Abstract
Metabolic reprogramming is a hallmark of cancer. The changes in metabolism are adaptive to permit proliferation, survival, and eventually metastasis in a harsh environment. Stable isotope-resolved metabolomics (SIRM) is an approach that uses advanced approaches of NMR and mass spectrometry to analyze the fate of individual atoms from stable isotope-enriched precursors to products to deduce metabolic pathways and networks. The approach can be applied to a wide range of biological systems, including human subjects. This review focuses on the applications of SIRM to cancer metabolism and its use in understanding drug actions.
Keywords: cancer, isotopic tracer, mass spectrometry (MS), metabolism, metabolomics, nuclear magnetic resonance (NMR)
Introduction
Metabolism is the collection of predominantly enzyme-catalyzed biochemical transformations that meet the growth and survival demands of an organism. For nearly a century, scientists have documented profound metabolic changes that occur in tumors (1, 2). One of the earliest insights into cancer metabolism came when Otto Warburg noted increased uptake and fermentation of glucose (Glc)3 to lactate in tumors relative to surrounding tissue, even in the presence of ample oxygen (2). Clinical cancer diagnosis exploits this “Warburg effect” by measuring uptake of the Glc analogue 18F-deoxyglucose using positron emission tomography (PET), as the metabolic demands of differentiated, non-proliferating tissues generally require far less Glc than tumors (3).
Oncoproteins and tumor suppressors are well-established regulators of metabolism, and mutations or dysregulated expression can lead to the altered metabolic phenotypes observed in many cancers (4, 5). Inactivating mutations in enzymes such as fumarate hydratase (FH) or succinate dehydrogenase (SDH) induce pathophysiological accumulation of substrates that inhibit other critical enzyme functions, whereas less common gain-of-function mutations such as in isocitrate dehydrogenases (IDH) produce metabolites that directly deregulate cellular processes (6). Additionally, cancer cells frequently express fetal isoforms of metabolic enzymes that are subject to different regulatory mechanisms to provide growth advantages (7). Gaining a systematic understanding of the metabolic changes that occur during carcinogenesis can thus be exploited for therapeutic and diagnostic purposes.
Stable isotope-resolved metabolomics (SIRM) is a powerful approach developed by others and by us where isotopically enriched precursors such as [13C6]Glc are administered to a biological system, and the ensuing metabolic transformations are determined using appropriate analytic techniques to enable robust reconstruction of metabolic pathways (8–11). Stable isotopes are non-radioactive and in SIRM practice are identical chemically and functionally to the most abundant isotope of that element. Incorporating stable isotopes such as 2H, 13C, or 15N into biological precursors has long been used to trace their metabolism in living systems (12). SIRM is thus distinct from non-tracer-based metabolomics profiling for statistical models. Here, we review SIRM applications and demonstrate how it is used to study cancer metabolism.
Analytical tools for SIRM
Mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy are the analytical tools of choice for SIRM studies. After isotopically labeling a system and extracting the metabolites, the number of heavy atoms (isotopologues) and their positions (isotopomers) are determined for each metabolite in crude extracts (13). As 13C has a nominal mass of 1 dalton greater than that of 12C, MS can readily determine the number of 13C atoms incorporated. Gas and liquid chromatography (GC and LC) methods have been developed to resolve large numbers of metabolites, which are often used orthogonally with MS detection. Ultra-high-resolution MS, defined here as m/z mass-resolving power of ≥350,000 (m/z 400), of which all current models are Fourier-transform MS (UHR-FTMS), can resolve all non-isomeric metabolites without chromatography (10, 14). At the sufficiently high resolution of UHR-FTMS, the neutron m/z differences for metabolites containing 13C, 15N, or 2H can all be distinguished, allowing for multiplexed labeling experiments. However, NMR is better suited for determining positional labeling, e.g. to distinguish the label of [13C1]lactate at carbon 1, 2, or 3, which are identical in molecular mass but have distinct NMR spectral properties. 13C is magnetically active, whereas 12C is silent thus enabling NMR to distinguish different positions of enrichment either by direct 13C detection or by indirect detection of the attached protons (15). Advances in 1D and 2D NMR have enabled isotopomer analysis in complex biological samples without fractionation (8). Coupling NMR and UHR-FTMS with an atom-resolved metabolic database and tracing tools can provide robust and unprecedented reconstruction of interconnected pathways and networks (8, 10, 16, 17).
Tracer selection for interrogation of metabolic pathways
Glycolysis
Glycolysis is the metabolic pathway of NAD+-dependent oxidation of Glc to pyruvate to generate ATP and glycolytic intermediates. [13C6]Glc is a commonly used tracer for measuring glycolysis as it is relatively inexpensive and will label all glycolytic carbons derived from Glc, and it enables tracing of glycolysis-derived precursors in other pathways. For example, [13C6]Glc produces the glycolytic intermediate 3-[13C3]phosphoglycerate, which is a substrate for purine synthesis via the Ser-Gly one-carbon pathway (18). However, due to ambiguities in resolving [13C3]lactate production from [13C6]Glc because of pathways other than glycolysis, Glc tracers labeled at 13C1-2 or 13C1-3 can provide better estimation of glycolytic flux (19).
Pentose phosphate pathway
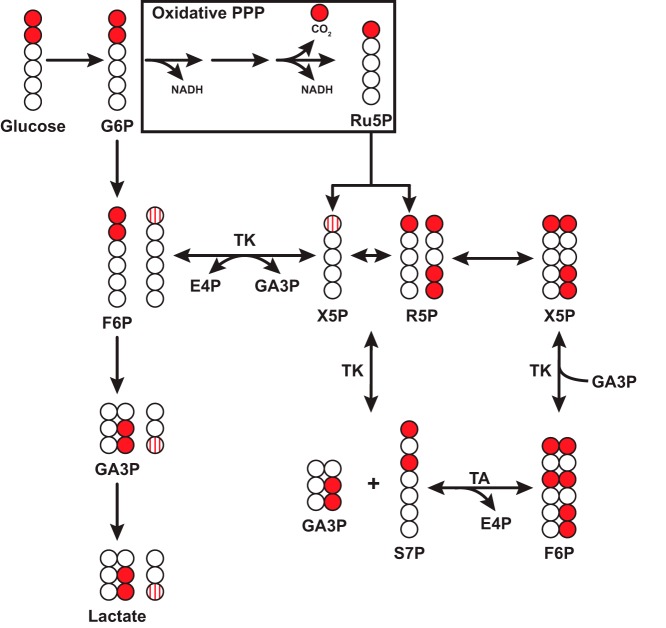
The pentose phosphate pathway (PPP) is an alternative route for Glc metabolism, yielding ribose 5-phosphate (R5P) for nucleotide biosynthesis and NADPH for fatty acid biosynthesis and decomposition of peroxides (20). The choice of tracer for analysis depends on the research question as LC-MS or ion chromatography-MS can detect most of the PPP intermediates (21, 22). [13C6]Glc provides maximal labeling of PPP intermediates as all carbons of the PPP precursors will contain labels. However, [13C2-1,2]Glc can distinguish the oxidative and non-oxidative branches of the PPP (Fig. 1) (23, 24). R5P isotopologues containing more than one label reflect activity of both the oxidative and non-oxidative branches of PPP as additional 13C label incorporation into R5P can only be derived through transaldolase and transketolase activity in the non-oxidative PPP branch (25). Furthermore, the ratio of [13C1-3]lactate to [13C2-2,3]lactate following labeling with [13C2-1,2]Glc has been used to estimate PPP activity in vivo (26).
Figure 1.
Production of [13C-1]ribose or [13C3-1,4,5]ribose in the oxidative or non-oxidative PPP from [13C2-1,2]glucose. Example scheme depicting generation of singly labeled ribose from the oxidative PPP or triply labeled ribose from the non-oxidative PPP. Glucose 6-phosphate (G6P) enters the oxidative pentose phosphate pathway to produce M + 1 ribulose 5-phosphate (Ru5P), which enters the non-oxidative branch to produce xylulose 5-phosphate (X5P) and ribose 5-phosphate (R5P). Transketolase (TK) transfers the first two carbons of X5P to R5P to produce [13C2-1,3]sedoheptulose 7-phosphate (S7P). Transaldolase (TA) transfers the first three carbons from S7P to glyceraldehyde 3-phosphate (GA3P), which can be derived from glycolysis (arrow) to produce F6P. TK then transfers two carbons from F6P to produce X5P and subsequently R5P. Note that F6P derived from X5P via TK can re-enter glycolysis to produce [13C1-3]GA3P and subsequently [13C1-3]lactate. Striped circles indicate labeled carbons derived from non-oxidative PPP. Other isotopomers are possible and are not shown for simplicity. E4P, erythrose 4-phosphate.
Krebs cycle and anaplerosis
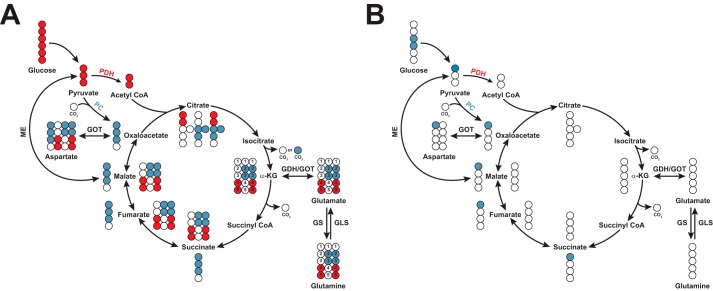
The Krebs cycle is a central metabolic hub that integrates carbohydrate, lipid, and amino acid metabolism. Glc, glutamine (Gln), and fatty acids are major carbon sources that feed into the Krebs cycle in cancer (27). [13C]Glc and -Gln are both commonly used tracers to label Krebs cycle intermediates via entry as pyruvate or α-ketoglutarate (α-KG), respectively, producing distinct labeling patterns. For example, pyruvate dehydrogenase (PDH) generates [13C2]acetyl-CoA, which condenses with oxaloacetate (OAA) to form [13C2]- or [13C4]citrate, depending on the first or second round of the Krebs cycle (Fig. 2A) (28). In contrast, pyruvate carboxylase (PC) carboxylates [13C3]pyruvate with unlabeled CO2, generating [13C3]OAA, which condenses with either unlabeled or [13C2]acetyl-CoA to generate [13C3]- or [13C5]citrate, respectively (Fig. 2A) (29, 30). Carbons from [13C2-3,4]Glc (or [13C1-1]pyruvate) will only enter the Krebs cycle through PC as these carbons are lost as CO2 in the PDH reaction (Fig. 2B) (31, 32).
Figure 2.
13C distribution in first turn of the Krebs cycle with [13C3]- or [lsqb]13C1-1]pyruvate. A, [13C3]pyruvate enters the Krebs cycle either through pyruvate carboxylase (PC, blue circles) or pyruvate dehydrogenase (PDH, red circles). Colored circles indicate 13C label for selected metabolites. PDH-derived and PC-derived metabolites are represented in the inner and outer circle, respectively. PC notably produces M + 3 citrate, malate, and fumarate during the first turn of the Krebs cycle, whereas PDH notably produces M + 2 citrate. [13C2-2,3]Glu and [13C2-3,4]Glu isotopomers are associated with PC and PDH activity, respectively. B, labels from [13C-1]pyruvate (blue circle, derived from [13C2-3,4]glucose) enter the Krebs cycle exclusively through PC as the labeled carbon is lost in the PDH reaction. Other isotopomers are possible but not shown. IDH, isocitrate dehydrogenase; GDH, glutamate dehydrogenase; GS, glutamine synthetase; GLS, glutaminase; ME, malic enzyme; GOT, glutamic-oxaloacetic transaminase.
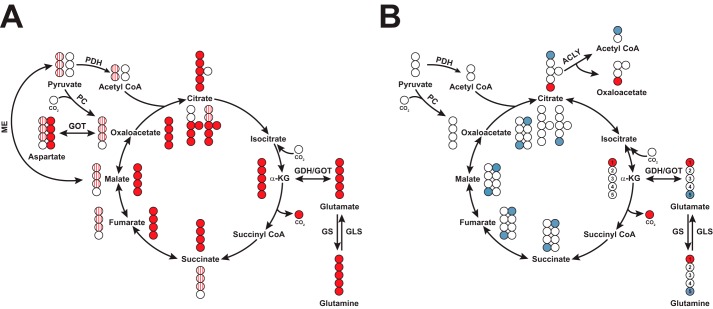
The choice of Gln tracer depends on the need to resolve Gln metabolism via oxidative or reductive pathways in the Krebs cycle. Gln undergoes glutaminolysis and enters the Krebs cycle via α-ketoglutarate (α-KG) as another anaplerotic input to the Krebs cycle (Fig. 3A). [13C5]Gln-derived α-KG is then oxidized to [13C4]succinate, -fumarate, -malate, and -OAA through the first turn of the Krebs cycle or reduced to [13C5]citrate through IDH-catalyzed reductive carboxylation to subsequently generate [13C3]OAA, -malate, and -fumarate (33). [13C1-1]Gln distinguishes between the two pathways because labeled products result from reductive carboxylation as the 13C-1 label is lost as CO2 via the α-KG dehydrogenase reaction (Fig. 3B). Similarly, [13C1-5]Gln is used to measure reductive carboxylation by tracing incorporation of Gln into fatty acids as most of the label is lost during oxidation through multiple turns of the Krebs cycle. Furthermore, [15N]Gln allows tracing of Gln into nitrogenous compounds such as amino acids and nucleotides (34). The uses for common 15N and 13C tracers are summarized in Table 1. Use of 2H in tracers requires very careful consideration of enzyme mechanisms and chemical exchange under mostly unknown in situ physical-chemical conditions.
Figure 3.
13C distribution in first turn of the Krebs cycle with [13C5]- or [13C-1,5]glutamine. A, [13C5]glutamine (red circles) enters the cycle and distributes as M + 4 isotopologues through oxidative glutaminolysis (inside of cycle) or M + 5 citrate through reductive carboxylation (outside of cycle). Striped dots represent labeled carbons from [13C3]pyruvate produced via malic enzyme re-entering the Krebs cycle. For simplicity, not all isotopologues are shown, and the re-entry of labeled pyruvate into the Krebs cycle could produce the same isotopologues as in Fig. 2A. B, labels from [13C-1]Gln (red circles) are lost following decarboxylation of C-1 by α-KG dehydrogenase. The 13C label is retained in citrate through the reverse reaction of IDH but is not incorporated into fatty acids. The label from [13C-5]Gln (blue circle) is retained through glutaminolysis but is only incorporated into fatty acids via reductive carboxylation.
Table 1.
Common stable-isotope tracers and their uses
| Tracer | Labeled pathway | Ref. |
|---|---|---|
| [U-13C]Glucose | Glycolysis, PPP, Krebs cycle, hexosamine, nucleotide, lipid synthesis | 22, 30 |
| [1,2-13C]Glucose | Non-oxidative versus oxidative PPP | 23, 24 |
| [3,4-13C]Glucose | PC anaplerosis | 31, 32 |
| [13C/15N]Glucose | Glutaminolysis, nucleotide biosynthesis, Krebs cycle, and fatty acid synthesis | 30, 39, 58 |
| 13C-Labeled fatty acids | β-Oxidation, fatty acid synthesis | 97 |
| [13C]Serine | Serine metabolism, one-carbon metabolism, lipid synthesis | 98 |
| [13C]Glycerol | Lipid synthesis, gluconeogenesis-pentose cycle interactions | 99, 100 |
SIRM profiling of cancer systems can reveal novel metabolic reprogramming and therapeutic targets
A major goal of SIRM is to delineate the differences in substrate utilization and subsequent metabolic transformations between cancerous and non-cancerous tissues. To this end, multiple cancer biological models have been used in SIRM studies, including 2D and 3D cell culture, animal tumor models derived spontaneously from defined oncogenic lesions or from implanted tissue/cells, and human tumors analyzed in vivo or ex vivo following surgical resection (35). Regardless of the model, most tumors and derived cells profiled by SIRM recapitulate Warburg's original findings and disprove the canard that tumor cells necessarily have dysfunctional mitochondria (36) by demonstrating that Glc-derived 13C incorporation into the Krebs cycle can be enhanced in cancerous versus paired non-cancerous tissues (37–39). Thus, SIRM profiling is excellently suited for uncovering novel aspects of cancer metabolism in model systems and directly in human subjects.
Proliferating cells shunt Glc into pathways other than glycolysis and the Krebs cycle, and SIRM has confirmed increases of Glc flux into these pathways. For example, enhanced non-oxidative and oxidative PPP activity has been reported for pancreatic (23) and renal (40) cancers, respectively, using a [13C2-1,2] Glc tracer. Many cancer cells also divert Glc from glycolysis into de novo serine synthesis and vary considerably in their de novo serine biosynthetic capacity (18). The wide range in de novo serine biosynthesis may correlate with tumorigenic potential because the Glc to serine flux increases as cells become progressively more tumorigenic (41). Glc carbon may also be diverted from glycolysis for other anabolic purposes: dihydroxyacetone phosphate is a precursor for glycerol 3-phosphate used for glycerolipid synthesis (42), and fructose 6-phosphate (F6P) is the precursor for the hexosamine pathway critical for protein glycosylation and intracellular regulation (43, 44). Collectively, these reactions indicate that in many proliferating cells the fraction of Glc consumed that is converted to lactate is much less than 100%.
Many cancer cells use anaplerosis to replenish intermediates that exit the Krebs cycle to support anabolic and/or anti-oxidative demands during cell proliferation (45). For example, PC-mediated carboxylation of pyruvate to OAA is active in several cancers. In a non-small cell lung cancer (NSCLC) patient study, patients receiving a bolus injection of [13C6]Glc prior to surgery showed enhanced production of the PC-associated metabolites [13C3]Asp, [13C3]malate, [13C3]citrate, and [13C2-2,3]Glu in cancerous versus surrounding non-cancerous lung tissues (30, 46). Elevated PC activity was also observed in mouse orthotopic glioblastoma (GBM) tumors after administering [13C2-3,4]Glc (47).
Cell line studies with [13C2-1,2]Glc suggest that pyruvate flux through PC and PDH is compartmentalized. Comparison of [13C2-2,3]Glu (PC) to [13C2-4,5]Glu (PDH) enrichment in Jurkat T leukemia cells yields a ratio of about 0.4 suggesting PDH flux is dominant in this line (25). However, the PC/PDH flux ratio determined from [13C1-2]Glu to [13C1-4]Glu enrichment is 30 times higher than the flux calculated from the doubly labeled Glu species. These singly labeled isotopomers may arise when C-1 of Glc is lost as CO2 in the oxidative PPP and re-enters glycolysis as fructose 6-phosphate generated by the non-oxidative PPP. This SIRM example demonstrates how cells with high PPP flux may preferentially utilize the PC pathway to meet the demands for nucleotide synthesis to support cell division.
Gln is another major anaplerotic source for Krebs cycle replenishment and serves as a precursor in numerous metabolic pathways (48). Many cancer cells appear to require Gln in some capacity, depending on the tumor type and oncogenic lesions (49). Cells that require Gln have characteristic 13C-labeling patterns with [13C6]Glc and [13C5]Gln tracers. For example, Gln-dependent U2OS osteosarcoma cells only produce [13C2]citrate whereas Gln-independent MCF7 breast adenocarcinoma cells also generate [13C6]citrate from [13C6]Glc (50). In contrast, all 13C-isotopologues of citrate are made from [13C5]Gln in U2OS cells confirming Gln anaplerosis in Gln-dependent cells (50). These cell line differences in Gln dependence are clinically relevant when studying cancer-initiating cells (CICs), which are undifferentiated cancer cells with self-renewal capacity and are highly resistant to conventional treatments (51). In a study comparing CICs with paired, differentiated cells from the same GBM tumor, CICs were much more resistant to Gln deprivation than their differentiated controls (52). This may be related to a higher capacity for [15N]Gln synthesis from 15NH4 in CICs than differentiated cells (52).
Given the heterogeneity in Gln requirements between cancer cell lines and even cells within the same tumor, several studies have profiled tumors in vivo to establish Glc and Gln dependences. Cell lines derived from KRASG12D;TP53−/−-induced mouse adenocarcinomas avidly consume Gln and are sensitive to glutaminase (GLS) inhibitors in vitro (53). However, xenografts from these cells were insensitive to GLS inhibition. Similarly, 13C labeling was higher in mouse orthotopic human GBM tumors following injection of [13C]Glc compared with [13C]Gln, suggesting that these tumors use less Gln in vivo (53). Tumors resected following [13C6]Glc infusion produce [13C3]citrate, -malate, and -aspartate, which is consistent with active PC, suggesting that both routes of anaplerosis are utilized by tumors depending on nutrient availability (30, 37, 46). These studies also suggest that metabolic alterations occur during adaptation to cell culture and/or that the tumor microenvironment substantially influences tumor metabolism (53).
Cancer cells synthesize lipids de novo to meet growth requirements (54), and we now appreciate that Gln carbons can fuel citrate and subsequent lipid biosynthesis through oxidative glutaminolysis or reductive carboxylation in the Krebs cycle. In the oxidative pathway, Gln-derived OAA condenses with acetyl-CoA to produce citrate as precursor to fatty acid biosynthesis. Alternatively, malic enzyme can convert Gln-derived malate to pyruvate, which then re-enters the Krebs cycle and generates acetyl-CoA via PDH (Fig. 2A). Certain conditions, such as hypoxia, inhibit PDH activity leading to reduced acetyl-CoA production from pyruvate (55). These conditions favor carboxylation of Gln-derived α-KG by the reverse reaction of IDH to generate citrate (Fig. 3B) (56, 57). For example, Gln-dependent GBM cells produce [13C4]fumarate, -malate, and -citrate when traced with [13C5]Gln under normoxia. However, hypoxia induces a shift to [13C5]citrate synthesis (56). [13C1-1]Gln tracing has confirmed reductive carboxylation in cell lines derived from lung, breast, melanoma, and colon cancers (57) and from cells with mitochondrial deficiencies in FH (58) or SDH (59). Reductive carboxylation also occurs in in vivo as evidenced by [13C]citrate production in mouse tumors following injection of [13C1-1]Gln (60).
Nutrient availability may significantly contribute to the metabolic plasticity in tumors. When mice bearing NSCLC tumors were injected with [13C1-2]lactate, 13C1-isotopologues of glycolytic and Krebs cycle intermediates were detected in these tumors suggesting lactate-derived pyruvate entered the Krebs cycle or the gluconeogenic pathway (37). NSCLC and breast cancer cells consume [13C3]lactate or [13C5]Gln to produce 13C-labeled glycolytic intermediates and [13C]serine through the key gluconeogenic enzyme phosphoenolpyruvate carboxykinase 2, and gluconeogenic activity is enhanced under Glc withdrawal (61, 62). Tumors can also consume acetate, and in mice bearing human orthotopic GBM tumors, infusion of [13C2,1,2]acetate produces [13C2,4,5]Glu suggesting that exogenous acetate can fuel the Krebs cycle (63). [13C2,1,2]Acetate oxidation has also been detected in human GBM tumors after resection, and GBMs consume [11C]acetate in vivo as measured by PET (63).
SIRM can elucidate downstream effects of oncogene activation
SIRM has been used to characterize some of the key metabolic alterations resulting from oncogenic lesions.
RAS
RAS proto-oncogenes are among the most frequently mutated oncogenes in human cancers (64). Oncogenic KRAS decouples glycolysis and the Krebs cycle by increasing Glc uptake and conversion to lactate while decreasing Glc entry into the Krebs cycle in fibroblasts (65, 66) and in cells derived from mice with KRAS-driven spontaneous tumor formation (53). Tracing by [13C2-1,2]Glc shows that oncogenic KRAS increases flux into the PPP, presumably to fuel nucleotide biosynthesis (21, 66). KRAS-driven cancer cells also require Gln for proliferation, although this requirement depends largely on extracellular environment (53) or specific mutation (67). Oncogenic KRAS increases production of [13C4]aspartate from [13C5]Gln (65), which can be transported into the cytoplasm and converted to OAA via cytosolic aspartate transaminase (68). The subsequent conversion to malate and pyruvate by malate dehydrogenase and malic enzyme increases NADPH to maintain cellular redox state. In contrast, HRASG12V transformation of immortalized epithelial cells increases [13C]Glc entry into the Krebs cycle and sensitizes the cells to mitochondrial inhibitors (69). These studies suggest that the RAS family members may transform cells by differentially reprogramming metabolic pathways.
MYC
The MYC transcription factor is frequently overexpressed in human cancers (70) and regulates the expression of many metabolic enzymes (71). Mice transgenic for MYC overexpression in liver develop sporadic hepatocellular carcinomas (72) that show increased [13C]Glc uptake and subsequent production of [13C]lactate relative to normal adjacent tissue due to glycolytic gene induction by MYC (49), which agrees with the effects of MYC overexpression in B cells (73). In carcinomas, MYC also drives increased [13C]Gln uptake and conversion to [13C]Glu by inducing glutamine transporters and GLS expression, leading to increased 13C labeling of Krebs cycle intermediates and increased Gln incorporation in lactate (4, 49, 74). In a MYC-driven hepatocellular carcinoma model using hyperpolarized [13C]pyruvate, [13C]lactate was the main product, but in pre-cancerous regions [13C]alanine was produced, a phenotype not found in normal or tumor tissues (75). Thus, SIRM led to identification of alanine aminotransferase as a potential early marker of MYC-induced carcinogenesis. Interestingly, hepatocellular carcinoma models driven by a different oncogene, MET, failed to phenocopy the MYC-driven metabolic perturbations suggesting that metabolic reprogramming differs based on oncogenic lesions (49).
Retinoblastoma protein
The retinoblastoma protein (pRb) is a tumor suppressor that restricts passage from G1 into S phase during the cell cycle, and pRb is functionally inactivated in most cancers (76). Unlike oncogenic KRAS, increased [13C]Glc uptake and lactate production was not observed in tissues derived from RB knock-out mice or RB-silenced epithelial cells (77). Instead, loss of pRb enhances Glc entry into the Krebs cycle, as evidenced by an increase in PDH-mediated production of [13C2]citrate (77). Loss of pRb more profoundly impacts Gln metabolism as Gln uptake and conversion to Glu were activated in mouse embryonic fibroblasts derived from RB knock-out mice (78). The most striking effect of pRb knock-out is increased 13C labeling of glutathione derived from [13C5]Gln (78, 79). These results suggest that the metabolic role of pRb may be to divert Gln carbons to anti-oxidative pathways that control reactive oxygen species accumulation during cell cycle progression.
Applications of SIRM in drug development
Many clinically successful drugs or promising drug candidates may benefit from SIRM analyses to gain insights into their molecular mechanisms. For example, SIRM implicated enhanced glycolysis and PC-mediated Krebs cycle activity in the mechanism of the anti-manic depression drug lithium on neurons and astrocytes (28). Likewise, selenium agents perturbed Krebs cycle activity and attenuated lipid biosynthesis in lung cancer cells, and these alterations were related to an activation of the AMP–AMP kinase pathway as was determined by SIRM and microarray experiments (80). Anti-cancer target discovery is one of the most promising translational applications for SIRM, and inhibitors of several targets have been developed and are showing promise in preclinical models (81). These inhibitors can also potentiate the efficacy of conventional chemotherapeutics (82).
Metabolic enzymes are frequently mutated in certain cancers such as SDH in gastrointestinal, renal cell carcinomas, and paraganglioma tumors (83), IDH in GBM (84), and FH in renal cell cancers (85). These mutations often result in the production of excess “oncometabolites” that promote activation of proliferative transcription factors and regulate DNA methylation (6). Tracer studies show that SDH-null cells produce citrate predominantly through reductive carboxylation of Gln and accumulate Gln-derived succinate, a product inhibitor of the α-KG-dependent dioxygenases, while relying on PC for anaplerosis and synthesis of aspartate to support nucleotide biosynthesis required for proliferation (59, 86, 87). Likewise, IDH1 mutant cells increased conversion of [13C1-2]Glc to PC-derived [13C1-3]Glu, which implicates the importance of PC anaplerosis for proliferation of these cells (88). More intriguingly, PC expression and in vivo activity were enhanced in human NSCLC tumors relative to adjacent non-cancerous lung tissues, and silencing PC expression in lung cancer lines slowed tumor growth in mouse xenografts (8, 53). Together, these results suggest that PC may be a viable drug target for certain types of cancer. Furthermore, [13C]Gln profiling of WT versus FH-null mice revealed a dramatic increase in [13C4]arginosuccinate, a urea cycle intermediate that is normally converted to fumarate and arginine by arginosuccinate lyase (ASL) (89). This accumulation of labeled arginosuccinate can be attributed to elevated fumarate levels in FH-null cells to drive the reverse reaction of ASL and deplete the cells of arginine, rendering them auxotrophic for arginine. Enzymatic depletion of arginine prevented cell proliferation, thus demonstrating the utility of targeting this system in FH-deficient cells (89).
SIRM may also be applied to cancer therapeutics by investigating the mechanisms of drug resistance. For example, imatinib treatment of chronic myeloid leukemia cells induces a shift from glycolysis to mitochondrial metabolism, a phenotype associated with non-cancerous tissues (90, 91). Imatinib-resistant cells display no such decrease in glycolysis in response to drug treatment. In addition, cells switched from the oxidative to non-oxidative PPP for ribose production as they became resistant to imatinib (91). Thus, screening cells from patients for these metabolic avoidance traits may predict their sensitivity to imatinib treatment.
As discussed previously, cell-based tracer experiments may not always reflect in vivo tumor metabolism, and target discovery in more relevant model systems is necessary (35). In a study comparing pancreatic CIC spheres with adherent, differentiated cells from the same tumor, the CICs showed less Glc flux into lactate and ribose via glycolysis and the PPP, respectively, and more flux into the Krebs cycle (92). Likewise, CICs were more susceptible to mitochondrial inhibitors than differentiated cells, and resistant clones were shown to have a mixed phenotype. By genetically and pharmacologically manipulating factors that regulate mitochondrial biogenesis, resistant CICs became sensitive to mitochondrial inhibitors (92). Thus, understanding the metabolic basis of chemotherapy resistance in relevant model systems will certainly facilitate more robust target identification for improving therapeutic outcomes.
Concluding remarks/future directions
SIRM profiling has unveiled the complex and plastic nature of cancer metabolism. Tumors are highly adaptable to changing nutrient supplies, which creates conceptual and technical research challenges that may not be met with commonly used model systems. For example, recent studies have demonstrated the importance of the microenvironment for tumor metabolism either through immune modulation (22) or through direct regulation by stromal cells (93). Our group has demonstrated that ex vivo cultured tumor slices from primary tumors are viable metabolically and immunologically with the maintenance of the 3D tumor architecture and microenvironment (22, 30, 39, 94). These ex vivo slices provide the unique opportunity to study the effects of environmental variants or drug treatment on metabolic networks directly in human tumors without systemic complications. As such, this new approach can circumvent the long-standing concerns for cell or animal models in recapitulating human tumor metabolism.
SIRM highly complements other “omic” profiling technologies already being practiced clinically. By gaining insights into metabolic dysfunction due to cancer development or drug interventions, SIRM-generated knowledge can be integrated with genomic and proteomic information to achieve systems biochemical insights in both model systems and individual human patients. SIRM can further promote discovery of mechanism-based biomarkers for early detection and prediction of drug responses. Metabolite profiling for statistical modeling has identified potential biomarkers for cancers such as lung (95) and cervix (96), but the etiology of these biomarkers is mostly unknown. SIRM investigations can unambiguously trace the pathway(s) from which such biomarkers are derived and accelerate the mapping of specific pathway dysregulations derived from other 'omics information. This knowledge is pivotal for translating biomarker candidates into clinical practices. We believe the future of cancer research and treatment will greatly benefit from SIRM as more researchers adapt this technology and contribute to the growing atlas of reprogrammed cancer metabolism.
This work was supported in part by National Institutes of Health Grants T32CA165990 (to R. C. B.), 1R01ES022191-01 (to T. W. M. F. and R. M. H.), and 1P01CA163223-01A1 (to A. N. L. and T. W. M. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Glc
- glucose
- α-KG
- α-ketoglutarate
- ASL
- arginosuccinate lyase
- CIC
- cancer-initiating cell
- F6P
- fructose 6-phosphate
- FH
- fumarate hydratase
- GBM
- glioblastoma
- GLS
- glutaminase
- IDH
- isocitrate dehydrogenase
- OAA
- oxaloacetate
- PDH
- pyruvate dehydrogenase
- PC
- pyruvate carboxylase
- PET
- positron emission tomography
- PPP
- pentose phosphate pathway
- pRB
- retinoblastoma protein
- R5P
- ribulose 5-phosphate
- SIRM
- stable isotope-resolved metabolomics
- SDH
- succinate dehydrogenase
- NSCLC
- non-small cell lung cancer
- UHR-FTMS
- ultra-high-resolution Fourier-transform MS.
References
- 1. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Warburg O. (1923) Versuche an überlebendem carcinomgewebe (methoden). Biochem. Zeitschr. 142, 317–333 [Google Scholar]
- 3. Kelloff G. J., Hoffman J. M., Johnson B., Scher H. I., Siegel B. A., Cheng E. Y., Cheson B. D., O'shaughnessy J., Guyton K. Z., Mankoff D. A., Shankar L., Larson S. M., Sigman C. C., Schilsky R. L., and Sullivan D. C. (2005) Progress and promise of FDG-PET imaging for cancer patient management and oncologic drug development. Clin. Cancer Res. 11, 2785–2808 [DOI] [PubMed] [Google Scholar]
- 4. Wise D. R., DeBerardinis R. J., Mancuso A., Sayed N., Zhang X. Y., Pfeiffer H. K., Nissim I., Daikhin E., Yudkoff M., McMahon S. B., and Thompson C. B. (2008) Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U.S.A. 105, 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gottlieb E., and Tomlinson I. P. (2005) Mitochondrial tumour suppressors: a genetic and biochemical update. Nat. Rev. Cancer 5, 857–866 [DOI] [PubMed] [Google Scholar]
- 6. Yang M., Soga T., and Pollard P. J. (2013) Oncometabolites: linking altered metabolism with cancer. J. Clin. Invest. 123, 3652–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mathupala S. P., Ko Y. H., and Pedersen P. L. (2010) The pivotal roles of mitochondria in cancer: Warburg and beyond and encouraging prospects for effective therapies. Biochim. Biophys. Acta 1797, 1225–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fan T. W., Lorkiewicz P. K., Sellers K., Moseley H. N., Higashi R. M., and Lane A. N. (2012) Stable isotope-resolved metabolomics and applications for drug development. Pharmacol. Ther. 133, 366–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fan T. W., Tan J., McKinney M. M., and Lane A. N. (2012) Stable isotope resolved metabolomics analysis of ribonucleotide and RNA metabolism in human lung cancer cells. Metabolomics 8, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Higashi R. M., Fan T. W., Lorkiewicz P. K., Moseley H. N., and Lane A. N. (2014) Stable isotope-labeled tracers for metabolic pathway elucidation by GC-MS and FT-MS. Methods Mol. Biol. 1198, 147–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boros L. G., Brackett D. J., and Harrigan G. G. (2003) Metabolic biomarker and kinase drug target discovery in cancer using stable isotope-based dynamic metabolic profiling (SIDMAP). Curr. Cancer Drug Targets 3, 445–453 [DOI] [PubMed] [Google Scholar]
- 12. Schoenheimer R., and Rittenberg D. (1935) Deuterium as an indicator in the study of intermediary metabolism: III. The role of the fat tissues. J. Biol. Chem. 111, 175–181 [DOI] [PubMed] [Google Scholar]
- 13. Lane A. N., Fan T. W., and Higashi R. M. (2008) Isotopomer-based metabolomic analysis by NMR and mass spectrometry. Methods Cell Biol. 84, 541–588 [DOI] [PubMed] [Google Scholar]
- 14. Lorkiewicz P., Higashi R. M., Lane A. N., and Fan T. W. (2012) High information throughput analysis of nucleotides and their isotopically enriched isotopologues by direct-infusion FTICR-MS. Metabolomics 8, 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fan T. W., and Lane A. N. (2011) NMR-based stable isotope resolved metabolomics in systems biochemistry. J. Biomol. NMR 49, 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fan T. W., and Lane A. N. (2008) Structure-based profiling of metabolites and isotopomers by NMR. Prog. Nucl. Magn. Reson. Spectrosc. 52, 69–117 [Google Scholar]
- 17. Fan T. W., and Lane A. N. (2016) Applications of NMR spectroscopy to systems biochemistry. Prog. Nucl. Magn. Reson. Spectrosc. 92, 18–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DeNicola G. M., Chen P. H., Mullarky E., Sudderth J. A., Hu Z., Wu D., Tang H., Xie Y., Asara J. M., Huffman K. E., Wistuba I. I., Minna J. D., DeBerardinis R. J., and Cantley L. C. (2015) NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 47, 1475–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Metallo C. M., Walther J. L., and Stephanopoulos G. (2009) Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J. Biotechnol. 144, 167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patra K. C., and Hay N. (2014) The pentose phosphate pathway and cancer. Trends Biochem. Sci. 39, 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ying H., Kimmelman A. C., Lyssiotis C. A., Hua S., Chu G. C., Fletcher-Sananikone E., Locasale J. W., Son J., Zhang H., Coloff J. L., Yan H., Wang W., Chen S., Viale A., Zheng H., et al. (2012) Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fan T. W., Warmoes M. O., Sun Q., Song H., Turchan-Cholewo J., Martin J. T., Mahan A., Higashi R. M., and Lane A. N. (2016) Distinctly perturbed metabolic networks underlie differential tumor tissue damages induced by immune modulator β-glucan in a two-case ex vivo non-small-cell lung cancer study. Cold Spring Harb. Mol. Case Stud. 2, a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boros L. G., Lerner M. R., Morgan D. L., Taylor S. L., Smith B. J., Postier R. G., and Brackett D. J. (2005) [1,2-13C2]-d-glucose profiles of the serum, liver, pancreas, and DMBA-induced pancreatic tumors of rats. Pancreas 31, 337–343 [DOI] [PubMed] [Google Scholar]
- 24. Lee W. N., Boros L. G., Puigjaner J., Bassilian S., Lim S., and Cascante M. (1998) Mass isotopomer study of the nonoxidative pathways of the pentose cycle with [1,2-13C2]glucose. Am. J. Physiol. 274, E843–E851 [DOI] [PubMed] [Google Scholar]
- 25. Miccheli A., Tomassini A., Puccetti C., Valerio M., Peluso G., Tuccillo F., Calvani M., Manetti C., and Conti F. (2006) Metabolic profiling by 13C-NMR spectroscopy: [1,2-13C2]glucose reveals a heterogeneous metabolism in human leukemia T cells. Biochimie 88, 437–448 [DOI] [PubMed] [Google Scholar]
- 26. Jalloh I., Carpenter K. L., Grice P., Howe D. J., Mason A., Gallagher C. N., Helmy A., Murphy M. P., Menon D. K., Carpenter T. A., Pickard J. D., and Hutchinson P. J. (2015) Glycolysis and the pentose phosphate pathway after human traumatic brain injury: microdialysis studies using 1,2-13C2 glucose. J. Cereb. Blood Flow Metab. 35, 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guppy M., Leedman P., Zu X., and Russell V. (2002) Contribution by different fuels and metabolic pathways to the total ATP turnover of proliferating MCF-7 breast cancer cells. Biochem. J. 364, 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fan T. W., Yuan P., Lane A. N., Higashi R. M., Wang Y., Hamidi A. B., Zhou R., Guitart X., Chen G., Manji H. K., and Kaddurah-Daouk R. (2010) Stable isotope-resolved metabolomic analysis of lithium effects on glial-neuronal metabolism and interactions. Metabolomics 6, 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buescher J. M., Antoniewicz M. R., Boros L. G., Burgess S. C., Brunengraber H., Clish C. B., DeBerardinis R. J., Feron O., Frezza C., Ghesquiere B., Gottlieb E., Hiller K., Jones R. G., Kamphorst J. J., Kibbey R. G., et al. (2015) A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 34, 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sellers K., Fox M. P., Bousamra M. 2nd, Slone S. P., Higashi R. M., Miller D. M., Wang Y., Yan J., Yuneva M. O., Deshpande R., Lane A. N., and Fan T. W. (2015) Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J. Clin. Invest. 125, 687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng T., Sudderth J., Yang C., Mullen A. R., Jin E. S., Matés J. M., and DeBerardinis R. J. (2011) Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl. Acad. Sci. U.S.A. 108, 8674–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crown S. B., Ahn W. S., and Antoniewicz M. R. (2012) Rational design of 13C-labeling experiments for metabolic flux analysis in mammalian cells. BMC Syst. Biol. 6, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J., Ahn W. S., Gameiro P. A., Keibler M. A., Zhang Z., and Stephanopoulos G. (2014) 13C isotope-assisted methods for quantifying glutamine metabolism in cancer cells. Methods Enzymol. 542, 369–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hiller K., Metallo C. M., Kelleher J. K., and Stephanopoulos G. (2010) Nontargeted elucidation of metabolic pathways using stable-isotope tracers and mass spectrometry. Anal. Chem. 82, 6621–6628 [DOI] [PubMed] [Google Scholar]
- 35. Lane A. N., Higashi R. M., and Fan T. W. (2016) Preclinical models for interrogating drug action in human cancers using stable isotope resolved metabolomics (SIRM). Metabolomics 12, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Warburg O. (1956) On respiratory impairment in cancer cells. Science 124, 269–270 [PubMed] [Google Scholar]
- 37. Hensley C. T., Faubert B., Yuan Q., Lev-Cohain N., Jin E., Kim J., Jiang L., Ko B., Skelton R., Loudat L., Wodzak M., Klimko C., McMillan E., Butt Y., Ni M., et al. (2016) Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maher E. A., Marin-Valencia I., Bachoo R. M., Mashimo T., Raisanen J., Hatanpaa K. J., Jindal A., Jeffrey F. M., Choi C., Madden C., Mathews D., Pascual J. M., Mickey B. E., Malloy C. R., and DeBerardinis R. J. (2012) Metabolism of [U-13C]glucose in human brain tumors in vivo. NMR Biomed. 25, 1234–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xie H., Hanai J., Ren J. G., Kats L., Burgess K., Bhargava P., Signoretti S., Billiard J., Duffy K. J., Grant A., Wang X., Lorkiewicz P. K., Schatzman S., Bousamra M. 2nd, Lane A. N., et al. (2014) Targeting lactate dehydrogenase-A inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 19, 795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang Y., Lane A. N., Ricketts C. J., Sourbier C., Wei M. H., Shuch B., Pike L., Wu M., Rouault T. A., Boros L. G., Fan T. W., and Linehan W. M. (2013) Metabolic reprogramming for producing energy and reducing power in fumarate hydratase null cells from hereditary leiomyomatosis renal cell carcinoma. PLoS ONE 8, e72179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Richardson A. D., Yang C., Osterman A., and Smith J. W. (2008) Central carbon metabolism in the progression of mammary carcinoma. Breast Cancer Res. Treat. 110, 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lane A. N., Fan T. W., Xie Z., Moseley H. N., and Higashi R. M. (2009) Stable isotope analysis of lipid biosynthesis by high resolution mass spectrometry and NMR. Anal. Chim. Acta 651, 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moseley H. N., Lane A. N., Belshoff A. C., Higashi R. M., and Fan T. W. (2011) Non-steady state modeling of UDP-GlcNAc biosynthesis is enabled by stable isotope resolved metabolomics (SIRM). BMC Biol. 9, 3721627825 [Google Scholar]
- 44. Wei S., Liu L., Zhang J., Bowers J., Gowda G. A., Seeger H., Fehm T., Neubauer H. J., Vogel U., Clare S. E., and Raftery D. (2013) Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer. Mol. Oncol. 7, 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Owen O. E., Kalhan S. C., and Hanson R. W. (2002) The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 277, 30409–30412 [DOI] [PubMed] [Google Scholar]
- 46. Fan T. W., Lane A. N., Higashi R. M., Farag M. A., Gao H., Bousamra M., and Miller D. M. (2009) Altered regulation of metabolic pathways in human lung cancer discerned by 13C stable isotope-resolved metabolomics (SIRM). Mol. Cancer 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marin-Valencia I., Yang C., Mashimo T., Cho S., Baek H., Yang X. L., Rajagopalan K. N., Maddie M., Vemireddy V., Zhao Z., Cai L., Good L., Tu B. P., Hatanpaa K. J., Mickey B. E., et al. (2012) Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 15, 827–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. DeBerardinis R. J., and Cheng T. (2010) Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 29, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yuneva M. O., Fan T. W., Allen T. D., Higashi R. M., Ferraris D. V., Tsukamoto T., Matés J. M., Alonso F. J., Wang C., Seo Y., Chen X., and Bishop J. M. (2012) The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 15, 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cetinbas N. M., Sudderth J., Harris R. C., Cebeci A., Negri G. L., Yılmaz Ö. H., DeBerardinis R. J., and Sorensen P. H. (2016) Glucose-dependent anaplerosis in cancer cells is required for cellular redox balance in the absence of glutamine. Sci. Rep. 6, 32606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hermann P. C., Huber S. L., Herrler T., Aicher A., Ellwart J. W., Guba M., Bruns C. J., and Heeschen C. (2007) Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 1, 313–323 [DOI] [PubMed] [Google Scholar]
- 52. Tardito S., Oudin A., Ahmed S. U., Fack F., Keunen O., Zheng L., Miletic H., Sakariassen P. Ø., Weinstock A., Wagner A., Lindsay S. L., Hock A. K., Barnett S. C., Ruppin E., Mørkve S. H., et al. (2015) Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat. Cell Biol. 17, 1556–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Davidson S. M., Papagiannakopoulos T., Olenchock B. A., Heyman J. E., Keibler M. A., Luengo A., Bauer M. R., Jha A. K., O'Brien J. P., Pierce K. A., Gui D. Y., Sullivan L. B., Wasylenko T. M., Subbaraj L., Chin C. R., et al. (2016) Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 23, 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mashima T., Seimiya H., and Tsuruo T. (2009) De novo fatty-acid synthesis and related pathways as molecular targets for cancer therapy. Br. J. Cancer 100, 1369–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim J. W., Tchernyshyov I., Semenza G. L., and Dang C. V. (2006) HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 [DOI] [PubMed] [Google Scholar]
- 56. Wise D. R., Ward P. S., Shay J. E., Cross J. R., Gruber J. J., Sachdeva U. M., Platt J. M., DeMatteo R. G., Simon M. C., and Thompson C. B. (2011) Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. U.S.A. 108, 19611–19616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Metallo C. M., Gameiro P. A., Bell E. L., Mattaini K. R., Yang J., Hiller K., Jewell C. M., Johnson Z. R., Irvine D. J., Guarente L., Kelleher J. K., Vander Heiden M. G., Iliopoulos O., and Stephanopoulos G. (2011) Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mullen A. R., Wheaton W. W., Jin E. S., Chen P. H., Sullivan L. B., Cheng T., Yang Y., Linehan W. M., Chandel N. S., and DeBerardinis R. J. (2011) Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lussey-Lepoutre C., Hollinshead K. E., Ludwig C., Menara M., Morin A., Castro-Vega L. J., Parker S. J., Janin M., Martinelli C., Ottolenghi C., Metallo C., Gimenez-Roqueplo A. P., Favier J., and Tennant D. A. (2015) Loss of succinate dehydrogenase activity results in dependency on pyruvate carboxylation for cellular anabolism. Nat. Commun. 6, 8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gameiro P. A., Yang J., Metelo A. M., Pérez-Carro R., Baker R., Wang Z., Arreola A., Rathmell W. K., Olumi A., López-Larrubia P., Stephanopoulos G., and Iliopoulos O. (2013) In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 17, 372–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vincent E. E., Sergushichev A., Griss T., Gingras M. C., Samborska B., Ntimbane T., Coelho P. P., Blagih J., Raissi T. C., Choinière L., Bridon G., Loginicheva E., Flynn B. R., Thomas E. C., Tavaré J. M., et al. (2015) Mitochondrial phosphoenolpyruvate carboxykinase regulates metabolic adaptation and enables glucose-independent tumor growth. Mol. Cell 60, 195–207 [DOI] [PubMed] [Google Scholar]
- 62. Leithner K., Hrzenjak A., Trötzmüller M., Moustafa T., Köfeler H. C., Wohlkoenig C., Stacher E., Lindenmann J., Harris A. L., Olschewski A., and Olschewski H. (2015) PCK2 activation mediates an adaptive response to glucose depletion in lung cancer. Oncogene 34, 1044–1050 [DOI] [PubMed] [Google Scholar]
- 63. Mashimo T., Pichumani K., Vemireddy V., Hatanpaa K. J., Singh D. K., Sirasanagandla S., Nannepaga S., Piccirillo S. G., Kovacs Z., Foong C., Huang Z., Barnett S., Mickey B. E., DeBerardinis R. J., Tu B. P., et al. (2014) Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell 159, 1603–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bos J. L. (1989) ras oncogenes in human cancer: a review. Cancer Res. 49, 4682–4689 [PubMed] [Google Scholar]
- 65. Gaglio D., Metallo C. M., Gameiro P. A., Hiller K., Danna L. S., Balestrieri C., Alberghina L., Stephanopoulos G., and Chiaradonna F. (2011) Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Syst. Biol. 7, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vizan P., Boros L. G., Figueras A., Capella G., Mangues R., Bassilian S., Lim S., Lee W. N., and Cascante M. (2005) K-ras codon-specific mutations produce distinctive metabolic phenotypes in NIH3T3 mice [corrected] fibroblasts. Cancer Res. 65, 5512–5515 [DOI] [PubMed] [Google Scholar]
- 67. Brunelli L., Caiola E., Marabese M., Broggini M., and Pastorelli R. (2014) Capturing the metabolomic diversity of KRAS mutants in non-small-cell lung cancer cells. Oncotarget 5, 4722–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Son J., Lyssiotis C. A., Ying H., Wang X., Hua S., Ligorio M., Perera R. M., Ferrone C. R., Mullarky E., Shyh-Chang N., Kang Y., Fleming J. B., Bardeesy N., Asara J. M., Haigis M. C., et al. (2013) Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Telang S., Lane A. N., Nelson K. K., Arumugam S., and Chesney J. (2007) The oncoprotein H-RasV12 increases mitochondrial metabolism. Mol. Cancer 6, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dang C. V. (2013) MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb. Perspect. Med. 3, a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zeller K. I., Zhao X., Lee C. W., Chiu K. P., Yao F., Yustein J. T., Ooi H. S., Orlov Y. L., Shahab A., Yong H. C., Fu Y., Weng Z., Kuznetsov V. A., Sung W. K., Ruan Y., et al. (2006) Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl. Acad. Sci. U.S.A. 103, 17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shachaf C. M., Kopelman A. M., Arvanitis C., Karlsson A., Beer S., Mandl S., Bachmann M. H., Borowsky A. D., Ruebner B., Cardiff R. D., Yang Q., Bishop J. M., Contag C. H., and Felsher D. W. (2004) MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 431, 1112–1117 [DOI] [PubMed] [Google Scholar]
- 73. Le A., Lane A. N., Hamaker M., Bose S., Gouw A., Barbi J., Tsukamoto T., Rojas C. J., Slusher B. S., Zhang H., Zimmerman L. J., Liebler D. C., Slebos R. J., Lorkiewicz P. K., Higashi R. M., Fan T. W., and Dang C. V. (2012) Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 15, 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gao P., Tchernyshyov I., Chang T. C., Lee Y. S., Kita K., Ochi T., Zeller K. I., De Marzo A. M., Van Eyk J. E., Mendell J. T., and Dang C. V. (2009) c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hu S., Balakrishnan A., Bok R. A., Anderton B., Larson P. E., Nelson S. J., Kurhanewicz J., Vigneron D. B., and Goga A. (2011) 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab. 14, 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu H., Dibling B., Spike B., Dirlam A., and Macleod K. (2004) New roles for the RB tumor suppressor protein. Curr. Opin. Genet. Dev. 14, 55–64 [DOI] [PubMed] [Google Scholar]
- 77. Nicolay B. N., Danielian P. S., Kottakis F., Lapek J. D. Jr., Sanidas I., Miles W. O., Dehnad M., Tschöp K., Gierut J. J., Manning A. L., Morris R., Haigis K., Bardeesy N., Lees J. A., Haas W., and Dyson N. J. (2015) Proteomic analysis of pRb loss highlights a signature of decreased mitochondrial oxidative phosphorylation. Genes Dev. 29, 1875–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Reynolds M. R., Lane A. N., Robertson B., Kemp S., Liu Y., Hill B. G., Dean D. C., and Clem B. F. (2014) Control of glutamine metabolism by the tumor suppressor Rb. Oncogene 33, 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nicolay B. N., Gameiro P. A., Tschöp K., Korenjak M., Heilmann A. M., Asara J. M., Stephanopoulos G., Iliopoulos O., and Dyson N. J. (2013) Loss of RBF1 changes glutamine catabolism. Genes Dev. 27, 182–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fan T. W., Bandura L. L., Higashi R. M., and Lane A. N. (2005) Metabolomics–edited transcriptomics analysis of Se anticancer action in human lung cancer cells. Metabolomics 1, 325–339 [Google Scholar]
- 81. Vander Heiden M. G. (2011) Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 10, 671–684 [DOI] [PubMed] [Google Scholar]
- 82. Svensson R. U., Parker S. J., Eichner L. J., Kolar M. J., Wallace M., Brun S. N., Lombardo P. S., Van Nostrand J. L., Hutchins A., Vera L., Gerken L., Greenwood J., Bhat S., Harriman G., Westlin W. F., et al. (2016) Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat. Med. 22, 1108–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gimenez-Roqueplo A. P., Favier J., Rustin P., Mourad J. J., Plouin P. F., Corvol P., Rötig A., and Jeunemaitre X. (2001) The R22X mutation of the SDHD gene in hereditary paraganglioma abolishes the enzymatic activity of complex II in the mitochondrial respiratory chain and activates the hypoxia pathway. Am. J. Hum. Genet. 69, 1186–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Parsons D. W., Jones S., Zhang X., Lin J. C., Leary R. J., Angenendt P., Mankoo P., Carter H., Siu I. M., Gallia G. L., Olivi A., McLendon R., Rasheed B. A., Keir S., Nikolskaya T., et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tomlinson I. P., Alam N. A., Rowan A. J., Barclay E., Jaeger E. E., Kelsell D., Leigh I., Gorman P., Lamlum H., Rahman S., Roylance R. R., Olpin S., Bevan S., Barker K., Hearle N., et al. (2002) Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 30, 406–410 [DOI] [PubMed] [Google Scholar]
- 86. Cardaci S., Zheng L., MacKay G., van den Broek N. J., MacKenzie E. D., Nixon C., Stevenson D., Tumanov S., Bulusu V., Kamphorst J. J., Vazquez A., Fleming S., Schiavi F., Kalna G., Blyth K., Strathdee D., and Gottlieb E. (2015) Pyruvate carboxylation enables growth of SDH-deficient cells by supporting aspartate biosynthesis. Nat. Cell Biol. 17, 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Saxena N., Maio N., Crooks D. R., Ricketts C. J., Yang Y., Wei M.-H., Fan T. W.-M., Lane A. N., Sourbier C., Rouault T. A., and Linehan W. M. (2016) SDHB-deficient cancers: the role of mutations that impair iron sulfur cluster delivery. J. Natl. Cancer Inst. 108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Izquierdo-Garcia J. L., Cai L. M., Chaumeil M. M., Eriksson P., Robinson A. E., Pieper R. O., Phillips J. J., and Ronen S. M. (2014) Glioma cells with the IDH1 mutation modulate metabolic fractional flux through pyruvate carboxylase. PLoS ONE 9, e108289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zheng L., Mackenzie E. D., Karim S. A., Hedley A., Blyth K., Kalna G., Watson D. G., Szlosarek P., Frezza C., and Gottlieb E. (2013) Reversed argininosuccinate lyase activity in fumarate hydratase-deficient cancer cells. Cancer Metab. 1, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Klawitter J., Kominsky D. J., Brown J. L., Klawitter J., Christians U., Leibfritz D., Melo J. V., Eckhardt S. G., and Serkova N. J. (2009) Metabolic characteristics of imatinib resistance in chronic myeloid leukaemia cells. Br. J. Pharmacol. 158, 588–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Serkova N., and Boros L. G. (2005) Detection of resistance to imatinib by metabolic profiling: clinical and drug development implications. Am. J. Pharmacogenomics 5, 293–302 [DOI] [PubMed] [Google Scholar]
- 92. Sancho P., Burgos-Ramos E., Tavera A., Bou Kheir T., Jagust P., Schoenhals M., Barneda D., Sellers K., Campos-Olivas R., Graña O., Viera C. R., Yuneva M., Sainz B. Jr., and Heeschen C. (2015) MYC/PGC-1α balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 22, 590–605 [DOI] [PubMed] [Google Scholar]
- 93. Zhao H., Yang L., Baddour J., Achreja A., Bernard V., Moss T., Marini J. C., Tudawe T., Seviour E. G., San Lucas F. A., Alvarez H., Gupta S., Maiti S. N., Cooper L., Peehl D., et al. (2016) Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 5, e10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fan T. W., Lane A. N., and Higashi R. M. (2016) Stable isotope resolved metabolomics studies in ex vivo tissue slices. Bio. Protoc. 6, e1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Vargas A. J., and Harris C. C. (2016) Biomarker development in the precision medicine era: lung cancer as a case study. Nat. Rev. Cancer 16, 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hou Y., Yin M., Sun F., Zhang T., Zhou X., Li H., Zheng J., Chen X., Li C., Ning X., Lou G., and Li K. (2014) A metabolomics approach for predicting the response to neoadjuvant chemotherapy in cervical cancer patients. Mol. Biosyst. 10, 2126–2133 [DOI] [PubMed] [Google Scholar]
- 97. Kasumov T., Adams J. E., Bian F., David F., Thomas K. R., Jobbins K. A., Minkler P. E., Hoppel C. L., and Brunengraber H. (2005) Probing peroxisomal βoxidation and the labelling of acetyl-CoA proxies with [1-13C]octanoate and [3-13C]octanoate in the perfused rat liver. Biochem. J. 389, 397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cowin G. J., Willgoss D. A., Bartley J., and Endre Z. H. (1996) Serine isotopomer analysis by 13C-NMR defines glycine-serine interconversion in situ in the renal proximal tubule. Biochim. Biophys. Acta 1310, 32–40 [DOI] [PubMed] [Google Scholar]
- 99. Qi J., Lang W., Geisler J. G., Wang P., Petrounia I., Mai S., Smith C., Askari H., Struble G. T., Williams R., Bhanot S., Monia B. P., Bayoumy S., Grant E., Caldwell G. W., et al. (2012) The use of stable isotope-labeled glycerol and oleic acid to differentiate the hepatic functions of DGAT1 and -2. J. Lipid Res. 53, 1106–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kurland I. J., Alcivar A., Bassilian S., and Lee W. N. (2000) Loss of [13C]glycerol carbon via the pentose cycle. Implications for gluconeogenesis measurement by mass isotoper distribution analysis. J. Biol. Chem. 275, 36787–36793 [DOI] [PubMed] [Google Scholar]