Abstract
Purpose
Tumor-derived cell-free DNA (cfDNA) from urine of patients with cancer offers non-invasive biologic material for detection of cancer-related molecular abnormalities such as mutations in Exon 2 of KRAS.
Experimental Design
A quantitative, mutation-enrichment next-generation sequencing test for detecting KRASG12/G13 mutations in urine cfDNA was developed and results were compared to clinical testing of archival tumor tissue and plasma cfDNA from patients with advanced cancer.
Results
With 90–110 mL of urine, the KRASG12/G13 cfDNA test had an analytical sensitivity of 0.002%–0.006% mutant copies in wild-type background. In 71 patients, the concordance between urine cfDNA and tumor was 73% (sensitivity, 63%; specificity, 96%) for all patients and 89% (sensitivity, 80%; specificity, 100%) for patients with urine samples of 90–110 mL. Patients had significantly fewer KRASG12/G13 copies in urine cfDNA during systemic therapy than at baseline or disease progression (P=0.002). Compared with no changes or increases in urine cfDNA KRASG12/G13 copies during therapy, decreases in these measures were associated with longer median time to treatment failure (P=0.03).
Conclusions
A quantitative, mutation-enrichment next-generation sequencing test for detecting KRASG12/G13 mutations in urine cfDNA had good concordance with testing of archival tumor tissue. Changes in mutated urine cfDNA were associated with time to treatment failure.
Keywords: Cell-free DNA, KRAS mutation, liquid biopsy, next-generation sequencing, cancer, urine, plasma
INTRODUCTION
Detecting molecular alterations can provide guidance for personalized cancer therapy in patients with melanoma, non-small cell lung cancer (NSCLC), colorectal cancer, and other cancers (1–5). KRAS mutations are associated with poor prognosis in diverse cancer types and with lack of benefit from anti–epidermal growth factor receptor (EGFR) targeted monoclonal antibodies in colorectal cancer (3, 6–8). Currently, oncogenic alterations such as KRAS mutations are assessed in archival tumor tissue, but the tissue availability is often a limiting factor that precludes molecular analysis (9, 10). In addition, mutation assessment of primary tumor tissue or an isolated metastasis does not necessarily reflect the genetic make-up of metastatic disease owing to tumor heterogeneity (11–13). Different oncogenic mutations occur in different areas of a primary tumor, and the mutation statuses of the primary tumor and distant metastases are discrepant in approximately 20–30% of cases (12, 14). In addition, translational studies in EGFR-mutated NSCLC suggest that cancer genotype can change over time; for example, Sequist et al. demonstrated in a group of 37 patients with EGFR-mutant NSCLC who had pre-treatment and post-progression tumor biopsies that some mutations can occur and disappear over time (15). Tumor cells undergoing apoptosis or necrosis release small fragments of cell-free (cf) DNA, which can be identified in blood, urine, and other biologic materials and offers an alternative source of material for genomic testing (16). Unlike performing tissue biopsies, obtaining samples of urine or plasma cfDNA is less invasive, with less risk to patients at a lower cost, and can be repeated at different times and provide valuable information about genetic changes that occur during the disease evolution. In colorectal cancer, sensitive techniques such as BEAMing (beads, emulsion, amplification, magnetics) polymerase chain reaction (PCR), droplet digital PCR, and next-generation sequencing (NGS) detected low-frequency clones with KRAS mutations in plasma cell-free DNA (cfDNA) not detected by standard clinical molecular testing, and these clones ultimately led to resistance to EGFR antibodies (17–20).
Preliminary data suggest that molecular testing of urine cfDNA is feasible in patients with advanced cancers (10, 21, 22). The purpose of this study was to develop and validate molecular detection and quantification of exon 2 KRAS mutations (KRASG12/G13) in urine and plasma cfDNA specimens from patients with advanced cancers and determine whether this approach has acceptable concordance, sensitivity, and specificity with conventional clinical testing of archival tumor samples. In addition, this study sought to determine whether changes in KRASG12/G13 copy numbers in urine or plasma cfDNA are correlated with treatment outcomes.
METHODS
Patients
Patients with progressing advanced cancers and known KRAS mutation statuses from conventional clinical testing of their archival formalin-fixed, paraffin-embedded (FFPE) tumor tissue specimens (described in the Supplementary Methods) treated at The University of Texas MD Anderson, Niguarda Cancer Center, and the University of Southern California Norris Comprehensive Cancer Center were enrolled for urine and plasma collection from December 2012 to November 2015. Patients had the option of providing longitudinally collected samples during the course of their therapy. The study was conducted in accordance with the approval of the participating institutions’ Institutional Review Boards and/or with the guidelines of their Ethical Committees.
Sample Collection and Processing
Urine and plasma samples for cfDNA isolation were collected at the time of disease progression before treatment initiation and, if feasible, repeatedly during subsequent therapy. The recommended urine collection volume was 90–110 mL; however, amounts as small as 10 mL were also accepted. Urine samples were collected in 120-mL containers supplemented with preservative and stored at −70°C. For cfDNA extraction, urine was concentrated to 4 mL using Vivacell 100 concentrators (Sartorius Corp, Bohemia, NY) and incubated with 700 μL of Q-sepharose Fast Flow quaternary ammonium resin (GE Healthcare, Pittsburg, PA). Tubes were spun to collect sepharose and bound DNA. The pellet was resuspended in a buffer containing guanidinium hydrochloride and isopropanol, and the eluted DNA was collected as a flow-through using polypropylene chromatography columns (BioRad Laboratories, Irvine, CA). The DNA was further purified using QiaQuick columns (Qiagen, Germany).
At MD Anderson and Niguarda Cancer Center, whole blood was collected in ethylenediaminetetraacetic acid–containing tubes and centrifuged and spun twice within 2 hours to yield plasma. At the University of Southern California, blood was collected in Cell-Free DNA BCT tubes (Streck, Omaha, NE), which allow storage for up to 2 weeks. The QIAamp Circulating Nucleic Acid kit (Qiagen, Valencia, CA) was used to isolate cfDNA from 1.5–4 mL of plasma according to the manufacturer’s instructions.
KRAS Mutation Analysis in cfDNA
We developed a new workflow to create an assay capable of detecting a low abundance of KRASG12/G13 mutations (≤ 0.01% in the wild-type [wt] DNA background) in short, highly fragmented urine cfDNA (Supplementary Fig. S1). The urine cfDNA extraction method was designed to preferentially isolate low-molecular-weight (< 400 bp) fragments of cfDNA. Quantitative analysis of 7 common mutations (G12A, G12C, G12D, G12R, G12S, G12V, and G13D) in codons 12 or 13 of exon 2 of the KRAS gene (KRASG12/G13 mutations) was performed using a mutation-enrichment PCR coupled with NGS (Trovagene, San Diego, CA). An ultra-short footprint PCR assay (gene-specific footprint 31 bp; overall amplicon length of 75 bp) was used to amplify highly degraded cfDNA KRASG12/G13 fragments. The PCR amplification utilized a preferential enrichment of KRASG12/G13-mutant cfDNA by using oligonucleotides complementary to wt KRAS DNA to block annealing of the PCR primers and to suppress the amplification of wt KRAS (Supplementary Fig. S2). PCR primers contained a 3′ gene-specific sequence and a 5′ common sequence that was used in the subsequent sample-barcoding step. The PCR enrichment cycling conditions utilized an initial 98°C denaturation step followed by an assay-specific 5 cycles of pre-amplification PCR and 30 cycles of mutation-enrichment PCR. Custom DNA sequencing libraries were constructed and indexed using the Access Array System for Illumina Sequencing Systems (Fluidigm, San Francisco, CA). The indexed libraries were pooled, diluted to equimolar amounts with buffer and the 5% PhiX Control library, and sequenced on an Illumina MiSeq platform at a high depth (~200,000 reads) using 150-V3 sequencing kits (Illumina, San Diego, CA). Primary image analysis, secondary base calling, and data quality assessment were performed on the MiSeq instrument using RTAv1.18.54 and MiSeq Reporter v2.6.2.3 software. The analysis output (FASTQ files) from the runs was processed using custom sequencing reads counting and variant calling algorithms to tally the sums of total target gene reads (wt KRAS or mutant KRAS reads) that passed predetermined sequence quality criteria (qscore ≥ 20). A custom quantification algorithm was developed to accurately determine the absolute number of mutant DNA molecules in the source cfDNA sample. The algorithm quantifies the mutational copy number by incorporating into each sequencing run a corresponding reference sample set with known copy numbers for each of the seven most common KRASG12/G13 mutations. Sequencing results from this reference sample set is used to generate standard curves and the mutant copy number from the source cfDNA sample is calculated by interpolation. Results are standardized to a 100,000 Genome Equivalents (GEq).
The KRASG12/13 mutation detection was determined as the number of KRAS mutations detected above a pre-defined cutpoint which were specific for each of the seven KRAS mutations assessed. The pre-defined cutpoint for each KRAS mutation was calculated as the copy number obtained from the mean plus three standard deviations of non-specific signal (copy number) established by analyzing urine cfDNA samples from 150 healthy volunteers and 24 patients with wt KRASG12/G13 metastatic cancer (by tumor tissue analysis). Similarly, assay cut-offs for plasma were established by analyzing plasma cfDNA samples from a separate cohort of 40 healthy volunteers and 80 patients with wt KRASG12/G13 metastatic cancer (by tumor tissue analysis). Detection cut-offs were standardized to 100,000 GEq.
Statistical Analysis
Concordance between the mutation analyses of urine cfDNA, plasma cfDNA, and archival tumor specimens was calculated using a kappa coefficient. Overall survival (OS) was defined as the time from the date of study entry to the date of death or last follow-up. Time to treatment failure (TTF) was defined as the time from the date of systemic therapy initiation to the date of removal from the treatment. The Kaplan-Meier method was used to estimate OS and TTF, and a log-rank test was used to compare OS and TTF among patient subgroups. Cox proportional hazards regression models were fit to assess the association between patient characteristics and OS or TTF. The Spearman rank coefficient was used to assess correlations. All tests were 2-sided, and P values < 0.05 were considered statistically significant. All statistical analyses were performed with the GraphPad (GraphPad Software, Inc., La Jolla, CA) or SPSS 23 (SPSS, Chicago, IL) software programs.
RESULTS
Performance of the Assay in Detecting KRASG12/G13 Mutations in Urine cfDNA
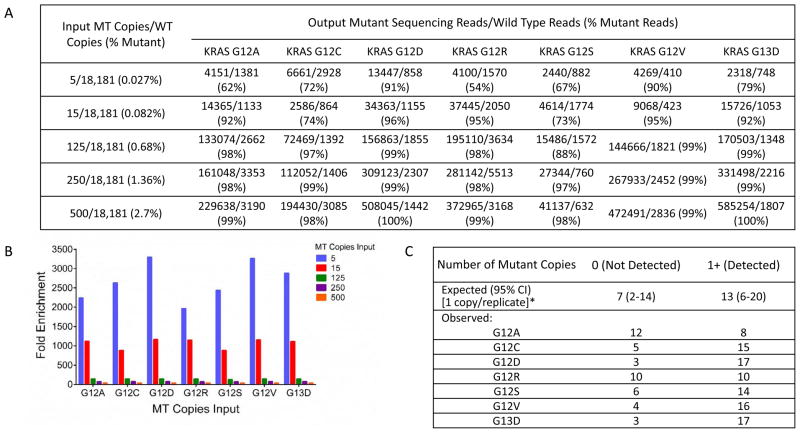
The performance of mutation-enrichment PCR coupled with NGS for the detection of KRASG12/G13 mutations in urine cfDNA was investigated by assessing fold mutation enrichment, lower limit of detection, and assay reproducibility in urine. Fold enrichment was assessed by spiking 5–500 copies of mutant DNA into 18,181 GEq of wt DNA (0.027%–2.7%). For the 7 most common KRASG12/G13 variants, 2,000- to 3,370-fold enrichment of mutant KRASG12/G13 fragments was obtained for an input of 5 copies of KRASG12/13 mutant DNA within 60 ng (18,181 GEq) of wt DNA (Fig. 1A and 1B). The resulting sequencing libraries comprised 69.5%–99.7% mutant reads, thus enabling sensitive mutation detection by NGS (Fig. 1A). Resulting fold-enrichment for KRASG12/G13-mutant fragments increased inversely with decreasing amount of mutant copies in the wt background (Fig. 1B).
Figure 1.
Mutation-enrichment next-generation sequencing (NGS) platform for the analysis of cell-free DNA from urine and plasma. A. Comparison between the input ratio of mutant/wild-type (wt) KRASG12/G13 copies and the output ratio of mutant/wt KRASG12/G13 sequencing reads for 5–500 input mutant copies of the 7 most common KRASG12/G13 variants diluted in 60 ng (~18,180 genome equivalents) of wt DNA (mutation abundance, 0.0275–2.75%). The output sequencing reads are the means of 18 replicates from 6 independent NGS dilution series experiments performed on 3 different days by 2 operators on 2 MiSeq instruments. B. Fold enrichment was calculated as the percent of input mutant KRASG12/G13 molecules divided by the percent of output mutant KRASG12/G13 sequencing reads in A. C. Verification of the analytical sensitivity (lower limit of detection, 1) of the KRASG12/G13 mutation-enrichment NGS assay. A DNA blend with 20 mutant copies in a background of ~363,620 wt genome equivalents (0.006%) was prepared and distributed over 20 wells to achieve a target concentration of 1 mutant copy/18,181 genome equivalents per well. Following mutation-enrichment NGS, the observed distribution frequency of the counts of 0 or ≥1 copies across 20 replicates was compared to theoretical Poisson expectations (95% confidence intervals [CIs]).
When quantifying rare DNA fragments, the frequency distribution of the number of DNA molecules that will be present in each PCR tube upon repeated measurements can be predicted by the Poisson distribution. Herein, the lower limit of detection was defined as the lowest number of copies for which the frequency distribution of the copy number events upon repeated measurements fell within the 95% confidence interval (CI) of expected frequency distribution determined by Poisson statistics. For lower limit of detection verification, 20–80 repeated measurements were performed on a single multiplexed NGS run for a target spike-in level of 1 mutant KRASG12/G13 copy within 18,181 GEq (60 ng) of wt KRAS DNA or for a target spike-in level of 2 mutant KRASG12/G13 copies within 100,000 GEq (330 ng) of wt KRAS DNA. Replicates were subjected to mutation-enrichment NGS analysis. The observed distribution of positive and negative hits in our experiments matched the theoretical hit rate of an ideal Poisson distribution for these replicates, confirming 1 copy detection sensitivity of the KRASG12/G13 assay in the background of 18,181 wt GEq (0.006%; Fig. 1C) and 2 copies detection sensitivity in a background of 100,000 wt GEq (0.002%; Supplementary Table S1).
The reproducibility of quantitative KRASG12/G13 mutations detection was analyzed using urine samples from patients with advanced cancers. Two to three cups (each 90–120 mL) of urine were obtained at a single time point from 3 patients with tumor biopsy specimens positive for KRASG12/G13 mutations. Intra-patient reproducibility of the urine KRASG12/G13 testing, calculated as the coefficient variation percent (CV%) for repeat measurements, varied from 2.3% to 19.6%. The average inter-patient reproducibility (CV%) was 9.7% (Table 1).
Table 1.
Reproducibility of the detection of KRASG12/G13 mutations in urine cell-free DNA from patients with advanced cancer. Two to three urine cups (each 90–120 mL) were collected at a single time point from 3 patients with known KRAS mutational status in tumor biopsies. Following urine extraction, cfDNA was assayed by mutation-enrichment NGS. Intra- and inter-patient reproducibility was calculated as CV%.
| Patient, Replicate | KRAS Variant | KRASG12/G13 Copies | CV% | Average CV% |
|---|---|---|---|---|
| 1, 1 | G12S | 18.29 | 2.3 | 9.7 |
| 1, 2 | 17.81 | |||
| 1, 3 | 18.66 | |||
| 2, 1 | G13D | 195.02 | 7.0 | |
| 2, 2 | 176.57 | |||
| 3, 1 | G12D | 10.43 | 19.6 | |
| 3, 2 | 7.26 | |||
| 3, 3 | 7.91 |
Abbreviation: CV%, coefficient variation percent.
Concordance, Sensitivity and Specificity of KRASG12/13 Mutation Detection in Urine cfDNA Compared to Tumor
This blinded study with prospectively collected liquid biopsy samples enrolled 71 patients with diverse advanced cancers and archival formalin-fixed paraffin-embedded (FFPE) tumor specimens with known KRASG12/G13 mutation status (Table 2). The patients’ median age was 59 years (range, 36–85 years). Most patients were white (n=51; 72%) and male (n=38; 54%). The most common tumor type was colorectal cancer (n=56; 79%), followed by breast cancer (n=4; 6%) and NSCLC (n=3; 4%). The median time from tissue to urine sampling was 23.0 months (range, 0.7–91.3 months), and the median time from tissue to plasma sampling was 16.9 months (range, 0.9–80.2 months). The median amount of cfDNA isolated per 1 mL of urine was 9.1 ng (range, 0.2–2057.0 ng) and that isolated per 1 mL of plasma was 18 ng (range, 3.1–605.4 ng).
Table 2.
Characteristics of 71 patients enrolled in the study.
| Characteristic | No. of Patients (%)* |
|---|---|
| Median age (range), years | 59 (36–85) |
| Gender | |
| Male | 38 (54) |
| Female | 33 (46) |
| Ethnicity | |
| Caucasian | 51 (72) |
| Hispanic | 12 (17) |
| African American | 5 (7) |
| Asian | 3 (4) |
| Cancer type | |
| Colorectal cancer | 56 (79) |
| Breast cancer | 4 (6) |
| Non-small cell lung cancer | 3 (4) |
| Pancreatic cancer | 2 (<3) |
| Ovarian cancer | 2 (<3) |
| Other cancers | 4 (6) |
| KRAS status in the tissue | |
| G12C | 7 (10) |
| G12D | 24 (34) |
| G12R | 2 (3) |
| G12S | 6 (8) |
| G12V | 6 (8) |
| G13D | 3 (4) |
| Wild-type | 23 (32) |
| KRAS status in urine cfDNA | |
| G12C | 4 (6) |
| G12D | 17 (24) |
| G12R | 1 (<1) |
| G12S | 4 (6) |
| G12V | 3 (4) |
| G13D | 2 (<3) |
| Wild-type | 40 (56) |
| KRAS status in plasma cfDNA (N=33) | |
| G12C | 2 (6) |
| G12D | 12 (36) |
| G12S | 2 (6) |
| G12V | 3 (9) |
| G13D | 3 (9) |
| Wild-type | 11 (33) |
Unless otherwise indicated.
Of the 71 patients, 49 (69%) had archival tumor specimens with KRASG12/G13 mutations, and 31 (44%) had detectable KRASG12/G13 mutations in urine cfDNA. There was overall concordance in KRASG12/G13 mutation status between urine cfDNA and tumor specimens in 52 cases (73%; kappa, 0.49; standard error [SE], 0.09; 95% confidence interval [CI], 0.31–0.66). The urine cfDNA test had a sensitivity of 63% (95% CI, 0.47–0.76), specificity of 96% (95% CI, 0.78–1.00), and positive predictive value (PPV) of 97% (95% CI, 0.83–1.00; Table 3; Supplementary Table S2).
Table 3.
Concordance assessment of KRASG12/G13 mutations in formalin-fixed, paraffin-embedded (FFPE) tumor tissue and urine cell-free DNA (cfDNA) from patients with advanced cancers.
| Concordance for urine samples
collected before systemic therapy tested for
KRASG12/G13 mutations versus FFPE tumor
samples tested in the clinical laboratory | ||
| Number of patients, N=71 | KRASG12/G13 Mutation in Tumor | KRASG12/G13 Wild-Type in Tumor |
| KRASG12/G13 mutation in cfDNA, no. of patients | 30 | 1 |
| KRASG12/G13 wild-type in cfDNA, no. of patients | 18 | 22 |
| Observed concordance | 52 (73%); kappa, 0.49; SE, 0.09; 95% CI, 0.31–0.66 | |
| Sensitivity | 63% (95% CI, 0.47–0.76) | |
| Specificity | 96% (95% CI, 0.78–1.00) | |
| Positive predictive value | 97% (95% CI, 0.83–1.00) | |
| Concordance for urine samples
(> 50 mL of urine) collected before systemic therapy tested for
KRASG12/G13 mutations versus FFPE tumor
samples tested in the clinical laboratory | ||
| Number of patients, N=43 | KRASG12/G13 Mutation in Tumor | KRASG12/G13 Wild-Type in Tumor |
| KRASG12/G13 mutation in cfDNA, no. of patients | 19 | 0 |
| KRASG12/G13 wild-type in cfDNA, no. of patients | 10 | 14 |
| Observed concordance | 33 (77%); kappa, 0.55; SE, 0.11; 95% CI, 0.34–0.77 | |
| Sensitivity | 66% (95% CI, 0.46–0.82) | |
| Specificity | 100% (95% CI, 0.77–1.00) | |
| Positive predictive value | 100% (95% CI, 0.82–1.00) | |
| Concordance for urine samples
(90–110 mL of urine) collected before systemic therapy tested
for KRASG12/G13 mutations versus FFPE tumor
samples tested in the clinical laboratory | ||
| Number of patients, N=19 | KRASG12/G13 Mutation in Tumor | KRASG12/G13 Wild-Type in Tumor |
| KRASG12/G13 mutation in cfDNA, no. of patients | 8 | 0 |
| KRASG12/G13 wild-type in cfDNA, no. of patients | 2 | 9 |
| Observed concordance | 17 (89%); kappa, 0.79; SE, 0.14; 95% CI, 0.52–1.00 | |
| Sensitivity | 80% (95% CI, 0.44–0.97) | |
| Specificity | 100% (95% CI, 0.66–1.00) | |
| Positive predictive value | 100% (95% CI, 0.63–1.00) | |
Although the recommended volume for urine specimen collection was 90–110 mL, urine specimens with smaller volumes were also collected (median, 60 mL; range, 20–150 mL). Therefore, we investigated whether the collected amount of urine affected the concordance, sensitivity, and specificity of the urine cfDNA test. Among the 43 patients who had urine specimens of > 50 mL, there was overall concordance in KRASG12/G13 mutation status between urine cfDNA and tumor specimens in 33 cases (77%; kappa, 0.55; SE, 0.11; 95% CI, 0.34–0.77), and the urine cfDNA test had a sensitivity of 66% (95% CI, 0.46–0.82), specificity of 100% (95% CI, 0.77–1.00), and PPV of 100% (95% CI, 0.82–1.00; Table 3). Among the 19 patients who had urine specimens of 90–110 mL, there was overall concordance in KRASG12/G13 mutation status between cfDNA and tumor specimens in 17 cases (89%; kappa, 0.79; SE, 0.14; 95% CI, 0.52–1.00), and the urine cfDNA test had a sensitivity of 80% (95% CI, 0.44–0.97), specificity of 100% (95% CI, 0.66–1.00), and PPV of 100% (95% CI, 0.63–1.00; Table 3).
Of the 71 patients, 33 (46%) had simultaneous collection of plasma cfDNA and urine cfDNA. Among these 33 patients, there was overall concordance in KRASG12/G13 mutation status between plasma cfDNA and tumor specimens in 31 cases (94%; kappa, 0.86; SE, 0.10; 95% CI, 0.67–1.00). The plasma cfDNA test had a sensitivity of 92% (95% CI, 0.73–0.99), specificity of 100% (95% CI, 0.66–1.00), and PPV of 100% (95% CI, 0.85–1.00; Table 4; Supplementary Table S2). In addition, there was overall concordance in KRASG12/G13 mutation status between urine cfDNA and plasma cfDNA specimens in 22 cases (67%; kappa, 0.35; SE, 0.15; 95% CI, 0.07–0.64). Using plasma as the reference, the urine cfDNA test (10–110 mL) had a sensitivity of 59% (95% CI, 0.36–0.79), specificity of 82% (95% CI, 0.48–0.98), and PPV of 87% (95% CI, 0.60–0.98; Table 4; Supplementary Table S2).
Table 4.
Concordance assessment of KRASG12/G13 mutations in plasma cell-free DNA (cfDNA) and formalin-fixed, paraffin-embedded (FFPE) tumor tissue or urine cfDNA from patients with advanced cancers.
| Concordance for plasma samples
collected before systemic therapy tested for
KRASG12/G13 mutations versus FFPE tumor
samples tested in the clinical laboratory | ||
| Number of patients, N=33 | KRASG12/G13 Mutation in Tumor | KRASG12/G13 Wild-Type in Tumor |
| KRASG12/G13 mutation in plasma, no. of patients | 22 | 0 |
| KRASG12/G13 wild-type in plasma, no. of patients | 2 | 9 |
| Observed concordance | 31 (94%); kappa, 0.86; SE, 0.10; 95% CI, 0.67–1.00 | |
| Sensitivity | 92% (95% CI, 0.73–0.99) | |
| Specificity | 100% (95% CI, 0.66–1.00) | |
| Positive predictive value | 100% (95% CI, 0.85–1.00) | |
| Concordance for plasma and
urine samples collected before systemic therapy tested for
KRASG12/G13 mutations | ||
| Number of patients, N=33 | KRASG12/G13 mutation in plasma | KRASG12/G13 wild-type in plasma |
| KRASG12/G13 mutation in urine, no. of patients | 13 | 2 |
| KRASG12/G13 wild-type in urine, no. of patients | 9 | 9 |
| Observed concordance | 22 (67%); kappa, 0.35; SE, 0.15; 95% CI, 0.07–0.64 | |
| Sensitivity | 59% (95% CI, 0.36–0.79) | |
| Specificity | 82% (95% CI, 0.48–0.98) | |
| Positive predictive value | 87% (95% CI, 0.60–0.98) | |
KRASG12/G13-Mutant Copy Number and cfDNA Concentration and Survival
To determine whether the number of KRASG12/G13-mutant copies in urine cfDNA was associated with OS, we first divided the 71 patients into 2 groups: those with < 26.3 KRASG12/G13-mutant copies and those with ≥ 26.3 KRASG12/G13-mutant copies. The threshold was selected based on a 5% trimmed mean value of KRASG12/G13-mutant cfDNA. This was deemed to be appropriate as the median percentage of KRASG12/G13-mutant cfDNA was 0% because 40 of the 71 patients had no KRASG12/G13 mutations in urine cfDNA. The median OS duration of the 57 patients with < 26.3 KRASG12/G13-mutant copies (11.1 months; 95% CI, 7.5–14.7 months) and that of the 14 patients with ≥ 26.3 of KRASG12/G13-mutant copies (16.5 months; 95% CI, 5.3–27.7 months) did not differ significantly (P = 0.63; Supplementary Fig. S3A). Similarly, again using a threshold selected based on a 5% trimmed mean, we found that the median OS duration of the 23 patients with < 198.8 KRASG12/G13-mutant copies in plasma cfDNA (18.7 months; 95% CI, 3.5–33.9 months) and that of the 10 patients with ≥ 198.8 KRASG12/G13-mutant copies in plasma cfDNA (12.6 months; 95% CI, 11.6–13.4 months) did not differ significantly (P = 0.90; Supplementary Fig. S3B).
We next analyzed whether cfDNA concentrations in urine or plasma were associated with OS using thresholds selected based on median values. For the 69 of 71 patients for whom urine cfDNA data were available, the median OS duration of the 35 patients with < 9.1 ng of cfDNA/mL (13.0 months; 95% CI, 7.2–18.8 months) and that of the 34 patients with ≥ 9.1 ng of cfDNA/mL (11.1 months; 95% CI, 7.4–14.8 months) did not differ significantly (P = 0.31; Supplementary Fig. S4A). Similarly, for the 33 patients for whom plasma cfDNA data were available, the median OS duration of the 16 patients with < 18.0 ng of cfDNA/mL (12.6 months; 95% CI, 5.9–19.2 months) and that of the 17 patients with ≥ 18 ng of cfDNA/mL (20.6 months; 95% CI, 5.9–35.3 months) did not differ significantly (P = 0.19; Supplementary Fig. S4B).
Serial Monitoring for KRASG12/13 Mutations in the cfDNA of Cancer Patients on Therapy
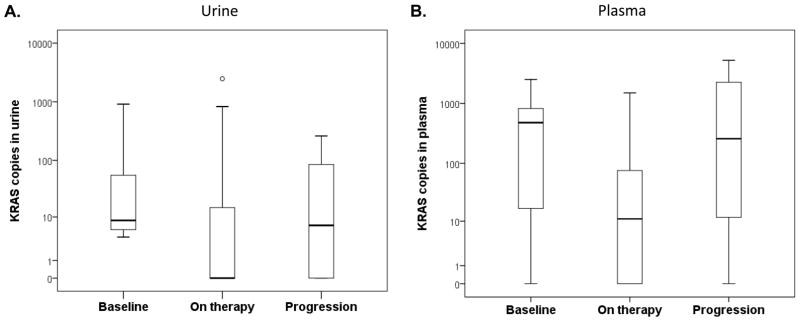
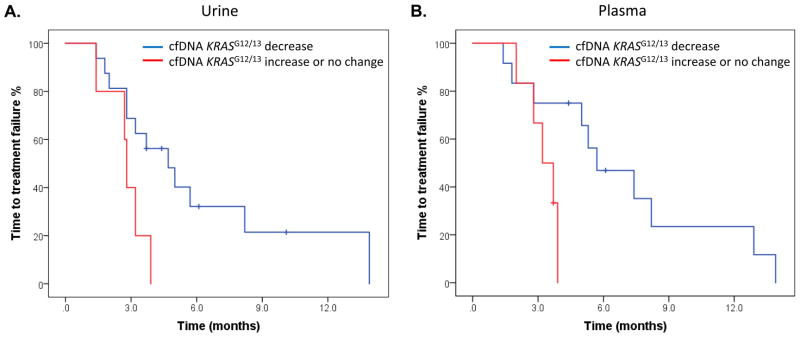
At least 2 (median, 6; range, 2–13) longitudinal serial urine collections were obtained before and during patients’ systemic therapy, which ranged from first-line therapies to experimental therapies after all standard treatment had failed, from 21 patients with KRASG12/G13 mutations in tumor tissue. Of these 21 patients, 17 (81%) had detectable KRASG12/G13 mutations in cfDNA in ≥ 1 urine specimen. The median KRASG12/G13 copy numbers in specimens collected at baseline (8.6), during therapy (0), and at disease progression (6.9) differed significantly (P = 0.002; Fig. 2A). The patients received 21 diverse systemic therapies (Supplementary Table S3). The best response to therapy (complete response [CR] or partial response [PR] or stable disease [SD] ≥ 6 months vs. SD < 6 months or progressive disease [PD]) on imaging per Response Evaluation Criteria in Solid Tumors (RECIST) was not associated with the best change in KRASG12/G13 copy numbers (median change percentage, −100% for patients with CR/PR/SD ≥ 6 months vs. −100% for patients with SD < 6 months/PD; P = 0.24) (23). Of the 21 therapies, 16 decreased the KRASG12/G13 copy numbers, and 5 caused no change or increased the KRASG12/G13 copy numbers. The median TTF of the patients with a decrease in KRASG12/G13 copy numbers (4.7 months; 95% CI, 2.6–6.8 months) was significantly longer than that of the patients with no change or an increase in copy numbers (2.8 months; 95% CI, 2.6–3.0 months; P = 0.03; Fig. 3A).
Figure 2.
A. The median KRASG12/G13 copy numbers in urine at baseline (8.6), on therapy (0), and at disease progression (6.9) differed significantly (P = 0.002). B. The median KRASG12/G13 copy numbers in plasma at baseline (488.5), during therapy (11.0), and at disease progression (258.6) also differed significantly (P < 0.001).
Figure 3.
Association between changes in cell-free DNA KRASG12/13 copies and time to treatment failure (TTF). A. The median TTF of patients with a decrease in KRASG12/G13 copy numbers in urine (4.7 months; 95% CI, 2.6–6.8 months; blue) was significantly longer than that of patients with no change or an increase in KRASG12/G13 copy numbers in urine (2.8 months; 95% CI, 2.6–3.0 months; red; P = 0.03). B. The median TTF of patients with a decrease in KRASG12/G13 copy numbers in plasma (5.7 months; 95% CI, 2.8–8.6 months; blue) was significantly longer than that of patients with no change or an increase in KRASG12/G13 copy numbers in plasma (3.2 months; 95% CI, 2.1–4.3 months; red; P = 0.04).
At least 2 (median, 5.5; range, 3–14) serial plasma collections were obtained before and during systemic therapy from 18 patients with KRASG12/G13 mutations in tumor tissue. All 18 patients had detectable KRASG12/G13 mutations in cfDNA in ≥ 1 plasma specimen. The median KRASG12/G13 copy numbers at baseline (488.5), during therapy (11.0), and at disease progression (258.6) differed significantly (P < 0.001; Fig. 2B). The patients received 20 diverse systemic therapies (Supplementary Table S3). The best response to therapy (CR, PR, or SD ≥ 6 months vs. SD < 6 months or PD) on imaging per RECIST showed a trend towards association with the best change in copy numbers (median change percentage, −100% for CR/PR/SD ≥ 6 months vs. −36% in SD < 6 months/PD; P = 0.09). Of the 18 therapies (2 therapies were excluded because of missing pre-treatment KRASG12/G13 copy number values), 12 decreased the KRASG12/G13 copy numbers, and 6 caused no change or increased KRASG12/G13 copy numbers. The median TTF of the patients with a decrease in KRASG12/G13 copy numbers (5.7 months; 95% CI, 2.8–8.6 months) was significantly longer than that of patients with no change or an increase in copy numbers (3.2 months; 95% CI, 2.1–4.3 months; P = 0.04; Fig. 3B).
DISCUSSION
Our findings demonstrate that mutation enrichment leads to an approximately 3,000-fold increase of the KRASG12/G13-mutant signal over the wt signal, which allows the detection of low-frequency mutant copies in samples of urine cfDNA. In a blinded study with prospectively collected samples, our assay using mutation-enrichment PCR coupled with NGS detected KRASG12/G13-mutant copies in urine cfDNA from patients with advanced cancers and had acceptable concordance (73–89%), sensitivity (63–80%), and specificity (96–100%) compared with the clinical testing of FFPE tumor tissue obtained at different times during routine care. The concordance increased with the amount of urine collected, which is ideally 90–110 mL. Furthermore, in a subset of patients for whom plasma cfDNA was available, we demonstrated excellent concordance of 94% with FFPE tumor tissue (sensitivity, 92%; specificity, 100%).
Although preliminary data on the molecular testing of urine cfDNA have been published, to our knowledge, ours is the first report of the development and laboratory and clinical validation of a urine cfDNA assay, whose concordance with testing of clinical samples appears to be similar to previously published data on plasma cfDNA (10, 21). One recent study demonstrated in a similar patient population that the testing of plasma cfDNA for KRASG12/G13 mutations with BEAMing PCR is concordant with the standard-of-care mutation analysis of FFPE primary or metastatic tumor in 83% of patients (24). A certain level of discordance can be anticipated if the tumor tissue and plasma are obtained at different times. Higgins et al. (25) found 100% concordance between testing plasma cfDNA with BEAMing PCR and testing simultaneously collected tumor tissue with conventional methods for PIK3CA mutations in a cohort of patients with advanced breast cancer. However, the concordance between the methods decreased to 79% in a cohort of patients whose tumor and plasma cfDNA samples were obtained at different times, which is consistent with our results. In another study of 100 patients with advanced colorectal cancer, droplet digital PCR detection of RAS mutations in plasma cfDNA was in concordance with archival tissue in 97% of cases (20). This rate was favorable compared with most other studies; however, the median time from tissue to plasma collection was only 43 days, which could explain the high concordance rate. In a phase III randomized trial of regorafenib vs. placebo, Tabernero et al. (26), using BEAMing PCR, showed concordant KRAS mutation status between plasma-derived cfDNA and archival tumor samples in 76% of tested patients with advanced colorectal cancer. Thierry et al. (27), using allele-specific quantitative PCR of plasma cfDNA and mutation detection in primary or metastatic tissue, demonstrated a 96% concordance for combined KRAS and BRAF mutation testing. Finally, Sacher et al. (28), in the only prospective study to date, demonstrated that digital droplet PCR detected KRASG12 mutations in the plasma cfDNA in 64% of patients with known KRASG12 mutations in the tumor. Compared with most of these previous studies’ findings, our concordance results for KRASG12/G13 mutations in urine cfDNA were similar, and those for KRASG12/G13 mutations in plasma cfDNA were favorable, despite the fact that the median times between archival tumor tissue collection and urine or plasma collection were relatively long (23.0 months and 16.9 months, respectively) and that fact that urine cfDNA is a far more challenging material because of its short fragments and low mutation allele frequencies (25–29). There is increasing evidence that the mutation analysis results for cfDNA are highly concordant with those for archival tumor tissue for concordantly, but not discordantly, collected samples, which may be explained by tumor biology, including tumor heterogeneity and evolution, and preanalytical factors such as inadequate specimen collection (28, 30). In addition, testing of urine cfDNA offers a completely non-invasive method and urine collection does not need to be done by a trained personnel, which can expand the use of molecular cfDNA testing.
In our study, we did not find any relationship between OS and KRASG12/G13 copy number values in urine or plasma cfDNA. An earlier study using BEAMing PCR to assess plasma cfDNA for KRASG12/G13 mutations in patients with advanced cancers found that a high amount of KRAS-mutant cfDNA was associated with shorter OS duration (4.8 months vs. 7.3 months; P = 0.008) (24). Another study that used the Idylla system to detect BRAFV600 mutations in plasma-derived cfDNA from patients with diverse advanced cancers showed that a higher percentage of BRAFV600-mutant cfDNA was associated with shorter OS (4.4 months vs. 10.7 months, P = 0.005) (31). Similarly, the phase III randomized trial of regorafenib vs. placebo showed that high baseline levels of KRAS-mutant cfDNA were associated with shorter OS durations in patients with advanced colorectal cancer (26). In other studies, higher amounts of KRAS-mutant cfDNA were associated with shorter OS durations in patients with advanced colorectal cancer treated with irinotecan and cetuximab and in patients with advanced NSCLC treated with carboplatin and vinorelbine (32, 33). Similarly, in a combined analysis of clinical trials of BRAF and MEK inhibitors in patients with advanced melanomas, a BRAFV600E mutation in cfDNA was associated with shorter OS duration (34). In contrast, in a study of patients with advanced NSCLC, those with EGFR exon 19 deletion in both the tissue and cfDNA had better survival than patients with EGFR exon 19 deletion in the tissue only (35). The results of our study may have been affected by the heterogeneity in the tumor types, setting of treatment administration (from first-line to third-line and higher, including clinical trials), and participating institutions and/or by its small sample sizes and large proportion of samples with less-than-optimal urine volumes; these factors may also explain some of the differences between our findings and those of previous studies. A larger prospective study to validate the clinical utility of KRAS mutation detection in the urine of patients with advanced colorectal cancer and it is association with treatment outcomes is ongoing.
Previous studies have investigated the use of detecting molecular aberrations in cfDNA to monitor response to cancer therapy (19, 21, 36–44). In the present study, we assessed serially collected urine and plasma cfDNA from patients treated with systemic therapies and found that the KRASG12/G13 copy numbers before therapy, during therapy, and at the time of disease progression differed significantly. We also found that patients with a decrease in KRASG12/G13 copy numbers in serially collected urine or plasma cfDNA during therapy had a longer median TTF compared with patients with no change or an increase in copy numbers (4.7 vs. 2.8 months, P = 0.03 for urine; 5.7 vs. 3.2 months, P = 0.04 for plasma). This observation is consistent with previously published data demonstrating that changes in plasma cfDNA can correspond with treatment outcomes (28, 29, 37–44). In particular, a study using the Idylla system to detect BRAFV600 mutations in plasma-derived cfDNA from patients with colorectal or other advanced cancers found that the median TTF of patients who received therapies associated with a decrease in BRAF-mutant cfDNA (10.3 months) was significantly longer than that of patients who received therapies associated with an increase or no change in BRAF-mutant cfDNA (7.4 months, P = 0.045) ((31). Overall, however, there is conflicting evidence that such changes in cfDNA can predict or at least correspond with treatment outcomes, and this issue will need to be investigated in future prospective studies.
Our study had several potential limitations. First, the amount of collected urine was suboptimal in many cases, which likely negatively impacted concordance and could have impacted serial monitoring analysis. Second, our study did not investigate if the timing of urine collection can impact results. Third, the sample size was limited. Fourth, we investigated only KRASG12/G13 mutations, which are clinically relevant to only a limited number of patients with certain tumor types. Finally, because of the heterogeneity in tumor types, systemic therapies and exploratory nature of the longitudinal analysis, the association between changes in mutant cfDNA and TTF needs to be validated in future prospective studies.
In summary, our study demonstrates that using mutation-enrichment PCR coupled with NGS to molecularly analyze urine cfDNA for the 7 most frequent hotspot KRASG12/G13 mutations is feasible and has good concordance with standard mutation testing of discordantly collected FFPE tumor tissue. Our results also suggest that the dynamics of KRASG12/G13-mutant copies in cfDNA corresponds with TTF. The clinical utility of cfDNA mutation testing is gaining increasing acceptance. Regulatory agencies in the United States and European Union have recently approved the use of an EGFR mutation plasma cfDNA test for advanced NSCLC when tissue is not available. The clinical utility of serial cfDNA testing is promising and should be further proven in future prospective clinical trials in which therapeutic interventions are tailored based on patients’ respective cfDNA mutation statuses.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
In patients with advanced cancers, mutation-enrichment next-generation sequencing detection of KRASG12/G13 mutations in urine cell-free DNA has good concordance with conventional clinical testing of archival tumor tissue, provided that the volume of collected urine is sufficient. Changes in mutated cell-free DNA correspond with time to treatment failure on systemic anticancer therapy.
Acknowledgments
Financial support: This study was supported by Trovagene, the Sidney Kimmel Foundation for Cancer Research, the Sheikh Khalifa Al Nahyan Ben Zayed Institute for Personalized Cancer Therapy, and the National Center for Advancing Translational Sciences (grant no. UL1 TR000371); by the National Institutes of Health through MD Anderson’s Cancer Center Support Grant (P30 CA016672) and the University of Southern California’s Cancer Center Support Grant (P30 CA014089); by the European Community’s Seventh Framework Programme under grant agreement no. 602901 MErCuRIC, European Community’s Seventh Framework Programme under grant agreement no. 635342-2 MoTriColor, IMI contract n. 115749 CANCER-ID, AIRC 2010 Special Program Molecular Clinical Oncology 5 per mille, Project n. 9970, Fondazione Piemontese per la Ricerca sul Cancro-ONLUS 5 per mille 2011 Ministero della Salute AIRC IG n. 16788, AIRC Special program 5xmille “Targeting resistances to molecular therapies in metastatic colorectal carcinomas,” European Community’s Seventh Framework Programme “Colon Therapies Research, COLTHERES”; and by Fondazione Oncologia Niguarda Onlus.
We thank Giran Cabrilo, Kiran Midwani, Rose Champ, and Debra Andrews for coordinating biospecimen collection and Latifa Hassaine and Benedetta Mussolin for assistance with urine analyses.
Footnotes
Conflict of interest: Filip Janku has research support from Novartis, Agios, Astellas, Deciphera, Symphogen, Piqur, Roche, BioMed Valley Discoveries, and Trovagene and is on the Scientific Advisory Boards of Deciphera and Guardant Health. Giulia Siravegna is a consultant for Trovagene. Federica Di Nicolantonio has research support from Trovagene. Alberto Bardelli has research support from Trovagene and is on the Scientific Advisory Boards of Horizon Discovery, Trovagene, and Biocartis.
References
- 1.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–67. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 2.Falchook GS, Long GV, Kurzrock R, Kim KB, Arkenau TH, Brown MP, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet. 2012;379:1893–901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–34. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 4.Janku F, Wheler JJ, Westin SN, Moulder SL, Naing A, Tsimberidou AM, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012;30:777–82. doi: 10.1200/JCO.2011.36.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:738–46. doi: 10.1016/S1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 6.Said R, Ye Y, Falchook GS, Janku F, Naing A, Zinner R, et al. Outcomes of patients with advanced cancer and KRAS mutations in phase I clinical trials. Oncotarget. 2014;5:8937–46. doi: 10.18632/oncotarget.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–39. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–8. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 9.Tsimberidou AM, Hong DS, Wheler JJ, Fu S, Piha-Paul S, Naing A, et al. Profile-related evidence to determine individualized cancer therapy (PREDICT): Preliminary results of the Personalized Phase I Clinical Trials program at MD Anderson Cancer Center. Proceedings of the 102nd Annual Meeting of the American Association for Cancer Research; 2011. p. 1287. [Google Scholar]
- 10.Janku F, Vibat CR, Kosco K, Holley VR, Cabrilo G, Meric-Bernstam F, et al. BRAF V600E mutations in urine and plasma cell-free DNA from patients with Erdheim-Chester disease. Oncotarget. 2014;5:3607–10. doi: 10.18632/oncotarget.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsimberidou AM, Iskander NG, Hong DS, Wheler JJ, Fu S, Piha-Paul SA, et al. Personalized medicine in a phase I clinical trials program: The M. D. Anderson Cancer Center Initiative. J Clin Oncol. 2011;29 doi: 10.1158/1078-0432.CCR-12-1627. abstr CRA2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupont Jensen J, Laenkholm AV, Knoop A, Ewertz M, Bandaru R, Weihua L, et al. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in Breast Cancer. Clin Cancer Res. 2010 doi: 10.1158/1078-0432.CCR-10-1133. [DOI] [PubMed] [Google Scholar]
- 13.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Angulo AM, Ferrer-Lozano J, Stemke-Hale K, Sahin A, Liu S, Barrera JA, et al. PI3K pathway mutations and PTEN levels in primary and metastatic breast cancer. Mol Cancer Ther. 2011;10:1093–101. doi: 10.1158/1535-7163.MCT-10-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polivka J, Jr, Pesta M, Janku F. Testing for oncogenic molecular aberrations in cell-free DNA-based liquid biopsies in the clinic: are we there yet? Expert review of molecular diagnostics. 2015;15:1631–44. doi: 10.1586/14737159.2015.1110021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morelli MP, Overman MJ, Dasari A, Kazmi SM, Mazard T, Vilar E, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015;26:731–6. doi: 10.1093/annonc/mdv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21:795–801. doi: 10.1038/nm.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyman DM, Diamond EL, Vibat CR, Hassaine L, Poole JC, Patel M, et al. Prospective blinded study of BRAFV600E mutation detection in cell-free DNA of patients with systemic histiocytic disorders. Cancer Discov. 2015;5:64–71. doi: 10.1158/2159-8290.CD-14-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reckamp KL, Melnikova VO, Karlovich C, Sequist LV, Camidge DR, Wakelee H, et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J Thorac Oncol. 2016 doi: 10.1016/j.jtho.2016.05.035. [DOI] [PubMed] [Google Scholar]
- 23.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 24.Janku F, Angenendt P, Tsimberidou AM, Fu S, Naing A, Falchook GS, et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies. Oncotarget. 2015;6:12809–21. doi: 10.18632/oncotarget.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. 2012;18:3462–9. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabernero J, Lenz HJ, Siena S, Sobrero A, Falcone A, Ychou M, et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015;16:937–48. doi: 10.1016/S1470-2045(15)00138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20:430–5. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 28.Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O’Connell A, Feeney N, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, et al. Serial Next-Generation Sequencing of Circulating Cell-Free DNA Evaluating Tumor Clone Response To Molecularly Targeted Drug Administration. Clin Cancer Res. 2015;21:4586–96. doi: 10.1158/1078-0432.CCR-15-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meric-Bernstam F, Frampton GM, Ferrer-Lozano J, Yelensky R, Perez-Fidalgo JA, Wang Y, et al. Concordance of genomic alterations between primary and recurrent breast cancer. Mol Cancer Ther. 2014;13:1382–9. doi: 10.1158/1535-7163.MCT-13-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janku F, Huang HJ, Claes B, Falchook GS, Fu S, Hong D, et al. BRAF Mutation Testing in Cell-Free DNA from the Plasma of Patients with Advanced Cancers Using a Rapid, Automated Molecular Diagnostics System. Mol Cancer Ther. 2016 doi: 10.1158/1535-7163.MCT-15-0712. [DOI] [PubMed] [Google Scholar]
- 32.Nygaard AD, Garm Spindler KL, Pallisgaard N, Andersen RF, Jakobsen A. The prognostic value of KRAS mutated plasma DNA in advanced non-small cell lung cancer. Lung Cancer. 2013;79:312–7. doi: 10.1016/j.lungcan.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Spindler KL, Pallisgaard N, Vogelius I, Jakobsen A. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res. 2012;18:1177–85. doi: 10.1158/1078-0432.CCR-11-0564. [DOI] [PubMed] [Google Scholar]
- 34.Santiago-Walker A, Gagnon R, Mazumdar J, Casey M, Long GV, Schadendorf D, et al. Correlation of BRAF Mutation Status in Circulating-Free DNA and Tumor and Association with Clinical Outcome across Four BRAFi and MEKi Clinical Trials. Clin Cancer Res. 2016;22:567–74. doi: 10.1158/1078-0432.CCR-15-0321. [DOI] [PubMed] [Google Scholar]
- 35.Karachaliou N, Mayo-de las Casas C, Queralt C, de Aguirre I, Melloni B, Cardenal F, et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol. 2015;1:149–57. doi: 10.1001/jamaoncol.2014.257. [DOI] [PubMed] [Google Scholar]
- 36.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 38.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Science translational medicine. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 39.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–12. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 40.Oxnard GR, Paweletz CP, Kuang Y, Mach SL, O’Connell A, Messineo MM, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20:1698–705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res. 2015;21:3196–203. doi: 10.1158/1078-0432.CCR-14-2594. [DOI] [PubMed] [Google Scholar]
- 42.Karlovich C, Goldman JW, Sun JM, Mann E, Sequist LV, Konopa K, et al. Assessment of EGFR Mutation Status in Matched Plasma and Tumor Tissue of NSCLC Patients from a Phase I Study of Rociletinib (CO-1686) Clin Cancer Res. 2016;22:2386–95. doi: 10.1158/1078-0432.CCR-15-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchetti A, Palma JF, Felicioni L, De Pas TM, Chiari R, Del Grammastro M, et al. Early Prediction of Response to Tyrosine Kinase Inhibitors by Quantification of EGFR Mutations in Plasma of NSCLC Patients. J Thorac Oncol. 2015;10:1437–43. doi: 10.1097/JTO.0000000000000643. [DOI] [PubMed] [Google Scholar]
- 44.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.