Abstract
Background
Capillary malformation is a cutaneous vascular anomaly that is present at birth, darkens over time, and can cause overgrowth of tissues beneath the stain. The lesion is caused by a somatic activating mutation in GNAQ. In a previous study we were unable to identify a GNAQ mutation in patients with a capillary malformation involving an overgrown lower extremity. We hypothesized that mutations in GNA11 or GNA14, genes closely related to GNAQ, also may cause capillary malformations.
Methods
Human capillary malformation tissue obtained from 8 patients that had tested negative for GNAQ mutations were studied. Lesions involved an extremity (n=7) or trunk (n=1). Droplet digital PCR (ddPCR) was used to detect GNA11 or GNA14 mutant cells (p.Arg183) in the specimens. Single molecule molecular inversion probe sequencing (smMIP-seq) was performed to search for other mutations in GNA11. Mutations were validated by sublconing and sequencing amplimers.
Results
We found a somatic GNA11 missense mutation (c.547C>T; p.Arg183Cys) in 3 patients with a diffuse capillary malformation of an extremity. Mutant allelic frequencies ranged from 0.3%–5.0%. GNA11 or GNA14 mutations were not found in 5 affected tissues or in unaffected tissues (white blood cell DNA).
Conculsions
GNA11 mutations are associated with extremity capillary malformations causing overgrowth. Pharmacotherapy that affects GNA11 signaling may prevent the progression of capillary malformations.
Keywords: capillary malformation, GNAQ, GNA11, vascular anomaly, extremity
INTRODUCTION
Capillary malformation is a congenital vascular lesion that affects the integument. Lesions darken, thicken, and cause disfigurement; adipose, muscle, and bone beneath the stain also can become overgrown. Somatic mutations in GNAQ (altering amino acid residue p.Arg183) result in constitutive activation of G-protein signaling and sporadic and syndromic (i.e., Sturge-Weber Syndrome) capillary malformations [1–3]. Mutations are enriched in the lesions’ endothelial cell compartment [3]. In a previous study we were unable to identify GNAQ mutations in a cohort of capillary malformations involving the lower extremity [3]. We hypothesized that somatic GNA11 mutations would also cause capillary malformations because GNAQ and GNA11 share > 90% amino acid identity, and mutations affecting a different amino acid residue p.Gln209 in either GNAQ or GNA11 cause congenital hemangiomas [4].
METHODS
The Committee on Clinical Investigation of Boston Children’s Hospital approved this study. Two biopsy specimens were collected as part of a research protocol while the remaining samples were obtained during a clinically-indicated procedure. Capillary malformation was diagnosed based on history, physical examination, and histopathology. Tissues were flash-frozen and placed in −80°C until further processing. Capillary malformation specimens from 8 patients that previously tested negative for a GNAQ mutation by droplet digital PCR (ddPCR) were included in this study. Because the most common GNAQ mutation in capillary malformation affects p.Arg183, we designed a ddPCR assay to screen for mutations in the equivalent codon of GNA11 or GNA14.
RESULTS
We found a somatic GNA11 missense mutation (c.547C>T; p.Arg183Cys) in specimens from 3 patients (Table 1, Fig. 1). Individuals had a phenotype consistent with diffuse capillary malformation with overgrowth [5]. We validated the presence of the mutation in affected tissue using an orthologous method by subcloning and sequencing amplimers from an independent PCR reaction performed on separately extracted DNA. In 2 patients with GNA11 mutations in affected tissue (participant 1, participant 5) we performed ddPCR on unaffected tissue (white blood cell DNA) and did not detect the mutant allele. We excluded false positive results, which could occur if our ddPCR assay had poor specificity, by not finding a GNA11 or GNA14 mutation in affected tissue from patients with capillary malformations caused by GNAQ mutations. To determine if other mutations in GNA11 account for the ddPCR-negative samples, we performed single molecule molecular inversion probe sequencing (smMIP-seq) [4] in 4 specimens that had sufficient DNA. No additional GNA11 mutations were identified.
Table 1.
GNA11 Mutation Detection in Capillary Malformation
| Patient | Age | Sex | Location | ddPCR | MIP-seq |
|---|---|---|---|---|---|
| 1 | 9 y/o | F | Lower extremity | 9/2871 (0.3%) | 1/78 (1%) |
| 2 | 9 y/o | F | Lower extremity | 1/2214 (0%)1 | 1/270 (0.4%) |
| 3 | 10 y/o | M | Lower extremity | 0/4306 (0%) | 0/146 (0%) |
| 4 | 14 y/o | F | Lower extremity | 0/3691 (0%) | 0/61 (0%) |
| 5 | 17 y/o | F | Lower extremity | 75/1557 (5%) | 5/94 (5%) |
| 6 | 21 y/o | F | Posterior Trunk | 1/7288 (0%)1 | 0/181 (0%) |
| 7 | 25 y/o | F | Lower extremity | 0/3815 (0%) | - |
| 8 | 27 y/o | M | Upper extremity | 90/3162 (3%) | - |
| Lower extremity | 21/4379 (0.5%) | - |
Abbreviations: F, female; M, male; ddPCR, droplet digital polymerase chain reaction; MIP-seq, molecular inversion probes sequencing.
ddPCR and MIP-seq columns indicate number of droplets and read depth, respectively. The rate of variant/total alleles is also depicted in the aforementioned columns. Dash (−) indicates the assay was not performed.
We considered a mutant allele frequency <1/000 to be background noise by ddPCR
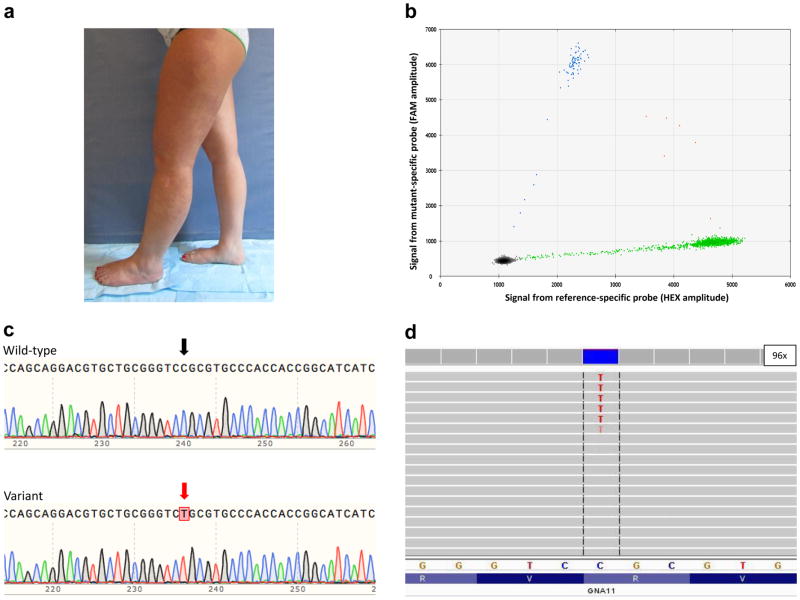
Figure 1.
GNA11 mutation in Participant 5. (a) Photograph depicting the patient’s capillary malformation and hypertrophy of her left lower extremity. (b) Results from a ddPCR reaction displaying the presence of the GNA11 c.547C>T (p.ArgR183Cys) mutation with a mutant allele frequency of ~5%. (c) Sanger sequencing of PCR-amplimer-subclones showing (Top) a clone with the wild-type sequence and (Bottom) another with the mutation at position 547 having a T (red arrow) instead of a C (black arrow). (d) Integrative Genomic Viewer screen-shot depicting MIP-seq coverage at the site of the mutation (c.547C>T) with ~5% mutant allele frequency. 96x indicates the depth of coverage at position 547.
DISCUSSION
The most common mutation found in capillary malformation is GNAQ p.Arg183Gln [1,3,6]. Other GNAQ somatic missense mutations affecting residue 183, p.Arg183Leu and p.Arg183Gly, also have been observed in patients with capillary malformation [3]. Here, we report a second gene responsible for this lesion, GNA11. Consistent with our finding, functional overlap or redundancy between GNAQ and GNA11 also have been observed in uveal melanoma [7], phakomatosis pigmentovascularis [8], melanomas associated with blue nevi [9], and congenital hemangiomas [4]. Although GNA14 shares ~80% amino acid identity with GNAQ and GNA11, and can cause vascular tumors [10], we did not identify GNA14 mutations in capillary malformations.
GNAQ and GNA11 R183 mutations partially inactivate guanosine triphosphatase (GTPase) activity, resulting in constitutive activation of the mitogen-activated protein (MAP) kinase pathway; Q209 mutations completely inactivate the enzyme [11]. These mutations affect vascular endothelial growth factor-2 (VEGF-2) function [12–14]. Because VEGF2 is critical for both vasculogenesis and angiogenesis, mutations affecting this protein likely affect the pathophysiology of different types of vascular lesions (i.e., capillary malformation, congenital hemangioma). The more severe Q209 inhibition of GTPase may explain the presence of this mutation only in tumors and not malformations [4,11].
Currently, we do not know whether there are phenotypic differences between patients with GNAQ and GNA11 p.Arg183 mutations. Among published reports, >95% of facial capillary malformations have a GNAQ p.Arg183 mutation [1,3,6]. In this study as well as a previous investigation [3], we have examined 10 extra-facial capillary malformations and have observed greater genetic heterogeneity: GNA11 p.Arg183Cys in 3 legs and 1 arm, GNAQ p.Arg183Leu in 1 leg, and neither GNA11 nor GNAQ in 4 legs and 1 trunk.
Our failure to find a GNA11, GNA14, or GNAQ mutation in several specimens could be because the mutant allele frequency was below our limit of detection. Intriguingly, participant 2 might be an example of this since her specimen had the GNA11 p.Arg183Cys allele detected by ddPCR and smMIP-seq, but at levels below our true-positive threshold. Alternatively, mutations in another gene may be responsible for causing capillary malformations. Our identification of GNA11 gain-of-function mutations in a subset of patients with capillary malformation indicate that drugs which suppress constitutive GNAQ or GNA11 signaling may have therapeutic value.
Acknowledgments
Research reported in this manuscript was supported by the National Institutes of Health Award NICHD-081004 (AKG), National Institutes of Health Award NICHD-082606 (AKG), National Institutes of Health Award HL127030 (JB, AKG), National Institutes of Health Award AR-064231 (MLW), the Translational Research Program Mid-Career Award Boston Children’s Hospital (AKG), and the Translational Neuroscience Center Pilot Study Award Boston Children’s Hospital (JB, AKG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors have no disclosures
References
- 1.Shirley MD, Tang H, Gallione CJ, et al. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013;368:1971–9. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakashima M, Miyajima M, Sugano H, et al. The somatic GNAQ mutation c.548G>A (p. R183Q) is consistently found in Sturge-Weber syndrome. J Hum Genet. 2014;59:691–3. doi: 10.1038/jhg.2014.95. [DOI] [PubMed] [Google Scholar]
- 3.Couto JA, Huang L, Vivero MP, et al. Endothelial Cells from Capillary Malformations Are Enriched for Somatic GNAQ Mutations. Plast Reconstr Surg. 2016;137:77e–82e. doi: 10.1097/PRS.0000000000001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayturk UM, Couto JA, Hann S, et al. Somatic Activating Mutations in GNAQ and GNA11 Are Associated with Congenital Hemangioma. Am J Hum Genet. 2016;98:789–95. doi: 10.1016/j.ajhg.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee MS, Liang MG, Mulliken JB. Diffuse capillary malformation with overgrowth: a clinical subtype of vascular anomalies with hypertrophy. J Am Acad Dermatol. 2013;69:589–94. doi: 10.1016/j.jaad.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 6.Lian CG, Sholl LM, Zakka LR, et al. Novel genetic mutations in a sporadic port-wine stain. JAMA Dermatol. 2014;150:1336–40. doi: 10.1001/jamadermatol.2014.1244. [DOI] [PubMed] [Google Scholar]
- 7.Metz CH, Scheulen M, Bornfeld N, Lohmann D, Zeschnigk M. Ultradeep sequencing detects GNAQ and GNA11 mutations in cell-free DNA from plasma of patients with uveal melanoma. Cancer Med. 2013;2:208–15. doi: 10.1002/cam4.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas AC, Zeng Z, Riviere JB, et al. Mosaic Activating Mutations in GNA11 and GNAQ Are Associated with Phakomatosis Pigmentovascularis and Extensive Dermal Melanocytosis. J Invest Dermatol. 2016;136:770–8. doi: 10.1016/j.jid.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa S, Byrne M, Pissaloux D, et al. Melanomas Associated With Blue Nevi or Mimicking Cellular Blue Nevi: Clinical, Pathologic, and Molecular Study of 11 Cases Displaying a High Frequency of GNA11 Mutations, BAP1 Expression Loss, and a Predilection for the Scalp. Am J Surg Pathol. 2016;40:368–77. doi: 10.1097/PAS.0000000000000568. [DOI] [PubMed] [Google Scholar]
- 10.Lim YH, Bacchiocchi A, Qiu J, et al. GNA14 Somatic Mutation Causes Congenital and Sporadic Vascular Tumors by MAPK Activation. Am J Hum Genet. 2016;99:443–50. doi: 10.1016/j.ajhg.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sivaraj KK, Li R, Albarran-Juarez J, et al. Endothelial Galphaq/11 is required for VEGF-induced vascular permeability and angiogenesis. Cardiovasc Res. 2015;108:171–80. doi: 10.1093/cvr/cvv216. [DOI] [PubMed] [Google Scholar]
- 13.Zeng H, Zhao D, Mukhopadhyay D. KDR stimulates endothelial cell migration through heterotrimeric G protein Gq/11-mediated activation of a small GTPase RhoA. J Biol Chem. 2002;277:46791–8. doi: 10.1074/jbc.M206133200. [DOI] [PubMed] [Google Scholar]
- 14.Zeng H, Zhao D, Yang S, Datta K, Mukhopadhyay D. Heterotrimeric G alpha q/G alpha 11 proteins function upstream of vascular endothelial growth factor (VEGF) receptor-2 (KDR) phosphorylation in vascular permeability factor/VEGF signaling. J Biol Chem. 2003;278:20738–45. doi: 10.1074/jbc.M209712200. [DOI] [PubMed] [Google Scholar]