Abstract
NMR spectroscopy is a powerful technique to study ribonucleic acids (RNAs) which are key players in a plethora of cellular processes. Although the NMR toolbox for structural studies of RNAs expanded during the last decades, they often remain challenging. Here, we show that solvent paramagnetic relaxation enhancements (sPRE) induced by the soluble, paramagnetic compound Gd(DTPA-BMA) provide a quantitative measure for RNA solvent accessibility and encode distance-to-surface information that correlates well with RNA structure and improves accuracy and convergence of RNA structure determination. Moreover, we show that sPRE data can be easily obtained for RNAs with any isotope labeling scheme and is advantageous regarding sample preparation, stability and recovery. sPRE data show a large dynamic range and reflect the global fold of the RNA suggesting that they are well suited to identify interaction surfaces, to score structural models and as restraints in RNA structure determination.
Introduction
In the last decades, ribonucleic acids (RNAs) have been found to be key regulators for numerous important cellular processes including transcription, translation, splicing, cell differentiation as well as cell death and proliferation1. Besides regulatory functions, RNAs are essential for catalysis in ribosomes and spliceosomes, sensing environmental parameters such as temperature or metabolite concentration, as well as storing the genomic information of RNA viruses. Understanding the underlying molecular mechanisms of these processes requires insights into the structure and dynamics at an atomic level. In the last decades, NMR spectroscopy has developed to a powerful technique to study RNA structure and dynamics2–8. However, technical challenges are caused by poor chemical shift dispersion leading to spectral overlap as well as by the low proton density and the low number of intramolecular interactions limiting the number of observable distance restraints that can be used to define RNA structure. Moreover, the RNA backbone is defined by several torsion angles, requiring a large set of restraints to obtain an accurate structural model. To overcome these limitations, different labeling strategies, including for example specific or uniformly 13C- and/or 15N-labeling and segmental labelling of RNAs were developed to reduce spectral complexity and signal overlap9–22. Large sets of restraints including NOE-based distances, torsion angles and orientational restraints derived from RDCs and J-couplings, respectively, as well as base pairing contacts obtained from J-couplings across hydrogen bonds are typically collected to provide a sufficient number of restraints for high-resolution models of RNAs2, 6, 23–26.
In previous NMR studies, solvent accessibility data were obtained by measuring the NOE between water and protein protons, but the data were found to be dominated by chemical exchange27, 28. Here, we demonstrate that solvent accessibility data derived from solvent paramagnetic relaxation enhancements (sPREs) provide valuable information to characterize the conformation of RNAs and will be useful to enable NMR studies of larger RNAs. sPRE data are a quantitative measure for solvent accessibility and thus readily provide distance-to-surface information29 (Fig. 1). Directly observing the solvent accessible areas is a promising approach not only for mapping interaction surfaces but also for structure determination, as has been shown for proteins30–32. Recently, computational protocols using sPRE data were developed for XplorNIH27 and the Rosetta framework33. The implementation and use of sPRE data has several advantages: i) no covalent labeling of RNAs is required, ii) the sample can be recovered, iii) complete chemical shift assignment is not required and iv) any type of NMR spectrum such as proton 1D, 1H, 15N HSQC or 1H, 13C HSQC experiments can be used. Direct measurements of solvent accessibility data of RNAs have been reported previously using the soluble compound TEMPOL34. While the correlation of TEMPOL-induced sPREs agreed with RNA structure for some parts, the data were limited to a qualitative validation of models. Moreover, unexpectedly large sPREs were found for certain nucleotides and it was later suggested that TEMPOL forms intermolecular hydrogen bonds leading to preferred binding to specific sites35. Here, sPRE data are obtained by titrating the RNA sample with Gd(DTPA-BMA), a soluble, well-characterized Gd3+ chelating paramagnetic contrast agent that was originally developed for MRI imaging31, 36.
Figure 1.
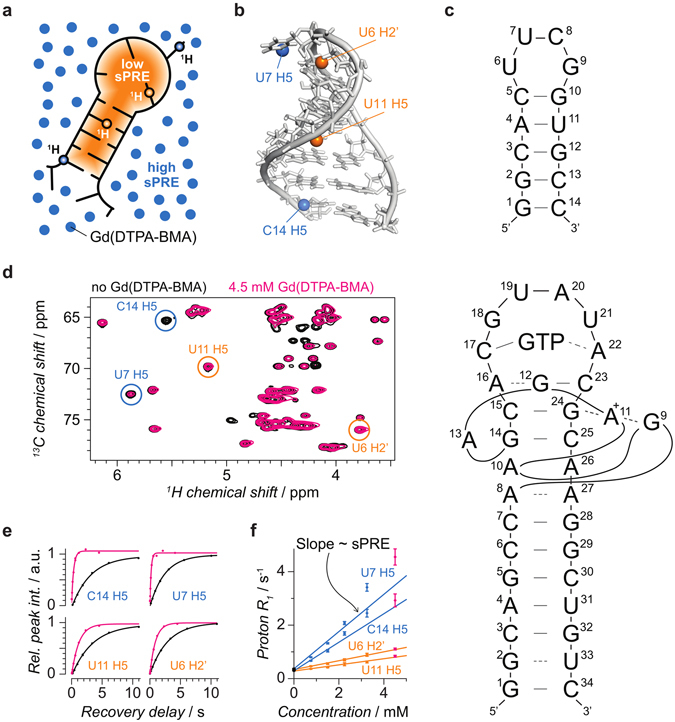
Concept of sPRE and exemplary sPRE data of the UUCG tetraloop. (a) sPRE data provide a quantitative measure for solvent accessibility and encode distance-to-surface information. sPRE data are acquired by titrating the RNA with the paramagnetic compound Gd(DTPA-BMA). (b) The NMR solution structure of the UUCG tetraloop (PDB code 2KOC) is shown and for illustration purpose, two solvent-exposed protons (blue spheres) and two buried protons (orange spheres) are highlighted. (c) Schematic representation of the UUCG tetraloop (top) and the GTP-bound GTP aptamer. (d) NMR spectra of the UUCG tetraloop are shown in the absence (black) and presence (magenta) of Gd(DTPA-BMA) with circles around the peaks corresponding to the protons shown in (b). (e) Quantitative sPRE data are obtained by measuring the longitudinal proton R 1 relaxation rate as a function of the concentration of Gd(DTPA-BMA). (f) Proton R 1 rates increase linearly with the concentration of the paramagnetic compound and the slopes correspond to the sPRE values of the respective resonances highlighted in (b).
Results
Gd(DTPA-BMA) provides quantitative solvent-accessibility data of RNAs
To first assess a potential binding to specific RNA moieties, fingerprint spectra in the absence and presence of the paramagnetic compound were recorded for two RNAs exhibiting distinct folds and structural elements. The resulting NMR spectra of the UUCG tetraloop37, 38 and the GTP class II aptamer39, 40 are shown in Supplementary Figures 1 and 2, respectively. As expected, the presence of Gd(DTPA-BMA) causes line-broadening of NMR signals. However, even in the presence of the paramagnetic compound, the vast majority of peaks are still well observable and the absence of chemical shift perturbations confirms the absence of specific binding of the compound to the RNA (Fig. 1d and Supplementary Figures 1 and 2). Thus Gd(DTPA-BMA) can be used as a paramagnetic probe that screens the accessible surface area of RNA molecules.
As unchelated lanthanide ions can efficiently hydrolyze RNA41, we checked for potential cleavage products of the RNAs in the presence of the Gd3+ compound. No degradation products were observed for any RNA studied and sPRE data obtained from a single RNA sample were reproducible even after several sPRE measurements, each followed by a removal of Gd(DTPA-BMA) (data not shown). The absence of phosphodiester hydrolysis can be explained by the very high chelating stability of DTPA-BMA36 and the presence of a slight excess of Ca2+-bound chelator.
The sPRE data were acquired by measuring proton longitudinal relaxation rates (1H-R 1) as a function of increasing concentrations of Gd(DTPA-BMA). As expected, 1H-R 1 rates were found to increase linearly with the concentration of Gd(DTPA-BMA) in the case of carbon-bound protons. For imino and amino protons, the increase of 1H-R 1 rates is larger at small concentrations of the paramagnetic compound (below about 1 mM), and becomes linear at higher concentrations (Supplementary Figure 3). This observation likely reflects an additional exchange contribution that mixes the relaxation mechanisms of water and RNA protons. Nevertheless, a quantitative measure for the solvent accessibility can be obtained for nitrogen-bound exchangeable protons by determining the minimum concentration of Gd(DTPA-BMA) for which a linear response is observed. Only titration points at this concentration or above are used to fit the linear sPRE model (0.5 mM or above for the UUCG tetraloop and 1.29 mM or above for the GTP aptamer; compare Supplementary Tables 1 and 2). For details about data acquisition and analysis, refer to the methods section in the supplementary information.
sPRE data correlate well with RNA structure
Next, we analyzed how sPRE data reflect the structural features of the RNAs studied. To this end, the NMR structures of the UUCG tetraloop and the GTP-bound GTP class II aptamer were used to predict the expected sPRE data using a previously published grid-based approach30, 31 (see method section for details). The experimental sPRE data correlate very well with the predicted data (Fig. 2a and Supplementary Figures 4a and 5). This demonstrates that sPRE data provide a quantitative measure of solvent-accessibility that correlates well with RNA structure. In particular, all buried protons of both RNAs show a small sPRE. Protons for which significant deviations between experimental and back-calculated sPREs are observed are found in loop regions or in terminal nucleotides. These regions are flexible and thus are expected to show a population-weighted average of the sPRE. The structural NMR ensembles used to predict the sPRE data represent the most stable conformations in solution and depend on the algorithm and experimental restraints used for the structure determination. Thus, the NMR ensembles do not necessarily reflect the equilibrium distribution of the different conformations of these solvent-exposed regions of the RNA. Since all conformations of the ensemble were equally weighted in the prediction of the sPRE data, the prediction is expected to deviate for dynamic regions of the RNA. It is further worth noting, that despite the compact fold of the UUCG tetraloop, the experimental sPRE data show a large dynamic range, which renders them a powerful source to characterize the structure of RNAs. The excellent correlation between experimental data and RNA structure in combination with a large dynamic range suggests sPRE data as promising parameters to probe RNA structures by NMR.
Figure 2.
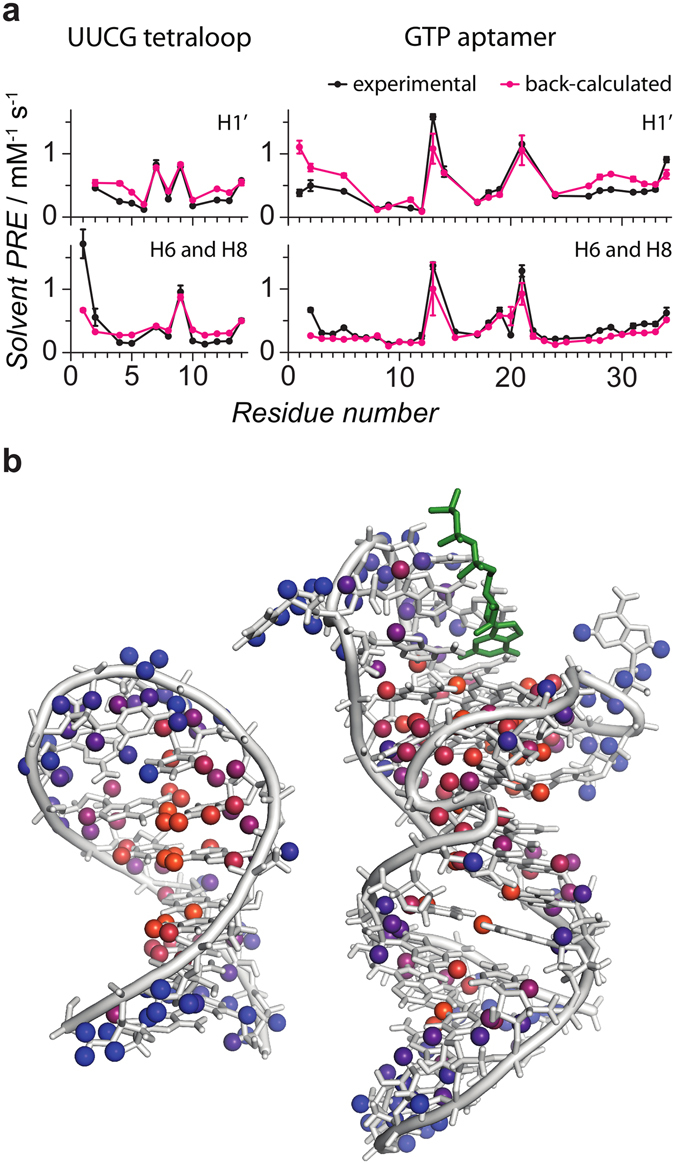
sPRE data correlates well with RNA structure. (a) Experimental sPRE data (black) are compared to predicted sPRE data based on NMR solution structures of the UUCG tetraloop (2KOC) and the GTP-bound aptamer (5LWJ). Data are shown for sugar protons (H1′) and aromatic protons (H6 and H8) as indicated. (b) NMR solution structures of the UUCG tetraloop37, 38 (2KOC, left) and the GTP-bound aptamer39, 40 (5LWJ, right, heavy atoms of GTP ligand shown in green) are shown. All protons for which a sPRE value was obtained are shown as spheres and colored according to the sPRE (blue corresponding to high and orange corresponding to low sPRE values).
As described above, significant deviations between predicted and measured sPRE data are observed for some terminal nucleotides. Among all 258 observed sPRE values (93 for the UUCG tetraloop and 165 for the GTP aptamer), only two protons that are located in non-terminal nucleotides show significant deviations (9Gua H1 and 6Ura H3 in the UUCG tetraloop). Both protons are located in the loop region of the RNA in close spatial proximity to each other (Supplementary Figure 4b) and the corresponding sPRE values are underestimated based on the NMR solution structure, independent of which structural model of the UUCG loop motif (PDB codes 1HLX, 1K2G, 1TLR, 1Z31, 2KHY, 2KOC, 2KZL, 2LHP, 2LUB and 2N6X) was used. Although the sPRE of these protons are expected to have an additional contribution due to chemical exchange with water, the sPRE values of 2.2 and 0.44 s−1mM−1 of 6Ura H3 and 9Gua H1, respectively, are significantly larger compared to the average of 0.079 ± 0.037 s−1mM−1 for all other imino protons. This suggests that conformational dynamics in this regions may cause the unusually high experimental sPREs. Indeed, recent molecular dynamics studies that included a large set of NMR relaxation data of the UUCG tetraloop, suggest the loop region to be more flexible than the RNA stem42–48. Moreover, intermediate structural states with the bases of the loop region experiencing higher solvent exposure were observed in these studies. This suggests that the increased sPREs are a result of increased flexibility and dynamics of the loop region. However, future studies are required to fully understand the usability of sPRE data to detect dynamics in RNAs.
sPRE data provide orthogonal restraints for RNA structure determination
Next, the potential of sPRE data to facilitate the structure determination of RNAs was investigated (Fig. 3). To this end, structural models of the UUCG tetraloop as well as the GTP-bound aptamer were computed using the XplorNIH framework27, 49–51 in combination with different sets of experimental NOE data (Supplementary Tables 3 and 4, Supplementary Figure 7). The results show that sPRE data significantly improves convergence and accuracy of the structure determination, in particular in cases where only limited sets of NOEs are available. For example, using only 258 experimental NOEs of the GTP-bound aptamer (30% of the all NOEs), the average RMSD of the obtained models was significantly reduced from 7.80 Å to 3.51 Å (Fig. 3b; Random set 2 in Supplementary Table 3). Next, the performance of the sPRE potential in combination with other experimental NMR-based restraints was investigated (Supplementary Table 5). The benchmark revealed that solvent accessibility data is an orthogonal restraint since its usage improves structure determination of the UUCG tetraloop in combination with NOE, RDC and torsion angle restraints. Strikingly, using only hydrogen bonds-derived restraints in combination with sPRE data, the structure of the UUCG tetraloop can be determined with an RMSD of 2.8 Å compared to the published NMR-based structure (Fig. 3a). Even sparse sPRE data sets, in some cases down to 25%, improve structural quality of the UUCG tetraloop (Supplementary Table 6).
Figure 3.
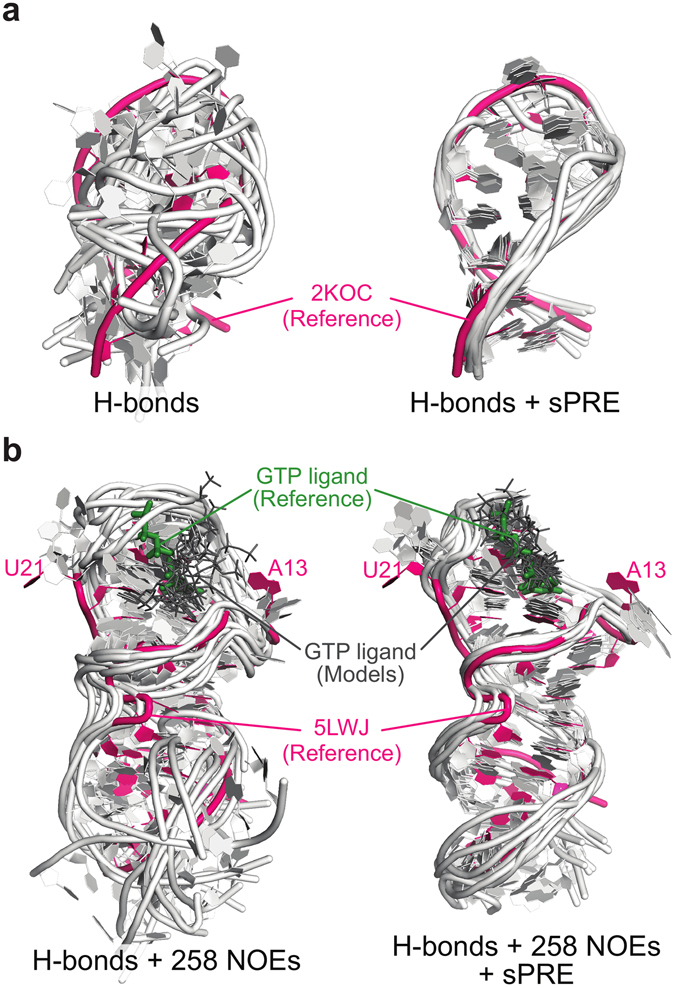
sPRE data improve structure determination of RNAs. Structural models of the UUCG tetraloop (a) and the GTP-bound GTP aptamer (b) were obtained without (left) and with (right) sPRE data using XplorNIH. The 10 best scored models (light gray) in terms of total energy were selected from a total of 200 models and aligned to the corresponding NMR structure (magenta). All restraints used in the computations are indicated below the respective models. In (b) heavy atoms of the GTP ligand are shown as sticks (Reference in green, computed models in dark gray) and the positions of the intrinsically flexible nucleotides A13 and U2140 are indicated.
Discussion
In summary, we show that sPRE data are efficient NMR observables to probe RNA structure. sPRE data obtained with the paramagnetic compound Gd(DTPA-BMA) yield quantitative information of solvent accessibility as they correlate well with RNA structure, provide structural information for both buried and surface-exposed atoms and can be used as restraints to drive molecular dynamics-based structure determination of RNAs. Since sPRE data reflect the global fold of a RNA they are well suited to identify tertiary contacts or map interaction surfaces with other molecules32, for example, in RNA-protein complexes. As the measurement of sPRE data does not require complete chemical shift assignments, surface accessibility restraints can be recorded for structural studies also in the case of large RNAs, in particular in combination with specific or segmental labelling, where it is difficult to obtain a comprehensive set of experimental restraints for structural analysis. Compared to proteins, several considerations have to be taken into account for sPRE-based studies of RNAs. First, different NMR pulse sequences might be needed, in particular for triple resonance experiments. Second, multiple data sets acquired for different RNA labelling schemes and different solvent conditions need to be combined. In order to compensate for minor differences between these data sets we have proposed an approach in which we normalize the sPRE data using the sPRE of water protons in each sample. Third, lower concentrations of Gd(DTPA-BMA) are used for RNAs compared to proteins due to the higher sPRE caused by the lack of large hydrophobic cores resulting in smaller distances to the RNA surface. Last, chemical exchange of protons with water is typically more efficient in RNAs and modulates sPREs of nitrogen-bound amino and imino protons. In these cases, only the region of linear response is used for quantitative analysis.
Our results show that sPRE-derived restraints are particularly promising to define the global fold of larger RNAs and provide valuable information for structure determination that is orthogonal to other NMR-based restraints in molecular dynamics-based approaches. Recently, accuracy of chemical-shift based RNA de novo structure prediction using the Rosetta framework was significantly enhanced by improving the chemical shift scoring function for RNAs52. Combining this improved score with a sampling algorithm driven by sPRE data33 and specific or segmental labeling could provide powerful tools for structural modeling of large RNAs based on NMR data in the future.
Methods
RNA sample preparation
A uniformly 13C, 15N labeled sample of the UUCG tetraloop (5′-GGCACUUCGGUGCC-3′) was purchased from Silantes (Munich, Germany). The NMR buffer contained 20 mM K2HPO4 and 0.4 mM EDTA and was adjusted to pH 6.4. Transcription and purification of the GTP class II aptamer in complex with GTP was been described previously39. The sample was measured in NMR buffer containing 25 mM KH2PO4, 25 mM KCl and 2 mM magnesium acetate. The buffer was adjusted to pH 6.3 and uniformly 13C, 15N labeled RNA was measured in the presence of a two-fold excess of unlabeled GTP. NMR assignments and a solution NMR structure of the UUCG tetraloop were obtained from literature37, 38 (PDB code 2KOC, BMRB 5705). For the GTP class II aptamer in complex with GTP, the recently reported NMR assignments and solution NMR structure were used39, 40 (PDB code 5LWJ, BMRB 25661).
NMR Experiments
NMR sPRE data were obtained by measuring proton R 1 relaxation rates as a function of the concentration of Gd(DTPA-BMA) using a saturation-recovery approach which has been described previously for protein applications30. Briefly, a 7.5 ms proton trim pulse followed by a gradient is used to dephase magnetization. Next, longitudinal magnetization is recovered during a recovery delay of variable length. The recovery delay is followed by a read-out spectrum, such as a 1H,15N or 1H,13C HSQC. This saturation-recovery scheme allows to use a short interscan delay (about 50 ms) which dramatically reduces experimental time compared to measuring proton R2 rates. Pseudo-3D experiments were acquired for 4 to 7 different concentrations of the paramagnetic compound. To recover the NMR RNA sample, the paramagnetic compound was first removed by dialysis against water using a membrane with a 1 kDa molecular weight cut-off and then, the sample was lyophilized and resuspended in the corresponding NMR buffer.
sPRE data for nitrogen-bound protons of the UUCG tetraloop were recorded with a 0.8 mM uniformly 13C, 15N labeled sample at 283 K on an Avance III Bruker 900 MHz NMR spectrometer equipped with a cryo-TXI probe head. Proton R 1 relaxation rates were measured in the absence and presence of 0.5, 1, 1.75, 2.75 and 4.0 mM Gd(DTPA-BMA) using 1H,15N HSQC-based pseudo-3D experiments. sPRE data for carbon-bound protons were acquired with the same sample at 298 K and R 1 relaxation rates were measured in the absence and presence of 0.75, 1.5, 2.25, 3.25 and 4.5 mM Gd(DTPA-BMA) using 1H,13C HSQC-based pseudo-3D experiments. Delay times and other NMR parameters for all experiments as well as the total NMR time are presented in Supplementary Table 1. To obtain sPRE data of the GTP aptamer in complex with GTP, samples containing about 150 µM of (13C, 15N)-GU, (13C, 15N)-C or (13C, 15N)-A labeled RNA and 300 µM unlabeled GTP were measured at 293 K on an Avance III Bruker 900 MHz NMR spectrometer equipped with a cryo-TXI probe head and an Oxford Instruments 600 MHz NMR spectrometer equipped with a Bruker Avance III console and a cryo-TCI probe head. 1H,13C and 1H,15N HSQC-based pseudo-3D experiments were used to acquire proton R 1 relaxation rates for 4 to 7 different concentrations of Gd(DTPA-BMA). Delay times and other NMR parameters for all experiments as well as the total NMR time are presented in Supplementary Table 2. For all RNA samples, amino and imino protons were measured in buffer containing 5 to 10% D2O whereas carbon-bound protons were acquired in buffer containing ≥99.990% D2O (Sigma-Aldrich). For every titration step of Gd(DTPA-BMA), solvent water R 1 was measured and the derived sPRE of the water solvent was used to normalize the RNA sPRE values. The normalized sPRE data were then rescaled by the mean sPRE of the solvent, obtained by averaging the sPRE of water protons in all relevant experiments. This procedure allows to combine sPRE measurements of different samples or different magnetic field strengths (e.g. in different buffers or with different labeling schemes; see Supplementary Figure 6). Furthermore, by plotting the solvent water R 1 against the concentration of Gd(DTPA-BMA) errors in pipetting can be detected and compensated accordingly if necessary.
In the case of nitrogen-bound protons, plotting the proton R 1 against the concentration of Gd(DTPA-BMA) revealed a non-linear correlation with rates at low concentrations of the paramagnetic compound being lower than expected according to a linear model. Consequently, the R 1 for low concentrations (0 mM in the case of the UUCG tetraloop, as well as 0 and 0.645 mM in the case of the GTP aptamer) were omitted in the computations of the sPRE values and only the linear correlation was used (compare Supplementary Table 1 and 2 as well as Supplementary Figure 3). In all cases, at least 4 different concentrations were used to derive the sPRE.
For large RNAs, specific isotope labeling schemes are required to reduce signal overlap9, 10, 53. In a simple and straight-forward approach, only one or two nucleotide types are enriched in NMR active nuclei. While this approach reduces spectral complexity, the measurement of sPREs with different samples might introduce systematic errors, due to pipetting errors. In addition, the usage of different magnetic field strengths affects the scaling of the sPRE data31. To ensure that sPRE data acquired on multiple samples and field strengths are consistent, the sPRE data sets are scaled by referencing to the sPRE of the water solvent in each sample. This referencing notably improves the correlation of calculated and experimental sPRE (Supplementary Figure 6). To normalize the sPRE data, the sPRE of water protons was acquired by measuring the R 1 rate of the water proton signal. The R 1 rate was measured using a pseudo 2D experiments with simple 1D proton read-out (the proton pulse length was set between 0.3 and 1 µs). The change of the R 1 rate of the water protons during the titration Gd(DTPA-BMA) with was used to obtain the sPRE of the water protons. The sPRE of the water solvent was then used to normalize the sPRE values of the RNA signals. The normalized sPRE values of different titration experiments (with different labeling schemes or at different field strengths) were then combined to generate a large sPRE data set for the corresponding RNA. To obtain an absolute quantity, the normalized sPRE data were then multiplied by the mean sPRE of the solvent, obtained by averaging the sPRE of water protons in all relevant experiments. This procedure allows to combine sPRE measurements of different samples or different magnetic field strengths (e.g. in different buffers or with different labeling schemes; see Supplementary Figure 6). Furthermore, by plotting the solvent water R 1 against the concentration of Gd(DTPA-BMA) errors in pipetting can be detected and compensated accordingly if necessary.
The measurement times for the collection of sPRE data can be quite long, as sPRE data are recorded as a series of proton R 1 relaxation experiments. Typical 1H-R 1 rates in the absence of Gd(DTPA-BMA) are relatively slow with an average 1H-R 1 rate of 0.23 s−1 (±0.10 s−1) at 900 MHz and 0.30 s−1 (±0.14 s−1) at 600 MHz. As a consequence, long delay times of several seconds (Supplementary Table 2) are required which in turn increase the overall experimental time. To overcome this drawback, the data sets of both RNAs were re-evaluated by determining the sPREs based on R 1 rates measured only in the presence of 0.75, 1.5, 3.25 and 4.5 mM Gd(DTPA-BMA) for the UUCG tetraloop and 0.8, 1.6, 3.2 and 4 mM for the GTP aptamer. Notably, the obtained sPRE data are fully consistent with those derived from a full set of measurements, but the optimized acquisition scheme reduces experimental time by about 40%. It should be noted, that experimental time could be further reduced by applying selective saturation schemes that employ longitudinal relaxation-enhancing techniques as demonstrated in a previous study54.
NMR data analysis
NMR spectra were processed with the NMRpipe software package55. Peak positions from literature were transferred using CcpNmr Analysis56 and peak intensities of well-resolved peaks were obtained using the nmrglue Python package57. Peak intensities of the pseudo-3D experiments were fitted to an exponential recovery function according to equation (1):
| 1 |
where τ is the recovery delay, I(τ) is the peak intensity measured for the recovery delay τ, I 0 is the maximum peak intensity, R 1 is the longitudinal proton relaxation rate and A is the amplitude of the relaxation.
To estimate the error of the peak intensities ε, the recovery delay τ d was measured twice to obtain two intensity values I(τ d, I, 1) and I(τ d, I, 2), where i is the index of the peak. For every peak i, the difference of the duplicates was computed. The overall error of the pseudo-3D experiment ε pseudo-3D was then computed according to equation (2):
| 2 |
where N is the number of peaks, and is the average difference of the duplicates. The subtraction of accounts for systematic errors, and in all cases was significantly lower in magnitude than the differences of the duplicates .
The error of the peak intensities ε pseudo-3D was then used to estimate the error of the fitted proton relaxation rates ΔR 1. To this end, the experimental data was resampled using the following combined Monte Carlo-type bootstrapping approach: For every peak in a given pseudo-3D experiment, a set of delay-intensity data points, I(τ), was measured. This set is used to generate 1000 new random sets , each having 2.5 times as many data points as the original data set I(τ) (Recovery delays measured as duplicates are only counted as one). These sets were created by randomly selecting data points from the original data set I(τ) and allowing values to be selected multiple times (Increasing the size of the new sets by a factor of 2.5 allows to generate a more diverse ensemble of sets). For every set , the intensity values were randomly altered by adding a random number drawn from a normal distribution centered at 0 and with a standard deviation of ε pseudo-3D. Every set was then fitted according to equation (1) giving rise to 1000 different rates. The error of the proton relaxation rate ΔR 1 was computed as the standard deviation of all values . This procedure was repeated for every peak and every pseudo-3D experiment.
To obtain the sPRE value, the proton R 1 rates and the corresponding errors ΔR 1 were collected for all measured concentrations of Gd(DTPA-BMA) c para and a weighted linear regression using equation (3) was performed
| 3 |
where R 1(c para) is the proton R 1 measured at the concentration c para of the paramagnetic compound, the slope m sPRE corresponds to the sPRE and is the fitted proton R 1 in the absence of the paramagnetic compound. The errors ΔR 1 (obtained from the resampling approach described above) were used as weights in a weighted linear regression and the error of the sPRE value as well as the error of the relaxation rate were directly obtained from the weighted linear regression.
Prediction of sPRE Data
To predict sPRE data based on published NMR solution structure, a previously published grid-based approach was used31. Briefly, the structural model was placed in a regularly-spaced grid representing the uniformly distributed paramagnetic compound and the grid was built with a point-to-point distance of 0.1 Å and a minimum distance of 20 Å between the RNA model and the outer border of the grid. Next, grid points that overlap with the RNA model were removed assuming a molecular radius of 3.5 Å for the paramagnetic compound. To compute the sPRE for a given RNA proton , the distance-dependent paramagnetic effect29 was numerically integrated over all remaining grid points according to equation (4):
| 4 |
where i is the index of a proton of the RNA, j is the index of the grid point, d i, j is the distance between the i-th proton and the j-th grid point and c is an arbitrary constant to scale the sPRE values. Here, c was chosen such that the sum of all predicted sPRE values is equal to the sum of all experimental sPRE values.
Structure Determination Benchmark
To demonstrate the potential of sPRE data for the structure determination of RNAs, structural models of the UUCG tetraloop as well as the GTP class II aptamer in complex with GTP were computed using the XplorNIH 2.40 framework49, 50. The used protocol is based on the gb1_rdc example that is included in the XplorNIH package and was adjusted to fold the respective RNA during an annealing procedure starting from an extended structure. The initial temperature was set to 3500 and the simulation at this high temperature was stopped after reaching 1000 ps or 10,000 steps. The temperature was then reduced to a final temperature of 25 in steps of 12.5 degrees and simulations were run for at least 4 ps or 2000 steps at every temperature step. The annealing procedure was repeated to obtain 200 models. The protocol made use of the recently published torsion potential RNA-ff151 and the sPRE data was included in the structure determination using the nbTargetPot module27 of the framework. nbTargetPot is the scaling factor (or weight) of the sPRE potential. The value of the scaling factor mainly depends on the scaling factor of the other energy terms. The weight of the sPRE potential should not be proportional to the size of the RNA. For the mentioned benchmark, the weight of the sPRE potential was determined by testing different weights and the weight that produced the best models was used for all runs. To this end, one weight was determined for every RNA and the obtained weight was used for all computations of the corresponding RNA. The lower value of the UUCG tetraloop can be explained with the fact that the Xplor runs of the UUCG tetraloop included more energy functions than the ones of the GTP aptamer (in the case of the UUCG tetraloop, torsion angles and RDC, both obtained from PDB entry 2KOC, were used). The sPRE data sets of the UUCG tetraloop and GTP aptamer were filtered to remove all data points with an experimental error above 10% and all data points of nitrogen-bound protons. The nbTargetPot energy was initialized using the slope and intercept parameter as returned by the calibrate function of the nbTargetPotTools module in combination with the NMR structure of the corresponding RNA. The weight of the nbTargetPot energy was set to 1000 in the case of the UUCG tetraloop and 3000 for the GTP aptamer. For more details such as weight factors of all potentials, please refer to the python code of the protocols which are shown at the end of this document for both RNAs.
The annealing protocol was then used to benchmark the benefit of the sPRE data for structure prediction. To this end, structural models of the UUCG tetraloop and the GTP-bound aptamer were computed in the absence and presence of the sPRE potential and in combination with experimental restraints derived from hydrogen bond information and experimental NOEs (PDB entries 2KOC and 5LWJ)37, 38, 40. To simulate several NOE assignments, random experimental NOE subsets were created by randomly drawing a certain percentage of restraints from the full NOE set (compare Supplementary Tables 3 and 4). To account for the effect of the random selection process, at least 3 different random sets with the same number of NOE restrains were created and used to benchmark the impact of the sPRE data. It should be noted, that the randomly created NOE subsets not only account for different levels of assignments, but also simulate different qualities of NOE data as random subsets with the same number of NOE restraints perform differently in driving the structure determination to the correct fold. For every given set of restraints, 200 structure models were computed with and without the sPRE data. To quantify the impact of the sPRE data, the 20 best models of each run (10%, scored according to the total energy) were selected and the RMSD to the published NMR structures was computed. Computation of RMSD was performed on all carbon, nitrogen and phosphorus atoms in the case of the UUCG tetraloop. For the GTP-bound aptamer, the RMSD was computed using all carbon, nitrogen and phosphorus atoms of the GTP ligand and all non-terminal nucleotides except the intrinsically flexible nucleotides A13 and U2140.
A second benchmark was performed to address the performance of the sPRE potential in the absence of any other experimental restraint as well as in combination with other experimental NMR restraints, such hydrogen bonds, NOEs, RDCs and torsion angles. Experimental restraints for the UUCG tetraloop were obtained from PDB entry 2KOC37, 38. Two hundred structure models of the UUCG tetraloop were computed with and without the sPRE data for every restraint set (compare Supplementary Tables 5) and the 20 best scored models according to the total energy were selected. The average RMSD to the published NMR structure was computed for the top 20 models and used to quantify the impact of the sPRE data (computation of RMSD was performed on all carbon, nitrogen and phosphorus atoms).
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. All scripts used for data analysis and back-calculation of sPRE data are openly available (http://mbbc.medunigraz.at/forschung/forschungseinheiten-und-gruppen/forschungsgruppe-tobias-madl/software/).
Electronic supplementary material
Acknowledgements
This work was supported by the Integrative Metabolism Research Center Graz (to T.M.), the Austrian infrastructure programme 2016/2017 (to T.M.), BioTechMed/Graz (to T.M.), the Omics Center Graz (to T.M.), the Bavarian Ministry of Sciences, Research and the Arts (Bavarian Molecular Biosystems Research Network, to T.M.), the President’s International Fellowship Initiative of CAS (No. 2015VBB045, to T.M.), the National Natural Science Foundation of China (No. 31450110423, to T.M.), the Austrian Science Fund (FWF: P28854 and W1226-B18 to T.M.) as well as the Deutsche Forschungsgemeinschaft (DFG) with the grants Wo901/1-1 (to J.W.), SFB1035 (to M.S.), GRK1721 (to M.S.) and MA5703/1-1 (to T.M.).
Author Contributions
C.H. and T.M. designed the studies, established and tested the NMR experiments, and performed the prediction of sPRE data. J.G., A.W., J.W. and M.S. prepared the isotope labelled RNA samples. C.H. and T.M., C.H. and J.G. performed the NMR titrations. C.H. processed and fitted the NMR data, performed the structure prediction computations and analyzed the obtained models. C.H. and T.M. wrote the manuscript and all authors have given critical feedback and approved the final version of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Christoph Hartlmüller and Johannes C. Günther contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05821-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furtig B, Richter C, Wohnert J, Schwalbe H. NMR spectroscopy of RNA. Chembiochem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 3.Latham MP, Brown DJ, McCallum SA, Pardi A. NMR methods for studying the structure and dynamics of RNA. Chembiochem. 2005;6:1492–1505. doi: 10.1002/cbic.200500123. [DOI] [PubMed] [Google Scholar]
- 4.Bothe JR, et al. Characterizing RNA dynamics at atomic resolution using solution-state NMR spectroscopy. Nat Methods. 2011;8:919–931. doi: 10.1038/nmeth.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez C, Schubert M, Duss O, Ravindranathan S, Allain FH. Structure determination and dynamics of protein-RNA complexes by NMR spectroscopy. Prog Nucl Magn Reson Spectrosc. 2011;58:1–61. doi: 10.1016/j.pnmrs.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Wijmenga SS, van Buuren BNM. The use of NMR methods for conformational studies of nucleic acids. Progress in Nuclear Magnetic Resonance Spectroscopy. 1998;32:287–387. doi: 10.1016/S0079-6565(97)00023-X. [DOI] [Google Scholar]
- 7.Wu H, Finger LD, Feigon J. Structure determination of protein/RNA complexes by NMR. Methods Enzymol. 2005;394:525–545. doi: 10.1016/S0076-6879(05)94022-6. [DOI] [PubMed] [Google Scholar]
- 8.Varani G, Aboul-ela F, Allain FH. NMR investigation of RNA structure. Prog Nucl Magn Reson Spectrosc. 1996;29:51–127. doi: 10.1016/0079-6565(96)01028-X. [DOI] [Google Scholar]
- 9.Duss O, Lukavsky PJ, Allain FH. Isotope labeling and segmental labeling of larger RNAs for NMR structural studies. Adv Exp Med Biol. 2012;992:121–144. doi: 10.1007/978-94-007-4954-2_7. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, et al. Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature. 2015;522:368–372. doi: 10.1038/nature14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu K, Miyazaki Y, Summers MF. Isotope labeling strategies for NMR studies of RNA. J Biomol NMR. 2010;46:113–125. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 14.Hennig, M., Williamson, J. R., Brodsky, A. S. & Battiste, J. L. Recent advances in RNA structure determination by NMR. Curr Protoc Nucleic Acid ChemChapter 7, Unit 7 7, doi:10.1002/0471142700.nc0707s02 (2001). [DOI] [PubMed]
- 15.Wunderlich CH, et al. Synthesis of (6-(13)C)pyrimidine nucleotides as spin-labels for RNA dynamics. J Am Chem Soc. 2012;134:7558–7569. doi: 10.1021/ja302148g. [DOI] [PubMed] [Google Scholar]
- 16.Johnson JE, Jr., Julien KR, Hoogstraten CG. Alternate-site isotopic labeling of ribonucleotides for NMR studies of ribose conformational dynamics in RNA. J Biomol NMR. 2006;35:261–274. doi: 10.1007/s10858-006-9041-x. [DOI] [PubMed] [Google Scholar]
- 17.Alvarado LJ, et al. Regio-selective chemical-enzymatic synthesis of pyrimidine nucleotides facilitates RNA structure and dynamics studies. Chembiochem. 2014;15:1573–1577. doi: 10.1002/cbic.201402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longhini AP, et al. Chemo-enzymatic synthesis of site-specific isotopically labeled nucleotides for use in NMR resonance assignment, dynamics and structural characterizations. Nucleic Acids Res. 2016;44:e52. doi: 10.1093/nar/gkv1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wenter P, Reymond L, Auweter SD, Allain FH, Pitsch S. Short, synthetic and selectively 13C-labeled RNA sequences for the NMR structure determination of protein-RNA complexes. Nucleic Acids Res. 2006;34:e79. doi: 10.1093/nar/gkl427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tzakos AG, Easton LE, Lukavsky PJ. Complementary segmental labeling of large RNAs: economic preparation and simplified NMR spectra for measurement of more RDCs. J Am Chem Soc. 2006;128:13344–13345. doi: 10.1021/ja064807o. [DOI] [PubMed] [Google Scholar]
- 21.Xu J, Lapham J, Crothers DM. Determining RNA solution structure by segmental isotopic labeling and NMR: application to Caenorhabditis elegans spliced leader RNA 1. Proc Natl Acad Sci USA. 1996;93:44–48. doi: 10.1073/pnas.93.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelissen FH, et al. Multiple segmental and selective isotope labeling of large RNA for NMR structural studies. Nucleic Acids Res. 2008;36:e89. doi: 10.1093/nar/gkn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipsitz RS, Tjandra N. Residual dipolar couplings in NMR structure analysis. Annu Rev Biophys Biomol Struct. 2004;33:387–413. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]
- 25.Dallmann A, Sattler M. Detection of hydrogen bonds in dynamic regions of RNA by NMR spectroscopy. Curr Protoc Nucleic Acid Chem. 2014;59:7 22 21–19. doi: 10.1002/0471142700.nc0722s59. [DOI] [PubMed] [Google Scholar]
- 26.Grzesiek S, Cordier F, Jaravine V, Barfield M. Insights into biomolecular hydrogen bonds from hydrogen bond scalar couplings. Progress in Nuclear Magnetic Resonance Spectroscopy. 2004;45:275–300. doi: 10.1016/j.pnmrs.2004.08.001. [DOI] [Google Scholar]
- 27.Wang Y, Schwieters CD, Tjandra N. Parameterization of solvent-protein interaction and its use on NMR protein structure determination. J Magn Reson. 2012;221:76–84. doi: 10.1016/j.jmr.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Modig K, Liepinsh E, Otting G, Halle B. Dynamics of protein and peptide hydration. Journal of the American Chemical Society. 2004;126:102–114. doi: 10.1021/ja038325d. [DOI] [PubMed] [Google Scholar]
- 29.Hocking HG, Zangger K, Madl T. Studying the structure and dynamics of biomolecules by using soluble paramagnetic probes. Chemphyschem. 2013;14:3082–3094. doi: 10.1002/cphc.201300219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madl T, Bermel W, Zangger K. Use of relaxation enhancements in a paramagnetic environment for the structure determination of proteins using NMR spectroscopy. Angew Chem Int Ed Engl. 2009;48:8259–8262. doi: 10.1002/anie.200902561. [DOI] [PubMed] [Google Scholar]
- 31.Pintacuda G, Otting G. Identification of protein surfaces by NMR measurements with a pramagnetic Gd(III) chelate. J Am Chem Soc. 2002;124:372–373. doi: 10.1021/ja016985h. [DOI] [PubMed] [Google Scholar]
- 32.Madl T, Guttler T, Gorlich D, Sattler M. Structural analysis of large protein complexes using solvent paramagnetic relaxation enhancements. Angew Chem Int Ed Engl. 2011;50:3993–3997. doi: 10.1002/anie.201007168. [DOI] [PubMed] [Google Scholar]
- 33.Hartlmuller C, Gobl C, Madl T. Prediction of Protein Structure Using Surface Accessibility Data. Angew Chem Int Ed Engl. 2016;55:11970–11974. doi: 10.1002/anie.201604788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venditti V, Niccolai N, Butcher SE. Measuring the dynamic surface accessibility of RNA with the small paramagnetic molecule TEMPOL. Nucleic Acids Res. 2008;36:e20. doi: 10.1093/nar/gkm1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernini A, et al. NMR studies on the surface accessibility of the archaeal protein Sso7d by using TEMPOL and Gd(III)(DTPA-BMA) as paramagnetic probes. Biophys Chem. 2008;137:71–75. doi: 10.1016/j.bpc.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Gadolinium(III) Chelates as MRI Contrast Agents: Structure, Dynamics, and Applications. Chem Rev. 1999;99:2293–2352. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]
- 37.Furtig B, Richter C, Bermel W, Schwalbe H. New NMR experiments for RNA nucleobase resonance assignment and chemical shift analysis of an RNA UUCG tetraloop. J Biomol NMR. 2004;28:69–79. doi: 10.1023/B:JNMR.0000012863.63522.1f. [DOI] [PubMed] [Google Scholar]
- 38.Nozinovic S, Furtig B, Jonker HR, Richter C, Schwalbe H. High-resolution NMR structure of an RNA model system: the 14-mer cUUCGg tetraloop hairpin RNA. Nucleic Acids Res. 2010;38:683–694. doi: 10.1093/nar/gkp956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolter AC, et al. NMR resonance assignments for the class II GTP binding RNA aptamer in complex with GTP. Biomol NMR Assign. 2016;10:101–105. doi: 10.1007/s12104-015-9646-7. [DOI] [PubMed] [Google Scholar]
- 40.Wolter AC, et al. A Stably Protonated Adenine Nucleotide with a Highly Shifted pKa Value Stabilizes the Tertiary Structure of a GTP-Binding RNA Aptamer. Angewandte Chemie. 2016 doi: 10.1002/anie.201609184. [DOI] [PubMed] [Google Scholar]
- 41.Matsumura K, Komiyama M. Enormously fast RNA hydrolysis by lanthanide(III) ions under physiological conditions: eminent candidates for novel tools of biotechnology. J Biochem. 1997;122:387–394. doi: 10.1093/oxfordjournals.jbchem.a021765. [DOI] [PubMed] [Google Scholar]
- 42.Deng NJ, Cieplak P. Free energy profile of RNA hairpins: a molecular dynamics simulation study. Biophys J. 2010;98:627–636. doi: 10.1016/j.bpj.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giambasu GM, York DM, Case DA. Structural fidelity and NMR relaxation analysis in a prototype RNA hairpin. RNA. 2015;21:963–974. doi: 10.1261/rna.047357.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villa A, Widjajakusuma E, Stock G. Molecular dynamics simulation of the structure, dynamics, and thermostability of the RNA hairpins uCACGg and cUUCGg. J Phys Chem B. 2008;112:134–142. doi: 10.1021/jp0764337. [DOI] [PubMed] [Google Scholar]
- 45.Borkar AN, Vallurupalli P, Camilloni C, Kay LE, Vendruscolo M. Simultaneous NMR characterisation of multiple minima in the free energy landscape of an RNA UUCG tetraloop. Phys Chem Chem Phys. 2017;19:2797–2804. doi: 10.1039/C6CP08313G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vallurupalli P, Kay LE. A suite of 2H NMR spin relaxation experiments for the measurement of RNA dynamics. J Am Chem Soc. 2005;127:6893–6901. doi: 10.1021/ja0427799. [DOI] [PubMed] [Google Scholar]
- 47.Williams DJ, Hall KB. Experimental and computational studies of the G[UUCG]C RNA tetraloop. J Mol Biol. 2000;297:1045–1061. doi: 10.1006/jmbi.2000.3623. [DOI] [PubMed] [Google Scholar]
- 48.Akke M, Fiala R, Jiang F, Patel D, Palmer AG., 3rd Base dynamics in a UUCG tetraloop RNA hairpin characterized by 15N spin relaxation: correlations with structure and stability. RNA. 1997;3:702–709. [PMC free article] [PubMed] [Google Scholar]
- 49.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Progress in nuclear magnetic resonance spectroscopy. 2006;48:47–62. doi: 10.1016/j.pnmrs.2005.10.001. [DOI] [Google Scholar]
- 50.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. Journal of magnetic resonance. 2003;160:65–73. doi: 10.1016/S1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 51.Bermejo GA, Clore GM, Schwieters CD. Improving NMR Structures of RNA. Structure. 2016;24:806–815. doi: 10.1016/j.str.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sripakdeevong P, et al. Structure determination of noncanonical RNA motifs guided by (1)H NMR chemical shifts. Nat Methods. 2014;11:413–416. doi: 10.1038/nmeth.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dallmann A, et al. Site-Specific Isotope-Labeling of Inosine Phosphoramidites and NMR Analysis of an Inosine-Containing RNA Duplex. Chemistry. 2016 doi: 10.1002/chem.201602784. [DOI] [PubMed] [Google Scholar]
- 54.Farjon J, et al. Longitudinal-relaxation-enhanced NMR experiments for the study of nucleic acids in solution. Journal of the American Chemical Society. 2009;131:8571–8577. doi: 10.1021/ja901633y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 56.Vranken WF, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 57.Helmus JJ, Jaroniec CP. Nmrglue: an open source Python package for the analysis of multidimensional NMR data. J Biomol NMR. 2013;55:355–367. doi: 10.1007/s10858-013-9718-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. All scripts used for data analysis and back-calculation of sPRE data are openly available (http://mbbc.medunigraz.at/forschung/forschungseinheiten-und-gruppen/forschungsgruppe-tobias-madl/software/).


