W.H., Kelsey T.W. Human ovarian reserve from conception to the menopause. PLoS One. 2010;5:e8772. doi: 10.1371/journal.pone.0008772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Russanova V.R., Gravina S., Hartley S., Mullikin J.C., Ignezweski A., Graham J., Segars J.H., DeCherney A.H., Howard B.H. DNA methylome and transcriptome sequencing in human ovarian granulosa cells links age-related changes in gene expression to gene body methylation and 3′-end GC density. Oncotarget. 2015;6:3627–3643. doi: 10.18632/oncotarget.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller M.J., Muller F., Liao J., Zhang Y., Gu H., Bock C., Boyle P., Epstein C.B., Bernstein B.E., Lengauer T. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011;7:e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller M.J., Gu H., Muller F., Donaghey J., Tsai L.T., Kohlbacher O., De Jager P.L., Rosen E.D., Bennett D.A., Bernstein B.E. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.