Abstract
EglN prolyl hydroxylases, a family of oxygen-sensing enzymes, hydroxylate distinct proteins to modulate diverse physiopathological signals. Aberrant regulations of EglNs result in multiple human diseases, including cancer. Different from EglN1 which function largely depends on the role of hypoxia-induce factor alpha (HIFα) in tumors, the functional significance and the upstream regulatory mechanisms of EglN2, especially in prostate cancer setting, remain largely unclear. Here, we demonstrated that dysregulation of EglN2 facilitated prostate cancer growth both in cells and in vivo. Notably, EglN2 was identified highly expressed in human prostate cancer tissues. Mechanically, Cullin 3-based E3 ubiquitin ligase SPOP, a well-characterized tumor suppressor in prostate cancer, could recognize and destruct EglN2. Meanwhile, androgen receptor (AR), playing a pivotal role in progression and development of prostate cancer, could transcriptionally up-regulate EglN2. Pathologically, SPOP loss-of-function mutations or AR amplification, frequently occurring in prostate cancers, could significantly accumulate EglN2 abundance. Therefore, our study not only underlines an oncogenic role of EglN2 in prostate cancer, but also highlights SPOP as a tumor suppressor to down-regulate EglN2 in prostate cancer.
Keywords: EglN2, SPOP, Ubiquitination, Prostate cancer, Androgen receptor
Introduction
EglN prolyl hydroxylases recently have attracted more attention due to the increasing identifications of their substrates and complicated roles in tumorigenesis [1,2]. Functionally, all three EglNs share the biochemical functions to hydroxylate hypoxia-induce factor alpha (HIFα), which acts as a key regulator of hypoxia-induced genes [3–6]. Subsequently, the von Hippel-Lindau tumor suppressor protein pVHL, forming a complex with Cullin 2, Skp1 and Rbx1, recognizes and destructs the hydroxylated HIFα [7]. However, the biological functions of EglNs towards hydroxylated HIFα are quite different. Specifically, EglN1 (also termed PHD2) is the primary HIF prolyl hydroxylase under normal conditions, whereas EglN2 (also termed PHD1) and EglN3 (also termed PHD3) might be involved in HIF prolyl hydroxylation under specific conditions, such as hypoxia [8]. The accumulation of HIFα incorporates with HIFβ (ARNT) to modulate cohorts of downstream genes, resulting in governing cellular behaviors to adapt the environment changes [9–12].
In addition to HIFα, multiple specific substrates recently were identified to be hydroxylated by distinct EglNs, such as Akt and NDRG3 as the substrates of EglN1 [13,14], FOXO3a, CEP192 and IKBKB as the substrates of EglN2 [2,15,16], PKM2 and EPOR as the substrates of EglN3 [17,18]. Thus, the biological functions of these EglNs are mainly dependent on their substrates in a tissue- or cell-context dependent manner. For instance, EglN1 serves as a tumor suppressor in triple negative breast cancer (TNBC), where HIF1α is characterized as an oncogene [19]. However, EglN1 may work as an oncogene in glioma due to the characterized tumor suppressor role of HIF1α in glioma setting [20]. Interestingly, EglN2 is demonstrated as a potential oncogene by hydroxylating FOXO3a to induce cyclin D1 expression in breast cancer [1,2], as well as involves in drug resistance of colorectal cancer (CRC) [21]. However, EglN3 plays oncogenic functions to protect cell from apoptosis in both HIF-dependent and -independent manners [22,23]. To better understand the physiopathological functions of EglNs, it is of value to investigate the regulatory mechanisms of EglNs. As reported before, EglN3 could be transcriptionally regulated by HIF signals [24,25], and EglN1 and EglN2 could be induced by estrogen receptor (ER) [1,26]. However, the regulation and degradation of EglNs, especial EglN2, are not well evaluated in prostate cancer setting.
The ubiquitin-proteasome system governs a variety of biological processes and disease conditions, such as cell cycle, apoptosis and cancer [27]. Cullin-Ring complexes are the largest family in this system. Notably, SPOP, a Cullin 3-based E3 ubiquitin ligase, has been shown to participate in diverse cellular processes and plays tumor suppressive and oncogenic roles in prostate cancer and renal carcinoma by targeting different substrates for ubiqutination-mediated proteolysis, respectively [28–30]. Recently, SPOP has drawn more attention due to its frequent mutations occurring in prostate cancers [31], and its characterized downstream oncogenic targets including AR, TRIM24 and ERG [32–34]. Therefore, it is of great value to identify novel substrates of SPOP to further explore its functions as a tumor suppressor in prostate cancer. In this study, we reported that EglN2, but not EglN1 and EglN3, was recognized and destructed by SPOP in a degron-dependent manner, and the aberrant accumulation of EglN2, induced by amplification of AR or loss-of-function of SPOP, could facilitate prostate cancer growth.
Material and methods
Bioinformatics analysis
The correlation of EglN2 expression with the overall survival of tumor patients were analyzed via PrognoScan (www.prognoscan.org) [35] and Kaplan–Meier Plotter (Kmplot.com) [36] tools. The detail information of the databases was listed in the figure legends.
Cell culture and transfection
HeLa, HEK293, HEK293T, HEK293FT were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies, CA) containing 10% fetal bovine serum (FBS) (Gibico), 100 units of penicillin and 100 mg/ml streptomycin. Prostate cancer cell line PC3, RV1, C4-2 and LNCaP were maintained in RPMI1640 (Life Technologies, CA) supplemented with 10% FBS. Spop knockout and counterpart mouse embryonic fibroblasts (MEFs) as gifts were obtained from Dr. Nicholas Mitsiades (Baylor College of Medicine, Houston, TX). Cell transfection was performed using Lipofectamine and Plus reagents, as described previously [33]. For viral transduction experiments, lentivirus or retrovirus constructs, along with helper plasmids (i.e., GAG-pol and VSV-G) were co-transfected into HEK93FT cells as previously described [37]. Medium with progeny virus from transfected cells was collected every 24 h for 2 days, and then filtered with 0.45 μm filters (Millipore) and freshly used to infect cancer cells overnight in the presence of 4 μg/ml Polybrene (Sigma–Aldrich). After infection, the cells were selected with 1 μg/ml puromycin (Sigma–Aldrich) for 72 h to eliminate the uninfected cells before harvesting the whole cell lysates for the subsequent biochemical assays.
Plasmids construction
Constructs of Flag-EglN1, Flag-EglN2, Flag-EglN3 and sh-EglN2 were kind gifts from Dr. William Kaelin (Dana-Farber Cancer Institute, Boston, MA). pCMV-GST-SPOP, pGEX-4T-SPOP, pCDNA3-HA-SPOP, pCDNA3-HA-AR were described previously [33]. Myc-Cullins constructs and shRNA against Cullin3 were gifts from Dr. J. DeCaprio (Dana-Farber Cancer Institute, Boston, MA). Flag-KLHL2 was a gift from Dr. Shinichi Uchida (Tokyo Medical and Dental University, Tokyo, Japan). Flag-Keap1 and Flag-KLHL12 were purchased from Addgene. Various EglN2 and SPOP mutants were generated utilizing the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instruction. Details of plasmid constructions are provided upon request.
Antibodies
All antibodies were used at a 1:1000 dilution in 5% non-fat milk for western blot. Anti-EglN2 (NB100-310) and anti-EglN3 (NB100-139) antibodies were obtained from Novus biological. Anti-SPOP (16750-1-AP) antibody was purchased from Proteintech. Anti-EglN1 (4835), anti-Cullin 3 (2759), anti-GST (2625), anti-GFP (2956), anti-AR (5153) and anti-Myc-Tag (2278) antibodies were purchased from Cell Signaling. Polyclonal anti-HA (SC-805) antibody was purchased from Santa Cruz. Monoclonal anti-Flag (F-3165, clone M2), anti-Tubulin (T-5168), anti-Vinculin (V-4505) antibodies, anti-Flag agarose beads (A-2220), anti-HA agarose beads (A-2095), peroxidase-conjugated anti-mouse secondary antibody (A-4416) and peroxidase-conjugated anti-rabbit secondary antibody (A-4914) were purchased from Sigma. Monoclonal anti-HA (MMS-101P) antibody was purchased from Covance.
Immunoblots (IB) and immunoprecipitations (IP)
Cell lysates were collected in EBC buffer (50 mM Tris pH 7.5, 120 mM NaCl, 0.5% NP-40) supplemented with protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (phosphatase inhibitor cocktail set I and II, Calbiochem). The protein concentrations of lysates were measured by the Beckman Coulter DU-800 spectrophotometer using the Bio-Rad protein assay reagent. Same amounts of whole cell lysates were resolved by SDS-PAGE and immunoblotted with indicated antibodies. For immunoprecipitation, cells were treated with MG132 (10 μM) overnight after transfection 20 h, 1000 μg lysates were incubated with the indicated antibody (1–2 μg) for 3–4 h at 4°C followed by 1 h incubation with addition of carrier beads. Immunoprecipitants were washed five times with NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5% NP-40), then were resolved by SDS-PAGE and immunoblotted with indicated antibodies.
Immunohistochemistry (IHC) and human tissue microarray
The human tissue microarrays were purchased from Outdo Biotech Company (HProA100PG02, Shanghai, China). Total 100 cases of prostate tissues, including 3 cases of normal tissues, 2 cases of paracancerous tissues and 95 cases of prostate cancer tissues with clinical information, were arranged into one tissue array block and stained with human EglN2 (NB100-310, dilution 1:200) for IHC assay as previously reported [38]. Immuno-stained sections were scanned using panoramic viewer (Budapest, Hungary). The staining of cells was assessed according to the intensity (0 point: no; 1 point: weak; 2 points: moderate; 3 points: strong) and the proportion of positive cells (0 point: no; 1 point: 1–10%; 2 points: 11–50%; 3 points: 51–80%; 4 points: 81–100%). The slides were examined independently by two pathologists blinded to the clinical and the pathologic information.
RNA extraction and real-time PCR (q-PCR) analysis
Total RNA was extracted using RNeasy Mini Kit (Qiagen) according to the manufacturer’s instruction, and quantified by a spectrophotometer (NanoDrop ND-1000, Thermo Scientific). First-strand cDNA was synthesized according to iScript™ Reverse Transcription Supermix (Bio-rad) for RT-PCR. EglN2 primers were synthesized as previously reported [33]: EglN2-5′ 5-AACATCGAGCCACTCTTTGAC, EglN2-3′, 5-TCCTTGGCATCAAAATACC. GAPDH primers were used as intrinsic control as previous reported [33]. The cDNA templates and ABI Taqman Fast Universal PCR Master Mix (4352042, Life technologies) were mixed together and the real time PCR reactions were performed with the ABI-7500 Fast Real-time PCR system. Each experiment with triple-wells was repeated three times independently.
Purification of GST-tagged proteins
Recombinant GST-SPOP proteins were generated by transforming the BL21 (D3) E. coli strain with pGEX-SPOP or pGEX-4T (empty vector control). The bacterial cultures were grown at 37°C until an O.D. of 0.8, then was induced for 12–16 h using 0.1 mM IPTG at 16°C with vigorous shaking. Recombinant proteins were purified from harvested pellets. Pellets were re-suspended in 5 ml EBC buffer and sonicated to harvest supernatant, which was incubated with 50% Glutathione-sepharose slurry (GE) for 3 h at 4°C. The Glutathione beads were washed 3 times with PBS buffer (1 ml/wash) and stored at 4°C in PBS buffer or eluted with elution buffer. Recovery and yield of the proteins was confirmed by analyzing 10 μl of beads by coomassie blue staining, and quantified against BSA standards.
Protein degradation analysis and protein half-life studies
Cells cultured in 6-cm dishes were transfected with 0.1 μg Flag-EglNs, along with different doses of GST-SPOP (from 10 to 200 ng). For half-life studies, 100 μg/ml cycloheximide (CHX, Sigma–Aldrich) was added to the cells after 36 h of post transfection. At the indicated time points, cells were harvested and protein concentrations were measured. Total 30 μg of the indicated whole cell lysates were separated by SDS-PAGE and protein levels were measured by immunoblot analysis.
In vivo ubiquitination assays
His-ubiquitin along with Flag-EglN2 and CMV-GST-SPOP were transfected into cells. Thirty-six hours after transfection, cells were treated with MG132 (20 μM) for 6 h, and then were lysed in buffer A (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, and 10 mM imidazole [pH 8.0]) and subjected to sonicate. After high-speed centrifuged, the supernatants were incubated with nickel-beads (Ni-NTA) (Qiagen) for 3 h at room temperature. The products were washed twice with buffer A, twice with buffer A/TI (1 volume buffer A and 3 volumes buffer TI), and one time with buffer TI (25 mM Tris–HCl and 20 mM imidazole [pH 6. 8]). The pull-down proteins were resolved in 8% SDS-PAGE for immunoblot analysis.
MTS assays
1000 cells/well were seeded into 96-well plates and maintained in 100 μl DMEM containing 10% FBS. 20 μl CellTiter 96 AQueous One Solution Reagent (Promega) was added to each well at indicated time point. After incubating at 37°C for 1 h, the absorbance at 490 nm was measured and survival rate was quantified. Each experiment with triple-wells was repeated three times independently.
Colony formation and soft agar assays
Cells were seeded into 6-well plates (300 or 600 cells/well) and left for 8–12 days until formation of visible colonies. Colonies were washed with PBS and fixed with 10% acetic acid/10% methanol for 20 min, then stained with 0.4% crystal violet in 20% ethanol for 20 min. After staining, the plates were washed and air-dried, and colony numbers were counted. Three independent experiments were performed to generate the standard error of the difference (SED). The anchorage-independent cell growth assays were performed as described previously (34). Briefly, the assays were preformed using 6-well plates where the solid medium consists of two layers. The bottom layer contains 0.8% noble agar and the top layer contains 0.4% agar suspended with 1 × 104 or 3 × 104 cells. 500 μl complete DMEM medium was added every 7 days to keep the top layer moisture and 4 weeks later the cells were stained with iodonitrotetrazolium chloride (1 mg/ml) (sigma I10406) for colony visualization and counting. Three independent experiments were performed to generate the standard error of the difference (SED).
Mouse xenograft assays
Mouse xenograft assays were performed as described previously [39]. Briefly, 2 × 106 PC3 cells stably knockdown of EglN2 with different shRNAs were injected into the flank of nude mice (NCRNU-M-M from Taconic, 4–5 weeks of age). Tumor size was measured every three days with a caliper, and the tumor volume was determined with the formula: L × W2 × 0.52, where L is the longest diameter and W is the shortest diameter. After 12 days, mice were sacrificed and xenografted solid tumors were dissected, then tumor weights were measured and recorded post-necropsy.
Statistics
Differences between control and test conditions were evaluated by Student’s t test or one-way analysis of variance (ANOVA) test using the SPSS 11.5 Statistical Software. Values of p < 0.05 were considered statistically significant.
Results
EglN2 is aberrantly expresed in human prostate cancer tissues
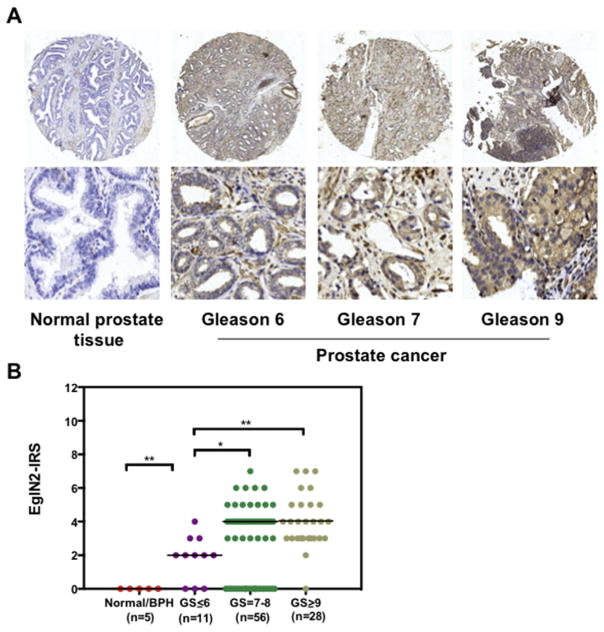
While EglN2, serving as a putative oncogene, participates in breast cancer cell proliferation [1] and colon cancer cell drug resistance [21], its physiological roles in other tumors are not well investigated yet. To this end, utilizing bioinformatics tools, such as Prognoscan [35] and Kappa–Meier plot [36], we revealed that high expression of EglN2 was markedly correlated with poor clinical outcome in diverse of cancer patients, including glioma, lung and gastric cancers (Fig. S1A–D). However, so far there is no any clues indicating the roles of EglN2 in prostate cancer setting. In order to uncover the potential function of EglN2 in prostate cancer, we initially assayed EglN2 protein levels in human prostate tissues by immunohistochemistry (IHC) assay, and found that EglN2 was aberrantly expressed in prostate cancers compared with normal or benign prostate tissues (Fig. 1A). Importantly, prostate cancers with high Gleason score (Gleason > 7) represented a higher expression of EglN2 (Fig. 1A and B), indicating that EglN2 is dysregulated in and positively correlated with advanced prostate cancers.
Fig. 1. EglN2 is aberrantly expressed in human prostate cancer tissues.
(A) Representative IHC staining images of EglN2 in prostate tissues. The magnification of upper and lower images is 10× and 200×, respectively. The EglN2 staining was assessed, calculated, plotted and analyzed in (B).*p < 0.05, **p < 0.01 (t test).
EglN2 promotes prostate cancer growth in cells and in vivo
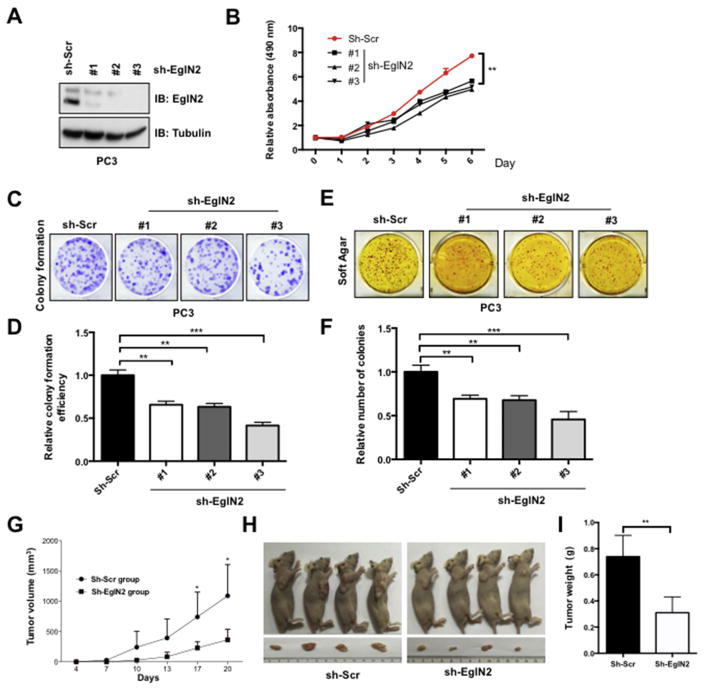
In order to evaluate the physiological roles of EglN2 in prostate cancer, we deleted endogenous EglN2 with independent sets of shRNA constructs in prostate cancer cell lines, including PC3 and RV1 (Fig. 2A and Fig. S2A). Consistent with the previous finding in breast cancer, deletion of EglN2 could significantly attenuate cell proliferation (Fig. 2B and Fig. S2B), colony formation (Fig. 2C and D, and Fig. S2C and D) and anchorage-independent growth (Fig. 2E and F) in prostate cancer. More importantly, EglN2 deficiency dramatically decreased PC3 cells tumor formation in xenograft mouse model (Fig. 2G–I). Conversely, stably expressed EglN2 could apparently enhance prostate cancer cell biological behaviors including proliferation (Fig. S3A,B,G and H), colony formation (Fig. S3C,D,I and J) and anchorage-independent growth (Fig. S3E and F). These findings together support the notion that dysregulation of EglN2 plays an important role in prostate tumorigenesis.
Fig. 2. Knockdown of endogenous EglN2 decreases prostate cancer growth.
(A) Immunoblot (IB) analysis of whole cell lysates (WCLs) derived from PC3 cells lentivirally infected with scramble (sh-Scr) or multiple independent EglN2 shRNAs (sh-EglN2). Infected cells were selected with puromycin (1 μg/ml) for 72 h before harvesting. (B) Cell viabilities were measured at indicated time point with MTS assays, and the relative cell viability was normalized and quantified to sh-Scr cells in the time of day 0 (Mean ± SD, n = 3). **p < 0.01 (t test). (C–F) Colony formation (C) and soft agar (E) assays were carried out with the cell line generated in (A). Relative colony numbers were further quantified for colony formation (D) and soft agar (F) (Mean ± SD, n = 3). **p < 0.01, ***p < 0.001 (t test). (G–I) Stable cell lines generated in (A) were used for xenograft assays, and tumor volume were monitored every three days (G). The tumors were dissected (H) and were weighed in (I) (mean ± SD, n = 6). *p < 0.05, **p < 0.01 (t test).
Cul3SPOP E3 ligase degrades EglN2
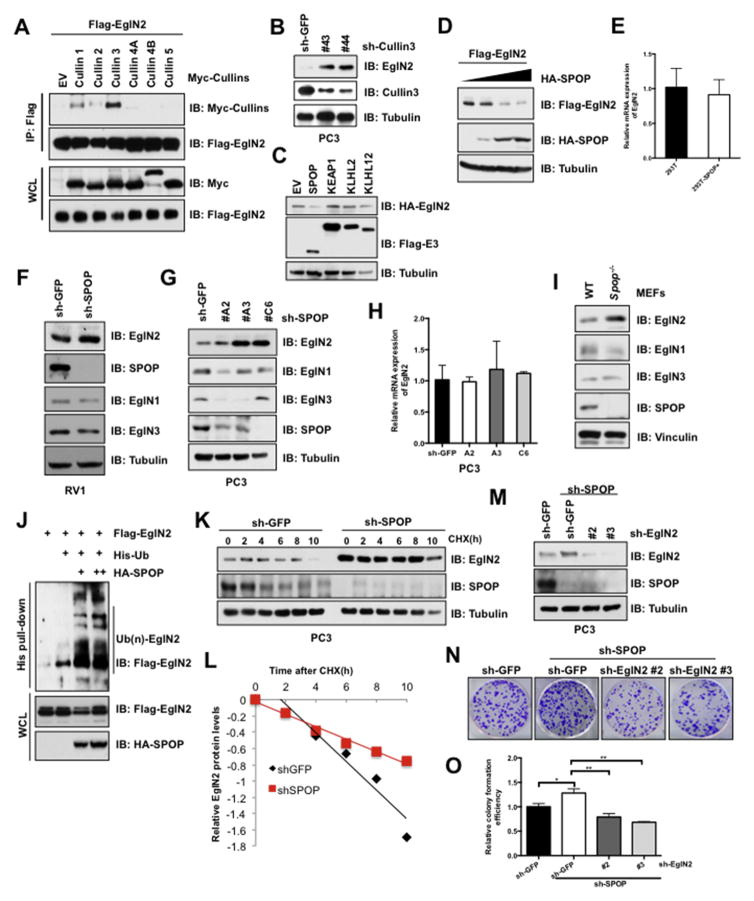
Next, to uncover the underlying regulatory mechanisms of EglN2 in prostate cancer, we initially try to identify the E3 ligase governing turnover of EglN2. Given the fact that Cullin-based E3 ligases composed of the largest family of E3 ligases, we then firstly screened a panel of Cullin scaffolding proteins to identify the potential E3 complex for EglN2. Interestingly, Cullin 3, to a lesser extent of Cullin 1, but not other Cullin family proteins, was validated to interact with EglN2 in cells (Fig. 3A). Consistently, deletion of endogenous Cullin 3 could largely increase EglN2 protein abundance (Fig. 3B). It is well-characterized that Cullin3-based BTB complexes including but not limited to SPOP, KEAP1, KLHL2 and KLHL12 [40]. However, only SPOP was observed to reduce EglN2 protein levels in a dose-dependent manner (Fig. 3C and D), instead of altering EglN2 mRNA levels (Fig. 3E). Consistent with these findings, deletion of SPOP in prostate cancer RV1 cells could increase the endogenous EglN2, but not EglN1 or EglN3, abundance (Fig. 3F). To further exclude the possibly indirect effect of SPOP-targeting other substrates, such as ER and AR [32,41], we revealed that SPOP-deficiency could largely increase EglN2 protein instead of mRNA levels in AR-null PC3 cells (Fig. 3G and H).
Fig. 3. Cul3SPOP E3 ligase complex interacts with and degrades EglN2.
(A) IB analysis of immunoprecipitates (IPs) and WCLs derived from 293T cells transfected with the indicated Cullin encoding constructs. Cells were treated with MG132 (10 μM) for 10 h before harvesting. (B) IB analysis of WCLs derived from PC3 cells lentivirally infected with the Cullin3 shRNA constructs. (C and D) IB analysis of WCLs derived from 293T cells transfected with indicated constructs. (E) Real-time PCR (qPCR) analysis to examine EglN2 mRNA levels in 293T cells transfected with indicated SPOP constructs (mean ± SD, n = 3). (F and G) IB analysis of WCLs derived from RV1 (F) and PC3 (G) cells lentivirally infected with the SPOP shRNA construct. (H) The mRNA levels of EglN2 in (G) were measured with qPCR (mean ± SD, n = 3). (I) IB analysis of WCLs derived from Spop−/− and counterpart MEFs. (J) IB analysis of WCLs and His pull-down products derived from 293T cells transfected with indicated constructs and treated with MG132 (10 μM) 10 h. (K and L) IB analysis of PC3 cells stably infected with the control and SPOP shRNA construct. Where indicated, Cycloheximide (CHX) (100 μg/ml) was added, and cells were harvested at the indicated time points. Relative EglN2 protein abundance in (K) was quantified by Image J and plotted in (L). (M) IB analysis of PC3 cells lentivirally infected with indicated constructs, and the resulting cells were performed colony formation assays (N). The relative colony numbers in (N) were further quantified in (O) (mean ± SD, n = 3). **p < 0.05, **p < 0.01 (t test).
In further support of SPOP as the upstream E3 ligase for EglN2, we observed that the endogenous EglN2, but not EglN1 and EglN3, were induced in Spop−/− MEFs compared with counterpart cells (Fig. 3I). In keeping with this finding, EglN2, but not EglN1 or EglN3, could be physically associated with mammalian expressed GST-SPOP (Fig. S4A), which was further validated by in vitro pull-down assays with the bacterially purified recombinant GST-SPOP (Fig. S4B). Conceivably, SPOP could promote EglN2 ubiqutination in a dose-dependent manner in cells (Fig. 3J), and subsequently deletion of SPOP could sustain the half-life of EglN2 (Fig. 3K and L). Conversely, ectopic expression of SPOP shortened the half-life of EglN2 (Fig. S4C and S4D). Importantly, deletion of EglN2 partially compromised SPOP knockdown-induced PC3 cell colony formation (Fig. 3M–O), indicating that negative regulation of EglN2 at least in part contributed to tumor suppressive functions of SPOP in prostate cancer.
SPOP induces EglN2 degradation in a degron-dependent manner
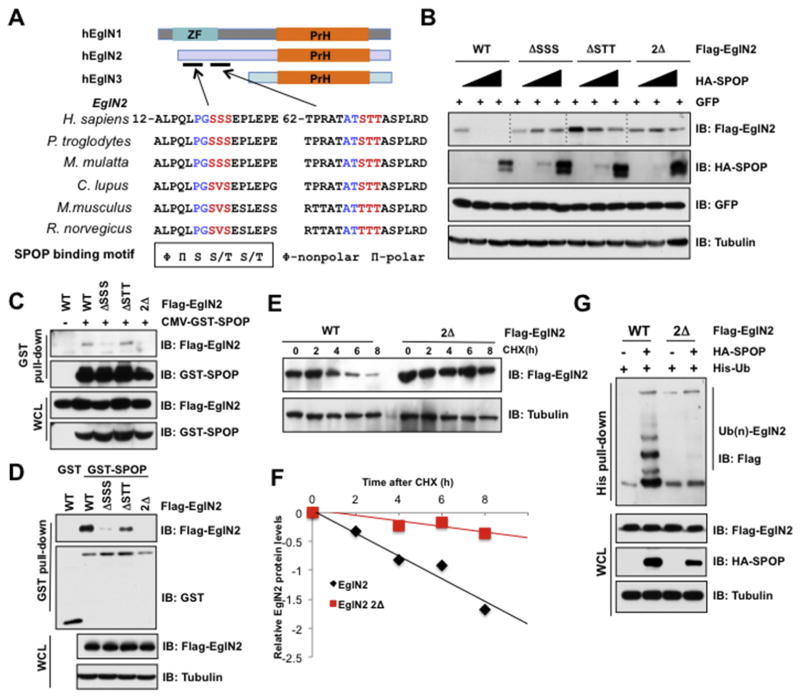
It is previously reported that majority of SPOP substrates share a consensus SPOP-binding motif as Φ-Π-S-S/T-S/T (Φ-nonpolar, Π-polar) [42]. Thus, we analyzed EglN protein sequence and found two putative SPOP binding degrons in the N-terminal of EglN2, but not in EglN1 and EglN3, which are evolutionarily conserved across different species (Fig. 4A). To reveal whether these degrons are indispensable for SPOP to reorganize EglN2, we deleted degron 1 (ΔSSS), and degron 2 (ΔSTT) of EglN2 individually or in combination (termed as 2Δ). The results showed that the deletion of degron 1, and to a lesser extent of degron 2, largely impaired SPOP-mediated degradation of EglN2 (Fig. 4B). Significantly, two degron deletions almost totally abolished the ability of SPOP to degrade EglN2 in cells (Fig. 4B). In keeping with these results, deletion of degron 1 individually, or in combination with degron 2, could attenuate the interaction of EglN2 with SPOP in cells or in vitro (Fig. 4C and D). These findings provide the evidence that degron 1 is the major functional degron responsible for SPOP physiological association with and degradation of EglN2. Biochemically, the half-life of EglN2-2Δ (deleted both degron 1 and 2) was significantly sustained compared with that of EglN2-WT (Fig. 4E and F). Moreover, the ubiquitination of EglN2-2Δ was diminished compared with EglN2-WT in cells (Fig. 4G). Together, these data suggest that SPOP mediates the ubiquitination and subsequent destruction of EglN2 in a degron-dependent manner.
Fig. 4. SPOP interacts with and degrades EglN2 in a degron-dependent manner.
(A) A schematic illustration of the domain structures of EglNs and the alignment of binding motif of SPOP in EglN2 across different species. (B) IB analysis of WCLs derived from 293T cells transfected with the indicated constructs. (C) IB analysis of WCLs and GST pull-down products derived from 293T cells transfected with the indicated plasmid. Cells were treated with MG132 (10 μM) for 10 h before harvesting. (D) GST pull-down assay was performed with bacterially purified recombinant GST-SPOP proteins (GST as negative control) and the WCLs derived from 293T cells transfected with indicated EglN2 mutants. (E and F) IB analysis of WCLs derived from 293T cells infected with the indicated constructs. Where indicated, Cycloheximide (CHX) (100 μg/ml) was added after transfection 30 h, and the resulting cells were harvested at the indicated time points. Relative EglN2 protein abundance in (E) was quantified by Image J and plotted in (F). (G) IB analysis of WCLs and His pull-down products derived from 293T cells transfected with constructs encoding the indicated proteins and treated with MG132 (10 μM) for 10 h before harvesting.
Patients-associated SPOP mutants are incapable to degrade EglN2
SPOP, as the major frequently mutated gene in prostate cancer, plays pivotal roles for prostate tumorigenesis and metastasis [43]. Most of the somatic mutations of SPOP, such as Y87C, F102C, W131G and F133V, occurred in MATH domain of SPOP, where is indispensable for SPOP binding with substrates (Fig. 5A) [30]. As an ubiquitin substrate of SPOP, EglN2 indeed disassociated with MATH-deleted SPOP (ΔMATH-SPOP) in cells (Fig. 5B). Conceivably, prostate cancer patients-associated SPOP mutants failed to interact with, and subsequently destruct EglN2 (Fig. 5C–E). Moreover, the ubiquitination of EglN2 was largely compromised by SPOP mutants compared with SPOP-WT (Fig. 5F). These data indicate that patient-associated SPOP mutants are deficient in promoting ubiquitination and destruction of EglN2, therefore partially providing a molecular mechanism to explain the aberrant accumulation of EglN2 in prostate cancer (Fig. 5G).
Fig. 5. SPOP mutants are impaired to interact with and degrade EglN2.
(A) A schematic illustration of SPOP domains and prostate cancer-associated mutations. (B) IB analysis of WCLs and IPs derived from 293T cells transfected with the indicated plasmids. The resulting cells were treated with MG132 (10 μM) for 10 h before harvesting. (C) In vitro GST pull-down assay demonstrated SPOP mutations impaired to interact with EglN2. (D) IB analysis of WCLs derived from 293T cells transfected with the indicated constructs. (E) IB analysis of LNCaP cells stably infected with indicated constructs. (F) IB analysis of WCLs and His pull-down products derived from 293T cells transfected with constructs encoding indicated proteins and treated with MG132 (10 μM) for 10 h before harvesting. (G) Schematic diagrams illustrate how SPOP regulates EglN2. Specifically, WT-SPOP could interact with and degrade EglN2, while loss-of-function mutations SPOP impaired its capability to destruct EglN2, importantly, SPOP could not recognize and destruct degrons deletion form of EglN2.
Androgen receptor transcriptionally up-regulates EglN2
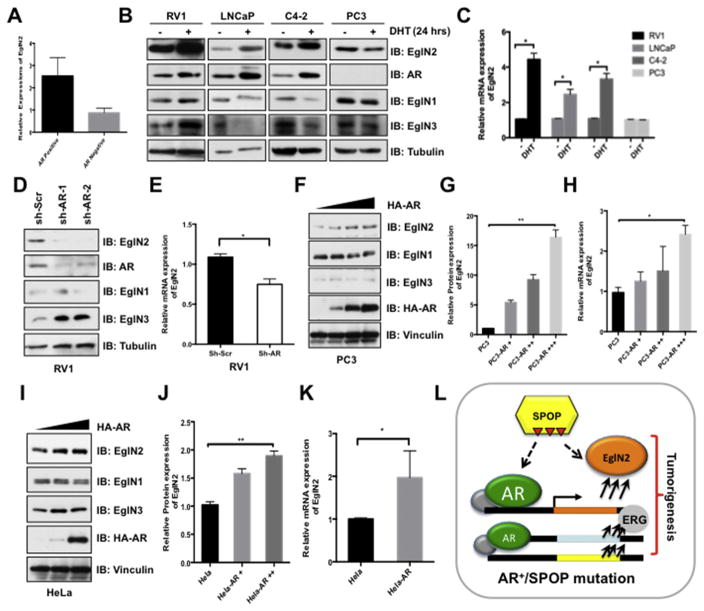
Since we have demonstrated that majority of prostate cancer tissues represented abnormal expression of EglN2 (Fig. 1), thus, there must be more common pathways involving in EglN2 regulation except loss-of-function mutations of SPOP in prostate cancers [43]. A previous report showed that estrogen receptor could transcriptionally regulate EglN2 expression in breast cancer setting [1]. So we explore a possible link between the hormone receptor AR and the regulation of EglN2 in prostate cancer setting. To this end, we analyzed the Gene Expression Omnibus (GEO) database and found that the EglN2 mRNA levels were much higher in AR-positive prostate cell lines compared with that of the AR negative cells (Fig. 6A). Strikingly, dihydrotestosterone (DHT) could strongly induce EglN2 expression in both protein and mRNA levels in AR positive (RV1, LNCaP and C4-2), but not in AR-negative (PC3) prostate cancer cells (Fig. 6B and C). Specifically, silencing endogenous AR in AR-positive prostate cancer cell line RV1 could markedly decrease EglN2 levels, with minor effect on the expression of EglN1 and EglN3 (Fig. 6D and E). In contrast, ectopic expression of AR in AR-negative prostate cancer cell line PC3 or a cervical cancer cell line HeLa increased EglN2 expression both in protein and mRNA levels, in a dose-dependent manner (Fig. 6F–K). These findings coherently support the notion that AR, a hallmark and critical effector of prostate cancer proliferation and development [44], could transcriptionally up-regulate EglN2, contributing to the aberrant expression of EglN2 in prostate cancers (Fig. 6L).
Fig. 6. Androgen Receptor (AR) transcriptionally up-regulates EglN2 expression.
(A) GEO (GSE4016) database analysis of relative EglN2 mRNA expression in AR-negative and AR-positive prostate cancer cells. (B) Prostate cancer RV1, LNCaP, C4-2 and PC3 cells were cultured in charcoal-treated medium for 72 h, then were stimulated with or without DHT (10 nM) for another 24 h before harvesting for IB analysis. (C) The mRNA levels of EglN2 in (B) were measured with qPCR (mean ± SD, n = 3). *p < 0.05 (t test). (D and E) IB analysis of WCLs derived from RV1 cells lentivirally infected with scramble (sh-Scr) or independent AR shRNAs (sh-AR). Infected cells were selected with puromycin (1 μg/ml) for 72 h before harvesting (D). The mRNA levels of EglN2 in (D) were measured with qPCR (E) (mean ± SD, n = 3). *p < 0.05 (t test). (F–K) IB analysis of WCLs derived from PC3 (F) and HeLa (I) cells transfected with indicated AR constructs. The relative EglN2 protein abundance in (F, I) was quantified by Image J and normalized with control cells in (G, J). Furthermore, the mRNA levels of EglN2 in (F, I) were measured with qPCR (H, K) (mean ± SD, n = 3). *p < 0.05 (t test). (L) Schematic diagrams illustrate the regulation of EglN2 in prostate cancer. Loss-of-function mutations in SPOP impaire its capable to destruct its downstream targets, including AR and EglN2, in turn, the accumulation of AR transcriptionally up-regulates EglN2. Ultimately, the elevated EglN2 cooperates with other oncogenic genes together contributing to prostate tumorigenesis.
Discussion
The primary function of EglNs is assigned to tightly regulate HIFα signals. In physio- or pathological conditions of lacking oxygen, inactive EglNs lead to the accumulation of HIFα, which in turn dramatically induces the expression of EglNs, especially the expression of EglN3 [45]. On the other hand, in VHL-deficient conditions, such as in clear cell renal carcinoma (ccRCC), the hydroxylated HIFα could not be destructed by the defective pVHL, leading to the increase of EglNs levels [46]. However, the regulation and degradation of EglNs in normal condition or other tissue-context, especially in prostate cancer are not well investigated. In this study, we demonstrated that EglN2, aberrantly expressed in prostate cancer patients, functions as an oncogene cooperating with other oncogenes to promote prostate tumor growth. Moreover, similarly to the regulation of EglN2 by estrogen in breast cancer cells [26], EglN2 might be also transcriptionally induced by AR in prostate cancer. Hence, in AR amplification prostate cancer cells, EglN2 expression is up-regulated and possibly synergizes with other AR target genes such as ERG, TRIM24 to facilitate prostate cancer initiation and progression (Fig. 6L).
Loss-of-function mutations of SPOP frequently occur in prostate cancers. In addition to AR, SPOP degrades multiple other oncogenic substrates, including but not limited to AR target proteins such as ERG and TRIM24 to exert its tumor suppressor role [32,34]. Here we show that SPOP also can degrade another AR target protein, EglN2, in a degron-dependent manner (Fig. 3), and EglN2 deletion could partially decrease SPOP deficiency-induced prostate cancer colony formation (Fig. 3M–O). Notably, the mutations of SPOP in prostate cancer patients, restrain its capability to degrade prostate oncoproteins, such as AR, TRIM24 and EglN2. At the same time, the accumulation of AR leads to amplification of its target genes transcriptionally, including EglN2, ERG and TRIM24, subsequently facilitate prostate cancer growth and progression (Fig. 6L). Together, in this study we not only highlight the oncogenic role of EglN2 in prostate cancer, but also reveal the upstream regulatory mechanisms of EglN2 by SPOP-mediated degradation and AR-mediated transcriptional up-regulation.
Supplementary Material
Acknowledgments
We thank Jinfang Zhang, Bin Wang and other Wei lab members for critical reading of the manuscript and helpful discussions. J.G. is an NRSA T32 trainee and supported by 5T32HL007893-17. W.W. is an LLS research scholar. This study was partially supported by the National Natural Science Foundation of China (81272491, 81572960).
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.canlet.2017.01.003.
Footnotes
Conflict of interest
All authors declare no conflict of interests.
References
- 1.Zhang Q, Gu J, Li L, Liu J, Luo B, Cheung HW, et al. Control of cyclin D1 and breast tumorigenesis by the EglN2 prolyl hydroxylase. Cancer Cell. 2009;16:413–424. doi: 10.1016/j.ccr.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng X, Zhai B, Koivunen P, Shin SJ, Lu G, Liu J, et al. Prolyl hydroxylation by EglN2 destabilizes FOXO3a by blocking its interaction with the USP9x deubiquitinase. Genes Dev. 2014;28:1429–1444. doi: 10.1101/gad.242131.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. HIF prolylhydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 5.Hirsila M, Koivunen P, Gunzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 6.Ivan M, Haberberger T, Gervasi DC, Michelson KS, Gunzler V, Kondo K, et al. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc Natl Acad Sci U S A. 2002;99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaelin WG., Jr Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb Symp Quant Biol. 2011;76:335–345. doi: 10.1101/sqb.2011.76.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science (New York, NY) 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 9.Semenza GL. HIF-1 and human disease: one highly involved factor. Genes Dev. 2000;14:1983–1991. [PubMed] [Google Scholar]
- 10.Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 11.Wenger RH. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J Off Publ Fed Am Soc Exp Biol. 2002;16:1151–1162. doi: 10.1096/fj.01-0944rev. [DOI] [PubMed] [Google Scholar]
- 12.Semenza GL. Targeting HIF-1 for cancer therapy. Nature Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 13.Lee DC, Sohn HA, Park ZY, Oh S, Kang YK, Lee KM, et al. A lactate-induced response to hypoxia. Cell. 2015;161:595–609. doi: 10.1016/j.cell.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Guo J, Chakraborty AA, Liu P, Gan W, Zheng X, Inuzuka H, et al. pVHL suppresses kinase activity of Akt in a proline-hydroxylation–dependent manner. Science (New York, NY) 2016;353:929–932. doi: 10.1126/science.aad5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moser SC, Bensaddek D, Ortmann B, Maure JF, Mudie S, Blow JJ, et al. PHD1 links cell-cycle progression to oxygen sensing through hydroxylation of the centrosomal protein Cep192. Dev Cell. 2013;26:381–392. doi: 10.1016/j.devcel.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cummins EP, Berra E, Comerford KM, Ginouves A, Fitzgerald KT, Seeballuck F, et al. Prolyl hydroxylase-1 negatively regulates IkappaB kinase-beta, giving insight into hypoxia-induced NFkappaB activity. Proc Natl Acad Sci U S A. 2006;103:18154–18159. doi: 10.1073/pnas.0602235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haase VH. Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev. 2013;27:41–53. doi: 10.1016/j.blre.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briggs KJ, Koivunen P, Cao S, Backus KM, Olenchock BA, Patel H, et al. Paracrine induction of HIF by glutamate in breast cancer: EglN1 senses cysteine. Cell. 2016;166:126–139. doi: 10.1016/j.cell.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henze AT, Riedel J, Diem T, Wenner J, Flamme I, Pouyseggur J, et al. Prolyl hydroxylases 2 and 3 act in gliomas as protective negative feedback regulators of hypoxia-inducible factors. Cancer Res. 2010;70:357–366. doi: 10.1158/0008-5472.CAN-09-1876. [DOI] [PubMed] [Google Scholar]
- 21.Deschoemaeker S, Di Conza G, Lilla S, Martin-Perez R, Mennerich D, Boon L, et al. PHD1 regulates p53-mediated colorectal cancer chemoresistance. EMBO Mol Med. 2015;7:1350–1365. doi: 10.15252/emmm.201505492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu J, Menzies K, Freeman RS, Taubman MB. EGLN3 prolyl hydroxylase regulates skeletal muscle differentiation and myogenin protein stability. J Biol Chem. 2007;282:12410–12418. doi: 10.1074/jbc.M608748200. [DOI] [PubMed] [Google Scholar]
- 23.Tennant DA, Gottlieb E. HIF prolyl hydroxylase-3 mediates alpha-ketoglutarate-induced apoptosis and tumor suppression. J Mol Med (Berlin, Germany) 2010;88:839–849. doi: 10.1007/s00109-010-0627-0. [DOI] [PubMed] [Google Scholar]
- 24.del Peso L, Castellanos MC, Temes E, Martin-Puig S, Cuevas Y, Olmos G, et al. The von Hippel Lindau/hypoxia-inducible factor (HIF) pathway regulates the transcription of the HIF-proline hydroxylase genes in response to low oxygen. J Biol Chem. 2003;278:48690–48695. doi: 10.1074/jbc.M308862200. [DOI] [PubMed] [Google Scholar]
- 25.Metzen E, Stiehl DP, Doege K, Marxsen JH, Hellwig-Burgel T, Jelkmann W. Regulation of the prolyl hydroxylase domain protein 2 (phd2/egln-1) gene: identification of a functional hypoxia-responsive element. Biochem J. 2005;387:711–717. doi: 10.1042/BJ20041736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mak P, Chang C, Pursell B, Mercurio AM. Estrogen receptor beta sustains epithelial differentiation by regulating prolyl hydroxylase 2 transcription. Proc Natl Acad Sci U S A. 2013;110:4708–4713. doi: 10.1073/pnas.1221654110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hershko A, Ciechanover A, Varshavsky A Basic medical research award. The ubiquitin system. Nat Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 28.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 30.Mani RS. The emerging role of speckle-type POZ protein (SPOP) in cancer development. Drug Discov Today. 2014;19:1498–1502. doi: 10.1016/j.drudis.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.An J, Wang C, Deng Y, Yu L, Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6:657–669. doi: 10.1016/j.celrep.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gan W, Dai X, Lunardi A, Li Z, Inuzuka H, Liu P, et al. SPOP promotes ubiquitination and degradation of the ERG oncoprotein to suppress prostate cancer progression. Mol Cell. 2015;59:917–930. doi: 10.1016/j.molcel.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Groner AC, Cato L, de Tribolet-Hardy J, Bernasocchi T, Janouskova H, Melchers D, et al. TRIM24 is an oncogenic transcriptional activator in prostate cancer. Cancer Cell. 2016;29:846–858. doi: 10.1016/j.ccell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorand RD, Nthale J, Myers JT, Barkauskas DS, Avril S, Chirieleison SM, et al. Cdk5 disruption attenuates tumor PD-L1 expression and promotes anti-tumor immunity. Science (New York, NY) 2016;353:399–403. doi: 10.1126/science.aae0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo J, Kim D, Gao J, Kurtyka C, Chen H, Yu C, et al. IKBKE is induced by STAT3 and tobacco carcinogen and determines chemosensitivity in non-small cell lung cancer. Oncogene. 2013;32:151–159. doi: 10.1038/onc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Inuzuka H, Gao D, Finley LW, Yang W, Wan L, Fukushima H, et al. Acetylation-dependent regulation of Skp2 function. Cell. 2012;150:179–193. doi: 10.1016/j.cell.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo JP, Shu SK, He L, Lee YC, Kruk PA, Grenman S, et al. Deregulation of IKBKE is associated with tumor progression, poor prognosis, and cisplatin resistance in ovarian cancer. Am J Pathol. 2009;175:324–333. doi: 10.2353/ajpath.2009.080767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L, Su B, Sun W, Li W, Luo M, Liu D, et al. Twist1 promotes radio-resistance in nasopharyngeal carcinoma. Oncotarget. 2016 doi: 10.18632/oncotarget.12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genschik P, Sumara I, Lechner E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J. 2013;32:2307–2320. doi: 10.1038/emboj.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang P, Gao K, Jin X, Ma J, Peng J, Wumaier R, et al. Endometrial cancer-associated mutants of SPOP are defective in regulating estrogen receptor-alpha protein turnover. Cell Death Dis. 2015;6:e1687. doi: 10.1038/cddis.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geng C, He B, Xu L, Barbieri CE, Eedunuri VK, Chew SA, et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci U S A. 2013;110:6997–7002. doi: 10.1073/pnas.1304502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 45.Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- 46.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.