Abstract
ETS transcription factor ETV2 / Etsrp functions as a key regulator of embryonic vascular development in multiple vertebrates. However, its role in pathological vascular development has not been previously investigated. To analyze its role in tumor angiogenesis, we utilized a zebrafish xenotransplantation model. Using a photoconvertible kdrl:NLS-KikGR line we demonstrated that all tumor vessels originate from the existing embryonic vasculature by the mechanism of angiogenesis. Xenotransplantation of mouse B16 melanoma cells resulted in a significant increase in expression of the ETS transcription factors etv2 and fli1b expression throughout the embryonic vasculature. etv2 null mutants which undergo significant recovery of embryonic angiogenesis during later developmental stages displayed a strong inhibition of tumor angiogenesis. We utilized highly specific and fully validated photoactivatable morpholinos to inhibit Etv2 function after embryonic vasculogenesis has completed. Inducible inhibition of Etv2 function resulted in a significant reduction of tumor angiogenesis and inhibition of tumor growth. Furthermore, inducible inhibition of Etv2 function in fli1b mutant embryos resulted in even stronger reduction in tumor angiogenesis and growth, demonstrating that Etv2 and Fli1b have a partially redundant requirement during tumor angiogenesis. These results demonstrate the requirement for Etv2 and Fli1b in tumor angiogenesis and suggest that inhibition of these ETS factors may present a novel strategy to inhibit tumor angiogenesis and reduce tumor growth.
Keywords: angiogenesis, ETS transcription factors, tumor, zebrafish, xenograft
INTRODUCTION
Angiogenesis plays a key role in tumor progression and metastatic spread. Most solid tumors secrete proangiogenic factors that induce vascular growth to supply the growing tumor with oxygen and nutrients. It is thought that solid tumors cannot grow beyond 1–2 mm without angiogenesis that supports tumor growth [1]. Angiogenesis also enables tumor cells to spread to distant sites. Therefore inhibition of angiogenesis is thought to be one of the most promising strategies to stop tumor growth and metastasis. However, in most cases angiogenic inhibitors are insufficient to stop the tumor growth completely. Resistance to antiangiogenic inhibitors is frequently developed and can even lead to acceleration of metastatic growth [2]. It is apparent that alternative strategies for inhibiting tumor induced angiogenesis are necessary.
The zebrafish is an ideal model system for both angiogenesis and tumor formation because they are optically transparent, inexpensive to maintain and breed, develop externally and are readily amenable to genetic and pharmacological screening [3,4]. Transplantation of mammalian tumor cells into zebrafish (xenotransplantation) has been successfully used to study tumor progression and angiogenesis both of which can be imaged at high resolution [3]. Both two-day old zebrafish embryos and adult zebrafish have been used to model tumor-induced angiogenesis [5,6]. The embryonic zebrafish tumor model allows much faster evaluation of tumor inhibition strategies in a higher number of embryos compared with the mouse model.
ETS transcription factors comprise a family of more than 20 different homologs that share an ETS winged helix-turn-helix DNA binding domain. Several members of the ETS transcription factor family are expressed in the embryonic vasculature and have distinct roles during vascular development [7]. We and others have previously demonstrated that the ETS domain transcription factor Etsrp/Etv2 functions as a key regulator of embryonic vascular development. Inhibition of Etv2 function in zebrafish embryos results in nearly complete loss of vasculature [8,9]. Etv2 in association with FoxC transcription factors can directly initiate expression of multiple endothelial specific genes including VegfR2, therefore in the absence of Etv2 function endothelial cells cannot respond to Vegf signaling [10]. Etv2 function is highly evolutionary conserved. Similar to zebrafish, mouse Etv2 is required for embryonic and yolk sac vasculature formation. Etv2 mouse knockout embryos die due to the absence of yolk sac blood islands and embryonic vasculature [11,12]. Besides its early role in vasculogenesis, Etv2 is also involved in embryonic angiogenesis where it functions redundantly with a related transcription factor fli1b. Inducible inhibition of Etv2 function using photoactivatable morpholinos in fli1b−/− mutant background resulted in the inhibition of embryonic angiogenesis while vasculogenesis was not affected [7]. Etv2 is also known to play role in regenerative angiogenesis [13].
While Etv2 plays a critical role in embryonic and regenerative vascular development, its role in tumor angiogenesis has not been previously investigated. In this study, we analyzed the role of Etv2 in tumor angiogenesis using zebrafish xenotransplantation model. We demonstrate that Etv2 functions partially redundantly with Fli1b during tumor angiogenesis. These results identify Etv2 and Fli1b as critical regulators of tumor angiogenesis and suggest an alternative strategy to inhibit tumor angiogenesis and reduce tumor growth.
MATERIALS AND METHODS
Fish Lines and Embryos
The following transgenic and mutant lines were used for experiments: Tg(fli1a:EGFP)y1 (abbreviated further as fli1a:GFP) [14], Tg(kdrl:EGFP)s843 (abbreviated further as kdrl:GFP) [15], fli1btpl50Gt [7], etsrpy11 [9], Tg(kdrl:NLS-KikGR)hsc7 [16] and AB wild type. Embryos were incubated at 28.5°C prior to the tumor transplantation and at 32°C after transplantation. Embryos were staged as described previously [17]. Embryos were treated with 0.003% 1-phenyl-2-thiourea (PTU) to inhibit pigment formation for analyses performed beyond 24 hours post fertilization (hpf). etv2y11/y11 (henceforth referred to as “etv2−/−”) mutants were identified at 24 hpf by the absence of intersomitic vessels and defective development of the axial vessels as previously reported [9].
Tumor and Control Cell Xenotransplantation
Fluorescently labeled murine B16-F10:dsRed melanoma cells were kindly donated by Chengjian Zhao (UCLA), while A673 Ewing sarcoma cell line was kindly donated by Dr. Timothy Cripe (Nationwide Children’s Hospital). 293T cells were originally obtained from the American Type Culture Collection (ATC). A673 cell line was validated by STR genotyping at Dr. Cripe’s laboratory, while B16 cells were validated by immunostaining of injected zebrafish embryos for specific markers S100 and melanA. Cell stocks were frozen in liquid nitrogen and periodically used to start new passages. To label A673 cells with EGFP, lentivirus was produced using the second generation system Lv203_EGFP (Genecopiea, Rockville, MD). The virus was mixed with 8μg/ml polybrene and used to infect A673 cells. At 48 hour after infection, cells were treated with 2μg/ml puromycine to select infected cells. Cells were maintained in DMEM (ATCC # 30-2002 containing 4 mM L-glutamine, 4500 mg/L glucose, 1 mM sodium pyruvate, and 1500 mg/L sodium bicarbonate) with 10% FBS and 100 U/ml each of penicillin and streptomycin. Cultures were harvested 2 days after passage when cells were at 70 – 80% confluence. Cells were washed with phosphate buffer saline (PBS), treated 5–10 minutes with trypsin (100U/ml), and trypsin was neutralized with DMEM medium with 10% FBS. Cells were collected by centrifugation for 5 min at 1,200 rpm, and suspended in 10 μl of DMEM medium with 10% FBS. Cells were transplanted immediately after collection as reported previously. Xenografts were performed by laterally mounting 2 dpf zebrafish embryos for injection in 0.6% low melting temperature agarose containing 125 mg/l of Tricaine (Sigma, cat# A-5040). Injections were performed using a borosilicate glass injection needle with a 50 – 100 μm tip which was back-filled with cell suspension. A pneumatic microinjector (model PLI-100, Harvard Apparatus) set to deliver a 50 – 100 ms pulse at 10 PSI was used to inject a defined volume of cells (31 nl) across the yolk from the dorsal to ventral side and into the perivitelline space. Embryos were then removed from the agarose and raised at 32 °C.
kdrl:NLS-KikGR photoconversion
kdrl:NLS-KikGR-positive embryos were photoconverted at 48 hpf prior to xenotransplantation by exposing them for 15–30 seconds to the 405 nm wavelength under a compound microscope (Zeiss) equipped with 10x objective (NA=0.3).
Measurement of Tumor Growth and Vascularization and Statistical Analysis
Xenotransplanted zebrafish were imaged at 2–3 days after injection using an upright confocal microscope (Nikon A1) equipped with a 16x water immersion objective. Tumor volume was calculated based on the tumor surface area measured from the confocal Z-stack files using the Imaris software package (Bitplane). To calculate the tumor vessel length and the number of blood vessel branch points, vessels were traced manually and measurement points were selected in the Imaris software, which were then used to calculate the length of traced vessels. To account for the variability between separate experiments, all measured parameters within each experiment (tumor volume, blood vessel length and the number of branch points) were normalized to the average values of these parameters in control embryos injected with the tumor cells. Thus, for each experimental embryo, the ratio for each parameter over the average value obtained in the control embryos was calculated. Each experiment was repeated at least twice and 20–60 embryos per experimental group were analyzed in each experiment. Unpaired Student’s t-test was used to calculate p values between the analyzed groups.
Etv2 Caged Morpholino Injections and Photoactivation
Caged etv2 morpholino solution was prepared as described previously [18]. Briefly, etv2 caging strand (Supernova Life Sciences) designed against etv2 MO2 [8] was mixed in excess with etv2 MO2 at final concentrations of 500 μM and 37.5 μM, respectively, in nuclease-free water and protected from ambient light. The mixture was denatured for 30 minutes at 70°C and allowed to anneal overnight at 4°C. A total of 2 nanoliters of the caged-MO solution was injected into zebrafish embryos at the 1-cell stage. Injections were performed in a room equipped with yellow filters over all overhead and microscope lights to prevent premature uncaging of caged etv2 morpholino. Dishes containing injected embryos were wrapped in aluminum foil to minimize background uncaging until they were uncaged at 24 hpf by exposure to 365 nm UV light for 30 minutes. Embryos were then injected with B16-F10:dsRed melanoma cells as described above.
Whole-mount In Situ Hybridization (WISH)
DIG-UTP labeled riboprobes were synthesized using T3, T7, or SP6 RNA polymerases (ThermoFisher). In situ hybridization was performed as described [19] using a previously reported antisense etv2 probe [20]. Processed embryos were cleared in BBA (2:1 benzyl benzoate: benzyl alcohol) or RIMS (57% Histodenz, 0.2% Tween 20, 0.02% sodium azide in 0.02M sodium phosphate buffer, pH=7.4) for imaging of tumor vessels as needed. Z-stacks of images were captured using an AxioImager compound microscope (Carl Zeiss Inc., USA) equipped with a Plan-Neofluar 10X/0.3 NA microscope objective (Carl Zeiss Inc., USA) and an AxioCam ICC3 color camera (Carl Zeiss Inc., USA). Extended focus images were produced using AxioVision 4.6 software (Carl Zeiss Inc., USA). Image levels and color balance were adjusted using Adobe Photoshop CS5.
Real time qPCR
Pools of 20–25 embryos were frozen on dry ice at 24hpf. Embryos were homogenized in lysis buffer using a 23-gauge needle and extraction of RNA from the embryos was carried out using the RNAqueous 4-PCR kit (Ambion). Quantification of purified RNA was carried out using a Nanodrop spectrophotometer machine. Next, cDNA synthesis was performed using SuperScript® VILO cDNA Synthesis Kit or Superscript III cDNA synthesis kit (Invitrogen). Quantitative real-time PCR (qRT-PCR) was performed using PowerUp™ SYBR™ Green Master Mix (Applied Biosystems) in a StepOnePlus™ Real-Time PCR System. Quantification was performed using the relative standard curve method; controls/DMSO treatment were assigned a value of 1 and experimental values were computed by the software based on the CT values, relative to the control standard curve. For each experiment, 2–3 replicates were performed with duplicates in each run and ef1α was used as an endogenous control. The relative quantity of cDNA in each sample of each gene was normalized to the value of ef1α. Data was analyzed using StepOne™ Software-version 2.3 (Life Technologies).
The following primers were used:
ef1α (TCACCCTGGGAGTGAAACAGC) and (ACTTGCAGGCGATGTGAGCAG),
etv2 (GAGCTGTTGCACAAAGGTCA) and (CAGAGAGGGACGAGGTTCTG),
fli1b (GACCAAAGTGCACGGCAAACGC) and (TGTTCAAGTGAGTGTGAGTGCTGG),
kdrl (CCATCATCCATTTGTGGAGG) and (GAGGATGAGGGTGTCACCGAC).
Whole-mount Immunofluorescence Staining
Embryos were fixed at selected stages in 4% paraformaldehyde (PFA) in PBS at room temperature for 4 hrs. The fixed embryos were then dehydrated through an ascending series of ethanol washes followed by rehydration through a descending ethanol series and finally into distilled water. Samples were permeabilized for an hour in acetone at −20°C, then in distilled water and PBD buffer (0.2% Tween, 5% Triton X-100 in PBS). Tissues were transferred to 0.1M maleic acid buffer and blocked for 30 minutes at room temperature (Roche, #1-096-176) prior to overnight incubation at +4°C in 1:500 polyclonal rabbit anti-human S100 (Dako, #Z0311) or 1:50 monoclonal mouse anti-human MelanA (Dako, #M7196). After six washes in PBD buffer, the samples were incubated overnight at 4°C in 1:1000 dilutions of goat anti-rabbit Alexa Fluor 647 or anti-mouse 647 secondary antibodies (ThermoFisher). Samples were then washed in TNT buffer and mounted in the Vectashield Antifade Mounting Medium with DAPI (Vector Labs, #H-1200) for imaging.
Whole transcriptome analysis by RNA-seq
Control uninjected fli1a:GFP embryos and B16 melanoma transplanted control fli1a:GFP embryos were frozen at 3 dpf (1 dpi) for RNA-seq analysis. Two biological replicates of 20 embryos each were analyzed in each experimental group. RNA was purified using RNAqueous Total RNA isolation kit (ThermoFisher). Library preparation and next generation sequencing was performed at the CCHMC Core Facility using Illumina HiSeq2500 (75 read length, single sided, 20M reads per sample). Quality check of the RNA-SEQ reads was performed using Fastqc [http://www.bioinformatics.babraham.ac.uk/projects/fastqc/]. Reads with low quality, adapter content and over-represented sequences were trimmed using trimmomatic [http://www.usadellab.org/cms/?page=trimmomatic]. We mapped and quantified the trimmed RNA-SEQ reads using RSEM to latest Zebrafish genome assembly GRCz10 [http://deweylab.github.io/RSEM/] for each sample at default thresholds. We performed Differential Expression using CSBB’s [https://github.com/skygenomics/CSBB-v1.0] DifferentialExpression module which uses RUV-Seq [https://www.bioconductor.org/packages/3.3/bioc/vignettes/RUVSeq/inst/doc/RUVSeq.pdf]. Functional and Pathway enrichment was performed using ToppGene [https://toppgene.cchmc.org/].
RESULTS
Xenotransplantation of mammalian tumor cells effectively induces tumor angiogenesis in the zebrafish model
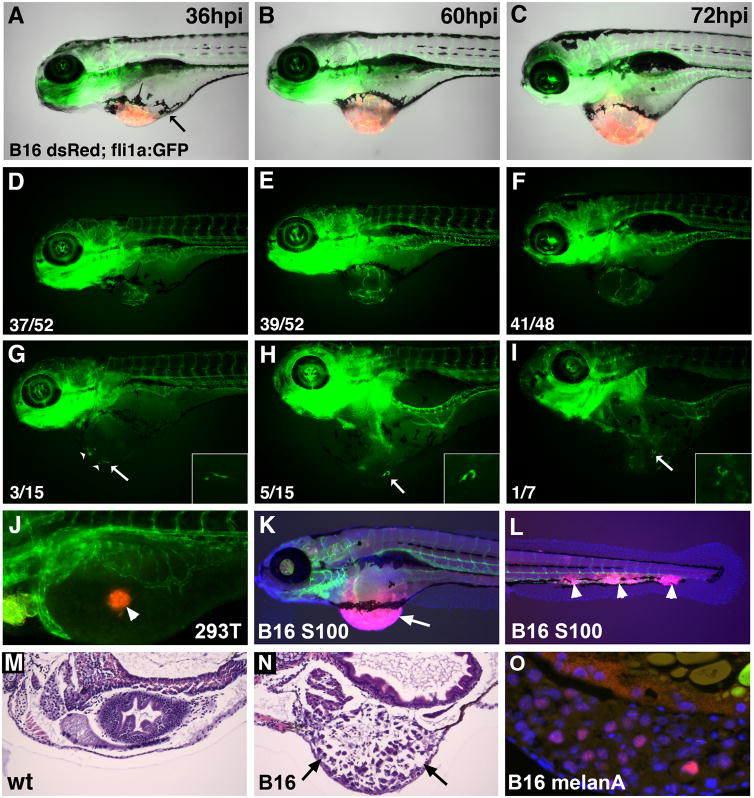
Xenotransplantation of mouse melanoma B16 tumor cells into zebrafish embryos was performed to test if Etv2 function is associated with tumor angiogenesis. B16 tumor cells expressing fluorescent dsRed protein were transplanted at 2 days-post-fertilization (dpf) into the perivitelline space of fli1a:GFP or kdrl:GFP transgenic zebrafish embryos which express GFP in vascular endothelial cells. Just 36 hours after injection (hpi), fli1a:GFP expressing cells were observed migrating from the adjacent embryonic vasculature toward the dsRed labeled tumor cells (Fig. 1A,D). At 60–72 hpi, extensive tumor angiogenesis and a measurable increase in tumor size was apparent (Fig. 1B,C,E,F). Intriguingly, individual migrating vascular endothelial cells were observed in a subset of embryos with tumors (Fig. 1G–I). No ectopic angiogenesis was evident in embryos transplanted with control epithelial kidney 293T cells (Fig. 1J). The hematoxylin-eosin (H&E) sections and immunostaining of neoplastic tissues with S100 and melanA antigens, specific to B16 melanoma, further confirmed the presence of B16 tumors in zebrafish embryos (Fig. 1K–O). In addition to the main tumor present at the injection site, metastasis to distant sites was observed (Fig. 1L). Thus melanoma B16 transplantation effectively induces tumor angiogenesis in the zebrafish model.
Figure 1. Induction of tumor growth and angiogenesis in zebrafish embryos transplanted with murine B16 melanoma cells.
(A–F) Lateral views of the same Tg(fli1a:GFP) vascular endothelial reporter embryo with murine B16-F10:dsRed xenograft imaged from 36 hpi through 72 hpi. The number of zebrafish with tumors (numerator) versus the total number injected (denominator) are indicated. (A–C) Merged dsRed, GFP and brightfield channels; (D–F) GFP channel.
(A) Tumors are partially demarcated by a ring of pigmented cells at 36 hpi (arrow) and roughly double in size over the next 24 – 36 hpi (B,C). Angiogenic sprouts extending from adjacent embryonic vasculature are evident by 36 hpi (D) and continued elaboration and branching of these vessels occurs with added tumor growth (E,F).
(G–I) Stray GFP-positive endothelial cells were evident in a subset of tumors (arrows). The number of embryos showing stray endothelial cells (numerator) versus the total number imaged (denominator) are shown. (G) Multiple GFP+ endothelial cells evident at 36 hpi (arrowheads). These cells extend, branch and persist through 72 hpi (I). Insets are magnified views of the cell marked with an arrow.
(J) No ectopic angiogenesis is observed after the transplantation of DiI-labeled control 293T cells (arrowhead).
(K,L) Immunofluorescent staining against S100 to identify melanoma cells (pink, arrow, K) and vascular endothelial kdrl:GFP at 6 dpf. Metastases along tail vasculature are apparent in (L) (arrowheads).
(M,N) Hematoxylin and eosin staining of histological sections in control (M) and melanoma B16 transplanted embryo (N) at 6 dpf. Note the tumor tissue present in (N, arrows). (O) Immunostaining of zebrafish tumor sections at 6 dpf for melanA, a marker for melanoma (pink, nuclei are stained blue with DAPI).
Tumor vasculature is derived from existing embryonic blood vessels
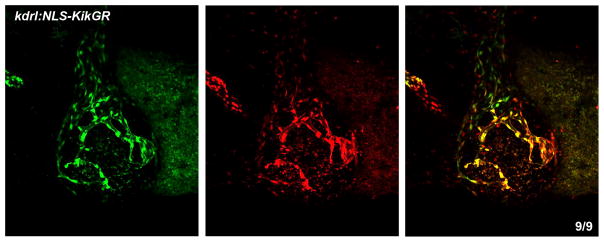
All tumor vasculature may be derived from the existing embryonic vasculature by tumor induced angiogenesis. Alternatively, some tumor vasculature may be derived by vasculogenesis de novo. To determine if vasculogenesis contributes to the formation of tumor vasculature in the zebrafish xenotransplantation model, xenotransplantation was performed in the kdrl:NLS-KikGR transgenic embryos which express green to red photoconvertible protein in vascular endothelial cells. The embryos were subjected to global photoconversion at 2 dpf, labeling existing endothelial cells in red, followed by the xenotransplantation of unlabeled melanoma B16 cells. All endothelial cells derived from the cells that expressed kdrl:NLS-KikGR at the time of photoconversion would show red fluorescence in addition to green fluorescence of newly synthesized KikGR, while any new endothelial cells that formed by vasculogenesis after photoconversion would only show green fluorescence. At 4 dpf (2 dpi) all tumor blood vessels displayed both green and red fluorescence (Fig. 2), indicating that they originated from the existing embryonic vasculature and did not form by vasculogenesis de novo.
Figure 2. Tumor blood vessels originate from the existing embryonic vasculature.
kdrl:NLS-KikGR embryos were photoconverted (green to red) immediately after B16 (non-fluorescent) tumor cell transplantation at 2 dpf. KikGR fluorescence was imaged at 2 dpi (4 dpf) in green (left) and red channels (middle). Right panel, merged image. All blood vessels in the tumor were positive for red and green expression. The number of embryos that have red positive cells over the total number of embryos is shown in lower right. Anterior is to the left, dorsal is up.
Expression of etv2 and fli1b is greatly upregulated during tumor angiogenesis
During normal development, etv2 expression within embryonic vasculature is highest prior to 24 hpf and is greatly diminished by 2–4 dpf [21,22]. To determine if etv2 expression is upregulated during tumor angiogenesis, we performed in situ hybridization and qPCR in B16-melanoma injected embryos. At 1 dpi, strong upregulation of etv2 expression throughout embryonic vasculature was evident in B16-injected embryos (Fig. 3A,B). Greatly increased etv2 expression was evident not only in the growing tumor blood vessels but also throughout the embryonic vasculature and the endocardium (Fig. 3C,D). Time course analysis of etv2 expression at 2–24 hours-post-injection indicated that etv2 was first upregulated as early as 4 hpi within the embryonic vasculature, initially in the posterior half of the embryo, and by 8 hpi was present throughout the entire embryonic vasculature (Fig. 3E–H). Expression of a related ETS transcription factor fli1b was also upregulated in melanoma-transplanted embryos (Fig. 3I,J). In contrast, there was no systemic upregulation of kdrl expression, a homolog of Vegf receptor Vegfr2/Flk1 in the embryonic vasculature. Rather, kdrl expression was induced in the tumor blood vessels in addition to its normal expression within the embryonic vasculature (Fig. 3K,L). At later stages of tumor growth (7 dpi), upregulated etv2 expression was observed in a subset of tumor vasculature, while kdrl expression was apparent in all tumor vessels (Fig. 3M–P). Induction of etv2 and fli1b expression at 1 dpi was also confirmed by qPCR using RNA purified from whole B16-injected and control embryos, while kdrl did not show such upregulation (Fig. 3Q).
Figure 3. etv2 and fli1b expression is induced during tumor angiogenesis.
(A–P) In situ hybridization analysis of etv2 (A–H, M,N), fli1b (I,J) and kdrl (K,L,O,P) expression in B16 melanoma injected embryos compared to uninjected controls.
(A,B) etv2 expression is induced throughout the embryonic vasculature (arrows) at 1 day post injection (dpi) in a B16 injected embryo (B), compared to uninjected control (A). White arrow marks the tumor injection site.
(C,D) Magnified images showing etv2 expression in tumor vasculature above the yolk (purple staining, arrows) and the endocardium (D) at 1 dpi.
(E–H) Time-course analysis of etv2 expression in B16 injected embryos at 2–24 hpi. Note the strong induction of etv2 expression in the main axial vasculature (arrows) from 4 hpi through 24 hpi. White arrow marks the tumor injection site.
(I–L) fli1b expression is upregulated throughout the embryonic vasculature at 1 dpi in melanoma injected embryos (J, arrows) while kdrl expression is primarily induced at the tumor injection site (L).
(M–P) Upregulated etv2 expression is restricted largely to the subset of the tumor vasculature at 7 dpi (N, arrows), while expanded kdrl expression corresponds to the entire tumor angiogenic network (P, arrows).
(Q) qPCR analysis of etv2, fli1b and kdrl expression in B16 tumor and control 293T cell injected embryos at 1 dpi (3 dpf) relative to uninjected controls. Note the significant induction of etv2 and fli1b expression in tumor injected embryos, while kdrl expression is not significantly affected. 293T cell injection did not significantly affect expression of any of the markers. (R) qPCR analysis of etv2 and fli1b expression in B16 transplanted embryos treated with control 0.1% DMSO solution or pan-Vegf inhibitor SU5416. Note that etv2 and fli1b expression is increased in xenotransplanted DMSO treated embryos as compared to sibling uninjected controls (p<0.01), while SU5416 treatment did not reduce etv2 or fli1b expression. Although SU5416 treated embryos showed greater upregulation of etv2 expression compared to control DMSO embryos in the replicate samples (p=0.04), this is likely to be an artifact due to a large variation in the level of etv2 induction observed among individual B16 transplanted embryos in separate experiments (see Fig. 3Q, for example).
These data suggest that the tumor secretes a pro-angiogenic factor which results in the systemic induction of etv2 and fli1b expression throughout the embryonic vasculature. To test if Vegf signaling is responsible for the upregulation of etv2 and fli1b expression, we treated B16-transplanted embryos immediately after xenotransplantation with 2 μM of Vegfr inhibitor SU5416. However, inhibition of Vegfr signaling did not affect the upregulation of etv2 expression in B16 melanoma injected embryos at 1 dpi (Fig. 3R). Higher concentrations of SU5416 resulted in the inhibition of embryonic angiogenesis, therefore it was not possible to evaluate their specific effect on tumor angiogenesis. This result suggests that etv2 and fli1b upregulation in response to tumor xenotransplantation is independent of Vegf signaling.
To confirm these results and to identify signaling pathways misregulated in tumor injected embryos, we performed global transcriptome analysis at 3 dpf (1 dpi) using RNA-seq. 675 genes were significantly upregulated in B16 melanoma injected embryos compared to uninjected sibling embryos (>2-fold upregulation, p<0.05, FDR corrected). The top 50 upregulated genes are listed in Table 1, and the complete list of differentially expressed genes is provided in Suppl. Table 1. Pathway analysis revealed significant enrichment in the pathways associated with extracellular matrix remodeling, cell migration and vascular development (Table 2). Significantly upregulated genes included metalloproteases mmp9 and mmp13, involved in extracellular matrix remodeling [23]. Expression of ETS factors etv2, fli1b as well as erg was significantly upregulated in the tumor injected embryos, consistent with the results from qPCR and in situ hybridization analysis. Intriguingly, genes associated with the innate immune response including neutrophil and macrophage specific markers myeloperoxidase mpx and lcp1 were upregulated [24,25]. We did not observe systemic upregulation of many other endothelial markers, however, likely because angiogenesis was limited to the tumor formation site. Thus RNA-seq analysis confirms specific upregulation of ETS transcription factors including etv2 and fli1b in tumor injected embryos.
Table 1. Top 50 genes upregulated in B16 melanoma injected embryos compared to control uninjected embryos as analyzed by RNA-seq at 3 dpf (1 dpi).
Expression is shown in TPM (transcripts per million).
| Gene | Annotated gene name | Log Fold Change | p value, FDR | Expression WT | Expression, B16 injected |
|---|---|---|---|---|---|
| si:ch73-368j24.6 | novel histone H3 protein | 7.99 | 1.31E-53 | 0.06 | 22.34 |
| CU459186.3 | novel miRNA | 7.50 | 4.47E-13 | 0.00 | 25.03 |
| zgc:172290 | natterin-like protein | 7.15 | 2.15E-10 | 0.02 | 1.15 |
| zgc:110286 | novel protein similar to vertebrate ADP-ribosylation factor 4a (ARF4A) | 5.92 | 8.73E-26 | 0.14 | 4.79 |
| arf4b | ADP-ribosylation factor 4b | 5.15 | 2.36E-59 | 0.51 | 15.12 |
| rhcga | Rh family, C glycoprotein a | 3.89 | 1.67E-30 | 1.50 | 13.59 |
| mmp9 | matrix metallopeptidase 9 | 3.83 | 4.61E-67 | 0.79 | 10.93 |
| hbz | hemoglobin zeta | 3.75 | 6.87E-34 | 2.70 | 33.77 |
| isg15 | ISG15 ubiquitin-like modifier | 3.69 | 2.79E-22 | 0.36 | 5.06 |
| si:dkey-23a13.17 | novel protein similar to vertebrate histone cluster 1, H1e (HIST1H1E) | 3.66 | 2.01E-06 | 0.24 | 1.97 |
| mmp13a | matrix metallopeptidase 13a | 3.48 | 6.59E-44 | 1.04 | 10.92 |
| si:ch1073-165f9.2 | ADP-ribosylation factor 1-like | 3.40 | 2.44E-22 | 0.86 | 8.32 |
| si:dkey-6n3.3 | lincRNA | 3.39 | 1.60E-14 | 0.63 | 6.78 |
| ccl34a.4 | chemokine (C-C motif) ligand 34a, duplicate 4 | 3.24 | 3.99E-27 | 2.05 | 18.47 |
| fosl1a | FOS-like antigen 1a | 3.21 | 2.53E-37 | 0.65 | 5.53 |
| scpp8 | secretory calcium-binding phosphoprotein 8 | 3.18 | 4.71E-11 | 0.23 | 2.39 |
| zgc:174719 | uncharacterized protein LOC100137112 | 3.07 | 6.84E-05 | 0.26 | 1.55 |
| si:dkey-119g10.4 | lincRNA | 3.01 | 1.66E-10 | 0.61 | 5.72 |
| si:dkey-83k24.5 | similar to UDP-galactose translocator | 3.00 | 1.68E-38 | 2.26 | 18.59 |
| NPC2 | Niemann-Pick disease, type C2 | 2.98 | 5.79E-10 | 0.83 | 6.03 |
| si:dkey-19a16.5 | SLAM family member 5-like | 2.89 | 2.76E-06 | 0.31 | 1.47 |
| si:ch211-194m7.8 | olfactomedin-4-like | 2.82 | 5.30E-09 | 0.18 | 1.71 |
| tnfaip2b | tumor necrosis factor, alpha-induced protein 2b | 2.81 | 9.77E-12 | 0.27 | 1.70 |
| ccl35.1 | chemokine (C-C motif) ligand 35, duplicate 1 | 2.80 | 3.50E-05 | 0.21 | 1.76 |
| si:ch211-226h7.3 | novel GP2-like protein | 2.76 | 1.34E-05 | 0.05 | 0.45 |
| cxcl8b.1 | chemokine (C-X-C motif) ligand 8b, duplicate 1 | 2.74 | 5.14E-10 | 0.73 | 5.14 |
| HACD4 | 3-hydroxyacyl-CoA dehydratase 4 | 2.74 | 4.53E-09 | 0.49 | 3.00 |
| si:dkey-97i18.5 | novel transcript | 2.74 | 4.98E-12 | 4.09 | 25.77 |
| si:ch211-13o20.1 | novel transcript | 2.69 | 2.91E-05 | 0.28 | 1.57 |
| si:ch211-284o19.8 | similar to vertebrate glutamate receptor, ionotropic, N-methyl D-asparate-associated protein 1 (glutamate binding) (GRINA) | 2.69 | 1.82E-08 | 0.37 | 2.11 |
| BX005284.1 | similar to LINE-1 reverse transcriptase | 2.68 | 1.25E-05 | 0.08 | 0.60 |
| CU019646.2 | E3 ubiquitin-protein ligase RNF182-like | 2.67 | 1.59E-32 | 2.61 | 16.73 |
| cbx7a | chromobox homolog 7a | 2.63 | 1.32E-28 | 7.52 | 45.09 |
| BX072532.1 | uncharacterized protein LOC108179104 | 2.61 | 2.48E-04 | 0.07 | 0.37 |
| si:dkey-102g19.3 | urokinase plasminogen activator surface receptor-like | 2.56 | 2.52E-08 | 0.50 | 3.42 |
| BX469901.1 | novel pseudogene | 2.54 | 2.60E-09 | 1.98 | 14.00 |
| si:ch211-194m7.5 | similar to vertebrate olfactomedin 4 (OLFM4) | 2.50 | 1.36E-06 | 0.36 | 2.03 |
| vaspa | vasodilator-stimulated phosphoprotein a | 2.46 | 3.14E-04 | 0.09 | 0.44 |
| si:dkey-9i5.2 | uncharacterized protein LOC101885950 | 2.43 | 1.61E-04 | 0.50 | 1.83 |
| lyz | lysozyme | 2.43 | 9.39E-29 | 9.82 | 53.10 |
| CABZ01075274.1 | NACHT, LRR and PYD domains-containing protein (NLRC3)-like | 2.42 | 7.62E-05 | 0.09 | 0.61 |
| adamts8a | ADAM metallopeptidase with thrombospondin type 1 motif, 8a | 2.41 | 8.93E-10 | 0.28 | 1.41 |
| mcm6l | MCM6 minichromosome maintenance deficient 6, like | 2.40 | 8.31E-05 | 0.12 | 0.58 |
| epm2a | epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) | 2.40 | 2.15E-04 | 0.29 | 1.48 |
| gcga | glucagon a | 2.38 | 7.57E-25 | 1.76 | 9.39 |
| zgc:172260 | novel ETX_MTX2 domain containing protein | 2.38 | 5.48E-05 | 0.19 | 1.34 |
| si:ch211-266c8.1 | novel NLR C nacht domain containing protein | 2.34 | 1.61E-04 | 0.09 | 0.48 |
| prl | prolactin | 2.34 | 1.02E-28 | 2.64 | 13.38 |
| cxcr4a | chemokine (C-X-C motif) receptor 4a | 2.34 | 1.76E-22 | 1.55 | 7.60 |
| bcl3 | B-cell CLL/lymphoma 3 | 2.33 | 4.46E-04 | 0.08 | 0.29 |
| Other selected genes | |||||
| mpx | myeloid-specific peroxidase | 2.31 | 1.61E-23 | 1.52 | 7.15 |
| etv2 | ets variant 2 | 0.93 | 6.16E-04 | 2.75 | 5.23 |
| fli1b | Fli-1 proto-oncogene, ETS transcription factor b | 0.58 | 1.74E-02 | 2.60 | 3.98 |
Table 2.
Top 20 pathways upregulated in B16 melanoma injected embryos as compared with control uninjected embryos at 3 dpf (1dpi)
| ID | Name | q-value FDR B&H |
|---|---|---|
| GO:0030198 | extracellular matrix organization | 4.76E-23 |
| GO:0043062 | extracellular structure organization | 4.76E-23 |
| GO:0072359 | circulatory system development | 3.38E-19 |
| GO:0072358 | cardiovascular system development | 3.38E-19 |
| GO:0001944 | vasculature development | 3.88E-18 |
| GO:0001568 | blood vessel development | 1.62E-17 |
| GO:0051270 | regulation of cellular component movement | 2.14E-17 |
| GO:0040012 | regulation of locomotion | 9.30E-17 |
| GO:2000145 | regulation of cell motility | 9.30E-17 |
| GO:0030334 | regulation of cell migration | 9.30E-17 |
| GO:0048514 | blood vessel morphogenesis | 2.39E-16 |
| GO:0006915 | apoptotic process | 4.34E-16 |
| GO:0012501 | programmed cell death | 5.05E-16 |
| GO:0007155 | cell adhesion | 9.24E-16 |
| GO:0022610 | biological adhesion | 1.94E-15 |
| GO:0048870 | cell motility | 2.69E-15 |
| GO:0051674 | localization of cell | 2.69E-15 |
| GO:0040011 | locomotion | 2.69E-15 |
| GO:0048646 | anatomical structure formation involved in morphogenesis | 2.69E-15 |
| GO:0016477 | cell migration | 2.69E-15 |
Etv2 function is required for tumor angiogenesis
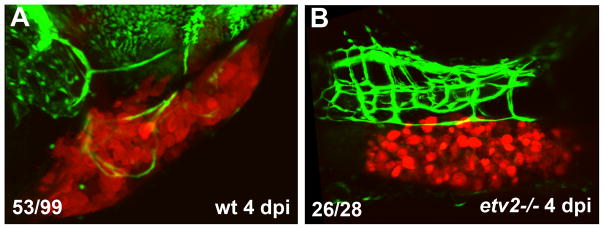
To test if Etv2 function is required for tumor angiogenesis, we analyzed tumor angiogenesis in the null etv2y11 mutants [9]. Although etv2 mutants show severe defects in early embryonic vasculogenesis and angiogenesis prior to 24 hpf, they undergo a partial recovery of embryonic angiogenesis and show considerable formation of vascular network at later stages [9,22]. In contrast to wild-type embryos, which showed induction of tumor angiogenesis at 4 dpi, etv2 mutants exhibited complete or nearly complete absence of tumor angiogenesis, while embryonic vasculature was still present (Fig. 4).
Figure 4. Tumor angiogenesis is inhibited in etv2−/− mutant embryos.
B16-dsRed melanoma cells were transplanted into fli1a:GFP; etv2−/− embryos and their wild-type siblings at 2 dpf, and tumor angiogenesis was analyzed at 4 dpi. Note that etv2−/− embryos exhibit absence of tumor angiogenesis as compared with their wild-type siblings (which represent a mixture of wt and etv2+/− heterozygous embryos). The number of embryos with pronounced tumor angiogenesis is shown in (A) (the others did not show extensive tumor angiogenesis), while the number of embryos with absent tumor angiogenesis is shown in (B).
Because etv2 mutants show defective early vasculogenesis, it is possible that the observed defects in tumor angiogenesis are a secondary consequence of mispatterned embryonic vasculature. Therefore we utilized an inducible knockdown approach using photoactivatable morpholinos to inhibit Etv2 function at a later stage. We have previously designed and validated this approach which utilizes a caging etv2 strand hybridized to a standard Etv2 morpholino (MO) [18,26]. Upon UV irradiation, the caging strand is cleaved and releases the Etv2 MO which can then inhibit gene function. As we previously reported [18], uncaging Etv2 MO at 4–5 hpf resulted in the specific inhibition of embryonic vasculogenesis and the absence of angiogenic sprouting at 24 hpf, observed in fli1a:GFP transgenic reporter line (Suppl. Fig. S1). The resulting phenotype was similar to or slightly weaker than the etv2 null mutant phenotype, and no other morphological defects were present in caged Etv2 MO injected embryos. As reported previously [18], Etv2 MO photoactivation at 24 hpf resulted in no apparent vascular or any other morphological defects, when analyzed either shortly after photoactivation (Suppl. Fig. S1C,G) or at later stages (data not shown). Caged Etv2 MO-injected embryos, which were not subjected to photoactivation, were also completely normal (Suppl. Fig. S1D,H). We have previously reported an expected loss of vascular marker expression in Etv2 MO injected embryos that were photoactivated early (4–5 hpf), while no defects were observed in the embryos which were not photoactivated [18]. These results, together with our previous validation of caged Etv2 MO, argue that Etv2 MO photoactivation results in a specific inhibition of Etv2 function.
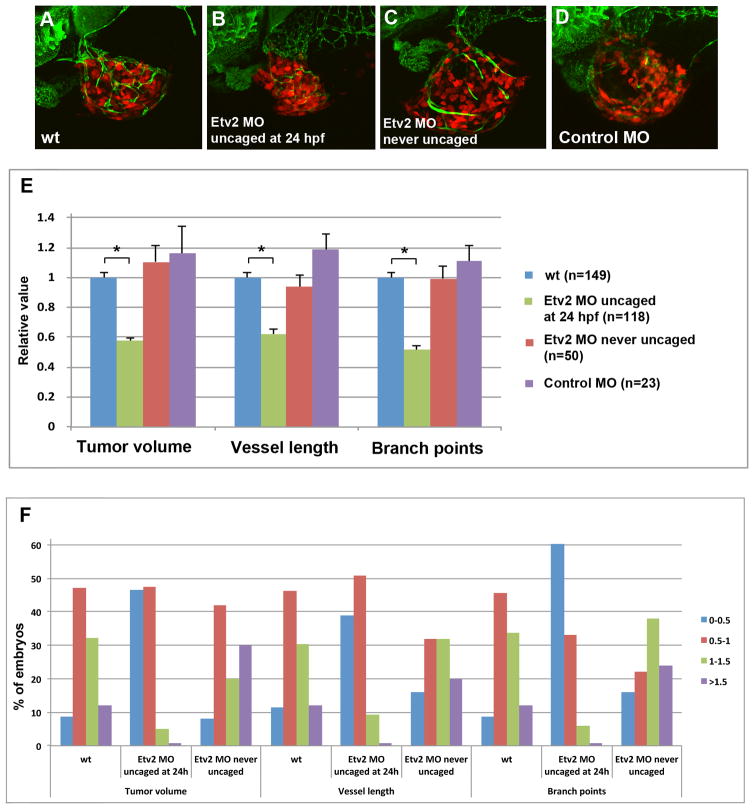
To test if Etv2 function is required for tumor angiogenesis, we injected embryos with caged Etv2 MO, photoactivated Etv2 MO at 24 hpf, and transplanted melanoma cells into the perivitelline space at 48 hpf. The embryos were then imaged using confocal microscopy at 5 dpf (Fig. 5A–D) to analyze the total length of blood vessels in the tumor, the number of branch points and the tumor volume. To minimize variability between replicate experiments, all measurements in each experiment were normalized relative to the mean values in the control embryos, which were not injected with the Etv2 morpholino (see Materials and Methods). Thus, the ratio of either tumor volume, blood vessel length or the number of branch points in each embryo over the mean corresponding value in the control embryos, was calculated. The tumor volume, tumor blood vessel length and the number of branch points in tumor vessels were significantly reduced in caged Etv2 MO injected embryos, which were photoactivated at 24 hpf, compared to control embryos which were either not injected with caged Etv2 MO or injected with the MO but not subjected to photoactivation (Fig. 5E). Injection of a control MO, hybridized to caged Etv2 strand, followed by photoactivation at 24 hpf, had no significant effect on the tumor volume, blood vessel length or vessel branch points (Fig. 5D,E). We also analyzed the distribution of embryos with small tumors (relative normalized volume less than 0.5), small to moderate (volume 0.5–1), moderate to large (1–1.5) and large (more than 1.5). The majority of wild-type control embryos (79.2%) had moderate size tumors (0.5–1.5 relative volume) (Fig. 5F). While only 8.7% of control embryos had small tumors (<0.5), the percentage of Etv2 knockdown embryos with small tumors was greatly increased (46.6%, Fig. 5F). Similarly, only 11.4% and 8.7% of control embryos had small blood vessel length and few branch points, respectively (less than 0.5 of relative value), while 39.0% and 60.2% of Etv2 knockdown embryos displayed reduced blood vessel length and the number of branch points (Fig. 5F). These results show that the knockdown of Etv2 function inhibited angiogenesis in melanoma induced tumors and reduced tumor growth.
Figure 5. Inhibition of Etv2 function using photoactivatable MO results in the reduction of xenotransplanted B16 tumor growth and tumor angiogenesis.
(A–D). Confocal images of dsRed labeled tumor cells and fli1a:GFP blood vessels at 5 dpf (3 dpi) in a control embryo not injected with MO (A), an embryo injected with caged Etv2 MO and uncaged at 24 hpf (B), an embryo injected with caged Etv2 MO which was not subjected to uncaging (C), and an embryo injected with a control caged MO which included a standard control MO hybridized to Etv2 caging strand, photoactivated at 24 hpf (D). Note the reduced tumor angiogenesis and tumor growth in (B).
(E) Quantification of the tumor volume, tumor blood vessel length and the number of tumor blood vessel branch points at 5 dpf (3 dpi). To account for the variation in tumor and blood vessel growth between different experiments, the absolute values were normalized in each experiment to wild-type fli1a:GFP embryos which were not injected with MO. Each experiment was repeated at least twice and the total number of embryos is shown. Note a highly significant reduction (asterisk) in the tumor volume (p=2.4 × 10−17), tumor blood vessel length (p=3.2×10−13), and the number of branch points (p=3.4×10−18) in Etv2 MO embryos uncaged at 24 hpf (green bars) compared to wild-type control embryos (blue bars). Unpaired Student’s t-test was used to calculate p values; ±SEM is shown.
(F) Distribution of embryos with different ranges of the relative tumor volume, blood vessel length or the number of branch points. The percentage of embryos in each group is shown. All values are relative to the tumor injected wild-type embryos, which have been normalized to 1 in each experiment. Note that the percentage of embryos with small tumors (<0.5) is greatly increased in caged Etv2 MO / uncaged at 24 hpf group (blue bar) compared to wt or never uncaged embryos, while a percentage of embryos with large tumors (1–1.5 and >1.5, green and purple bars) is greatly reduced. Similarly, the percentage of embryos with short vessel length and few branch points (<0.5, relative value) is greatly increased among caged Etv2 MO / uncaged at 24 hpf embryos, while the percentage of embryos with large vessel length and many branch points (1–1.5 and >1.5) is greatly reduced.
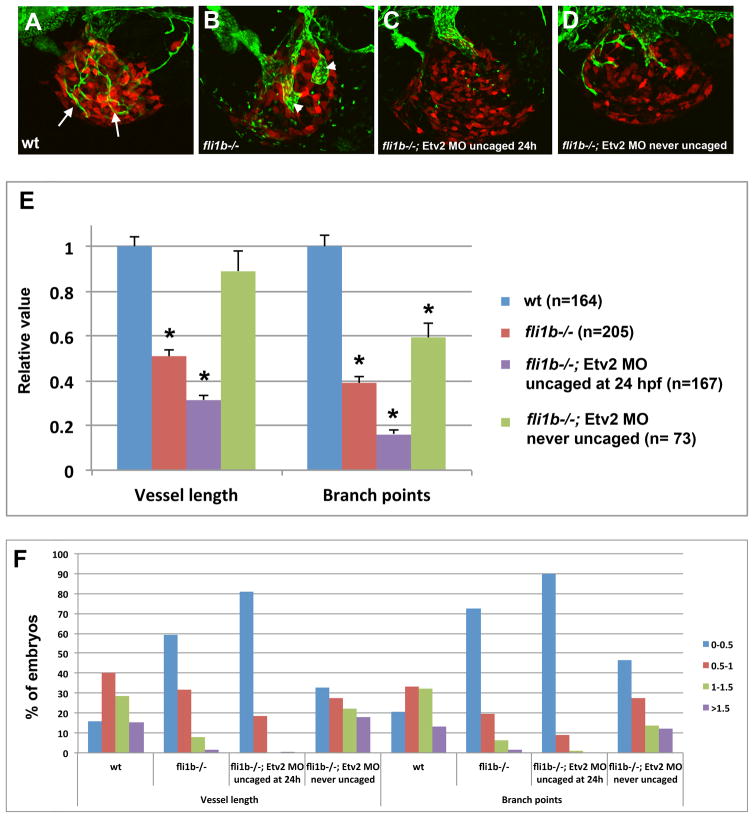
Fli1b and Etv2 function redundantly during tumor angiogenesis
Despite these results, Etv2 knockdown embryos displayed only partial inhibition of tumor angiogenesis. We had previously demonstrated that Etv2 and Fli1b function redundantly during embryonic angiogenesis [22]. To test for the functional redundancy between Etv2 and Fli1b in tumor angiogenesis, we analyzed tumor angiogenesis in fli1b−/− mutants and double Etv2 MO; fli1b−/− knockdown embryos. fli1b mutants have been previously generated by Tol2 transposase-mediated insertional mutagenesis which resulted in the integration of a gene trap construct containing a splice acceptor site and GFP reporter into the fli1b locus [22]. This insertion results in robust fli1b promoter driven GFP expression in the vasculature with less than 5% of endogenous fli1b transcript remaining in fli1b−/− embryos. Nevertheless, fli1b−/− embryos are viable and do not exhibit any apparent defects in embryonic vascular development [22]. B16 melanoma cells were transplanted into fli1b−/− embryos and control wild-type fli1a:GFP embryos at 48 hpf as described earlier. When analyzed at 4 dpf (2 dpi), fli1b−/− embryos showed 49% reduction in tumor blood vessel length and 61% reduction in branch points (Fig. 6A,B,E). The remaining tumor vessels were often greatly dilated and had few connections to other blood vessels. Enlarged vessels may be the remains of the common cardinal vein (CCV), which failed to remodel in melanoma cell injected fli1b−/− embryos. To examine the redundancy between Etv2 and Fli1b, we injected caged Etv2 MO into fli1b−/− mutants, followed by the photoactivation of Etv2 MO at 24 hpf. This resulted in even greater reduction in tumor angiogenesis; double knockdown embryos had only 31% of average tumor vessel length and 16% of tumor vessel branch points remaining as compared to the control embryos. (Fig. 6C,E). Although patterning of embryonic vasculature in the double mutants was not significantly affected, many embryos developed pericardial edema by 5 dpf which prevented us from measuring tumor growth (tumor angiogenesis was analyzed at 4 dpf when tumor growth in many embryos is not clearly pronounced at this stage yet). The cause of pericardial edema needs further investigation but it may be a consequence of additional functional requirement of Etv2 and Fli1b in the embryonic vasculature beyond 24 hpf. Control fli1b−/− embryos injected with Etv2 caged MO and not subjected to photoactivation showed only minor reduction in tumor vessel growth and branch point number (Fig. 6D,E), which is likely caused by incomplete inhibition of Etv2 MO by the caging strand. As we previously demonstrated, there is some inhibition of Etv2 function in caged MO injected embryos in the absence of photoactivation, but it is not sufficient to cause an embryonic phenotype [18]. As expected, the fraction of embryos with short tumor vessel length and few branch points (less than 0.5 of relative values as compared to wt embryos) was greatly increased in fli1b−/− mutants and further increased in fli1b−/−; Etv2 MO embryos uncaged at 24 hpf. In contrast, the percentage of embryos with relative values of >1 for vessel length and the number of branch points was greatly reduced in fli1b−/− mutants and almost absent among fli1b−/−; Etv2 MO embryos (Fig. 6F). These results argue that Etv2 and Fli1b function redundantly during tumor angiogenesis.
Figure 6. Combined inhibition of Etv2 and Fli1b function results in great reduction of tumor and blood vessel growth.
(A–D) Confocal images of B16 dsRed-labeled melanoma tumors transplanted into fli1a:GFP wild-type (A) and fli1b−/− embryos (B–D) at 4 dpf (2 dpi). fli1b−/− embryos were either not injected with MO (B), injected with caged Etv2 MO / photoactivated at 24 hpf (C) or injected with caged Etv2 MO and not subjected for photoactivation (D). Note GFP-positive thin tumor blood vessels in (A), and the absence of thin tumor blood vessels in (B) while only wide enlarged blood vessels are observed which may be the remnants of the common cardinal vein that failed to remodel. (C) Tumor vessels are completely or nearly completely absent in (C). Note that there was a variation in vessel width among fli1b−/− embryos, and dilated vessels were not apparent in every embryo (such as the one in (D)).
(E) Quantification of the tumor blood vessel length and the number of tumor vessel branch points at 4 dpf (2 dpi). Absolute values were normalized to the corresponding values in wild-type fli1a:GFP embryos. Note the significant reduction in tumor vessel length (p=1.4×10−27 for wt versus fli1b−/−, and p=8.6 × 10−36 for wt versus Etv2 MO; fli1b−/−) and the number of branch points (p=2.6×10−30 and 2.6×10−41, respectively, asterisks). Unpaired Student’s t-test was used to calculate p values; ±SEM is shown.
(F) Distribution of embryos with different ranges of the relative blood vessel length or the number of branch points. A percentage of embryos in each group is shown. All values are relative to the tumor injected wild-type embryos, which have been normalized to 1 in each experiment. Note that the percentages of embryos with short vessels and few branch points (<0.5, relative value, blue bars) are greatly increased among fli1b−/− embryos, and even more increased among Etv2 MO; fli1b−/− embryos, while the percentages of embryos with large vessel length and many branch points (1–1.5 and >1.5, green and purple bars) are greatly reduced.
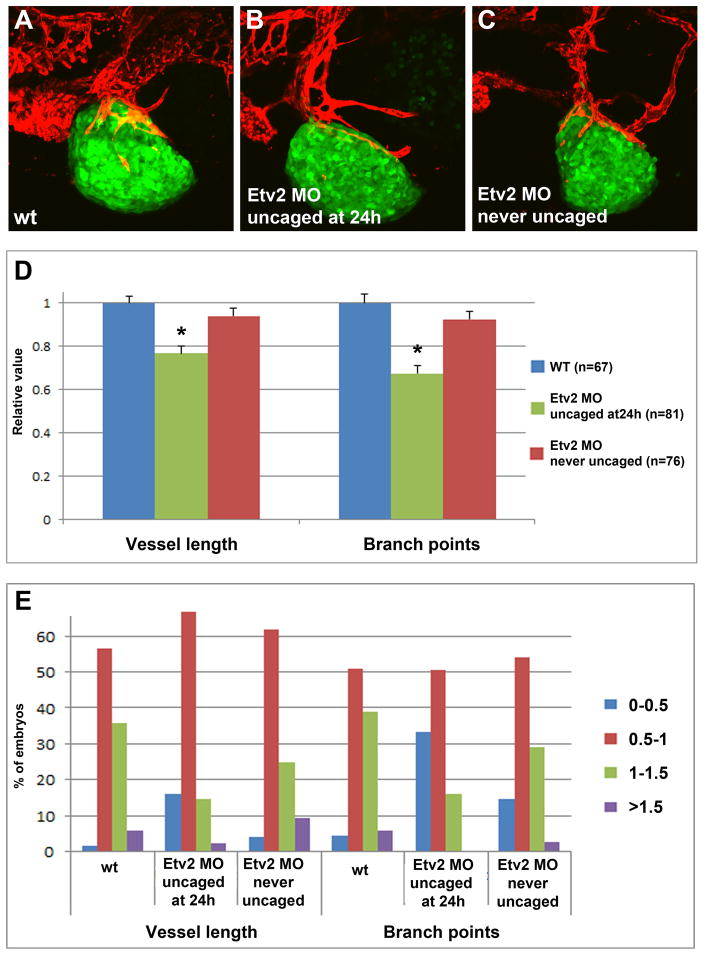
Etv2 knockdown inhibits tumor angiogenesis in an A673 Ewing Sarcoma xenograft model
To test if a similar inhibition of tumor angiogenesis was observed with other tumor cell lines, we used A673 Ewing Sarcoma tumor cells that had been transfected with a GFP reporter. The cells were transplanted at 2 dpf into caged Etv2 MO-injected and control kdrl:mCherry transgenic zebrafish embryos, followed by imaging analysis at 4 dpf (2 dpi). These cells formed very dense tumors which absorbed deep fluorescence, therefore only superficially located vessels were quantified. Nevertheless, there was a significant reduction in the tumor blood vessel length and the branch point number in Etv2 inhibited embryos compared to control uninjected embryos and the embryos that were injected with caged MO but never uncaged (Fig. 7A–D). The percentage of embryos which displayed shorter blood vessel length (<0.5 of the relative length and branch points) was greatly increased among Etv2 knockdown embryos (Fig. 7E). These results argue that Etv2 inhibition reduces tumor angiogenesis in different types of tumor lines.
Figure 7. Inhibition of Etv2 function results in reduced tumor angiogenesis in zebrafish embryos transplanted with A673 Ewing Sarcoma fluorescent cells.
(A–C) Confocal images of GFP-expressing A673 cells which were transplanted into kdrl:mCherry zebrafish embryos at 2 dpf. Imaging was performed at 4 dpf (2 dpi). (A) Control uninjected embryo; (B) Etv2 caged MO injected embryo uncaged at 24 hpf; (C) Etv2 caged MO injected embryo which was not subjected to photoactivation.
(D) Quantification of tumor blood vessel length and the number of branch points in tumor vasculature at 4 dpf (2 dpi). All values are relative to the tumor injected wild-type embryos, which have been normalized to 1 in each experiment. Note a significant reduction (asterisks) in the tumor blood vessel length (p=0.047) and branch points (p=3.4×10−9) in caged Etv2 MO injected embryos, photoactivated at 24 hpf compared to uninjected controls. Unpaired Student’s t-test was used to calculate p values; ±SEM is shown.
(E) Distribution of embryos with different ranges of relative blood vessel length or the number of branch points. A percentage of embryos in each group is shown. Note that the percentage of embryos with short vessels and few branch points (<0.5, relative value, red bars) is increased among Etv2 MO embryos, while the percentage of embryos with large vessel length and many branch points (1–1.5 and >1.5, green and purple bars) is greatly reduced.
DISCUSSION
Our results demonstrate that Etv2 is required for tumor angiogenesis and it functions partially redundantly with a related ETS transcription factor Fli1b. etv2 expression was significantly upregulated in zebrafish embryos after xenotransplantation. Knockdown of Etv2 function using photoactivatable morpholinos resulted in a significant reduction of tumor angiogenesis and reduction in tumor growth. Furthermore, the combined knockdown of Etv2 and Fli1b resulted in even greater effect and nearly complete inhibition of tumor angiogenesis without significantly affecting embryonic vascular development.
It has been widely accepted that the majority of tumor vasculature originates from existing host vessels and forms through the mechanism of angiogenesis [27,28]. However, vasculogenesis has also been implicated in tumor growth, and new blood vessels may arise from the circulating endothelial progenitor cells or via the process of vascular mimicry [29,30]. Our data obtained using kdrl:NLS-KikGR line argue that in the zebrafish xenotransplantation model all tumor vessels originate from the existing embryonic blood vessels, and do not originate via de novo vasculogenesis. We cannot, however, exclude contribution of vascular mimicry in this model, since only zebrafish derived vasculature is visualized using kdrl:NLS-KikGR fluorescence, while vascular mimicry would involve mouse melanoma derived vasculature.
During normal development, etv2 embryonic expression is downregulated after 24 hpf and remains localized to only a few selected vessels such as the pharyngeal arches [8,9,21]. It is remarkable that etv2 expression throughout the embryonic vasculature is upregulated within as little as 4 hpf after injecting mouse tumor cells. It is likely that this systemic upregulation is a response to tumor secreted factors that may enter circulation. It has been previously reported that VEGF can upregulate etv2 expression during embryonic vascular development [31]. However, treatment with VEGF inhibitor SU5416 failed to inhibit tumor induced upregulation of etv2 expression, suggesting that other tumor-secreted factors may induce etv2 expression. Nevertheless, our treatments were limited to only lower doses of SU5416 since higher doses resulted in the retraction or apoptosis of embryonic vasculature. Therefore, we cannot completely exclude VEGF as a factor which upregulates etv2 expression during tumor angiogenesis. Additional pathways including BMP, FGF and Wnt have been implicated in regulating etv2 expression [12] and may also be involved in inducing etv2 expression during tumor angiogenesis. Because etv2 expression can be upregulated in a VEGF independent manner, this could be one of the reasons why VEGF inhibition therapies are only partially effective in inhibiting tumor growth. It is possible that a combinatorial VEGF and Etv2 inhibition may result in a synergistic effect in reducing tumor angiogenesis, which could be examined in the future studies.
We have previously demonstrated that Etv2 and Fli1b function redundantly in embryonic angiogenesis [22]. Inducible Etv2 knockdown starting at the 18-somite stage using photoactivatable morpholinos in fli1b−/− background resulted in the inhibition of embryonic angiogenesis. Yet, as we observe in this study, inhibition of Etv2 function starting at 24 hpf stage even in fli1b mutant background did not significantly affect embryonic angiogenesis. It is likely that additional vascular specific ETS factors such as fli1a, ets1 and erg may compensate at later stages for the absence of Etv2 and Fli1b. Intriguingly, knockdown of Etv2 and Fli1b greatly inhibited tumor angiogenesis without affecting embryonic angiogenesis. Even fli1b−/− single mutants displayed reduction in tumor angiogenesis although they have no apparent defects in embryonic angiogenesis. A likely explanation is that tumor vessels grow fast and require greater levels of ETS factors as opposed to the slower growth of embryonic vessels which is observed at later developmental stages. It is also possible that expression of other ETS factors such as fli1a, ets1 and others is not induced as quickly in tumor vessels compared to etv2 and fli1b, therefore they cannot compensate for the absence of Etv2 and Fli1b function. It is likely that additional ETS factors may also play roles in tumor angiogenesis. In fact, many studies have implicated Ets1 in tumor angiogenesis [32,33].
Despite the significant knockdown of Etv2 and nearly complete loss in Fli1b function, there was still a partial tumor angiogenesis observed in Etv2 MO; fli1b−/− embryos. Etv2 knockdown is only partial and the MO effect becomes even weaker at later stages due to dilution and excretion, therefore it is likely that the remaining Etv2 protein is sufficient for partial formation of tumor vasculature. Alternatively, other ETS factors may compensate for the absence of Etv2 function.
Intriguingly, while etv2 was upregulated throughout the entire embryonic vasculature, induction of angiogenesis was only observed in the vicinity of the tumor. It is likely that additional local factors including Vegf are required to induce directional growth of blood vessels towards the tumor. During embryonic angiogenesis, Etv2 directly upregulates expression of multiple genes associated with vascular development [34]. It is likely that it functions in a similar way during tumor angiogenesis, upregulating expression of Vegf receptor Vegfr2 homologs as well as many other vascular specific genes. Although induction of kdrl expression was not observed throughout the entire embryonic vasculature and was restricted to the site of active tumor angiogenesis in tumor transplanted embryos, differently from etv2 and fli1b expression, it is possible that Etv2 functions in combination with a local signal such as VEGF to induce expression of Vegfr2 and other vascular endothelial markers at the tumor injection site. Further experiments are needed to clarify the mechanism of Etv2 function during tumor angiogenesis.
In summary, our results identify Etv2 and Fli1b as novel critical regulators of tumor angiogenesis. Because Etv2 and Fli1 function in embryonic vascular development is highly conserved among different vertebrates, it is likely that their role in tumor angiogenesis is also evolutionarily conserved. Therefore inhibition of Etv2 and Fli1b function may provide a novel strategy to inhibit tumor angiogenesis and reduce tumor growth.
Supplementary Material
Acknowledgments
We thank Timothy Cripe for providing A673 tumor cell line, and Chengjian Zhao for providing B16 melanoma cells. We thank Matthew Kofron and Mike Muntifering at the CCHMC confocal core for their help with the imaging and analysis (the confocal core is supported by the award from the National Institutes of Health 1S10RR029406). The research was supported by the awards from the National Institutes of Health (R01 HL107369), Ohio Cancer Research Associates and Cancer-Free Kids to S.S. and NIH 5T32HL00752 (M.P.C. / J. Whitsett).
References
- 1.Figg WD, Folkman J. Angiogenesis: an integrative approach from science to medicine. Springer; New York: 2008. [Google Scholar]
- 2.Ferrara N. Pathways mediating VEGF-independent tumor angiogenesis. Cytokine Growth Factor Rev. 2009 doi: 10.1016/j.cytogfr.2009.11.003. S1359-6101(09)00110-5 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Taylor AM, Zon LI. Zebrafish tumor assays: the state of transplantation. Zebrafish. 2009;6(4):339–346. doi: 10.1089/zeb.2009.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoletov K, Montel V, Lester RD, Gonias SL, Klemke R. High-resolution imaging of the dynamic tumor cell vascular interface in transparent zebrafish. Proc Natl Acad Sci U S A. 2007;104(44):17406–17411. doi: 10.1073/pnas.0703446104. 0703446104 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicoli S, Presta M. The zebrafish/tumor xenograft angiogenesis assay. Nature protocols. 2007;2(11):2918–2923. doi: 10.1038/nprot.2007.412. [DOI] [PubMed] [Google Scholar]
- 6.Zhao C, Wang X, Zhao Y, Li Z, Lin S, Wei Y, Yang H. A novel xenograft model in zebrafish for high-resolution investigating dynamics of neovascularization in tumors. PLoS One. 2011;6(7):e21768. doi: 10.1371/journal.pone.0021768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig MP, Sumanas S. ETS transcription factors in embryonic vascular development. Angiogenesis. 2016;19(3):275–285. doi: 10.1007/s10456-016-9511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4(1):e10. doi: 10.1371/journal.pbio.0040010. 05-PLBI-RA-0577R4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, Weinstein BM. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303(2):772–783. doi: 10.1016/j.ydbio.2006.10.030. S0012-1606(06)01323-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, Pennacchio LA, Dubchak I, Krieg PA, Stainier DY, Black BL. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135(6):1053–1064. doi: 10.1016/j.cell.2008.10.049. S0092-8674(08)01387-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferdous A, Caprioli A, Iacovino M, Martin CM, Morris J, Richardson JA, Latif S, Hammer RE, Harvey RP, Olson EN, Kyba M, Garry DJ. Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc Natl Acad Sci U S A. 2009;106(3):814–819. doi: 10.1073/pnas.0807583106. 0807583106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, Janknecht R, Lim DS, Choi K. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2(5):497–507. doi: 10.1016/j.stem.2008.03.008. S1934-5909(08)00120-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park C, Lee TJ, Bhang SH, Liu F, Nakamura R, Oladipupo SS, Pitha-Rowe I, Capoccia B, Choi HS, Kim TM, Urao N, Ushio-Fukai M, Lee D, Miyoshi H, Kim BS, Lim DS, Apte RS, Ornitz DM, Choi K. Injury-Mediated Vascular Regeneration Requires Endothelial ER71/ETV2. Arterioscler Thromb Vasc Biol. 2016;36(1):86–96. doi: 10.1161/ATVBAHA.115.306430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248(2):307–318. doi: 10.1006/dbio.2002.0711. S0012160602907116 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132(23):5199–5209. doi: 10.1242/dev.02087. dev.02087 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Lazic S, Scott IC. Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Dev Biol. 2011;354(1):123–133. doi: 10.1016/j.ydbio.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 17.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 18.Kohli V, Schumacher JA, Desai SP, Rehn K, Sumanas S. Arterial and venous progenitors of the major axial vessels originate at distinct locations. Developmental cell. 2013;25(2):196–206. doi: 10.1016/j.devcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jowett T. Analysis of protein and gene expression. Methods Cell Biol. 1999;59:63–85. doi: 10.1016/s0091-679x(08)61821-x. [DOI] [PubMed] [Google Scholar]
- 20.Sumanas S, Jorniak T, Lin S. Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants. Blood. 2005;106(2):534–541. doi: 10.1182/blood-2004-12-4653. 2004-12-4653 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore JC, Sheppard-Tindell S, Shestopalov IA, Yamazoe S, Chen JK, Lawson ND. Post-transcriptional mechanisms contribute to Etv2 repression during vascular development. Dev Biol. 2013;384(1):128–140. doi: 10.1016/j.ydbio.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craig MP, Grajevskaja V, Liao HK, Balciuniene J, Ekker SC, Park JS, Essner JJ, Balciunas D, Sumanas S. Etv2 and fli1b function together as key regulators of vasculogenesis and angiogenesis. Arterioscler Thromb Vasc Biol. 2015;35(4):865–876. doi: 10.1161/ATVBAHA.114.304768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh D, Srivastava SK, Chaudhuri TK, Upadhyay G. Multifaceted role of matrix metalloproteinases (MMPs) Front Mol Biosci. 2015;2:19. doi: 10.3389/fmolb.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood. 2001;98(10):3087–3096. doi: 10.1182/blood.v98.10.3087. [DOI] [PubMed] [Google Scholar]
- 25.Liu F, Wen Z. Cloning and expression pattern of the lysozyme C gene in zebrafish. Mechanisms of development. 2002;113(1):69–72. doi: 10.1016/s0925-4773(01)00658-x. [DOI] [PubMed] [Google Scholar]
- 26.Tomasini AJ, Schuler AD, Zebala JA, Mayer AN. PhotoMorphs: a novel light-activated reagent for controlling gene expression in zebrafish. Genesis. 2009;47(11):736–743. doi: 10.1002/dvg.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishna Priya S, Nagare RP, Sneha VS, Sidhanth C, Bindhya S, Manasa P, Ganesan TS. Tumour angiogenesis-Origin of blood vessels. Int J Cancer. 2016;139(4):729–735. doi: 10.1002/ijc.30067. [DOI] [PubMed] [Google Scholar]
- 28.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285(21):1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 29.Marcola M, Rodrigues CE. Endothelial progenitor cells in tumor angiogenesis: another brick in the wall. Stem Cells Int. 2015;2015:832649. doi: 10.1155/2015/832649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Huang J, Yao WY, Ben QW, Chen DF, He XY, Li L, Yuan YZ. The origins of vacularization in tumors. Front Biosci (Landmark Ed) 2012;17:2559–2565. doi: 10.2741/4071. [DOI] [PubMed] [Google Scholar]
- 31.Rasmussen TL, Shi X, Wallis A, Kweon J, Zirbes KM, Koyano-Nakagawa N, Garry DJ. VEGF/Flk1 signaling cascade transactivates Etv2 gene expression. PloS one. 2012;7(11):e50103. doi: 10.1371/journal.pone.0050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakano T, Abe M, Tanaka K, Shineha R, Satomi S, Sato Y. Angiogenesis inhibition by transdominant mutant Ets-1. J Cell Physiol. 2000;184(2):255–262. doi: 10.1002/1097-4652(200008)184:2<255::AID-JCP14>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 33.Wernert N, Raes MB, Lassalle P, Dehouck MP, Gosselin B, Vandenbunder B, Stehelin D. c-ets1 proto-oncogene is a transcription factor expressed in endothelial cells during tumor vascularization and other forms of angiogenesis in humans. Am J Pathol. 1992;140(1):119–127. [PMC free article] [PubMed] [Google Scholar]
- 34.Sumanas S, Choi K. ETS Transcription Factor ETV2/ER71/Etsrp in Hematopoietic and Vascular Development. Curr Top Dev Biol. 2016;118:77–111. doi: 10.1016/bs.ctdb.2016.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.