Abstract
PURPOSE
The purpose of our study was to assess whether a model combining clinical factors, MR imaging features, and genomics would better predict overall survival of patients with glioblastoma (GBM) than either individual data type.
METHODS
The study was conducted leveraging the Cancer Genome Atlas (TCGA) effort supported by the National Institutes of Health. Six neuroradiologists reviewed MRI images from The Cancer Imaging Archive (http://cancerimagingarchive.net) of 102 GBM patients using the VASARI scoring system. The patients’ clinical and genetic data were obtained from the TCGA website (http://www.cancergenome.nih.gov/). Patient outcome was measured in terms of overall survival time. The association between different categories of biomarkers and survival was evaluated using Cox analysis.
RESULTS
The features that were significantly associated with survival were: 1) clinical factors: chemotherapy; 2) imaging: proportion of tumor contrast enhancement on MRI, and 3) genomics: HRAS copy number variation. The combination of these three biomarkers resulted in an incremental increase in the strength of prediction of survival, with the model that included clinical, imaging, and genetic variables having the highest predictive accuracy (area under the curve 0.679 ± 0.068, Akaike’s information criterion 566.7, p < 0.001).
CONCLUSION
A combination of clinical factors, imaging features, and HRAS copy number variation best predicts survival of patients with GBM.
Introduction
Recent research in glioblastoma (GBM) treatment has focused on identification of biomarkers that may predict patient outcome, and may consequently impact therapeutic decisions through selection of more aggressive therapies for tumors with worse prognosis. The impact of clinical factors on patient outcome [1–4] and the correlation between MR imaging features of GBM and survival [5–10] have been previously studied. Similarly, the association between genomic biomarkers and patient outcome has received growing attention, in particular with the recent introduction of anti-angiogenetic drugs [11–15]. Only few studies so far have attempted to integrate clinical factors, imaging biomarkers (usually not evaluated across observers in a standardized fashion), and tumor gene expression data into a statistical model that would potentially provide a robust predictor of patient outcome than each individual type of data.[16] The purpose of our study was to assess whether such a model combining clinical factors, MR imaging features (assessed using standardized semantic imaging features across multiple observers), and genomics would predict patient survival more reliably than any individual data type.
Materials and Methods
Study Data
Our study was conducted leveraging the TCGA project (http://www.cancergenome.nih.gov/) of the National Cancer Institute (NCI) and National Human Genome Research Institute (NHGRI), which aims to catalogue gene mutations associated with cancer. One hundred and two patients with pathology proven GBM were included in this study. Their MR imaging studies were made available through The Cancer Imaging Archive (http://www.cancerimagingarchive.net). Clinical data were obtained from the Open Access Data Tier of the TCGA website, and Health Insurance Portability and Accountability Act of 1996 (HIPAA) de-identified clinical data.
Gene expression, mutation and copy number data were obtained from the TCGA data portal (https://tcga-data.nci.nih.gov). Three sets of genetic metadata were separately considered for the current study:
-
1)
Genes with p-value <0.05 in univariate COX model analysis of gene expression and overall survival were assessed further with Gene Set Enrichment Analysis (GSEA). GSEA is a method to evaluate the expression of genes in the context of pathways. A significant pathway means that gene expression in these pathways is significantly different compared to a random gene set. We used a hypergeometric test to detect significant biological pathways.
-
2)
The mutation status of specific genes of interest identified in previous work (EGFR, ERBB2, IDH1, NF1, PDGFRA, PIK3CA, PIK3R1, PTEN, RB1 and TP53) was also evaluated [17].
-
3)
As part of a copy number analysis, raw data were first converted into an ordinal ranking (homozygous deletion, heterozygous deletion, wild type, low level amplification, high level amplification), as described by the Computational Biology Center at Memorial Sloan-Kettering Cancer Center (http://www.cbioportal.org/public-portal/) prior to Cox survival analysis.
Imaging Review
Six neuroradiologists from the University of Virginia Health System, Thomas Jefferson University Hospital, Emory University, MD Anderson Cancer Center and the National Institute of Health, independently reviewed the MR images of 102 patients, using ClearCanvas workstations, which allow visualization as well as annotation and markup of DICOM images (customized version of ClearCanvas to support the capture of markups in Annotation and Image Markup (AIM) format, available at https://wiki.nci.nih.gov/x/z4×3Ag). The VASARI feature scoring system for human gliomas (https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project, was employed for the interpretation of the MR images to ensure interobserver consistency. The VASARI scoring system (Table 1) includes 30 semantic descriptors of imaging features of brain tumors clustered by categories pertaining to lesion location, morphology of the lesion substance, morphology of the lesion margin, alterations in the vicinity of the lesion, and extent of tumor resection [7, 18]. For each feature, the scoring system incorporates discrete qualitative, semi-quantitative, or quantitative values, appropriate to the nature of the feature assessed.
Table 1. Correlation between Vasari imaging features and survival time on univariate Cox analysis.
In univariate Cox model analyses, four VASARI features were associated with shorter survival time with an unadjusted p-value < 0.05: higher proportion of enhancing tumor (p=0.009), higher T1/FLAIR ratio (p=0.022), and tumor location (p=0.029) and side (p=0.049). Only the proportion of tumor enhancing (bolded) was significant in the multiple clinical feature model.
| Vasari Imaging Features | Values in our study population (counts) |
Coefficient | Hazard Ratio |
Standard error |
P value |
||
|---|---|---|---|---|---|---|---|
|
| |||||||
| F1 | Tumor Location | 1=Frontal | 32/102 | −0.609 | 0.544 | 0.278 | 0.029 |
| 2=Temporal | 45/102 | ||||||
| 3=Insular | 3/102 | ||||||
| 4=Parietal | 13/102 | ||||||
| 5=Occipital | 9/102 | ||||||
|
| |||||||
| F2 | Side of Tumor Epicenter | 1=Right | 52/102 | −0.525 | 0.592 | 0.267 | 0.049 |
| 2=Center/Bilateral | 0/102 | ||||||
| 3=Left | 50/102 | ||||||
|
| |||||||
| F3 | Eloquent Brain Involved | 1=None | 60/102 | −0.479 | 0.62 | 0.352 | 0.174 |
| 2=Speech motor | 7/102 | ||||||
| 3=Speech receptive | 14/102 | ||||||
| 4=Motor | 12/102 | ||||||
| 5=Vision | 9/102 | ||||||
|
| |||||||
| F4 | Enhancement Quality | 1=None | 0/102 | 0.5961 | 1.815153 | 0.596921 | 0.31792 |
| 2=Mild/Minimal | 6/102 | ||||||
| 3=Marked/Avid | 96/102 | ||||||
|
| |||||||
| F5 | Proportion Enhancing | 1=Not Available | 0/102 | −0.637 | 1.891 | 0.243 | 0.009 |
| 2=None (0%) | 0/102 | ||||||
| 3=<5% | 9/102 | ||||||
| 4=6–33% | 71/102 | ||||||
| 5=34–67% | 21/102 | ||||||
| 6=68–95% | 5/102 | ||||||
| 7=>95% | 0/102 | ||||||
| 8=All (100%) | 0/102 | ||||||
|
| |||||||
| F6 | Proportion Non-Enhancing | 1=Not Available | 0/102 | −0.155 | 0.857 | 0.173 | 0.372 |
| 2=None (0%) | 18/102 | ||||||
| 3=<5% | 33/102 | ||||||
| 4=6–33% | 29/102 | ||||||
| 5=34–67% | 12/102 | ||||||
| 6=68–95% | 10/102 | ||||||
| 7=>95% | 0/102 | ||||||
| 8=All (100%) | 0/102 | ||||||
|
| |||||||
| F7 | Proportion Necrosis | 1=Not Available | 0/102 | 0.171 | 1.187 | 0.254 | 0.50 |
| 2=None (0%) | 3/102 | ||||||
| 3=<5% | 18/102 | ||||||
| 4=6–33% | 41/102 | ||||||
| 5=34–67% | 30/102 | ||||||
| 6=68–95% | 10/102 | ||||||
| 7=>95% | 0/102 | ||||||
| 8=All (100%) | 0/102 | ||||||
|
| |||||||
| F8 | Cysts | 1=No | 97/102 | −0.490 | 0.613 | 0.721 | 0.497 |
| 2=Yes | 5/102 | ||||||
|
| |||||||
| F9 | Multifocal or Multicentric | 1=Not Available | 0/102 | −0.546 | 0.579 | 0.443 | 0.218 |
| 2=Focal | 94/102 | ||||||
| 3=Multifocal | 5/102 | ||||||
| 4=Multicentric | 1/102 | ||||||
| 5=Gliomatosis | 2/102 | ||||||
|
| |||||||
| F10 | T1/FLAIR ratio | 1=Expansive (T1~FLAIR) | 64/102 | −0.529 | 1.697 | 0.23 | 0.022 |
| 2=Mixed (T1<FLAIR) | 8/102 | ||||||
| 3=Infiltrative (T1<<FLAIR) | 30/102 | ||||||
|
| |||||||
| F11 | Thickness of enhancing margin | 1=Not Available | 0/102 | 0.596 | 1.815 | 0.597 | 0.318 |
| 2=None | 0/102 | ||||||
| 3=Thin | 7/102 | ||||||
| 4=Thick/solid | 95/102 | ||||||
|
| |||||||
| F13 | Definition of the Non-Enhancing Margin | 1=Not Available | 0/102 | −0.179 | 0.836 | 0.435 | 0.681 |
| 2=Smooth | 13/102 | ||||||
| 3=Irregular | 92/102 | ||||||
|
| |||||||
| F14 | Proportion of Edema | 1=Not Available | 0/102 | −0.236 | 0.79 | 0.206 | 0.251 |
| 2=None (0%) | 1/102 | ||||||
| 3=<5% | 19/102 | ||||||
| 4=6–33% | 32/102 | ||||||
| 5=34–67% | 48/102 | ||||||
| 6=68–95% | 2/102 | ||||||
| 7=>95% | 0/102 | ||||||
| 8=All (100%) | 0/102 | ||||||
|
| |||||||
| F16 | Hemorrhage | 1=No | 68/102 | 0.045 | 1.046 | 0.279 | 0.872 |
| 2=Yes | 34/102 | ||||||
|
| |||||||
| F17 | Diffusion | 1=No image | 0/102 | −0.273 | 0.761 | 0.405 | 0.5 |
| 2=Facilitated | 49/102 | ||||||
| 3=Restricted | 11/102 | ||||||
| 4=Neither/equivocal/both | 42/102 | ||||||
|
| |||||||
| F18 | Pial Invasion | 1=No | 60/102 | −0.179 | 0.836 | 0.274 | 0.512 |
| 2=Yes | 42/102 | ||||||
|
| |||||||
| F19 | Ependymal Invasion | 1=No | 41/102 | 0.263 | 1.301 | 0.27 | 0.329 |
| 2=Yes | 61/102 | ||||||
|
| |||||||
| F20 | Cortical Involvement | 1=No | 10/102 | −0.569 | 0.566 | 0.367 | 0.122 |
| 2=Yes | 92/102 | ||||||
|
| |||||||
| F21 | Deep White Matter Invasion | 1=No | 42/102 | −0.527 | 0.59 | 0.274 | 0.054 |
| 2=Yes | 60/102 | ||||||
|
| |||||||
| F22 | Nonenhancing Tumor Crosses Midline | 1=Not Available | 0/102 | 0.318 | 1.374 | 0.331 | 0.337 |
| 2=No | 80/102 | ||||||
| 3=Yes | 22/102 | ||||||
|
| |||||||
| F23 | Enhancing Tumor Crosses Midline | 1=Not Available | 0/102 | 1.007 | 2.738 | 0.529 | 0.057 |
| 2=No | 92/102 | ||||||
| 3=Yes | 10/102 | ||||||
|
| |||||||
| F24 | Satellites | 1=No | 77/102 | 0.13 | 1.139 | 0.327 | 0.691 |
| 2=Yes | 25/102 | ||||||
|
| |||||||
| F25 | Calvarial Remodeling | 1=No | 100/102 | −0.139 | 0.871 | 0.724 | 0.848 |
| 2=Yes | 2/102 | ||||||
|
| |||||||
| F29 & F30 | Lesion Size | Major Axis Length | 76.80598 (20.60491) | 0.007 | 1.007 | 0.007 | 0.283 |
| Minor Axis Length | 49.89373 (12.32175) | −0.001 | 0.999 | 0.012 | 0.934 | ||
| Mean(SD) | |||||||
Statistical Analysis
The 102 study patients were split into a training set of 68 patients and a separate testing set of 34 patients using a stratified sampling method. Stratified sampling was selected because of the small sample size and the skewed survival distribution, and because it ensures an approximately equal survival distribution of subjects between the training and testing data. The split data are also more representative of the population than the random sampling. Samples were sorted by survival in an ascending order. For every three samples, the first two samples were chosen as the training dataset and the third sample was selected as the testing dataset. Models were created in the training dataset and validated in the testing dataset.
Associations between survival (outcome) and different individual categories of predictors including clinical variables (Table 2), imaging features (Table 1), tumor gene expression, mutation, and copy number variation (Table 3) were assessed using univariate Cox regression models. Clinical, genetic, and imaging data were checked for co-linearity. Non-collinear, predicting variables shown as significant in the univariate Cox regression models were retained and included in several multivariate Cox regression models incorporating the clinical, imaging and/or genomic datasets using stepwise covariate model-building strategies. The models were compared with Akaike’s Information Criterion (AIC) to evaluate the model fitness, with the baseline model with clinical data only used as reference for comparisons.[19] A lower AIC value indicates a better model. AUC (area under curve) analysis was also used to measure the accuracy of prediction. A higher AUC value indicates that the model is better at predicting survival. The model using only clinical data was used as the reference. Statistical models were corrected for multiple comparisons using false discovery rate (FDR) procedure.
Table 2. Correlation between clinical features and survival time on univariate Cox analysis.
In univariate Cox model analyses, three clinical variables were correlated with survival with an unadjusted p-value < 0.05. These included radiation therapy (p<0.001), chemotherapy (p<0.001), and age at initial diagnosis (p=0.021). Only chemotherapy (bolded) was significant in the multiple clinical feature model.
| Clinical variables |
Values in our study population (mean±standard deviation or counts) |
Coefficient | Hazard ratio |
Standard error |
P value |
|---|---|---|---|---|---|
| Age at initial pathological diagnosis [years] | 57.7±14.6 | 0.023 | 1.023 | 0.010 | 0.021 |
| Gender | 66 males, 36 females | 0.275 | 1.317 | 0.276 | 0.318 |
| Ethnicity | 8 hispanic or latino; 94 non- hispanic or latino | 1.302 | 3.676 | 1.033 | 0.208 |
| Race | 83 caucasian, 6 african- americans, 5 asians | 0.446 | 1.562 | 0.391 | 0.255 |
| Radiation therapy | 96 yes, 6 no | −3.298 | 0.037 | 0.634 | <0.001 |
| Chemotherapy | 88 yes, 14 no | −1.311 | 0.270 | 0.339 | <0.001 |
| Hormonotherapy | 1 yes, 101 no | −0.049 | 0.952 | 1.014 | 0.962 |
| Immunotherapy | 2 yes, 100 no | −0.019 | 1.019 | 0.724 | 0.979 |
Table 3. Correlation between tumor gene expression and survival on Cox analysis.
-
1)FSHB, FSHR, LHCGR, TSHB, TSHR, PRSS2 genes in Neuroactive ligand-receptor interaction (p=0.012);
-
2)LDHAL6A, LDHAL6B, LDHA genes in Cysteine metabolism (p=0.030); and
-
3)EFNA5 and EPHA5 genes in Ephrin A reverse signaling (p=0.009).
| Type of genetic analysis |
Genes | Coefficient | Hazard ratio |
Standard error |
P value |
|---|---|---|---|---|---|
|
| |||||
| Copy Number | PIK3R1 | 1.588 | 4.896 | 0.566 | 0.005 |
|
| |||||
| AKT1 | 2.602 | 13.492 | 1.155 | 0.024 | |
|
| |||||
| HRAS | 1.125 | 3.080 | 0.623 | 0.071 | |
|
| |||||
| Mutation | PDGFRA | 2.463 | 11.741 | 1.118 | 0.028 |
|
| |||||
| NF1 | −0.713 | 0.490 | 0.480 | 0.082 | |
|
| |||||
| Gene Set Enrichment Analysis (GSEA) | FSHB | 1.311 | 3.709 | 0.636 | 0.039 |
| FSHR | 0.812 | 2.253 | 0.446 | 0.069 | |
| LHCGR | 1.022 | 2.779 | 0.431 | 0.017 | |
| TSHB | 0.787 | 2.198 | 0.460 | 0.087 | |
| TSHR | −0.370 | 0.689 | 0.282 | 0.188 | |
| PRSS2 | 0.463 | 1.589 | 0.255 | 0.069 | |
| (neuroactive ligand-receptor interaction pathway) | |||||
|
| |||||
| LDHAL6A | −0.854 | 0.425 | 0.642 | 0.183 | |
| LDHAL6B LDHA | −1.303 | 0.271 | 0.601 | 0.030 | |
| (cysteine metabolism pathway) | 0.356 | 0.356 | 0.163 | 0.029 | |
|
| |||||
| EFNA5 | −0.723 | 0.485 | 0.390 | 0.063 | |
| EPHA5 | 0.190 | 1.210 | 0.130 | 0.144 | |
| (Ephrin A reverse signalling pathway) | |||||
Results
Study Population Demographics
The clinical and imaging characteristics of our 102 patients are reported in Tables 2 and 1, respectively. We calculated the association of 17,787 genes with expression data, 10 genes with mutation data and 14 genes with copy number data. The median survival was 324.0 days (25 percentile: 144.5, 75 percentile: 578.8, minimum: 6.0, maximum: 1757.0)
Correlation between clinical variables and survival (Table 2)
Univariate Cox model analyses demonstrated that three clinical variables were significantly correlated to survival with an unadjusted p-value < 0.05. These included radiation therapy (p<0.001), chemotherapy (p<0.001), and age at initial diagnosis (p=0.021). The 6 patients who did not receive radiation therapy had a median survival of 66 days compared to median survival of 389 days in the 96 patients with radiation therapy. The 14 patients who did not receive chemotherapy had a median survival of 132 days compared to the median survival of 454 days in the 88 patients with chemotherapy. An increased age at the initial pathological diagnosis was associated with a shorter survival (p=0.021). All clinical features were used to build survival models in a stepwise fashion. Only chemotherapy was significant in the multiple clinical feature model.
Correlation between VASARI imaging features and survival (Table 1)
Univariate Cox model analyses showed that four VASARI features were associated with a shorter survival time with an unadjusted p-value < 0.05: higher proportion of enhancing tumor (p=0.009), a higher T1/FLAIR ratio (p=0.022), and tumor location (p=0.029) and side (p=0.049). The T1/FLAIR ratio measures the ratio between the size of the portion of the tumor that is hypointense on T1-weighted images in relation to the extent of FLAIR signal abnormality corresponding to the tumor. A higher T1/FLAIR ratio indicates a more infiltrative tumor. Tumor location in the frontal lobe (p=0.029) and tumors on the right side (p=0.049) correlated with a longer survival. Only the proportion of enhancing tumor was significant in the multiple clinical feature model (Figure 1).
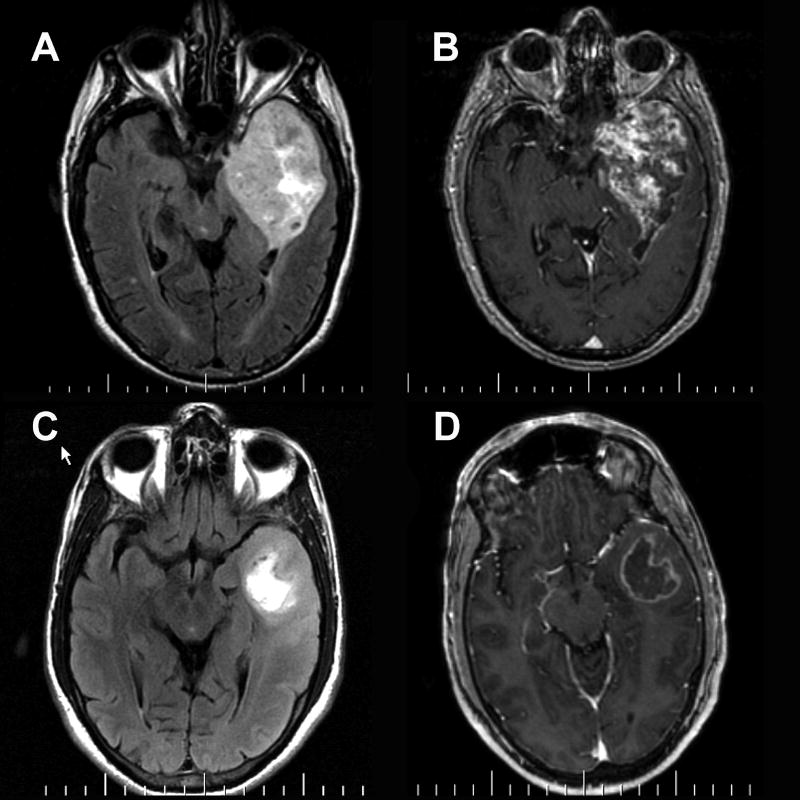
Figure 1.
(A,C) FLAIR and (B,D) post-contrast T1-weighted images in two patients with left temporal GBM. (A,B) In patient #1, enhancement involves nearly the entirety of the tumor. Patient #1 survived 82 days after the MRI scan. (C,D) In patient #2, enhancement involves only a small fraction of the tumor, basically a rim around the central necrosis. Patient #1 survived 467 days after the MRI scan.
Correlation between genomics and survival (Table 3)
Cox model analyses revealed that three genes with copy number variations, PIK3R1 (p=0.005), AKT1 (p=0.024), and HRAS (p=0.071), and two genes with mutation, PDGFRA (p=0.028) and NF1 (p=0.082) were associated with survival. However, none of these genes had FDR (false discovery rate) adjusted p-value < 0.050.[20]
We used a Cox model analysis to investigate association between survival and tumor gene expression. A total of 724 genes were associated with survival with an adjusted p-value<0.05 in the training dataset; however, none of the genes associated with survival passed the FDR test. Therefore, we carried out a Gene Set Enrichment Analysis (GSEA) of the 724 genes in order to select genes in significant biological pathways. This analysis revealed three gene categories associated with survival with an unadjusted p-value <0.050. These included:
-
1)
FSHB, FSHR, LHCGR, TSHB, TSHR, PRSS2 genes in Neuroactive ligand-receptor interaction;
-
2)
LDHAL6A, LDHAL6B, LDHA genes in Cysteine metabolism, and
-
3)
EFNA5 and EPHA5 genes in Ephrin A reverse signaling.
Only HRAS copy number variation was significantly associated with survival in the multiple genetic feature model.
Multivariate Cox analysis of the association between combined biomarkers and survival (Table 4)
Table 4. Multivariate analysis of the correlation between combined biomarkers and survival adjusted by age, race, and gender.
The model with smaller AIC is a better fit model. The model with larger AUC has better prediction power. Df indicated the variables used in the model building. P values (<0.05) showed that there is a significant difference when comparing the combined models to the baseline clinical model. The model that best predicted survival (highest AUC, smallest AIC) included clinical and imaging data and genomics.
| Model | Variables Included in Model |
df | AIC | AUC mean ± standard deviation |
P value |
|---|---|---|---|---|---|
| Clinical Data | Chemotherapy | 3 | 744.0 | 0.575 ± 0.029 | Not applicable |
| Clinical Data and Genomics | Chemotherapy HRAS | 4 | 569.9 | 0.560 ± 0.063 | < 0.001 |
| Clinical and Imaging Data | Chemotherapy, Proportion Enhancing | 4 | 739.8 | 0.589 ± 0.030 | 0.012 |
| Clinical and Imaging Data and Genomics | Chemotherapy Proportion Enhancing and HRAS | 5 | 566.7 | 0.679 ± 0.068 | < 0.001 |
Stepwise multivariate Cox’s models were used to assess the association between overall survival time and the clinical data, VASARI imaging features and genomic variations shown as statistically significant in the above-mentioned Cox analyses. Models were evaluated with AIC values for the model fitness and AUC values for the model prediction power. The clinical variable “chemotherapy”, the imaging variable “portion of the tumor enhancing” and HRAS copy number variation are the only variables that remained significant in the multivariate analysis. The AIC value for the clinical data only model was 744.0 and the one for the clinical and imaging data was 739.8. The AIC value for the clinical and genomic data model was smaller, 569.9, indicating a better model. The AIC value for the clinical, imaging and genomic data was the smallest, 566.7, indicating the best model. AUC analysis led to the same conclusion, supporting the concept that the model including clinical, imaging and genomic data is the best predictor for survival. As designed, we use the testing data to validate the model, AUC analysisled to the same conclusion with the combined model with AUC=0.679 ± 0.068 comparing to AUC values for Chemotherapy = 0.575 ± 0.029, Chemotherapy and Proportion Enhancing = 0.589 ± 0.030, Chemotherapy and HRAS = 0.560 ± 0.063. That supports the notion that the model including clinical, imaging and genomic data is the best predictor for survival.
Discussion
Accurate classification of gliomas is important as the treatment modalities are substantially different between WHO grade III and IV tumors. Patients with GBM have the worst prognosis with a median survival of approximately 12 months despite advances in surgery, radiation therapy, and chemotherapy.[21] The current histopathology-based classification system for gliomas is complicated by considerable interobserver variability [22]. Moreover, the existence of different subclasses with varying response to treatment within similar histologic grades of glioma has been shown.[22–24] Therefore, a classification using more reliable biomarkers has been postulated as a critical element to improve treatment selection and outcome.
The association of clinical variables with the outcome of GBM patients has been extensively studied in the literature. Among established independent predictors of poor outcome are older age (greater than 60 years), lower Karnofsky Performance Score (KPS), and incomplete tumor resection[1, 3, 4]. Barker et al. [4] also found in a population of 301 GBM patients that a more extensive surgical resection was associated with a better response to radiation therapy. Our univariate analysis demonstrated that three clinical features are associated with survival with a p value <0.05, including radiation therapy, chemotherapy, and age at initial diagnosis. Multivariate analysis only retained chemotherapy as significantly associated with improved survival.
Several studies have evaluated the prognostic value of the imaging features of GBM. In a recent study of 393 GBM patients by Chaichana et al. [5], periventricular location of the tumor was associated with worse survival. A retrospective study by Murakami et al.[6] of 79 GBM patients demonstrated that lower ADC values within the tumor correlated with shorter survival. A study by Pope et al.[7] of 15 imaging features of high grade gliomas, including 41 grade III gliomas and 110 GBMs, demonstrated that the absence of enhancing tumor, the absence of edema and the absence of satellite or multifocal lesions correlated with a doubling of the median survival. In a series of 141 grade III and IV gliomas, Steltzer et al [9] showed that involvement of the corpus callosum is a poor prognostic sign. In our study, we assessed the prognostic value of a systematic scoring system for the MR imaging features of GBM. Features that were associated with poor outcome on the basis of univariate analyses included the proportion of tumor enhancing, and a higher T1/FLAIR ratio of the tumoral signal abnormality, which corresponds to a more infiltrative tumor. These results are consistent as they include features that likely denote more aggressive tumor behavior. The association of tumoral involvement of the visual cortex with a worse prognosis possibly relates to the fact that this location precludes complete surgical resection. Location of the tumor in the frontal lobe and in the right cerebral hemisphere correlated with longer survival. This may reflect that frontal lobe resection and right hemispheric lesion resection in particular can usually be more extensive than for other locations, as the majority of patients are left hemisphere dominant. The multivariate analysis showed that only the proportion of tumor enhancing significantly correlated with a worse survival. This is concordant with a well-established imaging criterion that associates enhancement with more aggressive tumor behavior [7, 16]. Likewise, surgical biopsies and resections primarily target the enhancing portion of the tumor with the intent to sample and/or resect the higher grade component [25].
A number of genes among hundreds of currently identified tumor expression genes have been shown to have a predictive value in terms of GBM patient survival [10, 12]. The Cancer Genome Atlas Network recently cataloged recurrent genomic abnormalities in GBM, which allowed a robust gene expression-based molecular classification of GBM into 4 subtypes: proneural, neural, classical, and mesenchymal [11]. Response to aggressive therapy appears to differ by subtype, with the greatest benefit in the classical subtype and no benefit in the proneural subtype. Our multivariate genomic analysis demonstrated that only HRAS copy number variation correlated with poorer survival. The HRAS gene is located on the short (p) arm of chromosome 11. It encodes for a GTPase enzyme also referred to as transforming protein p21, which is involved in regulating cell division in response to growth factor stimulation. HRAS is a proto-oncogene, and HRAS mutations are encountered in bladder, kidney and thyroid cancers (http://ghr.nlm.nih.gov/gene/HRAS). Our finding of HRAS being prognostic of outcome in GBM patients is concordant with several prior reports that demonstrated the causative role of HRAS mutation in the induction of GBM [26] and its association with shorter survival [12]. This gene is not among those used by Verhaak et al. in the tumor gene-expression based classification of GBM[11] and likely plays a role in the genesis of GBM across all subtypes.
The incremental addition of clinical (chemotherapy), imaging (proportion of tumor enhancing) and genomic (HRAS) biomarkers resulted in an incremental increase in the strength of their association with survival. The best model was the one that included all three types of biomarkers. These results are promising as they suggest that the combination of imaging and genomic variables with conventional clinical variables adds significant value in terms of prognosticating outcome.
We acknowledge several limitations to our study. The most significant one is the lack of complete clinical information if our study patients. For instance, Karnofsky Performance Score, percentage of resection, details of the chemotherapy, etc were not available and could not be incorporated in the statistical analysis.
In conclusion, our study demonstrates that a subset of clinical variables, VASARI imaging features, and tumor gene expression, considered in combination with each other, correlate significantly with GBM patients’ survival. These results need to be confirmed in a larger database including more complete clinical information.
Abbreviation Key
- ADC
Average Diffusion Coefficient
- AIC
Akaike’s Information Criterion
- AUC
Area Under the Curve
- GBM
Glioblastoma
- GSEA
Gene Set Enrichment Analysis
- KPS
Karnofsky Performance Score
- NCI
National Cancer Institute
- NHGRI
National Human Genome Research Institute
- TCGA
The Cancer Genome Atlas
- TCIA
The Cancer Imaging Archive (http://cancerimagingarchive.net)
- VASARI
Visually AcesSAble Rembrandt Images (https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project)
- WHO
World Health Organization
References
- 1.Verger E, Valduvieco I, Caral L, Pujol T, Ribalta T, Vinolas N, Boget T, Oleaga L, Blanco Y, Graus F. Does gender matter in glioblastoma? Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2011;13(10):737–741. doi: 10.1007/s12094-011-0725-7. [DOI] [PubMed] [Google Scholar]
- 2.Marina O, Suh JH, Reddy CA, Barnett GH, Vogelbaum MA, Peereboom DM, Stevens GH, Elinzano H, Chao ST. Treatment outcomes for patients with glioblastoma multiforme and a low Karnofsky Performance Scale score on presentation to a tertiary care institution. Clinical article. Journal of neurosurgery. 2011;115(2):220–229. doi: 10.3171/2011.3.JNS10495. [DOI] [PubMed] [Google Scholar]
- 3.Allahdini F, Amirjamshidi A, Reza-Zarei M, Abdollahi M. Evaluating the prognostic factors effective on the outcome of patients with glioblastoma multiformis: does maximal resection of the tumor lengthen the median survival? World neurosurgery. 2010;73(2):128–134. doi: 10.1016/j.wneu.2009.06.001. discussion e116. [DOI] [PubMed] [Google Scholar]
- 4.Barker FG, 2nd, Chang SM, Larson DA, Sneed PK, Wara WM, Wilson CB, Prados MD. Age and radiation response in glioblastoma multiforme. Neurosurgery. 2001;49(6):1288–1297. doi: 10.1097/00006123-200112000-00002. discussion 1297’1288. [DOI] [PubMed] [Google Scholar]
- 5.Chaichana K, Parker S, Olivi A, Quinones-Hinojosa A. A proposed classification system that projects outcomes based on preoperative variables for adult patients with glioblastoma multiforme. Journal of neurosurgery. 2010;112(5):997–1004. doi: 10.3171/2009.9.JNS09805. [DOI] [PubMed] [Google Scholar]
- 6.Murakami R, Sugahara T, Nakamura H, Hirai T, Kitajima M, Hayashida Y, Baba Y, Oya N, Kuratsu J, Yamashita Y. Malignant supratentorial astrocytoma treated with postoperative radiation therapyprognostic value of pretreatment quantitative diffusion-weighted MR imaging. Radiology. 2007;243(2):493–499. doi: 10.1148/radiol.2432060450. [DOI] [PubMed] [Google Scholar]
- 7.Pope WB, Sayre J, Perlina A, Villablanca JP, Mischel PS, Cloughesy TF. MR imaging correlates of survival in patients with high-grade gliomas. AJNR American journal of neuroradiology. 2005;26(10):2466–2474. [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Jin H, Lu Y, Oh J, Chang S, Nelson SJ. Identification of MRI and 1H MRSI parameters that may predict survival for patients with malignant gliomas. NMR in biomedicine. 2004;17(1):10–20. doi: 10.1002/nbm.858. [DOI] [PubMed] [Google Scholar]
- 9.Steltzer KJ, Sauve KI, Spence AM, Griffin TW, Berger MS. Corpus callosum involvement as a prognostic factor for patients with high-grade astrocytoma. International journal of radiation oncology, biology, physics. 1997;38(1):27–30. doi: 10.1016/s0360-3016(96)00632-3. [DOI] [PubMed] [Google Scholar]
- 10.Gutman DA, Cooper LA, Hwang SN, Holder CA, Gao J, Aurora TD, Dunn WD, Jr, Scarpace L, Mikkelsen T, Jain R, et al. MR Imaging Predictors of Molecular Profile and Survival: Multi-institutional Study of the TCGA Glioblastoma Data Set. Radiology. 2013 doi: 10.1148/radiol.13120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serao NV, Delfino KR, Southey BR, Beever JE, Rodriguez-Zas SL. Cell cycle and aging, morphogenesis, and response to stimuli genes are individualized biomarkers of glioblastoma progression and survival. BMC medical genomics. 2011;4:49. doi: 10.1186/1755-8794-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung CS, Unterberg AW, Hartmann C. Diagnostic markers for glioblastoma. Histology and histopathology. 2011;26(10):1327–1341. doi: 10.14670/HH-26.1327. [DOI] [PubMed] [Google Scholar]
- 14.Parsa AT. Functional validation confirms genomic phenotypes of glioblastoma with implications for targeted therapy. World neurosurgery. 2011;75(5–6):573–574. doi: 10.1016/j.wneu.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 15.Brennan C. Genomic profiles of glioma. Current neurology and neuroscience reports. 2011;11(3):291–297. doi: 10.1007/s11910-011-0198-7. [DOI] [PubMed] [Google Scholar]
- 16.Zinn PO, Sathyan P, Mahajan B, Bruyere J, Hegi M, Majumder S, Colen RR. A novel volume-age-KPS (VAK) glioblastoma classification identifies a prognostic cognate microRNA-gene signature. PloS one. 2012;7(8):e41522. doi: 10.1371/journal.pone.0041522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Agarwal A, Kumar S, Narang J, Schultz L, Mikkelsen T, Wang S, Siddiqui S, Poptani H, Jain R. Morphologic MRI features, diffusion tensor imaging and radiation dosimetric analysis to differentiate pseudo-progression from early tumor progression. Journal of neuro-oncology. 2013 doi: 10.1007/s11060-013-1070-1. [DOI] [PubMed] [Google Scholar]
- 19.Akaike H. A Bayesian Analysis of the Minimum AIC Procedure. In: Parzen E, Tanabe K, Kitagawa G, editors. Selected Papers of Hirotugu Akaike. Springer; New York: 1998. pp. 275–280. [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the False Discovery RateA Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 21.Johnson DR, O’Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. Journal of neuro-oncology. 2012;107(2):359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 22.Shirai K, Suzuki Y, Okamoto M, Wakatsuki M, Noda SE, Takahashi T, Ishiuchi S, Hasegawa M, Nakazato Y, Nakano T. Influence of histological subtype on survival after combined therapy of surgery and radiation in WHO grade 3 glioma. Journal of radiation research. 2010;51(5):589–594. doi: 10.1269/jrr.10055. [DOI] [PubMed] [Google Scholar]
- 23.Robertson T, Koszyca B, Gonzales M. Overview, recent advances in neuropathology. Part 1: Central nervous system tumours. Pathology. 2011;43(2):88–92. doi: 10.1097/PAT.0b013e3283426e86. [DOI] [PubMed] [Google Scholar]
- 24.Jain R, Poisson L, Narang J, Gutman D, Scarpace L, Hwang SN, Holder C, Wintermark M, Colen RR, Kirby J, et al. Genomic Mapping and Survival Prediction in Glioblastoma: Molecular Subclassification Strengthened by Hemodynamic Imaging Biomarkers. Radiology. 2012 doi: 10.1148/radiol.12120846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moliterno JA, Patel TR, Piepmeier JM. Neurosurgical approach. Cancer journal. 2012;18(1):20–25. doi: 10.1097/PPO.0b013e3183243f6e3. [DOI] [PubMed] [Google Scholar]
- 26.Kondo T. Mouse induced glioma-initiating cell models and therapeutic targets. Anti-cancer agents in medicinal chemistry. 2010;10(6):471–480. doi: 10.2174/1871520611009060471. [DOI] [PubMed] [Google Scholar]