Abstract
Taking into account the limits of current genotyping methodologies, we have established a versatile direct PCR method on intact microtissue samples without prior DNA isolation. A simple and standard protocol was developed and validated on a wide range of living organisms including bacterial and fungal strains, plant species and human samples. This allows reliable amplification of target genomic DNA fragment directly from source material using minimal amount of tissue which makes DNA purification irrelevant for a number of biological applications. The direct PCR technique established here represents an excellent alternative to traditional amplification methods used for real-time detection. Since this approach was efficiently and universally applied for high-throughput molecular screening, its implementation will offer new insights for several investigations in human health, biomedical diagnosis, plant biotechnology, as well as in applied environmental and food microbiology.
Keywords: Direct PCR, Wide range amplification, DNA extraction, Molecular screening and genotyping, Biomedical diagnosis, Molecular detection
Introduction
Polymerase chain reaction (PCR) is known as a routine molecular biology technique widely used in various fields and for several purposes. Scarcely any invention has altered biological science so radically in such a short period as the PCR. Such reaction requires purified nucleic acid as template for target DNA amplification. Commonly, DNA template must be first extracted and then purified using commercial kits or lab’s protocols. Large number of procedures for isolation of highly purified DNA from a big range of bacteria, fungi, animal and plant species was reported (Ahmed et al. 2009). These methods include usually treatment with proteinase K in the presence of SDS and/or NaOH for alkaline lysis of cell membranes. Nucleic DNA thus liberated is subjected frequently to phenol–chloroform purification, ethanol precipitation and subsequent resuspension of DNA pellet in nuclease-free water. Such procedures are often laborious and time-consuming, with handling of toxic organic solvents, especially when we are used to manage a large number of DNA preparations. One of the major limitations has been the inability to apply PCR directly to tissue samples. The main cause appears to be the presence of inhibitors in samples that has been the focus of much of the published literature (Watson and Blackwell 2000). The direct amplification could be achieved if PCR-inhibitors may be avoided.
Direct PCR is rising as one of the most useful molecular techniques applied in many fields of biological research and diagnostic. During the last years, few data have been reported on direct amplification without prior DNA isolation. So far, this approach has been applied already to bacterial colony (Eszik et al. 2016; Pathmanathan et al. 2003), yeasts and fungi (AlShahni et al. 2009; Ben-Amar et al. 2012), plant and algal species (Rogers and Parkes 1999; Sharma et al. 2012), animal tissues (Shokralla et al. 2010; Werblow et al. 2016) and human whole blood (Cascella et al. 2015; Nishimura et al. 2000). Most of described protocols frequently require alkaline lysis and often need high performance DNA Taq polymerases (AlShahni et al. 2009; Cascella et al. 2015). Different procedures were used, but to our knowledge no standard protocol has been provided.
We describe here a versatile, reliable and standardized direct PCR-based approach to amplify genomic DNA derived from eukaryotic and prokaryotic organisms using limited amount of samples as crude templates. This technique provides considerable benefits of speed, robustness and efficiency as compared with conventional PCR as well as with other specific direct PCR methods.
Materials and methods
Biological material
Biological material used in the present study includes microorganisms (bacteria, fungi), plant leaf tissue, human samples, as well as soil, wastewater and food samples. Four bacterial strains were used and cultured overnight on LB medium, respectively, at 28 °C for Agrobacterium tumefaciens and at 37 °C for the others strains (Escherichia coli, Pseudomonas aeruginosa and Bacillus subtilis). Individual colony from each strain was used for further amplification. Fungal isolates included in this study (Aspergillus carbonarius, Penicillium verrucosum, Botrytis cinerea and Fusarium culmorum) were grown on PDA medium at 25 °C for 4–5 days until mycelium completely covered the agar surface. All bacteria and fungi used as microbiological specimens for dir-PCR assays belong to the microorganism collection of the laboratory.
Wastewater (residual irrigation water) and soil (1:2 sand, 1:2 peat-based potting mix, Blocking substrate, Klasmann-Deilmann GmbH, Germany) taken as environmental samples, along with food samples (strawberry fruit, milk and bread) were heat sterilized by autoclaving at 120 °C for 24 min to ensure their microorganism decontamination. These samples were further artificially contaminated by F. culmorum as described previously (Ben-Amar et al. 2012) and incubated at 25 °C for 4 days before being used directly for PCR-based detection of fungi without prior DNA isolation.
Small amount of leaf disc tissue (~5 mm2, corresponding to 10–15 mg) were collected from different plant species (tobacco Nicotiana benthamiana, tomato Solanum lycopersicum, potato Ipomea batatas, cactus Opuntia ficus-indica, and grapevine Vitis vinifera) and used as direct template for PCR. Human blood (1 mm2 of dried spot on Whatman paper), hair (2–5 mm long), skin (1 mm2) and buccal cell samples were also tested as templates for direct PCR assays.
DNA purification
Bacterial DNA isolation was performed using Wizard® Genomic DNA purification system (Promega, USA) according to the manufacturer’s instructions. Fungal genomic DNA was prepared using TNE solution (10 mM Tris-HCl pH 8, 1 mM EDTA, 100 mM NaCl) combined with 2% SDS treatment and phenol–chloroform purification as described previously (Ben-Amar et al. 2012). Plant DNA extraction was achieved using the MasterPure™ Plant Leaf DNA purification kit (Epicentre® Biotechnologies, USA). Human DNA was extracted from fresh blood with a DNA Isolation Kit for cells and tissue (Roche, Germany). All purified genomic DNA were adjusted to the concentration of 100 ng/μl and used as positive controls for PCR.
Sample preparation for direct PCR
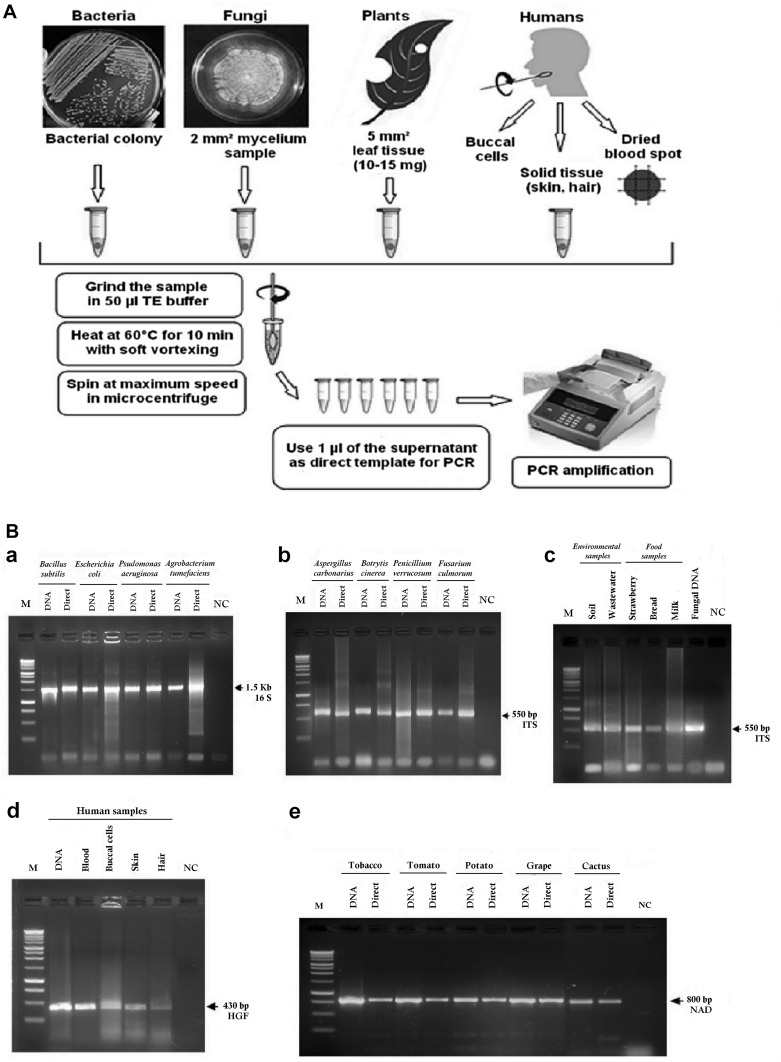
For direct PCR assays, bacterial colony was taken directly with sterile tip from LB agar plate into 1.5-ml microfuge tube and subjected to a brief heat shock (60 °C for 10 min followed by 2 min on ice). In the same way, a small amount (~2 mm2) of fungal mycelium was scratched from Petri dish and added to the PCR mix. For plant leaf discs as well as human tissue samples (skin, blood, hair root, buccal cells), we take as small sample as possible (~1–2 mm2) that is subjected to slight grinding in 50 μl of TE buffer with soft vortexing following incubation for 10 min at 60 °C. One microliter of this mixture was used directly for PCR (Fig. 1A). For environmental and food samples intended for fungal detection assay, small aliquots (approximately 10 mg of solid or 1 μl of liquid sample) of soil, polluted water, contaminated food were transferred in the same manner into PCR mix.
Fig. 1.
Direct PCR protocol universally adapted to living organism-derived samples including plants, animals, bacteria and fungi. A Flow chart presenting the direct PCR technique. B Amplification of constitutive genes (16S, ITS, HGF, NAD) from direct templates without DNA purification, respectively, using bacterial colonies (a), fungal mycelia, environmental samples and food products (b, c), human tissues (d) and plant leaf discs (e). Conventional PCR using purified genomic DNA (DNA) was performed versus direct PCR from crude templates (direct). DNA purified genomic DNA was presented as a positive control, M 1 Kb DNA ladder, NC negative control
PCR amplification
Oligonucleotide primers used for in the present study are listed in Table 1. Amplification reactions were performed in 20 μl containing 4 μl 5x reaction buffer, 1.6 μl of MgCl2 25 mM, 0.25 μl of dNTPs mix 10 mM, 0.25 μl of each primer 10 μM, 1 unit of GoTaq® DNA polymerase (Promega). One microliter of sample dilution mixture was used for direct PCR. Conventional PCR was performed using 100 ng of purified genomic DNA template as positive control. All PCR was achieved in GeneAmp® PCR system 9700 cycler (Applied Biosystems) using an initial 5 min denaturation at 94 °C followed by 35 cycles of 30 s denaturation at 94 °C, 45 s annealing at Tm (Table 1), 30 s extension at 72 °C, followed by final extension of 7 min at 72 °C. Amplified products (10 μl) were loaded on a 1.2% (w/v) agarose gel with TAE buffer and stained with ethidium bromide before revelation under UV light. PCR products were sequenced by the same primer set as the PCR amplification using ABI-PRISM 3130 Genetic Analyser Sequencer (Applied Biosystems) and obtained DNA sequences were analyzed with Clustal X software. PCR was repeated at least five times during optimization step to ensure reproducibility.
Table 1.
Primer sequences used for PCR amplification assays
| Primer pairs | Target sequence 5′–3′ | Target gene | Tm | Amplicon size | Specificity | Source |
|---|---|---|---|---|---|---|
| 16S-F 16S-R | AGAGTTTGATCCTGGCTCAG ACGGCTACCTTGTTACGACTT | 16-S ribosomal DNA (16 S rDNA) | 58 °C | ~1500 bp | Bacterial strains | Turner et al. (1999) |
| ITS-1 ITS-4 | TCCGTAGGTGAACCTGCGG TCCTCCGCTTATTGATATGC | Internal transcribed spacer (ITS) | 58 °C | ~550 bp | Fungal isolates | White et al. (1990) |
| HGF-F HGF-R | GCCTCCCCAACCATTCCCTTA TCACGGATTTCTGTTGTGTTTC | Human growth factor (HGF) | 60 °C | ~430 bp | Human tissues | Tomida and Saito (2004) |
| NAD-F NAD-R | ATCACCGAACCTGCACTCAGGAA TAGCCCGACCGTAGTGATGTTAA | Nicotinamide adenine dinucleotide (NAD) | 68 °C | ~800 bp | Plant species | Mannerlöf and Tenning (1997) |
Results and discussion
We investigated the PCR amplification directly from minute amount of samples without undergoing DNA isolation process. The direct PCR was carried out using universal primers and standard cycling conditions. A standard protocol was established using different kinds of templates (bacterial colony, mycelium, leaf disc, human tissues, cells and blood) as summarized in Fig. 1A.
The present work provides a simplified approach as untreated samples are used directly for PCR eliminating additional steps of cell cultivation and recovery or DNA extraction. The identity of all the obtained PCR products was confirmed by sequencing. The direct PCR was applied to several investigation areas including food control and environmental microbiology, human health and genetic disease diagnosis, agriculture and plant biotechnology.
Direct PCR in fundamental and applied microbiology
The used method provides a suitable mean to detect bacteria strains and fungi from the laboratory collection. The 16S primers yielded a 1500 bp fragment corresponding to bacterial ribosomal DNA amplified directly from single bacterial colony (Fig. 1B-a). While, for fungi, the amplification of the internal transcribed spacer (ITS1-5.8S-ITS2) of ribosomal DNA (rDNA) gave a 550 bp PCR product (Fig. 1B-b). This approach was also applicable either for spores or hyphal cells (data not shown). The technique is helpful as it allows a prior diagnosis of these microorganisms and will permit to save time to undergo microbiological assays dealing especially with identification purposes. In the present work, we targeted constitutive genes to detect bacteria and fungi.
The use of specific primers designed to target particular genes will permit the direct identification of microorganism specimens without requiring classic microbiological identification or DNA extraction methods. The direct PCR analysis was previously reported for detection of toxin-producing E. coli (Fode-Vaughan et al. 2003) using specific primers of functional genes. AlShahni et al. (2009) described a direct colony PCR of several medically important fungi while only 30% of the fungal strains analyzed showed PCR products when using standard PCR reagents. However, these methods required a long incubation period in lysis reagent or a cell fixation step using paraformaldehyde under saline buffer, which is labor and time-consuming. Our improved strategy provides a rapid amplification with neither serial dilution, nor extended incubation which reduces significantly template preparation time and also the cost of PCR.
Direct PCR in environmental samples and food industry
As soil or water may be contaminated with microorganisms, the direct PCR allows the identification of potential pathogens and provides a rapid tool to evaluate the risk assessment. The soil has been considered for a long time as an inhibitor-containing substrate which resulted to complete failure of the PCR (Watson and Blackwell 2000). Our data resulted in a high-quality yield with clear band profile of PCR products observed after agarose gel electrophoresis (Fig. 1B-c). Besides fungal detection on environmental samples as soil or water, processed food products (milk, bread) could be also a potential vector of fungi when they are improperly stored. Our method permitted to obtain PCR product targeting ITS region of rDNA with expected size of 550 bp indicating the presence of fungal, avoiding the PCR inhibiting-substances in food matrix (Fig. 1B-c). Among these, proteinases and calcium ions have been identified in milk as a source of PCR inhibition (Bickley et al. 1996). Recent attempts for real time PCR-based direct quantification of Aspergillus were reported (Chong et al. 2015).
Our procedure do not require prior microbiological culture step and could be also used to detect potential pathogens in food products which present a useful tool for controlling their quality during chain processing. Other studies in food toxicology intended to examine the presence of Salmonella in food (Pathmanathan et al. 2003) and E. coli O157:H7 responsible for toxin release in milk (Fode-Vaughan et al. 2003). In the same context, we previously established a specific PCR-based procedure to detect cereal contamination with trichothecene-producing Fusarium using functional Tri5 primers (Ben-Amar et al. 2012). The direct PCR-based method described here and broadly tested with different bacterial and fungal strains proved to be a reliable way of detecting the pathogen in food samples in a fast and inexpensive manner.
Direct PCR in human health and biomedical diagnosis
For medical purposes, we focus to investigate the direct PCR on human tissue samples to avoid the time-consuming process of genomic DNA extraction. Primers for the human hepatocyte growth factor HGF-tagged gene were used to amplify a 430 bp fragment. Results showed successful amplifications of the target gene using a variety of human samples such as: dried blood, buccal cells, skin, and hair root (Fig. 1B-d). Only few attempts briefly described PCR amplification directly from whole blood (Del Valle Mendoza et al. 2014), buccal cell swabs (Kovačević-Grujičić et al. 2012), and hair root (Hayashida et al. 2009); while they failed to establish truly an available direct PCR system for different tissues intended for routine practice. In contrast, we achieved an easy dir-PCR handling protocol using human samples described above and amplification occurred regardless of target tissue (Fig. 1B-d). Recent developed direct PCR assays enabling human HLA-genotyping showed that DNA extraction and amplification were combined in a single step, while no DNA isolation was performed in our trials. Interestingly, our designed approach was proven to be compatible with classic available Taq DNA polymerase that was feasible before only when using high performance Hot Start polymerases and several commercially specie or tissue-specific PCR amplification kits as shown in recent reports (Cascella et al. 2015; Gouveia et al. 2015; Gray et al. 2014). Furthermore, this method is fast, cost-saving omitting the complicated DNA extraction steps and minimizing the risk of DNA contamination during reagent handling. Besides its effectiveness, the technique will also accelerate PCR applications for medical purposes by eliminating the time and effort for nucleic acid extraction.
The direct PCR prevents the loss of trace samples in the DNA purification process which provides to the described technique a variety of applications in forensic science, preventive medicine and mutation/gene-based disease detection (Cascella et al. 2015; Gouveia et al. 2015). It could also facilitate the development of high-throughput PCR for large-scale diagnostic.
Direct PCR in plant biotechnology and agriculture
The nicotinamide adenine dinucleotide gene (NAD) was used in the present work as constitutive gene to establish the direct PCR from leaf discs of various plant species (Fig. 1B-e). From each species, 5–10 samples were tested to assess the reliability of the method using primer set of a conserved NAD gene. The efficiency of the dir-PCR was found to be similar as purified DNA used as positive control which makes DNA isolation inappropriate and approves the effectiveness of the described method, while previous report found some variability in PCR amplification (Rogers and Parkes 1999). We further developed the direct PCR on different types of plant material (nature and origin of the explant, developmental stage, etc.) to be standardized on a big range of templates (data not shown).
Leaf tissues of all plant species tested yielded positive results and showed successful amplification of target DNA sequence without adding chelating agents such as Chelex, EDTA or DMSO that have been often considered necessary elsewhere in last published methods (Cao et al. 2009) and led otherwise to the failure of PCR. Our described plant’s direct PCR protocol did not require specific treatments needed to remove PCR-inhibitors including polysaccharides and phenolic compounds known as commonly present in most templates and mainly expected for possible arrest of PCR amplification.
Our protocol involves a short incubation time in Tris–EDTA buffer sufficient to neutralize harmful effect of potential inhibitors in the PCR mix and, therefore, it may probably reduce the sensitivity of standard Taq DNA polymerase to these PCR inhibitory components. Also, heat-treated specimens were intended for easier release of their DNA with less harmful PCR-inhibitors as suggested previously (AlShahni et al. 2009; Ben-Amar et al. 2012). The inhibitor-containing substrate could be avoided using small amount of sample and especially a minute volume of the grinding sample mixture as PCR template as reported by Sharma et al. (2012). During this step, inhibitors may be largely diluted that common Taq polymerases will be non-vulnerable to the presence of these components in trace in the PCR mix which did not inhibit anymore the amplification.
Therefore, following this simplified procedure, we could efficiently and successfully amplify a DNA fragment from direct template eliminating consequently all extra-cost of DNA purification kits and reagents particularly in developing countries. The direct PCR method could be also applied to detect transgenes in transgenic plants, fruits and seeds which is suitable as a useful tool for biosafety applications as well as rapid large-scale screening of transformants. In fact the method is expected to be powerful, quite easy and relatively rapid especially when we have to handle a large number of samples.
Conclusion
We have developed an easy, fast and reliable method for direct PCR assay from a variety of living organisms including bacteria, fungi, plants, human tissues as well as environmental samples using a commercially available Taq DNA polymerase. Small amount of each sample was feasible to successfully amplify constitutive genes by direct PCR independently from target templates making the DNA purification irrelevant. Implementation of such procedure has diverse applications in research and applied molecular biology for many purposes such as environmental and food microbiology, biomedical diagnosis, forensic science, and agriculture biotechnology.
Acknowledgements
We are grateful to Dr. Andrea Devlin for proofreading this manuscript and Prof. Michael Florian Mette (Leibniz Institute of Plant Genetics and Crop Plant Research, IPK-Gatersleben, Germany) for helpful discussions. This work was supported by the Tunisian Ministry of Higher Education and Scientific Research.
Author contribution
ABA designed the work, developed the direct PCR technology, analyzed data and wrote the body of the paper; SO helped in the experimental work, microbiological assays and data analysis. AM co-ordinated the project together with ABA. All authors reviewed, edited and approved the final version of the manuscript.
Compliance with ethical standards
Ethical statement
We confirm that all experiments are approved by the committee and research studies conducted here using human samples. Experiments were performed in accordance with the relevant government’s regulatory guidelines and regulations.
References
- Ahmed I, Islam M, Arshad W, Mannan A, Ahmad W, Mirza B. High-quality plant DNA extraction for PCR: an easy approach. J Appl Genet. 2009;50(2):105–107. doi: 10.1007/BF03195661. [DOI] [PubMed] [Google Scholar]
- AlShahni MM, Makimura K, Yamada T, Satoh K, Ishihara Y, Takatori K, Sawada T. Direct colony PCR of several medically important fungi using Ampdirect®Plus. Jpn J Infect Dis. 2009;62:164–167. [PubMed] [Google Scholar]
- Ben-Amar A, Oueslati S, Ghorbel A, Mliki A. Prediction and early detection of mycotoxigenic Fusarium culmorum in wheat samples by direct PCR-based procedure. Food Control. 2012;23:506–510. doi: 10.1016/j.foodcont.2011.08.021. [DOI] [Google Scholar]
- Bickley J, Short JK, McDowell DG, Parkes HC. Polymerase chain reaction (PCR) detection of Listeria monocytogenes in diluted milk and reversal of PCR inhibition caused by calcium ions. Lett Appl Microbiol. 1996;22:153–158. doi: 10.1111/j.1472-765X.1996.tb01131.x. [DOI] [PubMed] [Google Scholar]
- Cao M, Fu Y, Guo Y, Pan J. Chlamydomonas (Chlorophyceae) colony PCR. Protoplasma. 2009;235:107–110. doi: 10.1007/s00709-009-0036-9. [DOI] [PubMed] [Google Scholar]
- Cascella R, Strafella C, Ragazzo M, Zampatti S, Borgiani P, Gambardella S, Pirazzoli A, Novelli G, Giardina E. Direct PCR: a new pharmacogenetic approach for the inexpensive testing of HLA-B* 57:01. Pharmacogn J. 2015;15:196–200. doi: 10.1038/tpj.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong GLM, Van De Sande WWJ, Dingemans GJH, Gaajetaan GR, Vonk AG, Hayette MP, Van Tegelen DWE, Simons GFM, Rijnders BJA. Validation of a new Aspergillus real-time PCR assay for direct detection of Aspergillus and azole resistance of Aspergillus fumigatus on bronchoalveolar lavage fluid. J Clin Microbiol. 2015;53(3):868–874. doi: 10.1128/JCM.03216-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle Mendoza J, Silva Caso W, Tinco Valdez C, Pons MJ, Del Valle LJ, Casabona Oré V, Champin Michelena D, Bazán Mayra J, Zavaleta Gavidea V, Vargas M, Ruiz J. Diagnosis of Carrion’s disease by direct blood PCR in thin blood smear negative samples. PLos One. 2014;9(3):e92283. doi: 10.1371/journal.pone.0092283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eszik I, Lantos I, Önder K, Somogyvári F, Burián K, Endrész V, Virok DP. High dynamic range detection of Chlamydia trachomatis growth by direct quantitative PCR of the infected cells. J Microbiol Methods. 2016;120(1):15–22. doi: 10.1016/j.mimet.2015.11.010. [DOI] [PubMed] [Google Scholar]
- Fode-Vaughan KA, Maki JS, Benson JA, Collins P. Direct PCR detection of Escherichia coli O157:H7. Lett Appl Microbiol. 2003;37:239–243. doi: 10.1046/j.1472-765X.2003.01386.x. [DOI] [PubMed] [Google Scholar]
- Gouveia N, Brito P, Serra A, Balsa F, Andrade L, Bento MS, Cunha P, Bogas V, Lopes V, Porto MJ. Direct amplification of reference samples with Globalfiler® PCR amplification kit. Forensic Sci Int. 2015 [Google Scholar]
- Gray K, Crowle D, Scott P. Direct amplification of casework bloodstains using the Promega PowerPlex® 21PCR amplification system. Forensic Sci Int. 2014;12:86–92. doi: 10.1016/j.fsigen.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Hayashida M, Iwao-Koizumi K, Murata S, Kinoshita K. Single-tube genotyping from a human hair root by direct PCR. Anal Sci. 2009;25:1487–1489. doi: 10.2116/analsci.25.1487. [DOI] [PubMed] [Google Scholar]
- Kovačević-Grujičić N, Davidivic S, Takic D, Mojsin M, Stevanovic M. Direct PCR amplification of the HVSI region in mitochondrial DNA from buccal cell swabs. Arch Biol Sci Belgrade. 2012;64(3):851–858. doi: 10.2298/ABS1203851G. [DOI] [Google Scholar]
- Mannerlöf M, Tenning P. Screening transgenic plants by multiplex PCR. Plant Mol Biol Rep. 1997;15:38–45. doi: 10.1007/BF02772111. [DOI] [Google Scholar]
- Nishimura N, Nakayama T, Tonoike H, Kojima K, Kato S. Direct polymerase chain reaction from whole blood without DNA isolation. Ann Clin Biochem. 2000;37:674–680. doi: 10.1258/0004563001899726. [DOI] [PubMed] [Google Scholar]
- Pathmanathan SG, Cardona-Castro N, Sa´nchez-Jime´nez MM, Correa-Ochoa MM, Puthucheary SD, Thong KL. Simple and rapid detection of Salmonella strains by direct PCR amplification of the hilA gene. J Med Microbiol. 2003;52:773–778. doi: 10.1099/jmm.0.05188-0. [DOI] [PubMed] [Google Scholar]
- Rogers HI, Parkes HC. Direct PCR amplification from leaf discs. Plant Sci. 1999;143:183–186. doi: 10.1016/S0168-9452(99)00048-5. [DOI] [Google Scholar]
- Sharma R, Kumar V, Mohapatra T, Khandelwal V, Vyas GK. A simple and non destructive method of direct-PCR for plant systems. J Plant Biol. 2012;55:114–122. doi: 10.1007/s12374-011-9191-6. [DOI] [Google Scholar]
- Shokralla S, Singer GAC, Hajibabaei M. Direct PCR amplification and sequencing of specimens’ DNA from preservative ethanol. Biotechniques. 2010;48(3):233–234. doi: 10.2144/000113362. [DOI] [PubMed] [Google Scholar]
- Tomida M, Saito T. The human hepatocyte growth factor (HGF) gene is transcriptionally activated by leukemia inhibitory factor through the stat binding element. Oncogene. 2004;23(3):679–686. doi: 10.1038/sj.onc.1207190. [DOI] [PubMed] [Google Scholar]
- Turner S, Pryer KM, Miao VPW, Palmer JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. 1999;46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- Watson RJ, Blackwell B. Purification and characterization of a common soil component which inhibit the polymerase chain reaction. Can J Microbiol. 2000;46:633–642. doi: 10.1139/w00-043. [DOI] [PubMed] [Google Scholar]
- Werblow A, Flechl E, Klimpel S, Zittra C, Lebl K, Kieser K, Laciny A, Silbermayr K, Melaun C, Fuehrer HP. Direct PCR of indigenous and invasive mosquito species: a time- and cost-effective technique of mosquito barcoding. Med Vet Entomol. 2016;30(1):8–13. doi: 10.1111/mve.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee SJ, Taylor W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and application. San Diego: Academic Press; 1990. [Google Scholar]