Abstract
Single-walled carbon nanotubes are of interest in biomedicine for imaging and molecular sensing applications, and as shuttles for various cargos such as chemotherapeutic drugs, peptides, proteins, and oligonucleotides. Carbon nanotube surface chemistry can be modulated for subcellular targeting while preserving photoluminescence for label-free visualization in complex biological environments, making them attractive materials for such studies. The cell nucleus is a potential target for many pathologies including cancer and infectious diseases. Understanding mechanisms of nanomaterial delivery to the nucleus may facilitate diagnostics, drug development, and gene editing tools. Currently, there are no systematic studies to understand how these nanomaterials gain access to the nucleus. Herein, we developed carbon nanotube-based hybrid material which elucidated a distinct mechanism of nuclear translocation of a nanomaterial in cultured cells. We developed a nuclear-targeted nanotube via cloaking photoluminescent single-walled carbon nanotubes in a guanidinium-functionalized helical polycarbodiimide. We found that the nuclear entry of the nanotubes was mediated by the import receptor importin β without the aid of importin α and not by the more common importin α/β pathway. Additionally, the nanotube photoluminescence exhibited distinct red-shifting upon entry to the nucleus, potentially functioning as a reporter of the importin β-mediated nuclear transport process. This work delineates a non-canonical mechanism for nanomaterial delivery to the nucleus and provides a reporter for the study of nucleus-related pathologies.
Keywords: helical polymer, hyperspectral imaging, cervical cancer, near-infrared sensor, biosensor, polycation, nuclear pore complex
The cell nucleus, nuclear transport receptors, and the nuclear pore complex (NPC) are potential therapeutic targets for diverse pathologies including cancer and infectious diseases.1 The targeted delivery of exogenous macromolecular materials such as transcription factors and nucleic acids for gene therapy to the nucleus is currently inefficient2 and challenging to engineer in part because macromolecule entry into the nucleus is tightly regulated by the NPCs,3 which are typically 20 – 50 nm in diameter4 and allow transit of import cargoes presenting active nuclear localization signals (NLSs)—commonly arginine- and lysine-rich short peptide sequences derived from nuclear and viral proteins.5
Several strategies have been applied, with mixed success, to localize macromolecules within the cell nucleus. One such strategy involves conjugating canonical NLS peptides to nanomaterials.6–8 Guanidine, the functional side chain in arginine, is a small moiety that allows integration into molecular probes with minimal structural perturbations and has been widely incorporated in synthetic drugs and drug-carriers, including polymers and nanoparticles, to improve cell uptake.9 Guanidine-functionalized oligomers and small polymers have been found to translocate to the cell nucleus in cultured cells,10 making guanidine a potential class of non-canonical nuclear localization signals to facilitate nuclear delivery. Non-canonical NLSs may enhance stability; obviate problems with the in vivo use of peptide-based targeting moieties, which are susceptible to enzymatic degradation and immunogenic responses. Currently, the mechanisms by which non-peptide based synthetic nanomaterials are trafficked to the cell nucleus, including guanidine-functionalized materials, are unknown.
Nucleo-cytoplasmic transport of macromolecules often involves karyopherin β (importin β), a major nuclear import receptor that acts as a chaperone for highly charged nuclear proteins.11 Structural analyses show that importin β has a large degree of flexibility that it uses to import cargoes directly, or more commonly through adaptor proteins such as importin α.12 Most nucleoproteins incorporate canonical NLS peptide sequence which binds to adaptor proteins to form a complex.13 The most common (canonical) mechanism14 involves a heterodimer, importin α/β, which binds to NLS-bearing cargoes resulting in translocation of the importin/cargo complex through the NPC.15 Although, some proteins and viruses have been found to translocate from the cytoplasm to the nucleus facilitated by importin β without the aid of importin α,16,17 this non-canonical mechanism of nucleocytoplasmic transport is relatively underexplored. Importin β is overexpressed in cervical, ovarian, and esophageal cancers,18 among others, and its function is also involved in the internalization of viruses,17 making it a potential therapeutic target in cancers and viral infections.
Single-walled carbon nanotubes (SWCNTs) are multifaceted nanomaterials with mechanical, photophysical and thermal properties which can function as nanocarriers, bioimaging tools, and molecular sensors.19,20 SWCNTs exhibit photostable, intrinsic photoluminescence in the near infrared (NIR) spectral window21 where there is minimal absorption, autofluorescence, and scattering of light in biological specimens.22 Therefore, SWCNTs are promising for the study of cellular transport processes. Importantly, nanotube photoluminescence also undergoes spectral changes mediated by redox,23,24 solvent dielectric,25,26 and electrostatic charge,27 potentiating nanotube-based optical sensors28 in complex biological environments. A large surface area provides opportunities for multivalent functionalization for subcellular targeting and molecular loading for targeted delivery.29 Nanotubes have been used to deliver various molecular cargos including chemotherapeutic drugs, oligonucleotides, peptides, and proteins inside live cells.19 Recent studies have demonstrated that surface functionalization can modulate nanotube physicochemical behavior and biological interactions, including cytotoxicity.22 Carbon nanotubes have been targeted via nuclear protein fragments containing NLS peptide sequences,30 and in their absence, observed to localize to the nucleus in certain cases.31 As with other nanomaterials, the mechanisms underlying the nuclear transport of carbon nanotubes remain elusive.32
In our previous work, we developed polycarbodiimide polymer derivatives with functional moieties that can effectively cloak and suspend SWCNTs in aqueous media, resulting in photoluminescent complexes that are stable under ambient conditions for several months.33 Here, we developed functional derivatives in this class of materials and presented a polycarbodiimide-nanotube complex that functions as a nanoscale probe to efficiently target the cell nucleus, discern its mechanism of nuclear transit, and report the mechanism via an optical response. Although carbon nanotubes have been made to localize in the nucleus, their mechanisms of nuclear translocation and optical response upon entering the nucleus were unexplored. We found that our probe, a photoluminescent single-walled carbon nanotube encapsulated in a multimeric guanidine-functionalized helical polycarbodiimide, translocated to the cell nucleus through the nuclear pore complex (NPC) via a distinct mechanism mediated by the importin β transport receptor without the aid of importin α. Upon entering the nucleus, the probe exhibited a distinct red-shift in its emission wavelength, thus functioning as a sensor for nuclear localization via importin β-mediated translocation through the NPC, potentiating an optical reporter for this cellular process.
Results and Discussion
We synthesized helical polycarbodiimide polymers33 to non-covalently suspend single-walled carbon nanotubes in aqueous media. The polymers contained phenyl groups to promote adhesion to the graphitic carbon sidewall of the nanotube via π-π and hydrophobic interactions and either (1) guanidine or (2) amine side chains to mimic arginine and lysine, respectively (Fig. 1a, Supporting Fig. S1). Such amino acids are common in cell-penetrating and NLS peptide sequences.5 The polymer-SWCNT complexes were water soluble and stable under ambient conditions without visual aggregation over several months. Atomic force microscopy (AFM) images suggested that the polymers ‘cloak’ the nanotubes, (Supporting Fig. S2) similarly to ssDNA.34 The polycarbodiimide-nanotube complexes exhibited spectral features (Fig. 1b, 1c) characteristic of SWCNTs produced by the HiPco method21 and positive surface potentials consistent with the charge of the polymers (Supporting Fig. S3).
Figure 1. Cationic helical polycarbodiimide-cloaked single-walled carbon nanotubes.
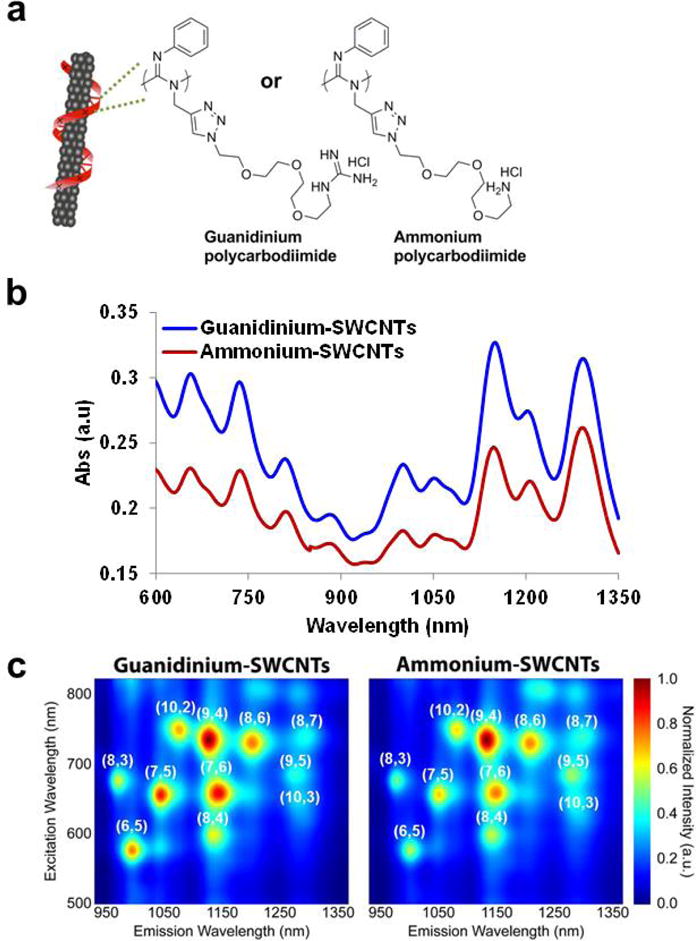
a, Chemical structures of two polycarbodiimide polymers used for non-covalent encapsulation of the carbon nanotubes. b, Vis-NIR absorbance spectra of aqueous suspensions of guanidinium and ammonium-nanotube complexes. c, Normalized photoluminescence excitation-emission (PL) plots of guanidinium and ammonium-nanotube complexes.
Human cervical cancer cell lines, HeLa and SiHa, were interrogated with the polycarbodiimide-nanotube complexes. Under standard cell culture conditions, the guanidinium-nanotube complexes exhibited substantial uptake and preferential accumulation in the cell nucleus (Fig. 2a and Supporting Fig. S4). Approximately 92% of SiHa cell nuclei and 84% of HeLa cell nuclei were positive for NIR emission from the nanotubes. Under the same conditions, the ammonium-nanotube complexes also exhibited high cell uptake but minimal nuclear localization (Fig. 2b). Ammonium functional moieties are known to be less efficient in facilitating endosomal escape of nanomaterials compared to guanidine functional groups, likely contributing to the reduced nuclear localization of ammonium-SWCNTs.35 The nuclear translocation of the guanidinium polycarbodiimide-functionalized nanotubes over the ammonium functionalization apparently mimicked behavior seen with peptides.36
Figure 2. Sub-cellular localization of polycarbodiimide-single-walled carbon nanotube complexes.
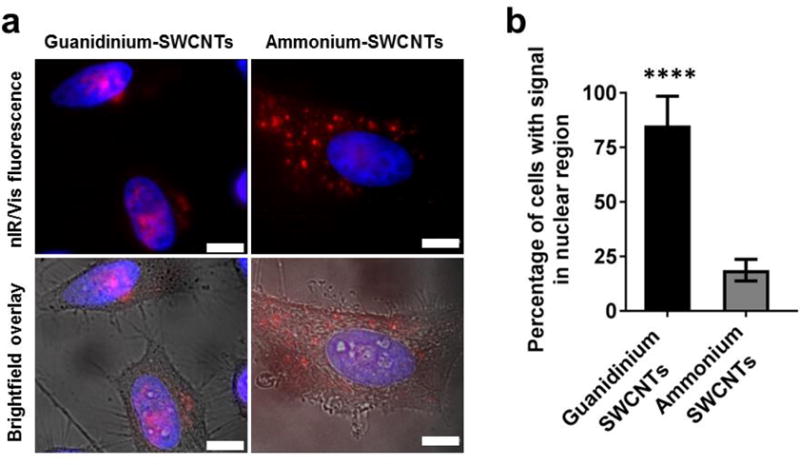
a, Fluorescence and brightfield overlay images of polymer-nanotube complexes in HeLa cells/nuclei. Red = carbon nanotube near-infrared emission, blue = Hoechst 33258 nuclear stain. b, Quantification of nuclear localization of guanidinium-polymer complexes and ammonium-polymer complexes. Graph represents mean ± standard deviation from 3 replicate experiments. Scale bars = 10 μm. ****P<0.0001 (P<0.05 significant).
Cells treated with the guanidinium-nanotube complexes were monitored via near-infrared microscopy to evaluate the timing of nuclear localization. After incubation, the guanidinium-nanotubes associated with the cell membranes, appearing as puncta (Supporting Fig. S5). After approximately 4 h, the nanotubes began to localize in the cell nucleus, and accumulation continued over 24 h. The guanidinium-nanotube complexes became motionless upon nuclear localization, whereas the ammonium-nanotube complexes were mobile (Movies S1–S3).
As proteoglycans are the primary receptors on cell surfaces for many cationic macromolecular cargos, including polyarginine derivatives,37 we investigated whether guanidinium-nanotube complexes would associate with proteoglycans on the cell surface to facilitate internalization. We deactivated proteoglycans by chemical treatment with sodium chlorate38 prior to addition of guanidinium nanotube complexes to the cells. The treatment distinctly reduced the uptake of the complexes (Supporting Fig. S6), suggesting that proteoglycans mediated their uptake.
We assessed whether nanotubes gain entry to the nucleus via NPCs by inhibiting this pathway with wheat germ agglutinin (WGA). Inside cells, WGA binds to carbohydrates on nuclear pore complex proteins and crosslinks nucleoporins, limiting entry of macromolecules via the NPCs.39 We pretreated the cells with WGA to block NPCs before administering guanidinium-nanotube complexes to cells incubated in WGA-free cell culture medium. Under these conditions, the nuclear translocation of nanotubes was attenuated by 92% compared to control cells that did not receive WGA treatments (Fig. 3a, 3b), suggesting that the nanotubes entered the nucleus via NPCs.
Figure 3. Mechanistic studies of nanoprobe nuclear translocation.
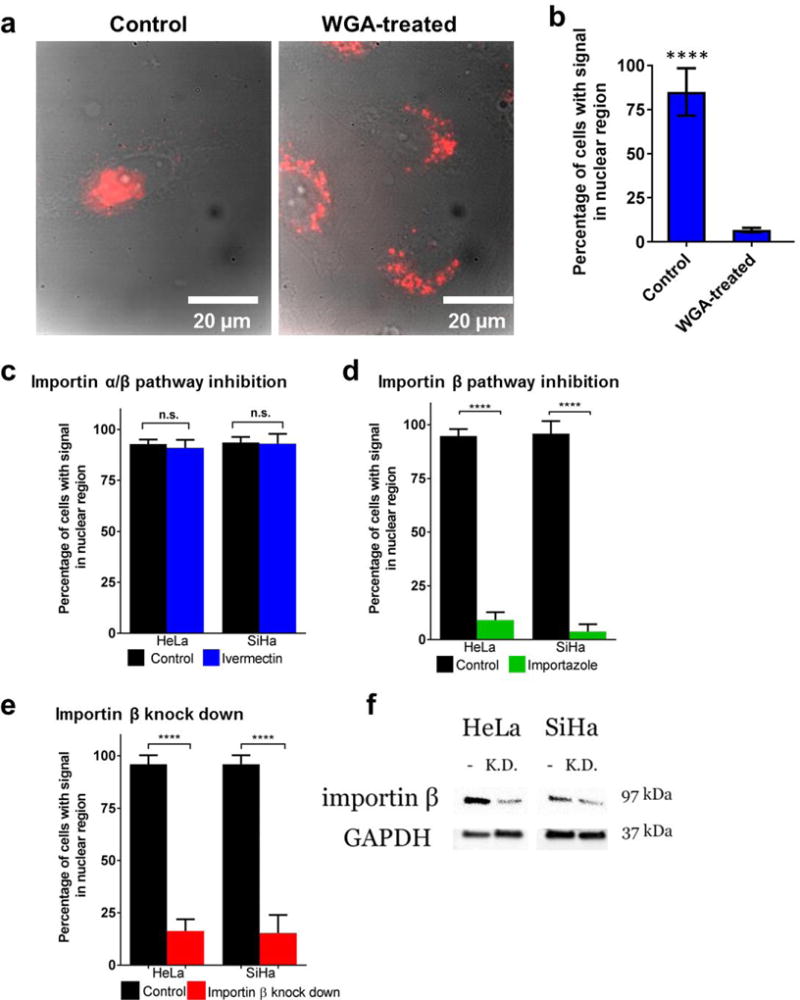
a, Near-infrared emission/brightfield overlay images of HeLa cells upon treatment with wheat germ agglutinin (WGA) and guanidinium-nanotube complexes. b, Graph quantifying nuclear localization from the experiment. c, Nuclear localization of nanotubes in cells treated with ivermectin, a pharmacological inhibitor of importin α/β-mediated nuclear import, quantified from NIR fluorescence images. d, Nanotube localization in cells treated with importazole, an inhibitor of importin β-mediated nuclear import. e, Nanotube localization in cells after knockdown of importin β via shRNA transfection. f, Western blot analysis of the shRNA-treated cells. Graphs represent mean ± standard deviation from 3 replicate experiments. ****P<0.0001 (P<0.05 significant).
Inside cells, the nanotubes presumably escape the endolysosomal pathway, most likely by membrane disruption in endosomes/lysosomes due to guanidine-rich sequences.35 As certain materials that cause such disruption often lead to cytotoxicity,40 cell viability was evaluated upon treatment with the guanidinium-nanotube complexes. The assays found that the complexes conferred no discernable cytotoxicities (Supporting Fig. S7).
To determine whether nanotube translocation followed the canonical importa α/β pathway, we treated cells with ivermectin, a pharmacological inhibitor of importa α/β-mediated nuclear transport.41 Nuclear entry was not significantly blocked in ivermectin treated cells compared to control cells (Fig. 3c and Supporting Fig. S8). Ruling out the importin α/β pathway, we then explored whether importa β alone resulted in nuclear translocation of the nanotubes. Treatment of the cells with importazole, an inhibitor of importin β-mediated nuclear translocation,42 caused a significant reduction in nuclear localization (Fig.3d and Supporting Fig. S9). We further confirmed this mechanism by knocking down importin β expression. Cells were transfected with shRNA against importin β before administering the nanotube complexes. The knockdown resulted in attenuated nuclear localization of the nanotubes by 83–84% compared to cells that were transfected with empty vector, according to imaging studies and consistent with the pharmacological inhibition experiment (Fig. 3e and Supporting Fig. S10). Importin β knockdown was confirmed by western blot analysis (Fig. 3f). These results imply that guanidinium-nanotube nuclear import is mediated by importin β without the aid of importin α.
Based on our findings, we propose a model for nuclear translocation of the guanidinium-nanotube complexes (Fig. 4). The complexes enter the cells via endocytosis facilitated by proteoglycans. The complexes then escape the endolysosomal pathway to enter the cytosol, where the nanotubes interact with importin β. The importin β then facilitates guanidinium-nanotube translocation via the nuclear pore complex, without the aid of importin α.
Figure 4. Model for proposed mechanism of nanoprobe translocation to the nucleus.
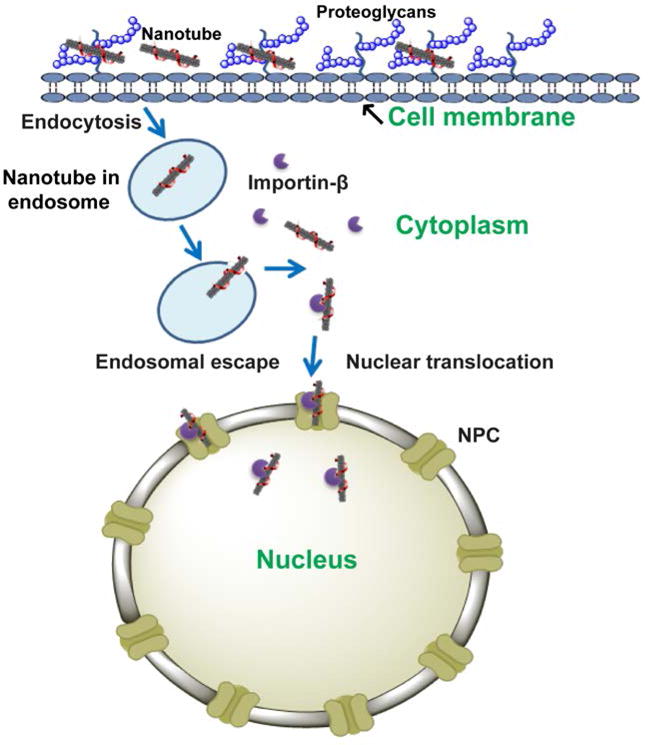
The nanotube complex enters the cell via endocytosis upon binding to proteoglycans on the cell membrane. The nanotube then escapes the nucleus and binds to importin β. The importin β-nanotube complex then transits the nuclear membrane via the NPC.
We assessed the optical response of the guanidinium-nanotube complexes in the nuclear environment. Using near-infrared hyperspectral microscopy,43 we collected spectral information from the nanotube complexes in the live cells and spatially resolved the spectral data. Upon measuring the nanotube emission, focusing on the (9,4) species, we found that the emission was red-shifted approximately 11 nm in the nucleus, compared with nanotubes in aqueous solution (Supporting Fig. S11). Upon interrogation with WGA and importazole to inhibit nuclear localization, the emission wavelength did not red shift and remained similar to the values in solution (Fig. 5a, b). The reagents themselves, WGA and importazole, did not cause significant changes in nanotube emission (Supporting Fig. S12).
Figure 5. Nanotube photoluminescence response on nuclear localization and imaging via hyperspectral microscopy.
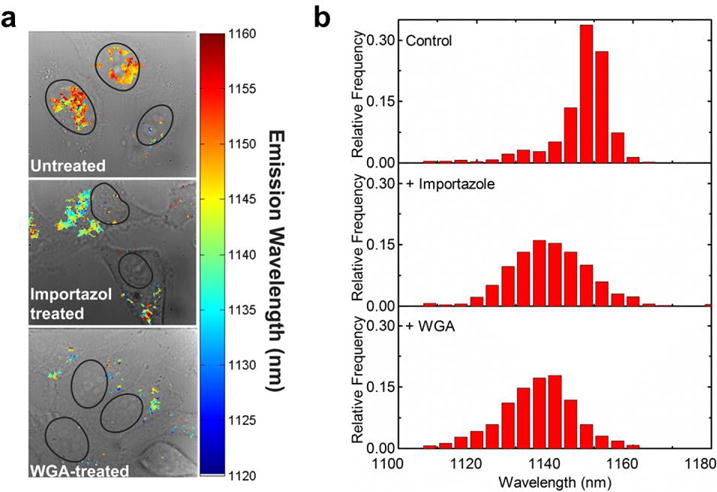
a, Hyperspectral maps of guanidinium-nanotube complex emission in live HeLa cells, overlaid on the brightfield images. Nuclear outlines are shown as black lines. b, Histograms of peak emission wavelength from the hyperspectral data, obtained from three replicate experiments.
We hypothesized that the highly charged character of chromatin may play a role in modulating the emission of the carbon nanotubes upon entry to the nucleus. Previous work found that carbon nanotubes, once in the nucleus, can directly interact with DNA.30 We also recently discovered that increased anionic charge due to polyelectrolytic biomacromolecules in the local environment of carbon nanotubes can result in distinct red-shifting of nanotube emission.27 To test whether DNA, found predominantly in the nucleus, could potentially cause a similar modulation of the optical bandgap, we introduced the guanidinium-nanotube complexes to salmon sperm DNA in solution. The DNA resulted in red-shifting of the nanotube emission by approximately 6 nm (Supporting Fig. S12). To attempt to recapitulate the magnitude of the shift in nuclei, the mixture of salmon DNA and nanotube complexes was centrifuged, yielding a pellet. The emission wavelength from the pellet was 12 nm red-shifted as compared to the emission from nanotube complexes in the absence of DNA. The resulting emission wavelength of 1148 nm in this condition was comparable to that in cell nuclei. This experiment suggests that the interaction of the guanidinium-nanotube complexes with DNA in the nucleus may mediate the spectral response. In the case where the aggregates were pelleted in vitro, it is currently unclear whether the increased nanotube contact with DNA or the aggregation itself caused an exacerbation of the red-shifting, and thus, further studies on the exact nature of this shift, and the nanotube-biomolecule interactions that cause it in the nucleus, are warranted.
Conclusions
In this work, we developed a nanoscale probe that targeted the cell nucleus via a distinct mechanism for carbon nanomaterial transport and reported that mechanism via an optical response. Using chemical inhibition, shRNA knockdown, and biochemical assays, we found that the nuclear translocation of the nanoprobe was mediated by the import receptor importin β and not by the more common importin α/β pathway. Upon entry to the nucleus, the probe emission wavelength was found to red-shift significantly. The photoluminescence shift was spatially mapped using NIR hyperspectral microscopy, resulting in quantitative images of nanotube optical bandgap modulation. The optical modulation in the nuclear environment constitutes a reporting event of the translocation mechanism and provides the optical reporter for this phenomenon. The work illustrates mechanistic evidence for the nuclear translocation of a synthetic nanomaterial through NPCs mediated specifically by importin β. The structural flexibility of importin β may be exploited for other nanomaterial delivery to nucleus especially in diseases where this import receptor is overexpressed.18 A probe for this process may have applications in drug delivery, in the screening/development of inhibitors of importin β and nucleo-cytoplasmic transport processes, and as a tool for investigations in cell and disease biology.
Experimental Section
Preparation of polymer-nanotube complexes
Polymer-nanotube complexes were prepared by introducing HiPco nanotubes (1 mg) to aqueous acidic solutions of either ammonium or guanidinium polycarbodiimide polymers (1 mL, 5 mg/mL water). The mixtures were probe sonicated for 20 min (750 W, 20 kHz, 40% Amplitude, SONICS VibraCell) at low temperature using a CoolRack M30 PF (BioCision), stored at −20 °C prior to use. Solutions were centrifuged (SORVALL Discovery 90SE, HITACHI) at 280,000 g for 30 min at room temperature. After centrifugation, ca. 80% of the supernatant was collected and filtered through a 100 kDa molecular weight cutoff centrifugal filter (Millipore Amicon) to remove excess polymer and acid. The washing step was repeated three more times before re-suspensing the nanotubes in water. Removal of acid from the suspension was confirmed with a litmus paper. Ultrapure water (18.2 mΩ) was used for all aqueous solutions. The suspensions were characterized by visible-near-infrared (VIS–NIR) absorbance, NIR fluorescence spectroscopies, zeta potential (surface charge) measurements, and atomic force microscopy.
Cell culture and nuclear import assay
HeLa cells were purchased from American Type Culture Collection (ATCC). SiHa cells were obtained from Antibody and Bioresource Core Facility at Memorial Sloan Kettering Cancer Center. STR analysis was performed for cell authentication. Cells were cultured in DMEM supplemented with 10% (V/V) heat inactivated FBS, 1% Penicillin Streptomycin, 1% glutamine, and 2.5% HEPES at 37 °C in humidified air containing 5% CO2. All reagents were purchased from Gibco (Life Technologies). Cells were trypsinized, washed, and plated onto a 35 mm glass bottom Petri dish (MatTek). Cells were used after 24 h at 70–80% confluence. Polymer-nanotube complexes were diluted to 3 mg/L in water to avoid potential agglomeration of cationic carbon nanotubes at high concentration in complete cell culture media. The resulting solutions were then added to cell culture medium containing serum to a final concentration of 0.2–2 mg/L of nanotubes. The cell culture medium was removed from the cells in the dishes and replaced with media containing polymer-nanotube complexes. The cells were incubated with the polymer-nanotube/media solutions for the time periods specified in the text, washed with PBS three times, and incubated in fresh cell culture media for up to 16 hours prior to imaging.
Live cell NIR imaging and hyperspectral microscopy
The near-infrared photoluminescence imaging and spectroscopy of the cells were performed using a hyperspectral microscope under 730 nm excitation and 0.2s exposure, as described.43 Broadband images and 3D stacks/overlays were collected using the same microscope without the hyperspectral functionalities. DAPI fluorescence images were collected using an X-cite 120Q lamp (Lumen Dynamics) with DAPI filter set and CCD camera (QIClick, QImaging) while near-infrared images were collected using a 2D InGaAs array. Images and movies were processed using ImageJ software (NIH) and Bitplane Imaris 8.0.2 software.
Nuclear pore complex blocking experiment
Cells were plated onto 35 mm glass bottom dishes as described and incubated overnight. Cells were incubated with fresh cell culture media containing wheat germ agglutinin (WGA) (50 μg/mL) at 37 °C for 30 min. Cells were washed three times with PBS and placed into fresh media. The cells were then treated with guanidinium polymer-nanotube complexes (0.2 mg/L) for 3h. The cells were washed 3x with PBS and, fresh media was added, and incubated for 16 h before imaging. Cells not treated with WGA were used as the control.
Pharmacologic inhibition of importin α/β and importin β mediated nuclear translocation
Cells were pre-incubated for 30 minutes with either importazole (50 μM), ivermectin (25 μM), or vehicle (DMSO) before introducing guanidinium-nanotube complexes (0.2 mg/L) for 3 h. The cells were washed with PBS and fresh media with importazole, ivermectin, or vehicle was added. The cells were then incubated for approximately 16 h before imaging.
Importin β knockdown by shRNA
Cells were seeded onto 35 mm Petri dishes and grown to 90% confluence in complete media. Cells were transfected following a protocol described in Nagy, V. & Watzele, M. Nature Methods, 2006 and using a mixture of 9 μL of FuGene HD (Promega) and 3 μL of either a plasmid for importin β knockdown (Mission shRNA plasmid DNA (human) against karyopherin (importin β1), or an empty plasmid (Mission pLKO 1-puro Vector control plasmid DNA). Cells were incubated in transfection media for 1 h then washed with PBS and placed onto fresh media. 24 hours after transfection, the cells were incubated with guanidinium-nanotube complexes (0.2 mg/L) for 30 minutes, the cells were then incubated for approximately 16 h before imaging.
Statistical analysis
Each experiment was performed in triplicate. Total numbers of cells examined for nuclear localization of guanidinium-nanotube complexes are as follows for HeLa cells: importazole treated cells, n = 95; vehicle control, n= 83; ivermectin-treated cells, n=84; vehicle control, n=91; shRNA knockdown, n=117; empty vector control, n=96. For SiHa cells: importazole treated cells, n=113; vehicle control, n= 131; ivermectin-treated cells, n=93; vehicle control, n=77; shRNA knockdown, n=100; empty vector control, n=111. In each case, the percentage of cells with NIR emission in the nuclear localization was calculated separately for three replicates. The significance was calculated using a two-tailed, unpaired t test.
Supplementary Material
Acknowledgments
This work was supported in part by the NIH New Innovator Award (DP2-HD075698), the Cancer Center Support Grant (P30 CA008748), the Anna Fuller Fund, the Louis V. Gerstner Jr. Young Investigator’s Fund, the Frank A. Howard Scholars Program, the Honorable Tina Brozman Foundation for Ovarian Cancer Research, Cycle for Survival, the Alan and Sandra Gerry Metastasis Research Initiative, Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center, the Imaging & Radiation Sciences Program, and the Center for Molecular Imaging and Nanotechnology of Memorial Sloan Kettering Cancer. J.B. was supported by the Tow Foundation Postdoctoral Fellowship, Center for Molecular Imaging and Nanotechnology at MSKCC. P. V. J. was supported by a an NIH National Cancer Institute T32 Training Grant (2T32CA062948-21). D.R. was supported by the American Cancer Society Roaring Fork Valley Postdoctoral Fellowship (PF-13-388801-TBG). The authors would like to thank V. Boyko for his help in image processing, and core facilities at Memorial Sloan Kettering Cancer Center: the Molecular Cytology Core Facility for AFM imaging, the Antibody and Bioresource Core Facility, and the Analytical Core Facility for nuclear magnetic resonance, FTIR, and high resolution mass spectroscopies.
Footnotes
Competing financial interests: None
Supporting Information
Additional experimental procedures, supporting figures, characterization data, and supporting movies. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Chahine MN, Pierce GN. Therapeutic Targeting of Nuclear Protein Import in Pathological Cell Conditions. Pharmacol Rev. 2009;61:358–372. doi: 10.1124/pr.108.000620. [DOI] [PubMed] [Google Scholar]
- 2.Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit JP. Progress in Developing Cationic Vectors for Non-Viral Systemic Gene Therapy Against Cancer. Biomaterials. 2008;29:3477–3496. doi: 10.1016/j.biomaterials.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 3.Sakiyama Y, Mazur A, Kapinos LE, Lim RY. Spatiotemporal Dynamics of the Nuclear Pore Complex Transport Barrier Resolved by High-Speed Atomic Force Microscopy. Nat Nanotechnol. 2016;11:719–723. doi: 10.1038/nnano.2016.62. [DOI] [PubMed] [Google Scholar]
- 4.Pouton CW, Wagstaff KM, Roth DM, Moseley GW, Jans DA. Targeted Delivery to the Nucleus. Adv Drug Deliver Rev. 2007;59:698–717. doi: 10.1016/j.addr.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin Alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kann NPaM. Nuclear Pore Complex Is Able to Transport Macromolecules with Diameters of 39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nitin N, LaConte L, Rhee WJ, Bao G. Tat Peptide Is Capable of Importing Large Nanoparticles Across Nuclear Membrane in Digitonin Permeabilized Cells. Ann Biomed Eng. 2009;37:2018–2027. doi: 10.1007/s10439-009-9768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma XW, Gong NQ, Zhong L, Sun JD, Liang XJ. Future of Nanotherapeutics: Targeting the Cellular Sub-Organelles. Biomaterials. 2016;97:10–21. doi: 10.1016/j.biomaterials.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 9.McKinlay CJ, Waymouth RM, Wender PA. Cell-Penetrating, Guanidinium-Rich Oligophosphoesters: Effective and Versatile Molecular Transporters for Drug and Probe Delivery. J Am Chem Soc. 2016;138:3510–3517. doi: 10.1021/jacs.5b13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim TI, Lee M, Kim SW. A Guanidinylated Bioreducible Polymer with High Nuclear Localization Ability for Gene Delivery Systems. Biomaterials. 2010;31:1798–1804. doi: 10.1016/j.biomaterials.2009.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harel A, Forbes DJ. Importin Beta: Conducting a Much Larger Cellular Symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Terry LJ, Shows EB, Wente SR. Crossing the Nuclear Envelope: Hierarchical Regulation of Nucleocytoplasmic Transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 13.Lott K, Cingolani G. The Importin Beta Binding Domain as a Master Regulator of Nucleocytoplasmic Transport. Bba-Mol Cell Res. 2011;1813:1578–1592. doi: 10.1016/j.bbamcr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kau TR, Way JC, Silver PA. Nuclear Transport and Cancer: From Mechanism to Intervention. Nat Rev Cancer. 2004;4:106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 15.Hill R, Cautain B, de Pedro N, Link W. Targeting Nucleocytoplasmic Transport in Cancer Therapy. Oncotarget. 2014;5:11–28. doi: 10.18632/oncotarget.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmeri D, Malim MH. Importin Beta Can Mediate the Nuclear Import of an Arginine-Rich Nuclear Localization Signal in the Absence of Importin Alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen S, Au S, Pante N. How Viruses Access the Nucleus. Bba-Mol Cell Res. 2011;1813:1634–1645. doi: 10.1016/j.bbamcr.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 18.van der Watt PJ, Chi A, Stelma T, Stowell C, Strydom E, Carden S, Angus L, Hadley K, Lang D, Wei W, Birrer MJ, Trent JO, Leaner VD. Targeting the Nuclear Import Receptor Kpnbeta1 As an Anticancer Therapeutic. Mol Cancer Ther. 2016;15:560–573. doi: 10.1158/1535-7163.MCT-15-0052. [DOI] [PubMed] [Google Scholar]
- 19.Serpell CJ, Kostarelos K, Davis BG. Can Carbon Nanotubes Deliver on Their Promise in Biology? Harnessing Unique Properties for Unparalleled Applications. Acs Central Sci. 2016;2:190–200. doi: 10.1021/acscentsci.6b00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernandez-Rivera M, Zaibaq NG, Wilson LJ. Toward Carbon Nanotube-Based Imaging Agents for the Clinic. Biomaterials. 2016;101:229–240. doi: 10.1016/j.biomaterials.2016.05.045. [DOI] [PubMed] [Google Scholar]
- 21.Bachilo SM, Strano MS, Kittrell C, Hauge RH, Smalley RE, Weisman RB. Structure-Assigned Optical Spectra of Single-Walled Carbon Nanotubes. Science. 2002;298:2361–2366. doi: 10.1126/science.1078727. [DOI] [PubMed] [Google Scholar]
- 22.Hong GS, Diao SO, Antaris AL, Dai HJ. Carbon Nanomaterials for Biological Imaging and Nanomedicinal Therapy. Chem Rev. 2015;115:10816–10906. doi: 10.1021/acs.chemrev.5b00008. [DOI] [PubMed] [Google Scholar]
- 23.Heller DA, Jin H, Martinez BM, Patel D, Miller BM, Yeung TK, Jena PV, Hobartner C, Ha T, Silverman SK, Strano MS. Multimodal Optical Sensing and Analyte Specificity Using Single-Walled Carbon Nanotubes. Nat Nanotechnol. 2009;4:114–120. doi: 10.1038/nnano.2008.369. [DOI] [PubMed] [Google Scholar]
- 24.Cognet L, Tsyboulski DA, Rocha JDR, Doyle CD, Tour JM, Weisman RB. Stepwise Quenching of Exciton Fluorescence in Carbon Nanotubes by Single-Molecule Reactions. Science. 2007;316:1465–1468. doi: 10.1126/science.1141316. [DOI] [PubMed] [Google Scholar]
- 25.Heller DA, Jeng ES, Yeung TK, Martinez BM, Moll AE, Gastala JB, Strano MS. Optical Detection of DNA Conformational Polymorphism on Single-Walled Carbon Nanotubes. Science. 2006;311:508–511. doi: 10.1126/science.1120792. [DOI] [PubMed] [Google Scholar]
- 26.Choi JH, Strano MS. Solvatochromism in Single-Walled Carbon Nanotubes. Appl Phys Lett. 2007;90:223114. [Google Scholar]
- 27.Roxbury D, Jena PV, Shamay Y, Horoszko CP, Heller DA. Cell Membrane Proteins Modulate the Carbon Nanotube Optical Bandgap via Surface Charge Accumulation. ACS Nano. 2016;10:499–506. doi: 10.1021/acsnano.5b05438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barone PW, Baik S, Heller DA, Strano MS. Near-Infrared Optical Sensors Based on Single-Walled Carbon Nanotubes. Nat Mater. 2005;4:86–92. doi: 10.1038/nmat1276. [DOI] [PubMed] [Google Scholar]
- 29.Serpell CJ, Rutte RN, Geraki K, Pach E, Martincic M, Kierkowicz M, De Munari S, Wals K, Raj R, Ballesteros B, Tobias G, Anthony DC, Davis BG. Carbon Nanotubes Allow Capture of Krypton, Barium and Lead for Multichannel Biological X-ray Fluorescence Imaging. Nat Commun. 2016;7:13118. doi: 10.1038/ncomms13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyer PD, Ganesh S, Qin Z, Holt BD, Buehler MJ, Islam MF, Dahl KN. Delivering Single-Walled Carbon Nanotubes to the Nucleus Using Engineered Nuclear Protein Domains. ACS applied materials & interfaces. 2016;8:3524–3534. doi: 10.1021/acsami.5b12602. [DOI] [PubMed] [Google Scholar]
- 31.Mu QX, Broughton DL, Yan B. Endosomal Leakage and Nuclear Translocation of Multiwalled Carbon Nanotubes: Developing a Model for Cell Uptake. Nano Lett. 2009;9:4370–4375. doi: 10.1021/nl902647x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pantarotto D, Briand JP, Prato M, Bianco A. Translocation of Bioactive Peptides Across Cell Membranes by Carbon Nanotubes. Chem Commun. 2004:16–17. doi: 10.1039/b311254c. [DOI] [PubMed] [Google Scholar]
- 33.Budhathoki-Uprety J, Jena PV, Roxbury D, Heller DA. Helical Polycarbodiimide Cloaking of Carbon Nanotubes Enables Inter-Nanotube Exciton Energy Transfer Modulation. J Am Chem Soc. 2014;136:15545–15550. doi: 10.1021/ja505529n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin H, Jeng ES, Heller DA, Jena PV, Kirmse R, Langowski J, Strano MS. Divalent Ion and Thermally Induced DNA Conformational Polymorphism on Single-Walled Carbon Nanotubes. Macromolecules. 2007;40:6731–6739. [Google Scholar]
- 35.El-Sayed A, Khalil IA, Kogure K, Futaki S, Harashima H. Octaarginine- and Octalysine-Modified Nanoparticles Have Different Modes of Endosomal Escape. J Biol Chem. 2008;283:23450–23461. doi: 10.1074/jbc.M709387200. [DOI] [PubMed] [Google Scholar]
- 36.Schroder T, Niemeier N, Afonin S, Ulrich AS, Krug HF, Brase S. Peptoidic Amino- and Guanidinium-Carrier Systems: Targeted Drug Delivery into the Cell Cytosol or the Nucleus. J Med Chem. 2008;51:376–379. doi: 10.1021/jm070603m. [DOI] [PubMed] [Google Scholar]
- 37.Fuchs SM, Raines RT. Pathway for Polyarginine Entry into Mammalian Cells. Biochemistry. 2004;43:2438–2444. doi: 10.1021/bi035933x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baeuerle PA, Huttner WB. Chlorate - a Potent Inhibitor of Protein Sulfation in Intact-Cells. Biochem Bioph Res Co. 1986;141:870–877. doi: 10.1016/s0006-291x(86)80253-4. [DOI] [PubMed] [Google Scholar]
- 39.Gillard M, Jia Z, Hou JJ, Song M, Gray PP, Munro TP, Monteiro MJ. Intracellular Trafficking Pathways for Nuclear Delivery of Plasmid DNA Complexed with Highly Efficient Endosome Escape Polymers. Biomacromolecules. 2014;15:3569–3576. doi: 10.1021/bm5008376. [DOI] [PubMed] [Google Scholar]
- 40.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A Two-Stage Poly(ethylenimine)-Mediated Cytotoxicity: Implications for Gene Transfer/Therapy. Mol Ther. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA. Ivermectin Is a Specific Inhibitor of Importin Alpha/Beta-Mediated Nuclear Import Able to Inhibit Replication of HIV-1 and Dengue Virus. Biochem J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M, Weis K, Heald R. Importazole, a Small Molecule Inhibitor of the Transport Receptor Importin-beta. ACS Chem Biol. 2011;6:700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roxbury D, Jena PV, Williams RM, Enyedi B, Niethammer P, Marcet S, Verhaegen M, Blais-Ouellette S, Heller DA. Hyperspectral Microscopy of Near-Infrared Fluorescence Enables 17-Chirality Carbon Nanotube Imaging. Sci Rep. 2015;5:14167. doi: 10.1038/srep14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


