Abstract
In total, 303 randomly selected clinical Mycobacterium tuberculosis (MTB) isolates from 303 patients (collected January to December 2012) in central Taiwan were examined. The major lineages found were Beijing (N = 114, 37.62%), Haarlem (N = 76, 25.08%) and East African–Indian (EAI) (N = 42, 13.86%). Notably, younger persons (≤30 years old) were 6.58 times more likely to be infected with a Beijing genotype compared to older persons (>70 years) (p < 0.05). Combining molecular typing methods and geographical information system (GIS) analysis, we uncovered a twofold higher incidence of Beijing strains in a hotspot area (33%) compared to non-hotspot areas (17%). By 24 MIRU-VNTR typing, persons in clustered groups were 1.96 times more likely to be infected with a Beijing strain compared with non-clustered persons, suggesting recent spread and emergence of MTB. Finally, we observed a trend in which TB incidence increased as the density/concentration of analyzed environmental factors increased, suggesting that environmental factors are associated with TB transmission; however, only population density was found to be significantly associated with increased risk of TB (p < 0.05). Molecular typing methods combined with spatial analysis suggest possible TB transmission. Early intervention to interrupt transmission may be most effective if targeted to hot zones of TB.
Introduction
Tuberculosis (TB), an airborne infectious disease caused by Mycobacterium tuberculosis (MTB), is one of the deadliest communicable diseases worldwide. As estimated by the World Health Organization in 2013, there were 9 million new TB cases and 1.5 million TB deaths globally1. Although Taiwan has a low to medium incidence of TB cases compared to all other countries, TB is still a significant public health threat, especially in high-risk groups and regions2, 3. Therefore, early intervention and improved investigation of the contacts of TB cases are critical to interrupt transmission. Zelner et al. suggested that targeting the highest burden TB areas might be more effective than blanket screening and treatment4. In past decades, the transmission dynamics of TB in social networks has been analyzed based on molecular epidemiological methods, including restriction fragment length polymorphism, spoligotyping, and MIRU-VNTR (mycobacterial interspersed repetitive units – variable number of tandem repeats) typing5–7. Recently, molecular epidemiological techniques combined with graphically based surveillance have been applied to identify areas with ongoing TB transmission and as a new strategy to limit the spread of TB4.
We have undertaken long-term surveillance of MTB in central Taiwan (including collecting information on MTB genotypes, drug resistance phenotypes, and TB patient demographic characteristics) to ascertain associations between transmission dynamics and risk factors. In our earlier studies, we applied molecular epidemiological methods to analyze hundreds of randomly chosen MTB clinical isolates from patients in northern, southern and eastern regions of Taiwan. Those studies demonstrated that the Beijing lineage is the predominant MTB strain in Taiwan3, 8–10. Our molecular analyses also showed that the East African–Indian (EAI) and Haarlem strains are particularly prevalent in southern and eastern Taiwan, respectively3, 9. However, the transmission dynamics of prevalent MTB strains in central Taiwan are not well understood.
The purpose of the present study was twofold. First, we sought to determine the genotypes of MTB isolates present in central Taiwan. Second, we sought to evaluate the factors associated with increased risk of TB by using molecular methods combined with geographical information system (GIS) analysis. Interestingly, Lai et al. and Smith et al. reported that fine particle (PM2.5) and traffic-related air pollution, including nitrogen dioxide, nitrogen oxide and carbon monoxide, were associated with elevated TB risk11, 12. Therefore, in this study we also investigated environmental factors. We further sought to identify possible sites of TB transmission by using hotspot analysis in combination with GIS and molecular typing techniques. Our spatial analysis revealed possible TB transmission areas, which we believe may be helpful in setting public health policies to limit the spread of TB4.
Materials and Methods
Study setting
This retrospective study was conducted at the National Health Research Institutes in Taiwan. The study included 303 MTB samples (from 303 patients) randomly selected from 795 isolates collected during January 2012 to December 2012 at the Changhua Christian Hospital (CCH). The mycobacterium laboratory of CCH is the major laboratory in Changhua providing TB testing services not only to Changhua Christian Healthcare System (CCHC comprises one medical center CCH and three branch hospitals distributed evenly at different locations of the county and covering both urban and rural areas) hospitals, but also to 27 health centers and other primary care clinics in each township of this county. All bacteria isolates were confirmed by routine microscopy, culture and positive nitrate and niacin tests. All isolated 303 MTB strains were then genotyped by spoligotyping and 24-locus MIRU-VNTR typing. This study was approved by the Human Ethics Committee of the National Health Research Institutes, Taiwan (Code: EC1010804-E-R1).
Genomic DNA extraction from mycobacterial cells
Mycobacterial genomic DNA extraction was performed as described previously9. Briefly, bacterial colonies were scraped from Lowenstein-Jensen medium and suspended in 100 μl distilled H2O. The samples were then incubated at 85 °C for 30 min. After centrifugation, the supernatant, containing DNA, was removed into a new tube and stored at −20 °C until use.
Spoligotyping and spoligotype analysis
Spoligotyping was performed according to the manufacturer’s instructions (Isogen Bioscience BV., Maarsen, The Netherlands). The new isolates were assigned by using the SUITVITWEB database13 and Spotclust14.
24-locus MIRU-VNTR typing
The 24 loci (MIRU 4, MIRU 26, MIRU 40, MIRU 10, MIRU 16, MIRU 31, Mtub04, ETR C, ETR A, Mtub30, Mtub39, QUB-4156, QUB-11b, Mtub21, QUB-26, MIRU 2, MIRU 23, MIRU 39, MIRU 20, MIRU 24, MIRU 27, Mtub29, ETR B, and Mtub34) were selected and analyzed as described previously15. The resulting genotyping pattern of each isolated strain was used to create a digit-allelic profile.
Drug susceptibility testing
Drug susceptibility tests were carried out according to the Clinical and Laboratory Standards Institute (CLSI) standard16. The tests were conducted by using the agar proportion method utilizing Middlebrook 7H10 agar supplemented individually with the following antibiotics: isoniazid (0.2 or 1 mg/L), rifampin (1 or 5 mg/L), ethambutol (5 or 10 mg/L), and streptomycin (5 or 10 mg/L).
Spatial analysis
Residential addresses of each participant at the time of diagnosis were geocoded by ArcGIS and Taiwan Geospatial One-Stop Portal (TGOS), the official website of the Information Center, Ministry of the Interior of Taiwan for geospatial data management. Three spatial analyses were conducted: (1) the Global Moran’s I statistic was applied to measure the spatial autocorrelation of TB outbreak at the township scale; (2) Kernel Density Estimation (KDE) was applied to identify the geographic hotspots of TB in the Chang-hua area; and (3) the clustered MTB strains were overlaid with several thematic maps, including ones depicting population density, fine particulate matter (PM2.5) concentration, and residential, commercial and industrial areas, to visually assess potential spatial trends among human activities, air pollutants, and distribution of clustered TB strains. Also, townships (with their numbers of TB cases or numbers of clustered strains) were examined for differences in population density and the magnitude of three air pollutants (PM2.5, NO2, and CO).
Supplementary Fig. 1A,B show the spatial distribution of all isolated TB cases for the year 2012 (N = 795), and the randomly selected MTB strains examined in this study (N = 303), respectively. The consistent spatial distributions in those two figures confirm the representative nature of the sampled TB subjects in the study area and their suitability for empirical analysis.
Kernel-density Estimation (KDE)
KDE is a tool for smoothing points in continuous space17. A smoothly curved surface is fitted over each point. The surface value is highest at the location of the point and diminishes with increasing distance from the point, reaching zero at the search radius distance from the point18. In our study, ArcMap GIS (version 10.2) and its Spatial Analyst Extension (ESRI Inc., Redland, CA) were used to generate the KDE maps with a search radius of 5 km. The distribution of MTB/clustered MTB strains was calculated using KDE methods.
Spatial variability of PM2.5
Hourly ambient PM2.5 concentrations were measured by the Taiwan Environmental Protection Administration from January 1, 2012 to December 31, 2012. We used automatic monitoring data, which were available from 71 stationary sites covering the Main Island. A modified ordinary kriging adopted from Liao et al. (2006) was applied to determine the spatial estimation of PM2.5 levels of the whole island, and then the extent of the study area was extracted. ArcMap GIS (version 10.2) and its Geostatistical Analysts Extension (ESRI Inc., Redland, CA) were utilized to construct the semivariogram. The median value of cross-validated R-squared was 0.7319.
Statistical analysis
Patient characteristics (gender, age) and the results of the sputum smear, chest X-ray, drug susceptibility test and MIRU-VNTR analysis were compared between Beijing- and non-Beijing-infected groups. Univariate regression analyses were performed to yield crude odds ratios and their 95% confidence intervals. Data management and analyses were carried out by using SAS/STAT software, version 9.3 for Windows (SAS Institute Inc., Cary, NC, USA).
Results
Demographic characteristics of TB patients
According to the TB registry database of the local health authority in Changhua county, a county with an intermediate incidence of TB (58.7 per 105 in 2012), there were 856 reported pulmonary TB cases in 2012, 692 of which were confirmed by the sputum culture. The CCH TB laboratory provides nearly 70% of all TB testing services in the whole county each year. Thus, the 303 specimens randomly selected from 795 isolates collected at the CCH may therefore be treated as a representative sample of the total culture-positive pulmonary TB in the community. In total, 303 randomly selected clinical isolates from 303 patients in central Taiwan who were diagnosed with culture-confirmed TB (January to December 2012) were subjected to spoligotyping and MIRU-VNTR typing. There were 2.6 times as many isolates from men as from women (male: 71.95%, 218; female: 28.05%, 85). The most prevalent genotype was Beijing, identified in 114 isolates (37.62%), followed by Haarlem (76; 25.08%), EAI (42; 13.86%), T (37; 12.21%), Latin American–Mediterranean (LAM) (7; 2.31), and Manu_ancestor (7; 2.3%) (Table 1). The Beijing lineage was particularly prevalent in the ≤30-year-old age group (77.78%), although the sample size was small; its prevalence in three older age groups (31–50, 51–70 and >70) ranged from about 31% to 44%. In contrast, the prevalence of the Haarlem lineage was lowest in the ≤30-year-old age group (about 11%) and tended to increase with patient age (to a high of nearly 28%) (Table 1).
Table 1.
Demographic characteristics of 303 tuberculosis patients from central Taiwan.
| Beijing | Haarlem | EAI | T | LAM | Manu_ancestor | Others | |
|---|---|---|---|---|---|---|---|
| No. of isolates* (%) | 114 (37.62) | 76 (25.08) | 42 (13.86) | 37 (12.21) | 7 (2.31) | 7 (2.31) | 20(6.60) |
| Gender | |||||||
| Male | 80 (36.70) | 51 (23.39) | 31 (14.22) | 33 (15.14) | 4 (1.83) | 4 (1.83) | 15 (6.88) |
| Female | 34 (40.00) | 25 (29.41) | 11 (12.94) | 4 (4.71) | 3 (3.53) | 3 (3.53) | 5 (5.88) |
| Age(years, mean/median) | 70/74.5 | 73/77.5 | 71/77.5 | 73/78 | 69/75 | 79.3/81 | 71/73.5 |
| Age group (years) | |||||||
| ≤30 | 7 (77.78) | 1 (11.11) | 1 (11.11) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| 31–50 | 10 (31.25) | 6 (18.75) | 5 (15.63) | 5 (15.63) | 2 (6.25) | 0 (0.00) | 4 (12.50) |
| 51–70 | 30 (44.12) | 19 (27.94) | 9 (13.24) | 6 (8.82) | 1 (1.47) | 0 (0.00) | 3 (4.41) |
| å 70 | 67 (34.54) | 50 (25.77) | 27 (13.92) | 26 (13.40) | 4 (2.06) | 7 (3.61) | 13 (6.70) |
*One sample was collected from each patient.
Characteristics of Beijing and non-Beijing strains isolated from TB patients
To better understand the clinical characteristics of patients infected with Beijing strains, the gender-, age-, sputum smear-, geographic location-, drug resistance and MIRU analysis-specific odds ratios were calculated (Table 2). There were no significant differences in gender, sputum smear, location and drug-resistance patterns between the Beijing and non-Beijing groups (Table 2). However, we found that persons in the youngest age group (≤30 years old) were 6.58 times more likely to be infected with a Beijing genotype compared with persons in the oldest age group (>70 years old) (Table 2). In addition, MIRU analysis revealed that patients in clustered groups were 1.96 times more likely to be infected with a Beijing strain compared with non-clustered patients, suggesting recent spread and emergence (Table 2).
Table 2.
Characteristics of tuberculosis patients associated with Mycobacterium tuberculosis Beijing and non-Beijing strains.
| Bacterial lineage | Crude odds ratio (95% CI) | ||
|---|---|---|---|
| Beijing; No. (%) | Non-Beijing; No. (%) | ||
| Gender | |||
| Male | 80 (70.18) | 138 (73.02) | 0.8696 (0.520–1.45) |
| Female | 34 (29.82) | 51 (26.98) | Reference |
| Age | |||
| ≤30 | 7 (6.14) | 2 (1.06) | 6.582 (1.330–32.570)* |
| 31–50 | 10 (8.77) | 22 (11.64) | 0.855 (0.383–1.910) |
| 51–70 | 30 (26.32) | 39 (20.63) | 1.45 (0.826–2.534) |
| >70 | 67 (58.77) | 126 (66.67) | Reference |
| Sputum smear | |||
| Positive | 62 (54.39) | 93 (49.21) | 1.31 (0.772–1.961) |
| Negative | 52 (45.61) | 96 (50.79) | Reference |
| Location | |||
| Pulmonary | 108 (94.74) | 183 (96.83) | Reference |
| Extra-pulmonary | 6 (5.26) | 6 (3.17) | 1.694 (0.533–5.385) |
| Antibiotic resistance pattern | |||
| All sensitive | 99 (86.84) | 163 (86.24) | Reference |
| Monoresistance | 2 (1.75) | 12 (6.35) | 0.2744 (0.006–1.252) |
| Polyresistance | 13 (11.40) | 13 (6.88) | 1.646 (0.735–3.695) |
| Multidrug resistance | 0 (0.00) | 1 (0.53) | — |
| MIRU analysis | |||
| Clustered | 36 (31.58) | 36 (19.05) | 1.962 (1.147–3.354)* |
| Non-clustered | 78 (68.42) | 153 (80.95) | Reference |
*Indicates that the OR is significant and p < 0.05.
Using GIS tools to identify hot zones of TB transmission
The genotypes and clustering characteristics of isolated MTB strains can be analyzed by MIRU-VNTR typing. However, the hotspot areas of ongoing TB transmission cannot be identified using molecular methods. Therefore, we combined molecular and GIS analyses to identify geographical areas with ongoing TB transmission.
Figure 1A shows the distribution of clustered MTB strains calculated using the Kernel Density Estimation (KDE) method. Two geographical hotspots of MTB strains (red areas) surrounded by several regions of moderately intensive clustering (yellow areas) were observed in the west-central townships of Chang-hua and Yuan-lin. Figure 1B shows the strain-clustering rate in each geographical region based on molecular methods without GIS information. We noted a twofold higher incidence of Beijing strains in the two hotspot areas (33%, 12/24) compared to non-hotspot regions (17%; 7/35) (p = 0.087).
Figure 1.
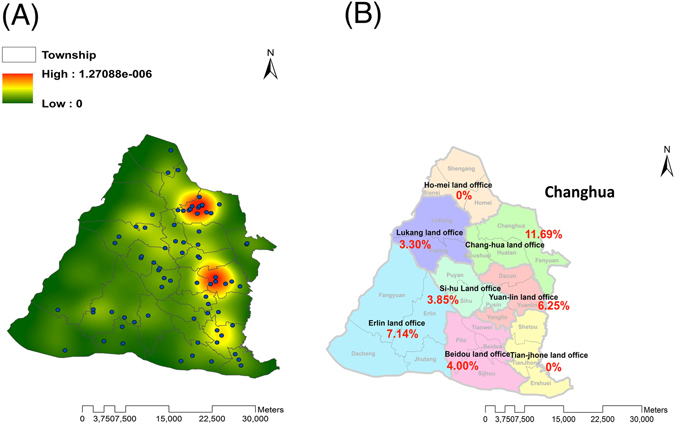
Spatial distribution of clustered MTB strains. (A) KDE maps were constructed based on GIS data representing the clustered MTB strain referral sites. (B) Distribution of clustered MTB strains was analyzed based on strain-clustering rates in each township. Esri ArcGIS 10.2 was used to create this figure (http://www.esri.com/software/arcgis).
Analysis of environmental risk factors associated with TB transmission
Lai et al. demonstrated that exposure to fine particles (PM2.5) and traffic-related air pollution (including nitrogen dioxide and carbon monoxide) was associated with increased risk of TB12. To investigate possible interactions between environmental factors and TB transmission, we examined the distribution of total TB cases/clustered strains associated with environmental factors, including population density, fine particulates, and human activity (Fig. 2).
Figure 2.
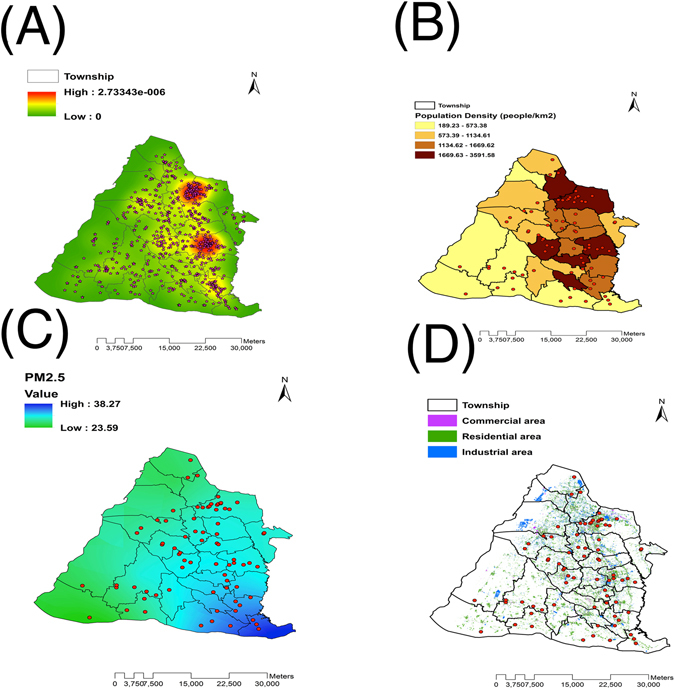
Distribution of Total MTB (A) and clustered strain (B, C, D) referral sites associated with (B) population density, (C) fine particle matter (PM2.5), and (D) human activity. Esri ArcGIS 10.2 was used to create this figure (http://www.esri.com/software/arcgis).
Spatial analysis of TB cases revealed that MTB strains (including clustered strains) tended to group in townships with high population densities and residential areas (Fig. 2A,B and D). To clarify associations between environmental factors and TB transmission, we performed the Kruskal-Wallis H test. First, townships were classified into three categories based on the number of TB cases. Interestingly, TB incidence increased as the density/concentration of each analyzed environmental factor increased, suggesting that environmental factors are associated with TB transmission (Table 3). Nevertheless, only population density was significantly associated with increased risk of TB (Table 3; p = 0.046). No significant association between clustered strains and environmental factors was found (data not shown).
Table 3.
Analysis of environmental factors for TB transmission by using the Kruskal-Wallis H test.
| TB case number | n | Mean rank | Chi-square statistic | p-value | |
|---|---|---|---|---|---|
| Population density | fewest | 9 | 8.56 | ||
| mid | 9 | 15 | 6.161 | 0. 046* | |
| most | 8 | 17.38 | |||
| PM2.5 | fewest | 9 | 12.56 | ||
| mid | 9 | 13.89 | 0.214 | 0.899 | |
| most | 8 | 14.13 | |||
| NO2 | fewest | 9 | 9.11 | ||
| mid | 9 | 15.61 | 4.549 | 0.103 | |
| most | 8 | 16.06 | |||
| CO | fewest | 9 | 11.56 | ||
| mid | 9 | 13.67 | 1.133 | 0.568 | |
| most | 8 | 15.50 |
*p < 0.05.
Discussion
To our knowledge, this is the first epidemiological study to combine GIS and molecular methods to investigate risk factors of TB transmission in Taiwan. Our results showed that younger persons (≤30 years old) were 6.58 times more likely to be infected with a Beijing genotype compared to older persons (>70 years old). Notably, hotspot regions have a twofold higher incidence of Beijing lineages (33%) compared to non-hotspot regions (17%). Based on the results of MIRU-VNTR typing, people in clustered groups were 1.96 times more likely to be infected with a Beijing strain compared with persons not in clustered groups. Taken together, our results suggest that targeting high-burden areas might be a good strategy to limit the spread of TB.
The Beijing genotype was the predominant genotype identified in central Taiwan, as well as in the two hotspot regions. This result coincides with those of previous studies in Taiwan and globally, in which Beijing strains, with their high transmissibility, are often associated with major TB outbreaks8, 10, 20–22. A recent study hypothesized that Beijing strains originated in Guangxi province in the south of China23, and they are the dominant MTB strains not only in China (up to 90%), but also in nearby countries, including South Korea (up to 97.1%), Japan (up to 80%%), Thailand (56%), Vietnam (up to 55.4%), Singapore (45.8%) and Myanmar (31.9%)21, 22, 24–30. The reasons for the success of Beijing strains remain controversial, but could include a variety of factors such as positive selection due to worldwide BCG vaccination coverage, population migration, and ineffective treatment, leading to disease transmission22, 31.
The patients in our cohort were, on average, older (70.0 years), and we found a trend of decreasing prevalence of Beijing genotype infections with age (Table 1). Moreover, Beijing is more common among young persons (≤30 years old; OR 6.58) and in clustered groups (OR 1.962), suggesting a tendency to spread in central Taiwan (Tables 1 and 2). In addition, as already noted, GIS results revealed a twofold higher incidence of Beijing lineages in hotspot areas (34%) compared to non-hotspot regions (17%), supporting increased transmission of Beijing strains in central Taiwan. The higher incidence of Beijing genotypes in hotspot areas may reflect the fact that young and middle-aged patients work and study in Chung-hua and Yuan-lin cities. These new results coincide with our previous findings in southern Taiwan and those reported by Buu et al. in Vietnam32, 33. In southern Taiwan, the percentage of Beijing-infected patients in young age groups increased monotonically from 3.9% in 2006 to 9.3% in 2008, but no such trend was observed for any non-Beijing genotype33. Apart from the Beijing genotype, the other major lineage found in central Taiwan was the Haarlem genotype (25.08%). Results of previous studies by our group revealed that the Haarlem lineage was predominant in eastern Taiwan, particularly in aborigines2, 3. In central Taiwan, most aborigines live in Nan-tou County, adjacent to Chang-hua city. A previous retrospective study conducted at Pulli Christian Hospital (located in Nan-tou County) demonstrated that Haarlem genotypes were the predominant MTB lineage2. Therefore, a high prevalence of Haarlem strains isolated in this study may be due to population migration and close interpersonal contact in those regions.
Interestingly, Lai et al. suggested that ambient air pollution is associated with increased risk of TB12. Similarly, Smith et al. suggested a potential association between long-term exposure to particulate matter and TB11. Notably, Rivas-Santiago et al. demonstrated that exposure of A549 cells, a human type II alveolar epithelial cell, to air pollution particulate matter induced cellular senescence, downregulation of beta-defensin (HBD-2 and HBD-3) expression, and impaired their ability to control MTB intracellular growth34. In the present study, associations between TB incidence and exposure to ambient air pollution, including fine particle matter (PM2.5), carbon monoxide and nitrogen dioxide, were evaluated. Our results coincide with those of previous studies, in which TB incidence increased as the density/concentration of the analyzed environmental factors increased; however, only population density was found to be significantly associated with increased risk of TB (p < 0.05). In developing countries, outdoor air pollution is a major problem and the people living in these regions experience long-term exposure to high levels of air pollutants. Taken together, these results suggest further investigation in the field of air pollution and TB control is warranted.
Our study has several limitations. The study was a retrospective one. Therefore, the effect on the results caused by missing data (e.g. host factors, including chronic diseases and genetics) is unknown. Also, the sample size for the oldest age group (>70 years) was small. We hope to initiate a prospective collection of patient information and MTB strains to further clarify associations between MTB transmission and risk factors such as environmental factors.
Despite these limitations, our study combined GIS and molecular epidemiological analyses to attempt to identify possible places of MTB transmission, with the idea that targeting high-burden areas might be an effective way to limit the spread of TB [2]. We observed a trend in which the numbers of TB cases increased as the density/concentration of the analyzed environmental factors increased. Taken together, our spatial analysis combined with molecular epidemiological methods has provided further insight into TB transmission dynamics in west-central Taiwan, and the approach should be considered as a new control policy in TB.
Electronic supplementary material
Acknowledgements
We thank the mycobacteriology laboratories of Changhua Christian Hospital for providing bacterial isolates and Chang-Hua County Public Health Bureau for its timely collection of data. All participants of this consortium are acknowledged for valuable discussions. This work was supported by grants from the National Health Research Institutes (IV-103-SP-06 and PH103-PP-05) (Zhunan, Taiwan) and the National Science Council (NSC 102-2320-B-400-006) (Taipei, Taiwan).
Author Contributions
Conceived and designed the experiments: Y.Y.C. and H.Y.D. Performed experiments: Y.Y.C., J.R.C., C.D.W., C.H.H., M.C.L., C.H.T. and M.S.L. Analyzed the data: Y.Y.C., Y.P.Y., S.J.Y. and I.J.S. Contributed to the writing of the manuscript: Y.Y.D. and H.Y.D. All authors reviewed and approved of the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05674-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO global tuberculosis control report 2016. Summary. Cent Eur J Public Health (2016). [PubMed]
- 2.Chen, Y. Y. et al. Molecular epidemiology of Mycobacterium tuberculosis in aboriginal peoples of Taiwan, 2006–2011. J Infect, doi:S0163-4453(13)00380-0 (2013). [DOI] [PubMed]
- 3.Chen YY, et al. Distinct modes of transmission of tuberculosis in aboriginal and non-aboriginal populations in Taiwan. PLoS One. 2014;9:e112633. doi: 10.1371/journal.pone.0112633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zelner JL, et al. Identifying Hotaspots of Multidrug-Resistant Tuberculosis Transmission Using Spatial and Molecular Genetic Data. J Infect Dis. 2016;213:287–294. doi: 10.1093/infdis/jiv387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Embden JD, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Supply, P. et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol44, 4498–4510, doi:JCM.01392-06 (2006). [DOI] [PMC free article] [PubMed]
- 7.Brudey, K. et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol6, 23, doi:1471-2180-6-23 (2006). [DOI] [PMC free article] [PubMed]
- 8.Dou HY, Chen YY, Kou SC, Su IJ. Prevalence of Mycobacterium tuberculosis strain genotypes in Taiwan reveals a close link to ethnic and population migration. J Formos Med Assoc. 2015;114:484–488. doi: 10.1016/j.jfma.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Chen YY, et al. Molecular epidemiology of tuberculosis in kaohsiung city located at southern taiwan, 2000-2008. PLoS One. 2015;10:e0117061. doi: 10.1371/journal.pone.0117061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dou HY, et al. Molecular epidemiology and evolutionary genetics of Mycobacterium tuberculosis in Taipei. BMC Infect Dis. 2008;8:170. doi: 10.1186/1471-2334-8-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith GS, Schoenbach VJ, Richardson DB, Gammon MD. Particulate air pollution and susceptibility to the development of pulmonary tuberculosis disease in North Carolina: an ecological study. Int J Environ Health Res. 2014;24:103–112. doi: 10.1080/09603123.2013.800959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai TC, et al. Ambient air pollution and risk of tuberculosis: a cohort study. Occupational and environmental medicine. 2016;73:56–61. doi: 10.1136/oemed-2015-102995. [DOI] [PubMed] [Google Scholar]
- 13.Kolk AH, et al. Detection of Mycobacterium tuberculosis in clinical samples by using polymerase chain reaction and a nonradioactive detection system. J Clin Microbiol. 1992;30:2567–2575. doi: 10.1128/jcm.30.10.2567-2575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demay C, et al. SITVITWEB–a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol. 2012;12:755–766. doi: 10.1016/j.meegid.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Chen YY, et al. Genetic diversity of the Mycobacterium tuberculosis East African-Indian family in three tropical Asian countries. J Microbiol Immunol Infect. 2015 doi: 10.1016/j.jmii.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 16.PA, W. Susceptibility testing of mycobacteria, nocardia and other aerobic actinomycetes. (National Committee for Clinical Laboratroy Standards, 2000). [PubMed]
- 17.Ribeiro FK, et al. Genotypic and Spatial Analysis of Mycobacterium tuberculosis Transmission in a High-Incidence Urban Setting. Clin Infect Dis. 2015;61:758–766. doi: 10.1093/cid/civ365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ArcGIS Desktop Help 10.2 Geostatistical Analyst. (Environmental Systems Research Institute (ESRI), 2014).
- 19.Pan, W. C. et al. Fine Particle Pollution, Alanine Transaminase, and Liver Cancer: A Taiwanese Prospective Cohort Study (REVEAL-HBV). J Natl Cancer Inst108, doi:10.1093/jnci/djv341 (2016). [DOI] [PubMed]
- 20.Chen YY, et al. Genetic diversity of the Mycobacterium tuberculosis Beijing family based on SNP and VNTR typing profiles in Asian countries. PLoS One. 2012;7:e39792. doi: 10.1371/journal.pone.0039792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mokrousov I. Mycobacterium tuberculosis phylogeography in the context of human migration and pathogen’s pathobiology: Insights from Beijing and Ural families. Tuberculosis (Edinb) 2015;95(Suppl 1):S167–176. doi: 10.1016/j.tube.2015.02.031. [DOI] [PubMed] [Google Scholar]
- 22.Mokrousov, I. et al. Origin and primary dispersal of the Mycobacterium tuberculosis Beijing genotype: clues from human phylogeography. Genome Res15, 1357–1364, doi:gr.3840605 (2005). [DOI] [PMC free article] [PubMed]
- 23.Wan K, et al. Investigation on Mycobacterium tuberculosis diversity in China and the origin of the Beijing clade. PLoS One. 2011;6:e29190. doi: 10.1371/journal.pone.0029190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wada T, Iwamoto T, Maeda S. Genetic diversity of the Mycobacterium tuberculosis Beijing family in East Asia revealed through refined population structure analysis. FEMS Microbiol Lett. 2009;291:35–43. doi: 10.1111/j.1574-6968.2008.01431.x. [DOI] [PubMed] [Google Scholar]
- 25.Shamputa IC, et al. Genetic diversity of Mycobacterium tuberculosis isolates from a tertiary care tuberculosis hospital in South Korea. J Clin Microbiol. 2010;48:387–394. doi: 10.1128/JCM.02167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim LK, et al. Molecular epidemiology of Mycobacterium tuberculosis complex in Singapore, 2006–2012. PLoS One. 2013;8:e84487. doi: 10.1371/journal.pone.0084487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen VA, et al. High prevalence of Beijing and EAI4-VNM genotypes among M. tuberculosis isolates in northern Vietnam: sampling effect, rural and urban disparities. PLoS One. 2012;7:e45553. doi: 10.1371/journal.pone.0045553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen VA, et al. Mycobacterium tuberculosis lineages and anti-tuberculosis drug resistance in reference hospitals across Viet Nam. BMC Microbiol. 2016;16:167. doi: 10.1186/s12866-016-0784-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phyu S, et al. Predominance of Mycobacterium tuberculosis EAI and Beijing lineages in Yangon, Myanmar. J Clin Microbiol. 2009;47:335–344. doi: 10.1128/JCM.01812-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srilohasin P, et al. Genetic diversity and dynamic distribution of Mycobacterium tuberculosis isolates causing pulmonary and extrapulmonary tuberculosis in Thailand. J Clin Microbiol. 2014;52:4267–4274. doi: 10.1128/JCM.01467-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurepina NE, et al. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber Lung Dis. 1998;79:31–42. doi: 10.1054/tuld.1998.0003. [DOI] [PubMed] [Google Scholar]
- 32.Buu TN, et al. The Beijing genotype is associated with young age and multidrug-resistant tuberculosis in rural Vietnam. Int J Tuberc Lung Dis. 2009;13:900–906. [PubMed] [Google Scholar]
- 33.Chang JR, et al. Clonal expansion of both modern and ancient genotypes of Mycobacterium tuberculosis in southern Taiwan. PLoS One. 2012;7:e43018. doi: 10.1371/journal.pone.0043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivas-Santiago CE, et al. Air pollution particulate matter alters antimycobacterial respiratory epithelium innate immunity. Infect Immun. 2015;83:2507–2517. doi: 10.1128/IAI.03018-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


