Abstract
Background: Mass cytometry (Cytometry by Time of Flight, CyTOF) allows single-cell characterization on the basis of specific metal-based cell markers. In addition, other metals in the mass range such as silver can be detected per cell. Bacteria are known to be sensible to silver and a protocol was developed to measure both the number of affected cells per population and the quantities of silver per cell.
Methods: For mass cytometry ruthenium red was used as a marker for all cells of a population while parallel application of cisplatin discriminated live from dead cells. Silver quantities per cell and frequencies of silver containing cells in a population were measured by mass cytometry. In addition, live/dead subpopulations were analyzed by flow cytometry and distinguished by cell sorting based on ruthenium red and propidium iodide double staining. Verification of the cells’ silver load was performed on the bulk level by using ICP-MS in combination with cell sorting. The protocol was developed by conveying both, fast and non-growing Pseudomonas putida cells as test organisms.
Results: A workflow for labeling bacteria in order to be analyzed by mass cytometry was developed. Three different parameters were tested: ruthenium red provided counts for all bacterial cells in a population while consecutively applied cisplatin marked the frequency of dead cells. Apparent population heterogeneity was detected by different frequencies of silver containing cells. Silver quantities per cell were also well measurable. Generally, AgNP-10 treatment caused higher frequencies of dead cells, higher frequencies of silver containing cells and higher per-cell silver quantities. Due to an assumed chemical equilibrium of free and bound silver ions live and dead cells were associated with silver in equal quantities and this preferably during exponential growth. With ICP-MS up to 1.5 fg silver per bacterial cell were detected.
Conclusion: An effective mass cytometry protocol was developed for the detection and quantification of silver in single bacterial cells of different physiological states. The silver quantities were generally heterogeneously distributed among cells in a population, the degree of which was dependent on micro-environmental conditions and on silver applied either in ion or nanoparticle-aggregated form.
Keywords: mass cytometry, metal-based cell marker, silver quantification in single cells, silver distribution, bacterial heterogeneity, silver nanoparticles
Introduction
Increasing use of antimicrobial commercial products amended with silver nanoparticles (AgNP) has caused concerns, and extensive study was directed on AgNP toxicity of causing generation of reactive oxygen species (Choi and Hu, 2008; Franci et al., 2015), cell growth inhibition (Ivask et al., 2014), or cell viability loss (Guo et al., 2017). The characterization of AgNP physicochemical behavior with the focus on their dissolution, aggregation, and transformation in biological environments became increasingly important (Fabrega et al., 2011). As a result of these efforts recent studies suggested that dissolved silver ions can be held responsible for the antimicrobial qualities of AgNP (Xiu et al., 2012). We confirmed these findings in an own study but revealed an additional particle effect supposedly caused by fast formation of huge AgNP-aggregates of about 500 nm in cell solutions and suggested their contribution to higher cell death ratios (Guo et al., 2017). The attachment of AgNP-aggregates to single bacterial cells was identified by TEM and SEM-EDX (Guo et al., 2017). Aside from TEM and SEM-EDX, other techniques have been used to visualize and identify metals in single human cells, e.g., AFM, SXFM (Cerchiaro et al., 2013). However, these microscopy techniques have limitations including random sample detection, and lack of quantitative information regarding cellular interactions with nanoparticles. Flow cytometry has been reported to characterize single cells in microbial populations with high-throughput by means of light scatter and fluorescent dyes (Müller and Nebe-von-Caron, 2010). Cellular AgNP uptake was conveyed by this method via changes of side scatter characteristics but only for eukaryotic cells (Lankoff et al., 2012; Zucker et al., 2013; Zhao and Ibuki, 2015). For bacterial cells side scatter did not reveal any AgNP uptake (for sizes of 10 and 30 nm; Guo et al., 2017). Instead, ICP-MS has been operated widely to quantify the contents of silver ions in bacterial cells (Swathy et al., 2014; Wakshlak et al., 2015). It is a destructive technique where entire cell suspensions are digested and injected which prevents differentiation of silver-affected cells from unaffected ones. The ICP-MS obtained average value from a highly heterogeneous cell population disregards cell subsets and diverse phenotypes that may be relevant to reveal causality of cell reaction and toxicity in response to silver.
A growing interest in single-cell analysis can be recognized and numerous analytical methods have been developed or improved. One of those technologies is mass cytometry, which couples mass spectrometry with single-cell analysis and was introduced as Cytometry by Time of Flight (CyTOF) (Bendall et al., 2011; Chang et al., 2017). CyTOF offers numerous potential advantages over fluorescence-based flow cytometry, e.g., overcoming the challenge of spectral overlap intrinsic to fluorescent dyes by using rare-earth-metal stable isotopes with little signal overlap. In addition, more than 35 simultaneously measured cellular parameters compared to 10 markers in fluorescence-based flow cytometry can be measured (Bjornson et al., 2013; Chattopadhyay et al., 2014). To date this promising technique is mainly used for human cells as an effective tool for drug development or improvement of therapeutic programs ranging from infectious disease to cancer (Bendall et al., 2011; Gavasso et al., 2016; Robinson and Mao, 2016; Baca et al., 2017; Baumgart et al., 2017). Also other inorganic nanoparticles have been lables (Vancaeyzeele et al., 2007; Lin et al., 2014; Tong et al., 2016; Schulz et al., 2017) but were in the focus of biodistribution experiments (Yang et al., 2017). Limited studies have been reported on microbial cells (Leipold et al., 2011; Miyashita et al., 2014), where, e.g., a combination of a metal-based membrane stain and lectins, conjugated to lanthanide-chelating polymers, was used to differentiate Escherichia coli cells based on their cell surface polysaccharides.
In this study, we tested the mass cytometry technology for discrimination of live/dead cell states and simultaneous quantification of silver in single bacterial cells. An earlier study (Guo et al., 2017) revealed random attachment of huge up to 500-nm-AgNP-aggregates to a limited number of cells in a population after few minutes treatment of cells with 10- and 30-nm AgNP at environmental relevant concentrations. A relation between viability states and increased quantities of silver ions in cells by those AgNP-aggregates was suggested. Because flow cytometry does not allow direct detection of these two events simultaneously, a mass cytometry workflow was developed for the purpose. Such data may be especially useful to link cell states and features with cell fate and thus to contribute to the development of models that implement immanent characteristics of an individual cell and its individual capacity to notice random, selective, and perhaps lethal influences from the environment.
Materials and Methods
Materials
Silver nitrate (AgNO3) (99.9%) and ruthenium red (RR) was purchased from Sigma–Aldrich (United States). AgNPs were provided by nanoComposix (United States) as aqueous suspensions [citrate coated, mass concentration (Ag) 0.02 mg/mL] of the size 10 nm (9.4 ± 1.7 nm, AgNP-10). Cis-Platinum (II) diamine dichloride (cisPt) was purchased from Enzo Life Sciences GmbH (Lörrach, Germany). Nitric acid (HNO3) was purchased from Merck (Germany). M12 medium and PBS compositions were shown in Supplementary Table S1. For washing purposes 18.2 MΩ⋅cm water (MilliQ, Germany) was used.
Bacterial Cultivation
Pseudomonas putida KT2440 was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Germany). Bacterial standard-growth was performed in M12 medium on a rotary shaker at 30°C and 170 rpm. The growth was monitored by optical density at λ = 600 nm (Spectra max Plus 384 photometer, Molecular Devices, Sunnyvale, CA, United States).
Bacterial Cultivation under Silver Treatment
An overnight pre-culture of P. putida KT2440 was incubated in M12 medium with an initial OD600 of 0.09 and grown for 72 h (30°C, 170 rpm). Either AgNP-10 (1.29 mg/L) or AgNO3 (0.19 mg/L) were implemented in the cultivations and chosen concentrations referred to the determined EC50 values from an earlier publication (Guo et al., 2017). Cultivations without silver treatment served as silver-ion negative control while application of AgNO3 served as silver-ion positive control. Cells were harvested at 0, 12, 48, and 72 h and treated separately according to the mass cytometry staining protocol (see below).
Determination of Cell Number
To analyze bacteria on the single cell level at the mass cytometer, a concentration of 5.0 × 105 cells/mL was required for each injection. Therefore, a fast and accurate cell counting method was required and for this a range of linear relationship between cell counts and OD600 was exploited. Cell counts were determined by a flow cytometer (Becton, Dickinson and Company, Franklin Lakes, NJ, United States) together with a calibrated suspension of microsphere standard (6.0 μm diameter microspheres at a concentration of 108 beads/mL in Milli-Q water containing 2 mM sodium azide, L34856, Thermo Fisher Scientific, Germany) for accurate cell count measurements. OD600 was analyzed by a spectrophotometer. All measurements were done in replicates and shown in Supplementary Figure S1.
All Cell Indicator for Mass Cytometry
To optimize RR staining for P. putida KT2440 populations, a stock solution of 1.3 mM of RR in PBS was prepared and stored at 4°C. Before use the solution was ultra-sonicated (Ultrasonic bath, Merck Eurolab, Darmstadt, Germany) for 10 min. 108 cells were treated with different concentrations of RR (0 to 0.33 μM) for 30 and 60 min. The final staining protocol requires 0.13 μM RR/108 cells/200 μL PBS and a staining time of 30 min at room temperature (RT). Details of staining optimization were shown in Supplementary Figure S2A.
Dead Cell Indicator for Mass Cytometry
To optimize cisPt staining for P. putida KT2440 populations, a stock solution of 25 mM of cisPt in dimethyl sulfoxide (Sigma–Aldrich, United States) was prepared and stored at -20°C. 108 cells were stained with different concentrations of cisPt (0 to 20 μM) and treated for 1, 5, 10, 30 min. The final staining protocol requires 5 μM cisPt/108 cells/1 mL PBS and a staining time of 10 min at RT. Details of staining optimization and calibration curves generated on the basis of RR and cisPt measurements were shown in Supplementary Figures S2B,C.
Dead Cell Indicator for Flow Cytometry
The fluorescent dye propidium iodide (PI) was used as dead cell indicator for P. putida KT2440. Staining optimization and calibration were published (Guo et al., 2017). The final protocol requires 2 μM PI/108 cells/1 mL PBS and a staining time of 2 min. For quantification of PI stained dead cells (PI+dead) of RR labeled P. putida KT2440, cells were double stained with RR (0.13 μM, 30 min), followed by PI (2 μM, 2 min) and analyzed by flow cytometry at log scale.
Workflow for Analysis of Bacterial Cells at the Mass Cytometer
Harvested cells were diluted to 108 cells/1 mL PBS and centrifuged at 3200 × g for 10 min. The pelleted cells were re-suspended in cisPt solution (5 μM, 10 min, RT). Subsequently, the cisPt stained dead cells (cisPt+dead) were washed (3200 × g, 10 min) twice with 3 and 1 mL PBS, respectively, to remove unbound cisPt. Finally, cells were stained with RR (0.13 μM, 30 min, RT), washed twice (3200 × g, 10 min) with 1 mL water to remove salts and unbound RR. For mass cytometry measurement the cell concentration was adjusted to 5.0 × 105 cells/mL in Milli-Q water. Four element calibration beads (Fluidigm, United States) were added 1:10 v/v before acquisition for later normalization (Finck et al., 2013).
Mass Cytometry
The CyTOF instrument (Fluidigm Corp, South Francisco, CA, United States) was tuned, calibrated, and cleaned on the daily base according to the manufacturer’s advice. Aqueous bacterial cell suspensions were acquired on a CyTOF instrument version 1 upgraded to the control software of v6.0.626 with a flow rate of 45 μL/min. Argon gas 5.0 was used to generate the plasma and for nebulizing the cell suspension. About 1.5 × 105 cells per sample were subsequently loaded onto a 450 μL sample loop and measured in a dual instrument mode with noise reduction turned on, a cell length range from 1 to 75 and with ‘on the fly’ processing. The initial raw data were processed into standard FCS file format, randomized and normalized by Software Helios version 6.5.358 (Fluidigm Corp, South San Francisco, CA, United States). Data analysis was performed with FlowJo (version 10) (TreeStar, Ashland, OR, United States). Monitoring was done by analyzing natural abundance isotopes of ruthenium (102Ru and 104Ru), platinum (195Pt) and silver (107Ag and 109Ag, 51.8 and 48.2%, respectively). Silver was measured in ionic form and resulting data were related to the 107Ag isotope.
Flow Cytometry and Cell Sorting
Cytometric measurements were performed with a BD Influx v7 Sorter USB, (Becton, Dickinson and Company, Franklin Lakes, NJ, United States) equipped with a blue 488-nm Sapphire OPS laser (400 mW, Coherent, Santa Clara, CA, United States). The 488-nm laser light was used for the analysis of the forward scatter (FSC, 488/10), the side scatter (SSC, 488/10, trigger signal), and the PI induced red fluorescence (616/23). The fluidic system was run at 33 psi using a 70-μm nozzle. The sheath fluid consisted of 0.5 × FACSFlow buffer (BD). For the optical calibration of the cytometer in the linear range, 1-μm blue fluorescent FluoSpheres (Molecular Probes, F-8815, Eugene, OR, United States) and 2-μm yellow-green fluorescent FluoSpheres (Thermo Fisher Scientific, F8827, Waltham, MA, United States) were used. For calibration in the log range, 0.5-μm UV Fluoresbrite Microspheres (Polysciences, 18339, Warrington, PA, United States) were applied. Sorting of PI unstained live (PI-live) and PI stained dead (PI+dead) cells into plastic tubes was performed at an event rate of 5.000/sec which corresponded to a sort rate of 700–1.500 cells/sec. The sort mode was 1.0 Drop Pure. To obtain sufficient cell amounts for the following mass cytometry and ICP-MS measurements, up to 3.0 × 106 cells were sorted for either PI-live or PI+dead cells. After sorting cells were pre-processed before CyTOF or ICP-MS analysis: sorted cells were transferred from plastic tubes into glass tubes and centrifuged at 3200 × g for 10 min, supernatant was removed and the cell pellets were stored at -20°C. Before mass cytometry measurements, cell pellets were washed once with 1 mL Milli-Q water by re-suspension in 1 mL Milli-Q water for injection.
ICP-MS Analysis
For bulk ICP-MS analysis, the sorted PI-live and PI+dead cell pellets were digested by addition of 50 μL HNO3 (60%, ultrapure) for a period of 15 min. Subsequently the solution was suspended in Milli-Q water dispensed with rhodium (103Rh) as internal standard [c (Rh) = 0.75 ng/mL] to a 1 mL solution. The solution was measured with an Inductively Coupled Plasma Mass Spectrometer (ICP-MS, Element XR, Thermo, Germany) equipped with a micro-concentric nebulizer (Micromist-100, Glass Expansion, Australia) consuming 135 μL sample per minute. Silver was measured and quantified in ionic form related to the 109Ag isotope.
Results and Discussion
Bacterial Labeling for Mass Cytometry Analysis
Cytometry by Time of Flight technology substitutes isotopes of transition elements and lanthanides for fluorescent dyes. The cells are directed into a narrow flow to be screened one by one and vaporized. Elements of the cells are atomized, ionized, and subsequently analyzed by a time-of-flight mass spectrometer. The measured signal of natural abundance isotopes of the rare-earth-metals ruthenium and platinum per cell from applied RR and cisPt provided the number of labeled cells and the isotopes’ quantity per cell. RR is a cationic reagent and has been widely used to locate acidic polysaccharide-like material (Fletcher and Floodgate, 1973; Fassel and Edmiston, 1999; Waller et al., 2004) and to visualize numerous ultra-structural details in and outside of cells (Lingens et al., 1985; Chatterjee et al., 2010; Perfumo et al., 2014). Because RR is easy to operate by addressing all cells in a population directly without pre-treatment (instead of, e.g., fixation, necessary for flow cytometric measurements; Günther et al., 2008) and its atomic mass is within the detectable mass range of mass cytometers, RR was used as an all cell indicator for the CyTOF workflow. However, the metal was not used in this context for bacteria before. The second dye used in this study, cisPt, is a readily available platinum based chemotherapeutic agent and reacts with protein nucleophiles, with which it can form covalent platinum–sulfur or sulfhydryl bonds (Fienberg et al., 2012). cispt enters cells with compromised cell membranes, where it non-specifically labels total cellular protein. The reagent can thus be used to discriminate live from dead cells which is well practiced for human cells (Bjornson et al., 2013; Nair et al., 2015). However, applications on bacteria haven’t been tested yet to our knowledge.
In our study, the Gram-negative bacterium P. putida KT2440 was used as a test organism to establish a protocol for RR and cisPt staining in order to detect all cells in a population and to differentiate live from dead cells. The RR stained bacteria were well detectable by the CyTOF and entirely identified within a cell gate plotted via the two most natural abundance ruthenium isotopes 102Ru/104Ru. Protein binding cisPt was used as second cell marker for dead cells and identified by the 195Pt isotope. For this combined marker group optimized staining conditions were tested with regard to dye concentrations and times of incubation and the trade-off between highest intensity and shortest staining time was chosen for the final workflow (Figure 1A and Supplementary Figure S2B). To verify the reliability of the cisPt marker different ratios of live and dead cells (70% ethanol treated, 20 min, RT) were prepared and measured at CyTOF. In parallel the same ratios were analyzed by using the fluorescent marker PI (binding on nucleic acids) for dead cells and flow cytometry. While flow cytometry mirrored the precise ratios, CyTOF gave lower dead cell counts (Supplementary Figure S2C). Up to now the dye was only applied to cell-wall-free cells such as human cells effectively (Majonis et al., 2011; Fienberg et al., 2012). Bacterial cells, however, have rigid cell walls, subsistent also in dead cells, forming a barrier which must be overcome by cisPt. It can be assumed that the limitation in the quantitative detection of dead cells in a population may be caused by this boundary. Nevertheless, despite the determination of dead cell numbers by cisPt in a population of P. putida KT2440 was lower in comparison to PI staining (Supplementary Figure S2C: kcisPt = 0.45, kPI = 0.98), the stable isotopes’ calibration of RR and cisPt proved to be useful cell markers. Standard-grown P. putida KT2440 cells (48 h) were represented in typical 2D-plots for RR staining (Figure 1B). The number of dead cells was 2.7% (Q2) measured by cisPt and mass cytometry (Figure 1C), and 4.1% (R2) measured by PI and flow cytometry (Figure 1D).
FIGURE 1.
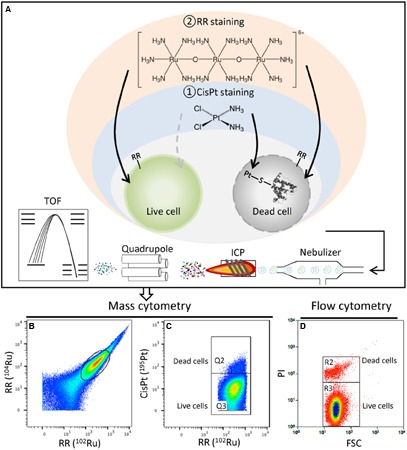
(A) CyTOF workflow for bacteria. Cells were first stained by cisPt for live/dead discrimination, followed by RR staining as an overall bacterial cell indicator. After washing, cells were subjected to the mass cytometer as a stream of single cells. In the plasma region elements of the cells are atomized and ionized. Ions of high-mass elements (i.e., ruthenium, platinum, and silver) enter selectively the TOF chamber and are separated according to their mass before detection. (B) CyTOF analysis of standard-grown and harvested P. putida KT2440 (48 h). CyTOF plots marked all events based on 102Ru/104Ru signal. (C) Dead cisPt+ (Q2) cells are distinguished from live cisPt– (Q3) cells. (D) Dead (R2) and live (R3) cells are distinguished by PI staining and flow cytometry.
Detection of Silver of Single Cells of P. putida
Pseudomonas putida KT2440 is known to react on various silver concentrations by changes in growth rates and live/dead cell ratios. The toxicity of silver was recently assumed to be caused by the ions only (Xiu et al., 2012; Giao et al., 2017; Juganson et al., 2017) but we detected an additional particle-related effect (Guo et al., 2017). We assumed that the typical fast aggregation of AgNPs to huge complexes in a nature-like environment within 30 min (Guo et al., 2017) might contribute to a further increase of dead cell counts due to their random attachment to bacterial cell surfaces and their thereby steady release of additional silver ions into those cells.
Thus, mass cytometry was involved in this study to clarify if the treatment of bacteria with either silver ions or AgNP (1) generates different frequencies of cells that contain silver which may cause different live/dead cell ratios and (2) produces disparate quantities of silver per cell suggesting that populations exposed to AgNP might contain cells with a higher silver load. To answer these questions P. putida KT2440 cells were treated with the respective EC50 concentrations of 1.29 mg/L for AgNP-10 and 0.19 mg/L for AgNO3 as the positive control. As determined before, the released ion concentrations were nearly similar because cell-free AgNP-10 dissolution kinetic curves inferred 0.08 mg/L silver ions released from AgNP-10 (1.29 mg/L) in comparison to 0.12 mg/L silver ions from AgNO3 (0.19 mg/L) over a time period of 72 h (Guo et al., 2017). Standard-grown P. putida KT2440 without silver treatment served as the negative control. The growth curves (Figure 2, bottom) showed a delayed lag-phase for the AgNP-10 treated bacteria and a slower growth rate (μ = 0.14/h) for the AgNO3 positive control in comparison to the negative control (μ = 0.18/h). CyTOF technology was used to mark all bacterial cells of those populations and to distinguish live from dead cells. The additional measured silver signal was used to determine the frequency of silver containing cells as well as the quantities of silver per cell via the intensity of the mass cytometric signal for silver (IAg).
FIGURE 2.
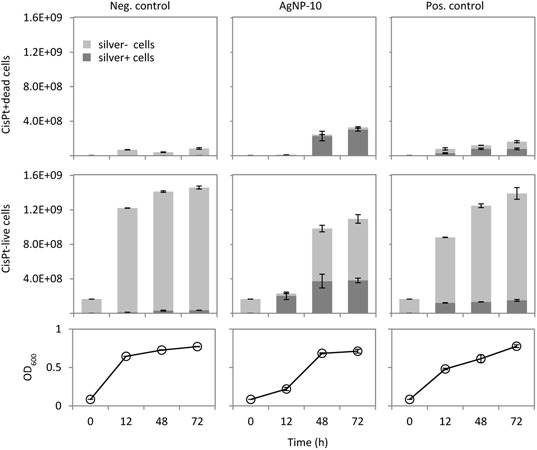
CyTOF measurements on cisPt-live/cisPt+dead cell discrimination and the number of silver containing cells in a population. P. putida KT2440 cells were cultivated in M12 medium with AgNP-10 (1.29 mg/L), with AgNO3 (0.19 mg/L) as positive control, and untreated as negative control. Growth curves were shown for 72 h by measuring OD600. Cells were harvested at 12, 48, 72 h and measured by CyTOF. Light gray bars showed cells without silver load, while dark gray bars showed silver containing cells. Standard errors were from replicate analysis.
After 72 h and in comparison to the negative control AgNP-10 treatment caused an increase of cisPt+dead cells by a factor of about 4 while the AgNO3 positive control showed a factor of 2 (Figure 2), indicating that the particles caused higher frequencies of dead cells. Further, the frequency of cells that contain silver (Figure 2, dark gray bars) was also higher for the AgNP-10 treatment in comparison to the positive control while cells of the untreated negative control were nearly not loaded with silver (Figure 2). After 72 h AgNP-10 treatment 92% of cisPt+dead cells and 35% of cisPt-live cells were loaded with silver. Instead, the AgNO3 positive control revealed 48% silver containing cells of the cisPt+dead and only 11% of the cisPt-live cells (Figure 2). The data show that AgNP-10 treatment caused the highest frequencies of dead cells in comparison to both controls after 72 h, and that almost all of the dead cells were loaded with silver. At the beginning of the exponential phase (12 h) where cells thrived at merely low numbers, live cells were still predominant but silver was loaded to any cell independent of its viability state. At longer exposure time the high frequency of silver loaded dead cells was almost consistent between 48 and 72 h. A similar tendency was observed within the larger fraction of surviving silver containing cells, indicating that the absorbed silver (ions) do not cause immediate cell death.
We also determined silver quantities per cell by measuring their IAg values which were different between AgNO3 positive control and AgNP-10 treated cells (Table 1). Overall, AgNP-10 treatment led to much higher silver quantities per cell in comparison to the positive control. Our data also suggest that the AgNP-aggregates, formed asap from AgNP-10, were not present at all or at least in their entirety due to the stringent washing procedures before CyTOF measurement otherwise we would expect an elevated silver signal in the 1D-plot of the silver channel (Supplementary Figure S3). But the silver ions released earlier by AgNP-aggregates and then bound by the cells were obviously responsible for the higher silver contents in AgNP-10 treated cells. In addition, for AgNP-10 treated cells we found nearly identical IAg values for both cisPt-live and cisPt+dead cells when cells grew exponentially (12 h: 18.3 ± 0.6 vs. 17.1 ± 1.1, Table 1) which was different for early and late stationary cells where the IAg values were higher for the cisPt+dead cells (e.g., for 72 h: 1.6 ± 0.1 vs. 10.3 ± 0.6). For AgNO3 treated cells the IAg values per cell were generally much lower and increased finally to only 3.0 ± 0.3 after 72 h for cisPt+dead cells. Thus, silver quantities per cell were higher under AgNP-10 treatment with the highest IAg values determined for cells from exponential phase independent if they were dead or alive.
Table 1.
The silver quantities per cell via the intensity of the mass cytometric signal for silver (IAg).
| Time (h) | Treatment | IAg |
|
|---|---|---|---|
| cispt-live cells | cispt+dead cells | ||
| 12 | Pos. control | 0.5 ± 0.0 | 0.9 ± 0.1 |
| AgNP-10 | 18.3 ± 0.6 | 17.1 ± 1.1 | |
| 48 | Pos. control | 0.3 ± 0.0 | 3.4 ± 0.3 |
| AgNP-10 | 2.1 ± 0.0 | 11.0 ± 0.5 | |
| 72 | Pos. control | 0.3 ± 0.0 | 3.0 ± 0.3 |
| AgNP-10 | 1.6 ± 0.1 | 10.3 ± 0.6 | |
P. putida KT2440 cells were cultivated in M12 medium with AgNP-10 (1.29 mg/L) and with AgNO3 (0.19 mg/L) as positive control for 72 h. Cells were harvested at 12, 48, 72 h and measured by CyTOF. Median values for intensities of 107Ag (IAg) in cisPt-live and cisPt+dead cells were shown. Examples of CyTOF plots were in Supplementary Figure S3.
The data of both the calculated frequencies in cell numbers and the silver quantities per cell follow the same trend. AgNP-10 treatment cause higher frequencies of dead cells, higher frequencies of silver affected cells and higher per-cell silver quantities whereby live and dead cells load silver in equal quantities and this preferably during exponential growth.
Independent from this, the population showed heterogeneous cell states with respect to silver quantities which might simply be caused by chemical equilibria of available free silver ions. Although we were not able to distinguish a silver signal related to cell-attached AgNP-aggregates or to distinguish aggregate-dissolved silver ions from free silver ions with CyTOF technology we assume that by a fast process an equilibrium is established and leads to the silver ions’ equal distribution among cells. When the equilibrium changes in favor of the cells, because of increasing cell numbers during growth, these mostly new live cells were found free of silver. This observation was made also with AgNO3 treated cells. It can also be assumed that a continuous uptake of silver ions by the cells may influence the equilibrium between AgNP-aggregate bound and unbound silver ions in the medium and cause further dissolution of silver ions from the aggregates (Yue et al., 2017).
The equal silver quantities in both live and dead exponential grown cells were verified by sorting identical numbers (3 × 106 cells each) of PI unstained/PI stained cells, from which an aliquot of cells of each sample were injected into the mass cytometer and 106 cells of each sample was used for ICP-MS measurements. As before, the cells were treated by AgNP-10 at EC50 dosage of 1.29 mg/L. From CyTOF analysis we found analogous silver quantities per cell among PI-live and PI+dead cells with IAg = 21.6 and IAg = 17.9, respectively. The equal silver quantities per cell were by tendency confirmed by the ICP-MS results with 1.5 fg and 1.1 fg silver per PI-live and PI+dead cells, respectively. Overall, the equal silver quantity distribution among live and dead cells suggested that random AgNP-aggregate attachment is not only affecting and toxic to an appended cell but also to every other cell in its vicinity.
Conclusion
The primary objective of this study was to establish a mass cytometry method for the analysis of bacteria exemplary taking the Gram-negative bacterium P. putida KT2440. Our data show that by using RR, mass cytometry is able to detect all bacteria in a population, and that, by combining RR with cisPt, the method differentiates live from dead bacterial cells. The panel was extended by including natural abundance silver isotopes in order to measure the silver level in a single bacterial cell.
In Guo et al. (2017), PI uptake was correlated with silver-cell toxicity. This proof, however, was indirect. Now, mass cytometry was not only able to confirm the data of the earlier study by a direct measurement of silver per cell but provided further findings. AgNP-10 treatment caused a two times higher number of dead cells in comparison to AgNO3 treatment after 72 h cultivation and three times higher numbers of cells with silver load. The per-cell silver quantities were also higher in AgNP-10 treated cells. Therefore, mass cytometry clearly supported the earlier discussion of a particle effect in addition to the toxicity of silver ions. The origin of this additional silver can be assumed to come from the AgNP-aggregates. In addition, the heterogenic distribution of silver ions was verified but seems to be independent on live/dead cell states but rather on existing chemical equilibria in the environmental vicinity. Thus, the action of the silver is clearly dependent on complex effects. Prediction of toxicant behavior to cells in natural environments will be even more complex due to the presence of further affecting parameters and can probably only be predicted when models implement equilibria of chemical components and their bioavailability and adjoin such data with the heterogeneous state or feature of single cells.
Author Contributions
YG designed and conducted the experiments, evaluated the data and wrote the paper. SB analyzed samples for mass cytometry, helped to evaluate the data, and contributed to writing. H-JS analyzed samples for ICP-MS, helped to evaluate the data, and contributed to writing. HH contributed to writing. SM designed the experiments, evaluated the data and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Graduate School “Leipzig School of Natural Sciences-Building with Molecules and Nano-objects (BuildMoNa)”, funded by the European Social Fund (ESF, project number R313H044), the European Regional Development Funds (EFRE – Europe funds Saxony, 100192205), and the German Federal Ministry of Education and Research (BMBF) within the framework of the Med research and funding concept (sysINFLAME, grant # 01ZX1306B). We thank Thomas Hübschmann for advice and bacterial flow cytometric measurements at Flow Cytometry Group in Helmholtz Centre for Environmental Research (Leipzig, Germany). We also thank Toralf Kaiser for cell counts measuring at Flow Cytometry Core Facility in German Rheumatism Research Center (Berlin, Germany).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01326/full#supplementary-material
References
- Baca Q., Cosma A., Nolan G., Gaudilliere B. (2017). The road ahead: implementing mass cytometry in clinical studies, one cell at a time: editorial. Cytometry B Clin. Cytom. 92 10–11. 10.1002/cyto.b.21497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart S., Peddinghaus A., Schulte-Wrede U., Mei H. E., Grützkau A. (2017). OMIP-034: comprehensive immune phenotyping of human peripheral leukocytes by mass cytometry for monitoring immunomodulatory therapies. Cytometry A 91 34–38. 10.1002/cyto.a.22894 [DOI] [PubMed] [Google Scholar]
- Bendall S. C., Simonds E. F., Qiu P., El-ad D. A., Krutzik P. O., Finck R., et al. (2011). Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332 687–696. 10.1126/science.1198704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson Z. B., Nolan G. P., Fantl W. J. (2013). Single-cell mass cytometry for analysis of immune system functional states. Curr. Opin. Immunol. 25 484–494. 10.1016/j.coi.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchiaro G., Manieri T. M., Bertuchi F. R. (2013). Analytical methods for copper, zinc and iron quantification in mammalian cells. Metallomics 5 1336–1345. 10.1039/c3mt00136a [DOI] [PubMed] [Google Scholar]
- Chang Q., Ornatsky O. I., Siddiqui I., Loboda A., Baranov V. I., Hedley D. W. (2017). Imaging mass cytometry. Cytometry A 91 160–169. 10.1002/cyto.a.23053 [DOI] [PubMed] [Google Scholar]
- Chatterjee A., Carpentieri A., Ratner D. M., Bullitt E., Costello C. E., Robbins P. W., et al. (2010). Giardia cyst wall protein 1 is a lectin that binds to curled fibrils of the GalNAc homopolymer. PLoS Pathog. 6:e1001059 10.1371/journal.ppat.1001059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay P. K., Gierahn T. M., Roederer M., Love J. C. (2014). Single-cell technologies for monitoring immune systems. Nat. Immunol. 15 128–135. 10.1038/ni.2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi O., Hu Z. (2008). Size dependent and reactive oxygen species related nanosilver toxicity to nitrifying bacteria. Environ. Sci. Technol. 42 4583–4588. 10.1021/es703238h [DOI] [PubMed] [Google Scholar]
- Fabrega J., Luoma S. N., Tyler C. R., Galloway T. S., Lead J. R. (2011). Silver nanoparticles: behaviour and effects in the aquatic environment. Environ. Int. 37 517–531. 10.1016/j.envint.2010.10.012 [DOI] [PubMed] [Google Scholar]
- Fassel T. A., Edmiston C. E., Jr. (1999). Ruthenium red and the bacterial glycocalyx. Biotech. Histochem. 74 194–212. 10.3109/10520299909047974 [DOI] [PubMed] [Google Scholar]
- Fienberg H. G., Simonds E. F., Fantl W. J., Nolan G. P., Bodenmiller B. (2012). A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A 81A 467–475. 10.1002/cyto.a.22067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck R., Simonds E. F., Jager A., Krishnaswamy S., Sachs K., Fantl W., et al. (2013). Normalization of mass cytometry data with bead standards. Cytometry A 83A 483–494. 10.1002/cyto.a.22271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher M., Floodgate G. D. (1973). An electron-microscopic demonstration of an acidic polysaccharide involved in the adhesion of a marine bacterium to solid surfaces. Microbiology 74 325–334. 10.1099/00221287-74-2-325 [DOI] [Google Scholar]
- Franci G., Falanga A., Galdiero S., Palomba L., Rai M., Morelli G., et al. (2015). Silver nanoparticles as potential antibacterial agents. Molecules 20 8856–8874. 10.3390/molecules20058856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavasso S., Gullaksen S.-E., Skavland J., Gjertsen B. T. (2016). Single-cell proteomics: potential implications for cancer diagnostics. Expert Rev. Mol. Diagn. 16 579–589. 10.1586/14737159.2016.1156531 [DOI] [PubMed] [Google Scholar]
- Giao N. T., Limpiyakorn T., Kunapongkiti P., Thuptimdang P., Siripattanakul-Ratpukdi S. (2017). Influence of silver nanoparticles and liberated silver ions on nitrifying sludge: ammonia oxidation inhibitory kinetics and mechanism. Environ. Sci. Pollut. Res. 24 9229–9240. 10.1007/s11356-017-8561-0 [DOI] [PubMed] [Google Scholar]
- Günther S., Hübschmann T., Rudolf M., Eschenhagen M., Röske I., Harms H., et al. (2008). Fixation procedures for flow cytometric analysis of environmental bacteria. J. Microbiol. Methods 75 127–134. 10.1016/j.mimet.2008.05.017 [DOI] [PubMed] [Google Scholar]
- Guo Y., Stärk H. J., Hause G., Schmidt M., Harms H., Wick L. Y., et al. (2017). Heterogenic response of prokaryotes toward silver nanoparticles and ions is facilitated by phenotypes and attachment of silver aggregates to cell surfaces. Cytometry A 10.1002/cyto.a.23055 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Ivask A., ElBadawy A., Kaweeteerawat C., Boren D., Fischer H., Ji Z., et al. (2014). Toxicity mechanisms in Escherichia coli vary for silver nanoparticles and differ from ionic silver. ACS Nano 8 374–386. 10.1021/nn4044047 [DOI] [PubMed] [Google Scholar]
- Juganson K., Mortimer M., Ivask A., Pucciarelli S., Miceli C., Orupõld K., et al. (2017). Mechanisms of toxic action of silver nanoparticles in the protozoan Tetrahymena thermophila: from gene expression to phenotypic events. Environ. Pollut. 225 481–489. 10.1016/j.envpol.2017.03.013 [DOI] [PubMed] [Google Scholar]
- Lankoff A., Sandberg W. J., Wegierek-Ciuk A., Lisowska H., Refsnes M., Sartowska B., et al. (2012). The effect of agglomeration state of silver and titanium dioxide nanoparticles on cellular response of HepG2, A549 and THP-1 cells. Toxicol. Lett. 208 197–213. 10.1016/j.toxlet.2011.11.006 [DOI] [PubMed] [Google Scholar]
- Leipold M. D., Ornatsky O., Baranov V., Whitfield C., Nitz M. (2011). Development of mass cytometry methods for bacterial discrimination. Anal. Biochem. 419 1–8. 10.1016/j.ab.2011.07.035 [DOI] [PubMed] [Google Scholar]
- Lin W., Hou Y., Lu Y., Abdelrahman A. I., Cao P., Zhao G., et al. (2014). A high-sensitivity lanthanide nanoparticle reporter for mass cytometry: tests on microgels as a proxy for cells. Langmuir 30 3142–3153. 10.1021/la403627p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingens F., Blecher R., Blecher H., Blobel F., Eberspächer J., Fröhner C., et al. (1985). Phenylobacterium immobile gen. nov., sp. nov., a gram-negative bacterium that degrades the herbicide chloridazon. Int. J. Syst. Evol. Microbiol. 35 26–39. 10.1099/00207713-35-1-26 [DOI] [Google Scholar]
- Majonis D., Ornatsky O., Kinach R., Winnik M. A. (2011). Curious results with palladium- and platinum-carrying polymers in mass cytometry bioassays and an unexpected application as a dead cell stain. Biomacromolecules 12 3997–4010. 10.1021/bm201011t [DOI] [PubMed] [Google Scholar]
- Miyashita S., Groombridge A. S., Fujii S., Minoda A., Takatsu A., Hioki A., et al. (2014). Highly efficient single-cell analysis of microbial cells by time-resolved inductively coupled plasma mass spectrometry. J. Anal. At. Spectrom. 29 1598–1606. 10.1039/C4JA00040D [DOI] [Google Scholar]
- Müller S., Nebe-von-Caron G. (2010). Functional single-cell analyses: flow cytometry and cell sorting of microbial populations and communities. FEMS Microbiol. Rev. 34 554–587. 10.1111/j.1574-6976.2010.00214.x [DOI] [PubMed] [Google Scholar]
- Nair N., Mei H. E., Chen S.-Y., Hale M., Nolan G. P., Maecker H. T., et al. (2015). Mass cytometry as a platform for the discovery of cellular biomarkers to guide effective rheumatic disease therapy. Arthritis Res. Ther. 17 127 10.1186/s13075-015-0644-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfumo A., Elsaesser A., Littmann S., Foster R. A., Kuypers M. M. M., Cockell C. S., et al. (2014). Epifluorescence, SEM, TEM and nanoSIMS image analysis of the cold phenotype of Clostridium psychrophilum at subzero temperatures. FEMS Microbiol. Ecol. 90 869–882. 10.1111/1574-6941.12443 [DOI] [PubMed] [Google Scholar]
- Robinson W. H., Mao R. (2016). Biomarkers to guide clinical therapeutics in rheumatology? Curr. Opin. Rheumatol. 28 168–175. 10.1097/BOR.0000000000000250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A. R., Stanislawiak S., Baumgart S., Grützkau A., Mei H. E. (2017). Silver nanoparticles for the detection of cell surface antigens in mass cytometry. Cytometry A 91 25–33. 10.1002/cyto.a.22904 [DOI] [PubMed] [Google Scholar]
- Swathy J. R., Sankar M. U., Chaudhary A., Aigal S., Anshup and Pradeep. T. (2014). Antimicrobial silver: an unprecedented anion effect. Sci. Rep. 4:7161 10.1038/srep07161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L., Lu E., Pichaandi J., Zhao G., Winnik M. A. (2016). Synthesis of uniform NaLnF 4 (Ln: Sm to Ho) nanoparticles for mass cytometry. J. Phys. Chem. C 120 6269–6280. 10.1021/acs.jpcc.6b00570 [DOI] [Google Scholar]
- Vancaeyzeele C., Ornatsky O., Baranov V., Shen L., Abdelrahman A., Winnik M. A. (2007). Lanthanide-containing polymer nanoparticles for biological tagging applications: nonspecific endocytosis and cell adhesion. J. Am. Chem. Soc. 129 13653–13660. 10.1021/ja073970w [DOI] [PubMed] [Google Scholar]
- Wakshlak R. B.-K., Pedahzur R., Avnir D. (2015). Antibacterial activity of silver-killed bacteria: the “zombies” effect. Sci. Rep. 5:9555 10.1038/srep09555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller L. N., Fox N., Fox K. F., Fox A., Price R. L. (2004). Ruthenium red staining for ultrastructural visualization of a glycoprotein layer surrounding the spore of Bacillus anthracis and Bacillus subtilis. J. Microbiol. Methods 58 23–30. 10.1016/j.mimet.2004.02.012 [DOI] [PubMed] [Google Scholar]
- Xiu Z., Zhang Q., Puppala H. L., Colvin V. L., Alvarez P. J. J. (2012). Negligible particle-specific antibacterial activity of silver nanoparticles. Nano Lett. 12 4271–4275. 10.1021/nl301934w [DOI] [PubMed] [Google Scholar]
- Yang Y.-S. S., Atukorale P. U., Moynihan K. D., Bekdemir A., Rakhra K., Tang L., et al. (2017). High-throughput quantitation of inorganic nanoparticle biodistribution at the single-cell level using mass cytometry. Nat. Commun. 8:14069 10.1038/ncomms14069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Li X., Sigg L., Suter M. J.-F., Pillai S., Behra R., et al. (2017). Interaction of silver nanoparticles with algae and fish cells: a side by side comparison. J. Nanobiotechnol. 15:16 10.1186/s12951-017-0254-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Ibuki Y. (2015). Evaluating the toxicity of silver nanoparticles by detecting phosphorylation of histone H3 in combination with flow cytometry side-scattered light. Environ. Sci. Technol. 49 5003–5012. 10.1021/acs.est.5b00542 [DOI] [PubMed] [Google Scholar]
- Zucker R. M., Daniel K. M., Massaro E. J., Karafas S. J., Degn L. L., Boyes W. K. (2013). Detection of silver nanoparticles in cells by flow cytometry using light scatter and far-red fluorescence. Cytometry A 83 962–972. 10.1002/cyto.a.22342 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


