Abstract
RAS and its downstream cascades transmit cellular signals resulting in increased transcription of genes involved in cell growth and division. Protein-coding gene targets of RAS signaling have been characterized extensively, but long non-coding RNAs (lncRNA) regulated by these processes have not. Using a custom-designed lncRNA microarray, we identified the lncRNA Orilnc1 as a genetic target of RAS that is critical for RAS oncogenicity. Orilnc1 expression was regulated by RAS-RAF-MEK-ERK signaling via the transcription factor AP1. Orilnc1 was highly expressed in BRAF-mutant cancers such as melanoma. Silencing of Orilnc1 blocked tumor cell proliferation and growth in vitro and in vivo. Additionally, Orilnc1 blockade reduced expression of Cyclin E1 and induced G1/S cell cycle arrest in tumor cells. Taken together, our results identify Orilnc1 as a novel, non-protein mediator of RAS/RAF activation which may serve as a therapeutic target in RAS/RAF-driven cancers.
INTRODUCTION
High-throughput sequencing studies have demonstrated that up to 70% of the human genome is transcribed into RNA (1, 2). Long non-coding RNAs (lncRNAs) are operationally defined as transcripts larger than 200 nt that do not appear to have protein-coding potential (2–11). Like protein-coding genes, lncRNA genes contain introns and exons, with fewer exons, transcripts are processed by the same spliceosome machinery and their transcription is subject to the same histone modification-mediated regulation (2–11). In addition, lncRNA genes exhibit greater cross-species diversity suggesting weaker selective evolutionary constraints. LncRNAs are generally in low abundance but often expressed in a strikingly cell type- and tissue-specific (12–14), and in many cases, primate-specific (1, 2). Recently lncRNAs have been revealed to play important roles during cancer initiation and progression (2–11). At a cellular level, lncRNAs have been implicated in cell proliferation, differentiation, migration, and apoptosis. At the molecular level, lncRNAs interact with proteins and other molecules to function as scaffolds or guides to regulate protein-protein or protein-DNA/RNA interactions, as decoys to bind proteins or miRNAs, and as enhancers to influence gene transcription, when transcribed from enhancer regions or their neighboring loci. Most importantly, lncRNAs are highly deregulated in tumors (15, 16).
Three related RAS proteins (HRAS, KRAS and NRAS) belong to the small GTPase protein family, and function as binary molecular switches that control intracellular signaling networks in normal cells (17–21). They are ubiquitously expressed in nearly all cell lineages and organs in humans. Mutant RAS is constitutively activated, promotes uncontrollable growth and serves as a central oncogene in a third of human cancers, including a high percentage of pancreatic, lung, and colorectal cancers, as well as acute lymphocytic leukemia and melanoma (17–21). RAS activates several signal transduction pathways, such as the mitogen-activated protein (MAP) kinase and the phosphatidylinositide 3-kinase (PI3K) cascades. Many genes coding for proteins in these downstream pathways are also highly altered in human cancers. For example, the BRAF serine/threonine-protein kinase gene is mutated in about half of patients with melanoma (22–27). RAS and its downstream signaling cascades transmit cellular signals and result in the transcription of a large number of genes involved in cell growth and division. During the last two decades, many protein-coding genes have been identified as RAS-regulated genes, which mediate the functions of RAS in proliferation, cell death and other cellular processes (17–21). Characterization of these downstream genes has greatly contributed to development of novel therapeutic methods to target the RAS pathway. However, most studies have focused on protein-coding genes. The discovery of non-coding RNAs, such as small non-coding RNAs - microRNA (miRNA), has dramatically changed our understanding of RAS signaling. For example, RAS activation leads to repression of miR-143/145, which regulate KRAS and RREB1, and functions as a feed-forward mechanism that potentiates RAS signaling (28). However, research on long non-coding transcripts regulated by the RAS signal cascades is still in its infancy. In the present study, using a custom-designed lncRNA microarray, we identified a novel lncRNA (oncogenic RAS-induced lncRNA 1, Orilnc1), which is regulated by the RAS-RAF-MEK-ERK pathway and is required for cell proliferation in RAS/BRAF-driven human cancers.
MATERIALS AND METHODS
Cell lines
Cell lines MDA-MB-231, MDA-MB-435, MCF10A, IMR90, SK-MEL-2 and LOX-IMVI were purchased from ATCC. The other melanoma cell lines (Table S1) were received from a Melanoma Cell Line Collection Bank (MCLCB) at the Melanoma Research Center, Wistar Institute. Each cell line was tested and authenticated by ATCC or MCLCB, based on morphological, cytogenetic, and DNA profile analysis such as short tandem repeat fragment analysis for characterization and authentication of cell lines. The cells were tested for mycoplasma at ATCC and MCLCB, and were not tested again in the author’s laboratory. Cell lines were obtained directly from ATCC or MCLCB. The cell lines were received within the last 3 years, propagated, expanded, and frozen immediately into 18 aliquots after arrival. The cells were passaged in the author’s laboratory for fewer than 6 months after receipt, and their identity was not formally verified for this study. The cells from our frozen stock of author’s laboratory were used within 5 to 10 passages and not exceeding a period of 3 months.
Custom-designed lncRNA microarray
A 60-mer oligonucleotide microarray with a total of 14,262 probes for 2,965 lincRNAs (an average of 5 probes for each lincRNA), was custom manufactured using Agilent’s SurePrint oligo technology. Probes for 11,081 protein coding genes were also included in the microarray platform. Total RNA was converted to cDNA using random primers. Labeling and hybridization were performed by the Molecular Profiling Facility according to manufacturer’s instruction.
Microarray analysis
The normalized microarray data were managed and analyzed by GeneSpring (Agilent), microarray software suite 4, BRB-ArrayTools version 3.6, and GenePattern. Gene Expression Omnibus access number: GSE90044.
Lentiviral transduction
pCDH-KRAS [G12D] and pCDH-Ctrl for KRAS were generated for the expression in IMR90 cells. The pCDH empty vector was purchased from System Biosciences. Lentiviral shRNAs targeting KRAS, NRAS, BRAF and Orilnc1 were constructed. pLKO.1 empty vector was purchased from OpenBiosystms. Non-target shRNA (SHC002, Sigma) was used as control. Lentiviral vector and packaging vectors were transfected into the packaging cell line 293T (ATCC) using the FuGENE6 Transfection Reagent (Promega). The media was changed 8 hours post-transfection and the media containing lentivirus was collected 48 hours later. Tumor cells were infected with lentivirus in the presence of 8 μg/ml of polybrene (Sigma). Overexpress and knockdown efficiency were detected 72 hours after infection. The shRNA sequences are listed below:
| shKRAS-F | 5′-CCGGGACGAATATGATCCAACAATACTCGAGTATTGTTGGATCATATTCGTCTTTTTG-3′ |
| shKRAS-R | 5′-AATTCAAAAAGACGAATATGATCCAACAATACTCGAGTATTGTTGGATCATATTCGTC-3′ |
| shBRAF-F | 5′-CCGGTTGGTTGGGACACTGATATTTCTCGAGAAATATCAGTGTCCCAACCAATTTTTG-3′ |
| shBRAF-R | 5′-AATTCAAAAATTGGTTGGGACACTGATATTTCTCGAGAAATATCAGTGTCCCAACCAA-3′ |
| shNRAS-F | 5′-CCGGCACAACTTTGACGTAATTAAACTCGAGTTTAATTACGTCAAAGTTGTGTTTTTG-3′ |
| shNRAS-R | 5′-AATTCAAAAACACAACTTTGACGTAATTAAACTCGAGTTTAATTACGTCAAAGTTGTG-3′ |
| shOrilnc1-1F | 5′-CCGGCTAGGGCAGTGGTTCTCAAATCTCGAGATTTGAGAACCACTGCCCTAGTTTTTG-3′ |
| shOrilnc1-1R | 5′-AATTCAAAAACTAGGGCAGTGGTTCTCAAATCTCGAGATTTGAGAACCACTGCCCTAG-3′ |
| shOrilnc1-2F | 5′-CCGGGATTGATCAAAGGGCTCTTAACTCGAGTTAAGAGCCCTTTGATCAATCTTTTTG-3′ |
| shOrilnc1-2R | 5′-AATTCAAAAAGATTGATCAAAGGGCTCTTAACTCGAGTTAAGAGCCCTTTGATCAATC-3′ |
RNA isolation and Quantitative real time RT-PCR (qRT-PCR)
Total RNA was extracted using TRIzol Reagent (Invitrogen) and reverse-transcribed using High Capacity RNA-to-cDNA Kit (Applied Biosystems) under conditions provided by the supplier. cDNA was quantified by qRT-PCR using ABI ViiA 7 System (Applied Biosystems). PCR was performed using SYBR Green reagents (Applied Biosystems) according to manufacturer’s instructions. GAPDH was used as an internal control. Primers used for qRT-RCR are listed below:
| qRT-PCR primer | 5′ | 3′ |
| OTTHUMT00000046024 | GGGTGAGGTACAGGCTCAGT | GCAGCACTCTGCTCACATTC |
| OTTHUMT00000046025 | CCAGCTGTGAAAGGACTGCT | TCCCACCAACTCTTGGTACTG |
| OTTHUMT00000091113 | CAGCAGGTGGATATGGGACT | ACAAGGGAGCGCTTTATCCT |
| OTTHUMT00000256182 | CATCTGGAAAGGGGACAACT | GCATACAAGACCCAGGAACG |
| OTTHUMT00000326402 | TTTGGGAGGCCTTACTTCTG | TCACAGTCATGGCCTTGGTA |
| OTTHUMT00000328267 | GCTGGAGTGAGGAAGCTGAC | GGCGCTCCTAATTCTTGTTG |
| BMP2 | TGGACGCTCTTTCAATGGAC | GGAAGCAGCAACGCTAGAAG |
| SMAD6 | ACCTCCCTACTCTCGGCTGT | GCAGTGATGAGGGAGTTGGT |
| KRAS | CAAGAGTGCCTTGACGATACAG | CCAAGAGACAGGTTTCTCCATC |
| NRAS | ATACAAAACAAGCCCACGAACT | AAAAGCATCTTCAACACCCTGT |
| BRAF | ACACGCCAAGTCAATCATCC | CCCACTCCATCGAGATTTCA |
Reporter Assay
The ~3000bp promoter region of Orilnc1 and the different truncations of this promoter region were cloned into pGL3 luciferase reporter vector. Cells were plated on a 24-well plate. Reporter plasmid was transfected using FuGENE6 transfection reagent alone or with mutant KRAS or mutant BRAF. At 48 hours after transfection, cells were harvested and reporter assays were performed using a dual luciferase reporter assay system (Promega) according to the manufacturer’s instructions. Renilla and firefly luciferase activities were measured using a Fluoroskan Ascent FL fluorometer (Thermo Fisher Scientific). The primers for cloning were listed as following:
| Promoter2k F | 5′-CGG GGTACC CTAAGACGTGACTTGCTCTTCC-3′ |
| Promoter3k F | 5′-CGG GGTACC CCTCTCTTGGATGGGTGAAA-3′ |
| Promoter R | 5′-CCG CTCGAG CACCAGATTCAAATATGCAGAAG-3′ |
| PM-AP-1 | 5′-CCT GTC CAG ATA ATC ACC AGT GAT CAT CCT GTA TAA ATG-3′ |
| 5′-CAT TTA TAC AGG ATG ATC ACT GGT GAT TAT CTG GAC AGG-3′ | |
| PM-AP-2 | 5′-CCT GTC CAG ATA ATC CAC TAT GAT CAT CCT GTA TAA ATG-3′ |
| 5′-CAT TTA TAC AGG ATG ATC ATA GTG GAT TAT CTG GAC AGG-3′ |
ChIP assay
Cells were cross-linked in 1% formaldehyde at room temperature for 10min. Crosslinking was terminated by the addition of 125mM (final concentration) glycine. Cells were incubated with glycine at room temperature for 5min with gentle mixing. Cells were then washed with ice-cold PBS, scraped and resuspended in PBS. After being washed with PBS, cell pellets were lysed by cell lysis buffer (10mM Tris-HCl (pH 7.5), 10mM NaCl, 3mM MgCl2, 0.5% IGEPAL, 1mM PMSF), keep the lystate on ice for 10min, centrifuge 5000rpm for 5min at 4°C to pellet the nuclei. The nuclei pellets were lysed by nuclear lysis buffer (50mM Tris-HCl (pH 8.0), 10mM EDTA, 1% SDS, 1mM PMSF and proteinase inhibitor cocktail) then diluted by IP dilution buffer (16.7mM Tris-HCl(pH 8.0), 1.2mM EDTA, 1.1% Triton X-100, 167mM NaCl, 0.01% SDS). Nuclear lysates were sonicated and the debris was removed by centrifugation. c-Jun antibody or IgG antibody (Cell Signaling) were mixed with clear nuclear lysates for immunoprecipitation. DNA was purified from the precipitates then quantified using real-time PCR. Primers are listed below:
| ChIP-AP-1 | 5′-GGGATCCCTGCCTACGTAAA-3′ |
| 5′-TGCTAGCCAACAATTGAACG-3′ | |
| ChIP-AP-2 | 5′-TTGTTGGCTAGCAACCTGTG-3′ |
| 5′-TCTTGGAAGAGGAGGAGGAG-3′ | |
| ChIP-AP-3 | 5′-AGTTCTGAGGCTATCTCCTCCT-3′ |
| 5′-CCTCCAAACAACATTGGCTTA-3′ | |
| ChIP-AP-4 | 5′-AGCCAATGTTGTTTGGAGGT-3′ |
| 5′-CAGCTCCAATGCACAAAAAG-3′ |
Cell viability assay
Cells were seeded in 96-well plates. 24 hours later, MTT assays (in 96-well plate) were performed at different time points (24hr, 48hr, 72hr, 96hr). The MTT assay was conducted with the Cell Proliferation Kit (I) (Roche) according to the manufacturer’s instructions. The resulting colored solution was quantified using an ELx800 Absorbance Microplate Reader (BioTek) at 570 nm with a reference wavelength of 630 nm.
Soft agar assay
The bottom layer was prepared with 0.8% agarose (Invitrogen) solution in culture medium into 6-well plate. Allow the gel to be set for 20min. Cells (2.5 × 103) were resuspended in 0.4% top agarose solution (in culture medium) then add on top of the bottom agarose in 6-well plates carefully. The plates were incubated at 37 °C with 5% CO2 until colonies were formed. Cells were fed 1–2 times per week with cell culture medium. After 2 to 3 weeks, cell colonies were stained by crystal violet (Sigma) and counted under a microscope.
Xenograft model in vivo
Six to eight week old female nude mice (Harlan Laboratories) were used for xenograft assay. Control shRNA and two different Orilnc1 shRNAs were stably expressed in MDA-MB-231 cells and 1205Lu cells. These stable cells were trypsinized and suspended in PBS. A total volume of 0.1 ml PBS containing 1205Lu cells (1×106) or MDA-MB-231 cells (1.5×106) were injected subcutaneously into the mouse flank respectively. Approximately 12 and 15 days later, tumors were detectable and tumor size was measured by a vernier caliper. Tumor volumes were calculated by the formula V= ½ L×W2, where L is the length (longest dimension) and W is the width (shortest dimension). The animal study protocol was reviewed and approved by the institutional animal care and use committee (IACUC) of the University of Pennsylvania.
Protein Isolation and western blot
Cells were lysed in mammalian protein extraction reagent (Pierce). After quantification using a BCA protein assay kit (Pierce), total protein was separated by SDS-PAGE under denaturing conditions, and transferred to PVDF membranes (Millipore). Membranes were blocked in 5% non-fat milk (Bio-Rad) and then incubated with anti-Cyclin E1(Cell Signaling) antibody, followed by incubation with an anti-rabbit secondary antibody conjugated with horseradish peroxidase (GE healthcare life sciences) together with HRP-conjugated primary antibody for beta-actin (Sigma). Immunoreactive proteins were visualized using the LumiGLO chemiluminescent substrate (Cell Signaling).
RPPA Sample Preparation
The cells were collected and washed twice in ice-cold PBS. Cell pellets were lysed in mammalian protein extraction reagent (Pierce) with protease inhibitor cocktails (Sigma) and PhosStop (Roche) for 30 min on ice. Then the cell lysate was centrifuged at 14,000 rpm for 20 minutes at 4 °C and the supernatant was collected. Protein concentration was determined by BCA assay. The protein concentration was adjusted to 1–1.5 ug/ul. Cell lysate was mixed with 4 × SDS sample buffer without bromophenol blue, the boiled the samples for 5 minutes. Each sample has three biological replicates. Samples were sent to the RPPA Core Facility at MD Anderson Cancer Center for RPPA analysis.
Cell cycle assay
Cell cycle was analyzed using in situ cell proliferation kit FLUOS (Roche) according to manufacturer’s instruction. Briefly, cells were labeled with Brdu 40min before trypsinization. Cells were rinsed with PBS then fixed by adding into 70% ice-cold ethanol. Fixed cells were incubated with 4M HCl at room temperature for 30min then washed with PBS. Then the cells were incubated with anti-Brdu-FLUOS antibody at room temperature for 45min. Wash cells with PBS and resuspend cells in 0.1ml PBS. Cell suspensions were incubated with 7-AAD for 5 min and immediately analyzed by flow-cytometry. The data were analyzed by FlowJo (Tree Star).
Statistical analysis
Statistical analysis was performed using SPSS and SAS software. The two-tailed Student’s t test was used in our analyses. All results were expressed as mean ± SD, and p<0.05 indicates significance.
RESULTS
The oncogenic RAS-regulated lncRNAs were initially identified by a custom-designed microarray
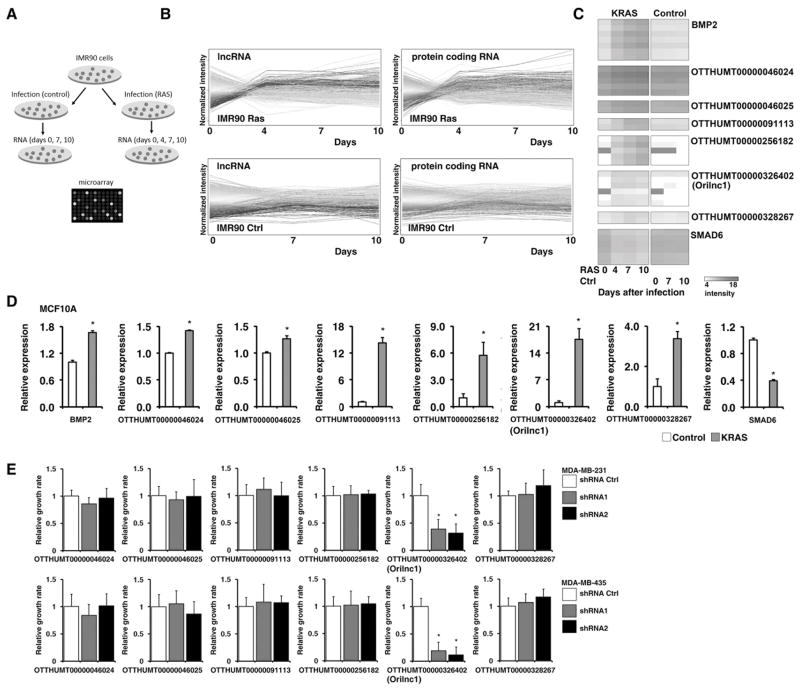
To explore oncogenic RAS-induced transcriptional changes of lncRNA, we analyzed the transcriptional profiles of IMR90, an immortalized human diploid fibroblast cell line, in response to oncogenic RAS activation. KRASG12D- expressing or control lentiviruses were used to infect IMR90 cells, and total RNAs were harvested from cells at different time points (0, 4, 7, and 10 days for KRAS cells and 0, 7, and 10 days for control cells, Figure 1A). To characterize the kinetic transcriptional profiles of lncRNA and protein-coding genes (PCGs) in IMR90 cells expressing either KRAS or the vector control, we designed a custom 60-mer oligonucleotide microarray platform that contains probes recognizing 2,965 lncRNA and 11,081 protein-coding genes (29) (Table S2). We found that KRAS expression induced a significant transcriptional change in both lncRNAs and PCGs (Figure 1B). To identify KRAS target genes, we first identified all genes that were consistently upregulated in response to KRAS expression. Consistent up-regulation was defined as expression ≥ 1.5 fold higher or lower at Day 4 and ≥ 2 fold at Day 7 and 10, compared to that at Day 0. Genes whose expression were also altered in the control cells over the course of the experiment were excluded from the analysis. In summary, 115 (3.9%) lncRNAs and 481 (4.3%) PCGs were found to be regulated by oncogenic RAS activation in IMR90 cells (Table S3). Genome mapping indicated that like KRAS-induced PCGs, there is no specific chromosomal preference of the KRAS-induced lncRNAs (Figure S1A).
Figure 1. The oncogenic RAS-regulated lncRNAs were identified by a custom-designed microarray.
A. The schematic of the IMR90 cells infected with control or KRAS lentivirus. B. The kinetic profiles of the expression of lncRNAs and PCGs in IMR90 cells expressing oncogenic KRASD12G (upper panels) and control vectors (lower panels). Normalized intensity (y-axis) was plotted against days post infection (x-axis). Each line represents a gene. C. Relative expression of BMP2, SMAD6, and the six selected lncRNA genes, measured by qRT-PCR, in control (white bars) and KRAS expressing (orange bars) IMR90 cells at different time points after lentiviral infection. D. Relative expression of BMP2, SMAD6, and the six selected lncRNA genes, measured by qRT-PCR, in MCF10A cells expressing KRAS (orange bars) and vector control (white bars) 8 days after lentiviral infection. E. Six RAS-regulated lncRNAs were knocked down by shRNAs in MDA-MB-231 (upper panel) and MDA-MB-435 (lower panel) cell lines. The cell proliferation after lncRNA knockdown was analyzed by a MTT assay.
Our custom-designed array platform identified many previously characterized KRAS regulated PCGs, such as bone morphogenetic protein 2 (BMP2) and mothers against decapentaplegic homolog 6 (SMAD6), which were found to be significantly up-and down-regulated, respectively, in response to KRAS expression in IMR90 cells (Figure 1C). This confirmed that our custom-designed array platform was a robust approach to identify genes regulated by RAS. To further validate our assay, we conducted qRT-PCR analysis to determine the RNA expression of BMP2, and SMAD6, as well as six selected RAS-regulated lncRNAs in IMR90 cells infected with KRAS or control lentivirus. The qRT-PCR results verified that BMP2 and the six lncRNAs were significantly induced by KRAS expression in a time-dependent manner, whereas the expression of SMAD6 was significantly decreased (Figure S1B). These results were further corroborated in immortalized mammary epithelial MCF10A cells. Expression of BMP2 and the six lncRNAs were significantly induced by KRASD12G expression, while that of SMAD6 was significantly decreased (Figure 1D). In summary, using a customized microarray platform, we have initially identified 115 lncRNAs whose expression is regulated by RAS activation in IMR90 cells. To identify the RAS-regulated lncRNA which is functionally involved in tumorigenesis, we knocked down six lncRNA candidates showed in Figure 1C by specific short hairpin RNAs (shRNAs) in MDA-MB-231 and MDA-MB-435 cells (for each lncRNA, two independent shRNAs were designed). One of the RAS-induced lncRNAs, OTTHUMT00000326402 (named as oncogenic RAS-induced lncRNA 1, Orilnc1), showed strong effort on cell proliferation after shRNA knockdown (Figure 1E). Therefore, we decided to choose this lncRNA for our further study. We analyzed Orilnc1 expression in MCF10A cells treated with EGF or IGF-1 by qRT-PCR. EGF or IGF-1 stimulation did not significantly increase Orilnc1 expression in MCF10 cells which expressed wild-type RAS and RAF (Figure S1C) suggesting that the up-regulation of Orilnc1 might be an oncogenic growth promoting effect. Next, we knocked down Orilnc1 expression in MCF10A cells, and cell growth was measured by cell counting. There was no significant growth inhibition in MCF10A cells (Figure S1D). Taken together, our results suggest that suppression of Orilnc1 does not affect normal epithelial cell growth.
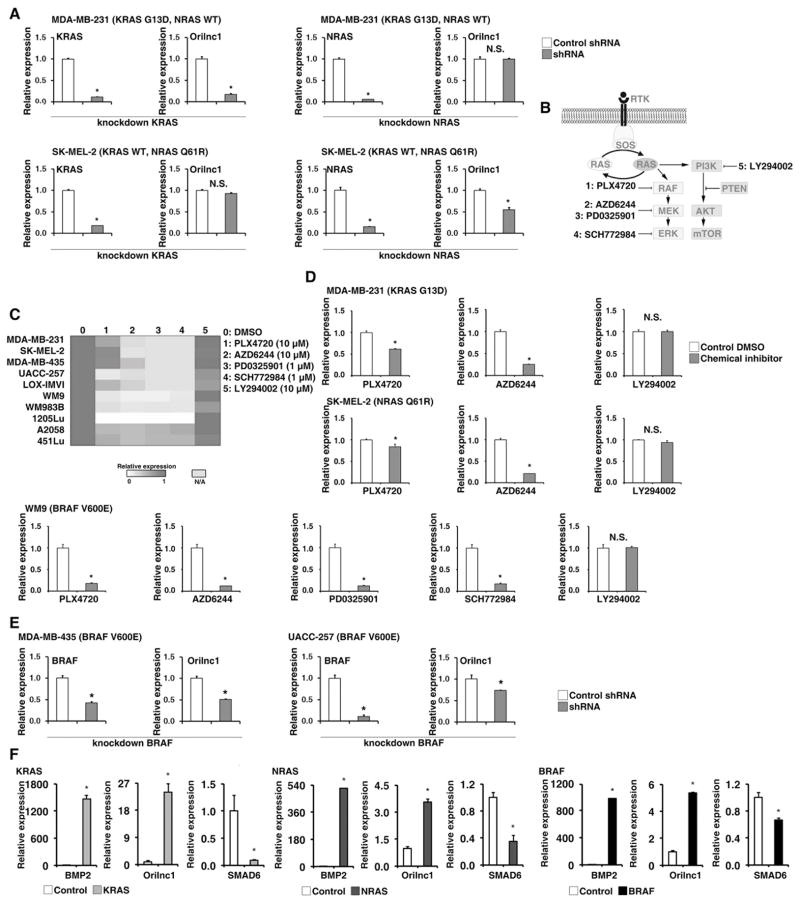
The expression of Orilnc1 is regulated by the RAS-RAF-MEK-ERK signaling cascade
To explore transcriptional regulation of Orilnc1 by endogenous oncogenic RAS proteins we examined the expression of Orilnc1 in MDA-MB-231 and SK-MEL-2 cells, which carry a KRAS and an NRAS mutation, respectively. KRAS specific shRNAs caused a significant decrease of expression of Orilnc1 in MDA-MB-231 cells (KRAS mutation and wild-type NRAS), but not in SK-MEL-2 cells (wild-type KRAS and mutant NRAS) (Figure 2A). NRAS- specific shRNAs caused a significant decrease of expression of Orilnc1 in SK-MEK-2 but not MDA-MB-231 cells (Figure 2A). These data further functionally demonstrated that Orilnc1 expression is dependent on activation of oncogenic RAS, and both KRAS and NRAS can regulate its expression in cancer cells. Because the RAS-RAF-MEK-ERK and the PI3K/AKT pathways can be activated by oncogenic RAS, downstream pathways were mapped with small molecule inhibitors (Figure 2B) and Orilnc1 expression was examined (Figure 2C). Inhibition of PI3K with LY294002 caused a moderate decrease of Orilnc1 expression, whereas inhibition of BRAF, MEK and ERK caused a significant decrease in Orilnc1 expression (Figure 2C and 2D), suggesting that RAS-RAF-MEK-ERK signaling is important in regulating Orilnc1 expression. To confirm this finding, we knocked down BRAF expression in two melanoma cell lines that carry the BRAFV600E mutation and found that both lines expressing BRAF specific shRNA have significantly lower Orilnc1 expression than control cells (Figure 2E).
Figure 2. The expression of Orilnc1 (OTTHUMT00000326402) is regulated by the RAS-RAF-MEK-ERK signaling cascade.
A. Relative expression of Orilnc1 in MDA-MB-231 (upper panels, with KRAS mutation) and SK-MEL-2 (lower panels, with NRAS mutation) cells expressing shRNAs specific to KRAS or NRAS. The expression levels of Orilnc1 in control and shRNA-RAS cells were determined by qRT-PCR. The knockdown efficiency of KRAS (or NRAS) shRNA was also confirmed by qRT-PCR. B. Schematic diagram of RAS downstream signaling pathways. Small molecule inhibitors to specific proteins in each pathway are illustrated. C. Heatmap of Orilnc1 expression in multiple cell lines treated with different inhibitors targeting downstream pathways of RAS activation. The expression of Orilnc1 was determined by qRT-PCR. Red: high expression; white: low expression: gray: no treatment was available in certain cell lines. D. Quantitative data of relative expression of Orilnc1 in MDA-MB-231, SK-MEL-2, and WM9 cells treated with different RAS pathway inhibitors. D presents the same data shown in C by a quantitative bar graph. E. Relative expression of Orilnc1 in MDA-MB-435 and UACC-257 expressing control and BRAF specific shRNAs. The expression of Orilnc1 in control and shRNA-BRAF cells was determined by qRT-PCR. The knockdown efficiency of BRAF shRNA was also confirmed by qRT-PCR. F. Activation of RAS or BRAF in melanocytes was able to significantly up-regulate Orilnc1 expression. Relative expression of BMP2, SMAD6, and Orilnc1, measured by qRT-PCR, in FOM226-1 (a skin epidermal melanocyte) expressing KRASG12D, NRASQ61R or BRAFV600E (red bars) and vector control (white bars) 7~10 days after lentiviral infection.
To test if activated oncogenic RAS or BRAF expression can induce the expression of Orilnc1 in melanocytes, we over-expressed KRASD12G, NRASQ61R or BRAFV600E in FOM226-1, a skin epidermal melanocyte. Expression of Orilnc1 as well as two control genes, BMP2 and SMAD6, were examined by qRT-PCR. Orilnc1 was detectable in FOM226-1 cells, although at a relatively low level compared to melanoma cell lines. Upon RAS or BRAF activation, BMP2 and Orilnc1 expression were significantly induced, while SMAD6 was significantly decreased (Figure 2F). Together, these results strongly suggest that the expression of Orilnc1 is regulated by activity of RAS-RAF-MEK-ERK pathway.
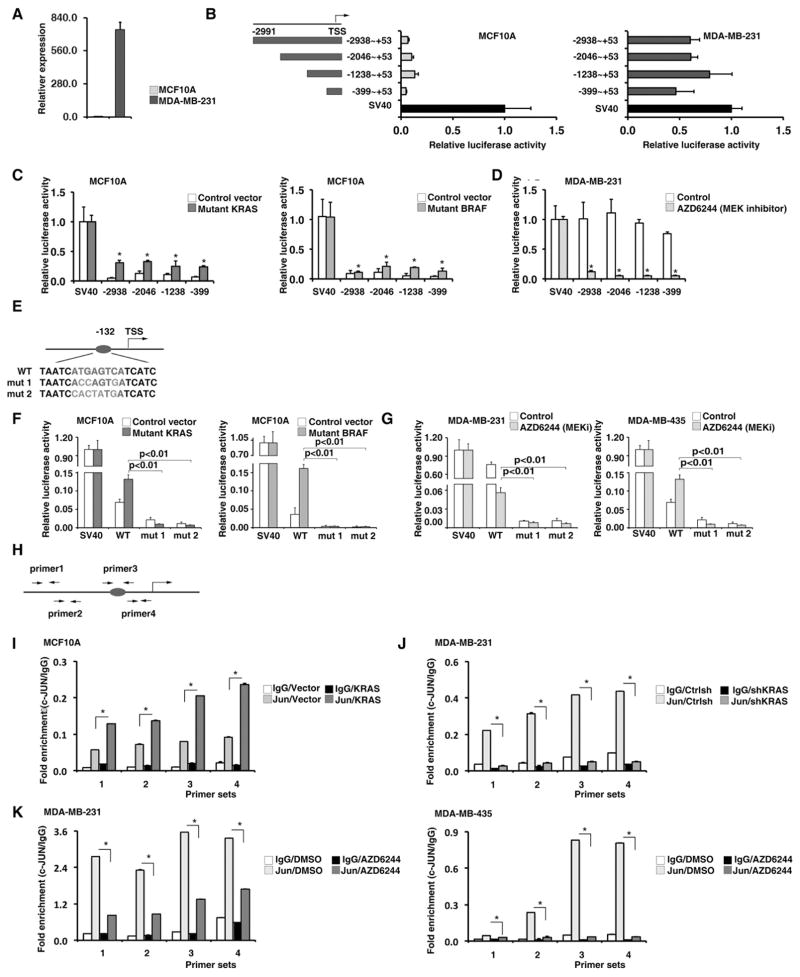
The RAS-RAF-MEK-ERK induced expression of Orilnc1 is mediated by transcriptional factor AP1
We next defined transcriptional regulation of Orilnc1 mediated by the RAS-RAF-MEK-ERK pathway. First, we compared endogenous levels of Orilnc1 of MCF10A cells, which express wild-type RAS and RAF, and MDA-MB-231 cells, which express mutant KRAS. As expected, there was a dramatically higher expression of Orilnc1 in MDA-MB-231 cells than in MCF10A cells (Figure 3A). We cloned the promoter region of Orilnc1 (-2938 to +53) and generated a series of 5′ truncations (-2938 to +53; -2046 to +53; -1238 to +53; and -399 to +53), inserted these fragments into the pGL3-basic luciferase vector and transfected the constructs into MCF10A and MDA-MB-231 cells, respectively. In parallel, the pGL3 luciferase vector containing the SV40 promoter was transfected into these cell lines to serve as a positive control. As shown in Figure 3B, in MCF10A cells, luciferase activity under the control of the Orilnc1 promoter constructs was much lower than under the SV40 promoter, while in MDA-MB-231 cells the luciferase activity under the Orilnc1 and SV40 promoter constructs were comparable (Figure 3B). This confirms that the difference of endogenous Orilnc1 expression in these two cell lines is mediated via the promoter.
Figure 3. The RAS-RAF-MEK-ERK induced expression of Orilnc1 is mediated by transcriptional factor AP1.
A. The expression of endogenous Orilnc1 in MCF10 (RAS and BRAF wide type) and MDA-MB-231 (KRAS mutated) cells by qRT-PCR. B. Luciferase reporter assay assessing the promoter activities of different Orilnc1 promoter fragments in MCF10A and MDA-MB-231 cells. TSS, transcription start site; the numbers ranges (i.e.−2938 to +53) are the location of each fragment relative to the TSS. The activity of SV40 promoter was set as 1 in each cell line. C. Luciferase reporter assay assessing the promoter activities of different Orilnc1 promoter fragments in MCF10A expressing KRASG12D, BRAFV600E or the control vector. Open bar, cells expressing control vector; red or orange bar, cells expressing mutant KRAS or BRAF, respectively. D. Luciferase reporter assay assessing the promoter activities of different Orilnc1 promoter fragments in MDA-MB-231 cells treated with MEK inhibitor AZD6244 or vehicle. Open bar, cells treated with vehicle; green bar, cells treated with AZD6244. E. An illustration of AP1 binding site at −132 (red indicates the consensus motif) and the sequences of the two AP1 mutations (blue indicates the mutant nucleotides). F. Luciferase reporter assay assessing the promoter activities of Orilnc1 core promoter construct and its AP1 mutant counterparts in MCF10A expressing KRASG12D, BRAFV600E or the control vector. Open bar, cells expressing control vector; red or orange bar, cells expressing mutant KRAS or BRAF, respectively. G. Luciferase reporter assay assessing the promoter activities of Orilnc1 core promoter construct and its AP1 mutant counterparts in MDA-MB-231 (left, KRASG13D, breast) and MDA-MB-435 (right, BRAFV600E, melanoma) cells treated with MEK inhibitor AZD6244 or vehicle. Open bar, cells treated with vehicle; green bar, cells treated with AZD6244. H. The locations of the primer pairs used in the ChIP assay. I. The promoter occupancy of c-Jun measured with four pairs of primers in MCF10A cells expressing control and mutant KRAS. J. The promoter occupancy measured with four pairs of primers in MDA-MB-231 cells expressing control or shRNA targeting KRAS. K. The promoter occupancy measured with four pairs of primers in MDA-MB-231 and MDA-MB-435 cells treated with vehicle or AZD6244.
To further map the location of the regulatory region of the RAS signal on the Orilnc1 promoter, we activated the RAS-RAF-MEK-ERK pathway in MCF10A cells by expressing mutant KRAS or mutant BRAF. Compared to control MCF10A cells, all four Orilnc1 promoter constructs in cells expressing KRASG12D or BRAFV600E demonstrated significantly higher luciferase activities (Figure 3C). In contrast, the MEK inhibitor, AZD6244, caused significant decrease in the activity of these Orilnc1 promoter constructs in MDA-MB-231 cells (Figure 3D). The similar response of all of the promoter constructs to RAS-RAF-MEK-ERK pathway activity suggests that regulation of this pathway resides between nt −399 and +53 from the transcription start site, the shortest fragment, which we refer to as the core promoter.
In silico transcriptional factor binding motif analysis revealed that a consensus AP1 binding site was present at −132 bp from the transcription start site of Orilnc1 (Figure 3E). To determine if this AP1 site is involved in RAS/BRAF-mediated regulation of Orilnc1 transcription, we generated two mutations of this AP1 binding motif and cloned the mutant core promoter into the pGL3-basic luciferase vector. In MCF10A cells, the promoter activities of the mutant constructs were significantly lower than that of the wild-type construct (Figure 3F). While KRASG12D or BRAFV600E expression induced the activity of the wild-type construct, they had no effect on the mutant constructs (Figure 3E). In MDA-MB-231 cells (KRAS mutant), while AZD6244 treatment significantly reduced the activity of the wild type construct, the mutations markedly decreased the promoter activities. Similar observations were also made in MDA-MB-435 cells, which express mutated BRAF (Figure 3G).
To determine whether AP1 (c-Jun) directly binds to the promoter region of Orilnc1, we designed four pairs of AP1 binding region specific primers (Figure 3H) and performed chromatin immunoprecipitation (ChIP) assays using anti-c-Jun specific antibodies. In all three cell lines tested, the core promoter region was detected in the c-Jun co-precipitate complex (Figure 3I to 3K). In MCF10A cells, ectopic KRASG12D expression significantly enhanced the association between the promoter region and c-Jun (Figure 3I). In MDA-MB-231 cells, knocking down endogenous mutated KRAS significantly reduced the amount of Orilnc1 promoter associated with c-Jun (Figure 3J). Similarly, AZD6244 treatment caused decreases of the amount of Orilnc1 promoter in c-Jun complexes in both MDA-MB-231 and MDA-MB-435 cell lines (Figure 3K). Together, our data demonstrates that the transcription of Orilnc1, regulated by RAS-RAF-MEK-ERK pathway, is mediated by AP1.
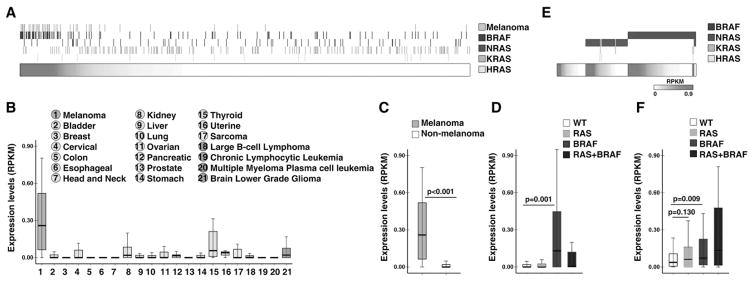
Orilnc1 is highly expressed in BRAF mutant melanoma
To investigate the clinical significance of Orilnc1 expression in human cancer, we used RNA-sequencing profiles (RNA-seq) from the Cancer Cell Line Encyclopedia (CCLE) project (30). The transcriptomic and genomic profiles as well as tumor lineage annotation of a total of 934 human tumor cell lines, which originated from 21 different cancer types including melanoma, epithelial, hematopoietic, and neurological cancers, were analyzed. Expression of Orilnc1 RNA in these cell lines was correlated with tissue origin. Orilnc1 was detectable in 35.9% (335/934) of cell lines examined, and interestingly, most melanoma cell lines (84.6%, 44/52) express detectable levels of Orilnc1 (Figure 4A). To further confirm these findings, we calculated the expression of Orilnc1 among the different tissue origins (n=21) of the 934 cell lines. Indeed, Orilnc1 expression in melanoma is remarkably higher than that of other cancer types (Figure 4B). While Orilnc1 RNA was expressed in thyroid and kidney cancer lines, the levels were much lower than in melanoma (Figure 4B). As such, melanoma cell lines have a significantly higher level of Orilnc1 than non-melanoma cell lines (p<0.001, Figure 4C).
Figure 4. Orilnc1 is highly expressed in melanoma cells with mutant BRAF.
A. Heatmap of Orilnc1 expression in 934 CCLE cell lines (horizontal bar) and their RAS/BRAF mutation status/tissue origins (vertical bars). Each column represents a cell line, and was ranked based on Orilnc1 expression. Red density represents Orilnc1 expression level, and white non-detectable. The rows above the horizontal bar represent the RAS/BRAF mutation status and tissue origins. Yellow: the tissue origin; dark blue: BRAF status; green: NRAS status; blue: KRAS status; and light blue: HRAS status. The presence of a vertical bar indicates the cell line has the corresponding mutation or is a melanoma cell line. B. Box plot of Orilnc1 expression in CCLE cell lines grouped by tumor types. The tumor types are color-coded based on their tissue origin: melanoma is orange; epithelia cancers are green; sarcoma is yellow; hematopoietic cancers are purple; and neurological cancers are blue. C. Box plot of Orilnc1 expression in melanoma and non-melanoma cell lines of CCLE. D. Box plot of Orilnc1 expression in cell lines grouped by RAS/BRAF status. E. Heatmap of Orilnc1 expression in TCGA melanoma specimens (horizontal bar) and their RAS/BRAF mutation status (vertical bars). Each column represents a melanoma patient specimen, and was ranked based on Orilnc1 expression in each subgroup. Red density represents Orilnc1 expression level, and white non-detectable. The rows above the horizontal bar represent the RAS/BRAF mutation status. Dark blue: BRAF status; green: NRAS status; blue: KRAS status; and light blue: HRAS status. The presence of a vertical bar indicates the cell line has the corresponding mutation. F. Box plot of Orilnc1 expression in TCGA melanoma specimen grouped by RAS/BRAF status.
Next, we characterized the correlations of Orilnc1 expression with the genomic status of the genes in the RAS-associated pathways, such as RAS (KRAS, NRAS, and HRAS), BRAF, PIK3CA, PTEN and AKT. Notably, we found that the cell lines expressing high level of Orilnc1 often harbor BRAF mutations and there was no significant correlation with other genomic alterations of RAS pathway genes including RAS itself (Figure 4A). To confirm this result, we divided the 934 cell lines into four groups by their RAS/BRAF status: wild type group that has no RAS or BRAF mutations; RAS group has at least one RAS mutation but no BRAF mutations; BRAF group has BRAF mutations but no RAS mutations; RAS plus BRAF group has both BRAF and RAS mutations. Consistent with our observation in Figure 4A, the cell lines with BRAF mutations have significantly higher Orilnc1 expression than wild type cell lines. To further test if the cell-line based observation is also true in primary tumor specimens, we obtained genomic and RNA-seq profiles of melanoma specimens from TCGA (27) and grouped the tumors in the same way as with the cell lines (Figure 4E to 4F). Consistently, we found that a large percentage (76.8%, 202/263) of melanoma specimens express Orilnc1 and tumors with BRAF mutations have significantly higher Orilnc1 expression than wild type tumors (Figure 4E to 4F). Together, using RNA-seq profiles from cancer cell lines (CCLE) and tumor specimens (TCGA), we demonstrated that Orilnc1 is highly expressed in melanoma tumor cells harboring BRAF mutations, and activation of RAS/BRAF in melanocytes was able to significantly up-regulate Orilnc1 expression.
Knockdown of Orilnc1 expression reduces cancer cell growth in vitro and in vivo
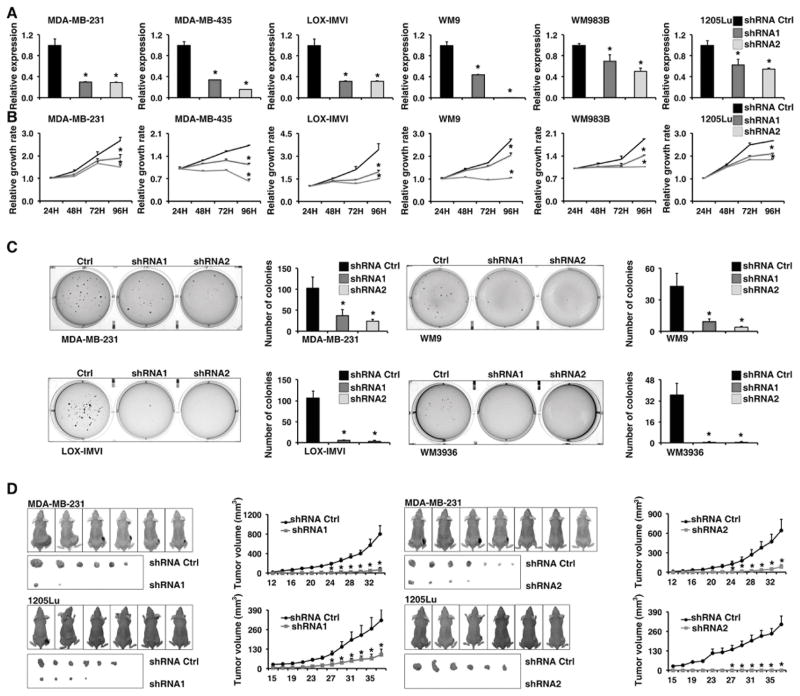
To examine the biological function of Orilnc1 in human cancer, we chose a panel of melanoma cell lines with different genomic alterations of RAS/BRAF as well as a non-melanoma cancer cell line, MDA-MB-231, which harbors mutant KRAS and expresses high level of Orilnc1. First, we suppressed Orilnc1 expression by two independent shRNA sequences to minimize the possibility of observing off-target effects. qRT-PCR analysis demonstrated that both shRNA vectors were able to significantly reduce the endogenous Orilnc1 expression in these cancer cell lines (Figure 5A). Notably, we consistently found that the expression of Orilnc1-specfic hairpins significantly reduced growth rates in all cell lines tested by MTT assay (Figure 5B). Moreover, down-regulating Orilnc1 expression also significantly reduced anchorage-independent growth in the four cell lines tested by soft agar assay in this study (Figure 5C). Finally, we injected MDA-MB-231- and 1205Lu cells-expressing control and Orilnc1-specific hairpins into nude mice and found that the expression of the Orilnc1-specific hairpins significantly suppressed tumor growth in vivo in both cell lines tested in our study (Figure 5D). Taken together, our in vitro and in vivo experiments demonstrated that blocking Orilnc1 expression significantly blocked tumor cell proliferation and growth.
Figure 5. Knockdown of Orilnc1 expression reduces cancer cell growth in vitro and in vivo.
A. Relative expression of Orilnc1 in cells expressing control and Orilnc1 shRNAs in a panel of melanoma cell lines and a non-melanoma cancer cell, MDA-MB-231 (Breast; KRASG13D). B. Growth curves of cancer cell lines transfected with control or Orilnc1 shRNAs. C. Soft-agar assay with cells expressing control and Orilnc1 shRNAs. D. In vivo xenograft tumor growth curves of MDA-MB-231 (upper panel) and 1205Lu (lower panel) cells expressing control and Orilnc1 shRNAs.
Knockdown of Orilnc1 represses the expression of Cyclin E1 and induced G1/S arrest
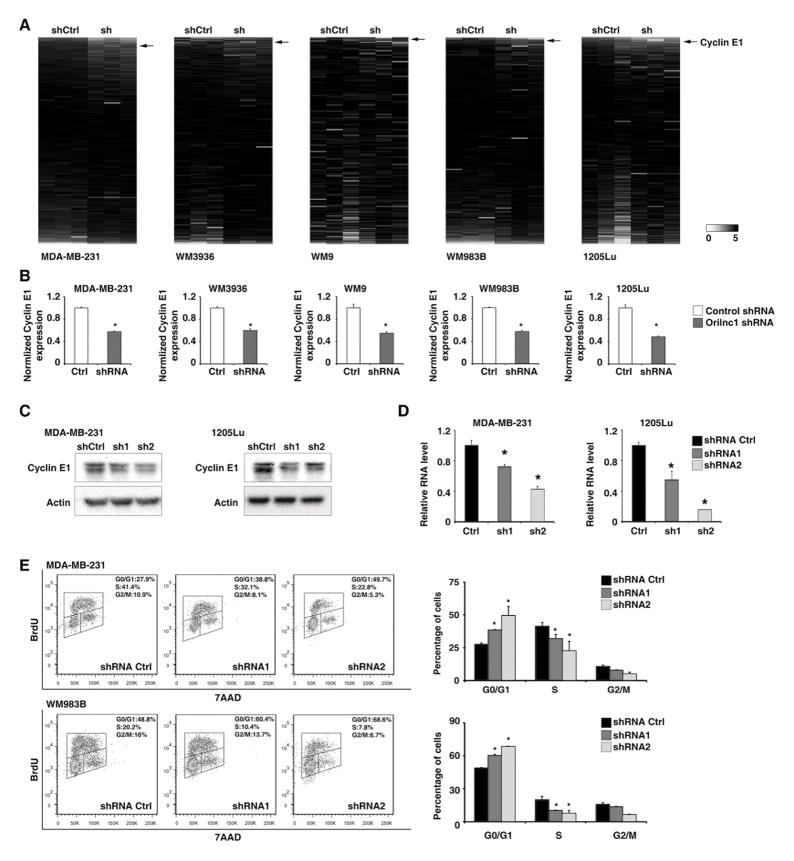
To further characterize the molecular mechanism by which Orilnc1 promotes cancer cell proliferation, we profiled the protein expression changes of cells, in which Orilnc1 was stably knocked down by shRNA, compared to control cells, by reverse phase protein array (RPPA) analysis (Figure 6A and Table S4). Interestingly, in all five cancer cell lines tested in our study, the cell cycle regulated protein, Cyclin E1, was significantly reduced in the Orilnc1 knockdown cells compared to the control cells (Figure 6A and 6B). This result was further validated by two independent shRNAs using Western blots in MBA-MD-231 and 1205Lu cells (Figure 6C). Furthermore, using a qRT-PCR assay, we found that inhibition of Orilnc1 not only reduced Cyclin E1 protein expression but also significantly decreased its mRNA expression (Figure 6D). Cyclin E1 forms a complex with and functions as a regulatory subunit of CDK2, whose activity is required for cell cycle G1/S transition. Cyclin E1 accumulates at the G1-S phase boundary and is degraded as cells progress through S phase. Therefore, we hypothesized that Orilnc1 may regulate cell cycle via regulating Cyclin E1 expression. To this end, we examined if knocking down Orilnc1 leads to the same G1/S cell-cycle arrest phenotype often seen in cells with Cyclin E1 inhibition. Using a BrdU incorporation assay, we found that knocking down Orilnc1 expression by shRNAs significantly induced G1/S arrest in both MDA-MB-231 and WM983B cell lines (Figure 6E). This is in agreement with our previous observation that Orilnc1 knockdown significantly blocked cell proliferation (Figure 5). Taken together, our results indicated that Orilnc1 exerts its function on cell growth at least in part via regulating Cyclin E1 expression.
Figure 6. Knockdown of Orilnc1 represses the expression of Cyclin E1 and induced G1/S arrest.
A. Heatmap of the protein expression that was analyzed by a RPPA assay in cells introduced control or Orilnc1 shRNA (n=3 each condition). The proteins (antibodies) were ranked according to the magnitude of the fold change in each given cell line. Red and green indicate up- and down- regulation, respectively. The black arrow indicates Cyclin E1 antibody. B. The quantitative result of the Cyclin E1 expression of the RPPA assay. C. Western blot of Cyclin E1 in cells expressing control or Orilnc1 shRNAs. D. qRT-PCR of CCNE1 in cells expressing control or Orilnc1 shRNAs. E. Cell-cycle profiles of cells expressing control or Orilnc1 shRNAs.
DISCUSSION
RAS signaling is one of the most studied biological pathways and plays important roles in tumorigenesis and cancer progression (17–21). The RAS pathway controls a wide range of cellular processes, including cell proliferation, differentiation, migration, and cell death, through transcriptional regulation. While many RAS-regulated protein coding genes have been identified, little is known on how RAS regulates the transcription of non-coding RNAs (17–21), which comprise up to 70% of the human genome (1, 2). In the current study, we analyzed lncRNA expression profiles in response to RAS activation with a customized array. This RAS-regulated lncRNA profile can be a valuable resource to further understand RAS mediated lncRNA regulation. Furthermore, with emerging evidence indicating lncRNA as a new regulator for cancer, the lncRNA profile we report here will help to further elucidate the mechanisms underlying RAS-mediated tumorigenesis and progression. One of the RAS- induced lncRNAs identified through our customized array is Orilnc1. Our study revealed that Orilnc1 is required for cell growth in cancer cell lines with an activated RAS-RAF pathway. An AP1 consensus site is present in Orilnc1 promoter region, which provides a molecular explanation to our observation that the transcription of Orilnc1 is induced by the RAF-MEK-ERK pathway downstream of RAS. Furthermore, we demonstrated that Orilnc1 can regulate Cyclin E1 and inhibition of Orilnc1can induce the same phenotype as Cyclin E1 knock down. These observations together suggest that Orilnc1 may be an important non-protein effector in RAS-RAF signaling pathway through regulation of Cyclin E1.
Consistent with the observation that Orilnc1 is induced by RAS pathway activation in cell lines, Orilnc1 is highly expressed in melanomas carrying BRAF mutations, which are detected in approximately 50% of melanomas (22–27). Though many BRAF inhibitors have been developed, BRAF inhibitor resistance emerges in most patients (22–27). A significant amount of effort has been devoted to elucidate the mechanism underlying BRAF inhibitor resistance. Here we have demonstrated that repressing Orilnc1 expression inhibited the growth of tumor cells with BRAF mutations. These observations warrant further investigation on the non-coding effectors of BRAF and suggest that Orilnc1 may functionally contribute to resistance to BRAF inhibition.
Most recently, many cancer associated lncRNAs have been successfully identified in human cancers. For example, SPRY4-IT1(31, 32), BANCR (33), DRAIC/PCAT29 (34), CASC15 (35), MIR31HG (36), SPRIGHTLY (37), RMEL3 (38), SLNCR1 (39) and SAMMSON (40) have been proposed to serve as novel oncogenes or tumor suppressor genes in melanoma. The discovery of lncRNA cancer driver will contribute to cancer diagnosis and treatment. Several features of lncRNAs make them good candidates as biomarkers: they often form secondary structures, which make them stable in body fluid; their levels are deregulated in diseased tissue and their deregulation is often disease/tissue specific; and they are easy to detect and quantify. In fact, the Food and Drug Administration has approved a method of detecting PCA3, a prostate cancer-associated lncRNA found in urine, as a non-invasive prostate cancer diagnosis tool (41). Given that Orilnc1 level is higher in tumors with BRAF mutations than other tumors, we reasoned that Orilnc1 may serve as a non-invasive biomarker for certain melanomas. Further investigations will be needed to test if Orilnc1 expression in patients can predict response to treatment with BRAF inhibitors. Recent advances in oligonucleotide/nanoparticle technology have made targeting non-coding genes a possible therapeutic approach (3–11). Several RNAi-based therapies are under clinical evaluation. Our study here demonstrated that suppression of Orilnc1 expression decreased the growth of xenografts. This result provided proof-of-concept evidence for developing Orilnc1 specific RNA-based therapy for BRAF-mutant cancer. In conclusion, our study provided a useful resource for studying RAS-regulation of lncRNA, elucidated a novel molecular mechanism for RAS-mediated cancer cell proliferation, and demonstrated that Orilnc1 has the potential to serve as biomarker and therapeutic target for cancer, especially melanoma.
Supplementary Material
Acknowledgments
This work was supported, in whole or in part, by the NIH P50CA174523 (M. Herlyn and L. Zhang), P01CA114046 (M. Herlyn), R01CA142776 (L. Zhang), R01CA190415 (L. Zhang), U54HG008100 (G.B. Mills), P50CA083638 (G.B. Mills), the Basser Center for BRCA (L. Zhang) and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (M. Herlyn). D. Zhang and L. Yang were supported by the China Scholarship Council.
We thank the TCGA and CCLE project teams.
Footnotes
No potential conflicts of interest were disclosed.
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews Genetics. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 4.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154:1190–3. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–52. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends in cancer. 2015;1:93–109. doi: 10.1016/j.trecan.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, Slack FJ, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34:5003–11. doi: 10.1038/onc.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome research. 2006;16:11–9. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:716–21. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nature genetics. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer cell. 2015;28:529–40. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nature reviews Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 18.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nature reviews Molecular cell biology. 2008;9:517–31. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nature reviews Cancer. 2011;11:761–74. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nature reviews Drug discovery. 2014;13:928–42. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 21.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nature reviews Drug discovery. 2014;13:828–51. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo JA, Fisher DE. The melanoma revolution: from UV carcinogenesis to a new era in therapeutics. Science (New York, NY) 2014;346:945–9. doi: 10.1126/science.1253735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribas A, Flaherty KT. BRAF targeted therapy changes the treatment paradigm in melanoma. Nature reviews Clinical oncology. 2011;8:426–33. doi: 10.1038/nrclinonc.2011.69. [DOI] [PubMed] [Google Scholar]
- 24.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes & development. 2012;26:1131–55. doi: 10.1101/gad.191999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibney GT, Messina JL, Fedorenko IV, Sondak VK, Smalley KS. Paradoxical oncogenesis--the long-term effects of BRAF inhibition in melanoma. Nature reviews Clinical oncology. 2013;10:390–9. doi: 10.1038/nrclinonc.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nature reviews Cancer. 2014;14:455–67. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas N. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, et al. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes & development. 2010;24:2754–9. doi: 10.1101/gad.1950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Y, Gholamin S, Schubert S, Willardson MI, Lee A, Bandopadhayay P, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med. 2014;20:732–40. doi: 10.1038/nm.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–62. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 32.Mazar J, Zhao W, Khalil AM, Lee B, Shelley J, Govindarajan SS, et al. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget. 2014;5:8959–69. doi: 10.18632/oncotarget.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22:1006–14. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakurai K, Reon BJ, Anaya J, Dutta A. The lncRNA DRAIC/PCAT29 Locus Constitutes a Tumor-Suppressive Nexus. Mol Cancer Res. 2015;13:828–38. doi: 10.1158/1541-7786.MCR-15-0016-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lessard L, Liu M, Marzese DM, Wang H, Chong K, Kawas N, et al. The CASC15 Long Intergenic Noncoding RNA Locus Is Involved in Melanoma Progression and Phenotype Switching. The Journal of investigative dermatology. 2015;135:2464–74. doi: 10.1038/jid.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montes M, Nielsen MM, Maglieri G, Jacobsen A, Hojfeldt J, Agrawal-Singh S, et al. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nature communications. 2015;6:6967. doi: 10.1038/ncomms7967. [DOI] [PubMed] [Google Scholar]
- 37.Zhao W, Mazar J, Lee B, Sawada J, Li JL, Shelley J, et al. The long non-coding RNA SPRIGHTLY regulates cell proliferation in primary human melanocytes. The Journal of investigative dermatology. 2016 doi: 10.1016/j.jid.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goedert L, Pereira CG, Roszik J, Placa JR, Cardoso C, Chen G, et al. RMEL3, a novel BRAFV600E-associated long noncoding RNA, is required for MAPK and PI3K signaling in melanoma. Oncotarget. 2016 doi: 10.18632/oncotarget.9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ, et al. The lncRNA SLNCR1 Mediates Melanoma Invasion through a Conserved SRA1-like Region. Cell Rep. 2016;15:2025–37. doi: 10.1016/j.celrep.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531:518–22. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 41.Lee GL, Dobi A, Srivastava S. Prostate cancer: diagnostic performance of the PCA3 urine test. Nature reviews Urology. 2011;8:123–4. doi: 10.1038/nrurol.2011.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.