Abstract
The aim of this study was to investigate the effects of dietary supplementation with two alternatives to antibiotics (Candida tropicalis and mulberry leaf flavonoids) on intestinal microbiota of preweaned calves challenged with Escherichia coli K99. Sixty Holstein calves were randomly assigned to 5 treatments: fed a basal diet (N-CON); fed a basal diet and challenged with E.coli K99 (P-CON); fed a basal diet supplemented with C.tropicalis (CT), mulberry leaf flavonoids (MLF), and the combination of the two additives (CM), respectively, and challenged with E.coli K99. The MLF and CM groups had significantly higher average daily grain and feed efficiency, and significantly lower fecal scores compared with the P-CON group after E. coli K99 challenge. The supplementation groups increased the relative abundance, at the phylum level, of Bacteroidetes and Proteobacteria, whereas at the genus level, they increased the relative abundance of Prevotella, Lactobacillus, and Enterococcus. Quantitative PCR revealed that the CT, MLF, and CM groups had significantly lower copy numbers of E.coli K99 compared with the P-CON group. The CT, MLF, and CM treatments reduce days of diarrhea, improve intestinal health, and beneficially manipulate the intestinal microbiota in preweaned calves.
Introduction
Neonatal calf diarrhea is a serious health and welfare problem on dairy farms, with the resulting high mortality and morbidity contributing to considerable economic losses worldwide in the cattle industry1–3. Enterotoxigenic Escherichia coli expressing K99 fimbriae and heat-stable type Ia (STa) toxin is one of the major pathogens associated with neonatal calf diarrhea4–6. The K99 fimbrial adhesins promote attachment and colonization of bacterial cells to the surface of epithelial cells of the small intestines, while the STa toxin damages the epithelial cells and disrupts fluid homeostasis, resulting in fluid and electrolyte hypersecretion that leads to watery diarrhea, dehydration, and acidosis in neonatal calves7–9.
Although antibiotics are given therapeutically after scours is observed, concerns have been raised regarding microbial resistance to antibiotics and increasing passage of laws banning the use of antibiotics in livestock production throughout the world. Therefore, alternatives to antibiotics for prevention and treatment of neonatal calf diarrhea are urgently needed to maintain the health of livestock. Recently recognized alternatives include yeasts and flavonoid-containing plant extracts, which are showing beneficial effects on animal intestinal health in an increasing number of studies10–13.
Candida tropicalis, a yeast of the Candida genus, is considered an important inhabitant of the healthy animal gut, and it can commonly be isolated from the gastrointestinal tracts of humans14, bovines15, birds16, and fish17. Previous ruminal fermentation studies demonstrated that C. tropicalis stimulated total and cellulolytic microbial populations, increased gas production, and activated in vitro ruminal fermentation, indicating its excellent potential for use as a feed additive in ruminants15, 18, 19. Long et al. reported C. tropicalis stimulated lactate uptake by Selenomonas ruminantium and increased the production of acetate and propionate and the ratio of propionate to acetate20. C. tropicalis isolated from some fish gastrointestinal tracts increased phytase and tannase production, as well as crude protein, lipid, and mineral contents, and reduced the antinutritional factors in different plant feedstuff17, 21.
Plant-derived flavonoids, such as those extracted from mulberry (Morus alba) leaves, have also shown health-promoting properties due to the alteration of the expression and activity of key enzymes in lipid and carbohydrate metabolism22, 23, induction of protective effects against hydroxyl and superoxide radical damage24, antimicrobial activity25, 26, antiparasitic activity27, and antioxidant activity24, 28, among others. Previous studies have shown that flavonoids isolated from the leaves of many different plants exhibit antimicrobial activity against Gram-negative and Gram-positive bacteria and fungi in in vitro antimicrobial assays29–34. Phytochemical investigations have shown that lipophilic flavonoids exert their antimicrobial activities through their ability to penetrate biological membranes35. Omosa et al.26 demonstrated that the antibacterial activity depended on the relative positions of the hydroxyl and methoxy groups on the flavone skeleton, and that a methoxy group at the C-3 position in the flavone skeleton was associated with good activity against E. coli. Several studies have demonstrated that probiotic yeasts might have inhibitory activity against specific pathogens36–38, but very few experimental and clinical trials have examined C. tropicalis as a possible probiotic. A considerable number of studies have shown that flavonoids isolated from different plants demonstrate antimicrobial activity, but most of these have been in vitro investigations. The aim of the present study was therefore to examine the effects of dietary supplementation with C. tropicalis and mulberry leaf flavonoids, singly or in combination, on the intestinal bacterial community composition in preweaned calves challenged with E. coli K99.
Results
Growth performance of calves
In order to determine the effects of C. tropicalis, mulberry leaf flavonoids, and their combination on growth performance of calves before and after E. coli K99 challenge, the average daily grain (ADG), dry matter intake (DMI), and feed efficiency were analyzed (Table 1). Before E. coli K99 challenge, the ADG of N-CON, P-CON, CT, MLF, and CM groups were 0.60, 0.59, 0.62, 0.69, and 0.70 kg/d, respectively. The ADG and DMI had no significant difference among groups, but the feed efficiency of calves in the MLF group was significantly higher than that in the control groups. After E. coli K99 challenge, the ADG of N-CON, P-CON, CT, MLF, and CM groups were 0.89, 0.56, 0.62, 0.85, and 0.92 kg/d, respectively. The ADG and feed efficiency of calves in the MLF and CM groups were significantly higher than that in the P-CON and CT groups, but had no significant difference compared with that in the N-CON group. The DMI had no significant difference among groups.
Table 1.
Effects of Candida tropicalis, mulberry leaf flavonoids, and their combination on growth performance of calves.
| Item | Treatments | P | ||||
|---|---|---|---|---|---|---|
| N-CON | P-CON | CT | MLF | CM | ||
| 28–56 d (before challenge) | ||||||
| Average daily gain, kg/d | 0.60 ± 0.11 | 0.59 ± 0.12 | 0.62 ± 0.11 | 0.69 ± 0.08 | 0.70 ± 0.11 | 0.29 |
| Dry matter intake, kg/d | 1.17 ± 0.10 | 1.16 ± 0.11 | 1.13 ± 0.09 | 1.14 ± 0.09 | 1.23 ± 0.12 | 0.85 |
| Feed efficiency1, % | 51.28 ± 3.67b | 51.30 ± 3.30b | 54.87 ± 4.51ab | 60.53 ± 3.46a | 56.91 ± 4.63ab | 0.02 |
| 57–63 d (after challenge) | ||||||
| Average daily gain, kg/d | 0.89 ± 0.11a | 0.56 ± 0.12b | 0.62 ± 0.14b | 0.85 ± 0.13a | 0.92 ± 0.11a | <0.0001 |
| Dry matter intake, kg/d | 1.43 ± 0.12 | 1.38 ± 0.16 | 1.43 ± 0.10 | 1.40 ± 0.18 | 1.47 ± 0.14 | 0.65 |
| Feed efficiency% | 62.40 ± 6.94a | 40.95 ± 7.35b | 43.29 ± 8.76b | 61.34 ± 7.52a | 62.51 ± 7.30a | <0.0001 |
1Feed efficiency = (Average daily gain (kg/d)/Dry matter intake (kg/d)) × 100%. Values are mean ± SD. a,bValues in the same row with different superscripts differ significantly (P < 0.05).
Fecal scores across different treatments
In order to observe whether dietary supplementation with the two alternatives to antibiotics would have effects on prevention of diarrhea in preweaned calves challenged with E. coli, fecal scores were performed after E. coli K99 challenge. Calves from the P-CON group suffered from diarrhea on the first day after the E. coli K99 challenge, and all the other calves challenged with E. coli K99 experienced diarrhea on the second day. However, calves fed C. tropicalis (CT), mulberry leaf flavonoids (MLF), or the combination (CM) had lower fecal scores and experienced fewer days with fecal score >2 than that in the P-CON group (Fig. 1). Fecal scores of the MLF and CM groups were significantly lower (P < 0.05) when compared with the P-CON group on d 2–5 after the E. coli K99 challenge. Dietary supplementation with C. tropicalis and mulberry leaf flavonoids, singly or in combination, improved fecal scores and reduced the number of days with mild or watery diarrhea.
Figure 1.
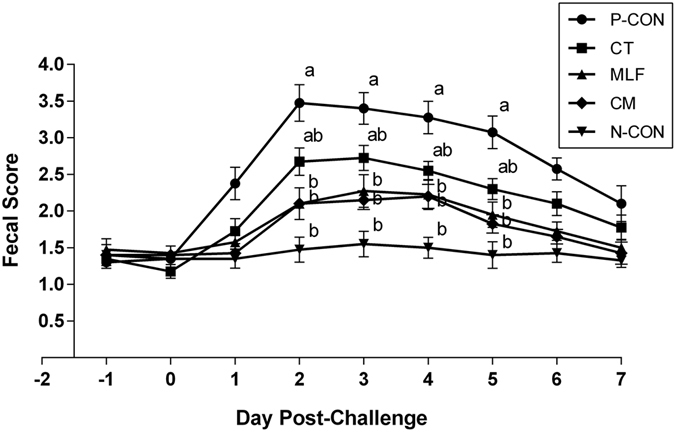
The effect of feeding Candida tropicalis and mulberry leaf flavonoids, singly or in combination, on fecal scores of dairy calves challenged with E. coli K99. N-CON: fed a basal diet and not challenged with E.coli K99; P-CON: fed a basal diet and challenged with E.coli K99; CT, MLF, and CM: fed a basal diet supplemented with C.tropicalis, mulberry leaf flavonoids, and the combination of the two additives, respectively, and challenged with E.coli K99. The X-axis denotes the day post-challenge and the Y-axis the fecal score. Values are mean ± SD. a,bValues in the same column with different superscripts differ significantly (P < 0.05).
Sequencing depth and alpha diversity
Illumina MiSeq sequencing analysis of the 24 jejunum digesta samples of preweaned calves generated a total of 1017,001 trimmed reads with an average of 42,622 ± 12,268 reads per sample after data filtering, quality control, and removal of primers, chimeras, and low-confidence singletons. For further analyses, all reads were classified into 512 operational taxonomic units (OTUs) based on ≥ 97% nucleotide sequence identity between reads.
We assessed whether our sequencing depth provided sufficient diversity coverage to accurately describe the bacterial composition of each group by generating sample-based rarefaction curves for each group (Supplementary Fig. S1). The results indicated a sufficient sequencing depth for the samples from different groups. The OTU numbers of N-CON, P-CON, CT, MLF, and CM were 154, 208, 217, 189, and 289, respectively. The CM group had significantly higher OTU numbers compared with other groups (P < 0.05). The community diversity (Shannon index) differed significantly (P < 0.05) between the CM group and other groups in preweaned calves (Fig. 2).
Figure 2.
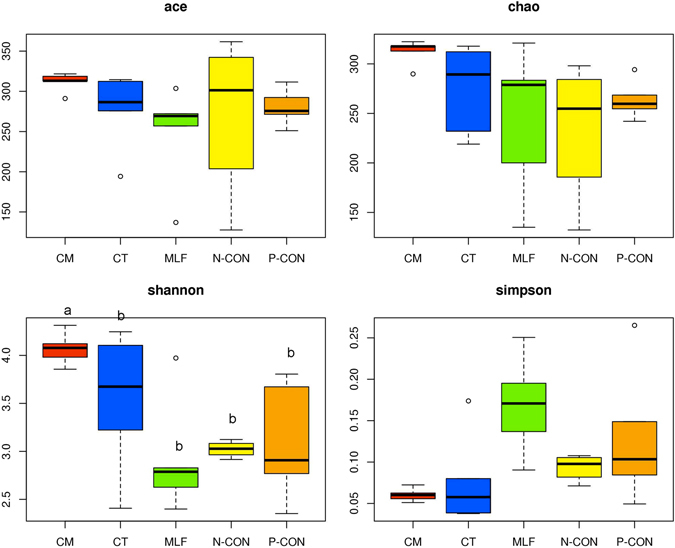
Community richness estimates (ACE and Chao1) and diversity indices (Shannon and Simpson) for different treatments. a,bBoxes with different superscripts differ significantly (P < 0.05).
Gut bacterial composition across different treatments
At the phylum level, 15 phyla were identified in the samples from the preweaned calf guts, and these were dominated by Firmicutes, Actinobacteria, Proteobacteria, and Bacteroidetes, regardless of treatment group (Fig. 3 and Table 2). However, the relative abundance of these predominant phyla varied considerably among the different calf groups. The Firmicutes dominated in all treatment groups (Table 2). The phylum Actinobacteria was abundant in the samples taken from the N-CON and P-CON groups when compared with the MLF, CT and CM groups, and this phylum was significantly lower (P < 0.05) in the MLF group than in the N-CON and P-CON groups. The phyla Bacteroidetes and Proteobacteria were abundant in samples taken from the MLF, CT, and CM groups when compared with the N-CON and P-CON groups, and the two phyla were more abundant in the MLF and CM groups (P < 0.05) than in the other groups. Among other minor phyla, the TM7 and Verrucomicrobia were more prominent in the MLF, CT and CM groups, while the Tenericutes and Synergistetes were more prominent in the N-CON and P-CON groups.
Figure 3.
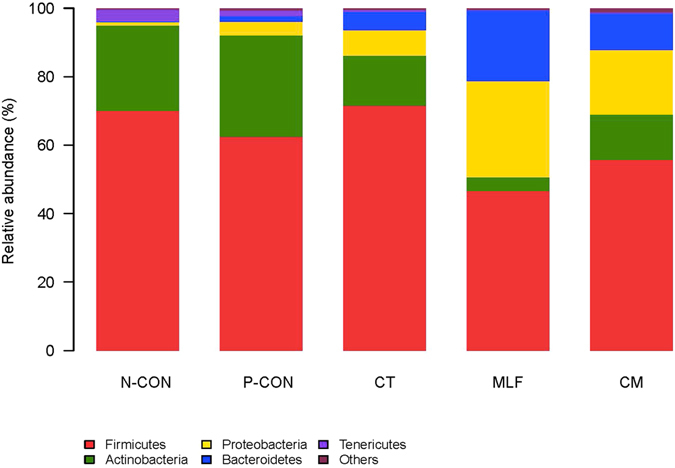
Phylum level composition. Color-coded bar plot showing the relative abundances of the five most abundant phyla across different groups.
Table 2.
Phylum-level composition of the jejunum digesta among different groups.
| Phylum | Relative abundance (%) | P | ||||
|---|---|---|---|---|---|---|
| N-CON | P-CON | CT | MLF | CM | ||
| Acidobacteria | 0.00 ± 0.00b | 0.03 ± 0.02b | 0.02 ± 0.02b | 0.04 ± 0.03b | 0.13 ± 0.08a | 0.0103 |
| Actinobacteria | 24.86 ± 2.75a | 29.59 ± 16.18a | 14.53 ± 12.38ab | 4.07 ± 2.30b | 13.23 ± 10.67ab | 0.0112 |
| Bacteroidetes | 0.36 ± 0.23c | 1.65 ± 1.51c | 5.37 ± 4.47bc | 20.62 ± 17.28a | 10.82 ± 2.87ab | 0.0022 |
| Chloroflexi | 0.02 ± 0.00 | 0.07 ± 0.05 | 0.03 ± 0.02 | 0.06 ± 0.04 | 0.15 ± 0.12 | 0.1822 |
| Firmicutes | 70.07 ± 7.78 | 62.54 ± 15.51 | 71.59 ± 9.20 | 46.59 ± 28.78 | 55.70 ± 6.42 | 0.1798 |
| Proteobacteria | 0.99 ± 0.30c | 3.97 ± 3.29bc | 7.43 ± 4.54b | 28.10 ± 14.77a | 18.85 ± 2.62a | <0.0001 |
| Synergistetes | 0.24 ± 0.18 | 0.19 ± 0.13 | 0.05 ± 0.02 | 0.06 ± 0.04 | 0.01 ± 0.00 | 0.4400 |
| Tenericutes | 3.36 ± 2.18 | 1.69 ± 0.84 | 0.62 ± 0.57 | 0.16 ± 0.16 | 0.28 ± 0.11 | 0.2553 |
| TM7 | 0.05 ± 0.02 | 0.07 ± 0.03 | 0.21 ± 0.14 | 0.08 ± 0.06 | 0.28 ± 0.19 | 0.1281 |
| Verrucomicrobia | 0.02 ± 0.02b | 0.05 ± 0.05b | 0.04 ± 0.03b | 0.07 ± 0.06b | 0.22 ± 0.09a | 0.0468 |
Values are mean ± SD. a,bValues in the same row with different superscripts differ significantly (P < 0.05).
At the genus level, 185 genera belonging to the 15 phyla were detected in the samples. In total, 25 most abundant shared genera with a relative abundance ≥ 0.1% (Table 3) were present in all samples across different groups, but their relative abundance levels were markedly different among the different treatment groups (Fig. 4 and Table 3). The Prevotella, Enterococcus, and Lactobacillus made up the main bacterial species in MLF group, with Prevotella and Lactobacillus having the highest relative abundances in this group. The Enterococcus and Pseudomonas were the two most abundant genera in the samples from the CM group.
Table 3.
Shared genera with a relative abundance ≥ 0.1% in all samples among different groups.
| Phylum | Genus | Relative abundance (%) | P | ||||
|---|---|---|---|---|---|---|---|
| N-CON | P-CON | CT | MLF | CM | |||
| Actinobacteria | Arthrobacter | 0.11 ± 0.05 | 0.30 ± 0.16 | 0.41 ± 0.33 | 0.23 ± 0.25 | 0.55 ± 0.07 | 0.0945 |
| Atopobium | 4.83 ± 2.31 | 2.27 ± 1.15 | 3.45 ± 2.88 | 0.79 ± 0.56 | 0.33 ± 0.13 | 0.0072 | |
| Bifidobacterium | 0.40 ± 0.19 | 0.76 ± 0.74 | 0.51 ± 0.40 | 0.54 ± 0.32 | 0.45 ± 0.45 | 0.9593 | |
| Corynebacterium | 0.32 ± 0.28 | 0.36 ± 0.12 | 0.34 ± 0.16 | 0.17 ± 0.23 | 0.49 ± 0.25 | 0.3389 | |
| Olsenella | 16.24 ± 6.21a | 22.93 ± 14.68a | 10.50 ± 9.04ab | 2.02 ± 1.93b | 9.72 ± 8.05ab | 0.0341 | |
| Bacteroidetes | Alistipes | 0.12 ± 0.07b | 0.29 ± 0.27b | 1.11 ± 0.83ab | 1.03 ± 0.78ab | 1.89 ± 0.89a | 0.0640 |
| Prevotella | 0.05 ± 0.03b | 0.06 ± 0.05b | 2.06 ± 2.02b | 15.22 ± 7.31a | 3.23 ± 2.87b | 0.0039 | |
| Myroides | 0.08 ± 0.04 | 0.23 ± 0.13 | 0.20 ± 0.18 | 0.20 ± 0.19 | 0.34 ± 0.16 | 0.4611 | |
| Firmicutes | Acetitomaculum | 2.05 ± 1.94 | 4.07 ± 3.69 | 0.71 ± 0.43 | 0.56 ± 0.20 | 0.65 ± 0.10 | 0.0682 |
| Acidaminococcus | 0.70 ± 0.47 | 0.32 ± 0.21 | 0.99 ± 0.68 | 1.60 ± 0.82 | 1.18 ± 0.92 | 0.8772 | |
| Bacillus | 1.43 ± 0.39 | 3.58 ± 2.61 | 5.03 ± 3.63 | 2.02 ± 1.51 | 5.55 ± 2.72 | 0.2204 | |
| Butyrivibrio | 0.93 ± 0.52 | 1.54 ± 1.32 | 1.17 ± 1.19 | 0.74 ± 0.66 | 1.05 ± 1.17 | 0.8966 | |
| Dialister | 2.14 ± 1.76 | 1.78 ± 0.88 | 0.62 ± 0.59 | 2.10 ± 1.25 | 1.02 ± 0.90 | 0.6678 | |
| Enterococcus | 0.35 ± 0.14b | 1.42 ± 1.21b | 3.63 ± 2.12b | 8.28 ± 6.47b | 13.83 ± 6.37a | 0.0209 | |
| Howardella | 0.54 ± 0.27a | 0.34 ± 0.28ab | 0.16 ± 0.09b | 0.22 ± 0.26ab | 0.07 ± 0.05b | 0.0501 | |
| Lactobacillus | 0.30 ± 0.25b | 0.73 ± 0.47b | 12.70 ± 6.43a | 13.14 ± 7.26a | 0.16 ± 0.07b | 0.0121 | |
| Lactococcus | 0.45 ± 0.13b | 1.21 ± 0.83ab | 2.06 ± 1.54ab | 0.94 ± 0.40ab | 2.52 ± 0.92a | 0.0931 | |
| Megasphaera | 11.08 ± 9.95 | 12.58 ± 9.10 | 10.32 ± 8.07 | 2.48 ± 2.36 | 5.04 ± 3.57 | 0.2282 | |
| Mitsuokella | 3.23 ± 2.34 | 0.93 ± 0.72 | 2.08 ± 1.07 | 1.14 ± 0.68 | 0.41 ± 0.39 | 0.3383 | |
| Pseudoramibacter | 0.43 ± 0.23ab | 0.85 ± 0.76a | 0.49 ± 0.17ab | 0.11 ± 0.05b | 0.16 ± 0.14b | 0.0451 | |
| Roseburia | 0.26 ± 0.18 | 2.12 ± 1.79 | 0.33 ± 0.29 | 0.33 ± 0.28 | 0.28 ± 0.15 | 0.7039 | |
| Ruminococcus | 0.27 ± 0.25 | 0.11 ± 0.03 | 0.25 ± 0.14 | 0.17 ± 0.12 | 0.41 ± 0.16 | 0.2007 | |
| Syntrophococcus | 6.03 ± 5.03a | 0.78 ± 0.17b | 0.68 ± 0.28b | 0.21 ± 0.11b | 0.27 ± 0.21b | 0.0002 | |
| Proteobacteria | Pseudomonas | 0.64 ± 0.16b | 1.77 ± 1.32b | 3.42 ± 0.36a | 2.60 ± 2.45ab | 5.94 ± 1.28a | 0.0006 |
| Psychrobacter | 0.11 ± 0.09 | 0.17 ± 0.06 | 0.26 ± 0.22 | 0.10 ± 0.08 | 0.42 ± 0.27 | 0.2002 | |
Values are mean ± SD. a,bValues in the same row with different superscripts differ significantly (P < 0.05).
Figure 4.
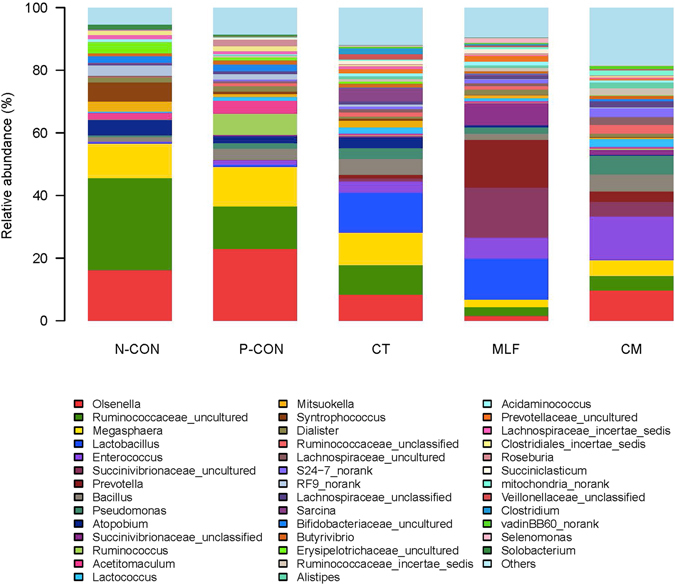
Genus level composition. Color-coded bar plot showing the relative abundances of different genera across different groups.
Quantification of total bacteria, E.coli K99, and the four selected bacterial species
We used absolute quantitative real-time PCR to investigate the quantification of total bacteria, E.coli K99, and four selected bacterial species that showed statistically significant in sequencing results (Fig. 5). The copy numbers of total bacteria and of the genus Prevotella were significantly higher in MLF group than in other groups. The genus Enterococcus had significantly higher copy numbers in CM group than in P-CON and CT groups. The genus Lactobacillus had significantly higher copy numbers in MLF group than in N-CON, P-CON, and CM groups. The copy numbers of Pseudomonas were no significant difference across different treatments. The relative abundances of the four bacterial species calculated by quantitative real-time PCR (Supplementary Table S1) had the similar statistical differences with that calculated by Illumina MiSeq sequencing. No E. coli K99 was detected in the digesta samples taken from the N-CON group. The copy numbers of the E.coli K99 were significantly higher in P-CON group than that in CT, MLF, and CM groups (Fig. 5).
Figure 5.
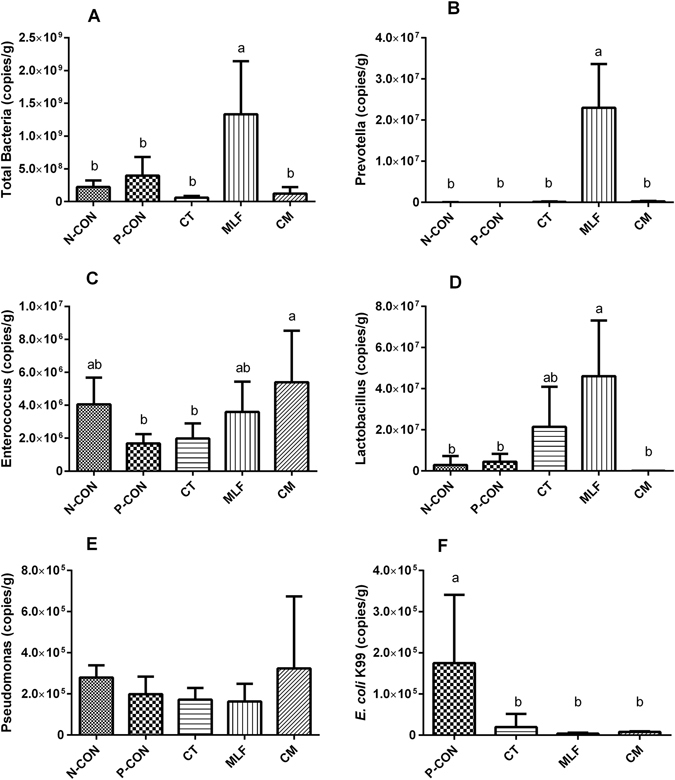
The copy numbers of selected bacterial species and of E.coli K99 in the jejunum digesta. A to F represent the copy numbers per gram of total bacteria, Prevotella, Enterococcus, Lactobacillus, Pseudomonas, and E.coli K99, respectively. In figure F, no E. coli K99 was detected in N-CON group. Values are mean ± SD. a,bBars with different superscripts differ significantly (P < 0.05).
Discussion
A role in disease has been reported for the intestinal microbiota, whereby gut microbes function as a key interface between host and environment and some bacteria protect the host from pathogens that cause infectious diarrhea39. The use of antibiotics has already been reported to affect intestinal microbiota profiles in humans40 and swine41. Evidence is also emerging that antibiotic usage in animal production may contribute to the antibiotic resistance of human pathogens42. Alternatives to antibiotics for the prevention and treatment of disease in young calves are continuously being evaluated and are urgently required in order to minimize the need for antibiotics43. This need prompted the present analysis of the effects of two potential antibiotic alternatives–C. tropicalis and mulberry leaf flavonoids, supplied singly or in combination–on the intestinal bacterial community composition in preweaned calves challenged with E. coli K99.
In the present study, the MLF and CM groups had significantly higher ADG and feed efficiency, and significantly lower fecal scores compared with the P-CON group after E. coli K99 challenge. Diet supplementation with C. tropicalis and mulberry leaf flavonoids, singly or in combination, significantly increased the relative abundance of Lactobacillus and Enterococcus, which belong to lactic acid producing bacteria (LAB). Many studies indicated that diet supplemented with LAB could improve weight gain and feed efficiency, and reduced diarrhea incidence44–46. Furthermore, the structure of flavonoids is similar to estradiol, which can regulate the secretion of growth hormone by the hypothalamus-pituitary hormone axis47. Growth hormone directly accelerates protein synthesis or stimulates insulin-like growth factors 1 to promote muscle tissue growth and body weight gain48.
Dietary supplementation with C. tropicalis and mulberry leaf flavonoids, singly or in combination, improved fecal scores and reduced the number of days with mild or watery diarrhea, suggesting protective effects of both supplements in calves with a high risk of morbidity. Previous studies showed that a yeast culture supplement decreased the risk of diarrhea due to E. coli, because the E. coli adhered to the oligosaccharides present in the yeast cell walls rather than attaching to and invading the intestinal cells49, 50. This might be the reason that the copy number of E. coli K99 in CT group was significantly lower than that in the P-CON group. A considerable number of studies have demonstrated that flavonoids isolated from the leaves of many different plants show good antimicrobial activity against E. coli 26, 31, 33, 34. The antimicrobial activity of flavonoids is apparently due to their ability to penetrate biological membranes35. The antimicrobial activity of flavonoids might be one of the reasons that the MLF group had significantly lower copy number of E. coli K99 compared with the P-CON group. As mentioned above, feeding C. tropicalis, mulberry leaf flavonoids, or their combination increased the relative abundance of LAB in the calf gut. The LAB can adhere to the intestinal tract and produce lactate and acetate, which reduces the attachment and colonization of E. coli on the surface of intestinal epithelial cells44, 46. The increasing abundance of LAB in the calf gut might be another reason that the CT, MLF, and CM groups had significantly lower copy number of E. coli K99 compared with the P-CON group. The observed increase in LAB abundance would be expected to improve gut health and may explain the benefits seen in fecal scores and diarrhea observed in this study when calves were fed either of the two supplements or their combination.
The community diversity index (Fig. 2) and OTU number significantly increased in the CM group compared with all other groups, suggesting that the combination of C. tropicalis and mulberry leaf flavonoid supplements increased the intestinal community diversity in preweaned calves, whereas each supplement on its own did not have this effect. As mentioned above, flavonoids isolated from the leaves of many plants exhibit antimicrobial activity29–34, which means flavonoids might decrease the community diversity. However, in the current study, dietary supplementation with the combination of C. tropicalis and mulberry leaf flavonoid increased the intestinal community diversity. The reason might be the synergistic effect of C. tropicalis and mulberry leaf flavonoid. The action mechanism needs to be further studied in future.
The OTU number and community diversity of the gastrointestinal tracts of the calves in the present study were lower than previously reported in some studies. One reason might be that the previous studies examined weaned calves or adult cows, and that the species richness and diversity in the gut increases with age51–53. The gradual increase in consumption of large amounts of different solid feeds might be one reason for an age-dependent increase in bacterial diversity54, 55. Another reason might be that the digesta samples were collected from different gastrointestinal tract regions in different studies. Malmuthuge et al. reported that the highest number of OTUs and the greatest bacterial community diversity indices were observed in the rumen, followed by the large intestine (cecum and colon), and then the small intestine (jejunum and ileum)56. The rumen and large intestine are regarded as fermentation tanks for microbial fermentation of indigestible dietary substrates, and the retention time of digesta is longer in the rumen and large intestine than in the small intestine, which would facilitate the growth of a more complex bacterial community56.
The dominant phyla found in all groups were Firmicutes, Actinobacteria, Proteobacteria, and Bacteroidetes. The groups shared 113 genera, and the 25 most abundant shared genera, with a relative abundance ≥ 0.1%, were present in all samples across the different groups. These dominant phyla and shared genera represented the core microbiome of the calves of this age, irrespective of the treatments. However, the relative abundance of the phyla and genera from this shared community varied considerably among the groups.
In the present study, Firmicutes showed the highest overall relative abundance and dominated in all the treatment groups. Our findings are consistent with the studies of Oikonomou et al. and Malmuthuge et al., who detected a significantly higher relative abundance of Firmicutes in the gut digesta of preweaned Holstein calves53, 56. Malmuthuge et al. implied that Firmicutes tended to more readily colonize the small intestinal digesta of preweaned calves, while Bacteroidetes tended to more readily colonize the rumen and large intestinal contents56. The regional variations in the gastrointestinal tract of preweaned calves revealed differences in the dominant bacterial community56.
The LABs, such as Lactobacillus and Enterococcus, have known beneficial effects on feed efficiency and animal health46, and both Lactobacillus and Enterococcus have been used as direct-fed microbials for a long time. Studies have reported that direct feeding of calves or dairy cows with microbials consisting of Lactobacillus and Enterococcus prevented declines in ruminal pH and decreased the risk of metabolic acidosis. These effects occurred due to facilitation of the growth of ruminal microorganisms adapted to the presence of lactic acid and by stimulation of the utilization of lactic acid by lactic acid utilizing bacteria46, 57, 58. Calves fed Lactobacillus and Enterococcus also showed an improved abundance of ruminal cellulolytic bacteria, such as Butyrivibrio fibriosolvens and Eubacterium ruminantium 58. In our study, the relative abundance of Lactobacillus increased in the CT and MLF groups, and the relative abundance of Enterococcus increased in the CT, MLF, and CM groups.
The phylum Bacteroidetes was significantly more abundant in the MLF and CM groups, and especially in the MLF group, when compared to the N-CON and P-CON groups. Bacteroidetes was composed mainly of the genera Alistipes, Myroides, and Prevotella in all groups, and the genus Prevotella was significantly more abundant in the MLF group, accounting for up to 15.22% of the total reads. The Prevotella genus contains several ruminal species that are capable of utilizing starches, other non-cellulosic polysaccharides, and simple sugars as energy sources59, and this genus was more abundant in the CT, MLF, and CM groups than in the N-CON and P-CON groups.
Previous studies have reported that the Bacteroidetes found in the rumens of preweaned calves fed whole milk or milk replacer contain more Bacteroides than Prevotella 56, 60, whereas the rumens of adult cattle contain almost exclusively Prevotella 51. A recent study showed that the relative abundance of Bacteroides in the feces of adult cattle was negatively associated with a high fiber diet54. The diet of preweaned calves, which mainly includes whole milk or milk replacer, is rich in protein, fat, and sugar, whereas the adult cattle’s diet is composed mainly of plant fiber. In the present study, the small intestinal digesta of the 9-week-old preweaned calves contained more Prevotella than Bacteroides and the diet of the calves consisted of milk replacer, calf starter, and hay, with the calf starter and hay dominating, suggesting that, during calf development, increased fiber ingestion and decreased milk consumption decreases the relative abundance of Bacteroides 55.
Other phyla, such as the Actinobacteria and Proteobacteria, were also found in high proportion in all treatment groups. The relative abundance of Actinobacteria decreased and Proteobacteria increased in the MLF, CT, and CM groups, and especially in the MLF group, when compared with the N-CON and P-CON groups, indicating that the dietary supplements helped to encourage the growth of Proteobacteria and to inhibit the growth of Actinobacteria in the preweaned calves.
Conclusions
The results presented here provide new information regarding the effects of dietary supplementation with two alternatives to antibiotics (C. tropicalis and mulberry leaf flavonoids), and their combination on the intestinal microbiota in preweaned calves challenged with E. coli K99. The MLF and CM groups had significantly higher ADG and feed efficiency compared with the P-CON group after E. coli K99 challenge. Dietary supplementation with the two alternatives to antibiotics, singly or in combination, improved fecal scores and reduced days with mild or watery diarrhea. Dietary supplementation with the combination of C. tropicalis and mulberry leaf flavonoids significantly increased the number of OTUs and the community diversity. Dietary supplementation with the two alternatives to antibiotics, singly or in combination, increased the relative abundance, at the phylum level, of Bacteroidetes and Proteobacteria and decreased the relative abundance of Actinobacteria, while at the genus level, this supplementation increased the relative abundance of Prevotella, Lactobacillus, and Enterococcus. Quantitative real-time PCR revealed that dietary supplementation with mulberry leaf flavonoids significantly increased the copy numbers of total bacteria and of the genera Prevotella and Lactobacillus in jejunum digesta. The CT, MLF, and CM groups had significantly lower copy numbers of E.coli K99 compared with the P-CON group. Our results establish a strong foundation for evaluating the potential of C. tropicalis and mulberry leaf flavonoids as feed additives for the reduction of diarrhea and improvement of intestinal health in preweaned calves challenged with E. coli K99.
Materials and Methods
Animal experiment and sample collection
The experiments were approved by the Animal Ethics Committee of the Chinese Academy of Agricultural Sciences, Beijing, China. All methods were performed in accordance with the relevant standard operating procedures approved by the above mentioned ethics committee.
Sixty newborn Holstein bull calves with body weight 40 ± 2.0 kg were purchased from a commercial dairy farm, fed colostrum within 2 h after birth and for the first 3 d of life. The calves were then housed individually in 1.6 m × 3.6 m pens with wood shavings for bedding at the experimental farm of Chinese Academy of Agricultural Sciences and fed with commercial pasteurized whole milk twice a day until d 21. Over the following week, the pasteurized whole milk was gradually replaced with milk replacer (1:7 w/v; Table 4), which was then fed twice daily at a total amount corresponding to 10% of the calves’ body weight until d 64. All calves had ad libitum access to calf starter (Table 4) and hay from d 4 to 64. Clean fresh water was offered free choice daily throughout the study.
Table 4.
Ingredients and chemical composition of starter and milk replacer.
| Item | Starter (%) | Milk replacer (%) |
|---|---|---|
| Ingredient | ||
| Corn | 20.00 | |
| Extruded corn | 22.90 | |
| Soybean meal | 20.00 | |
| Extruded soybean | 18.00 | |
| Dried whey | 5.00 | |
| Wheat bran | 10.00 | |
| Calcium hydrogen phosphate | 0.80 | |
| Limestone | 1.80 | |
| Salt | 0.50 | |
| Premix1 | 1.00 | |
| Chemical composition | ||
| DM2 | 85.36 | 95.36 |
| OM | 92.21 | 94.85 |
| CP | 19.08 | 24.27 |
| Ether extract | 2.21 | 12.85 |
| NDF | 18.59 | 4.02 |
| ADF | 10.65 | 2.11 |
| Calcium | 1.09 | 1.07 |
| Phosphorous | 0.47 | 0.48 |
| Gross energy, MJ/kg | 15.45 | 19.86 |
1Premix was manufactured by the Precision Animal Nutrition Research Centre, Beijing, China. Premix provided per kilogram of concentrate: vitamin A, 15,000 IU; vitamin D, 5,000 IU; vitamin E, 50 mg; Fe, 90 mg; Cu, 12.5 mg; Mn, 30 mg; Zn, 90 mg; Se, 0.3 mg; I, 1.0 mg. 2DM = dry matter; OM = organic matter; CP = crude protein; NDF = neutral detergent fiber; ADF = acid detergent fiber.
At 28 d of age, 60 calves were randomly assigned to 5 treatments with 12 calves each based on BW and date of birth. The treatments were as follows: 1) negative control (N-CON) treatment: fed a basal diet and not challenged with E. coli K99; 2) positive control (P-CON): fed a basal diet and orally challenged with E. coli K99 (30 mL; 1 × 109 CFU/mL); 3) C. tropicalis treatment (CT): fed a basal diet supplemented daily with C. tropicalis (5.0 × 109 CFU/g; 1 g/calf) and then orally challenged with E. coli K99 (30 mL; 1.0 × 109 CFU/mL); 4) mulberry leaf flavonoid treatment (MLF): fed a basal diet supplemented daily with mulberry leaf flavonoids (5.0%, w/w; 3 g/calf) and then orally challenged with E. coli K99 (30 mL; 1.0 × 109 CFU/mL); and 5) combined C. tropicalis and mulberry leaf flavonoid treatment (CM): fed a basal diet supplemented daily with both C. tropicalis (5.0 × 109 CFU/g; 1 g/calf) and mulberry leaf flavonoids (5.0%, w/w; 3 g/calf) and then orally challenged with E. coli K99 (30 mL; 1.0 × 109 CFU/mL). The basal diet (Table 4) was free of antibiotics or other additives. Experimental treatments were applied from d 28 to 64, with the oral challenges with E. coli K99 carried out on d 57.
The body weight of each calf was recorded at d 28, 56, and 64 of age. Feed intake and fecal scores of each calf was recorded daily. Five calves were selected from each group and euthanized at 64 d of age. Jejunum digesta samples were collected from the middle of the jejunum and at the same site consistently for all the animals. Jejunum digesta samples were snap frozen in liquid nitrogen and kept at −80 °C until further analysis. The flow scheme of this trial was shown in Fig. 6.
Figure 6.
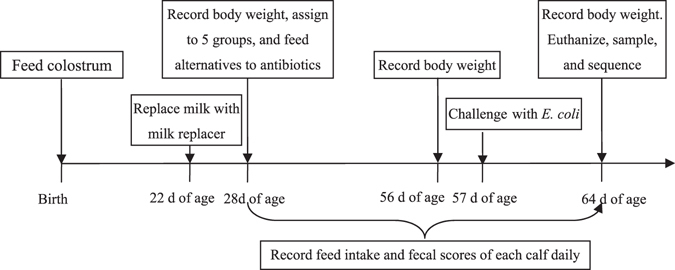
The flow scheme of this study. Calves were fed colostrum within 2 h after birth. At 22 d of age, whole milk was gradually replaced with milk replacer, which was then fed twice daily until d 64. At d 28, 56, and 64 of age, body weight of each calf was recorded. From d 28 to 64 of age, feed intake and fecal scores were recorded. At 28 d of age, calves were randomly assigned to 5 treatments and treated with alternatives to antibiotics. At 57 d of age, E. coli K99 challenge were carried out. At 64 d of age, calves were euthanized and jejunum digesta samples were collected and sequenced.
Additives preparation and oral challenge
A commercial CT probiotic (5.0 × 109 CFU/g) was purchased from Beijing Huanong Biological Engineering Co., Ltd., Beijing, China. Commercial mulberry (Morus alba Linn.) leaf extracts were purchased from Xi’an Feida Biotechnology Co. Ltd., Xi’an, China. The mulberry leaf extracts were vacuum-dried and contained 5.0% flavonoids (w/w). The CT and MLF were mixed with the milk replacer and administered with the morning feeding.
The E. coli K99 strain was obtained from China Veterinary Culture Collection Center. The viability of the culture was ensured by growing it aerobically in Luria Bertani broth for 24 h at 37 °C with shaking (120 rpm). A growth curve was constructed for E. coli K99 to determine the appropriate incubation time required to reach the target challenge dosage of approximately 1.0 × 109 CFU/mL. E. coli K99 (30 mL, 1.0 × 109 CFU/mL) was mixed with milk replacer and fed to the calves on d 57.
Fecal consistency scoring
Fecal consistency scoring was performed daily before the morning milk feeding using a 1 to 4 scale, as described by Magalhaes et al.50. Briefly, fecal consistency was scored as 1 when firm, 2 when soft or of moderate consistency, 3 when runny or mild diarrhea, and 4 when watery and profuse diarrhea. Daily fecal scores were generated for individual calves for statistical analyses. Calves with fecal score >2 were used for analysis of incidence of diarrhea.
DNA extraction, PCR amplification, and Illumina miseq sequencing
Microbial DNA was extracted from jejunum digesta samples using the OMEGA E.Z.N.A.® digesta DNA Kit (Omega Bio-tek, Norcross, GA, USA), according to the manufacturer’s protocols. The quality and quantity of the DNA were measured using an ND1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA). The V3–V4 regions of the bacterial 16S ribosomal RNA gene were amplified by PCR (95 °C for 3 min, followed by 27 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s and a final extension at 72 °C for 10 min) using primers 338 F 5′-barcode-ACTCCTACGGGAGGCAGCAG-3′ and 806 R 5′-barcode-GGACTACHVGGGTWTCTAAT-3′61, where barcode is an eight-base sequence unique to each sample. PCR reactions were performed in triplicate 20 μl mixtures containing 4 μl 5×FastPfu Buffer, 2 μl 2.5 mM dNTPs, 0.8 μl of each primer (5 μM), 0.4 μl FastPfu Polymerase, and 10 ng template DNA.
Amplicons were extracted from 2% agarose gels, purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer’s instructions, and quantified using the QuantiFluor™ -ST system (Promega, Madison, WI, USA). Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 300 bp) on an Illumina MiSeq PE300 platform (Illumina, Inc., San Diego, CA, USA) according to the standard protocols.
Processing of sequencing data
After raw FASTQ files demultiplexed, sequences were filtered using Trimmomatic62 following the criteria: (i) the 300-bp reads were truncated at any site receiving an average quality score of < 20 over a 50-bp sliding window, and truncated reads shorter than 50 bp were discarded; (ii) reads containing any mismatch in the barcode region, two or more nucleotide mismatches in the primer sequence, or ambiguous characters were removed; and (iii) only sequences with an overlap of > 10 bp and < 10% mismatches were assembled and reads that could not be assembled were discarded. The assembled sequences were then trimmed of primers and barcodes. Chimeric sequences were identified and removed using usearch63. After quality control, the assembled sequences were assigned to operational taxonomic units (OTUs) at a 97% identity threshold using UPARSE64. Alpha diversity index, including ACE, Chao1, Shannon, and Simpson were calculated by normalizing the number of reads in all samples to 7668 sequences using mothur65. Rarefaction curves were analyzed with mothur and plotted using R. The taxonomy of each 16S rRNA gene sequence was assigned against the SILVA bacteria alignment database66 using RDP classifier67 with a confidence threshold of 70%. Sequences were aligned against the PyNAST68, and a phylogenetic tree was built using FastTree69. The sequencing data obtained in this study were deposited in the NCBI Sequence Read Archive (SRA) under accession numbers SRR5406973 to SRR5406996.
Real-time PCR
Absolute quantitative real-time PCR analysis was performed to estimate the copy numbers of the total bacteria, E. coli K99, and four selected bacterial species in the jejunum digesta using the amplification primers shown in Supplementary Table S2. Briefly, a standard curve was generated for the total bacterial gene, E.coli K99, and each individual bacterial strain selected using universal primers. Real-time PCR was performed in a 20 μl reaction mixture containing 10 μl 2 × SG Green qPCR Mix (SinoGene, Beijing, China), 0.5 μl of each primer (10 μM), 0.5 μl 10 ng DNA templates, and 8.5 μl nuclease-free water. Amplification involved one holding cycle at 95 °C for 10 min for initial denaturation and then 40 cycles at 95 °C for 20 s for denaturation followed by annealing at 60 °C for 30 s and extension at 72 °C for 20 s. The copy numbers of the total bacteria, E.coli K99, and four bacterial species per gram of jejunum digesta were then calculated. The relative abundances of four bacterial species were calculated by dividing the gene copy number of each bacterial species by the gene copy number of total bacteria.
Statistical analyses
The body weight and feed intake of calves, the alpha diversity indices, and the quantification of total bacteria, E. coli K99, and the four selected bacterial species were analyzed by one-way ANOVA using SAS (version 9.2; SAS Institute Inc., Cary, NC, USA). Statistical differences among the means of the treatments were compared using the Duncan’s Multiple Range Test. The relative abundances of communities do not fit normal distribution and arcsine transformation function were performed before analyses. The transformed data of the abundances of communities at the phylum and genus levels were analyzed by one-way ANOVA using SAS. Daily fecal scores were analyzed by ANOVA using the MIXED procedure of SAS, fitting a Poisson distribution50 and square root transformation function with repeated measures for count data. The MIXED procedure model included the fixed effects of treatment and day, interaction between treatment and day, and the random effect of the individual nested within treatment. Treatment differences with P < 0.05 were considered statistically significant, and 0.05 ≤ P < 0.10 was designated as a tendency.
Electronic supplementary material
Acknowledgements
This study was supported by the Beijing Dairy Industry Innovation Consortium of Agriculture Research System (Beijing, China) and Fundamental Research Funds for Central Non-profit Scientific Institution.
Author Contributions
This study was designed by Q.Y.D. and Y.T. Experimental research was carried out by Y.L.B. and C.T.Y. Data analysis was performed by Y.L.B., Y.L.B. wrote the manuscripts. All authors approved the manuscript before submission.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05376-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qiyu Diao, Email: diaoqiyu@caas.cn.
Yan Tu, Email: tuyan@caas.cn.
References
- 1.Bazeley K. Investigation of Diarrhoea in the Neonatal Calf. IN PRACTICE. 2003;25:152. doi: 10.1136/inpract.25.3.152. [DOI] [Google Scholar]
- 2.Kim WI, Kim W, Liu SY, Kinyon JM, Yoon KJ. Development of a Panel of Multiplex Real-Time Polymerase Chain Reaction Assays for Simultaneous Detection of Major Agents Causing Calf Diarrhea in Feces. J Vet Diagn Invest. 2010;22:509–517. doi: 10.1177/104063871002200403. [DOI] [PubMed] [Google Scholar]
- 3.Zafar MA, et al. Evaluation of Relative Resuscitative Effects of Hypertonic and Isotonic Saline Solutions as an Adjunct to Ceftiofur HCl in Bovine Neonatal Diarrhea Associated with Escherichia coli. Int J Agric Biol. 2015;17:953–960. doi: 10.17957/IJAB/15.0014. [DOI] [Google Scholar]
- 4.Shams Z, et al. Detection of Enterotoxigenic K99 (F5) and F41 from Fecal Sample of Calves by Molecular and Serological Methods. Comp Clin Pathol. 2012;21:475–478. doi: 10.1007/s00580-010-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolenda, R., Burdukiewicz, M. & Schierack, P. A Systematic Review and Meta-Analysis of the Epidemiology of Pathogenic Escherichia coli of Calves and the Role of Calves as Reservoirs for Human Pathogenic E. Coli. Front Cell Infect Mi5, (2015). [DOI] [PMC free article] [PubMed]
- 6.Ok M, et al. The Studies on the Aetiology of Diarrhoea in Neonatal Calves and Determination of Virulence Gene Markers of Escherichia coli Strains by Multiplex PCR. Zoonoses Public Hlth. 2009;56:94–101. doi: 10.1111/j.1863-2378.2008.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagy B, Fekete PZ. Enterotoxigenic Escherichia coli in Veterinary Medicine. Int J Med Microbiol. 2005;295:443–454. doi: 10.1016/j.ijmm.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Nagy B, Fekete PZ. Enterotoxigenic Escherichia coli in Veterinary Medicine. Int J Med Microbiol. 2005;295:443–454. doi: 10.1016/j.ijmm.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Acres SD. Enterotoxigenic Escherichia coli Infections in Newborn Calves; A Review. J Dairy Sci. 1985;68:229–256. doi: 10.3168/jds.S0022-0302(85)80814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaghoubi SMJ, Ghorbani GR, Rahmani HR, Nikkhah A. Growth, Weaning Performance and Blood Indicators of Humoral Immunity in Holstein Calves Fed Supplemental Flavonoids. J Anim Physiol An N. 2008;92:456–462. doi: 10.1111/j.1439-0396.2007.00734.x. [DOI] [PubMed] [Google Scholar]
- 11.Finck DN, et al. Yeast Supplementation Alters the Performance and Health Status of Receiving Cattle. Prof Anim Scient. 2014;30:333–341. [Google Scholar]
- 12.Uyeno Y, Shigemori S, Shimosato T. Effect of Probiotics/Prebiotics on Cattle Health and Productivity. Microbes Environ. 2015;30:126–132. doi: 10.1264/jsme2.ME14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maciej J, et al. Short Communication: Effects of Oral Flavonoid Supplementation on the Metabolic and Antioxidative Status of Newborn Dairy Calves. J Dairy Sci. 2016;99:805–811. doi: 10.3168/jds.2015-9906. [DOI] [PubMed] [Google Scholar]
- 14.Hallen-Adams HE, Kachman SD, Kim J, Legge RM, Martinez I. Fungi Inhabiting the Healthy Human Gastrointestinal Tract: A Diverse and Dynamic Community. Fungal Ecol. 2015;15:9–17. doi: 10.1016/j.funeco.2015.01.006. [DOI] [Google Scholar]
- 15.Marrero Y, et al. Morphological, Biochemical and Molecular Identification of the Yeast Levica 25: A Potential Ruminal Microbial Additive. Global Veterinaria. 2011;7:60–65. [Google Scholar]
- 16.Costa Sidrim JJ, et al. Candida Species Isolated from the Gastrointestinal Tract of Cockatiels (Nymphicus Hollandicus): In Vitro Antifungal Susceptibility Profile and Phospholipase Activity. Vet Microbiol. 2010;145:324–328. doi: 10.1016/j.vetmic.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Mandal S, Ghosh K. Isolation of Tannase-Producing Microbiota from the Gastrointestinal Tracts of some Freshwater Fish. J Appl Ichthyol. 2013;29:145–153. doi: 10.1111/j.1439-0426.2012.02054.x. [DOI] [Google Scholar]
- 18.Marrero Y, et al. Identification of Levica Yeasts as a Potential Ruminal Microbial Additive. Czech J Anim Sci. 2013;58:460–469. [Google Scholar]
- 19.Marrero Y, Castillo Y, Ruiz O, Burrola E, Angulo C. Feeding of Yeast (Candida Spp.) Improves in Vitro Ruminal Fermentation of Fibrous Substrates. J Integr Agr. 2015;14:514–519. doi: 10.1016/S2095-3119(14)60830-3. [DOI] [Google Scholar]
- 20.Long M, et al. Effect of Different Yeasts on Selenomonas Ruminantium Utilizing Lactate in Vitro. Indian J Anim Res. 2013;47:126–131. [Google Scholar]
- 21.Paramita D, Koushik G. Evaluation of Phytase Production by Candida Tropicalis Isolated from Fish Gut and Subsequent Bio-Processing of Groundnut Oil Cake under Solid State Fermentation. J Microbiol Biotech Food Sci. 2014;3:470–476. [Google Scholar]
- 22.Kobayashi Y, Miyazawa M, Kamei A, Abe K, Kojima T. Ameliorative Effects of Mulberry (Morus alba L.) Leaves on Hyperlipidemia in Rats Fed a High-Fat Diet: Induction of Fatty Acid Oxidation, Inhibition of Lipogenesis, and Suppression of Oxidative Stress. Biosci Biotech Bioch. 2010;74:2385–2395. doi: 10.1271/bbb.100392. [DOI] [PubMed] [Google Scholar]
- 23.Stoldt A, et al. Effects of a 6-Wk Intraduodenal Supplementation with Quercetin on Energy Metabolism and Indicators of Liver Damage in Periparturient Dairy Cows. J Dairy Sci. 2015;98:4509–4520. doi: 10.3168/jds.2014-9053. [DOI] [PubMed] [Google Scholar]
- 24.Cavia-Saiz M, et al. Antioxidant Properties, Radical Scavenging Activity and Biomolecule Protection Capacity of Flavonoid Naringenin and its Glycoside Naringin: A Comparative Study. J Sci Food Agr. 2010;90:1238–1244. doi: 10.1002/jsfa.3959. [DOI] [PubMed] [Google Scholar]
- 25.Moreira GMB, Matsumoto LS, Silva RMG, Domingues PF, Mello-Peixoto ECT. Antibacterial Activity of the Hydroalcoholic Extract of Punica Granatum Linn. on Staphylococcus Spp. Isolated from Bovine Milk. Pesqui Vet Brasil. 2014;34:626–632. doi: 10.1590/S0100-736X2014000700003. [DOI] [Google Scholar]
- 26.Omosa LK, et al. Antimicrobial Flavonoids and Diterpenoids from Dodonaea Angustifolia. S Afr J Bot. 2014;91:58–62. doi: 10.1016/j.sajb.2013.11.012. [DOI] [Google Scholar]
- 27.Madzimure J, et al. Acaricidal Efficacy Against Cattle Ticks and Acute Oral Toxicity of Lippia Javanica (Burm F.) Spreng. Trop Anim Health Pro. 2011;43:481–489. doi: 10.1007/s11250-010-9720-1. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh D, et al. Black Tea Extract: A Supplementary Antioxidant in Radiation-Induced Damage to DNA and Normal Lymphocytes. J Environ Pathol Tox. 2012;31:155–166. doi: 10.1615/JEnvironPatholToxicolOncol.v31.i2.70. [DOI] [PubMed] [Google Scholar]
- 29.Pinzon LC, Uy MM, Sze KH, Wang M, Chu IK. Isolation and Characterization of Antimicrobial, Anti-Inflammatory and Chemopreventive Flavones From Premna Odorata Blanco. J Med Plants Res. 2011;5:2729–2735. [Google Scholar]
- 30.Chandrashekar C, Kulkarni VR. Isolation Characterization and Antimicrobial Activity of Annona Squamosa Leaf. J Pharm Res. 2011;4:1831–1832. [Google Scholar]
- 31.Diab Y, Atalla K, Elbanna K. Antimicrobial Screening of some Egyptian Plants and Active Flavones from Lagerstroemia Indica Leaves. Drug discoveries & therapeutics. 2012;6:212–217. [PubMed] [Google Scholar]
- 32.Ngo Mbing J, et al. New Flavonoids C-glycosides from Rhabdophyllum Arnoldianum. Nat Prod Res. 2014;28:539–544. doi: 10.1080/14786419.2014.883393. [DOI] [PubMed] [Google Scholar]
- 33.Okwu DE, Nnamdi FU. Two Novel Flavonoids From Bryophyllum Pinnatum and their Antimicrobial Activity. J Chem Pharm Res. 2011;3:1–10. [Google Scholar]
- 34.Bagla, V. P., McGaw, L. J., Elgorashi, E. E. & Eloff, J. N. Antimicrobial Activity, Toxicity and Selectivity Index of Two Biflavonoids and a Flavone Isolated from Podocarpus Henkelii (Podocarpaceae) Leaves. BMC Complem Altern M14, (2014). [DOI] [PMC free article] [PubMed]
- 35.Harborne JB. Toxins of Plant–Fungal Interactions. Handbook of Natural Toxins. Plant and Fungal Toxins. New York: Mercel Dekker; 1983. p. 743. [Google Scholar]
- 36.Generoso SV, et al. Saccharomyces Cerevisiae Strain UFMG 905 Protects Against Bacterial Translocation, Preserves Gut Barrier Integrity and Stimulates the Immune System in a Murine Intestinal Obstruction Model. Arch Microbiol. 2010;192:477–484. doi: 10.1007/s00203-010-0574-8. [DOI] [PubMed] [Google Scholar]
- 37.Franca RC, et al. Pichia Pastoris X-33 Has Probiotic Properties with Remarkable Antibacterial Activity Against Salmonella Typhimurium. Appl Microbiol Biot. 2015;99:7953–7961. doi: 10.1007/s00253-015-6696-9. [DOI] [PubMed] [Google Scholar]
- 38.Tiago FCP, et al. Physiological Characterization of non-Saccharomyces Yeasts from Agro-Industrial and Environmental Origins with Possible Probiotic Function. World J Microb Biot. 2009;25:657–666. doi: 10.1007/s11274-008-9934-9. [DOI] [Google Scholar]
- 39.O’Hara AM, Shanahan F. The Gut Flora as a Forgotten Organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Claesson MJ, et al. Composition, Variability, and Temporal Stability of the Intestinal Microbiota of the Elderly. P Natl Acad Sci USA. 2011;1081:4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Looft T, et al. In-Feed Antibiotic Effects on the Swine Intestinal Microbiome. P Natl Acad Sci USA. 2012;109:1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fey PD, et al. Ceftriaxone-Resistant Salmonella Infection Acquired by a Child From Cattle. New Engl J Med. 2000;342:1242–1249. doi: 10.1056/NEJM200004273421703. [DOI] [PubMed] [Google Scholar]
- 43.Heinrichs AJ, Jones CM, Heinrichs BS. Effects of Mannan Oligosaccharide or Antibiotics in Neonatal Diets on Health and Growth of Dairy Calves. J Dairy Sci. 2003;86:4064–4069. doi: 10.3168/jds.S0022-0302(03)74018-1. [DOI] [PubMed] [Google Scholar]
- 44.Frizzo LS, et al. Lactic Acid Bacteria to Improve Growth Performance in Young Calves Fed Milk Replacer and Spray-Dried Whey Powder. Anim Feed Sci Tech. 2010;157:159–167. doi: 10.1016/j.anifeedsci.2010.03.005. [DOI] [Google Scholar]
- 45.Abe F, Ishibashi N, Shimamura S. Effect of Administration of Bifidobacteria and Lactic Acid Bacteria to Newborn Calves and Piglets. J Dairy Sci. 1995;78:2838–2846. doi: 10.3168/jds.S0022-0302(95)76914-4. [DOI] [PubMed] [Google Scholar]
- 46.Seo JK, et al. Direct-Fed Microbials for Ruminant Animals. Asian Austral J Anim. 2010;23:1657–1667. doi: 10.5713/ajas.2010.r.08. [DOI] [Google Scholar]
- 47.Miksicek RJ. Commonly Occurring Plant Flavonoids Have Estrogenic Activity. Mol Pharmacol. 1993;44:37–43. [PubMed] [Google Scholar]
- 48.Wang Z, Guo Y, Yuan J, Zhang B. Effect of Dietary Beta-1,3/1,6-Glucan Supplementation on Growth Performance, Immune Response and Plasma Prostaglandin E-2, Growth Hormone and Ghrelin in Weanling Piglets. Asian Austral J Anim. 2008;21:707–714. doi: 10.5713/ajas.2008.70559. [DOI] [Google Scholar]
- 49.White LA, Newman MC, Cromwell GL, Lindemann MD. Brewers Dried Yeast as a Source of Mannan Oligosaccharides for Weanling Pigs. J Anim Sci. 2002;80:2619–2628. doi: 10.2527/2002.80102619x. [DOI] [PubMed] [Google Scholar]
- 50.Magalhaes VJA, et al. Effect of Feeding Yeast Culture on Performance, Health, and Immunocompetence of Dairy Calves. J Dairy Sci. 2008;91:1497–1509. doi: 10.3168/jds.2007-0582. [DOI] [PubMed] [Google Scholar]
- 51.Jami E, Israel A, Kotser A, Mizrahi I. Exploring the Bovine Rumen Bacterial Community From Birth to Adulthood. ISME J. 2013;7:1069–1079. doi: 10.1038/ismej.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edrington TS, et al. Development of Colonic Microflora as Assessed by Pyrosequencing in Dairy Calves Fed Waste Milk. J Dairy Sci. 2012;95:4519–4525. doi: 10.3168/jds.2011-5119. [DOI] [PubMed] [Google Scholar]
- 53.Oikonomou, G. et al. Fecal Microbial Diversity in Pre-Weaned Dairy Calves as Described by Pyrosequencing of Metagenomic 16S rDNA. Associations of Faecalibacterium Species with Health and Growth. PLOS ONE8, (2013). [DOI] [PMC free article] [PubMed]
- 54.Kim M, et al. Investigation of Bacterial Diversity in the Feces of Cattle Fed Different Diets. J Anim Sci. 2014;92:683–694. doi: 10.2527/jas.2013-6841. [DOI] [PubMed] [Google Scholar]
- 55.Klein-Joebstl, D. et al. Pyrosequencing Reveals Diverse Fecal Microbiota in Simmental Calves During Early Development. Front Microbiol5, (2014). [DOI] [PMC free article] [PubMed]
- 56.Malmuthuge N, Griebel PJ, Guan LL. Taxonomic Identification of Commensal Bacteria Associated with the Mucosa and Digesta throughout the Gastrointestinal Tracts of Preweaned Calves. Appl Environ Microb. 2014;80:2021–2028. doi: 10.1128/AEM.03864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nocek JE, Kautz WP, Leedle J, Allman JG. Ruminal Supplementation of Direct-Fed Microbials on Diurnal pH Variation and in Situ Digestion in Dairy Cattle. J Dairy Sci. 2002;85:429–433. doi: 10.3168/jds.S0022-0302(02)74091-5. [DOI] [PubMed] [Google Scholar]
- 58.Qadis AQ, et al. Effects of a Bacteria-Based Probiotic on Ruminal pH, Volatile Fatty Acids and Bacteria Flora of Holstein Calves. J Vet Med Sci. 2014;76:877–885. doi: 10.1292/jvms.14-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Purushe J, et al. Comparative Genome Analysis of Prevotella Ruminicola and Prevotella Bryantii: Insights into their Environmental Niche. Microb Ecol. 2010;60:721–729. doi: 10.1007/s00248-010-9692-8. [DOI] [PubMed] [Google Scholar]
- 60.Li RW, Connor EE, Li C, Baldwin RL, Sparks ME. Characterization of the Rumen Microbiota of Pre-Ruminant Calves Using Metagenomic Tools. Environ Microbiol. 2012;14:129–139. doi: 10.1111/j.1462-2920.2011.02543.x. [DOI] [PubMed] [Google Scholar]
- 61.Dennis KL, et al. Adenomatous Polyps are Driven by Microbe-Instigated Focal Inflammation and are Controlled by IL-10-Producing T Cells. Cancer Res. 2013;73:5905–5913. doi: 10.1158/0008-5472.CAN-13-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolger AM, Lohse M, Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. BIOINFORMATICS. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edgar RC. Search and Clustering Orders of Magnitude Faster than BLAST. BIOINFORMATICS. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 64.Edgar RC. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat Methods. 2013;10:996. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 65.Schloss PD, et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl Environ Microb. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quast C, et al. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microb. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeSantis TZ, et al. NAST: A Multiple Sequence Alignment Server for Comparative Analysis of 16S rRNA Genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price MN, Dehal PS, Arkin AP. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


