Abstract
A large population of proliferative stem cells (neoblasts) is required for physiological tissue homeostasis and post-injury regeneration in planarians. Recent studies indicate that survival of a few neoblasts after sublethal irradiation results in the clonal expansion of the surviving stem cells and the eventual restoration of tissue homeostasis and regenerative capacity. Yet, the precise mechanisms regulating the population dynamics of neoblasts remain largely unknown. Here, we uncovered a central role for Epidermal Growth Factor (EGF) signaling during in vivo neoblast expansion mediated by Smed-egfr-3 (egfr-3) and its putative ligand Smed-neuregulin-7 (nrg-7). Furthermore, the EGFR-3 protein localizes asymmetrically on the cytoplasmic membrane of neoblasts and the ratio of asymmetric to symmetric cell divisions decreases significantly in egfr-3(RNAi) worms. Our results not only provide the first molecular evidence of asymmetric stem cell divisions in planarians, but also demonstrate that EGF signaling likely functions as an essential regulator of neoblast clonal expansion.
Keywords: extracellular signaling, epidermal growth factor, sublethal irradiation, neoblast regeneration, stem cell repopulation, regeneration
Graphical abstract
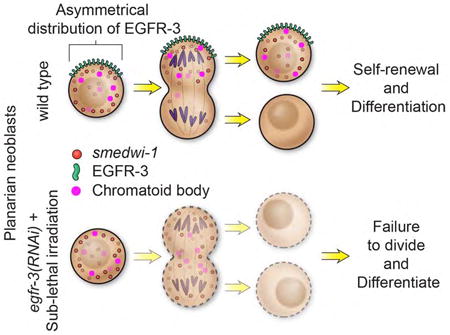
Stem cell repopulation can be achieved from even a single pluripotent stem cell in planarian flatworms. Lei et al. show that EGF signaling is required for stem cell repopulation in Schmidtea mediterranea, which is regulated through proper control of symmetric versus asymmetric cell division.
Introduction
The ability of planarians to restore their stem cells after substantial damage is in contrast to mammals, which lack a robust capacity to replenish a variety of stem cell pools after ablation by chemo- or radiotherapy (Biteau et al., 2011; Miyajima et al., 2014; Vermeulen and Snippert, 2014). Self-renewal and the production of differentiated progeny are key stem cell attributes required to sustain the function of many adult tissues (Hsu and Fuchs, 2012). Understanding how planarian stem cells regulate these processes during homeostasis and in response to injury has important implications for regenerative medicine and for developing effective cancer therapies.
Planarians harbor a remarkable capacity to regenerate complete animals from small tissue fragments (Morgan, 1900; Sánchez Alvarado and Newmark, 1998). This ability derives, in part, from an abundant population of adult stem cells collectively known as neoblasts (Baguñà et al., 1989; Reddien and Sánchez Alvarado, 2004). In asexually reproducing planarians, neoblasts are the only known proliferating cells (Newmark and Sánchez Alvarado, 2000). As such, it is possible to completely or partially ablate these cells by exposing animals to ionizing radiation (Bardeen and Baetjer, 1904; Guedelhoefer and Sánchez Alvarado, 2012; Reddien et al., 2005; Reddien and Sánchez Alvarado, 2004; Wolff and Dubois, 1948). Interestingly, these two experimental paradigms have helped demonstrate that the entire population of planarian stem cells can be fully reconstituted from a single pluripotent neoblast presently termed “cNeoblast” (Salvetti et al., 2009; Wagner et al., 2012; Wagner et al., 2011). Therefore, the robust stem cell regulation occurring in planarians combined with methodologies to effectively visualize neoblasts at whole organism resolution makes these animals a powerful in vivo model system to study the molecular mechanisms underpinning adult stem cell repopulation and the dynamics of stem cell populations under normal and aberrant conditions.
Extracellular signals usually associated with stem cell niches such as Epidermal Growth Factor (EGF), Fibroblast Growth Factor (FGF), Vascular Endothelial Growth Factor (VEGF), Wnt, Notch, and Transforming Growth Factor beta (TGF-β) are known to play prominent roles in regulating stem cell homeostasis and repopulation in vertebrate and invertebrate species (Hogan et al., 2014; Hsu and Fuchs, 2012; Kotton and Morrisey, 2014; Mendelson and Frenette, 2014; Shi and Garry, 2006). In many organisms, these extracellular signals affect cell proliferation by regulating both the frequency of cell division (Alberts et al., 2002) and/or the type of daughter cells produced by modulating whether symmetric or asymmetric cell divisions take place (Morrison and Kimble, 2006; Neumuller and Knoblich, 2009). However, the roles that developmental signaling pathways play in neoblast population dynamics remain unclear.
In planarians, it is known that Wnt/β-catenin and Hedgehog signaling are required for establishing anterior-posterior polarity during homeostasis and tissue regeneration (Adell et al., 2009; Gurley et al., 2008; Iglesias et al., 2008; Kobayashi et al., 2007; Petersen and Reddien, 2008, 2009; Reddien et al., 2007; Rink et al., 2009). TGF-β signaling is essential for maintenance and regeneration of dorsal-ventral and medial-lateral axes (Gaviño and Reddien, 2011; Molina et al., 2007; Reddien et al., 2007), as well as for sensing signals related to wound and/or missing tissues (Gaviño et al., 2013; Roberts-Galbraith and Newmark, 2013; Yazawa et al., 2009). FGF signaling is required for brain patterning (Cebrià et al., 2002) and tissue homeostasis (Wagner et al., 2012). EGF and Insulin signaling pathways are crucial for maintenance of the neoblast population during homeostasis and regeneration (Fraguas et al., 2011; Miller and Newmark, 2012). However, little is known about the potential functions these signaling pathways may play in regulating neoblast population dynamics, and no roles have been reported for any of these pathways in modulating neoblast repopulation after challenge by sublethal irradiation. For example, a previous RNAi screen aimed at identifying modulators of stem cell proliferation and differentiation did not include these pathways except for FGF (Wagner et al., 2012). Additionally, even though both symmetric and asymmetric cell divisions within the planarian stem cell pool have long been hypothesized to occur, no direct evidence demonstrating these two phenomena has yet been put forward (Coward, 1974; Reddien, 2013; Rink, 2013; Zhu et al., 2015).
In this study, we aimed to determine whether the EGF, FGF, Insulin, VEGF, TGF-β, Wnt/β-catenin, Hedgehog, and/or Notch signaling pathways play a role in regulating the proliferation dynamics of planarian stem cells. By taking advantage of dsRNA-mediated gene knockdown (Newmark et al., 2003; Reddien et al., 2005) and a colony expansion assay (Wagner et al., 2012), we performed a screen to test whether abrogation of signaling pathways would have an effect on neoblast expansion after sub-lethal irradiation. We found that EGFR-3 and a putative planarian EGF ligand, NEUREGULIN-7 are required for neoblast repopulation. In addition, a candidate approach and RNA-sequencing analysis revealed EGFR-3 downstream factors, including lkb1, ampk, and a DNA damage response gene rad54b to be are required for neoblast repopulation following sublethal irradiation. We also provide evidence for the existence of asymmetric cell division in planarian stem cells and show that asymmetric cell division and early progeny differentiation during neoblast repopulation is blocked in egfr-3(RNAi) worms. We propose that EGF signaling plays a central role in regulating asymmetric cell division and cell fate decision during neoblast repopulation.
Experimental Procedures
Planarian culture and irradiation treatment
Asexual Schmidtea mediterranea (strain CIW4) were maintained at 20°C as previously described (Newmark and Sánchez Alvarado, 2000). For all experiments, animals were starved for 7-14 days. A GammaCell 40 Exactor irradiator exposed animals to either 1,250 or 6,000 rads for sublethal and lethal irradiations, respectively.
Molecular cloning and RNAi feeding
cDNAs of all tested genes were cloned into a pPR-T4P vector as described previously (Gurley et al., 2008). RNAi food was prepared by adding 125 μL of liver paste (9 parts of liver to 1 part of water) into a bacterial pellet obtained from 50 mL overnight cultures. Unc22, a Caenorhabditis elegans gene without nucleotide sequence homology in planarians, was used as control RNAi. Animals in all RNAi experiments were fed 6 times with 3 days between feedings. 1,250 rads of irradiation or amputation was carried out 7 or 4 days after last feeding, respectively. Day 0 represents time of irradiation or amputation.
In situ hybridizations and antibody staining
In situ hybridizations were performed as previously described (King and Newmark, 2013; Pearson et al., 2009) with some modifications. Animals were fixed for 45 minutes in 4% formaldehyde, 0.5% Triton X-100, and 1 × PBS. To improve optical clarity after signal development, Sca/A2 (Hama et al., 2011) with 80% glycerol and 4 M urea was used for colorimetric whole mount in situ hybridization (WISH). 20% glycerol, 2.5% DABCO (Sigma-Aldrich, St. Louis, MO), and 4M urea Sca/A2 with was used for fluorescent WISH (FISH). Anti-phospho-Histone H3 (Ser10) (H3P) antibody (1:1,000, Abcam, Cambridge, UK; ab32107) and Alexa-conjugated goat anti-rabbit secondary antibodies (1:1,000, Abcam, Cambridge, UK; ab150086) were used to stain proliferating cells at the G2/M phase of the cell cycle.
Bromodeoxyuridine (BrdU) labeling
Animals were soaked in 25 mg/mL BrdU (Fluka, Sigma-Aldrich, St. Louis, MO) for 4 hours (van Wolfswinkel et al., 2014) and fixed at specified time points. In situ hybridization and BrdU antibody staining were performed as previously described (Thi-Kim Vu et al., 2015). BrdU was detected via a rat anti-BrdU antibody (1:1,000, Abcam, Cambridge, UK; ab6326).
TUNEL assay
Animals were fixed and stained with a TUNEL assay kit (Roche, Indianapolis, USA) as previously described (Pellettieri et al., 2010). TUNEL-stained specimens were imaged for quantification on a Perkin Elmer Ultraview spinning disk microscope.
Antibody generation
The anti-EGFR-3 antibody was generated by YenZym Antibodies, LLC. We used a peptide antigen derived from EGFR-3 amino acids 361-376 to immunize rabbits. Polyclonal antibodies were affinity purified with Hitrap NHS-activated HP column (GE Healthcare Life Sciences, Pittsburgh, USA).
Microscopy
Colorimetric WISH images were captured using a Zeiss SteREO Lumar stereoscope. Confocal images were captured using either a Zeiss LSM-700 or a Zeiss LSM-510. For quantification, tiled images of individual worms were acquired using a Perkin Elmer Ultraview spinning disk microscope, then stitched and spots counted as described previously (Adler et al., 2014).
Protein in vitro binding assay
Recombinant MYC-NRG-7-HIS was expressed in bacteria and recombinant FLAG-EGFR-3-HIS extracellular domain (residues 1 to 818 aa) was expressed in baculovirus. Proteins were affinity purified using standard methods with nickel affinity resin. To determine direct ligand/receptor interaction, NRG-7 and EGFR-3 were incubated along with Anti-c-Myc Agarose Affinity Gel (Sigma-Aldrich, St. Louis, MO) with 100ng/uL BSA in NETN buffer (250 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl pH 8.0, 0.5% NP-40, 10% glycerol, 1mM DTT). After 1 hour at 4°C, the affinity resin was washed 5 times with 15 volumes of NETN. Bound proteins were eluted with 2× urea-based Laemmli buffer (50mM Tris HCl pH 6.8, 1.6% SDS, 7% glycerol, 8M urea, 4% β-mercaptoethanol, 0.016% bromophenol blue). Samples were heated 10 minutes at 80°C and fractionated by SDS-PAGE for analysis.
Next generation RNAseq
Worms and sorted X1 cells were homogenized in TRIzol (Life Technologies, Grand Island, USA), and RNA isolated according to the manufacturer-supplied protocol. ∼100ng of RNA per sample were used for library generation using the Illumina TruSeq kit. Libraries were sequenced in 50bp single reads using an Illumina HiSeq 2500 sequencer. A set of 43,806 predicted S. mediterranea transcripts were used to analyze the RNAseq data (Robb et al., 2015). All sequencing data have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE84025.
Statistical analyses
Two-tailed Fisher's exact test was performed for statistical analyses of symmetric versus asymmetric cell divisions. All other statistical analyses were performed using Student's t test. p < 0.05 was considered a significant difference.
Results
egfr-3 is required for neoblast repopulation after sublethal irradiation
To assess the role of evolutionarily conserved extracellular signaling pathways in regulating stem cell repopulation, we first determined which of these candidate genes were expressed in neoblasts. Since neoblasts can be selectively eliminated within 24 hours following lethal irradiation (Reddien et al., 2005), we compared the expression of candidate genes in normal versus lethally irradiated planarians. We performed whole mount in situ hybridization (WISH) for the receptors of EGF (egfr-1, egfr-2, egfr-3), FGF (fgfr-1, fgfr-2, fgfr-3, fgfr-4), TGF-β (activinR-1, activinR-2), insulin (inr-1), VEGF (vegfr-1), and Hedgehog (ptc) pathways. We also assessed the expression of the transcriptional effectors of the canonical Wnt (β-catenin-1) and Notch (su(H)) pathways. After lethal irradiation (6,000 rads) and 1 day post irradiation (dpi), we detected a noticeable decrease in the expression levels of egfr-3, fgfr-1, fgfr-3, fgfr-4, and inr-1 (Figure S1A), suggesting expression of these genes in neoblasts. We then carried out double fluorescence in situ hybridization (FISH) with the neoblast-specific marker smedwi-1 to examine egfr-3, fgfr-1, fgfr-3, fgfr-4, and inr-1 co-expression. Indeed, the majority of the egfr-3+ (85%), fgfr-1+ (79%), fgfr-3+ (74%), fgfr-4+ (92%), and inr-1+ (100%) cells also expressed smedwi-1 (Figure S1B), consistent with previous reports (Fraguas et al., 2011; Miller and Newmark, 2012; Ogawa et al., 2002; Wagner et al., 2012). This suggests a functional role for the EGF, FGF and Insulin pathways in neoblasts.
Next, we confirmed previous observations on the dynamics of neoblast repopulation following sublethal (1,250 rads) irradiation (Wagner et al., 2012; Wagner et al., 2011), and defined a quantitative neoblast depletion and recovery assay that scores smedwi-1+ cells at 0, 1, 2, 3, 5, 7, 9, 11, and 14 dpi. We noted the total number of neoblasts decreased dramatically within the first 3 days (Figure 1A), followed by a significant increase at 7 dpi in control animals (Figure 1A and 1B, 14 dpi). To determine whether the candidate EGF, FGF, or Insulin receptors are required for neoblast repopulation, we reasoned that the number of neoblasts in both control and RNAi-treated animals should be comparable at 7 dpi, but significantly lower in RNAi-treated animals at 14 dpi (Figure 1C). Using this assay, we quantified the density of neoblasts in control versus RNAi-treated planarians at 7 and 14 dpi (Figure 1C).
Figure 1. egfr-3 is required for neoblast repopulation.
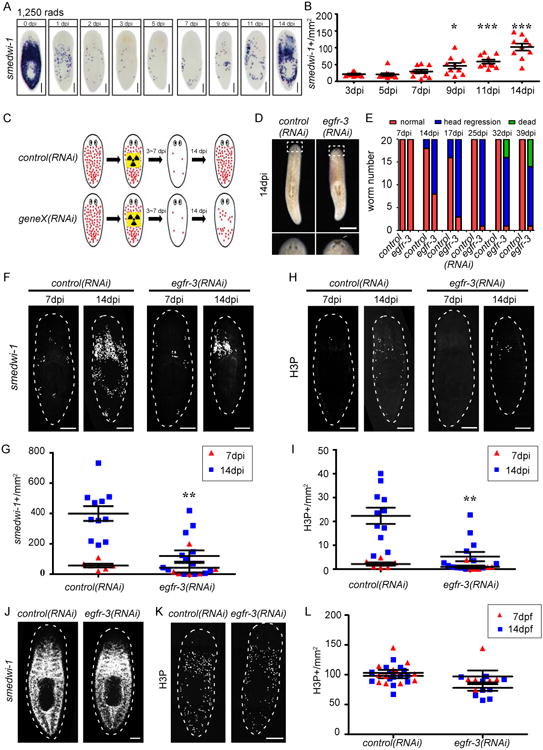
(A) smedwi-1 WISH of uninjured wild type planarians exposed to sublethal irradiation (1,250 rads) at indicated days post irradiation (dpi). (B) Quantification of smedwi-1+ cells/mm2 following sublethal irradiation in wild type planarians. Each dot= one animal. Error bars= S.E.M. *, p<0.05; ***, p<0.001 compared to 3 dpi. (C) Experimental strategy to identify gene(s) (geneX) required for neoblast repopulation following sublethal irradiation. Red dots represent smedwi-1+ neoblasts. (D) Live egfr-3(RNAi) planarians display head regression at 14 dpi following 1,250 rad irradiation. (E) Histogram of number of control(RNAi) vs. egfr-3(RNAi) planarians with head regression or death at indicated times. (F) control(RNAi) and egfr-3(RNAi) planarians at 7 and 14 dpi stained for smedwi-1 FISH. (G) Quantification of smedwi-1+ cells/mm2 in egfr-3(RNAi) planarians. **, p<0.01 vs. control(RNAi). (H) control(RNAi) and egfr-3(RNAi) planarians at 7 dpi and 14 dpi immunostained with anti-H3P antibody. (I) Quantification of H3P+ cells/mm2 in sublethally-irradiated egfr-3(RNAi) planarians. (J) Unirradiated control(RNAi) and egfr-3(RNAi) planarians stained for smedwi-1 FISH. (K) Unirradiated control(RNAi) and egfr-3(RNAi) planarians immunostained with anti-H3P antibody. (L) Quantification of H3P+ cells/mm2 in unirradiated egfr-3(RNAi) planarians compared to controls. Scale bars= 200 μm (A, D), 500 μm (F, H, J, K). See also Figures S1-S2.
Neither individual, nor combination knockdown of fgfr-1, fgfr-3, or fgfr-4 caused defects in neoblast repopulation after sublethal irradiation (Figure S1C-S1F). On the other hand, inr-1(RNAi) ablated the neoblast population in both unirradiated and sublethally irradiated planarians, and caused locomotion defects (Figure S2A-S2E, and S2K-S2M). When assayed by smedwi-1 FISH and immunostaining for mitotic cells using an anti-Phospho-Histone H3 (Ser10) antibody (anti-H3P), RNAi of insulin-like peptide 1 (ilp-1) (Figure S2F) (Miller and Newmark, 2012) yielded similar numbers of smedwi-1+ and H3P+ cells in unirradiated planarians compared to controls (Figure S2K-S2M), while in sublethal irradiations it resulted in slower neoblast repopulation (Figure S2G-S2J). In marked contrast, egfr-3(RNAi) animals displayed a density of neoblasts and mitotic cells similar to controls at 7 dpi, but significantly lower at 14 dpi (Figure 1F-1I). egfr-3(RNAi) planarians also showed head regression after 14 dpi and ultimately lysed, while control(RNAi) animals recovered (Figure 1D and 1E). Consistent with a previous report (Fraguas et al., 2011), no significant change in smedwi-1+ and H3P+ cell density was observed in unirradiated egfr-3(RNAi) planarians (Figure 1J-1L). Flow-cytometry analyses further confirmed that neoblasts did not decrease in numbers before sublethal irradiation in egfr-3(RNAi) planarians (Figure S1G).
Altogether, these data indicate that FGF signaling may not be required for neoblast repopulation. In contrast, insulin signaling facilitates both homeostatic neoblast population maintenance and expansion after irradiation damage, and additional insulin-like peptides may exist and function redundantly with ilp-1. Only egfr-3(RNAi) specifically prevented neoblast expansion without affecting homeostasis. We conclude that EGF signaling via egfr-3 is required for the expansion of neoblasts when their numbers are diminished by sublethal irradiation and that this role is distinct from that of other signaling molecules.
egfr-3 is required for cell proliferation during neoblast repopulation
Defects in neoblast repopulation can be caused by either slower proliferation or increased cell death. To distinguish between these two possibilities, we carried out BrdU labeling and TUNEL assays, respectively. Cells were labeled with BrdU for 4 hours in control and egfr-3(RNAi) at 7 dpi (Figure 2A). In controls, the number of BrdU+ cells increased gradually and doubled approximately 24 hours after labeling (Figure 2B and 2D). However, no obvious increase was observed in egfr-3(RNAi) (Figure 2C and 2D). Furthermore, smedwi-1+ cells increased in control(RNAi), but not in egfr-3(RNAi) (Figure 2E-2G). These results suggest that egfr-3 promotes cell proliferation during neoblast repopulation.
Figure 2. egfr-3(RNAi) affects cell proliferation but not cell apoptosis during neoblast repopulation.
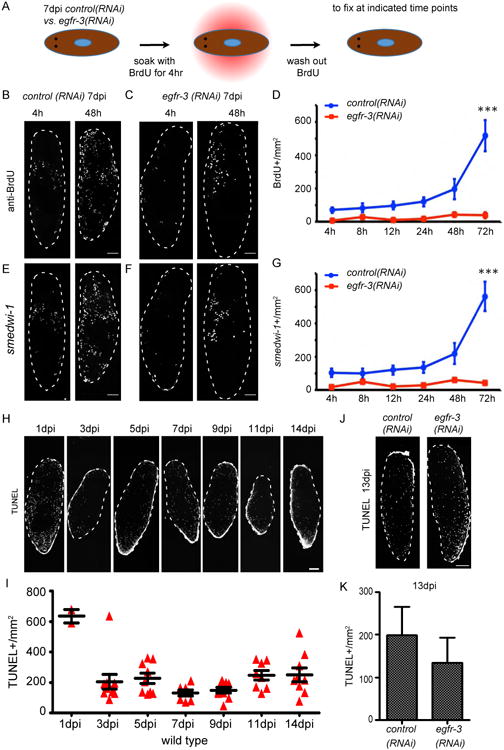
(A) Strategy for comparing cell proliferation in control(RNAi) and egfr-3(RNAi) animals at 7 dpi via BrdU pulse-chasE. (B and C) BrdU+ cells in control(RNAi) (B) and egfr-3(RNAi) (C) planarians at 4 and 48 hours after BrdU soaking. h, hour. Scale bars= 200 μm. (D) Quantification of BrdU+ cells/mm2 in egfr-3(RNAi) planarians vs. controls at 72 hours post chase. ***, p<0.001. (E and F) FISH for smedwi-1 shows neoblasts in control(RNAi) and egfr-3(RNAi) planarians at 4 and 48 hours after BrdU labeling. Scale bars= 200 μm. (G) Quantification of smedwi-1+ cells/mm2 in egfr-3(RNAi) planarians compared to controls at 72 hours post chase. ***, p<0.001. (H) TUNEL-positive-nuclei of cells in wild type planarians at indicated time points after irradiation with 1,250 rads. Scale bar= 200 μm. (I) Quantification of TUNEL+ nuclei/mm2 at indicated time points after 1,250 rads irradiation. (J) TUNEL staining of apoptotic cells in control(RNAi) and egfr-3(RNAi) planarians at 13 dpi. Scale bars= 500 μm (K) Quantification of TUNEL+ nuceli/mm2 in control(RNAi) and egfr-3(RNAi) planarians at 13 dpi. See also Figure S3.
To examine apoptosis, TUNEL assays were carried out in control(RNAi) and egfr-3(RNAi) animals after sublethal irradiation. In controls, there were a great number of apoptotic cells at 1 dpi, which was consistent with the observed neoblast depletion during this time (Figure 2H and 2I). After 3 dpi, the density of TUNEL+ cells leveled off in controls (Figure 2H and 2I). In egfr-3(RNAi) animals, the density of TUNEL+ cells was similar to controls at 13 dpi (Figure 2J and 2K) suggesting that egfr-3 may not drastically affect the amount of apoptosis during neoblast repopulation. Therefore, the egfr-3(RNAi) repopulation defects appear likely to be due to a failure in neoblast proliferation following sublethal irradiation.
Previous studies have shown that amputation induces neoblast proliferation (Newmark and Sánchez Alvarado, 2000; Wenemoser and Reddien, 2010). We therefore tested whether egfr-3 also functions in amputation-induced cell proliferation. Control and egfr-3(RNAi) planarians were amputated and immunostained with anti-H3P antibody staining to compare the number of proliferating cells. Consistent with a previous report (Fraguas et al., 2011), egfr-3(RNAi) animals displayed impaired regeneration at 3 days post amputation (dpa) (Figure S3A). At 6 hours post amputation (hpa), the first peak of amputation-induced hyperproliferation was slightly lower in egfr-3(RNAi) compared to controls (Figure S3B and S3C). In contrast, the second peak of hyperproliferation at 48 hpa was dramatically decreased in egfr-3(RNAi) animals (Figure S3B and S3C). These results suggest that egfr-3 is required for both phases of regeneration-associated hyperproliferation.
The putative EGF ligand neuregulin-7 also regulates neoblast repopulation
Since egfr-3 is a signaling molecule that appears to play a significant role in neoblast repopulation, we sought to identify its upstream ligands. Twelve putative EGF ligands were predicted to exist in the planarian genome based upon EGF domain and homology sequence comparisons (Figure S4A and S4B). Their expression patterns were examined by WISH in unirradiated and lethally irradiated planarians (6,000 rads, 1 dpi) (Figure 3A). None of the tested ligand-encoding genes were expressed in irradiation sensitive cells, suggesting they do not show enriched expression in neoblasts. To examine their function in neoblast repopulation, we screened the putative ligands in the sublethal irradiation assay following RNAi feeding (see Figure 1C for experimental strategy) and found that only neuregulin-7 (nrg-7) RNAi robustly impaired neoblast repopulation (Figure 3B-3E, S4C-S4I). Like egfr-3(RNAi), nrg-7(RNAi) did not affect the homeostasis of the neoblast population compared to controls (Figure 3F-3H), nor did it affect regeneration. As it was the case in egfr3(RNAi) animals, flow-cytometry analyses further confirmed that neoblasts did not decrease in number before sublethal irradiation in nrg-7(RNAi) planarians (Figure S4J) nor was recovery observed. To determine the cell types in which nrg-7 is expressed, we performed nrg-7 FISH with smedwi-1, epidermal progeny markers (early and late progeny prog-1 and agat), and differentiated cell markers (muscle: collagen; central nerve system: PC2). We found that nrg-7 was expressed in prog-1+ and PC2+ cells (Figure S4K). To test whether this molecule could interact with EGFR-3, we performed in vitro binding assays using purified recombinant EGFR-3 and NRG-7. Our analyses showed a direct interaction of NRG-7 and EGFR-3 in vitro (Figure 3I). Together, these data suggest that NRG-7 is a likely ligand of EGFR-3 required during neoblast repopulation.
Figure 3. nrg-7 is required for neoblast repopulation after sublethal irradiation.
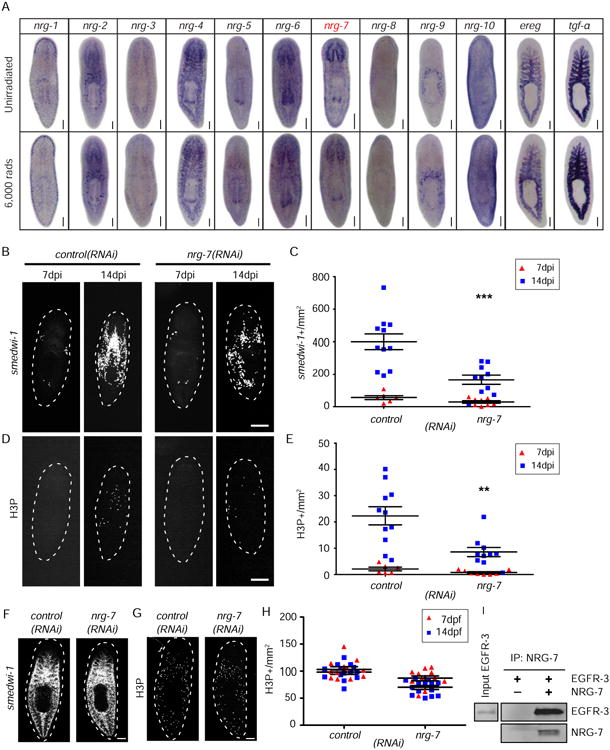
(A) WISH of putative egf ligands in unirradiated and 6,000 rads irradiated (1 dpi) planarians. (B) FISH for smedwi-1 in control(RNAi) and nrg-7(RNAi) planarians at 7 and 14 dpi. (C) Quantification of smedwi-1+ cells/mm2 in nrg-7(RNAi) animals. *, p<0.05. (D) Immunostaining with anti-H3P antibody in control(RNAi) and nrg-7(RNAi) planarians at 7 and 14 dpi. (E) Quantification of H3P+ cells/mm2 in nrg-7(RNAi) planarians. (F) FISH for smedwi-1 in control(RNAi) and nrg-7(RNAi) unirradiated planarians at 14 days post feeding. (G) Immunostaining with anti-H3P antibody in control(RNAi) and nrg-7(RNAi) unirradiated planarians. (H) Quantification of H3P+ cells/mm2 in nrg-7(RNAi) compared to controls. (I) Purified recombinant MYC-NRG-7-HIS (NRG-7) and the extracellular domain of FLAG-EGFR-3-HIS (EGFR-3) (residues 1 to 818 aa) were incubated together and NRG-7 was captured using anti-MYC affinity resin. EGFR-3 and NRG-7 are detected only in the experimental reaction containing NRG-7. Scale bars= 200 μm (A, B, D), 500 μm (F, G). See also Figure S4.
lkb1 and ampk function downstream of egfr-3 during neoblast repopulation
To better understand how egfr-3 regulates neoblast repopulation, we tested the function of downstream, conserved EGF pathway components erk, pi3k, stat, lkb1, and ampk. First, WISH was performed to examine their respective expression patterns in wild type and lethally irradiated animals (6,000 rads, 1 dpi). While none of these genes displayed obvious neoblast expression patterns, erk-1, lkb1, and ampk were noticeably decreased after 6,000 rads (Figure 4A, S5A, and S5B). Double FISH with smedwi-1 confirmed that erk-1, lkb1, and ampk were expressed in neoblasts (Figure 4B and S5C). To determine if any of these downstream effectors were required for neoblast repopulation, RNAi-treated animals were subjected to sublethal irradiation (1,250 rads). erk-1(RNAi) planarians displayed normal neoblast repopulation (Figure S5D-S5G). However, lkb1(RNAi) and ampk(RNAi) showed disrupted neoblast repopulation (Figure 4C-4F). Similar to egfr-3(RNAi), unirradiated lkb1(RNAi) and ampk(RNAi) planarians maintained normal levels of neoblasts and proliferating cells (Figure 4G-4I). These results suggest that egfr-3 regulates neoblast repopulation through the lkb1-ampk branch of the EGF signaling pathway; however, future biochemical work assessing pathway activation is still needed.
Figure 4. LKB1-AMPK signaling is required for neoblast repopulation.
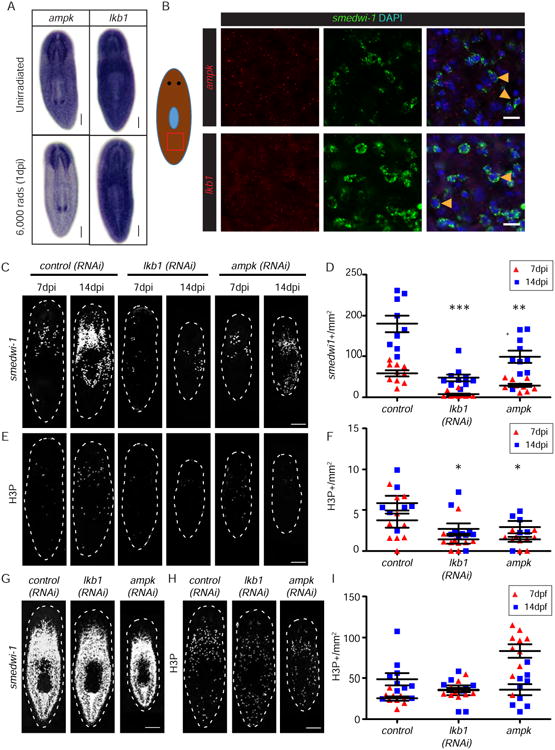
(A) WISH for ampk, and lkb1 in unirradiated and 6,000 rads irradiated planarians. (B) FISH for ampk and lkb1 (red) in smedwi-1+ neoblasts (green). (C) FISH for smedwi-1 in control(RNAi), ampk(RNAi), and lkb1(RNAi) animals at 7 and 14 dpi. (D) Quantification of smedwi-1+ cells/mm2 in ampk(RNAi) and lkb1(RNAi) planarians. *, p<0.05. (E) Immunostaining with anti-H3P antibody in control(RNAi), ampk(RNAi), and lkb1 (RNAi) planarians at 7 and 14 dpi. (F) Quantification of H3P+ cells/mm2 in ampk(RNAi) and lkb1(RNAi) planarians. (G) FISH for smedwi-1 in control(RNAi), ampk(RNAi), and lkb1(RNAi) unirradiated animals. (H) Immunostaining with anti-H3P antibody in control(RNAi), ampk(RNAi), and lkb1(RNAi) unirradiated animals. (I) Quantification of H3P+ cells/mm2 in ampk(RNAi) and lkb1(RNAi) compared to controls. Scale bars= 200 μm. See also Figure S5.
rad54b is likely an effector of egfr-3 signaling in neoblast repopulation
To further address how egfr-3 may regulate changes in gene expression during neoblast repopulation, we used an RNAseq approach to identify differentially expressed genes in control versus egfr-3(RNAi) planarians in whole animals during neoblast repopulation and in isolated neoblasts (X1 cells) from unirradiated animals (Figure 5A). During neoblast repopulation at 8 dpi and 10 dpi, 418 genes were downregulated in egfr-3(RNAi) animals compared to control planarians. Of these, 35 genes were also downregulated in X1 neoblasts isolated from egfr-3(RNAi) animals compared to X1s from controls (Figure 5B, Table S1, p value<0.05). To identify genes that may function specifically during neoblast repopulation, we focused on genes functioning in DNA damage response and transcription regulation. GO analysis was performed in the remaining 383 genes downregulated post sublethal irradiation (Figure 5B and S6). 48 genes were identified based on the terms of transcription factor complex or DNA binding that may function in DNA damage response and gene transcription regulation (Table S2). Because the decreased levels identified in RNAseq may be caused by fewer number of neoblasts in egfr-3(RNAi), WISH was carried out to compare the gene expression in neoblasts (Figure 5C). Candidate genes were cloned and WISH was performed to determine whether they were expressed in neoblasts. In the end, 27 genes were verified to be expressed in neoblasts (Figure 5D). Furthermore, FISH was performed to determine whether gene expression was affected in egfr-3(RNAi) compared to control(RNAi) animals. The data revealed that a DNA damage repair gene rad54b (Hiramoto et al., 1999) was significantly down-regulated in egfr-3(RNAi) compared to control(RNAi) animals (Figure 5E and 5F). Knockdown of rad54b impaired the dynamics of neoblast repopulation without affecting neoblast homeostasis (Figure 6). These results suggest that egfr-3 regulates the expression of rad54b during neoblast repopulation and functionally associates aspects of DNA repair with EGF signaling. Because EGFR and lkb1-ampk have been found associated with DNA damage repair in other systems (Chen and Nirodi, 2007; Sanli et al., 2014; Mahajan and Mahajan, 2015), the regulatory role of EGFR-3 in DNA damage is likely to be conserved in planarians in maintaining genome stability and ushering neoblast repopulation. Recently, another essential DNA damage repair gene rad51 in planarian was characterized to be required for DNA integrity, cell proliferation, and cell apoptosis during cell turnover (Peiris et al., 2016). Neoblasts could not recover in rad51(RNAi) animals after sublethal irradiation (Peiris et al., 2016), which is consistent with our study of rad54b, a rad51 functional partner. This suggest that DNA damage response and repair pathways are conserved in planarians, and future studies will explore the mechanism by which planarians maintain their genome stability through incessant cell proliferation.
Figure 5. RNAseq analysis of egfr-3(RNAi) in neoblast repopulation.
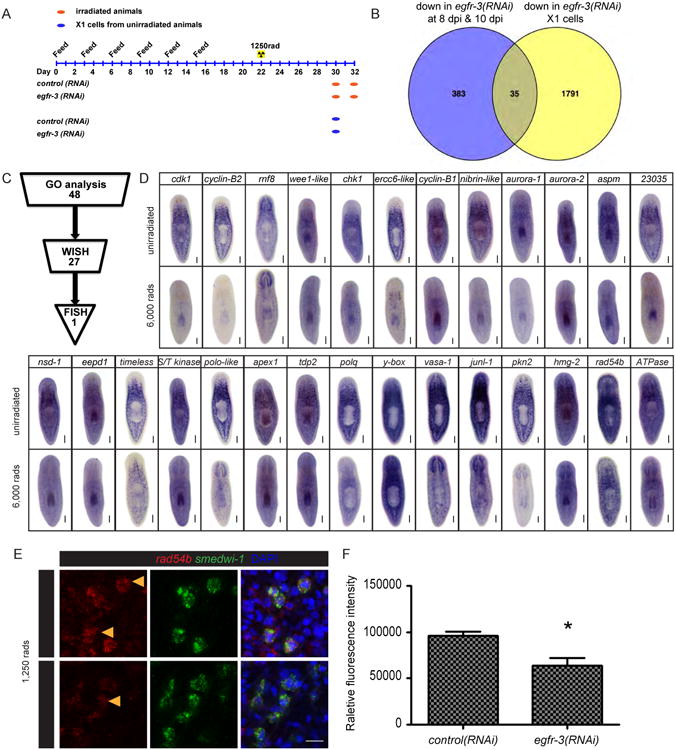
(A) Feeding schedule and sample collection of control(RNAi) and egfr-3(RNAi) planarians for RNAseq. (B) Venn diagram showing differentially expressed genes in egfr-3(RNAi) compared to control(RNAi) planarians at 8 dpi and 10 dpi, and in X1 cells of unirradiated animals. (C) Procedure for identifying the egfr-3 regulated gene including GO analysis, WISH, and FISH. (D) WISH of candidate genes in unirradiated and 6,000 rads irradiated planarians. Scale bars= 200 μm. (E) FISH for rad54b (red) in smedwi-1+ neoblasts (green). Scale bars= 20 μm. (F) Histogram showing rad54b fluorescence intensity in control(RNAi) and egfr-3(RNAi) planarians. See also Figures S6, Table S1, and Table S2.
Figure 6. rad54b are required for neoblast repopulation.
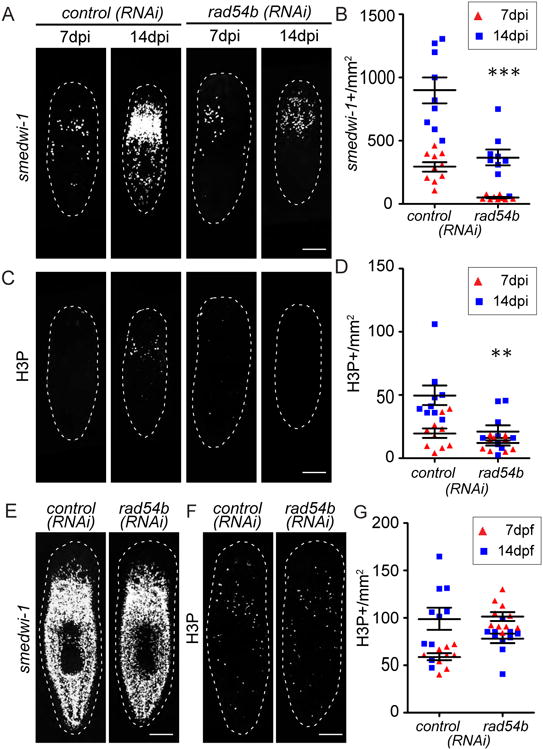
(A) FISH for smedwi-1 in control(RNAi) and rad54b(RNAi) planarians at 7 and 14 dpi. (B) Quantification of smedwi-1+ cells/mm2 in rad54b(RNAi) planarians. ***, p<0.001. (C) Immunostaining with anti-H3P antibody in control(RNAi) and rad54b(RNAi) planarians at 7 and 14 dpi. (D) Quantification of H3P+ cells/mm2 in rad54b(RNAi) planarians. **, p<0.01. (E) FISH for smedwi-1 in control(RNAi) and rad54b(RNAi) unirradiated animals. (F) Immunostaining with anti-H3P antibody in control(RNAi) and rad54b(RNAi) unirradiated animals. (G) Quantification of H3P+ cells/mm2 in rad54b(RNAi) compared to controls. Scale bar, 200 μm.
egfr-3 regulates asymmetric cell division in neoblast repopulation
To understand how egfr-3 regulated neoblast repopulation after sublethal irradiation, we sought to determine the subcellular localization of EGFR-3 protein in neoblasts. cDNA of egfr-3 was cloned from Schmidtea mediterranea and co-transfected along with EGFP-H1b, a nuclear marker, or Frizzled4-GFP, a cytoplasmic membrane marker, in 293T cells. Immunofluorescent staining showed clear cytoplasmic membrane localization of EGFR-3, consistent with its SMART structure prediction (Figure S7A-S7C). Next, an anti-EGFR-3 antibody was raised to determine the subcellular localization of EGFR-3 in vivo. Western blot of lysate from egfr-3 transfected cells and egfr-3(RNAi) planarians verified the specificity of the antibody (Figure S7D-S7I). Together with smedwi-1 in situ hybridization, it was clear that EGFR-3 was mainly expressed on the cytoplasmic membrane of neoblasts in control(RNAi) planarians (Figure 7A). As expected, EGFR-3 could not be detected in egfr-3(RNAi) neoblasts (Figure 7A). These results confirm that Smed-egfr-3 codes for a cytoplasmic membrane protein in planarian neoblasts.
Figure 7. Knockdown of egfr-3 reduces asymmetric cell division during neoblast repopulation after sublethal irradiation.
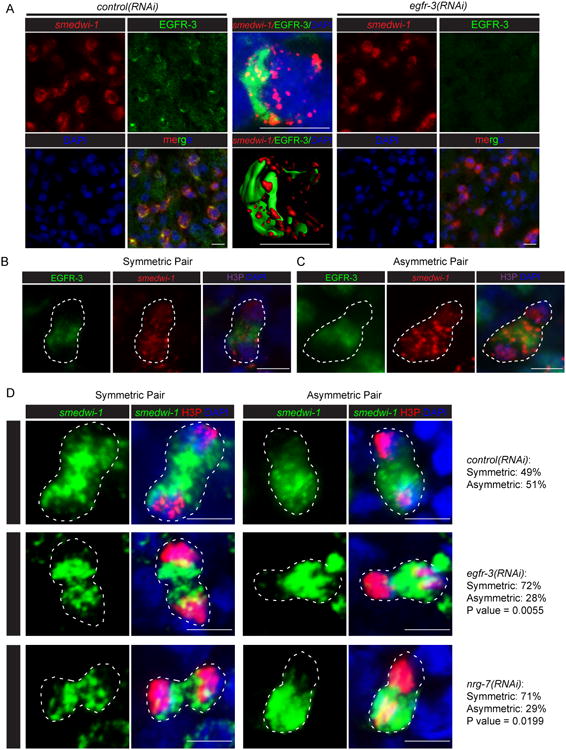
(A) Immunostaining with an antibody against EGFR-3 in control(RNAi) planarians. Egfr-3 knockdown eliminates the staining. (B) Representative images showing EGFR-3 and smedwi-1 distribution in symmetric cell division. (C) Representative images showing EGFR-3 and smedwi-1 distribution in asymmetric cell division. (D) Representative images and numbers of asymmetric vs. symmetric cell divisions in control(RNAi), egfr-3(RNAi), and nrg-7(RNAi) planarians at 14 dpi. Scale bar= 10 μm. p value calculated for egfr-3(RNAi) vs. control(RNAi) and nrg-7(RNAi) vs. control(RNAi), respectively, using Two tailed Fisher's exact test. See also Figures S7.
Further analysis of EGFR-3 subcellular localization uncovered an asymmetric distribution of this protein on the cytoplasmic membrane of neoblasts (Figure 7A). We hypothesized that egfr-3 likely regulates neoblast repopulation by controlling asymmetric versus symmetric cell division. To test this idea, we first examined the distribution of EGFR-3 in dividing cells and found that EGFR-3 distribution was associated with symmetric/asymmetric distribution of of smedwi-1 transcripts (Figure 7B and 7C). We next examined anti-H3P+ dividing cells at mitotic anaphase and telophase during homeostasis and neoblast repopulation and analyzed the symmetric and asymmetric cell division based on the distribution of smedwi-1 transcripts (Figure S7J-S7L). Both symmetric and asymmetric cell divisions were found in unirradiated and sublethally irradiated control(RNAi) animals (Figure 7D and S7M). To our surprise, asymmetric but not symmetric cell divisions were markedly decreased in sublethally irradiated egfr-3(RNAi) and nrg-7(RNAi) animals (Figure 7D, p value= 0.0055 and 0.0199, respectively), suggesting that egfr-3 promotes asymmetric cell division during neoblast repopulation. The direct consequence of symmetric and asymmetric cell division in neoblasts remains to be further characterized. However, a neoblast-specific organelle known as the chromatoid body, also displays symmetric or asymmetric distribution in dividing cell pairs (Figure 8A), which is consistent with smedwi-1 distribution. Asymmetric cell divisions shown with chromatoid bodies were also markedly decreased in sublethally irradiated egfr-3(RNAi) animals (Figure 8A, p value= 0.0449), suggesting disturbed cell fate regulation in egfr-3(RNAi) animals. A previous study reported that egfr-3 functions in cell differentiation during head regeneration after amputation (Fraguas et al., 2011). Therefore, we suspected that egfr-3 may also function in cell differentiation during neoblast repopulation. The density of prog-1+ cells was measured and the ratio of [prog-1+]/[smedwi-1+] at 14dpi was significantly lower in egfr-3(RNAi) compared to controls, suggesting a defect in prog-1+ cell differentiation in egfr-3(RNAi) planarians (Figure S8). The above results suggest that egfr-3 might regulate neoblast repopulation and their subsequent differentiation by controlling asymmetric cell division.
Figure 8. Asymmetric distribution of chromatoid body in dividing cells is impaired in egfr-3(RNAi) animals during neoblast repopulation after sublethal irradiation.
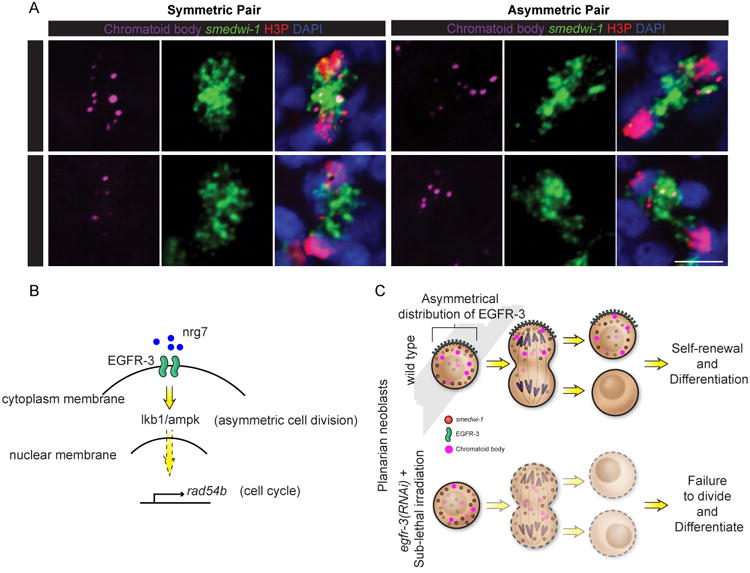
(A) Representative images and numbers of asymmetric vs. symmetric cell divisions observed in control(RNAi) and egfr-3(RNAi) planarians at 14dpi based upon distribution of chromatoid bodies (anti-Y-12+ antibody, purple), and smedwi-1 RNA (green). p value= 0.0449. Dividing cells are labeled with anti-H3P antibody (red), and nuclei are stained with DAPI (blue). Scale bar= 10 μm. (B) EGF signaling regulation of asymmetric cell division and cell cycle through nrg-7, egfr-3, lkb1/ampk, and rad54b during neoblast repopulation after sublethal irradiation. (C) Proposed model of EGF signaling in asymmetric cell division in planaria. See also Figures S8.
We need to emphasize that even though the ratio of asymmetric vs. symmetric cell division changed, the total number of mitotic events during neoblast repopulation also decreases in egfr-3(RNAi) animals (see Figure 1H and 1I). Although we can only speculate as to why the cells fail to expand under a symmetric cell division strategy, the most likely explanation may be that the necessary post-mitotic progeny fail to form and signals likely supporting the neoblasts proper made by these cells are absent. We have seen similar outcomes already for two different RNAi treatments in the past: smedwi-2 (Reddien et al., Science 2005) and p53 (Pearson et al, Development, 2009). Under both of these conditions, the neoblasts are initially not affected in their capacity to proliferate, but eventually as the neoblasts fail to generate normal post-mitotic progeny, the neoblast population becomes exhausted and disappears. As such, it is likely that a hypothetical differentiation factor may be normally distributed to the opposite poles leading to asymmetric cell division but that in egfr3(RNAi) treated animals such asymmetry does not occur. This idea is supported by the fact that we observed the distribution of the cytoplasmic chromatoid bodies was affected in egfr-3(RNAi) animals (Figure 8A). However, we cannot exclude the possibility that egfr-3 regulates both cell cycle and asymmetric cell division independently through lkb1/ampk and rad54b or other unidentified factors. Taken together, our studies have led us to propose a model in which egfr-3 regulates cell proliferation and asymmetric cell division during neoblast repopulation (Figure 8B and 8C).
Discussion
Planarians are a powerful system to study signaling regulation of stem cell expansion in vivo
How does the organism instruct stem cells to produce the correct types and numbers of cells to maintain normal tissue homeostasis? How do stem cells regulate the balance between self-renewal and the production of postmitotic division progeny? How do stem cells control their expansion during regeneration? The relatively large abundance of a constantly cycling population of pluripotent stem cells in planarians (Newmark and Sánchez Alvarado, 2000; Wagner et al., 2011) provides a robust and experimentally accessible in vivo setting in which to examine fundamental problems of proliferative dynamics.
In humans, resident stem cells have been shown to facilitate homeostasis in many adult tissues (Biteau et al., 2011). However, their population dynamics and contributions to regeneration after tissue damage remain challenging to study (van Es et al., 2012; Vaughan et al., 2015; Zuo et al., 2015). Animal model systems help overcome the difficulties by enabling the molecular and genetic dissection of stem cell biology. In particular, the abundance, experimental accessibility, and remarkable pluripotency of planarian neoblasts make this animal a tantalizing in vivo system for investigating fundamental questions of stem cell biology (Newmark and Sánchez Alvarado, 2000; Wagner et al., 2011). How, for instance, does the organism instruct stem cells to produce the correct types and numbers of cells during tissue homeostasis and regeneration? How do stem cells regulate the balance between self-renewal and the production of postmitotic division progeny? How do planarians achieve all these without developing stem-cell associated diseases observed in other animals, such as cancer?
By using sublethal irradiation to induce clonal expansion of planarian neoblasts (Wagner et al., 2011), we tested the role of conserved signaling pathways in regulating this process. We identified a specific role for the EGF pathway in modulating the expansion of neoblasts, further demonstrating the utility of planarians for studying the behavior and functions of adult stem cells in vivo.
The EGF signaling pathway regulates planarian stem cell expansion
In S. mediterranea, four EGFRs have been identified: egfr-1, egfr-2, egfr-3 and egfr-5. All these genes are expressed in differentiated tissues, but egfr-3 distinguishes itself by also being expressed in neoblasts (Fraguas et al., 2011; Rink et al., 2011). Interestingly, loss of egfr-3 expression affects cephalic regeneration and has been suggested to play a role in modulating neoblast functions (Fraguas et al., 2011). However, a direct role for egfr-3 in regulating neoblast proliferation had not been demonstrated, and RNAi screens aimed at identifying modulators of stem cell proliferation did not include this gene likely because its expression is not confined exclusively to neoblasts (Wagner et al., 2012). Because our study used a candidate approach to identify extracellular signals capable of modulating neoblast repopulation, egfr-3 was included as a candidate in our RNAi screen. Given that under normal, homeostatic conditions RNAi of either egfr-3 or its putative ligand nrg-7 did not result in detectable defects, our findings suggest that EGFR-3/NGR-7 are not required for the maintenance of neoblasts, but may instead have a specialized role in the expansion and/or self-renewal of stem cells.
Multiple cytokine sources regulate tissue homeostasis, regeneration, and neoblast repopulation
In most studied species, stem cell activities are greatly influenced by diverse cytokines or niche signals (Forbes and Rosenthal, 2014). Our finding that loss of egfr-3 and nrg-7 via RNAi resulted in a repopulation-specific neoblast deficiency indicates that a high degree of specialization of signaling molecules and their pathways must exist in planarians. Planarian muscle cells have recently been shown to serve as signaling centers capable of generating multiple factors for tissue regeneration and turnover (Witchley et al., 2013), and neuropeptides (Baguñà et al., 1989; Saló and Baguñà, 1986) and hedgehog (Hh) expressed in the nervous system (Rink et al., 2009; Yazawa et al., 2009) have been shown to affect neoblast division rates. Cells undergoing differentiation have also been reported to affect the maintenance and dynamics of the neoblast population (Tu et al., 2015; Zhu et al., 2015). Our finding that nrg-7 is expressed in prog-1+ cells and PC2+ neuron cells (Figure S4K), provides additional cytokine sources regulating neoblasts in planarians.
EGFR-3 signaling is required for asymmetric vs. symmetric cell division in neoblasts during neoblast repopulation
Adult stem cells rely on asymmetric cell division for both self-renewal and generation of differentiated progeny, a process that allows organisms to precisely maintain stem cell numbers and regulate somatic cell turnover (Inaba and Yamashita, 2012). While asymmetric cell division has long been proposed to exist in planarians (Reddien, 2013; Rink, 2013), there has been no direct evidence supporting its existence until now. As in tissue homeostasis, precise regulation of stem cell self-renewal and differentiation is also required for regeneration (Tanaka and Reddien, 2011). Hence, molecular heterogeneity of the stem cell population in planarians is to be expected, and has begun to be uncovered in recent studies (Hayashi et al., 2006; Lapan and Reddien, 2011; Pearson and Sánchez Alvarado, 2010; Tu et al., 2015; van Wolfswinkel et al., 2014). Interestingly, the complex molecular heterogeneity of planarian stem cells can be restored by the proliferation of the very few neoblasts survived after sublethal irradiation, and with some frequency by single transplanted neoblasts (Wagner et al., 2012; Wagner et al., 2011). These two experimental approaches have led to postulate the existence of a clonogenic or cNeoblast (Wagner et al., 2011). Furthermore, during the colony expansion following sublethal irradiation, a spliced leader RNA SL3 was found expressed in all neoblasts of small colonies (<6 neoblasts), but only expressed in subsets of neoblasts of bigger colonies (>8 neoblasts) (Rossi et al., 2013), supporting the idea that developmental mechanisms likely drive the molecular heterogeneity of the neoblast population (Reddien, 2013). It is not clear, however, whether asymmetric cell division functions in the generation of specialized neoblasts from cNeoblasts.
Here we show that smedwi-1 mRNA and chromatoid bodies display both symmetric and asymmetric distribution in dividing neoblasts during homeostasis (Figure 7D, S7M, and 8A). egfr-3(RNAi) did not noticeably alter the ∼1:1 ratio of symmetric to asymmetric cell divisions in unirradiated animals (Figure S7M). However, after sublethal irradiation asymmetric cell divisions were impaired in egfr-3(RNAi) animals (Figure 7D), drastically affecting neoblast repopulation and the production of a specialized lineage (Figure 1F-1I, and S8). This is the first functional demonstration for a role of asymmetric cell division in stem cell function in planarians. Altogether, we propose a model in which the observed asymmetric distribution of EGFR-3 (Figure 7A and 7B) is required for the proliferation and differentiation of neoblasts surviving from sub-lethal irradiation into lineage-specified neoblasts (Figure 8C). In the future, experiments aimed at determining the requirement of asymmetric cell division in the ontogeny of the observed neoblast heterogeneity in planarians will help shed light on how this remarkable population of adult stem cells regulates its dynamics.
The role of EGF signaling in regulating asymmetric stem cell division and repopulation is likely evolutionarily ancient
Asymmetric distribution of EGFR in stem cells was first described in mice, in which adult neural stem cells are regulated by the asymmetric distribution of EGFR (Sun et al., 2005). In the mouse brain, Egfr is required for both maintaining adult stem cells in the sub-ventricular zone (Suh et al., 2009) and generating CNS progenitor cells (Sun et al., 2005). egfr is also required in Drosophila for intestinal stem cell proliferation and midgut regeneration (Jiang et al., 2011), the maintenance of the germline niche architecture (Chen et al., 2013), and for the self-renewal and establishment of the cell polarity of epidermal follicle stem cells (Castanieto et al., 2014). The discovery in the Lophotrochozoa (e.g., planarians), a sister group to the Deuterostomes (e.g., vertebrates) and the Ecdysozoa (e.g., Drosophila) of the asymmetric distribution of EGFR-3 in stem cells (Figure 7A) and the role it plays in the asymmetric production of daughter cells (Figure 7B-7D), indicates an ancient evolutionary origin for these functions.
Such conservation may be intricately associated with the evolution of cell polarity in complex tissues. A recent study in Drosophila revealed that asymmetric EGFR distribution in follicle stem cells is required for the establishment of cell polarity via ERK and LKB1-AMPK and for the proper localization of the Par-PKC-Baz cell polarity complex, which disappeared when egfr was mutated (Castanieto et al., 2014). Our study also suggests a similar role for LKB1-AMPK. RNAi of these genes in sublethally irradiated animals affected neoblast repopulation in a manner that was indistinguishable from egfr-3(RNAi) treated animals. However, it remains to be determined whether EGFR-3 regulates LKB1-AMPK directly or indirectly. Moreover, lkb1 and ampk function in planarians may be pleiotropic, as suggested by the broad expression of these genes and the reduction in animal size observed in unirradiated animals during homeostasis (Figure 4). Exploring the possible roles of planarian egfr-3 in the conserved hierarchy of polarity establishment and maintenance, along with the identification and characterization of additional MAPKs will be the focus of future investigations.
Egfr-3 may be a regulator of genomic stability during neoblast expansion
The mechanisms by which the lifelong maintenance of the neoblast population in planarians is regulated and how these cells are kept from uncontrolled hyperproliferation and genomic instability remain unknown. Our analyses of the genomic output modulated by egfr-3 identified not only expected regulators of the cell cycle (e.g., cyclin b, cyclin dependent kinase 1), but also DNA damage response genes (rad54b, rnf8, p53, DNA lyase). The link between egfr-3, DNA damage repair and asymmetric cell divisions suggests the intriguing possibility that a detection/checkpoint mechanism for asymmetric cell division may normally occur during neoblast proliferation, which may have been exacerbated by the DNA damage introduced by sublethal irradiation. Therefore, we hypothesize that the DNA damage response and repair complex in neoblasts may be under EGF signaling regulation and may play an essential role in symmetric versus asymmetric cell divisions by maintaining planarian stem cell genomic stability, and mitigating normal cellular aging processes. Given our increasing ability to visualize DNA damage in planarian chromosomes (Xiang et al., 2014), future studies will aim to measure this process during neoblast proliferation and clonal expansion.
Supplementary Material
Highlights.
egfr-3 is required for neoblast repopulation after sublethal irradiation
The putative EGF ligand neuregulin-7 also regulates neoblast repopulation
lkb1, ampk and rad54B function downstream of egfr-3 during neoblast repopulation
egfr-3 regulates asymmetric cell division in neoblast repopulation
Acknowledgments
We thank all members of the Sánchez Alvarado lab, especially Eric Ross for bioinformatics assistance, Li-Chun Cheng, Carlos Guerrero Hernandez and Aurimas Gumbrys for technical help and probes, and Sarah Elliott, Erin Davies, Kim Tu, Elizabeth Duncan, Christopher Arnold, and Stephanie Nowotarski for comments on the manuscript. We also thank Mark Miller for the Illustration in Figure 8C; Xin Qian and all members of the Reptile & Aquatics, Molecular Biology, Cytometry, Microscopy, and Histology Core Facilities at the Stowers Institute for Medical Research for technical support. This work was supported by NIH R37GM057260 to A.S.A., who is Howard Hughes Medical Institute and a Stowers Institute for Medical Research investigator.
Footnotes
Author Contributions: Conceptualization, K.L. and A.S.A.; Methodology, K.L. and A.S.A.; Investigation, K.L., H.T.V., R.D.M.; Formal Analysis, K.L., H.T.V., R.D.M., S.A.M, C.W.S., R.A., and K.G.; Writing Original Draft, K.L. and A.S.A.; Writing, Review & Editing, K.L. and A.S.A.; Funding Acquisition, A.S.A.; Supervision, J.L.W. and A.S.A.
Conflicts of Interest: The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adell T, Saló E, Boutros M, Bartscherer K. Smed-Evi/Wntless is required for β-catenin-dependent and -independent processes during planarian regeneration. Development. 2009;136:905–910. doi: 10.1242/dev.033761. [DOI] [PubMed] [Google Scholar]
- Adler CE, Seidel CW, McKinney SA, Sánchez Alvarado A. Selective amputation of the pharynx identifies a FoxA-dependent regeneration program in planaria. Elife. 2014;3:e02238. doi: 10.7554/eLife.02238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J. Molecular Biology of the Cell. 4th. New York: Garland Science; 2002. Extracellular Control of Cell Division, Cell Growth, and Apoptosis. [Google Scholar]
- Baguñà J, Saló E, Auladell C. Regeneration and pattern formation in planarians. III. Evidence that neoblasts are totipotent stem cells and the source of blastema cells. Development. 1989;107:77–86. [Google Scholar]
- Baguñà J, Saló E, Romero R. Effects of activators and antagonists of the neuropeptides substance P and substance K on cell proliferation in planarians. Int J Dev Biol. 1989;33:261–266. [PubMed] [Google Scholar]
- Bardeen C, Baetjer F. The inhibitive action of the Roentgen rays on regeneration in planarians. Journal of Experimental Zoölogy. 1904;1:5. [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell. 2011;9:402–411. doi: 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanieto A, Johnston MJ, Nystul TG. EGFR signaling promotes self-renewal through the establishment of cell polarity in Drosophila follicle stem cells. Elife. 2014;3 doi: 10.7554/eLife.04437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrià F, Kobayashi C, Umesono Y, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Itoh M, Taira M, Sánchez Alvarado A, et al. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature. 2002;419:620–624. doi: 10.1038/nature01042. [DOI] [PubMed] [Google Scholar]
- Chen DJ, Nirodi CS. The epidermal growth factor receptor: a role in repair of radiation-induced DNA damage. Clin Cancer Res. 2007;13:6555–6560. doi: 10.1158/1078-0432.CCR-07-1610. [DOI] [PubMed] [Google Scholar]
- Chen H, Chen X, Zheng Y. The nuclear lamina regulates germline stem cell niche organization via modulation of EGFR signaling. Cell Stem Cell. 2013;13:73–86. doi: 10.1016/j.stem.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward SJ. Chromatoid bodies in somatic cells of the planarian: observations on their behavior during mitosis. Anat Rec. 1974;180:533–545. doi: 10.1002/ar.1091800312. [DOI] [PubMed] [Google Scholar]
- Forbes SJ, Rosenthal N. Preparing the ground for tissue regeneration: from mechanism to therapy. Nat Med. 2014;20:857–869. doi: 10.1038/nm.3653. [DOI] [PubMed] [Google Scholar]
- Fraguas S, Barberan S, Cebrià F. EGFR signaling regulates cell proliferation, differentiation and morphogenesis during planarian regeneration and homeostasis. Developmental biology. 2011;354:87–101. doi: 10.1016/j.ydbio.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Gaviño MA, Reddien PW. A Bmp/Admp regulatory circuit controls maintenance and regeneration of dorsal-ventral polarity in planarians. Curr Biol. 2011;21:294–299. doi: 10.1016/j.cub.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaviño MA, Wenemoser D, Wang IE, Reddien PW. Tissue absence initiates regeneration through Follistatin-mediated inhibition of Activin signaling. Elife. 2013;2:e00247. doi: 10.7554/eLife.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedelhoefer OC, Sánchez Alvarado A. Amputation induces stem cell mobilization to sites of injury during planarian regeneration. Development. 2012 doi: 10.1242/dev.082099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Rink JC, Sánchez Alvarado A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ. 2006;48:371–380. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- Hiramoto T, Nakanishi T, Sumiyoshi T, Fukuda T, Matsuura S, Tauchi H, Komatsu K, Shibasaki Y, Inui H, Watatani M, et al. Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene. 1999;18:3422–3426. doi: 10.1038/sj.onc.1202691. [DOI] [PubMed] [Google Scholar]
- Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, Niklason L, Calle E, Le A, Randell SH, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15:123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Fuchs E. A family business: stem cell progeny join the niche to regulate homeostasis. Nat Rev Mol Cell Biol. 2012;13:103–114. doi: 10.1038/nrm3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias M, Gomez-Skarmeta JL, Saló E, Adell T. Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development. 2008;135:1215–1221. doi: 10.1242/dev.020289. [DOI] [PubMed] [Google Scholar]
- Inaba M, Yamashita YM. Asymmetric stem cell division: precision for robustness. Cell stem cell. 2012;11:461–469. doi: 10.1016/j.stem.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Jiang H, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8:84–95. doi: 10.1016/j.stem.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RS, Newmark PA. In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev Biol. 2013;13:8. doi: 10.1186/1471-213X-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi C, Saito Y, Ogawa K, Agata K. Wnt signaling is required for antero-posterior patterning of the planarian brain. Dev Biol. 2007;306:714–724. doi: 10.1016/j.ydbio.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Kotton DN, Morrisey EE. Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med. 2014;20:822–832. doi: 10.1038/nm.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan SW, Reddien PW. dlx and sp6-9 Control optic cup regeneration in a prototypic eye. PLoS Genet. 2011;7:e1002226. doi: 10.1371/journal.pgen.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan K, Mahajan NP. Cross talk of tyrosine kinases with the DNA damage signaling pathways. Nucleic Acids Res. 2015;43:10588–10601. doi: 10.1093/nar/gkv1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med. 2014;20:833–846. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CM, Newmark PA. An insulin-like peptide regulates size and adult stem cells in planarians. Int J Dev Biol. 2012 doi: 10.1387/ijdb.113443cm. [DOI] [PubMed] [Google Scholar]
- Miyajima A, Tanaka M, Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell. 2014;14:561–574. doi: 10.1016/j.stem.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Molina MD, Saló E, Cebrià F. The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev Biol. 2007;311:79–94. doi: 10.1016/j.ydbio.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Morgan T. Regeneration in planarians. Archiv Entwick Mech. 1900;10:58–119. [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark PA, Reddien PW, Cebrià F, Sánchez Alvarado A. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc Natl Acad Sci U S A. 2003;100(1):11861–11865. doi: 10.1073/pnas.1834205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Kobayashi C, Hayashi T, Orii H, Watanabe K, Agata K. Planarian fibroblast growth factor receptor homologs expressed in stem cells and cephalic ganglions. Dev Growth Differ. 2002;44:191–204. doi: 10.1046/j.1440-169x.2002.00634.x. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Developmental dynamics: an official publication of the American Association of Anatomists. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Sánchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development. 2010;137:213–221. doi: 10.1242/dev.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sánchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Dev Biol. 2010;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris TH, Ramirez D, Barghouth PG, Ofoha U, Davidian D, Weckerle F, Oviedo NJ. Regional signals in the planarian body guide stem cell fate in the presence of DNA instability. Development. 2016 doi: 10.1242/dev.131318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Smed-βcatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. A wound-induced Wnt expression program controls planarian regeneration polarity. Proc Natl Acad Sci U S A. 2009;106:17061–17066. doi: 10.1073/pnas.0906823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW. Specialized progenitors and regeneration. Development. 2013;140:951–957. doi: 10.1242/dev.080499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Kicza AM, Sánchez Alvarado A. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development. 2007;134:4043–4051. doi: 10.1242/dev.007138. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Rink JC. Stem cell systems and regeneration in planaria. Dev Genes Evol. 2013;223:67–84. doi: 10.1007/s00427-012-0426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink JC, Gurley KA, Elliott SA, Sánchez Alvarado A. Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science. 2009;326:1406–1410. doi: 10.1126/science.1178712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink JC, Vu HT, Sánchez Alvarado A. The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development. 2011;138:3769–3780. doi: 10.1242/dev.066852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb SM, Gotting K, Ross E, Sánchez Alvarado A. SmedGD 2.0: The Schmidtea mediterranea genome database. Genesis. 2015;53:535–546. doi: 10.1002/dvg.22872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Newmark PA. Follistatin antagonizes activin signaling and acts with notum to direct planarian head regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1363–1368. doi: 10.1073/pnas.1214053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Ross EJ, Jack A, Sánchez Alvarado A. Molecular cloning and characterization of SL3: A stem cell-specific SL RNA from the planarian Schmidtea mediterranea. Gene. 2013 doi: 10.1016/j.gene.2013.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saló E, Baguñà J. Stimulation of cellular proliferation and differentiation in the intact and regenerating planarian Dugesia(G) tigrina by the neuropeptide substance P. J Exp Zool. 1986;237:129–135. doi: 10.1002/jez.1402370117. [DOI] [PubMed] [Google Scholar]
- Salvetti A, Rossi L, Bonuccelli L, Lena A, Pugliesi C, Rainaldi G, Evangelista M, Gremigni V. Adult stem cell plasticity: neoblast repopulation in non-lethally irradiated planarians. Developmental biology. 2009;328:305–314. doi: 10.1016/j.ydbio.2009.01.029. [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark P. The use of planarians to dissect the molecular basis of metazoan regeneration. Wound Repair Regen. 1998;6:413–420. doi: 10.1046/j.1524-475x.1998.60418.x. [DOI] [PubMed] [Google Scholar]
- Sanli T, Steinberg GR, Singh G, Tsakiridis T. AMP-activated protein kinase (AMPK) beyond metabolism: a novel genomic stress sensor participating in the DNA damage response pathway. Cancer Biol Ther. 2014;15:156–169. doi: 10.4161/cbt.26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Garry DJ. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006;20:1692–1708. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- Suh Y, Obernier K, Holzl-Wenig G, Mandl C, Herrmann A, Worner K, Eckstein V, Ciccolini F. Interaction between DLX2 and EGFR regulates proliferation and neurogenesis of SVZ precursors. Mol Cell Neurosci. 2009;42:308–314. doi: 10.1016/j.mcn.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Sun Y, Goderie SK, Temple S. Asymmetric distribution of EGFR receptor during mitosis generates diverse CNS progenitor cells. Neuron. 2005;45:873–886. doi: 10.1016/j.neuron.2005.01.045. [DOI] [PubMed] [Google Scholar]
- Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Dev Cell. 2011;21:172–185. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi-Kim Vu H, Rink JC, McKinney SA, McClain M, Lakshmanaperumal N, Alexander R, Sánchez Alvarado A. Stem cells and fluid flow drive cyst formation in an invertebrate excretory organ. Elife. 2015;4 doi: 10.7554/eLife.07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Cheng LC, Tk Vu H, Lange JJ, McKinney SA, Seidel CW, Sánchez Alvarado A. is a post-mitotic regulator of planarian epidermal differentiation. Elife. 2015;4 doi: 10.7554/eLife.10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Wagner DE, Reddien PW. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell. 2014;15:326–339. doi: 10.1016/j.stem.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517:621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L, Snippert HJ. Stem cell dynamics in homeostasis and cancer of the intestine. Nat Rev Cancer. 2014;14:468–480. doi: 10.1038/nrc3744. [DOI] [PubMed] [Google Scholar]
- Wagner DE, Ho JJ, Reddien PW. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell. 2012;10:299–311. doi: 10.1016/j.stem.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol. 2010;344:979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchley JN, Mayer M, Wagner DE, Owen JH, Reddien PW. Muscle Cells Provide Instructions for Planarian Regeneration. Cell Rep. 2013 doi: 10.1016/j.celrep.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff E, Dubois F. Sur la migration des cellules de régénération chez les planaires. Rev Suisse Zool. 1948;55:218–227. [Google Scholar]
- Xiang Y, Miller DE, Ross EJ, Sánchez Alvarado A, Hawley RS. Synaptonemal complex extension from clustered telomeres mediates full-length chromosome pairing in Schmidtea mediterranea. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E5159–5168. doi: 10.1073/pnas.1420287111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa S, Umesono Y, Hayashi T, Tarui H, Agata K. Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc Natl Acad Sci U S A. 2009;106:22329–22334. doi: 10.1073/pnas.0907464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SJ, Hallows SE, Currie KW, Xu C, Pearson BJ. A mex3 homolog is required for differentiation during planarian stem cell lineage development. Elife. 2015;4 doi: 10.7554/eLife.07025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, et al. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 2015;517:616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


