Abstract
There has been growing interest in disrupting bacterial virulence mechanisms as a form of infectious disease control through the use of ‘anti-infective’ drugs. Pseudomonas aeruginosa is an opportunistic pathogen noted for its intrinsic antibiotic resistance that causes serious infections requiring new therapeutic options. In this study, an analysis of the P. aeruginosa PAO1 deduced proteome was performed to identify pathogen-associated proteins. A computational screening approach was then used to discover drug repurposing opportunities, i.e. identifying approved drugs that bind and potentially disrupt the pathogen-associated protein targets. The selective oestrogen receptor modulator raloxifene, a drug currently used in the prevention of osteoporosis and/or invasive breast cancer in post-menopausal women, was predicted from this screen to bind P. aeruginosa PhzB2. PhzB2 is involved in production of the blue pigment pyocyanin produced via the phenazine biosynthesis pathway. Pyocyanin is toxic to eukaryotic cells and has been shown to play a role in infection in a mouse model, making it an attractive target for anti-infective drug discovery. Raloxifene was found to strongly attenuate P. aeruginosa virulence in a Caenorhabditis elegans model of infection. Treatment of P. aeruginosa wild-type strains PAO1 and PA14 with raloxifene resulted in a dose-dependent reduction in pyocyanin production in vitro; pyocyanin production and virulence were also reduced for a phzB2 insertion mutant. These results suggest that raloxifene may be suitable for further development as a therapeutic for P. aeruginosa infection and that such already approved drugs may be computationally screened and potentially repurposed as novel anti-infective/anti-virulence agents.
Keywords: Anti-virulence, Pseudomonas aeruginosa, Pyocyanin, Raloxifene
1. Introduction
Pseudomonas aeruginosa is an opportunistic, Gram-negative bacterial pathogen that causes infections in immunocompromised hosts, burn victims, individuals in intensive care and patients with cystic fibrosis (CF). The lungs of nearly all CF patients are chronically colonised by P. aeruginosa, which significantly reduces life expectancy and is the leading cause of morbidity and mortality for CF patients. The effectiveness of P. aeruginosa as a pathogen can be attributed to its arsenal of virulence mechanisms and its large metabolic capacity, including the ability to intrinsically resist antibiotics owing to its impermeable outer membrane, efflux capabilities, tendency to colonise surfaces in a biofilm form and ability to acquire and maintain antibiotic plasmids [1]. Novel approaches for the treatment of P. aeruginosa infection are urgently needed.
The concept of targeting virulence using anti-infective/anti-virulence drugs that ‘disarm’ pathogenic bacteria rather than kill them has garnered increasing attention in recent years. It is believed that such an approach places less selective pressure on bacteria to evolve new strategies for survival, thereby being more specific to pathogenic bacteria and subject to less selection for drug resistance [2]. To this end, in a previous analysis we identified pathogen-associated proteins having homologues only in pathogenic bacteria and not in non-pathogens [3]. Such proteins are more likely to have virulence-related functions.
The list of pathogen-associated proteins identified included components of the phenazine biosynthesis pathway. Strains of P. aeruginosa produce and secrete a variety of redox-active phenazine compounds, the most well studied being pyocyanin. Pyocyanin is responsible for the blue–green colour characteristic of Pseudomonas spp. and is considered both a virulence factor and a quorum sensing (QS) signalling molecule for P. aeruginosa [4]. QS-regulated virulence factors, and pyocyanin in particular, are active in lung infections associated with CF and produce numerous effects that are relevant to CF (reviewed in [4,5]). Pyocyanin modulates redox cycling and generates reactive oxygen species capable of causing significant oxidative stress, which in turn affects calcium homeostasis [4,5]. It inhibits cellular respiration, depletes intracellular cAMP and ATP levels, and thus affects chloride ion channels that are controlled by ATP-driven conformational changes [5]. Pyocyanin inhibits prostacyclin release and can inactivate human V-ATPases (involved in receptor-mediated endocytosis), α1-protease inhibitor (which modulates serine protease activity, including neutrophil elastase) and nitric oxide (which influences blood flow, blood pressure and immune functions) [5]. Pyocyanin also alters the host immune response in several ways to aid evasion of the immune system and establish chronic infection. Evidence suggests that pyocyanin could prevent the development of an effective T-cell response against P. aeruginosa and prevents the activation of monocytes and macrophages (through inhibition of cytokine production) [5]. High concentrations of pyocyanin in the sputum of CF patients suggest that this compound plays a role in pulmonary tissue damage observed with chronic lung infections, and early (in the growth curve) overexpression of QS-regulated virulence factors such as LasA, elastase and pyocyanin is common among populations of a highly successful CF epidemic strain [6]. Additional evidence demonstrates that pyocyanin significantly contributes to lung destruction during chronic P. aeruginosa infection of bronchiectasis airways in a mouse model and supports the hypothesis that pyocyanin contributes to the accelerated decline in lung function of CF and other bronchiectasis patients once they are infected with P. aeruginosa [7]. Pyocyanin biosynthesis is therefore an attractive target for anti-infective drug intervention.
In P. aeruginosa PAO1, pyocyanin is synthesised from chorismate in a complex series of intermediates by enzymes encoded by the homologous phzA1B1C1D1E1F1G1 and phzA2B2C2D2E2F2G2 operons (henceforth referred to as the phz operons) [8]. Two additional genes, phzS and phzM, convert phenazine-1-carboxylic acid to pyocyanin in the final steps of biosynthesis. The two phz operons are 98.3% identical at the DNA level, with almost all of the sequence divergence occurring in the first pair of genes, phzA1B1 and phzA2B2. Products of the phzA and phzB genes are also homologous (with ca. 67% amino acid identity).
The crystal structures for several components of the phenazine biosynthesis pathway have been solved, making it possible to use computational methods to help identify potential inhibitors. A promising yet underexploited avenue for discovering new antimicrobials is through drug repositioning, using existing drugs for new indications through identification of off-target interactions. Numerous screens to identify potential antimicrobials for P. aeruginosa infection using libraries of novel drug classes and bioactive molecules have been conducted, but screens using libraries of existing approved drugs have been limited despite the obvious benefits of drug repositioning, i.e. existing drugs already have a clinical history and therefore require less time and money to develop into a drug specific for treating a different disease. To our knowledge, only one study to repurpose known drugs for the treatment of P. aeruginosa infection has been reported [9].
Pseudomonas aeruginosa is a well studied model system for pathogenesis. Genome-wide essentiality studies have provided mutant strains for its non-essential gene repertoire—tools we can use to validate computationally predicted anti-infective targets [10–12]. There are also established infection models for P. aeruginosa, including the powerful and inexpensive Caenorhabditis elegans pathogenesis model. When P. aeruginosa strain PA14 is grown on low-salt media, it accumulates in the C. elegans intestine, killing the worms relatively slowly over the course of 2–3 days. This infection-like process referred to as ‘slow killing’ differs from ‘fast killing’ on high-osmolarity media, which involves the secretion of diffusible toxins and takes place over the course of several hours [13]. Many P. aeruginosa virulence-related genes required for mammalian infection have been shown to play a role in C. elegans killing, including proteins that regulate the transcription and export of virulence factors via the type III secretion machinery as well as proteins involved in QS [14]. Novel genes not previously known to be involved in virulence have also been identified in this manner [15].
Here we describe our application of drug repurposing methodology to identify potential anti-infective drug targets and drug leads for P. aeruginosa. The computational screen identified potential interactions between PhzB2 from the phenazine biosynthesis pathway and the selective oestrogen receptor modulator raloxifene. We present evidence that raloxifene attenuates P. aeruginosa virulence and promotes C. elegans survival. A pyocyanin quantification assay demonstrated that pigment production is reduced in the presence of raloxifene. These results suggest that raloxifene could be a novel source of anti-infective agents for the treatment of P. aeruginosa infection and, furthermore, that drug repurposing is a promising, efficient approach for novel antimicrobial drug discovery.
2. Materials and methods
2.1. Database of protein–drug complexed structures
Drug repurposing uses existing approved drugs for new indications by targeting a new site or sites of interest on a protein involved in a different disease. A database of all protein–drug complexed structures was generated by searching the RCSB Protein Data Bank (PDB) (http://www.pdb.org) for entries having a ligand bound to them and then cross-matching the ligand entries to the DrugBank database [16]. Ligands in DrugBank with fewer than 10 non-hydrogen atoms, or that were known to be additives from crystallographic experiments, were considered as ‘solvents’ and were removed from the data set. DrugBank ligands that were labelled as ‘approved’ but not considered ‘nutraceutical’ or ‘solvents’ were retained, resulting in 1059 ligands bound to proteins with structures available in the PDB. Additional filtering eliminated structures containing DNA/RNA ligands only, C-alpha atoms only, large ribosomal subunits, or where the ligand was a modified residue. In total, 232 drugs are represented across 940 PDB entries. The distribution is skewed towards drugs that are more commonly crystallised or better studied. Detailed information on the drugs and PDB entries can be found at http://funsite.sdsc.edu/drugome/TB/.
2.2. Drug repurposing analysis based on ligand binding site similarity to identify novel drug leads
The computational drug repurposing approach applied in this study was based on an approach used previously to repurpose an existing drug in treating drug-resistant tuberculosis (TB) [17], to reconstruct TB-drugome (a structural proteome-scale drug-target network in Mycobacterium tuberculosis), to reveal molecular mechanisms of anti-cancer activity of HIV protease inhibitors, and to identify side-effect profiles of cholesterylester transport protein inhibitors and oestrogen receptor modulators (reviewed in [18]). Briefly, it involves extracting the binding site of a commercially available drug from a three-dimensional structure or model of the target protein, identifying off-targets with similar binding sites across the P. aeruginosa structural proteome using SMAP software [19,20], and evaluating the atomic interactions between the putative off-targets and the drug using protein–ligand docking. A total of 20 targets (see Supplementary Table S1) were chosen from an initial set of 207 P. aeruginosa proteins with structures deposited in the PDB based on our classification of pathogen-associated genes [3], flux balance analysis with a genome-scale metabolic reconstruction of the strain [21] and the ligand binding site similarity network. These were further screened using a bidirectional BLAST analysis to the human genome to avoid potentially lethal side effects. The nine remaining targets were then subjected to the drug repurposing approach to compare their binding sites with those in the protein–drug complexed database described above. A P-value cut-off of 1e-4 for ligand binding site similarity was used to identify potential target–drug interactions. Docking was performed using Glide [22] to validate potential interactions, and the top binding poses were manually inspected for hydrophobic interactions and potential hydrogen bonds. This initial study did not consider closely related homologous structures in other organisms.
2.3. Bacterial strains and compounds/drugs for laboratory assays
The bacterial strains used in this study are listed in Table 1. Two wild-type P. aeruginosa strains were studied, PAO1 [23] and PA14 [24], plus phzB1 and phzB2 transposon insertion mutants for both strains PAO1 and PA14 [9,11]. Escherichia coli OP50 was used as a food source and non-pathogenic control for C. elegans infection studies. Raloxifene was purchased from Cayman Chemical (Ann Arbor, MI). Adenosine, chenodeoxycholic acid, dexamethasone, oestradiol, fusidic acid, spironolactone, trifluoperazine dihydrochloride, triiodothyronine (T3), trimethoprim and thyroxine were purchased from Sigma-Aldrich Canada (Oakville, ON, Canada). The stock solution of raloxifene was prepared with dimethyl sulphoxide and oestradiol with methanol.
Table 1.
Bacterial strains used in this study
| Strain | Phenotype | Reference |
|---|---|---|
| Escherichia coli OP50 | Uracil auxotroph with limited growth on NGM plates | [30] |
| Pseudomonas aeruginosa PA14 | Wild-type (highly virulent clinical isolate used as a reference strain; capable of infecting multiple diverse hosts) | [24] |
| PA14_phzB1 | PA14_09470 transposon mutant (mutant ID 24980) | [11] |
| PA14_phzB2 | PA14_39960 transposon mutant (mutant ID 48282) | [11] |
| PA14_lpxC | PA14_57260 transposon mutant (mutant ID 34855) | [11] |
| P. aeruginosa PAO1 | Wild-type (standard laboratory strain and genetic reference strain) | [23] |
| PAO1_phzB1 | PA4211 transposon mutant (mutant ID 22096) | [9] |
| PAO1_phzB2.1 | PA1900 transposon mutant (mutant ID 21450) | [9] |
| PAO1_phzB2.2 | PA1900 transposon mutant (mutant ID 52014) | [9] |
NGM, nematode growth medium.
2.4. Growth assays
Stock cultures of P. aeruginosa PAO1 and PA14 as well as the mutant strains were provided by Prof. Bob Hancock (University of British Columbia, Vancouver, Canada). A single colony of the bacterium was used to inoculate 5 mL of Luria–Bertani (LB) broth, which was then incubated overnight (<16 h) at 37 °C. A 0.3 mL aliquot was taken from the overnight culture and transferred to 30 mL of LB broth containing 0.1 mM of one of the approved drugs mentioned above (or in the case of raloxifene and oestradiol, 100 μg/mL). Growth at 37 °C was monitored by absorbance at 600 nm. Measurements were blanked with LB broth containing an equivalent amount of the drug being tested. Growth curves were performed a minimum of two times.
2.5. Infection model to evaluate mutants and drugs
Caenorhabditis elegans(wild-type Bristol N2) were maintained on nematode growth medium (NGM) plates by feeding on E. coli OP50 lawns. Pseudomonas aeruginosa PAO1, PA14 and transposon mutants were grown on LB culture medium. Infection of C. elegans (L4 stage) with PAO1, PA14 or transposon mutants was performed at 24 °C. Fresh NGM plates with 0.35% peptone were seeded with bacterial cultures (50 μL of overnight culture per 3.5 cm diameter plates). The cultures were incubated at 37 °C overnight and were then equilibrated at 24 °C for another 24 h. To minimise the confounding effects of progeny development, the NGM plates were supplemented with 5-fluorodeoxyuridine (100 μg/mL) 1 h before picking of worms. In each assay, 30–40 worms were transferred onto each plate. Worms were evaluated for viability for several days (up to 120 h). Worms were considered dead when they did not move and then did not respond to touch (moving worms were not touched to reduce stress on them). For drug assays, drugs were incorporated at 0.1 mM on low-osmolarity NGM inoculated with PAO1, PA14 or mutants. All experiments were performed at a minimum in triplicate.
2.6. Pyocyanin production
Production of the pyocyanin pigment was measured using a quantitative chemical assay. Absorbance at 520 nm in acidic solution was measured as described by Essar et al. [25]. Briefly, bacteria from 1 mL of culture were pelleted. The supernatant containing the pyocyanin was then extracted with 0.6 mL of chloroform, followed by extraction with 1 mL of 0.2 M HCl and measurement of the solution’s absorbance at 520 nm. The concentration of pyocyanin in the supernatant was expressed in μg/mL by multiplying the absorbance by 17.072.
3. Results
3.1. Computationally predicted interactions between approved drugs and Pseudomonas aeruginosa drug targets
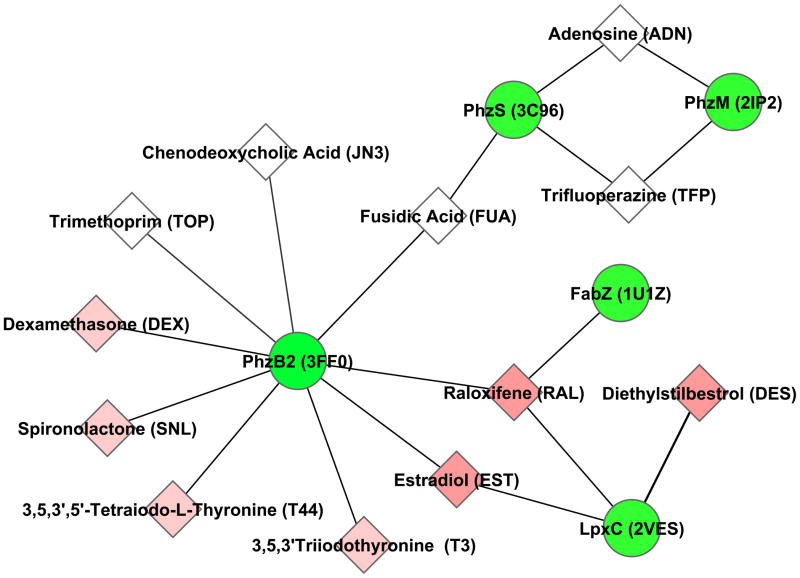
Using an efficient functional site search algorithm, the binding sites of US Food and Drug Administration (FDA)-approved drugs crystallised with their respective ligands were extracted and compared with binding pockets on the surface of nine target proteins, including the PhzB2, PhzM and PhzS members of the phenazine biosynthesis pathway. Within the network generated from this analysis (Fig. 1), PhzB2 met the criteria of being pathogen-associated (having homologues only in pathogenic bacteria and not in non-pathogens) and likely to be non-essential based on transposon mutagenesis studies [9–11], making it an attractive anti-infective drug target. PhzB1, the homologous protein in the alternate phz operon that produces pyocyanin, was not included in the drug repurposing analysis (and thus the drug network) because its crystal structure had not been experimentally determined. However, it is an equally attractive target because PhzB1 and PhzB2 are 91% identical at the amino acid level, with almost all of the variation occurring in the first 25 residues. These residues are excluded from the ligand binding pocket used in this analysis, making the predicted drug interactions just as likely for PhzB1 as they are for PhzB2.
Fig. 1.
Computational drug repurposing analysis predicts interactions between Pseudomonas aeruginosa phenazine biosynthesis proteins and known drugs. Green circles denote P. aeruginosa proteins labelled with gene name followed by Protein Data Bank structure identifier in brackets. Diamonds denote known drugs predicted to interact with the P. aeruginosa proteins (identified by name and heterogen molecule ID in brackets), with coloured nodes indicating hormones or hormone receptor antagonists (darker nodes indicate oestrogenic compounds). Edges in the graph represent significant binding pocket similarity (using the SMAP software at P ≤ 1e–4) to one or more existing approved drugs in the database of protein–drug complexed structures that was developed (see Section 2.2).
Thirteen drugs were predicted to bind to PhzB2. Each of these was docked to the PhzB2 protein model to estimate the relative protein–ligand binding affinities for each drug. Nine compounds were selected for further study based on their availability from drug vendors (see Supplementary Table S2). An additional two compounds predicted to bind to PhzS and PhzM (trifluoperazine dihydrochloride and adenosine) were also selected for testing (Fig. 1).
3.2. Effect of compounds on Pseudomonas aeruginosa viability in vitro
To confirm that none of the drugs tested were antimicrobial (as part of efforts to identify anti-infective drugs that would not kill the bacteria but rather disrupt virulence), the drugs were screened for their effect on growth of wild-type P. aeruginosa in vitro. All of the drugs tested had little to no effect on growth of wild-type P. aeruginosa PAO1 or PA14 growth in LB broth at 37 °C as measured at 600 nm (see Fig. 2C and Supplementary Fig. S1).
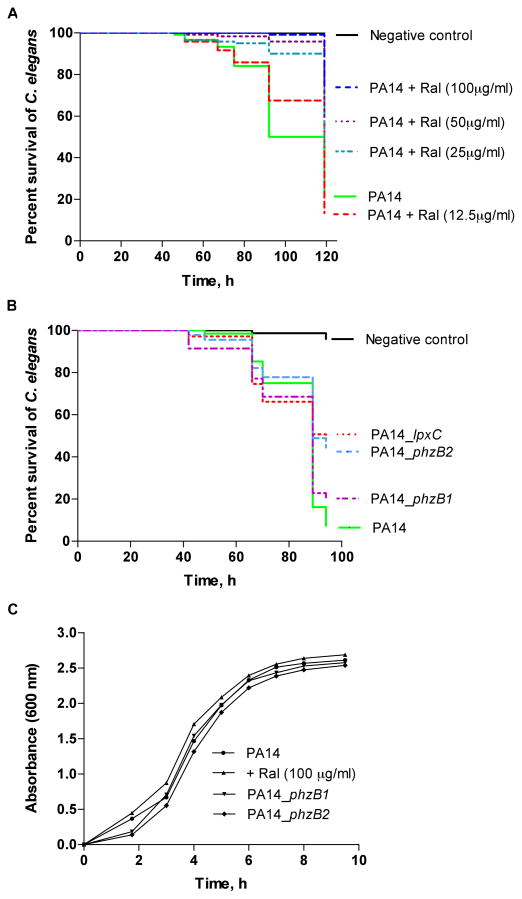
Fig. 2.
Raloxifene treatment and infection with phzB1/2 mutants attenuates killing of Caenorhabditis elegans by Pseudomonas aeruginosa PA14. (A) Survival of C. elegans following exposure to infection with P. aeruginosa PA14 plus raloxifene at varying doses. The negative control is non-pathogenic Escherichia coli OP50. (B) Survival of C. elegans following infection with P. aeruginosa transposon insertion mutants for phzB1, phzB2 and lpxC versus the wild-type PA14 positive control and non-pathogenic E. coli OP50 negative control. (C) Growth curve for P. aeruginosa PA14 cultures with and without the presence of raloxifene as well as for P. aeruginosa phzB1/2 insertion mutants. Cell density was measured at a wavelength of 600 nm. The results indicate that P. aeruginosa infection is attenuated by raloxifene, but not due to a direct effect on the viability of P. aeruginosa as measured in vitro.
3.3. Inhibition of Pseudomonas aeruginosa pathogenicity by raloxifene
The compounds were tested for attenuated P. aeruginosa PAO1 and PA14 virulence in the C. elegans infection model (Supplementary Fig. S1). The most notable attenuation of C. elegans killing was observed for raloxifene, which promoted C. elegans survival compared with controls lacking raloxifene without affecting in vitro growth (Fig. 2A, C). The effect was dose-dependent, with 75–95% survival observed at the highest raloxifene concentration of 100 μg/mL (or 0.21 mM) versus 20–25% survival when infected with wild-type PA14 with no raloxifene present (representative example shown in Fig. 2A). Raloxifene is an oral selective oestrogen receptor modulator that is used for the prevention of osteoporosis in post-menopausal women and for the reduction of risk of invasive breast cancer [26] and has not been previously reported to have anti-infective properties for treatment of any infectious disease.
Two other compounds attenuated P. aeruginosa virulence. Addition of triiodothyronine (T3), one of the two natural thyroid hormones, to the culture medium had little effect on P. aeruginosa PAO1 or PA14 growth in vitro but attenuated C. elegans killing (Supplementary Fig. S1) and reduced pyocyanin production (data not shown). Oestradiol, predicted to bind PhzB2 and LpxC, exhibited anti-infective properties but attenuated virulence to a lesser degree than either raloxifene or T3 (see Supplementary Fig. S2). Evaluation of the Glide docking results revealed that raloxifene binds to PhzB2 with a more favourable score than T3 or oestradiol (Supplementary Table S2), correlating with observations that raloxifene is a more effective inhibitor of P. aeruginosa virulence.
3.4. Treatment of Pseudomonas aeruginosa with raloxifene reduces pyocyanin production
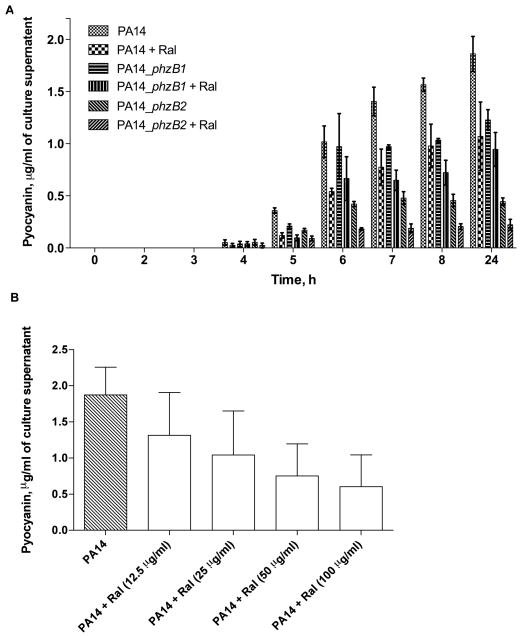
A quantitative chemical assay was used to measure the amount of pyocyanin produced by P. aeruginosa PA14 in the presence and absence of raloxifene. Fig. 3A shows a clear reduction in pyocyanin pigment production compared with control cultures 4–10 h after addition of raloxifene, supporting the possible inhibition of the phenazine biosynthesis pathway. The inhibition of pyocyanin occurred in a dose-dependent manner (Fig. 3B).
Fig. 3.
Wild-type Pseudomonas aeruginosa PA14 treated with raloxifene and PA14 phzB1/2 mutants show reduced pyocyanin biosynthesis. (A) Quantification of pyocyanin production (in acidic solution) in wild-type P. aeruginosa PA14 and P. aeruginosa phzB1/2 insertion mutants, with and without the addition of 100 μg/mL raloxifene. (B) Dose-dependent inhibition of pyocyanin biosynthesis for P. aeruginosa PA14 treated with raloxifene at the specified doses. Quantification was performed on overnight cultures.
The amount of pyocyanin produced after treatment with raloxifene was similar to that of a PA14 mutant lacking a functional copy of PhzB2 which results in disruption of the phenazine biosynthesis pathway. Pyocyanin production for the phzB2 insertion mutant was comparable with that of P. aeruginosa treated with 100 μg/mL raloxifene (Fig. 3A). Pyocyanin production for the PA14 phzB1 insertion mutant was slightly reduced.
Pyocyanin production was correlated with C. elegans survival. Worms infected with the phzB2 mutant survived longer than worms infected with the wild-type PA14 strain (Fig. 2B). The phzB1 mutant showed slightly reduced virulence for strain PA14. Analogous experiments were conducted for strain PAO1. Similar effects were observed for raloxifene treatment and the phzB2 mutant; however, the phzB1 mutant appeared to have a stronger effect on survival in strain PAO1 compared with PA14 (Fig. 2B and Supplementary Fig. S3).
4. Discussion
This study investigated the possibility of repurposing existing drugs to treat P. aeruginosa infections as well as targeting virulence rather than essential cellular functions through an initial computational screen. Raloxifene was found to strongly protect C. elegans against killing by P. aeruginosa PAO1 and PA14, with the effect likely due to inhibition of phenazine biosynthesis. Pyocyanin production is required for Pseudomonas pathogenesis in plants, and increased levels of pyocyanin have been observed in Pseudomonas–mouse burn and mouse lung infection models. These observations strongly suggest that pyocyanin is a conserved virulence factor with important roles in the success of P. aeruginosa to infect hosts of diverse evolutionary origin [12,27]. Thus, inhibition of pyocyanin synthesis in the C. elegans model is likely to be relevant to mammalian systems as well.
One concern might be that computational docking is known to have high false-positive rates; however, the similarity in the binding sites cannot be ignored. In any case, regardless of whether raloxifene binds to the exact location predicted using this approach, phenazine biosynthesis is clearly impacted by raloxifene treatment, and virulence in the C. elegans model is attenuated. Another concern might be that the effects observed are the result of interference with QS. Preliminary data suggest that this is not the case given that Pseudomonas quinolone signal (PQS), a key QS molecule that regulates the expression of a subset of genes belonging to the QS regulon, including the phz operon [28], is not impacted by the addition of raloxifene (Supplementary Fig. S4).
Of the 11 compounds initially selected for testing (9 with predicted PhzB2 interactions and 2 with predicted PhzM and PhzS interactions), 3 showed promise as anti-infective drug candidates—a success rate of 27%. This is significantly higher and more efficient than the 0.25% hit rate obtained from an untargeted screen for anti-infective agents for Enterococcus faecalis [29]. The failure of some of the computationally predicted compounds to attenuate virulence is not surprising given the biases present in selecting protein binding sites as well as subjectivity inherent in preparing molecules for molecular docking studies. However, it is notable that the computational methods employed in this study accelerated the ability to identify new drug leads for P. aeruginosa.
From a practical standpoint, this represents only the first stage in a process to eventually demonstrate efficacy for treatment of human CF-associated infections. For example, drug delivery mechanisms and dosage levels required to be effective in the microaerophilic/biofilm conditions that are relevant to CF remain to be determined. It should also be appreciated that potential anti-infective drugs, such as those identified here, represent treatments that would not directly kill the bacterium, but through ‘disarming’ them would allow the host immune system to potentially clear the infection without damage to the host occurring, or limit the ability of the bacterium to gain/maintain a foothold in the host environment. However, the lack of direct killing of the bacterium should reduce selective pressure for the bacterium to develop resistance to the drug, making such anti-infective drugs attractive as new, potentially quite sustainable options for infectious disease control. This study demonstrates that anti-infective activity can be found among existing collections of FDA-approved compounds, which may represent an untapped reservoir of drugs for infectious disease control.
Supplementary Material
Acknowledgments
Funding: This work was supported principally by the Simon Fraser University Community Trust and National Institute of Health GM078596, with additional partial support by the Cystic Fibrosis Foundation, Genome Canada, and the Natural Sciences and Engineering Research Council of Canada. SJHS is supported by a Canadian Cystic Fibrosis Foundation fellowship. FSLB is a Michael Smith Foundation for Health Research Senior Scholar.
Footnotes
Competing interests: None declared.
Ethical approval: Not required.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67:351–68. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- 2.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–8. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 3.Ho Sui SJ, Fedynak A, Hsiao WW, Langille MG, Brinkman FS. The association of virulence factors with genomic islands. PLoS One. 2009;4:e8094. doi: 10.1371/journal.pone.0008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau GW, Hassett DJ, Ran H, Kong F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med. 2004;10:599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Winstanley C, Fothergill JL. The role of quorum sensing in chronic cystic fibrosis Pseudomonas aeruginosa infections. FEMS Microbiol Lett. 2008;290:1–9. doi: 10.1111/j.1574-6968.2008.01394.x. [DOI] [PubMed] [Google Scholar]
- 6.Fothergill JL, Panagea S, Hart CA, Walshaw MJ, Pitt TL, Winstanley C. Widespread pyocyanin over-production among isolates of a cystic fibrosis epidemic strain. BMC Microbiol. 2007;7:45. doi: 10.1186/1471-2180-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldwell CC, Chen Y, Goetzmann HS, Hao Y, Borchers MT, Hassett DJ, et al. Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am J Pathol. 2009;175:2473–88. doi: 10.2353/ajpath.2009.090166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183:6454–65. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, Rybtke MT, Jakobsen TH, Hentzer M, Bjarnsholt T, Givskov M, et al. Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob Agents Chemother. 2009;53:2432–43. doi: 10.1128/AAC.01283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100:14339–44. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewenza S, Falsafi RK, Winsor G, Gooderham WJ, McPhee JB, Brinkman FS, et al. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 2005;15:583–9. doi: 10.1101/gr.3513905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A. 2006;103:2833–8. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa–Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 14.Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P.aeruginosavirulence factors. Proc Natl Acad Sci U S A. 1999;96:2408–13. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aballay A, Ausubel FM. Caenorhabditis elegans as a host for the study of host–pathogen interactions. Curr Opin Microbiol. 2002;5:97–101. doi: 10.1016/s1369-5274(02)00293-x. [DOI] [PubMed] [Google Scholar]
- 16.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–6. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinnings SL, Liu N, Buchmeier N, Tonge PJ, Xie L, Bourne PE. Drug discovery using chemical systems biology: repositioning the safe medicine Comtan to treat multi-drug and extensively drug resistant tuberculosis. PLoS Comput Biol. 2009;5:e1000423. doi: 10.1371/journal.pcbi.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie L, Xie L, Kinnings SL, Bourne PE. Novel computational approaches to polypharmacology as a means to define responses to individual drugs. Annu Rev Pharmacol Toxicol. 2012;52:361–79. doi: 10.1146/annurev-pharmtox-010611-134630. [DOI] [PubMed] [Google Scholar]
- 19.Xie L, Bourne PE. A robust and efficient algorithm for the shape description of protein structures and its application in predicting ligand binding sites. BMC Bioinformatics. 2007;8(Suppl 4):S9. doi: 10.1186/1471-2105-8-S4-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie L, Bourne PE. Detecting evolutionary relationships across existing fold space, using sequence order-independent profile–profile alignments. Proc Natl Acad Sci U S A. 2008;105:5441–6. doi: 10.1073/pnas.0704422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberhardt MA, Puchalka J, Fryer KE, Martins dos Santos VA, Papin JA. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J Bacteriol. 2008;190:2790–803. doi: 10.1128/JB.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 23.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–64. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 24.Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Essar DW, Eberly L, Hadero A, Crawford IP. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee WL, Chao HT, Cheng MH, Wang PH. Rationale for using raloxifene to prevent both osteoporosis and breast cancer in postmenopausal women. Maturitas. 2008;60:92–107. doi: 10.1016/j.maturitas.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Lau GW, Ran H, Kong F, Hassett DJ, Mavrodi D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect Immun. 2004;72:4275–8. doi: 10.1128/IAI.72.7.4275-4278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, et al. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A. 2004;101:1339–44. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moy TI, Conery AL, Larkins-Ford J, Wu G, Mazitschek R, Casadei G, et al. High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem Biol. 2009;4:527–33. doi: 10.1021/cb900084v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.