Abstract
Liver fibrosis is characterized by changes in tissue architecture and extracellular matrix composition. Liver fibrosis affects not only hepatocytes but also the non-parenchymal cells such as hepatic stellate cells (HSCs), which are essential for maintaining an intact liver structure and function. Transforming growth factor β1 (TGF-β1) is a multifunctional cytokine that induces liver fibrosis through activation of Smad signaling pathways. To improve a new therapeutic approach, synthetic TGF-β1/Smad oligodeoxynucleotide (ODN) was used to suppress both TGF-β1 expression and Smad transcription factor using a combination of antisense ODN and decoy ODN. The aims of this study are to investigate the anti-fibrotic effects of TGF-β1/Smad ODN on simultaneous suppressions of both Smad transcription factor and TGF-β1 mRNA expression in the hepatic fibrosis model in vitro and in vivo. Synthetic TGF-β1/Smad ODN effectively inhibits Smad binding activity and TGF-β1 expression. TGF-β1/Smad ODN attenuated the epithelial mesenchymal transition (EMT) and activation of HSCs in TGF-β1-induced AML12 and HSC-T6 cells. TGF-β1/Smad ODN prevented the fibrogenesis and deposition of collagen in CCl4-treated mouse model. Synthetic TGF-β1/Smad ODN demonstrates anti-fibrotic effects that are mediated by the suppression of fibrogenic protein and inflammatory cytokines. Therefore, synthetic TGF-β1/Smad ODN has substantial therapeutic feasibility for the treatment of liver fibrotic diseases.
Keywords: oligodeoxynucleotide, decoy, antisense, liver fibrosis, TGF-β1, Smad
Introduction
Liver fibrosis is a potentially reversible wound-healing response, which can be caused by chronic liver injuries including viral agents, alcoholic hepatitis, and autoimmune hepatitis.1 Fibrosis in the liver is characterized by an excess deposition of extracellular matrix (ECM) in and around injured liver tissues.2 The process of hepatic fibrosis involves multiple cellular and molecular mechanisms in most chronic liver diseases.3 Also, this process affects not only hepatocytes but also non-parenchymal cells such as hepatic stellate cells (HSCs) and liver myofibroblasts, which are essential for maintaining an intact liver structure and function.4 Hepatocytes can also transdifferentiate into mesenchymal cells via epithelial mesenchymal transition (EMT) and the deposition of collagen in the liver during chronic injury.5
EMT is a biological process in which tubular cells lose epithelial phenotypes, undergo morphological change, and acquire new characteristic features of mesenchymal properties.6 The development and progression of EMT requires specific micro-environmental conditions and the precise regulation of intracellular signals.7 The intercellular junction of epithelial cells are interfered or decreased by inhibition of adhesion molecules and tight junction protein such as E-cadherin and ZO-1.8 EMT also appears by morphological changes from epithelial to fibroblast-like with upregulation of mesenchymal markers protein, including fibronectin and vimentin.9 Injury to epithelial cells, which can be either hepatocytes or bile duct epithelium, elicits a cascade of injuries and cytokine release that contributes to the activation of resident HSCs.10
HSCs are nonparenchymal perisinusoidal cells situated in the subendothelial space between hepatocytes and sinusoidal endothelial cells.11, 12 These cells are characterized by the cytoplasmic storage of vitamin A in perinuclear cytoplasmic droplets.13 HSCs have emerged as the key cell type responsible for excess deposition of ECM during fibrogenesis.14 During hepatic fibrogenesis, HSCs are activated by inflammatory cytokines and growth factors such as transforming growth factor β1 (TGF-β1) and platelet-derived growth factor (PDGF) in a paracrine and autocrine manner. Proinflammatory cytokines are promoted during early hepatic inflammation and contributes to the activation of HSCs.15 Previous reports have investigated the crucial role of TGF-β1 in the activation of HSCs and fibrogenesis.16, 17 In response to liver injury, the quiescent HSCs undergo a complex transactivation process and differentiate into proliferating myofibroblast-like cells.18 Activated stellate cells evaluates the production of collagen type I and type III and α-SMA, which are the major cell types of matrix formation in damaged liver tissue leading to fibrosis.19 Choi et al.20 reported anti-fibrotic therapies for liver fibrosis that target both hepatocyte and HSCs.
The TGF-β1 family regulates a variety of cellular process, including proliferation, differentiation, apoptosis, and migration.17, 21 TGF-β1 is also the key mediator of liver fibrosis, promoting ECM production and tissue fibrosis.22 In response to injury, TGF-β1 commands cross talk between inflammatory, parenchymal, and myofibroblast cells.23 Although many other liver cells may generate TGF-β1, Kupffer cells and recruited macrophages centrally produce TGF-β1 in the fibrotic liver. Furthermore, TGF-β1 expression is also associated with morphological alterations, such as EMT in hepatocyte and changes in survival signaling pathway.24
TGF-β1 mediates its biological function through Smad signaling pathway.25 TGF-β1 signals through its type I and type II receptors, which have serine/threonine kinase activity.26 TGF-β1 exerts its effects by binding to the type II TGF-β receptor, which then recruits and activates the type I TGF-β receptor.27 The activated type I TGF-β receptor phosphorylates Smad2 and Smad3, which then form heteromeric complexes with Smad4.28 Smad complex translocate into the nucleus, where they bind to Smad binding element, which contains four base pairs, “CAGA.” This consensus sequence is present in the promoters regions of TGF-β1 and bind activated fibrogenic genes.29, 30 Moreover, Smad7 effectively prevents Smad2 and Smad3 interaction, efficiently preventing Smad signaling regulated protein expression.31 Abnormal TGF-β1 signaling has been implicated in a growing number of fibrotic and inflammatory conditions, including pulmonary fibrosis, liver cirrhosis, systemic sclerosis, and hypertrophic scars.32, 33, 34 Because TGF-β1 signaling has been postulated to play an important role in liver fibrosis, the effective blockade of TGF-β1/Smad signaling may provide a useful therapeutic tool for treating these diseases.
To improve a new therapeutic approach, synthetic TGF-β1/Smad oligodeoxynucleotide (ODN) was used to suppress both TGF-β1 expression and Smad transcription factor using a combination of antisense ODN and decoy ODN.35 Antisense ODN uses the selective impairment of protein synthesis in the cytoplasm through the use of antisense ODN sequences as its basis.36, 37 However, the decoy ODN technique is employed to block transcription factor activity through the use of a synthetic double-stranded ODN containing consensus sequences of DNA binding sites, which works at the pre-transcription level.38 Kemaladewi et al.39 investigated that TGF-β type I receptor antisense ODN mediates knockdown of TGF-β1 signaling cascades. Kim et al.40 found that chimeric decoy ODN affects renal interstitial fibrosis via the inhibition of both nuclear factor κB (NF-κB) and Sp1 transcription factors. In this study, synthetic TGF-β1/Smad ODN played a dual function, consisting of antisense ODN and decoy ODN. Furthermore, the modification of ODN’s composition to prolong ODN stability in vivo and development of a delivery system into liver tissues will be critical for the enhancement of potential therapeutic efficacy. This synthetic TGF-β1/Smad ODN strategy presents a novel method to target TGF-β signaling cascades with potential effects of fibrosis.
The aim of this study was to investigate the inhibitory effects of TGF-β1/Smad ODN on hepatocyte, activated HSCs, and hepatic fibrosis mouse models. TGF-β1/Smad ODN was designed to contain both TGF-β1 antisense ODN and Smad-binding sequences, which enhance the effective blockades of TGF-β1/Smad signaling. TGF-β1/Smad ODN was used to inhibit both mRNA expression of TGF-β1 and Smad transcription factors in the hepatocyte, activated HSCs, and CCl4-induced liver fibrosis mouse models. TGF-β1/Smad ODN also inhibits inflammatory response as well as fibrotic change associated with hepatic fibrogenesis in vitro and in vivo.
Results
Confirmation of FITC-Labeled TGF-β1/Smad ODN in AML12 and HSC-T6 Cells
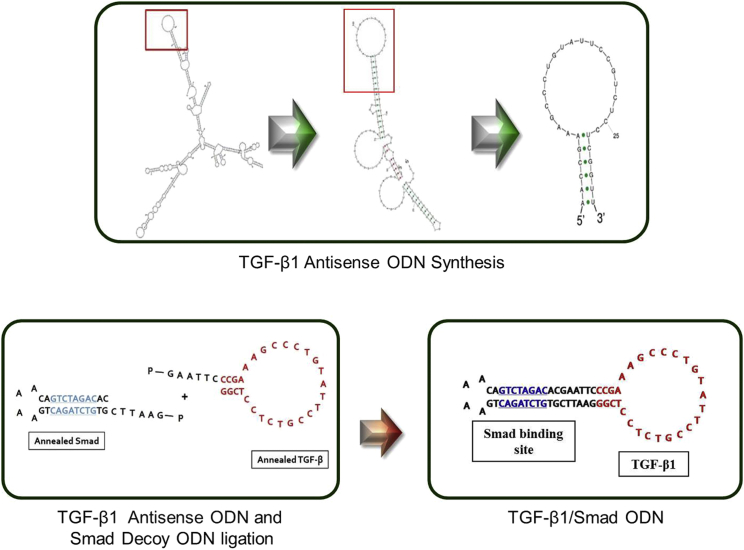
We first designed TGF-β1/Smad ODN based on both TGF-β1 antisense ODN and Smad decoy ODN. Target site of TGF-β1 mRNA sequence for TGF-β1 antisense ODN was selected the secondary structure by the S-fold program (Figure 1). Also, ring-type Smad decoy ODN synthesized double-stranded ODN containing the consensus Smad binding element (GTCTAGAC).
Figure 1.
Construction of Synthetic TGF-β1/Smad ODN
Target sites for TGF-β1 were selected via the sequential overlap simulation of secondary structures using the S-Fold program. TGF-β1/Smad ODN ligates the antisense TGF-β1 ODN and Smad decoy ODN.
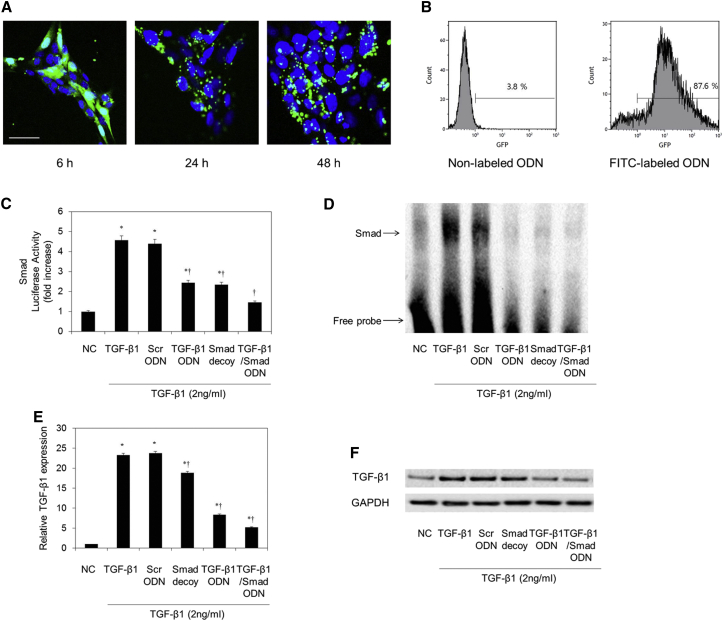
To determine the stability of synthetic TGF-β1/Smad ODN in cells, we measured the distribution of FITC-labeled TGF-β1/Smad ODN in AML12 and HSC-T6 cells using confocal microscopy (Figures S1A and 2A). The FITC-labeled TGF-β1/Smad ODN appeared strongly fluorescent in both cytoplasm and nucleus of AML12 and HSC-T6 cells after 6 hr of transfection. After 48 hr of FITC-labeled TGF-β1/Smad ODN transfection, fluorescence was detected in both cytoplasm and nucleus of AML12 and HSC-T6 cells. Furthermore, we determined the transfection efficiency of FITC-labeled TGF-β1/Smad ODN through flow cytometry in AML12 and HSC-T6 cells (Figures S1B and 2B). Flow cytometry analysis showed a high positive rate (89.2%, 87.6%) of fluorescence activity in respective AML12 and HSC-T6 cells. These results indicated that TGF-β1/Smad ODN was successfully transfected into AML12 and HSC-T6 cells.
Figure 2.
Confirmation of FITC-Labeled TGF-β1/Smad ODN in HSC-T6 Cells
(A) Fluorescence activity was detected in both cytoplasm and nucleus with FITC-labeled ODN deposition. Scale bar, 50 μm. (B) Transfection efficiency of FITC-labeled TGF-β1/Smad ODN through flow cytometry. Effect of Smad decoy, TGF-β1 antisense ODN, and TGF-β1/Smad ODN on Smad transcription activity and expression levels of TGF-β1 in HSC-T6 cells. (C–F) Luciferases activity in TGF-β1-treated HSC-T6 cells (C), electrophoretic mobility shift assay (EMSA) (D), real-time PCR (E), and western blot assay (F). *p < 0.05 versus normal control. †p < 0.05 versus TGF-β1-treated group.
TGF-β1/Smad ODN Regulated the Expression of TGF-β1 and Smad Binding Activity in HSC-T6 Cells
To confirm the function of TGF-β1/Smad ODN by interfering with Smad complex binding to DNA, we performed electrophoretic mobility shift assay (EMSA) with a Smad-specific DNA-binding activity. EMSA was performed with nuclear extracts of HSC-T6 cells that had been transfected with TGF-β1/Smad ODN to determine the effect of ODN on the TGF-β1-stimulated DNA-binding activity of Smad (Figure 2D). TGF-β1 (2 ng/mL) treatment increased DNA-binding activity of Smad in HSC-T6 cells. In contrast, TGF-β1/Smad ODN, TGF-β1 antisense ODN, and Smad decoy ODN suppressed the DNA-binding activity of Smad compared with Scr ODN in HSC-T6 cells. In addition, Smad-luciferase activity determined Smad transcriptional activity through a (CAGA)12-Luc reporter gene assay in HSC-T6 cells (Figure 2C). TGF-β1/Smad ODN, TGF-β1 antisense ODN, and Smad decoy ODN significantly inhibited TGF-β1-dependent gene transcriptional activity. Of these, TGF-β1/Smad ODN was the most efficient of the three ODNs.
Also, the effect of TGF-β1/Smad ODN on the inhibition of TGF-β1 expression levels was confirmed by RT-PCR and western blotting in HSC-T6 cells (Figures 2E and 2F). Transfection of TGF-β1/Smad ODN and TGF-β1 antisense ODN significantly decreases the activated TGF-β1 expression compared with Scr ODN group in mRNA and protein levels in HSC-T6 cells. In addition, efficiency of TGF-β1/Smad ODN was better than that of TGF-β1 antisense ODN on the silence of TGF-β1. Therefore, these results confirmed that transfection of TGF-β1/Smad ODN simultaneously inhibits the expression of TGF-β1 and Smad binding activity in HSC-T6 cells.
TGF-β1/Smad ODN Inhibited the Activated HSCs through the Regulation of Smad Signaling Pathway
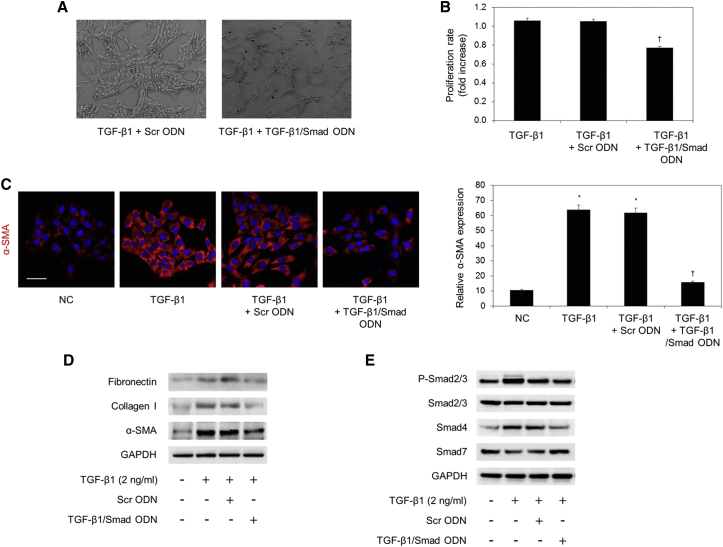
In chronic liver injury, the activation of quiescent HSCs into myofibroblast-like cells is a pivotal step for the progression of hepatic fibrosis.41 To determine the inhibitory effect of TGF-β1/Smad ODN on regulating TGF-β1-induced HSC activation, HSC-T6 cells were transfected with the TGF-β1/Smad ODN after TGF-β1 treatment. As shown in Figure 3A, TGF-β1/Smad ODN treatment inhibited the proliferation of HSC-T6 cells more than Scr ODN treatment. In addition, CCK-8 assay indicated that the viability of HSC-T6 cells after incubation with TGF-β1 and TGF-β1/Smad ODN was significantly decreased compared with the Scr ODN-transfected cells (Figure 3B). These results confirmed that TGF-β1/Smad ODN inhibits proliferation of activated HSCs.
Figure 3.
TGF-β1/Smad ODN Effectively Inhibits Activated HSCs by TGF-β1
(A) Morphology of TGF-β1-treated HSC-T6 cells with Scr ODN or TGF-β1/Smad ODN. (B) CCK-8 assay shows that TGF-β1/Smad ODN inhibits proliferation of activated HSC induced by TGF-β1. (C) Representative immunofluorescence images show the expression of α-SMA (red) and Hoechst 33342 (blue) in HSC-T6 cells. Scale bar, 50 μm. *p < 0.05 versus normal control. †p < 0.05 versus TGF-β1-treated group. (D and E) Effect of TGF-β1/Smad ODN on expressions of (D) fibrogenesis-related protein through (E) Smad signaling pathway in TGF-β1-treated HSC-T6 cells using western blot analysis.
To explore the influence of TGF-β1/Smad ODN on the activity of α-SMA, we examined the expression of α-SMA using immunofluorescence staining in HSC-T6 cells (Figure 3C). TGF-β1 treatment activates the expression of α-SMA (red), whereas TGF-β1/Smad ODN significantly decreased as compared to the Scr ODN. Additionally, western blot results show that the administration of TGF-β1+Scr ODN increased fibronectin, α-SMA, and collagen I expression, whereas TGF-β1/Smad ODN transfection downregulated fibronectin, α-SMA, and collagen I in HSC-T6 cells (Figure 3D).
To explore the effect of TGF-β1/Smad ODN, the expression of Smad proteins were determined by western blotting (Figure 3E). Smad2/3 phosphorylation was increased by TGF-β1 treatment with Scr ODN, but TGF-β1/Smad ODN diminished the level of Smad2/3 phosphorylation and Smad4 in HSC-T6 cells. In addition, Smad7 was efficient as an antagonist of TGF-β1/Smad signaling. Smad7 expression was increased by TGF-β1/Smad ODN treatment compared with Scr ODN in HSC-T6 cells. Therefore, these findings suggested that TGF-β1/Smad ODN affects regulating the proliferation of activated HSC-T6 and the expression of fibrosis-related protein via blocks of Smad signaling.
TGF-β1/Smad ODN Inhibited TGF-β1-Induced Fibrosis and EMT via Blocks of Smad Signaling Pathway in AML12 Cells
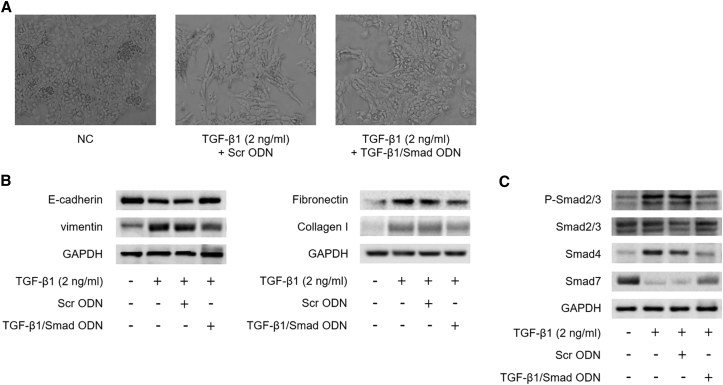
During liver fibrogenesis, hepatocytes lose their epithelial phenotype and acquire features of mesenchyme via EMT processes.42 To induce EMT, AML12 cells were incubated for 48 hr with TGF-β1 (2 ng/mL), which is the key profibrogenic cytokine in the liver EMT process. Cell morphology by TGF-β1 treatment was observed in time-lapse images taken at 0, 24, and 48 hr using a phase contrast microscope in AML12 cells (Figure S2A). AML12 cells appeared to have a typical polygonal epithelial morphology and tight arrangement. In response to 2 ng/mL TGF-β1, AML12 cells changed to a fibroblastic spindle-shaped morphology after 24 and 48 hr of treatment. We also determined the expression level of EMT marker through western blot assay in TGF-β1-induced AML12 cells (Figure S2B). E-cadherin is a prototypical epithelial cell marker that plays an important role in the maintenance of cellular integrity at the cell-cell junction. In addition, vimentin is an intermediate filament used to identify mesenchymal cells in EMT. Downregulation of E-cadherin, the well-known epithelial marker was observed during TGF-β1 treatment in time-dependent manner. On the other hand, TGF-β1 treatment upregulated vimentin expression in AML12 cells, indicating that hepatocytes acquired a mesenchymal phenotype.
To investigate the effects of TGF-β1/Smad ODN on fibrosis and EMT, we measured the TGF-β1-induced fibrosis-related gene and EMT marker protein expression by TGF-β1/Smad ODN in AML12 cells. AML12 cells morphology concurrently treated with TGF-β1 and TGF-β1/Smad ODN retained epithelial characteristics compared with Scr ODN (Figure 4A). When AML12 cells were exposed to 2 ng/mL TGF-β1, the expression level of E-cadherin was increased, whereas the expression of vimentin was decreased in TGF-β1/Smad ODN compared with Scr ODN treatment in TGF-β1-stimulated AML12 cells (Figure 4B). In addition, TGF-β1 and Scr ODN treatment increased fibronectin and collagen I expression, whereas TGF-β1/Smad ODN treatment downregulated fibronectin and collagen I in TGF-β1-treated AML12 cells.
Figure 4.
TGF-β1/Smad ODN Suppresses EMT and Fibrosis-Related Protein in TGF-β1-Induced AML12 Cells
(A) Effect of TGF-β1/Smad ODN on morphologic change of AML12 cells treated with TGF-β1. (B and C) Effect of TGF-β1/Smad ODN on expressions of EMT and fibrosis-related protein (B) through Smad signaling pathway (C) using western blot analysis
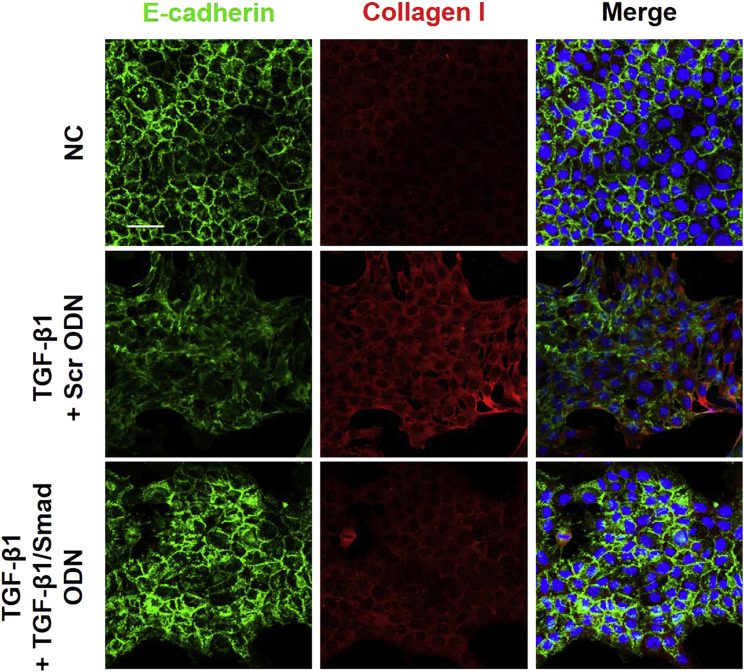
Additionally, immunofluorescence staining revealed that TGF-β1 treatment induced the suppression of E-cadherin expression and the induction of collagen I expression (Figure 5). Interestingly, TGF-β1/Smad ODN treatment resulted in the restoration of E-cadherin and the reduction of collagen I expression in TGF-β1-treated cells. Taken together, these data suggest that TGF-β1/Smad ODN suppresses fibrosis through the regulation of EMT in hepatocytes, which reveal the upregulation of E-cadherin expression and the downregulation of vimentin expression.
Figure 5.
Effect of TGF-β1/Smad ODN on E-cadherin and Collagen I Expression in TGF-β1-Treated AML12 Cells
Immunofluorescence double staining for E-cadherin (green) and collagen I (red) localization. Cells were counterstained with Hoechst 33342 (blue). Scale bar, 50 μm.
To assess the molecular mechanism of TGF-β1/Smad ODN in TGF-β1-induced AML12 cells, we examined its effects on liver fibrosis and matrix accumulation through regulation of TGF-β signaling pathway (Figure 4C). TGF-β-induced Smad2/3 phosphorylation was increased by Scr ODN treatment, whereas TGF-β1/Smad ODN decreased the Smad2/3 phosphorylation and Smad4 expression in TGF-β1-induced AML12 cells. TGF-β1/Smad ODN also regulated the expression of Smad7. Therefore, these results suggest that TGF-β1/Smad ODN suppresses fibrosis and EMT through the blocks of Smad signaling in TGF-β1-induced AML12 cells.
Inhibitory Effects of TGF-β1/Smad ODN on TGF-β1 Expression and Smad Binding Activity in Mouse Liver
To confirm the effective transfer of TGF-β1/Smad ODN in vivo, we measured the distribution of FITC-labeled TGF-β1/Smad ODN in mouse livers using confocal microscopy (Figure S3A). The FITC-labeled TGF-β1/Smad ODN appeared strongly fluorescent in both cytoplasm and nucleus of mouse liver. To determine the effect of TGF-β1/Smad ODN on Smad DNA-binding activity in CCl4-induced mouse model, EMSA was performed to analyze the transcriptional activity of Smad (Figure S3B). Smad binding activity was significantly increased in CCl4+Scr mice. On the other hand, administration of TGF-β1/Smad ODN significantly suppressed the activation of Smad in CCl4-induced mice.
The inhibitory effect of TGF-β1/Smad ODN on the expression of TGF-β1 was confirmed by RT-PCR and western blotting in CCl4-induced mouse model (Figures S3C and 3D). Administration of TGF-β1/Smad ODN significantly decreases the increased TGF-β1 expression compared with administration of Scr ODN at the mRNA and protein levels in CCl4-induced mice. Therefore, these results confirmed that transfection of TGF-β1/Smad ODN simultaneously inhibits the expression of TGF-β1 and Smad binding activity in CCl4-induced mouse models.
TGF-β1/Smad ODN Protected Liver Damage in CCl4-Induced Mouse Model
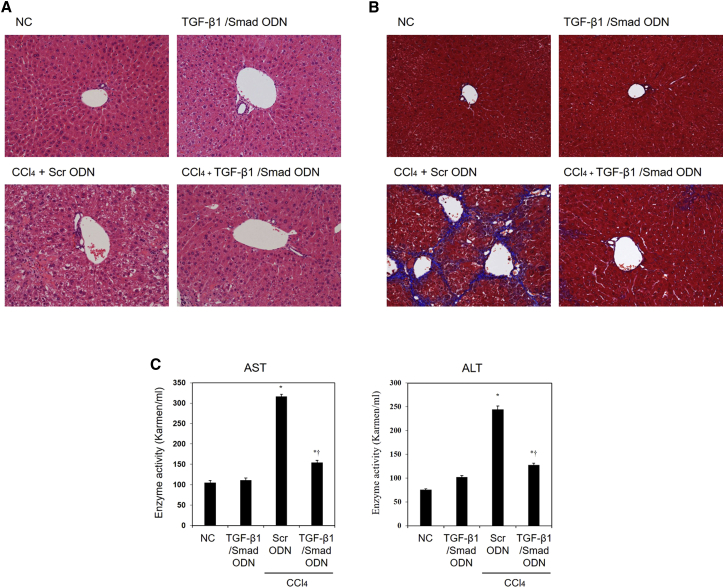
TGF-β1/Smad ODN was transferred biweekly during the CCl4-induced hepatotoxicity for 8 weeks. We histologically evaluated by H&E staining and Masson’s trichrome staining to determine the extent of hepatic fibrosis development in CCl4-treated mouse model (Figures 6A and 6B) Administration of CCl4 with Scr ODN induced a significant degree of liver injury, which was characterized by inflammatory cell infiltration and necrosis. In addition, chronic CCl4 treatment induced the development of collagen fibrils deposition, which was evidenced by trichrome staining. The CCl4-induced histological change and collagen deposition were markedly attenuated in TGF-β1/Smad ODN treatment, indicating that TGF-β1/Smad ODN plays a critical role in suppressing CCl4-induced chronic liver injury. Moreover, alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels, which were used as serum markers for liver damage, were obviously elevated in CCl4+Scr ODN mice, whereas the ALT and AST levels were significantly lower in CCl4+TGF-β1/Smad ODN administration (Figure 6C). Therefore, administration of TGF-β1/Smad ODN significantly reduced collagen deposition and mitigated the degree of liver fibrosis.
Figure 6.
TGF-β1/Smad ODN Suppresses CCl4-Induced Liver Damage and Collagen Accumulation
(A and B) Histological section of murine liver stained with (A) H&E stain, (B) Masson’s trichrome staining after 8 weeks of CCl4 administration. Histological examinations are performed at 200× magnification under light microscopy. (C) Serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were significantly decreased by TGF-β1/Smad ODN treatment compared with Scr ODN treatment in CCl4-induced mice. *p < 0.05 versus normal control. †p < 0.05 versus CCl4+Scr ODN-treated group.
TGF-β1/Smad ODN Attenuated Inflammation in CCl4-Induced Mouse Model
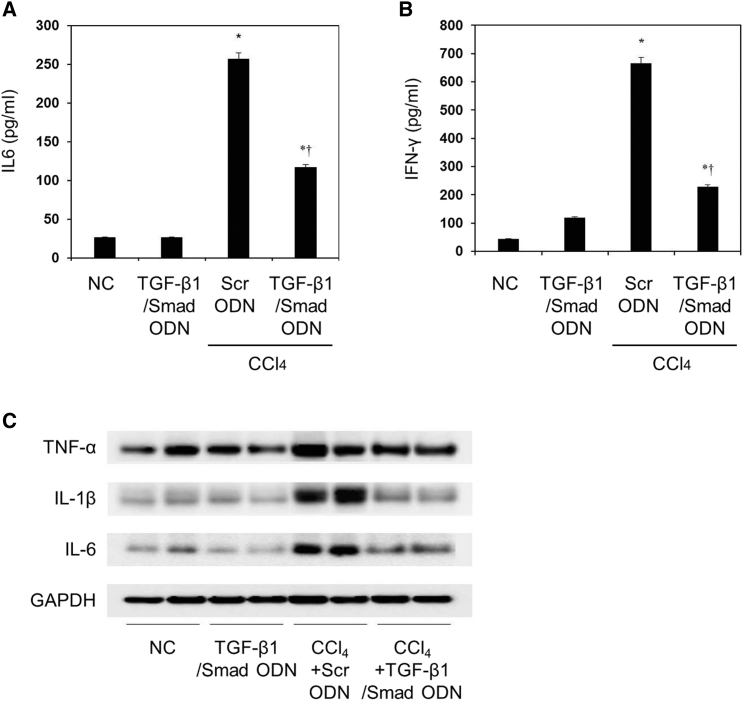
To investigate effects of TGF-β1/Smad ODN on the expression of inflammatory cytokines, we examined the levels of inflammatory cytokines on liver fibrosis using ELISA and western blot analysis. CCl4 administration increased the serum concentration of interleukin-6 (IL-6) and interferon γ (IFN-γ) in CCl4+Scr mice (Figures 7A and 7B). On the other hand, TGF-β1/Smad ODN treatment significantly inhibits the secretion of IL-6 and IFN-γ. Also, liver tissue from mice that received CCl4 administration showed an increased expression of tumor necrosis factor α (TNF-α), IL-1β, and IL-6 (Figure 7C). Treatment with TGF-β1/Smad ODN reduced the expressions of TNF-α, IL-1β, and IL-6 more than treatment with Scr ODN in CCl4-treated mice. These results showed that TGF-β1/Smad ODN significantly inhibits the secretion of inflammatory cytokine.
Figure 7.
TGF-β1/Smad ODN Significantly Inhibits the Pro-inflammatory Cytokine in CCl4-Induced Mouse Model
(A and B) ELISA results demonstrate that TGF-β1/Smad ODN significantly inhibits the pro-inflammatory cytokine in CCl4-induced mice: IL-6 (A) and IFN-γ (B) expression. *p < 0.05 versus normal control. †p< 0.05 versus the CCl4+Scr ODN-treated group. (C) Western blot results show that TGF-β1/Smad ODN inhibits expressions of TNF-α, IL-1β, and IL-6.
TGF-β1/Smad ODN Inhibited Fibrogenesis-Related Gene in CCl4-Induced Mouse Model
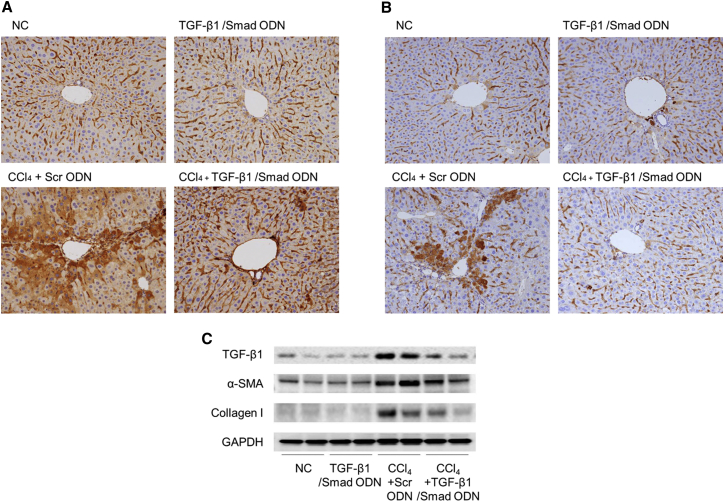
To investigate the anti-fibrotic effect of TGF-β1/Smad ODN in CCl4-induced mouse model, we examined its effects on liver fibrogenesis and ECM accumulation. An immunohistochemistry analysis study demonstrated that the accumulation of fibronectin-positive cells within the liver sinusoid was increased in CCl4+Scr ODN group compared with normal control (NC) group (Figure 8A). However, TGF-β1/Smad ODN markedly suppressed the expression of fibronectin in CCl4-induced mice. Administration of CCl4 with Scr ODN significantly increased the expression of FSP-1 in the fibrotic area (Figure 8B). However, TGF-β1/Smad ODN treatment diminishes the expression of FSP-1. Western blot analysis revealed an increased expression of TGF-β1, α-SMA, and collagen I in CCl4-induced Scr ODN treatment experimental groups that was greater than that of the NC group (Figure 8C). However, TGF-β1/Smad ODN treatment inhibited the expression of TGF-β1, α-SMA, and collagen I in CCl4-induced mice. Taken together, these results show that TGF-β1/Smad ODN has anti-fibrotic properties through the regulation of fibrosis-related gene and protein in CCl4-induced mice.
Figure 8.
TGF-β1/Smad ODN Significantly Inhibits the Fibrogenesis-Related Gene in CCl4-Induced Mouse Model
(A and B) Immunochemical staining results showed that TGF-β1/Smad ODN inhibits expression of fibronectin (A) and FSP-1 (B) in CCl4-induced mouse model. Histological examinations are performed at 200× magnification under light microscopy. (C) Western blot results show that TGF-β1/Smad ODN inhibits expressions of TGF-β1, α-SMA, and collagen I.
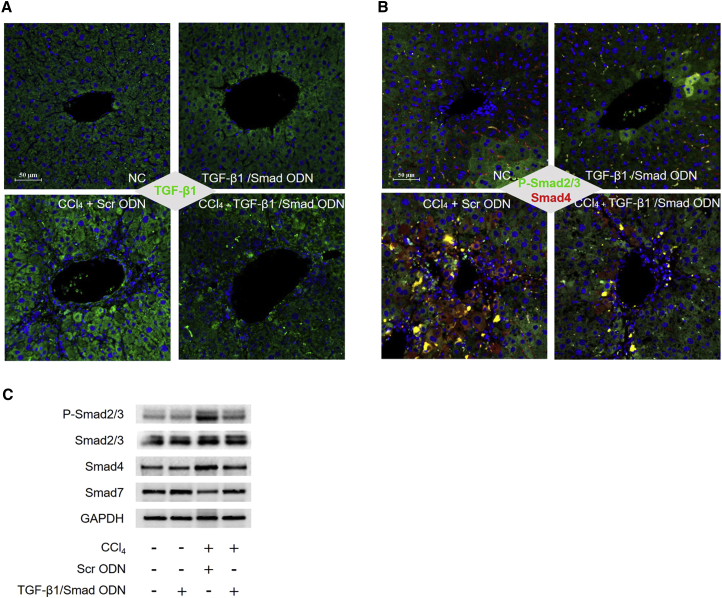
To assess the molecular mechanism of TGF-β1/Smad ODN in CCl4-induced mouse model, we measured the expression of TGF-β1, p-Smad2/3, and Smad 4 using western blot analysis. CCl4 administration with Scr ODN expressed upregulated p-Smad2/3 and Smad4 and downregulated Smad7. However, TGF-β1/Smad ODN treatment showed decrease of p-Smad2/3 and Smad4 expressions and increase of Smad7 expression in CCl4-induced mice (Figure 9C). Additionally, we examined the expression of TGF-β1, p-Smad2/3, and Smad 4 by immunofluorescence assay (Figures 9A and 9B). The liver of mice that received CCl4 administration with Scr ODN showed a higher expression of TGF-β1 (green), and TGF-β1/Smad ODN treatment suppressed the expression of TGF-β1. In addition, the expression of p-Smad2/3 (Green) and Smad4 (red) were expressed at cytosol and increased by CCl4 administration with Scr ODN group. In contrast, TGF-β1/Smad ODN decreased the expression of p-Smad2/3 and Smad4 in CCl4-induced mice. Based on these results, TGF-β1/Smad ODN might protect liver during CCl4 stimulation through the regulation of TGF-β1-stimulated Smad signaling pathway.
Figure 9.
TGF-β1/Smad ODN Significantly Inhibits the Smad Signaling Pathway in CCl4-Induced Mouse Model
(A and B) Immunofluorescence staining results showed that TGF-β1/Smad ODN inhibits expression of TGF-β1 (green) (A) and phosphorylation of Smad2/3 (green) and Smad4 (B) in CCl4-induced mice. Scale bar, 50 μm. (C) Western blot results showed that TGF-β1/Smad ODN inhibits expressions of phosphorylated Smad2/3, Smad4, and Smad7.
Discussion
Hepatic fibrosis is a complex pathophysiological process of numerous chronic liver diseases.43 This process is related to an inflammatory response and an increased deposition of ECM. If the hepatic injury persists, liver regeneration ultimately fails and hepatocytes accumulate with abundant ECM, including fibrillar collagen.15 For an effective treatment that targets the causes of liver fibrosis, it is necessary to systemically determine the therapeutic component based on the inhibition of initial molecule mechanism.
In this study, we investigated the effects of TGF-β1/Smad ODN on the simultaneous regulations of both Smad transcription factors and mRNA expression of TGF-β1 to induced hepatic fibrosis in vitro and in vivo. New techniques to inhibit target gene expression based on a short RNA and DNA strategy were provided due to recent progress in cellular and molecular research.35 Antisense ODNs were exploited to regulate the molecular process of gene expression through binding to target mRNA.44 Antisense ODN designed complementary nucleic acid fragments that specifically trigger via the selective ribonuclease H (RNase H) cleavage of the target mRNA in the nucleus.45 Several papers reported that antisense ODN can be employed as a novel therapeutic agents for various diseases.46, 47, 48, 49 Furthermore, a decoy ODN strategy blocks transcription factors of a specific gene, which can recognize their consensus binding sequences. Previous studies demonstrated that the effects of decoy ODNs significantly regulate transcription factor in several disorders.50, 51 Lee et al.52 reported the efficacy of chimeric decoy ODNs using NF-κB and Sp1 on atherosclerosis with immunological complication. In addition, chimeric decoy ODNs were more potent than the single transfection of NF-kB and ets decoy ODNs.53 Yuan et al.54 observed that dual AP-1 and Smad decoy ODN inhibited fibrosis associated with acute dermal wounds in mice through the inhibition of proinflammatory and antifibrotic effects.
HSCs are an initiating cell type of the disease and a major mechanistic contributor to the development of liver cirrhosis and fibrosis.55 Activated HSCs express myogenic genes acquiring a myofibroblast-like phenotype and start to secrete ECM components actively, including fibrillar collagens (collagen I and III).56 Moreover, HSCs are the main source of tissue inhibitors metalloproteinases (TIMPs), which may diminishes ECM degradation through the suppression of matrix metalloproteinases (MMPs) activities.57 Fang et al.58 investigated that TRPM7 channel regulates the expression of collagen and ECM protein such as MMPs and TIMPs in HSC-T6 cells. Furthermore, hepatocytes have been investigated as a source of TIMPs and other matrix modulators protein. Thus, they could play a role in processes of fibrogenesis and fibrosis regeneration.59 Fibrosis progresses by HSCs, and hepatocyte damage deteriorates.20 Damaged hepatocytes release reactive oxygen species (ROS) and fibrogenic mediators and lead to the recruitment of white blood cells by inflammatory cells.15 During fibrosis, hepatocytes undergo EMT and senescence due to telomere shortening. Consequently, hepatocytes lose their function and liver failure occurs.60 These interrelated events of fibrosis are orchestrated at the cellular level by numerous factors including cytokines and the pericellular matrix. Of these, TGF-β appears to be central, having been implicated in multiple factors of the injury response including fibrogenesis, growth regulation, growth dysregulation in cancer, and apoptosis.16
TGF-β1 signaling pathway modulates a wide range of cellular functions and has essential roles in fibrogenesis.61 TGF-β1 is also involved in collagen synthesis and chronic liver diseases. In CCl4-induced liver fibrosis, the expression of TGF-β1 and collagen type I, III, and IV was increased in the lipocytes, which are major sources of overproduction of matrix synthesis during hepatic fibrogenesis.62 Therefore, functional importance of TGF-β1 signaling in hepatic fibrosis has been demonstrated both in vivo and in vitro.39 TGF-β1 strategies were successfully used to counteract this process in different tissues, including liver, lung, and kidney.63 In this study, TGF-β1/Smad ODN inhibited the expression of TGF-β1 at the RNA and protein level.
Smad proteins, the first proteins to be identified by TGF-β1, play a central role in the transduction of receptor signals to target genes in the nucleus.22 Previous reports showed that Smad2 attenuates hepatocyte growth and dedifferentiation independent of TGF-β signaling.64 Moreover, Smad3 is required for TGF-β1-mediated Smad-containing DNA-binding complex formation in cultured HSCs.28 In Smad3 knockout mice isolated HSCs, collagen I expression reduced that additional signaling pathway may contribute to collagen gene expression.65 In addition, Xu et al.61 demonstrated that Smad3 and Smad4 are pro-fibrotic genes and that the deletion of Smad3 inhibits type I collagen expression and blocks EMT. In this study, EMSA revealed that Smad binding activity was blocked by Smad decoy ODN and TGF-β1/Smad ODN. The Smad-DNA complex was significantly decreased in the TGF-β1/Smad ODN-treated groups compared with Scr ODN-treated group. Besides, Smad7 forms a stable complex with activated TGF-β type I receptor and effectively prevented Smad2 and Smad3 interaction with the receptor and subsequent phosphorylation, thus efficiently preventing downstream signaling.66 Also, suppression of Smad7 was rescued by epigenetic modulation, which reduced α-SMA and collagen expression and ameliorated hepatic fibrosis.67 These results show that synthetic TGF-β1/Smad ODN regulates expression of Smad7 in hepatocytes and activated HSCs via TGF-β stimulation. Furthermore, pharmacological inhibition of TGF-β1/Smad ODN ameliorates liver fibrosis induced by chronic CCl4 exposure.
In fibrotic condition, activated fibroblasts and HSCs generate a deposition of collagen.1 The collagen deposits that accumulate during the progression of liver fibrosis are predominantly located in the generated fibrous septa, although some limited collagen deposition, primarily of collagen type IV, also occurs in the perisinusoidal space. A recent study reported that large amount of collagen production contributed to HSCs more significantly than hepatocyte and endothelial cells in vitro.55 Initiation of HSCs activation is associated with increases in several inflammatory cytokines including TNF-α, IL-1β, and IL-6, which modulate collagen expression.
Homeostatic inflammatory process control hemodynamic change, capability permeability, leukocyte migration into tissues, and secretion of inflammatory mediators.68 This homeostatic inflammation in the liver is tightly regulated, and the activation of inflammatory processes is intimately linked to mechanisms that resolve inflammation and promote tissue regeneration.69 The fibrotic process is regulated by inflammatory cytokines and growth factors, which are released by leukocytes that traffic to the damaged tissue.70 These cytokines include TNF-α, IL-6, PDGF, and TGF-β. Kupffer cells, resident tissue macrophages, are shown to synthesize pro-inflammatory cytokines such as IL-1, IL-6, and TNF-α upon activation due to phagocytosis or binding of activation-triggering compounds such as endotoxin.71 These cytokines lead to the activation and proliferation of HSCs, which are potent producers of extracellular matrix components, including α-SMA and collagen I.72 Our results showed that the expressions of TNF-α and IL-6 were increased by TGF-β treatment, whereas TGF-β1/Smad ODN treatment reduced a fibrotic process through the inhibition of TNF-α, IL-1β, and IL-6 production in vitro and in vivo.
In summary, this study demonstrated the feasibility of using TGF-β1/Smad ODN by blocking TGF-β/Smad signaling to prevent hepatic fibrosis in a hepatocyte, HSCs, and liver fibrosis mouse model. TGF-β1/Smad ODN attenuated the EMT and activation of HSCs in TGF-β1-induced AML12 and HSC-T6 cells. Moreover, TGF-β1/Smad ODN prevented the fibrogenesis and deposition of collagen in CCl4-treated mouse model. Given the successful inhibition of hepatic fibrosis using TGF-β1/Smad ODN in cells and mouse model, gene therapy targeted to suppress mRNA level of TGF-β1 and transcription activity of Smad simultaneously might provide a new therapeutic strategy to prevent liver fibrosis.
Materials and Methods
Construction of Ring-Type ODNs
The target sites for TGF-β1 were selected via the sequential overlapsimulation of secondary structures using the S-Fold program. Synthetic ODNs were synthesized on a macrogen. Synthetic ODN sequences were used as follows (the target site or consensus binding sequence is underlined): scrambled (Scr) ODN: 5′-GAATTCCCGAAGTGCCAAGTCCTCCTCTCCACGG-3′; 5′-GAATTCCAGGTACGGCAAAAAATTGCCGTACCTG-3′; TGF-β1 antisense ODN (target site): 5′-GAATTCCCGAAAGCCCTGTATTCCGTCTCCTCGG-3′; Smad decoy ODN (consensus sequence is underlined): 5′-GAATTCGTGTCTAGACTGAAAACAGTCTAGACAC-3′.
TGF-β1/Smad ODN and Scr ODN were annealed for 6 hr, while thetemperature was decreased from 80°C to 25°C. To obtain a covalent ligation for ring-type ODN, each ODN was mixed with T4 ligase (Takara Bio) and incubated for 18 hr at 16°C.
Cell Culture
AML12 cells, the murine hepatocyte cell line, were obtained from the American Type Culture Collection (ATCC). AML12 cells cultured in DMEM/F-12 medium containing 10% fetal bovine serum (FBS) and 1% antibiotics (GIBCO BRL; Life Technologies) supplemented with insulin-transferrin-selenium and dexamethasone (40 ng/mL) (Sigma). HSC-T6 cells, which were an immortalized rat HSC cell line, which had the stable phenotype and biochemical characters, was kindly provided by Dr. S.L. Friedman (Liver Center Laboratory, San Francisco General Hospital). HSC-T6 cells were cultured in DMEM containing 10% FBS and 1% antibiotics. Cells were cultured at 37°C in a humidified incubator under a 5% CO2 atmosphere. AML12 and HSC-T6 cells were seeded in a complete medium for 24 hr. Cells were replaced with fresh serum-free media containing 2 ng/mL TGF-β1. After 12 hr of TGF-β1 treatment, cells were transfected with ODN using Lipofectamine 2000 (Invitrogen).
Mouse Models for CCl4-Induced Liver Fibrosis
Male C57BL/6 mice (6 weeks old, 20–22 g; Samtako) were housed in a room with controlled humidity and temperature, and a 12-hr light-dark cycle. All experiments were performed in accordance with the ethical guidelines of Institutional Animal Care and Use Committee of the Catholic University of Daegu (EXP-IRB number: 2014-0001-CU-AEC-04-A). To examine the in vivo transfection efficiency of synthetic TGF-β1/Smad ODN, FITC-labeled TGF-β1/Smad ODN was injected into mice via the tail vein. The mice were killed 24 hr after injection. Liver tissues were frozen with OCT compound (Sakura Finetek Japan). Cryosections of liver, which were transferred with FITC-labeled TGF-β1/Smad ODN, were examined using fluorescence microscopy.
Chronic liver injuries were induced by intraperitoneal injections of CCl4 (2 mL/kg, dissolved in corn oil [1:3 ratio]) three times a week.42, 73 One week after the first CCl4 injection, either Scr ODN or TGF-β1/Smad ODN (10 μg) was transferred biweekly via the mouse tail vein, using an in vivo gene delivery system (Mirus Bio). Mice were killed 8 weeks after the first CCl4 injection.
EMSA
Nuclear extracts fractionation from cells and liver tissue of mouse was conducted using an NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo Scientific), which was operated according to the manufacturer’s instructions.
EMSA (Thermo Scientific) was performed to analyze the expres sion of Smad. ODNs containing the consensus Smad binding site (5′-AGTATGTCTAGACTGA-3′) were used as primer.
Luciferase Assay
To measure TGF-β1 signaling, we used a TGF-β1-sensitive reporter construct, 100 ng of (CAGA)12-Luc reporter, which encodes 12 copies of the CAGA canonical Smad DNA-binding sequence. Smad promoter activity was measured by cell-based analysis methods. Cells were lysed in 100 μL of reporter-lysis buffer (Promega). For luciferase measurement, 50 μL of the lysate was used and 50 μL for the β-galactosidase assay (normalization of the transfection efficiency). Assays were performed using kits from Promega in accordance with the instruction manuals.
Cell Viability Assay
HSC-T6 cells were plated in 24-well culture plates at 8 × 104 cells per well in culture medium and allowed to attach for 24 hr. Media were then discarded and replaced with 500 μL of new medium containing TGF-β1 (2 ng/mL) for 12 hr. After incubation of the TGF-β1, cells added for the transfection of Scr ODN or TGF-β1/Smad ODN using Lipofectamine 2000. After experimental treatment, 50 μL of WST-8 solution (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) was added to each well, and the HSC-T6 cells were incubated for additional 4 hr at 37°C. The absorbance was measured at 450 nm using a microplate reader.
ELISA
The blood of each mouse was collected and serum was separated. The concentrations (in picograms per milliliter) of IFN-γ and IL-6 in the supernatant of cultured cells and serum were measured using a commercially available Quantikine mouse IFN-γ and IL-6 ELISA kit (R&D Systems), in accordance with the manufacturer’s instructions. Reading of the absorbance at 450 nm was done using an ELISA reader (BMG Labtech).
Real-Time PCR
Total RNA was extracted from cells and liver tissues using TRIzol method (Invitrogen) according to the manufacturer’s protocol. RNA was reverse transcribed using the AccuPower RT PreMix kit (Bioneer). Real-time PCR was performed for initial denaturation of 10 min at 95°C followed by 45 cycles of amplification for 20 s at 95°C and 20 s at 60°C, and 20 s at 72°C using LightCycler Nano Instrument Real-Time PCR Systems (Roche Life Science) with a FastStart Essential DNA Green Master Kit (Roche). The cDNA was amplified by real-time PCR with the following primers: TGF-β1, 5′-GTGTGGAGCAACATGTGGAACTCTA-3′ (forward) and 5′-CGCTGAATCGAAAGCCCTGTA-3′ (reverse), and GAPDH, 5′-GACAACTTTGGCATCGTGGA-3′ (forward) and 5′-ATGCAGGGATGATGTTCTGG-3′ (reverse).
Western Blot Analysis
Cells were lysed in lysis buffer (Sigma). After incubation for 30 min on ice, lysate was centrifuged at 8,000 × g for 15 min at 4°C and the supernatant was retained. Protein samples were separated using SDS-PAGE with Bolt 4%–12% Bis-Tris gel (Invitrogen), transferred to nitrocellulose membrane (Millipore). Membranes were incubated with primary antibodies, and horseradish peroxidase (HRPO)-conjugated secondary antibodies were used for detection. GAPDH was used as a protein loading control. The enhanced chemiluminescence was captured using the ChemiDoc XRS+ system and quantified using Image Lab software (Bio-Rad). Primary antibodies used in this study were as follows: anti-p-Smad2/3 and anti-Smad2/3 (Cell Signaling), anti-Smad4, anti-Smad7, anti-IL-1β and anti-GAPDH (Santa Cruz), anti-collagen I and anti-TNF-α (Abcam), and anti-fibronectin, anti-E-cadherin, and anti-vimentin (BD Biosciences).
Histological and Immunohistochemical Staining
H&E, Masson’s Trichrome, and immunohistochemical staining were performed according to the described procedure.40 Sections were stained with H&E and Masson’s Trichrome. For immunohistochemical analysis, sections were incubated with anti-fibroblast specific protein (FSP)-1 (Abcam) and anti-fibronectin for 1 hr at 37°C, processed by an indirect immunoperoxidase technique using a commercial kit (DAKO). The slides were examined with an Eclipse 80i microscope (Nikon) and analyzed using iSolution DT software (IMT i-Solution).
Immunofluorescence Staining
Paraffin-embedded mouse liver sections (3-μm thickness) were prepared using a routine procedure. After blocking with 10% donkey serum for 30 min, the slides were immunostained with primary antibodies against collagen I, E-cadherin, p-Smad2/3, Smad4, and α-SMA. After washing, they were incubated with the secondary antibodies (Alexa Fluor 488 and/or Alexa Fluor 594) for 30 min at 37°C. Sections were then counterstained with Hoechst 33342. Stained slides were imaged using a NIKON A1+ confocal microscope (Nikon).
Statistical Analysis
All data represent at least three experiments and expressed as mean ± SD. ANOVA and paired or unpaired t test were performed for statistical analysis as appropriate. p < 0.05 was considered statistically significant.
Author Contributions
J.-Y.K. and K.-K.P. conceived the study and provided leadership. J.-Y.K., H.-J.A., W.-H.K., M-G.G., and H.G. performed the experiments. J.-Y.K. and K.-K.P. drafted the manuscript. J.-Y.K. and Y.-Y.P. performed data analysis. All authors discussed, revised, and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the National Research Foundation of Korea grant funded by the Korean Government (NRF-2015R1D1A1A01061026).
Footnotes
Supplemental Information includes three figures and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.06.022.
Supplemental Information
References
- 1.Lee U.E., Friedman S.L. Mechanisms of hepatic fibrogenesis. Best Pract. Res. Clin. Gastroenterol. 2011;25:195–206. doi: 10.1016/j.bpg.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman S.L. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman S.L. Seminars in medicine of the Beth Israel Hospital, Boston. The cellular basis of hepatic fibrosis. Mechanisms and treatment strategies. N. Engl. J. Med. 1993;328:1828–1835. doi: 10.1056/NEJM199306243282508. [DOI] [PubMed] [Google Scholar]
- 4.Weiler-Normann C., Herkel J., Lohse A.W. Mouse models of liver fibrosis. Z. Gastroenterol. 2007;45:43–50. doi: 10.1055/s-2006-927387. [DOI] [PubMed] [Google Scholar]
- 5.Copple B.L. Hypoxia stimulates hepatocyte epithelial to mesenchymal transition by hypoxia-inducible factor and transforming growth factor-beta-dependent mechanisms. Liver Int. 2010;30:669–682. doi: 10.1111/j.1478-3231.2010.02205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiery J.P., Sleeman J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 7.Zhe C., Yu F., Tian J., Zheng S. Trps1 regulates biliary epithelial-mesenchymal transition and has roles during biliary fibrosis in liver grafts: a preliminary study. PLoS One. 2015;10:e0123233. doi: 10.1371/journal.pone.0123233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee W.R., Kim K.H., An H.J., Kim J.Y., Lee S.J., Han S.M., Pak S.C., Park K.K. Apamin inhibits hepatic fibrosis through suppression of transforming growth factor β1-induced hepatocyte epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2014;450:195–201. doi: 10.1016/j.bbrc.2014.05.089. [DOI] [PubMed] [Google Scholar]
- 9.Shrestha N., Chand L., Han M.K., Lee S.O., Kim C.Y., Jeong Y.J. Glutamine inhibits CCl4 induced liver fibrosis in mice and TGF-beta1 mediated epithelial-mesenchymal transition in mouse hepatocytes. Food Chem. Toxicol. 2016;93:129–137. doi: 10.1016/j.fct.2016.04.024. [DOI] [PubMed] [Google Scholar]
- 10.Friedman S.L. Hepatic fibrosis: emerging therapies. Dig. Dis. 2015;33:504–507. doi: 10.1159/000374098. [DOI] [PubMed] [Google Scholar]
- 11.Greuter T., Shah V.H. Hepatic sinusoids in liver injury, inflammation, and fibrosis: new pathophysiological insights. J. Gastroenterol. 2016;51:511–519. doi: 10.1007/s00535-016-1190-4. [DOI] [PubMed] [Google Scholar]
- 12.Xu R., Zhang Z., Wang F.S. Liver fibrosis: mechanisms of immune-mediated liver injury. Cell. Mol. Immunol. 2012;9:296–301. doi: 10.1038/cmi.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman S.L. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye Z., Houssein H.S., Mahato R.I. Bioconjugation of oligonucleotides for treating liver fibrosis. Oligonucleotides. 2007;17:349–404. doi: 10.1089/oli.2007.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bataller R., Brenner D.A. Liver fibrosis. J. Clin. Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bissell D.M., Roulot D., George J. Transforming growth factor beta and the liver. Hepatology. 2001;34:859–867. doi: 10.1053/jhep.2001.28457. [DOI] [PubMed] [Google Scholar]
- 17.Cheng K., Yang N., Mahato R.I. TGF-beta1 gene silencing for treating liver fibrosis. Mol. Pharm. 2009;6:772–779. doi: 10.1021/mp9000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreira R.K. Hepatic stellate cells and liver fibrosis. Arch. Pathol. Lab. Med. 2007;131:1728–1734. doi: 10.5858/2007-131-1728-HSCALF. [DOI] [PubMed] [Google Scholar]
- 19.Safadi R., Friedman S.L. Hepatic fibrosis—role of hepatic stellate cell activation. MedGenMed. 2002;4:27. [PubMed] [Google Scholar]
- 20.Choi J.S., Kim J.K., Yang Y.J., Kim Y., Kim P., Park S.G., Cho E.Y., Lee D.H., Choi J.W. Identification of cromolyn sodium as an anti-fibrotic agent targeting both hepatocytes and hepatic stellate cells. Pharmacol. Res. 2015;102:176–183. doi: 10.1016/j.phrs.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Bauer M., Schuppan D. TGFbeta1 in liver fibrosis: time to change paradigms? FEBS Lett. 2001;502:1–3. doi: 10.1016/s0014-5793(01)02655-2. [DOI] [PubMed] [Google Scholar]
- 22.Bran G.M., Sommer U.J., Goessler U.R., Hörmann K., Riedel F., Sadick H. TGF-ß1 antisense impacts the SMAD signalling system in fibroblasts from keloid scars. Anticancer Res. 2010;30:3459–3463. [PubMed] [Google Scholar]
- 23.Cong M., Iwaisako K., Jiang C., Kisseleva T. Cell signals influencing hepatic fibrosis. Int. J. Hepatol. 2012;2012:158547. doi: 10.1155/2012/158547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y.L., Zhu R.T., Sun Y.L. Epithelial-mesenchymal transition in liver fibrosis. Biomed. Rep. 2016;4:269–274. doi: 10.3892/br.2016.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kisseleva T., Brenner D.A. Mechanisms of fibrogenesis. Exp. Biol. Med. (Maywood) 2008;233:109–122. doi: 10.3181/0707-MR-190. [DOI] [PubMed] [Google Scholar]
- 26.Wrana J.L., Attisano L., Wieser R., Ventura F., Massagué J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 27.Massagué J. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 28.Schnabl B., Kweon Y.O., Frederick J.P., Wang X.F., Rippe R.A., Brenner D.A. The role of Smad3 in mediating mouse hepatic stellate cell activation. Hepatology. 2001;34:89–100. doi: 10.1053/jhep.2001.25349. [DOI] [PubMed] [Google Scholar]
- 29.Miyazawa K., Shinozaki M., Hara T., Furuya T., Miyazono K. Two major Smad pathways in TGF-beta superfamily signalling. Genes Cells. 2002;7:1191–1204. doi: 10.1046/j.1365-2443.2002.00599.x. [DOI] [PubMed] [Google Scholar]
- 30.Schiller M., Javelaud D., Mauviel A. TGF-beta-induced SMAD signaling and gene regulation: consequences for extracellular matrix remodeling and wound healing. J. Dermatol. Sci. 2004;35:83–92. doi: 10.1016/j.jdermsci.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Nakao A., Afrakhte M., Morén A., Nakayama T., Christian J.L., Heuchel R., Itoh S., Kawabata M., Heldin N.E., Heldin C.H., ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 32.Castro S.V., Jimenez S.A. Biomarkers in systemic sclerosis. Biomarkers Med. 2010;4:133–147. doi: 10.2217/bmm.09.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gauglitz G.G., Korting H.C., Pavicic T., Ruzicka T., Jeschke M.G. Hypertrophic scarring and keloids: pathomechanisms and current and emerging treatment strategies. Mol. Med. 2011;17:113–125. doi: 10.2119/molmed.2009.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ismail M.H., Pinzani M. Reversal of liver fibrosis. Saudi J. Gastroenterol. 2009;15:72–79. doi: 10.4103/1319-3767.45072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim K.H., Park K.K. Small RNA- and DNA-based gene therapy for the treatment of liver cirrhosis, where we are? World J. Gastroenterol. 2014;20:14696–14705. doi: 10.3748/wjg.v20.i40.14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 37.Liu Q., Paroo Z. Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- 38.Mann M.J., Dzau V.J. Therapeutic applications of transcription factor decoy oligonucleotides. J. Clin. Invest. 2000;106:1071–1075. doi: 10.1172/JCI11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kemaladewi D.U., Pasteuning S., van der Meulen J.W., van Heiningen S.H., van Ommen G.J., Ten Dijke P., Aartsma-Rus A., ’t Hoen P.A., Hoogaars W.M. Targeting TGF-β signaling by antisense oligonucleotide-mediated knockdown of TGF-β type I receptor. Mol. Ther. Nucleic Acids. 2014;3:e156. doi: 10.1038/mtna.2014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim K.H., Park J.H., Lee W.R., Park J.S., Kim H.C., Park K.K. The inhibitory effect of chimeric decoy oligodeoxynucleotide against NF-κB and Sp1 in renal interstitial fibrosis. J. Mol. Med. (Berl.) 2013;91:573–586. doi: 10.1007/s00109-012-0972-2. [DOI] [PubMed] [Google Scholar]
- 41.Hellerbrand C. Hepatic stellate cells—the pericytes in the liver. Pflugers Arch. 2013;465:775–778. doi: 10.1007/s00424-012-1209-5. [DOI] [PubMed] [Google Scholar]
- 42.Kim K.H., Lee W.R., Kang Y.N., Chang Y.C., Park K.K. Inhibitory effect of nuclear factor-κB decoy oligodeoxynucleotide on liver fibrosis through regulation of the epithelial-mesenchymal transition. Hum. Gene Ther. 2014;25:721–729. doi: 10.1089/hum.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman S.L. Liver fibrosis—from bench to bedside. J. Hepatol. 2003;38(Suppl 1):S38–S53. doi: 10.1016/s0168-8278(02)00429-4. [DOI] [PubMed] [Google Scholar]
- 44.Wraight C.J., White P.J. Antisense oligonucleotides in cutaneous therapy. Pharmacol. Ther. 2001;90:89–104. doi: 10.1016/s0163-7258(01)00133-4. [DOI] [PubMed] [Google Scholar]
- 45.Sepp-Lorenzino L., Ruddy M. Challenges and opportunities for local and systemic delivery of siRNA and antisense oligonucleotides. Clin. Pharmacol. Ther. 2008;84:628–632. doi: 10.1038/clpt.2008.174. [DOI] [PubMed] [Google Scholar]
- 46.Li H., Quan J., Zhang M., Yung B.C., Cheng X., Liu Y., Lee Y.B., Ahn C.H., Kim D.J., Lee R.J. Lipid-albumin nanoparticles (LAN) for therapeutic delivery of antisense oligonucleotide against HIF-1α. Mol. Pharm. 2016;13:2555–2562. doi: 10.1021/acs.molpharmaceut.6b00363. [DOI] [PubMed] [Google Scholar]
- 47.Johns R.E., El-Sayed M.E., Bulmus V., Cuschieri J., Maier R., Hoffman A.S., Stayton P.S. Mechanistic analysis of macrophage response to IRAK-1 gene knockdown by a smart polymer-antisense oligonucleotide therapeutic. J. Biomater. Sci. Polym. Ed. 2008;19:1333–1346. doi: 10.1163/156856208786052326. [DOI] [PubMed] [Google Scholar]
- 48.Wang H., Hang J., Shi Z., Li M., Yu D., Kandimalla E.R., Agrawal S., Zhang R. Antisense oligonucleotide targeted to RIalpha subunit of cAMP-dependent protein kinase (GEM231) enhances therapeutic effectiveness of cancer chemotherapeutic agent irinotecan in nude mice bearing human cancer xenografts: in vivo synergistic activity, pharmacokinetics and host toxicity. Int. J. Oncol. 2002;21:73–80. [PubMed] [Google Scholar]
- 49.Murano M., Maemura K., Hirata I., Toshina K., Nishikawa T., Hamamoto N., Sasaki S., Saitoh O., Katsu K. Therapeutic effect of intracolonically administered nuclear factor kappa B (p65) antisense oligonucleotide on mouse dextran sulphate sodium (DSS)-induced colitis. Clin. Exp. Immunol. 2000;120:51–58. doi: 10.1046/j.1365-2249.2000.01183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao C.C., Ding X.Q., Ou Z.L., Liu C.F., Li P., Wang L., Zhu C.F. In vivo transfection of NF-kappaB decoy oligodeoxynucleotides attenuate renal ischemia/reperfusion injury in rats. Kidney Int. 2004;65:834–845. doi: 10.1111/j.1523-1755.2004.00463.x. [DOI] [PubMed] [Google Scholar]
- 51.Kim K.H., Lee E.S., Cha S.H., Park J.H., Park J.S., Chang Y.C., Park K.K. Transcriptional regulation of NF-kappaB by ring type decoy oligodeoxynucleotide in an animal model of nephropathy. Exp. Mol. Pathol. 2009;86:114–120. doi: 10.1016/j.yexmp.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 52.Lee W.R., Kim K.H., An H.J., Park Y.Y., Kim K.S., Lee C.K., Min B.K., Park K.K. Effects of chimeric decoy oligodeoxynucleotide in the regulation of transcription factors NF-κB and Sp1 in an animal model of atherosclerosis. Basic Clin. Pharmacol. Toxicol. 2013;112:236–243. doi: 10.1111/bcpt.12029. [DOI] [PubMed] [Google Scholar]
- 53.Aoki T., Kataoka H., Nishimura M., Ishibashi R., Morishita R., Miyamoto S. Regression of intracranial aneurysms by simultaneous inhibition of nuclear factor-κB and Ets with chimeric decoy oligodeoxynucleotide treatment. Neurosurgery. 2012;70:1534–1543. doi: 10.1227/NEU.0b013e318246a390. discussion 1543. [DOI] [PubMed] [Google Scholar]
- 54.Yuan H.F., Huang H., Li X.Y., Guo W., Xing W., Sun Z.Y., Liang H.P., Yu J., Chen D.F., Wang Z.G. A dual AP-1 and SMAD decoy ODN suppresses tissue fibrosis and scarring in mice. J. Invest. Dermatol. 2013;133:1080–1087. doi: 10.1038/jid.2012.443. [DOI] [PubMed] [Google Scholar]
- 55.Mederacke I., Hsu C.C., Troeger J.S., Huebener P., Mu X., Dapito D.H., Pradere J.P., Schwabe R.F. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat. Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Madsen D.H., Jürgensen H.J., Ingvarsen S., Melander M.C., Vainer B., Egerod K.L., Hald A., Rønø B., Madsen C.A., Bugge T.H. Endocytic collagen degradation: a novel mechanism involved in protection against liver fibrosis. J. Pathol. 2012;227:94–105. doi: 10.1002/path.3981. [DOI] [PubMed] [Google Scholar]
- 57.Uchinami H., Seki E., Brenner D.A., D’Armiento J. Loss of MMP 13 attenuates murine hepatic injury and fibrosis during cholestasis. Hepatology. 2006;44:420–429. doi: 10.1002/hep.21268. [DOI] [PubMed] [Google Scholar]
- 58.Fang L., Huang C., Meng X., Wu B., Ma T., Liu X., Zhu Q., Zhan S., Li J. TGF-β1-elevated TRPM7 channel regulates collagen expression in hepatic stellate cells via TGF-β1/Smad pathway. Toxicol. Appl. Pharmacol. 2014;280:335–344. doi: 10.1016/j.taap.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Calabro S.R., Maczurek A.E., Morgan A.J., Tu T., Wen V.W., Yee C., Mridha A., Lee M., d’Avigdor W., Locarnini S.A. Hepatocyte produced matrix metalloproteinases are regulated by CD147 in liver fibrogenesis. PLoS One. 2014;9:e90571. doi: 10.1371/journal.pone.0090571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iwaisako K., Brenner D.A., Kisseleva T. What’s new in liver fibrosis? The origin of myofibroblasts in liver fibrosis. J. Gastroenterol. Hepatol. 2012;27(Suppl 2):65–68. doi: 10.1111/j.1440-1746.2011.07002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu F., Liu C., Zhou D., Zhang L. TGF-beta/SMAD pathway and its regulation in hepatic fibrosis. J. Histochem. Cytochem. 2016;64:157–167. doi: 10.1369/0022155415627681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakatsukasa H., Nagy P., Evarts R.P., Hsia C.C., Marsden E., Thorgeirsson S.S. Cellular distribution of transforming growth factor-beta 1 and procollagen types I, III, and IV transcripts in carbon tetrachloride-induced rat liver fibrosis. J. Clin. Invest. 1990;85:1833–1843. doi: 10.1172/JCI114643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng Y.L., Xiong X.Z., Cheng N.S. Organ fibrosis inhibited by blocking transforming growth factor-β signaling via peroxisome proliferator-activated receptor γ agonists. Hepatobiliary Pancreat. Dis. Int. 2012;11:467–478. doi: 10.1016/s1499-3872(12)60210-0. [DOI] [PubMed] [Google Scholar]
- 64.Ju W., Ogawa A., Heyer J., Nierhof D., Yu L., Kucherlapati R., Shafritz D.A., Böttinger E.P. Deletion of Smad2 in mouse liver reveals novel functions in hepatocyte growth and differentiation. Mol. Cell. Biol. 2006;26:654–667. doi: 10.1128/MCB.26.2.654-667.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsukada S., Westwick J.K., Ikejima K., Sato N., Rippe R.A. SMAD and p38 MAPK signaling pathways independently regulate alpha1(I) collagen gene expression in unstimulated and transforming growth factor-beta-stimulated hepatic stellate cells. J. Biol. Chem. 2005;280:10055–10064. doi: 10.1074/jbc.M409381200. [DOI] [PubMed] [Google Scholar]
- 66.Heldin C.H., Miyazono K., ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 67.Dooley S., Hamzavi J., Breitkopf K., Wiercinska E., Said H.M., Lorenzen J., Ten Dijke P., Gressner A.M. Smad7 prevents activation of hepatic stellate cells and liver fibrosis in rats. Gastroenterology. 2003;125:178–191. doi: 10.1016/s0016-5085(03)00666-8. [DOI] [PubMed] [Google Scholar]
- 68.Tacke F., Luedde T., Trautwein C. Inflammatory pathways in liver homeostasis and liver injury. Clin. Rev. Allergy Immunol. 2009;36:4–12. doi: 10.1007/s12016-008-8091-0. [DOI] [PubMed] [Google Scholar]
- 69.Robinson M.W., Harmon C., O’Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cell. Mol. Immunol. 2016;13:267–276. doi: 10.1038/cmi.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borthwick L.A., Wynn T.A., Fisher A.J. Cytokine mediated tissue fibrosis. Biochim. Biophys. Acta. 2013;1832:1049–1060. doi: 10.1016/j.bbadis.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sica A., Invernizzi P., Mantovani A. Macrophage plasticity and polarization in liver homeostasis and pathology. Hepatology. 2014;59:2034–2042. doi: 10.1002/hep.26754. [DOI] [PubMed] [Google Scholar]
- 72.Weiskirchen R., Tacke F. Cellular and molecular functions of hepatic stellate cells in inflammatory responses and liver immunology. Hepatobiliary Surg. Nutr. 2014;3:344–363. doi: 10.3978/j.issn.2304-3881.2014.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park J.H., Jo J.H., Kim K.H., Kim S.J., Lee W.R., Park K.K., Park J.B. Antifibrotic effect through the regulation of transcription factor using ring type-Sp1 decoy oligodeoxynucleotide in carbon tetrachloride-induced liver fibrosis. J. Gene Med. 2009;11:824–833. doi: 10.1002/jgm.1355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.