Abstract
Prostate cancer (PCa) is the most common type of cancer and the second leading cause of cancer-related death in men. Despite extensive research, the molecular mechanisms underlying PCa initiation and progression remain unclear, and there is increasing need of better biomarkers that can distinguish indolent from aggressive and life-threatening disease. With the advent of advanced genomic technologies in the last decade, it became apparent that the human genome encodes tens of thousands non-protein-coding RNAs (ncRNAs) with yet to be discovered function. It is clear now that the majority of ncRNAs exhibit highly specific expression patterns restricted to certain tissues and organs or developmental stages and that the expression of many ncRNAs is altered in disease and cancer, including cancer of the prostate. Such ncRNAs can serve as important biomarkers for PCa diagnosis, prognosis, or prediction of therapy response. In this review, we give an overview of the different types of ncRNAs and their function, describe ncRNAs relevant for the diagnosis and prognosis of PCa, and present emerging new aspects of ncRNA research that may contribute to the future utilization of ncRNAs as clinically useful therapeutic targets.
Key Points
| Recent developments in expression-profiling technologies, combined with large-scale efforts to sequence patient samples, have drastically enhanced the discovery of disease-associated non-coding RNAs (ncRNAs). |
| ncRNAs are associated with disease progression in patients with prostate cancer. |
| ncRNAs are promising as diagnostic, prognostic, and predictive biomarkers in prostate cancer. |
| RNA-targeting drugs may provide the basis for precision cancer therapies. |
| Further investigations, large validation studies, and industrial partnerships are necessary for the successful translation of ncRNA research into clinical practice. |
Introduction
The Paradigm of Prostate Cancer (PCa): Current Needs and Demands
Prostate cancer (PCa) is the most common cancer and the second cause of cancer-related death among western men. PCa incidence is increasing mainly because of population ageing, increased awareness, and the widespread introduction of the prostate-specific antigen (PSA) test [1]. Current PCa diagnosis and “informed” clinical decisions involve digital rectal examination (DRE), serum PSA measurement, and multiple invasive tissue biopsies for histological grading. Yet, patient treatment is often hampered by invasive monitoring, risk of overtreatment after early diagnosis, or poor prediction of treatment response in advanced disease stages.
Such limitations in the clinical management of PCa arise from the pathological variability and molecular heterogeneity of prostate tumors. While some patients with indolent tumors live for up to 20 years after diagnosis, others die of metastatic cancer within 2–3 years. In addition, molecular polyclonality of the tumors of individual patents can cause intrinsic or acquired treatment resistance in late-stage disease [2–4]. As a result, patients with early-stage PCa are heavily overtreated [5] and often experience serious side effects, whereas optimal therapeutic sequence in late-stage PCa is yet to be established and must be balanced against toxicity, symptoms, and performance status [6]. The future advancement of tailored PCa treatment requires the development and introduction of a new generation of biomarkers that allow accurate early prognosis as well as appropriate monitoring and prediction of therapy response and resistance.
The genomic revolution led to the discovery of defining mutations and gene-expression profiles that accurately predict the progression and clinical course of many cancers. Genomic and transcriptomic studies identified many potential PCa tissue markers, including TMPRSS2-ERG, PSCA, BRCA1/2, PTEN, KLK2, AMACR, and TGFB. However, these markers hold little prognostic value and cannot accurately predict the course of disease [7–10]. Over the last few years, several new tests based on gene expression have been offered that are available from Clinical Laboratory Improvement Amendment (CLIA)-certified clinical laboratories (Decipher, Oncotype DX, and Prolaris) [11]. However, despite better prognostic performance, these tests remain dependent on invasive tissue sampling [12–14]. Recently, the so-called SelectMDx urine test was developed that determines urinary DLX1 and HOXC6 messenger RNA (mRNA) levels. This non-invasive test can be used to identify patients with aggressive or clinically significant PCa, including in men with low serum PSA values [15], and shows improved risk stratification for high-grade PCa and biopsy decision making when combined with traditional clinical risk factors [16]. This achievement clearly demonstrated the potential of RNA-based marker tests for the non-invasive diagnosis and prognosis of PCa and—together with the introduction of the PCA3 urine test based on the detection of a non-coding RNA (ncRNA) molecule—opened the doors for extensive exploration of the transcriptome as a new versatile field of marker and therapy research.
Definition and Types of Non-Coding RNAs (ncRNAs)
A most unexpected discovery that arose from the sequencing of the human genome was the finding that no more than 3% of human DNA appears to encode for protein. With the advancement of genomic technologies in the 2 decades that followed, it became apparent that over 80% of the genome is actively transcribed to different RNA products that are not translated to protein (known as ncRNAs) [17]
Based on size and the arbitrary cut-off of 200 nucleotides, ncRNAs are divided into two highly diverse groups: small ncRNAs (sncRNAs) and long ncRNAs (lncRNAs) (Table 1).
Table 1.
Types of non-coding RNAs
| ncRNA class | Common abbreviation | Approved gene symbola | Function |
|---|---|---|---|
| Small ncRNA | sncRNA | ||
| MicroRNA | miRNA | MIR | Regulation of gene expression |
| Transfer RNA | tRNA | Amino acid synthesis | |
| Spliceosomal RNA | U | RNU | Splicing |
| Small nucleolar RNA | snoRNA | Ribosome maturation by site-specific chemical modification; post-transcriptional modification of tRNAs and spliceosomal RNAs | |
| H/ACA box | SNORA | ||
| CD box | SNORD | ||
| Small Cajal body RNA | scaRNA | SCARNA | |
| Endogenous small interfering RNA | endo-siRNA | RNA interference; post-transcriptional gene silencing | |
| PiWi-interacting RNA | piRNA | PIRC | Transposon silencing |
| U7 small nuclear RNA | U7 | RNU7 | 3′-end maturation of histone pre-mRNA |
| 7SK RNA | 7SK | RN7SK | Activity regulation of positive transcription elongation factor b (P-TEFb) |
| 7SL RNA | SRP 7SL | RN7SL | RNA component of the SRP RNP |
| Small ILF3/NF90-associated RNA | SNAR | Binds interleukin enhancer binding factor ILF3/NF90 | |
| Ro-associated Y RNA | Y RNA | RNY | Component of the Ro RNP; proposed involvement in DNA replication |
| Vault RNA | VTRNA | Component of the vault RNP; proposed involvement in multidrug resistance | |
| Ribonuclease P RNA component H1 | RPPH1 | 5′-end processing of tRNA precursors | |
| RNA component of RNase MRP | RMRP | Maturation of precursor rRNAs; mitochondrial DNA replication | |
| Telomerase RNA | hTR | TERC | RNA component of human telomerase |
| Ribosomal RNA | |||
| Mitochondrial rRNA | mit-rRNA | MT-RNR1/2 | Protein synthesis |
| Genomic rRNA | rRNA | RN18S, N28S, RN5–8S, RN5S |
|
| Long ncRNA | lncRNA | ||
| Antisense transcripts | AS OS |
Reside on the opposite strand of protein-coding genes and intersect their exons | |
| Overlapping transcripts | OT | Contain a coding gene within an intron on the same strand | |
| Intronic transcripts | IT | Reside within introns of a coding gene but do not intersect any exons | |
| Host genes | HG | Primary hosts of small ncRNA genes nested within their introns | |
| Pseudogenes | P | Transcribed from genes that have lost their protein-coding potential | |
| Intergenic lncRNAs | lincRNA | LINC | Originate from protein non-coding genomic regions |
| Circular RNAs | circRNA | Form during splicing by chemical bonding of different exons | |
| Enhancer RNAs | eRNAs | Transcribed from genomic enhancer regions | |
| sno-related lncRNAs | sno-lncRNA | Flanked by snoRNAs instead of 5′-cap and 3′-poly(A) tail | |
ER endoplasmic reticulum, mRNA messenger RNA, ncRNA non-coding RNA, RNP ribonucleoprotein particle, SRP signal recognition particle
aHUGO Gene Nomenclature Committee (HGNC) (http://www.genenames.org/search)
Small ncRNAs (sncRNAs)
sncRNAs and particularly microRNAs (miRNAs) are extensively studied because of their function as gene regulators during development and disease. miRNAs are single-stranded RNA molecules 21–24 nucleotides in size and they complex with the Argonaute (AGO) family of proteins to form the so called RNA-induced silencing complex (RISC). In their canonical pathway, RISC-complexed miRNAs regulate the function of their target mRNAs at the post-transcriptional level, most often via mRNA degradation or translational repression [18]. It has been demonstrated that a subset of miRNAs have the ability to translocate back to the nucleus where they activate or repress transcriptional activity, possibly via the recruitment of chromatin-modifying factors to the target region (reviewed by Schwarzenbach et al. [19]). To date, 2588 human miRNAs originating from 1881 bona fide miRNA precursors have been described with high confidence, and strict criteria for their annotation have been set (miRBase v21; www.mirbase.org) [20].
Besides miRNAs, PiWi-interacting RNAs (piRNAs) and endogenous small interfering RNAs (endo-siRNAs) are also involved in post-transcriptional gene regulation and operate via interaction with AGO proteins and the RISC [21]. piRNAs are the largest class of sncRNAs, with over 30,000 unique sequences catalogued so far [22]. piRNAs function as post-transcriptional regulators of transposon silencing and indispensable guardians of genome stability in animal cells [23].
Other small RNAs such as small nucleolar RNAs (snoRNA) are involved in the biochemical maturation of ribosomal RNA and transfer RNAs (tRNAs) and in protein synthesis. It has long been thought that snoRNAs and tRNAs are house-keeping molecules. Nevertheless, recent genome-wide and cross-species studies demonstrate that snoRNAs exhibit strong tissue-specific expression patterns [24] and function beyond their canonical pathways. It was discovered that both snoRNAs and tRNAs give rise to even smaller RNA species referred to as snoRNA-derived RNAs (sdRNAs) and tRNA-derived fragments (tRFs) that operate in regulatory pathways different than those of their precursors [25, 26]. These small molecules form a new layer of dynamic and highly versatile small regulatory RNAs that are involved in alternative splicing, stress response, transcriptional regulation, and epigenetic control [27–29].
Genomic Abundance and Specific Expression of Long ncRNAs (lncRNAs)
The systematic annotation of human lncRNAs was initiated by the ENCODE [30] and GENCODE [31] projects, which aim to identify and annotate all functional elements in the human genome sequence. These projects clearly demonstrated that pervasive transcription of the human genome gives rise to many lncRNAs (Table 2). The latest release of the GENCODE database (version 25, GRCh38; www.gencodegenes.org/stats) counts more than 15,000 human lncRNA genes and over 14,000 pseudogenes, many of which produce multiple transcripts. Nevertheless, lncRNA gene annotations remain incomplete, and methods to define and annotate them still exhibit limitations regarding the full repertoire and dynamic range of transcript expression in the cell [32]. While the total number and function of many ncRNAs, particularly lncRNAs, is yet to be established, accumulating evidence demonstrates that ncRNAs interact with, regulate, or fine tune major pathways of cell growth, proliferation, differentiation, and survival. Subsequently, alterations in the functions of different ncRNAs are associated with developmental disorders, diseases, and cancer.
Table 2.
Long non-coding RNA characteristics
| GENCODE established lncRNA characteristics [31] |
|---|
| lncRNAs are independent transcriptional units that lack protein-coding potential. They are not unrecognized extensions of neighboring protein-coding genes |
| lncRNA genes can have an unusual exonic structure, but their processing, splicing signals, exon/intron length, and poly-adenylation are similar to those of protein-coding genes |
| lncRNA genes, when expressed, have the typical histone modifications associated with active transcription but show more tissue-specific and generally lower expression than protein-coding genes |
| Human lncRNAs are under weaker selective constraints than protein-coding genes, and many are primate specific |
| Some human lncRNA genes belong to evolutionary conserved families that evolve faster than protein-coding genes. In this process, sequence similarity seems to be preserved mainly in regions involved in secondary structure formation |
lncRNA long non-coding RNA
ncRNAs as Biomarkers for the Minimally Invasive Management of PCa
Several of the intrinsic properties of ncRNAs suggest that the non-coding transcriptome can be a useful source of disease and stage-specific biomarkers. For example, there are many more ncRNAs than mRNAs, hence the chance of finding a specific marker is higher. Furthermore, ncRNAs are the final gene product, and thus biologically relevant levels are measured. The highly specific tissue and/or disease expression of ncRNA can provide the high discriminative power required for a successful biomarker. ncRNAs, and RNAs in general, have uniform biochemical properties that make it easier to manufacture clinical assays. Finally, ncRNAs can be detected in body fluids, enabling the development of minimally invasive “liquid biopsy” assays.
Over the last decade, many efforts for ncRNA biomarker discovery have been initiated. These resulted in the identification of numerous disease-associated miRNAs, lncRNAs, and other non-coding transcripts. Platforms for the accurate quantification of ncRNA levels in tissue and body fluid specimens have evolved accordingly. In this section, we provide an overview of PCa-associated ncRNAs and ncRNA marker tests for PCa.
State of the Art: PCA3, A Clinically Approved ncRNA Marker Test
The very first PCa-associated ncRNA to be discovered was PCA3 (a.k.a. DD3, PCAT3), an lncRNA identified in 1999 via differential display analysis [33]. PCA3 is specifically expressed in prostate epithelial cells, and—compared with benign tissue—PCA3 is highly overexpressed in PCa and high-grade prostatic intra-epithelial neoplasia [33–35]. PCA3 is an antisense intronic lncRNA located in the tumor-suppressive protein-coding gene PRUNE2. Recently it was proposed that PCA3 controls PRUNE2 mRNA levels via the formation of a PRUNE2/PCA3 double-stranded RNA that undergoes adenosine deaminase, RNA specific (ADAR)-mediated adenosine-to-inosine RNA editing [36]. However, these findings conflict with the previously reported lack of correlation between PCA3 and PRUNE2 RNA expression in PCa tissue specimens [37], and the role of PCA3 in RNA editing remains uncertain. Knockdown studies of PCA3 demonstrated that cell viability of PCa cells was reduced and that the expression of androgen receptor (AR) target genes were altered [38]. PCA3 can also be found in the urine of men with PCa after DRE, and this finding has led to the development of the Progensa PCA3 test for PCa detection. Progensa PCA3 is a single-tube and isothermic amplification assay to quantify PCA3 levels in urine specimens [39, 40]. The Progensa® PCA3 (Hologic Inc.) test has been approved by the US FDA for clinical use to predict biopsy outcome in men aged ≥50 years with a suspicion of PCa based on serum PSA levels and/or DRE and/or one or more previous negative biopsies. Compared with serum PSA, the urinary PCA3 score (i.e., PCA3 RNA to KLK3 [=PSA] mRNA ratio) has improved sensitivity in the detection of PCa. Urinary PCA3 tests more accurately predict repeat biopsy outcomes and thus may lead to a reduction in the number of unnecessary prostate biopsies [9, 41–45]. In patients with clinically localized PCa, the urinary PCA3 score also shows correlation with tumor volume and may differentiate between low-volume/low-grade cancer and significant cancer [46–49]. Hence, it may be an independent risk factor for PCa. However, results on the prognostic value of PCA3 testing have been conflicting, as several independent studies failed to detect significant associations between PCA3 score and any prognostic parameter (including stage, Gleason score, tumor volume, or extra-prostatic extension; reviewed by Hessels and Schalken [50]). Therefore, PCA3 may not differentiate the aggressiveness of a tumor, hence biomarkers indicative of tumor aggressiveness are still an unmet need in PCa.
Candidate ncRNA Biomarkers
lncRNAs
Two major large-scale profiling studies to identify PCa-associated lncRNAs have yielded dozens of disease-associated transcripts [51, 52]. For example, PCa-associated transcript 1 (non-protein coding) (PCAT1) is a prostate-specific lncRNA that is upregulated in high-grade PCa (Gleason score ≥7), metastatic disease, and castration-resistant PCa (CRPC) [51, 53]. With respect to prognostic value, PCAT1 has a favorable expression pattern compared with PCA3, as the latter is over-expressed in almost all primary tumors and to a lesser extent in CRPC and metastatic lesions. Recently it has been described that PCAT1 regulates the DNA repair gene BRCA2 and thereby controls homologous recombination in vitro and sensitivity to poly ADP ribose polymerase (PARP)-1 inhibitors in vivo [53, 54]. Hence, PCAT1 also represents a promising biomarker to predict response to PARP1 inhibition, a cancer treatment now tested in clinical trials. Nevertheless, molecular tests and clinical trials using PCAT1 as a biomarker are yet to be reported. SCHLAP1 (second chromosome locus associated with prostate-1, a.k.a. PCAT11) is overexpressed in 25% of PCa. SCHLAP1 is associated with risk of biochemical recurrence, clinical progression, and PCa-specific death [53, 55, 56]. Therefore, SCHLAP1 is a promising biomarker for aggressive and advanced PCa, and SCHLAP1 assays are in development [53]. PCAT14 was shown to be a strong prognostic marker and—like SCHLAP1—has the ability to predict biochemical recurrence, clinical progression to systemic disease, and PCa–specific mortality. Furthermore, in a multivariate analysis, PCAT14 expression also predicted resistance to androgen-deprivation therapy (ADT) (p = 0.012) [57, 58]. The recent study by Bottcher et al. [52] identified another set of over 300 known (including SCHLAP1) and novel PCa-associated transcripts (EPCATs). When combined into an lncRNA panel, 11 EPCATs classified 80% of PCa samples correctly, while maintaining 100% specificity. This high specificity was confirmed for EPCAT176 (SCHLAP1) and EPCAT966 by in situ hybridization on extensive tissue microarrays, clearly demonstrating that EPCATs are candidate diagnostic biomarkers of PCa [52].
MALAT1 (metastasis-associated lung adenocarcinoma transcript 1, a.k.a. NEAT2) is an lncRNA associated with many types of cancer, including PCa. In PCa, MALAT1 over-expression is associated with indicators of poor prognosis [59]. The use of a recently developed urinary MALAT1 score model would prevent about one-third of unnecessary biopsies without missing any high-grade cancers [60]. PCGEM1 (prostate-specific transcript 1 [non-protein coding], a.k.a. PCAT9) is over-expressed in about 60% of PCa cases [61]. No efforts to develop PCGEM1-based biomarkers have been reported. Interestingly, PCGEM1 levels were found to be higher in PCa cells from African-American men than in those from Caucasian-American men; the mortality rate of PCa is highest in the former [62]. Hence, PCGEM1 may contribute to an aggressive tumor phenotype.
Several other promising PCa-associated lncRNAs have been identified, including CDKN2B-AS1 (a.k.a. ANRIL), PCAT6, PCAT7, PCAT18, PCGEM1, PTENP1, and SPRY4-IT1 [63]. These RNAs show elevated expression levels in (a subset of advanced) prostate tumors, but further preclinical and clinical evaluation of these lncRNAs is necessary to determine their clinical utility (Table 3).
Table 3.
Overview of prostate cancer-associated non-coding RNAs with (potential) clinical application
| ncRNA (HGNC symbol) | Alias(es) | Function | (Potential) application | Available test (company) | References |
|---|---|---|---|---|---|
| lncRNAs | |||||
| EPCAT176, 190, 234, 273, 522, 524, 602, 633, 709, 850, 966 | – | Enhance cell viability and function unknown | Diagnosis (EPCAT panel, n = 11); prognostic potential novel EPCATs unknown | [52] | |
| MALAT1 |
NEAT2,
LINC00047 |
Unclear; interacts with EZH2 | Diagnosis: prediction biopsy outcome. Prognosis: prediction biopsy GS, LN metastasis. | [59, 60] | |
| PCA3 | DD3, PCAT3 | Enhances cell viability; interacts with ADAR and downregulates PRUNE2 | Diagnosis: prediction (repeat) biopsy outcome | Progensa PCA3 (Hologic) | [9, 33–49] |
| PCAT1 | PCA1 | Promotes cell proliferation through MYC (inhibits BRCA2) | Prognosis: prediction – advanced disease. Theranostics: selection of patients for e.g. PARP inhibitors | [51, 53, 54] | |
| PCAT14 | – | Unknown | Diagnosis (combined with other markers); theranostic: prediction of ADT response | [57, 58] | |
| PCGEM1 |
PCAT9,
LINC00071 |
Inhibits apoptosis, promotes cell proliferation; interacts with activated AR | Role in CRPC? | [61, 62] | |
| SCHLAP1 |
PCAT11,
PCAT114, LINC00913 |
Promotes invasion and metastasis; interacts with SWI/SNF complex | Prognosis: prediction of BCR, metastasis and death from PCa (after primary radical treatment) | CLIA-certified test available | [53, 55] |
| Circulating miRNAs | |||||
| MIRLET7 MIR19A MIR19B MIR21 MIR200B MIR375 isomiRs of MIR21, MIR375 and MIR204 |
let-7 hsa-mir-19a hsa-mir-19b hsa-mir-21 hsa-mir-200b hsa-mir-375 |
Diagnosis; prediction of presence of cancer in blood and urine samples | [71, 73, 80, 161–164] | ||
| MIR141 MIR145 MIR200B MIR375 |
hsa-mir-141 hsa-mir-145 hsa-mir-200b hsa-mir-375 |
Prognosis; prediction of Gleason and/or metastasis in blood samples | [73, 161, 163, 165] | ||
ADT androgen-deprivation therapy, AR androgen receptor, BCR biochemical recurrence, CLIA Clinical Laboratory Improvement Amendment, CRPC castration-resistant prostate cancer, GS Gleason score, HGNC HUGO Gene Nomenclature Committee, LN lymph node, lncRNA long non-coding RNA, miRNA microRNA, ncRNA non-coding RNA, PARP poly ADP ribose polymerase, PCa prostate cancer, SWI/SNF switch/sucrose non-fermentable
MicroRNAs (miRNAs) and isoMiRs
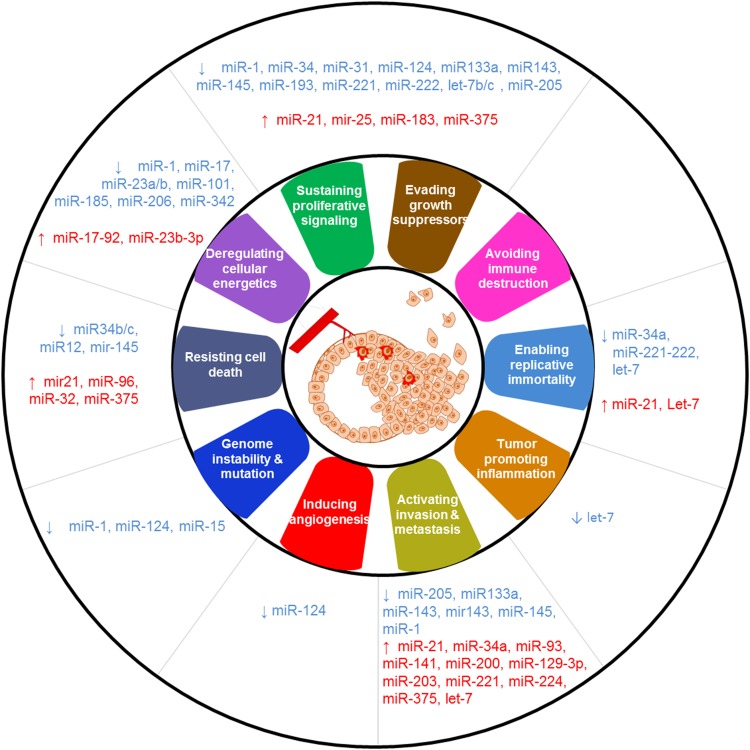
It is well established that the pattern of miRNA expression is highly tissue-type specific [64, 65]. Several miRNAs have been described for their relation and biomarker potential in PCa, at the level of diagnosis, prognosis, and therapy-response monitoring [66–69] (reviewed by Fabris et al. [70]). A main challenge in this process is the heterogeneous growth of cancer lesions throughout the prostate. In addition, differences in study designs between research groups, such as different profiling platforms and (often limited) sample sizes, can cause inconsistencies in the reported miRNAs. Nevertheless, a consensus list has emerged of miRNAs commonly deregulated in PCa. For example, deregulation of target genes of the Let-7 family influences cellular equilibrium in the prostate gland and promotes cancer development in PCa and several other cancers [71]; miR-25 regulates integrin expression, thereby preventing inhibition of cell growth and inducing metastasis [65, 66, 69, 72]; and miR-21 has a role in early PCa development and has been found upregulated in both tissue and blood samples as a circulating miRNA [73]. Many other miRNAs have been implicated in different aspects of PCa initiation, development, and progression (Fig. 1).
Fig. 1.
Many dysregulated microRNAs (miRNAs) affect the hallmarks of prostate cancer [70, 140, 158–160]
The development of next-generation sequencing (NGS) technologies for (small) ncRNAs has enabled the identification of novel miRNAs. Typically, a miRNA is annotated as one defined sequence. However, multiple length and/or sequence variants have been identified for many miRNAs via NGS [74]. Such variants are called isomiRs and are frequently found in most (if not all) tissue types. IsomiRs have biological function, for example, they can suppress mRNAs, though the modifications can also lead to binding of different mRNA targets or opposite strand selections because of changes in thermodynamic stability. The heterogeneity of a miRNA sequence length can arise from imprecise processing by endoribonucleases Dicer or Drosha [75]; by enzymatic post-transcriptional modifications, for example, by exoribonucleases (Nibbler and QUP); or by terminal uridylyl transferases (TUTs) and poly(A) polymerases (PAPs) [76]. Variations in miRNA sequences can be introduced via post-transcriptional editing by enzymes such as the ADAR proteins [77]. In breast cancer, isomiRs were found to be differentially expressed between healthy and cancer tissues, and were able to discriminate between different breast cancer subtypes [78, 79]. IsomiRs are not randomly distributed within tissues but are expressed in patterns that are more complex than initially thought [78]. IsomiRs can also be detected in both blood and urine [80]. In urinary extracellular vesicles (EVs) from patients with PCa, many isomiRs are differentially expressed between cancer samples and age-matched controls. Importantly, in accordance with findings in breast cancer, prostate tumors that exhibit deregulated expression of an miRNA simultaneously exhibit deregulated expression of isomiRs, which are derived from the same miRNA precursor. This has been demonstrated in PCa EVs isolated from urine, where isomiRs of miR-21, miR-375, and miR-204 are highly differentially expressed [80]. The actual biomarker potential of miRNAs and their isomiRs needs validation in larger clinical trials.
Other small RNAs as Potential Biomarkers of Disease
Besides the identification of isomiRs, ncRNA NGS technologies revealed the existence of many additional RNA fragments that are derived from sncRNAs. Although initially discarded as being RNA turnover artefacts, accumulating evidence suggests that smaller RNAs derived from snoRNA and tRNA are not just random degradation products but instead stable entities that may have functional activity in the normal cell and are deregulated in cancer [28]. In PCa, the small RNA transcriptome is dominated by sncRNAs other than miRNAs [69, 81]. Many of these sncRNAs can be classified as sdRNAs and tRFs. NGS data analysis on radical prostatectomy specimens demonstrates that sdRNAs and tRFs are upregulated in malignant tissue compared with normal adjacent prostate or benign prostate hyperplasia [69, 81, 82]. Interestingly, the expression of specific snoRNAs and sdRNAs (e.g., SNORD78 and the sdRNA derived from it, sd78) is already increased at the time of radical prostatectomy in subsets of patients with PCa who develop aggressive metastatic cancer years after surgery. This suggests that such sncRNAs have unique potential as a prognostic marker for aggressive metastatic PCa [81]. Besides snoRNA and sdRNA, tRFs can also be associated with high-grade, recurrent disease. For example, the expression levels of tRFs with opposing expression patterns derived from tRNAPheGAA and tRNALysCTT have been used to calculate an expression ratio that correctly discriminates high- from low-grade PCa and significantly associates with a shorter period to disease relapse in different cohorts [82]. The prognostic marker potential of sdRNAs or tRFs has been reported in not only PCa but also breast cancer [83–85], colorectal cancer [86], and lung cancer [87–89].
Pros and Cons of lncRNAs and sncRNAs in Their Utilization as Clinical Biomarkers
The detection of ncRNAs has the potential to improve the diagnosis and prognosis of PCa. Disease- and cell-type-specific expression, and the availability of relatively easy, sensitive, and quantitative detection methods for RNA make them extremely suitable cancer biomarkers. lncRNAs generally exhibit a major disease- and cell-type specificity, although a detailed expression analysis for each disease-associated lncRNA is necessary to rule out expression in non-cancerous conditions or other pathologies. The importance of the latter is demonstrated by the cancer-associated MALAT1 lncRNA, which also appears to be deregulated in benign conditions, such as cardiovascular diseases [90]. The disadvantage of lncRNAs is that they generally exhibit low to moderate levels of expression, which can have a negative impact on sensitive detection in clinical samples. Despite this potential pitfall, it has been proven that the PCA3 lncRNA, which is moderately expressed, can be detected robustly and sensitively even in <1 ml of post-DRE urine samples [39]. Many sncRNAs are expressed at higher levels than lncRNAs. Because of their size, sncRNAs are generally well protected from the action of native and contaminating nucleases, which ensures great stability even under different storage conditions. Nevertheless, often the specific function of different sncRNAs can affect tissue and cell-type specificity. In addition, the design of specific primers and oligonucleotide probes for amplification and detection of sncRNAs can be challenging for a number of reasons: their short length, which offers little room for design variation; their homology with other closely related sncRNA family members; and the expression of different isoforms (often differing by only one nucleotide residue). Nevertheless, new techniques under development (discussed in the following sections) address this challenge and promise increased sensitivity and specificity of detection. Whether the many natural post-transcriptional modifications made to sncRNAs significantly affect detection methods for RNA quantification is another field yet to be investigated.
Perspectives in ncRNA Detection and Monitoring
Liquid Biopsy
Definitive diagnosis of PCa currently depends on the histological verification of adenocarcinoma in prostate biopsy cores or on unexpected discovery in transurethral resection (TUR) specimens [1]. The collection of tissue biopsies from the prostate is a highly invasive procedure that requires multiple sampling and carries the risk of tumor cells not being reached, leading to unnecessary follow-up biopsies and increasing the risk of infection. Therefore, most recently developed and currently emerging diagnostic tools for PCa aim at minimally invasive approaches using the so-called liquid biopsies. Both cell-free RNA (cfRNA) and the RNA content of circulating tumor cells (CTCs) in peripheral blood are being studied extensively for their usefulness as early prognostic biomarkers of aggressive disease or as biomarkers of therapeutic response in late-stage metastatic cancer [91–93], as recently reviewed by Hegemann et al. [94] and Miyamoto and Lee [95].
Blood collection is considered a minimally invasive procedure that provides samples enriched in a variety of disease-associated markers, including proteins (i.e., PSA), DNA fragments, and RNA. Differentially expressed ncRNAs in serum and plasma have been studied mostly for the class of miRNAs [70]. In blood, miRNAs remain highly stable because AGO2 complexes or EVs protect them from the action of nucleases [96]. Various circulating miRNAs have been associated with PCa, including miR-21, miR-141, and miR-221 [97]. Levels of miR-375 and miR-141 have also been found to be substantially increased during progression to CRPC [98]. Nonetheless, blood is a very complex biofluid that contains ncRNAs secreted by virtually all tissues in the body, which can introduce high background and complicate downstream analysis. However, blood remains the biofluid of choice for follow-up after primary treatment and for monitoring therapeutic response in advanced disease.
EVs—a large family of diverse vesicles consisting of exosomes, oncosomes, microvesicles, apoptotic bodies, and many other variants of these—are an additional promising source of cancer-related RNA biomarkers [99–101]. When derived from the prostate, such vesicles are sometimes referred to as prostasomes. EVs are released in the extracellular environment by a large number of cell types and can be found in practically any biological fluid. Since the molecular content of EVs resembles the normal or malignant cells from which they originate, they can be used as a rich source of protein and RNA biomarkers. For example, EVs isolated from the peripheral blood of patients with advanced-stage PCa contain the disease and therapy resistance-associated androgen receptor variant 7 (ARV7) transcript and can be used as a marker of therapy resistance [102]. It has also been recently suggested that the elevated levels of PCa EVs could themselves have diagnostic value [103–105].
Urine is a less complex biofluid than blood in that it can be collected non-invasively (Fig. 2). Urine contains exfoliated prostate cells, EVs, and factors secreted by prostate cells, including ncRNAs, which can be used for diagnostic and prognostic testing. Preferably, urine should be collected from the first void after DRE as this enriches the level of prostatic components [103]. However, because of its invasive nature, DRE is not always accepted by patients and clinicians, and the necessity of DRE for urinary biomarker detection is still under debate [106]. Given the invasive nature of DRE, urine collection for PCa diagnostic testing is considered to be a minimally invasive procedure when performed after DRE. EVs detected in urine are highly enriched for small RNAs, including miRNAs and tRNA fragments [80]. Multiple studies have investigated the use of urine as a source of biomarkers for PCa. Urine sediment is used for the detection of PCA3, whereas the supernatant after low-speed centrifugation is more attractive for measuring ncRNA biomarkers present in EVs. As handling and storage influence the EV populations (e.g., exosomes, apoptotic bodies, ectosomes, and microvesicles), and hence the total extravesicular ncRNA profile, it is important to use rapid standardized collection and storage procedures that preserve EV and RNA integrity and prevent bacterial contamination. In addition, the Tamm-Horsfall protein (THP) can interfere with the isolation of urinary EVs at low temperatures [107]. Dithiothreitol (DTT) will release captured EVs from formed THP complexes, but it should be used with caution as it will also release protein–RNA complexes that contain other (ratios of) ncRNAs [108].
Fig. 2.
Different strategies for sample collection in the diagnosis and monitoring of prostate cancer. Definitive prostate cancer diagnosis is made after the histopathological evaluation of multiple core biopsies. The detection of molecular markers in blood is considered a minimally invasive approach and can be used to monitor disease progression and treatment response, e.g., by measuring prostate-specific antigen (PSA) protein levels. Urine collection is a non-invasive approach and can be used for the prediction of biopsy outcome with a suspicion of prostate cancer, e.g., via PCA3 test. cfDNA cell-fee DNA, cfRNA cell-free RNA, CTCs circulating tumor cells, EVs extracellular vesicles, RNPs ribonucleoprotein complexes
Small RNAs can be isolated and analyzed from urine or urine EVs without discriminating between their source. However, a selective capture of prostate- or even PCa-derived EVs from urine or blood could provide the most optimal sensitive and specific detection. This would allow a more specific isolation of ncRNAs of interest without a bulk of background ncRNAs from other cells from the urinary tract or—for blood—the entire body. Extensive efforts in the development and optimization of assays for the isolation, quantification, and characterization of cancer- or organ-specific EVs from body fluids are ongoing [109–123]. EV RNA isolation protocols and novel approaches to the detection of RNA from EV samples are also continuously improving [124–128].
Seminal fluid and post-ejaculate urethral washings (PEUW) have been suggested as yet another rich source of PCa biomarkers [129, 130]. These body fluids are enriched in prostate components by nature. Nevertheless, limitations associated with the collection of these fluids makes clinical implementation more challenging.
Technological Developments in RNA Detection
With the identification of new ncRNAs for the diagnosis of PCa, detection methods with improved sensitivity and specificity are essential. NGS is considered one of the most specific and sensitive detection platforms for discovery and profiling of ncRNAs because the entire sequence and abundance of different isoforms are detected in a quantitative manner. Nevertheless, standard clinical diagnostic testing requires other detection methods that are more affordable and robust when only limited amounts of starting material are available. Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) is the most common method for the quantification of lncRNAs. In the case of miRNAs, unambiguous detection by conventional qPCR can be challenging because of their small size, the high sequence homology between miRNA family members, and the simultaneous presence of multiple isoforms. Poly(A) addition and locked nucleic acid (LNA) modified primer extensions are commonly used for the quantification of miRNAs and other sncRNAs. Stem-loop PCR can also be used for the detection of sncRNA and is particularly useful for the discrimination of specific small RNA isoforms [131]. The absolute quantification of low abundant miRNAs by qPCR remains challenging. Digital PCR has been developed for the detection of nucleic acids present at very low levels [132], and droplet digital PCR (ddPCR) has been successfully used with TaqMan and LNA-based miRNA assays to measure miRNA expression in plasma [133]. Several alternative techniques that do not rely on amplification have been proposed, but the majority are still in developmental stages pending extensive validation [134–137]. NanoString’s nCounter analysis system is a promising, this is a recently developed technology for the detection of mRNAs, lncRNAs, and miRNAs [138]. This technology uses digital color labeling and barcoding to simultaneously measure the expression of multiple genes. NanoString promises very high precision and sensitivity with possible utilization in clinical applications [139].
Future Potential of ncRNAs as Therapeutic Targets in PCa and Emerging Technologies in ncRNA Research
ncRNA appeared rapidly as a novel class of molecules that control and tune major cellular processes. The deregulation of multiple types of ncRNAs in cancer and other diseases, with miRNAs as a prominent example, demonstrates their potential as viable markers of disease and provides new opportunities for the exploration of novel targeted therapies and the development of new therapeutic tools. Over the last 20 years, the biology of miRNAs has been extensively studied and the insights gained have led to the development of multiple therapeutic strategies targeting miRNAs in cancer, some of which are currently in clinical trials (recently reviewed by Shah et al. [140]). In contrast, a lot more is yet to be learned about the function, structural organization, and mechanistic action of lncRNAs and many small RNAs. Nevertheless, technologies that target ncRNA and RNA in general are well on their way and have also entered clinical trials. Several different therapeutic approaches to targeting lncRNAs are under investigation. It has been demonstrated that siRNAs, commonly used for the knockdown of protein-coding transcripts can also successfully reduce lncRNA expression [141, 142] and sensitize cells to appoptosis [143, 144]. Two major limitations associated with the use of siRNAs lie in (1) their double-stranded nature, which requires additional (lipophilic) agents for efficient delivery and (2) their dependence on RISC, which limits the number of possible ncRNA targets to those with cytoplasmic localization [145]. An alternative strategy for the targeting of ncRNA uses single-stranded synthetic antisense oligonucleotides (AONs or ASOs). AONs are generally designed with a chemically modified sugar-phosphate backbone, which makes them highly stable in tissue and resistant to the action of the majority of cellular nucleases [146]. The addition of LNAs in their sequence provides increased specificity toward the targeted ncRNA. AONs can be effective tools that correct errors in nuclear transcript processing such as exon skipping or inclusion. Gapmers are yet another type of synthetic AON that can correct for RNA gain-of-function effects or reduce the levels of oncogenic transcripts. Gapmers are chemically similar to standard AONs, but their sequence contains a stretch (gap) of unmodified DNA nucleotides in the middle. After binding, the DNA “gap” in the middle of the oligo forms a DNA/RNA heteroduplex with the targeted RNA, which recruits RNase H and causes the degradation of the targeted transcript. Gapmers that selectively target and destroy mRNAs by the induction of RNase H are currently being evaluated in phase III trials for STAT3 in different cancers and in a phase II trial for wild-type AR and AR variants in treatment-resistant metastatic CRPC [147, 148].
Cellular responses inflicted by the overexpression or knock-down/knock-out of specific ncRNAs do provide insight into their functional role. Ideally, these types of experiments would be reinforced by direct observation of the targeted RNA in its native, tissue, and cellular context. In cell biology, (fluorescent) imaging has been very fruitful in unraveling the function and action mechanism of proteins. Likewise, visualization technologies that could be used in functional analysis of ncRNAs are being developed. The detection of small RNAs in tissues is usually problematic because of their short size. Technologies that utilize probe oligonucleotides with LNA (LNA oligos) can overcome this by introducing increased stability of the RNA–LNA complex [149–152]. lncRNAs and mRNAs can be visualized in situ via a similar technique (RNAscope), which relies on the design of multiple probes and the major amplification of ISH signal for the detection and (quantitative) subcellular localization in cells and tissues [52, 153]. Both techniques can be applied on fresh-frozen or formalin-fixed, paraffin-embedded material but are largely dependent on the quality of fixation and sensitive to RNA degradation in archive samples. Visualization of RNA molecules in living cells would provide additional information on their spatio-temporal organization and mechanism of action. A pioneering and rapidly developing approach in this field resembles green fluorescent protein (GFP)-based imaging technology, where the protein of interest is tagged with GFP [154]. The RNA-labeling technique (termed Spinach or its improved version Broccoli) relies on an aptamer RNA structure, fused to the (nc)RNA of interest. When properly folded, the aptamer binds a green fluorophore, resulting in a fluorescently labelled RNA molecule, which can be monitored in living cells [154, 155]. This technique has been successfully used to demonstrate the translocation of 5S ribosomal RNA (rRNA) upon the induction of stress in eukaryotic cells; following additional development and optimization, the potential for use with ncRNAs is strong.
Conclusions and Perspectives
There is an urgent need for more accurate non-invasive tests for PCa diagnosis and to allow the stratification of patients with life-threatening disease. Because of the ease of collection, and the fact that prostate cells are directly released into the urethra through prostatic ducts, urine has become the first choice for diagnostic and prognostic non-invasive biomarker testing. Blood-based tests will be needed to monitor therapy response. Several biomarkers are promising because of their specificity for the disease in tissue; however, so far, only the PCA3 lncRNA is used as a urinary biomarker for PCa diagnosis in clinical practice.
This review highlights the tissue- and lineage-specific expression of several sncRNAs and lncRNAs, and discusses the potential of these ncRNAs for PCa diagnosis, prognosis, or disease monitoring. Now, validation of ncRNA biomarkers in large-scale multicenter studies using standardized quantification methods are needed to confirm their accuracy in detecting the stage of disease. In addition to well-designed validation trials, collaboration with industrial partners will be essential for successful development and positioning of any new test in the market [156].
Before an ncRNA-biomarker test can be used in clinic, several issues need to be considered. Some of the most important factors are the standardization of biofluid collection and storage, as well as consensus on adequate and standardized subsequent EV and ncRNA extraction procedures. Furthermore, normalization of ncRNA expression, especially in biofluids, remains a challenge. One way to overcome this problem is to use RNA expression ratios [11].
Validation of the large number of molecular markers that have been discovered and comparing their performance against the paltry number that have made it to clinical application is one part of the solution. Combining multiple biomarkers and ‘classical’ clinicopathological parameters is another. Markers need to be integrated into prediction models, preferably using marker expression levels as a continuous variable. Finally, the clinical consequences of using such models in clinical practice need to be evaluated [157], as exemplified by the SelectMDx test that can predict biopsy outcome [16].
Several questions remain to be addressed when investigating ncRNAs as targets for therapy. The overwhelming amount of newly discovered ncRNAs warrants the establishment of criteria to help narrow down the best target RNAs from a repertoire of tens of thousands. A better understanding of the domain organization and tertiary structure of lncRNAs is necessary to gain insight into the mechanistic basis of their function and overcome current obstacles and controversies in ncRNA research. The low conservation of large numbers of lncRNAs demands the development of new model systems that can assist the translation of ncRNA discoveries into therapies and diagnostic tools. Finally, further investigations, large validation studies, and industrial partnerships are necessary for the successful translation of ncRNA research into clinical practice.
Compliance with Ethical Standards
Conflicts of interest
IVB, MER, and ESM declare no conflict of interest. GWV is an inventor on the PCA3-related IP. The IP is owned by his employer, Radboud university medical center, which has licensed the technology and receives royalty payments.
Funding
This review was made possible through the collaboration of the authors in the “IMMPROVE” consortium (Innovative Measurements and Markers for Prostate Cancer Diagnosis and Prognosis using Extracellular Vesicles), which is sponsored by an Alpe d’HuZes grant of the Dutch Cancer Society (grant #EMCR2015-8022). Open access was funded as part of the COMPACT agreement between Springer and the Association of Dutch Universities and Academy Institutes.
Footnotes
All authors contributed equally.
References
- 1.Mottet N, Bellmunt J, Briers E, et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Arnhem: European Association of Urology; 2016. Updated March 2016. https://uroweb.org/guideline/prostate-cancer/. Accessed 29 Nov 2016.
- 2.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520(7547):353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maughan BL, Antonarakis ES. Enzalutamide in chemo-naive castration-resistant prostate cancer: effective for most but not for all. Asian J Androl. 2014;16(6):807–808. doi: 10.4103/1008-682X.137680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, van der Kwast T, et al. EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Update 2013. Eur Urol. 2014;65(1):124–137. doi: 10.1016/j.eururo.2013.09.046. [DOI] [PubMed] [Google Scholar]
- 6.Ritch CR, Cookson MS. Advances in the management of castration resistant prostate cancer. BMJ. 2016;355:i4405. doi: 10.1136/bmj.i4405. [DOI] [PubMed] [Google Scholar]
- 7.Hessels D, Smit FP, Verhaegh GW, Witjes JA, Cornel EB, Schalken JA. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin Cancer Res. 2007;13(17):5103–5108. doi: 10.1158/1078-0432.CCR-07-0700. [DOI] [PubMed] [Google Scholar]
- 8.Irshad S, Bansal M, Castillo-Martin M, Zheng T, Aytes A, Wenske S, et al. A molecular signature predictive of indolent prostate cancer. Sci Trans Med. 2013;5(202):202ra122. [DOI] [PMC free article] [PubMed]
- 9.van Gils MP, Hessels D, van Hooij O, Jannink SA, Peelen WP, Hanssen SL, et al. The time-resolved fluorescence-based PCA3 test on urinary sediments after digital rectal examination; a Dutch multicenter validation of the diagnostic performance. Clin Cancer Res. 2007;13(3):939–943. doi: 10.1158/1078-0432.CCR-06-2679. [DOI] [PubMed] [Google Scholar]
- 10.Velonas VM, Woo HH, dos Remedios CG, Assinder SJ. Current status of biomarkers for prostate cancer. Int J Mol Sci. 2013;14(6):11034–11060. doi: 10.3390/ijms140611034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sartori DA, Chan DW. Biomarkers in prostate cancer: what’s new? Curr Opin Oncol. 2014;26(3):259–264. doi: 10.1097/CCO.0000000000000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PloS One. 2013;8(6):e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knezevic D, Goddard AD, Natraj N, Cherbavaz DB, Clark-Langone KM, Snable J, et al. Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics. 2013;14. [DOI] [PMC free article] [PubMed]
- 14.Crawford ED, Scholz MC, Kar AJ, Fegan JE, Haregewoin A, Kaldate RR, et al. Cell cycle progression score and treatment decisions in prostate cancer: results from an ongoing registry. Curr Med Res Opin. 2014;30(6):1025–1031. doi: 10.1185/03007995.2014.899208. [DOI] [PubMed] [Google Scholar]
- 15.Leyten GH, Hessels D, Smit FP, Jannink SA, de Jong H, Melchers WJ, et al. Identification of a Candidate Gene Panel for the Early Diagnosis of Prostate Cancer. Clin Cancer Res. 2015;21(13):3061–3070. doi: 10.1158/1078-0432.CCR-14-3334. [DOI] [PubMed] [Google Scholar]
- 16.Van Neste L, Hendriks RJ, Dijkstra S, Trooskens G, Cornel EB, Jannink SA, et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker-Based Risk Score. Eur Urol. 2016. [DOI] [PubMed]
- 17.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec No 1:R17-29. [DOI] [PubMed]
- 18.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 20.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42 Database issue:D68-73. [DOI] [PMC free article] [PubMed]
- 21.Dallaire A, Simard MJ. The implication of microRNAs and endo-siRNAs in animal germline and early development. Dev Biol. 2016;416(1):18–25. doi: 10.1016/j.ydbio.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Zhang P, Si X, Skogerbo G, Wang J, Cui D, Li Y, et al. piRBase: a web resource assisting piRNA functional study. Database (Oxford). 2014;2014:bau110. [DOI] [PMC free article] [PubMed]
- 23.Moyano M, Stefani G. piRNA involvement in genome stability and human cancer. J Hematol Oncol. 2015;8:38. doi: 10.1186/s13045-015-0133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorjani H, Kehr S, Jedlinski DJ, Gumienny R, Hertel J, Stadler PF, et al. An updated human snoRNAome. Nucleic Acids Res. 2016;44(11):5068–5082. doi: 10.1093/nar/gkw386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15(7):1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23(22):2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupuis-Sandoval F, Poirier M, Scott MS. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA. 2015;6(4):381–397. doi: 10.1002/wrna.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA - Novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013;340(2):201–211. doi: 10.1016/j.canlet.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 29.Kumar P, Kuscu C, Dutta A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs) Trends Biochem Sci. 2016;41(8):679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lagarde J, Uszczynska-Ratajczak B, Santoyo-Lopez J, Gonzalez JM, Tapanari E, Mudge JM, et al. Extension of human lncRNA transcripts by RACE coupled with long-read high-throughput sequencing (RACE-Seq) Nat Commun. 2016;7:12339. doi: 10.1038/ncomms12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59(23):5975–5979. [PubMed] [Google Scholar]
- 34.de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62(9):2695–2698. [PubMed] [Google Scholar]
- 35.Popa I, Fradet Y, Beaudry G, Hovington H, Beaudry G, Tetu B. Identification of PCA3 (DD3) in prostatic carcinoma by in situ hybridization. Mod Pathol. 2007;20(11):1121–1127. doi: 10.1038/modpathol.3800963. [DOI] [PubMed] [Google Scholar]
- 36.Salameh A, Lee AK, Cardo-Vila M, Nunes DN, Efstathiou E, Staquicini FI, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci U S Am. 2015;112(27):8403–8408. doi: 10.1073/pnas.1507882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salagierski M, Verhaegh GW, Jannink SA, Smit FP, Hessels D, Schalken JA. Differential expression of PCA3 and its overlapping PRUNE2 transcript in prostate cancer. Prostate. 2010;70(1):70–78. doi: 10.1002/pros.21040. [DOI] [PubMed] [Google Scholar]
- 38.Ferreira LB, Palumbo A, de Mello KD, Sternberg C, Caetano MS, de Oliveira FL, et al. PCA3 noncoding RNA is involved in the control of prostate-cancer cell survival and modulates androgen receptor signaling. BMC Cancer. 2012;12:507. doi: 10.1186/1471-2407-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Groskopf J, Aubin SM, Deras IL, Blase A, Bodrug S, Clark C, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006;52(6):1089–1095. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 40.Hessels D, Klein Gunnewiek JM, van Oort I, Karthaus HF, van Leenders GJ, van Balken B, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44(1):8–15. (discussion -6). [DOI] [PubMed]
- 41.Haese A, de la Taille A, van Poppel H, Marberger M, Stenzl A, Mulders PF, et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur Urol. 2008;54(5):1081–1088. doi: 10.1016/j.eururo.2008.06.071. [DOI] [PubMed] [Google Scholar]
- 42.Marks LS, Fradet Y, Deras IL, Blase A, Mathis J, Aubin SM, et al. PCA3 molecular urine assay for prostate cancer in men undergoing repeat biopsy. Urology. 2007;69(3):532–535. doi: 10.1016/j.urology.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 43.Aubin SM, Reid J, Sarno MJ, Blase A, Aussie J, Rittenhouse H, et al. PCA3 molecular urine test for predicting repeat prostate biopsy outcome in populations at risk: validation in the placebo arm of the dutasteride REDUCE trial. J Urol. 2010;184(5):1947–1952. doi: 10.1016/j.juro.2010.06.098. [DOI] [PubMed] [Google Scholar]
- 44.Deras IL, Aubin SM, Blase A, Day JR, Koo S, Partin AW, et al. PCA3: a molecular urine assay for predicting prostate biopsy outcome. J Urol. 2008;179(4):1587–1592. doi: 10.1016/j.juro.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 45.Wei JT, Feng Z, Partin AW, Brown E, Thompson I, Sokoll L, et al. Can urinary PCA3 supplement PSA in the early detection of prostate cancer? J Clin Oncol. 2014;32(36):4066–4072. doi: 10.1200/JCO.2013.52.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakanishi H, Groskopf J, Fritsche HA, Bhadkamkar V, Blase A, Kumar SV, et al. PCA3 molecular urine assay correlates with prostate cancer tumor volume: implication in selecting candidates for active surveillance. J Urol. 2008;179(5):1804–1809. doi: 10.1016/j.juro.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Ploussard G, Durand X, Xylinas E, Moutereau S, Radulescu C, Forgue A, et al. Prostate cancer antigen 3 score accurately predicts tumour volume and might help in selecting prostate cancer patients for active surveillance. Eur Urol. 2011;59(3):422–429. doi: 10.1016/j.eururo.2010.11.044. [DOI] [PubMed] [Google Scholar]
- 48.Whitman EJ, Groskopf J, Ali A, Chen Y, Blase A, Furusato B, et al. PCA3 score before radical prostatectomy predicts extracapsular extension and tumor volume. J Urol. 2008;180(5):1975–1978. doi: 10.1016/j.juro.2008.07.060. [DOI] [PubMed] [Google Scholar]
- 49.van Poppel H, Haese A, Graefen M, de la Taille A, Irani J, de Reijke T, et al. The relationship between Prostate CAncer gene 3 (PCA3) and prostate cancer significance. BJU Int. 2012;109(3):360–366. doi: 10.1111/j.1464-410X.2011.10377.x. [DOI] [PubMed] [Google Scholar]
- 50.Hessels D, Schalken JA. Urinary biomarkers for prostate cancer: a review. Asian J Androl. 2013;15(3):333–339. doi: 10.1038/aja.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bottcher R, Hoogland AM, Dits N, Verhoef EI, Kweldam C, Waranecki P, et al. Novel long non-coding RNAs are specific diagnostic and prognostic markers for prostate cancer. Oncotarget. 2015;6(6):4036–4050. doi: 10.18632/oncotarget.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM, et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15(13):1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74(6):1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehra R, Udager AM, Ahearn TU, Cao X, Feng FY, Loda M, et al. Overexpression of the long non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur Urol. 2016;70(4):549–52. doi: 10.1016/j.eururo.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45(11):1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malik R, Zhang X, Shukla S, Niknafs YY, Zhao S, Feng FY, et al. Long noncoding RNA PCAT14/PRCAT104; A prognostic biomarker in prostate cancer. Cancer Res. 2016;76(6 Supplement):B39. doi: 10.1158/1538-7445.NONRNA15-B39. [DOI] [Google Scholar]
- 58.Shukla S, Zhang X, Niknafs YS, Xiao L, Mehra R, Cieslik M, et al. Identification and Validation of PCAT14 as Prognostic Biomarker in Prostate Cancer. Neoplasia. 2016;18(8):489–499. doi: 10.1016/j.neo.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013;190(6):2278–2287. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Wang F, Ren S, Chen R, Lu J, Shi X, Zhu Y, et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget. 2014;5(22):11091–11102. doi: 10.18632/oncotarget.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci USA. 2000;97(22):12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L, et al. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23(2):605–611. doi: 10.1038/sj.onc.1207069. [DOI] [PubMed] [Google Scholar]
- 63.Mouraviev V, Lee B, Patel V, Albala D, Johansen TE, Partin A, et al. Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis. 2016;19(1):14–20. doi: 10.1038/pcan.2015.48. [DOI] [PubMed] [Google Scholar]
- 64.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 65.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68(15):6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27(12):1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 68.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 69.Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Moller S, Trapman J, et al. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31(8):978–991. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 70.Fabris L, Ceder Y, Chinnaiyan AM, Jenster GW, Sorensen KD, Tomlins S, et al. The potential of MicroRNAs as prostate cancer biomarkers. Eur Urol. 2016;70(2):312–322. doi: 10.1016/j.eururo.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wagner S, Ngezahayo A, Murua Escobar H, Nolte I. Role of miRNA let-7 and its major targets in prostate cancer. Biomed Res Int. 2014;2014:376326. doi: 10.1155/2014/376326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zoni E, van der Horst G, van de Merbel AF, Chen L, Rane JK, Pelger RC, et al. miR-25 modulates invasiveness and dissemination of human prostate cancer cells via regulation of αv- and α6-integrin expression. Cancer Res. 2015;75(11):2326–2336. doi: 10.1158/0008-5472.CAN-14-2155. [DOI] [PubMed] [Google Scholar]
- 73.Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106(4):768–774. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 76.Boele J, Persson H, Shin JW, Ishizu Y, Newie IS, Sokilde R, et al. PAPD5-mediated 3’ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc Natl Acad Sci USA. 2014;111(31):11467–11472. doi: 10.1073/pnas.1317751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Telonis AG, Loher P, Jing Y, Londin E, Rigoutsos I. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 2015;43(19):9158–9175. doi: 10.1093/nar/gkv922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu X, Zeng R, Wu S, Zhong J, Yang L, Xu J. Comprehensive expression analysis of miRNA in breast cancer at the miRNA and isomiR levels. Gene. 2015;557(2):195–200. doi: 10.1016/j.gene.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 80.Koppers-Lalic D, Hackenberg M, de Menezes R, Misovic B, Wachalska M, Geldof A, et al. Noninvasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget. 2016;7(16):22566–22578. doi: 10.18632/oncotarget.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martens-Uzunova ES, Hoogstrate Y, Kalsbeek A, Pigmans B, Vredenbregt-van den Berg M, Dits N, et al. C/D-box snoRNA-derived RNA production is associated with malignant transformation and metastatic progression in prostate cancer. Oncotarget. 2015;6(19):17430–17444. doi: 10.18632/oncotarget.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Olvedy M, Scaravilli M, Hoogstrate Y, Visakorpi T, Jenster G, Martens-Uzunova ES. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget. 2016;7(17):24766–24777. doi: 10.18632/oncotarget.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krishnan P, Ghosh S, Wang B, Heyns M, Graham K, Mackey JR, et al. Profiling of small nucleolar RNAs by next generation sequencing: potential new players for breast cancer prognosis. PLoS One. 2016;11(9):e0162622. doi: 10.1371/journal.pone.0162622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krishnan P, Ghosh S, Wang B, Heyns M, Li D, Mackey JR, et al. Genome-wide profiling of transfer RNAs and their role as novel prognostic markers for breast cancer. Sci Rep. 2016;6:32843. doi: 10.1038/srep32843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37(21):7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okugawa Y, Toiyama Y, Toden S, Mitoma H, Nagasaka T, Tanaka K, et al. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut. 2017;66(1):107–17. doi: 10.1136/gutjnl-2015-309359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liao J, Yu L, Mei Y, Guarnera M, Shen J, Li R, et al. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer. 2010;9:198. doi: 10.1186/1476-4598-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mannoor K, Shen J, Liao J, Liu Z, Jiang F. Small nucleolar RNA signatures of lung tumor-initiating cells. Mol Cancer. 2014;13:104. doi: 10.1186/1476-4598-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gao L, Ma J, Mannoor K, Guarnera MA, Shetty A, Zhan M, et al. Genome-wide small nucleolar RNA expression analysis of lung cancer by next-generation deep sequencing. Int J Cancer. 2015;136(6):E623–E629. doi: 10.1002/ijc.29169. [DOI] [PubMed] [Google Scholar]
- 90.Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol. 2016;12(6):360–373. doi: 10.1038/nrneph.2016.51. [DOI] [PubMed] [Google Scholar]
- 91.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 92.Jiang R, Lu Y-T, Ho H, Li B, Chen J-F, Lin M, et al. A comparison of isolated circulating tumor cells and tissue biopsies using whole-genome sequencing in prostate cancer. Oncotarget. 2015;6(42):44781–44793. doi: 10.18632/oncotarget.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meyer CP, Pantel K, Tennstedt P, Stroelin P, Schlomm T, Heinzer H, et al. Limited prognostic value of preoperative circulating tumor cells for early biochemical recurrence in patients with localized prostate cancer. Urol Oncol. 2016;34(5):235.e11–235.e16. doi: 10.1016/j.urolonc.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 94.Hegemann M, Stenzl A, Bedke J, Chi KN, Black PC, Todenhöfer T. Liquid biopsy: ready to guide therapy in advanced prostate cancer? BJU Int. 2016;118(6):855–863. doi: 10.1111/bju.13586. [DOI] [PubMed] [Google Scholar]
- 95.Miyamoto DT, Lee RJ. Cell-free and circulating tumor cell–based biomarkers in men with metastatic prostate cancer: Tools for real-time precision medicine? Urol Oncol. 2016;34(11):490–501. doi: 10.1016/j.urolonc.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 96.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yaman Agaoglu F, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N, et al. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biol. 2011;32(3):583–588. doi: 10.1007/s13277-011-0154-9. [DOI] [PubMed] [Google Scholar]
- 98.Nguyen HC, Xie W, Yang M, Hsieh CL, Drouin S, Lee GS, et al. Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate. 2013;73(4):346–354. doi: 10.1002/pros.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Duijvesz D, Luider T, Bangma CH, Jenster G. Exosomes as biomarker treasure chests for prostate cancer. Eur Urol. 2011;59(5):823–831. doi: 10.1016/j.eururo.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 100.Crescitelli R, Lässer C, Szabó TG, Kittel A, Eldh M, Dianzani I, et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. 2013 doi: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Del Re M, Biasco E, Crucitta S, Derosa L, Rofi E, Orlandini C, et al. The detection of androgen receptor splice variant 7 in plasma-derived Exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur Urol. 2016 doi: 10.1016/j.eururo.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 103.Duijvesz D, Versluis CY, van der Fels CA, Vredenbregt-van den Berg MS, Leivo J, Peltola MT, et al. Immuno-based detection of extracellular vesicles in urine as diagnostic marker for prostate cancer. Int J Cancer. 2015;137(12):2869–2878. doi: 10.1002/ijc.29664. [DOI] [PubMed] [Google Scholar]
- 104.Park YH, Shin HW, Jung AR, Kwon OS, Choi Y-J, Park J, et al. Prostate-specific extracellular vesicles as a novel biomarker in human prostate cancer. Sci Rep. 2016;6:30386. doi: 10.1038/srep30386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cappello F, Logozzi M, Campanella C, Bavisotto CC, Marcilla A, Properzi F, et al. Exosome levels in human body fluids: a tumor marker by themselves? Eur J Pharm Sci. 2017;96:93–98. doi: 10.1016/j.ejps.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 106.Donovan MJ, Noerholm M, Bentink S, Belzer S, Skog J, O’Neill V, et al. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis. 2015;18(4):370–375. doi: 10.1038/pcan.2015.40. [DOI] [PubMed] [Google Scholar]
- 107.Kosanovic M, Jankovic M. Isolation of urinary extracellular vesicles from Tamm- Horsfall protein-depleted urine and their application in the development of a lectin-exosome-binding assay. Biotechniques. 2014;57(3):143–149. doi: 10.2144/000114208. [DOI] [PubMed] [Google Scholar]
- 108.Wachalska MK-LD, van Eijndhoven M, Pegtel DM, Geldof AA, Lipinska A, van Moorselaar RJ, Bijnsdorp IV. Protein complexes in urine interfere with extracellular vesicle biomarker studies. J Circulat Biomark. 2016;5:4. doi: 10.5772/62579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Royo F, Zuniga-Garcia P, Sanchez-Mosquera P, Egia A, Perez A, Loizaga A, et al. Different EV enrichment methods suitable for clinical settings yield different subpopulations of urinary extracellular vesicles from human samples. J Extracell Vesicles. 2016;5:29497. doi: 10.3402/jev.v5.29497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oliveira-Rodriguez M, Serrano-Pertierra E, Garcia AC, Lopez-Martin S, Yanez-Mo M, Cernuda-Morollon E, et al. Point-of-care detection of extracellular vesicles: Sensitivity optimization and multiple-target detection. Biosens Bioelectron. 2016;87:38–45. doi: 10.1016/j.bios.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 111.Lof L, Ebai T, Dubois L, Wik L, Ronquist KG, Nolander O, et al. Detecting individual extracellular vesicles using a multicolor in situ proximity ligation assay with flow cytometric readout. Sci Rep. 2016;6:34358. doi: 10.1038/srep34358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Erdbrugger U, Lannigan J. Analytical challenges of extracellular vesicle detection: a comparison of different techniques. Cytometry Part A. 2016;89(2):123–134. doi: 10.1002/cyto.a.22795. [DOI] [PubMed] [Google Scholar]
- 113.Taller D, Richards K, Slouka Z, Senapati S, Hill R, Go DB, et al. On-chip surface acoustic wave lysis and ion-exchange nanomembrane detection of exosomal RNA for pancreatic cancer study and diagnosis. Lab Chip. 2015;15(7):1656–1666. doi: 10.1039/C5LC00036J. [DOI] [PubMed] [Google Scholar]
- 114.Yoshioka Y, Kosaka N, Konishi Y, Ohta H, Okamoto H, Sonoda H, et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun. 2014;5:3591. doi: 10.1038/ncomms4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol. 2014;32(5):490–495. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Erdbrugger U, Rudy CK, Etter ME, Dryden KA, Yeager M, Klibanov AL, et al. Imaging flow cytometry elucidates limitations of microparticle analysis by conventional flow cytometry. Cytometry Part A. 2014;85(9):756–770. doi: 10.1002/cyto.a.22494. [DOI] [PubMed] [Google Scholar]
- 117.Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014;14(11):1891–1900. doi: 10.1039/C4LC00136B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maas SL, de Vrij J, van der Vlist EJ, Geragousian B, van Bloois L, Mastrobattista E, et al. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. J Control Rel. 2015;200:87–96. doi: 10.1016/j.jconrel.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Maas SLN, De Vrij J, Broekman MLD. Quantification and size-profiling of extracellular vesicles using tunable resistive pulse sensing. J Vis Exp. 2014;92:e51623. doi: 10.3791/51623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mehdiani A, Maier A, Pinto A, Barth M, Akhyari P, Lichtenberg A. An innovative method for exosome quantification and size measurement. J Vis Exp. 2015;95:50974. doi: 10.3791/50974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.van der Pol E, Coumans FA, Grootemaat AE, Gardiner C, Sargent IL, Harrison P, et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014;12(7):1182–1192. doi: 10.1111/jth.12602. [DOI] [PubMed] [Google Scholar]
- 122.Shih C-L, Chong K-Y, Hsu S-C, Chien H-J, Ma C-T, Chang JW-C, et al. Development of a magnetic bead-based method for the collection of circulating extracellular vesicles. N Biotechnol. 2016;33(1):116–122. doi: 10.1016/j.nbt.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 123.Puhka M, Nordberg ME, Valkonen S, Rannikko A, Kallioniemi O, Siljander P, et al. KeepEX, a simple dilution protocol for improving extracellular vesicle yields from urine. Eur J Pharm Sci. 2017;98:30–9. doi: 10.1016/j.ejps.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 124.Channavajjhala SK, Rossato M, Morandini F, Castagna A, Pizzolo F, Bazzoni F, et al. Optimizing the purification and analysis of miRNAs from urinary exosomes. Clin Chem Lab Med. 2014;52(3):345–354. doi: 10.1515/cclm-2013-0562. [DOI] [PubMed] [Google Scholar]
- 125.Goda T, Masuno K, Nishida J, Kosaka N, Ochiya T, Matsumoto A, et al. A label-free electrical detection of exosomal microRNAs using microelectrode array. Chem Commun (Camb) 2012;48(98):11942–11944. doi: 10.1039/c2cc36111f. [DOI] [PubMed] [Google Scholar]
- 126.Lee JH, Kim JA, Jeong S, Rhee WJ. Simultaneous and multiplexed detection of exosome microRNAs using molecular beacons. Biosens Bioelectron. 2016;86:202–210. doi: 10.1016/j.bios.2016.06.058. [DOI] [PubMed] [Google Scholar]
- 127.Lee JH, Kim JA, Kwon MH, Kang JY, Rhee WJ. In situ single step detection of exosome microRNA using molecular beacon. Biomaterials. 2015;54:116–125. doi: 10.1016/j.biomaterials.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 128.Yang J, Wei F, Schafer C, Wong DT. Detection of tumor cell-specific mRNA and protein in exosome-like microvesicles from blood and saliva. PLoS One. 2014;9(11):e110641. doi: 10.1371/journal.pone.0110641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roberts MJ, Richards RS, Chow CWK, Doi SAR, Schirra HJ, Buck M, et al. Prostate-based biofluids for the detection of prostate cancer: a comparative study of the diagnostic performance of cell-sourced RNA biomarkers. Prostate Int. 2016;4(3):97–102. doi: 10.1016/j.prnil.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Roberts MJ, Richards RS, Gardiner RA, Selth LA. Seminal fluid: a useful source of prostate cancer biomarkers? Biomark Med. 2015;9(2):77–80. doi: 10.2217/bmm.14.110. [DOI] [PubMed] [Google Scholar]
- 131.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10(10):1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ma J, Li N, Guarnera M, Jiang F. Quantification of plasma miRNAs by digital PCR for cancer diagnosis. Biomark Insights. 2013;8:127–136. doi: 10.4137/BMI.S13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Eissa S, Matboli M, Essawy NOE, Shehta M, Kotb YM. Rapid detection of urinary long non-coding RNA urothelial carcinoma associated one using a PCR-free nanoparticle-based assay. Biomarkers. 2015;20(3):212–217. doi: 10.3109/1354750X.2015.1062918. [DOI] [PubMed] [Google Scholar]
- 135.Degliangeli F, Pompa PP, Fiammengo R. Nanotechnology-based strategies for the detection and quantification of microRNA. Chemistry. 2014;20(31):9476–9492. doi: 10.1002/chem.201402649. [DOI] [PubMed] [Google Scholar]
- 136.Gu LQ, Wanunu M, Wang MX, McReynolds L, Wang Y. Detection of miRNAs with a nanopore single-molecule counter. Expert Rev Mol Diagn. 2012;12(6):573–584. doi: 10.1586/erm.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wegman DW, Ghasemi F, Khorshidi A, Yang BB, Liu SK, Yousef GM, et al. Highly-sensitive amplification-free analysis of multiple miRNAs by capillary electrophoresis. Anal Chem. 2015;87(2):1404–1410. doi: 10.1021/ac504406s. [DOI] [PubMed] [Google Scholar]
- 138.Malkov VA, Serikawa KA, Balantac N, Watters J, Geiss G, Mashadi-Hossein A, et al. Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter Assay System. BMC Res Notes. 2009;2:80. doi: 10.1186/1756-0500-2-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, Harbron CG, et al. Evaluating robustness and sensitivity of the nanostring technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res. 2015;75(13):2587–2593. doi: 10.1158/0008-5472.CAN-15-0262. [DOI] [PubMed] [Google Scholar]
- 140.Shah MY, Ferrajoli A, Sood AK, Lopez-Berestein G, Calin GA. microRNa therapeutics in cancer—an emerging concept. EBioMedicine. 2016;12:34–42. doi: 10.1016/j.ebiom.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 142.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43(7):621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488(7409):111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44(2):863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bennett CF, Baker BF, Pham N, Swayze E, Geary RS. Pharmacology of antisense drugs. Annu Rev Pharmacol Toxicol. 2017;57:81–105. doi: 10.1146/annurev-pharmtox-010716-104846. [DOI] [PubMed] [Google Scholar]
- 147.Yamamoto Y, Loriot Y, Beraldi E, Zhang F, Wyatt AW, Al Nakouzi N, et al. Generation 2.5 antisense oligonucleotides targeting the androgen receptor and its splice variants suppress enzalutamide-resistant prostate cancer cell growth. Clin Cancer Res. 2015;21(7):1675–1687. doi: 10.1158/1078-0432.CCR-14-1108. [DOI] [PubMed] [Google Scholar]
- 148.Hong D, Kurzrock R, Kim Y, Woessner R, Younes A, Nemunaitis J, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med. 2015;7(314):314ra185. doi: 10.1126/scitranslmed.aac5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lu J, Tsourkas A. Imaging individual microRNAs in single mammalian cells in situ. Nucleic Acids Res. 2009;37(14):e100. doi: 10.1093/nar/gkp482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12(2):187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2(6):1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- 152.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 153.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14(1):22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333(6042):642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Filonov GS, Moon JD, Svensen N, Jaffrey SR. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc. 2014;136(46):16299–16308. doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Poste G. Bring on the biomarkers. Nature. 2011;469(7329):156–157. doi: 10.1038/469156a. [DOI] [PubMed] [Google Scholar]
- 157.Vickers AJ. Markers for the early detection of prostate cancer: some principles for statistical reporting and interpretation. J Clin Oncol. 2014;32(36):4033–4034. doi: 10.1200/JCO.2014.58.5547. [DOI] [PubMed] [Google Scholar]
- 158.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 159.Ceder Y. Non-coding RNAs in prostate cancer: from discovery to clinical applications. Adv Exp Med Biol. 2016;886:155–170. doi: 10.1007/978-94-017-7417-8_8. [DOI] [PubMed] [Google Scholar]
- 160.Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59(5):671–681. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 161.Shen J, Hruby GW, McKiernan JM, Gurvich I, Lipsky MJ, Benson MC, et al. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate. 2012;72(13):1469–77. doi: 10.1002/pros.22499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mahn R, Heukamp LC, Rogenhofer S, von Ruecker A, Müller SC, Ellinger J. Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology. 2011;77(5):1265.e9–1265.e16. doi: 10.1016/j.urology.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 163.Kelly BD, Miller N, Sweeney KJ, Durkan GC, Rogers E, Walsh K, et al. A circulating microrna signature as a biomarker for prostate cancer in a high risk group. J Clin Med. 2015;4(7):1369–1379. doi: 10.3390/jcm4071369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stuopelytė K, Daniūnaitė K, Jankevičius F, Jarmalaitė S. Detection of miRNAs in urine of prostate cancer patients. Medicina. 2016;52(2):116–24. doi: 10.1016/j.medici.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 165.Brase JC, Johannes M, Schlomm T, Fälth M, Haese A, Steuber T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128(3):608–16. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]