Abstract
Rad52 is a highly conserved protein involved in the repair of DNA damage. Human RAD52 has been shown to mediate single-stranded DNA (ssDNA) and is synthetic lethal with mutations in other key recombination proteins. For this study, we used single-molecule imaging and ssDNA curtains to examine the binding interactions of human RAD52 with replication protein A (RPA)-coated ssDNA, and we monitored the fate of RAD52 during assembly of the presynaptic complex. We show that RAD52 binds tightly to the RPA-ssDNA complex and imparts an inhibitory effect on RPA turnover. We also found that during presynaptic complex assembly, most of the RPA and RAD52 was displaced from the ssDNA, but some RAD52-RPA-ssDNA complexes persisted as interspersed clusters surrounded by RAD51 filaments. Once assembled, the presence of RAD51 restricted formation of new RAD52-binding events, but additional RAD52 could bind once RAD51 dissociated from the ssDNA. Together, these results provide new insights into the behavior and dynamics of human RAD52 during presynaptic complex assembly and disassembly.
Keywords: DNA-binding protein, DNA recombination, DNA repair, microscopic imaging, single-molecule biophysics
Introduction
Homologous recombination (HR)2 is a conserved pathway for the repair of double-stranded breaks and is important for maintaining genomic integrity (1–8). HR involves the exchange of genetic information between homologous DNA molecules and performs crucial roles in the repair of double-stranded breaks (DSBs) (2, 3), the rescue of stalled replication forks (9, 10), meiosis (11–13), and break-induced replication (14–16). DSBs that are left unrepaired can lead to genetic instability and chromosomal rearrangements (1–4, 8). Defects in HR have been linked to many genetic diseases and cancer syndromes, such as Fanconi anemia and Bloom syndrome (3, 17–19).
Pairing of homologous sequences during recombination requires the action of ATP-dependent DNA recombinases, which are members of the Rad51/RecA family of proteins (20–22). These proteins form extended helical filaments on single-stranded DNA (ssDNA), and the resulting nucleoprotein filaments are referred to as the presynaptic complex (20–22). The presynaptic complex is a key intermediate in homologous recombination and is necessary to align and pair homologous DNA sequences (20–22).
During DSB repair, long 3′ ssDNA overhangs are generated by nucleases that resect the newly generated dsDNA end in the 5′ → 3′ direction (23–25). These ssDNA overhangs are first bound by replication protein A (RPA), which protects the ssDNA from nucleases and also helps remove secondary structures (26, 27). RPA can inhibit presynaptic complex assembly, but this inhibition is overcome by the action of mediator proteins (22, 28). Saccharomyces cerevisiae Rad52 is one of the most studied mediator proteins (29, 30). S. cerevisiae Rad52 is a ring-shaped oligomer (31) that helps load Rad51 on RPA-ssDNA (32, 33), binds tightly to ssDNA (31, 34), and promotes ssDNA annealing (35–37). Rad52 also participates in second-strand capture during the later stages of recombination and promotes the initiation of new DNA synthesis (38, 39). The profound importance of Rad52 to DNA repair in S. cerevisiae is highlighted by the extreme sensitivity of rad52 mutants to DNA-damaging agents (2, 40, 41).
Rad52 is highly conserved, and human RAD52 shares many biochemical traits with its yeast counterpart (42, 43). Like yeast Rad52, human RAD52 also forms ring-like structures (44–47), binds tightly to ssDNA, and promotes ssDNA annealing (47–51). However, human RAD52 does not have a known mediator activity (42, 43), and in striking contrast to S. cerevisiae, RAD52 deletions do not produce a strong phenotype in vertebrates (42, 43, 52, 53). These observations suggested that RAD52 was of lesser importance for DNA repair in higher organisms and that its functions had been overtaken by other proteins, such as BRCA2. However, RAD52 deficiencies are now known to be synthetically lethal with several key human recombination proteins, including BRCA1, BRCA2, PABL2, and the RAD51 paralog XRCC3 (54–56). In addition, recent studies have revealed an important role for mammalian RAD52 in promoting DNA synthesis during replication stress (57–59). These findings have led to a renewed interest in understanding how human RAD52 functions during DNA repair (42, 60) and have revealed RAD52 as an important target for anticancer therapeutics (60–64).
Despite the wealth of information available for S. cerevisiae Rad52, the exact role(s) human RAD52 in homologous recombination remains largely unclear. Here, we use two-color single-molecule imaging and ssDNA curtains to begin examining the dynamics of human RAD52, RPA, and RAD51 on ssDNA during presynaptic complex assembly. We show that RAD52 binds very tightly to RPA-ssDNA, and the presence of RAD52 restricts facilitated exchange of RPA, highlighting a potential regulatory role of RAD52 that is conserved from yeast to humans. Although most of the bound RAD52 and RPA is displaced upon addition of RAD51, we find that many small RAD52-RPA clusters remain embedded between longer RAD51 filaments. However, RAD52 appears incapable of directly associating with regions of ssDNA that are coated by RAD51, suggesting that RAD52 cannot bind to internal regions of the RAD51 filaments. Together, these data begin to provide new insights into the behavior of human RAD52 during presynaptic complex assembly.
Results
RAD52 binding to ssDNA curtains
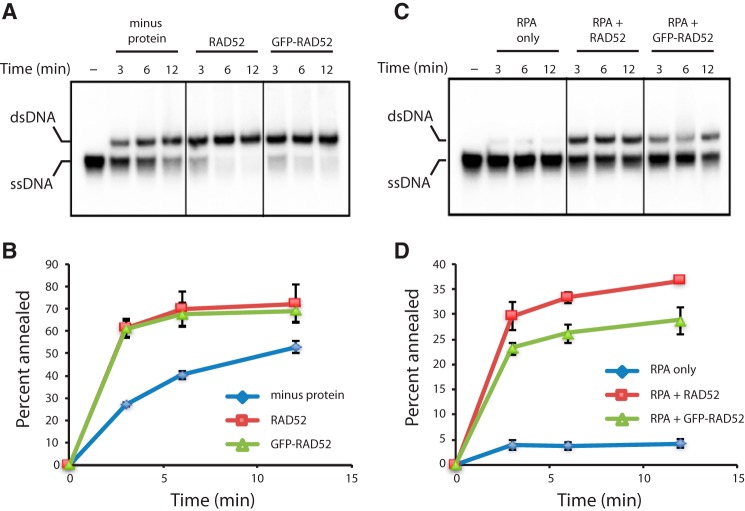
We have previously shown that S. cerevisiae Rad52 binds very tightly to ssDNA coated with yeast RPA (65). Given the high degree of conservation, we hypothesized that human RAD52 should behave similarly. For single molecule visualization, we expressed human RAD52 as fusion construct labeled at the N terminus with GFP. Previous studies have shown that GFP-tagged RAD52 is targeted to repair foci in vivo (66–69), and our bulk biochemical assays confirmed that GFP-RAD52 retained the ability promote strand annealing similar to unlabeled RAD52 (Fig. 1, A and B). GFP-RAD52 was also able to overcome RPA-mediated inhibition of ssDNA at levels comparable with those of unlabeled RAD52 (Fig. 1, C and D).
Figure 1.
PAGE assays for RAD52 ssDNA annealing activity. A and B, gel image of an ssDNA annealing by RAD52 and GFP-RAD52 in the absence of RPA (A) and corresponding quantitation analysis (B). C and D, ssDNA annealing by RAD52 and GFP-RAD52 in the presence of RPA (C) and corresponding quantitation analysis (D). Error bars represent standard deviation from three experiments.
We next used ssDNA curtains and human RPA-RFP to mimic the early stages of HR in which the processed 3′ ssDNA ends are initially coated with RPA (Fig. 2, A and B) (65, 70–73). When 50 pm GFP-RAD52 was injected into the sample chambers, we were able to readily detect binding of individual GFP-RAD52 complexes to both RFP-tagged RPA-ssDNA (Fig. 2B) and also to unlabeled RPA-ssDNA (Fig. 2C). At the low protein concentrations used in these reactions, the GFP-RAD52 complexes bound as discrete puncta to the RPA-ssDNA. However, when the concentration of the GFP-RAD52 injection was increased to 600 pm, the RAD52 complexes were able to coat the entire RPA-ssDNA molecules (Fig. 2E). Given these findings, subsequent experiments were conducted at 50 pm GFP-RAD52, unless stated otherwise.
Figure 2.
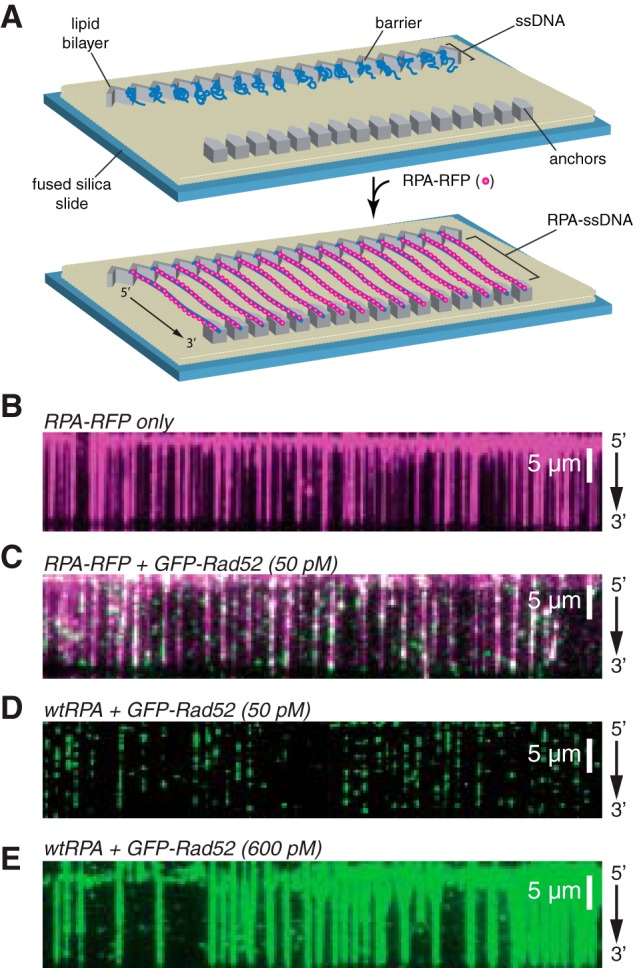
ssDNA curtain assay for RPA and RAD52 binding. A, schematic of double-tethered ssDNA curtains showing the nanofabricated patterns on the surface of a fused silica microscope slide. All hRPA-ssDNA molecules are anchored at both ends. 5′ anchoring is achieved with biotin-streptavidin-biotin linkage to the modified lipids in the bilayer and aligned at the zig-zag shaped chromium barriers. 3′ is anchored through nonspecific binding to the exposed chromium pedestals. B, wide-field image of RPA-RFP (magenta) bound to ssDNA in the absence of RAD52. C, wide-field image of RPA-RFP (magenta) curtain in the presence of 50 pm GFP-RAD52 (green). D, wide-field image of GFP-RAD52 (50 pm; green) bound to an wtRPA-ssDNA curtain (unlabeled). E, wide-field image of GFP-RAD52 (600 pm; green) bound to an wtRPA-ssDNA curtain (unlabeled).
RAD52 binds as discrete complexes
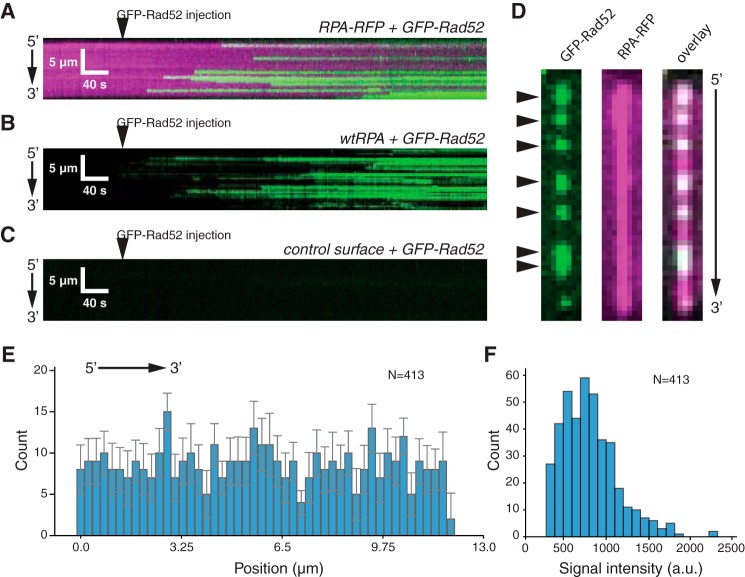
When viewed in real time, we could detect individual GFP-RAD52-binding events on ssDNA bound by either RPA-RFP (Fig. 3A) or bound by unlabeled RPA (Fig. 3B). In contrast to these results with RPA-ssDNA, we could detect no binding of GFP-RAD52 to a control surface that did not have a tethered ssDNA substrate (Fig. 3C), confirming that the GFP-RAD52-binding events only occurred on the RPA-ssDNA. We saw no evidence of either cooperative binding as would be evidenced by nucleation and spreading of GFP-RAD52 along the RPA-ssDNA. Instead, GFP-RAD52 binding always occurred as discrete events (Fig. 3, A and B). This finding is in contrast to our prior results with S. cerevisiae Rad52, which binds and appears to spread more extensively along the RPA-ssDNA complexes (65).
Figure 3.
RAD52-binding behavior. A, kymograph showing real time binding of GFP-RAD52 (50 pm; green) to an RPA-RFP-coated ssDNA molecule (magenta). B, kymograph showing binding of GFP-RAD52 (50 pm; green) to a wtRFP-coated ssDNA molecule (unlabeled). C, kymograph showing that GFP-RAD52 (50 pm; green) does not bind to a control surface within the sample chamber that lacks an ssDNA molecule. D, wide-field images of a single ssDNA molecule showing GFP-RAD52 complexes (green; highlighted with arrowheads), the RPA-RFP-coated ssDNA (magenta), and an overlaid image of the GFP-RAD52 and RFP-RFP signals. E, binding site distribution of GFP-RAD52 bound to the RPA-coated ssDNA molecules. Error bars represent standard deviation obtained through Bootstrap analysis of the data. F, histogram showing the intensity distribution of the RAD52-GFP complexes.
We next analyzed both the signal intensity and position distributions of the GFP-RAD52–binding interactions. When examined at the level of single ssDNA molecules, we could readily identify the locations of discrete GFP-RAD52 complexes that bound to the RPA-ssDNA (Fig. 3D). A binding position distribution histogram confirmed that there was no position or regional specificity for GFP-Rad52 binding to the RPA-ssDNA complexes (Fig. 3E). Interestingly, the bound GFP-RAD52 visually appeared to have relatively uniform fluorescence signal, which was confirmed by quantitation of the signal intensities for individual GFP-RAD52 complexes (Fig. 3F). The presence of a single prominent peak in this signal intensity distribution is consistent with the conclusion that GFP-RAD52 was behaving as a relatively well defined entity, rather than a distribution of complexes with vastly different numbers of GFP fluorophores.
RAD52 binds tightly to RPA-ssDNA
We next examined the stability of interactions observed between GFP-RAD52 and the RPA-ssDNA complexes. For this, we examined the GFP-RAD52 bound to RFP-tagged RPA-ssDNA over a total period of either 12 or 100 min (Fig. 4, A and B). In each of these experiments, the laser illumination was shuttered between each frame, and the frame acquisition rates were adjusted such that the total time during which the samples were exposed to laser illumination was identical for each of the two measurements, as previously described (71). From each of these measurements, we then plotted survival probability for the individual GFP-RAD52 complexes (Fig. 4, C and D). These experiments revealed a half-life of 29.4 ± 0.56 min for the 12 min observation and 256 ± 10.9 min for the 100 min measurements, and in both cases more than ∼72% of the GFP-RAD52 remained visible by the end of the measurements (Fig. 4, C and D). The similar slopes for these two graphs (Fig. 4, C and D) and the finding that the half-time varies in proportion to laser exposure time indicate that any loss of GFP-RAD52 during these observations could be attributed to photo-bleaching, whereas the complexes themselves are so stable that their binding lifetime must significantly exceed the 100-min maximum duration of our experiments. We conclude that human RAD52, like S. cerevisiae Rad52 (65), binds very tightly to RPA-ssDNA and does not dissociate appreciably for at least ≥2 h.
Figure 4.
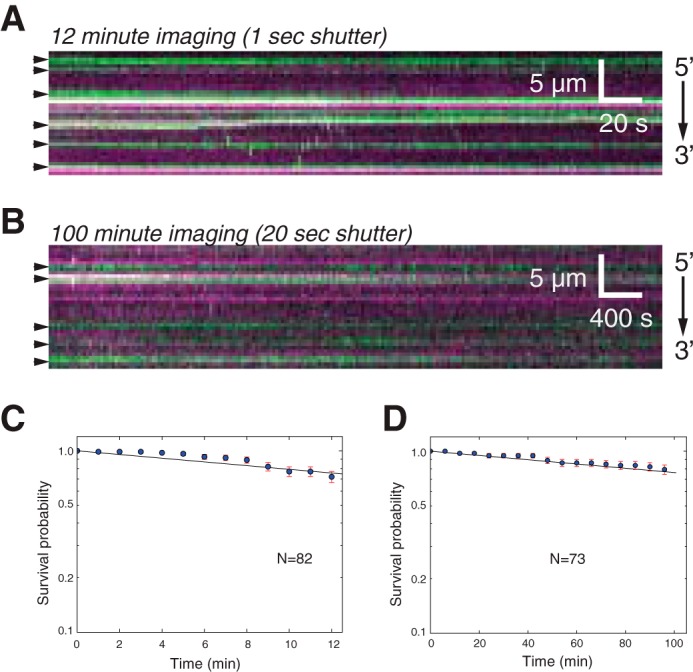
GFP-RAD52 binding lifetime. A and B, kymograms showing GFP-RAD52 (green) bound to RPA-RFP-ssDNA (magenta) and visualized for either 12 min (A) or visualized for 100 min (B), as indicated. For both experiments, the shuttering time was adjusted so that the total laser exposure time was identical for both experiments. C, survival probability analysis for RAD52 complexes viewed for the duration of the 12-min experiment. D, survival probability analysis for RAD52 complexes viewed for the duration of the 100-min experiment. The error bars represent standard deviation obtained through bootstrap analysis of the data.
RPA turnover in the presence of RAD52
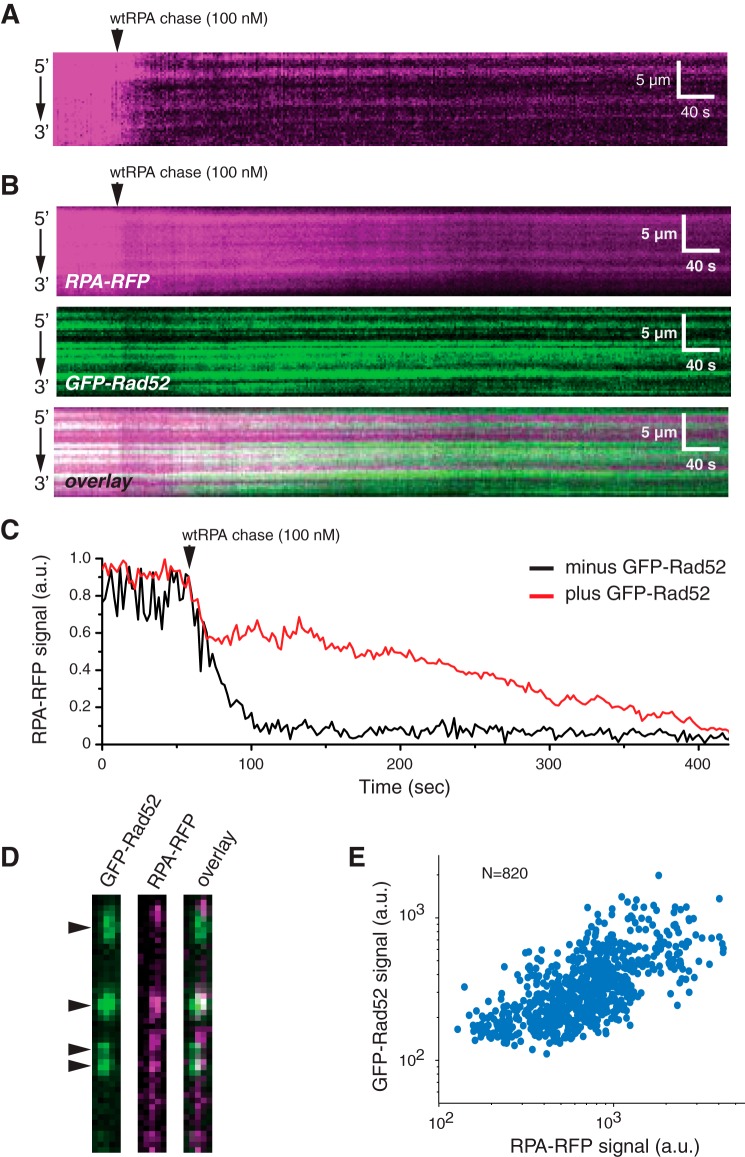
We have previously shown that yeast and human RPA can bind ssDNA very stably when free RPA is absent from solution (71). However, when free protein is present in solution, the bound RPA can undergo rapid exchange between free and bound states through a mechanism called facilitated dissociation (71). This mechanism involves the existence of microscopically dissociated states of the bound RPA, which lead to macroscopic dissociation only when free RPA is present to compete for the transiently exposed naked ssDNA (71, 74).
Interestingly, our previous results have shown that yeast Rad52 restricts the ability of RPA to undergo facilitated exchange (65). To determine whether human RAD52 acts similarly, we conducted RPA exchange experiments with two different substrates: (i) ssDNA-RPA (Fig. 5A) and (ii) ssDNA-RPA bound by RAD52 (Fig. 5B). In the first scenario, as expected, the bound RPA was rapidly exchanged by the free dark wtRPA, which was confirmed the sharp decline in overall RPA-RFP signal intensity (Fig. 5, A and C). In the second case, there was also a decrease in RPA-RFP signal intensity, but this reduction was smaller compared to the experiment without RAD52 (Fig. 5, B and C).
Figure 5.
RAD52 regulation of RPA turnover. A and B, kymograms showing RPA-RFP (magenta) turnover in the absence (A) and presence (B) of GFP-RAD52 (green). C, time-dependent changes in total integrated RPA-RFP fluorescence signal integrated over an entire ssDNA molecule in the presence and absence of GFP-RAD52 (green), as indicated, after chasing with 100 nm wtRPA. D, examples of side-by-side wide-field images showing the co-localization of GFP-RAD52 (green) and turnover-resistant RPA-RFP (magenta) after chasing with 100 nm wtRPA. E, scatter plot showing the correlation between the bound GFP-RAD52 and the turnover-resistant RPA-RFP.
We further studied the mechanism of this inhibition by examining the spatial relationship between the bound GFP-RAD52 complexes and the RPA-RFP that remained bound following the wtRPA chase. Inspection of this data revealed co-localization of bound GFP-RAD52 with exchange resistant RPA-RFP (Fig. 5D). This finding was also confirmed by correlation analysis, which revealed that the areas bound by exchange-resistant RPA-RFP were correlated with the presence of GFP-RAD52 complexes (Pearson's r = 0.66; P < 1 × 10−5; N = 820) (Fig. 5E). Together, these findings suggest that human RAD52 had an inhibitory effect on RPA-facilitated exchange.
RAD52 during presynaptic complex assembly
The role of human RAD52 in HR remains unknown. To help better understand its potential function(s), we next asked how GFP-RAD52 behaves during assembly of the RAD51 presynaptic complex. To determine the fate of GFP-RAD52 during presynaptic complex assembly, we first prepared single-tethered RFP-tagged RPA-ssDNA curtains, and we then injected 200 pm GFP-RAD52 into the sample chamber (Fig. 6A). Under these conditions, we observed extensive co-localization of GFP-RAD52 and RPA-RFP on the ssDNA (Fig. 6B). We used single-tethered curtains for these measurements so that we could observe the binding of GFP-RAD52 and RPA-RFP and also independently monitor the binding of wild-type (unlabeled) RAD51 based upon the observed changes in length of the ssDNA molecules, as previously described (65, 72).
Figure 6.
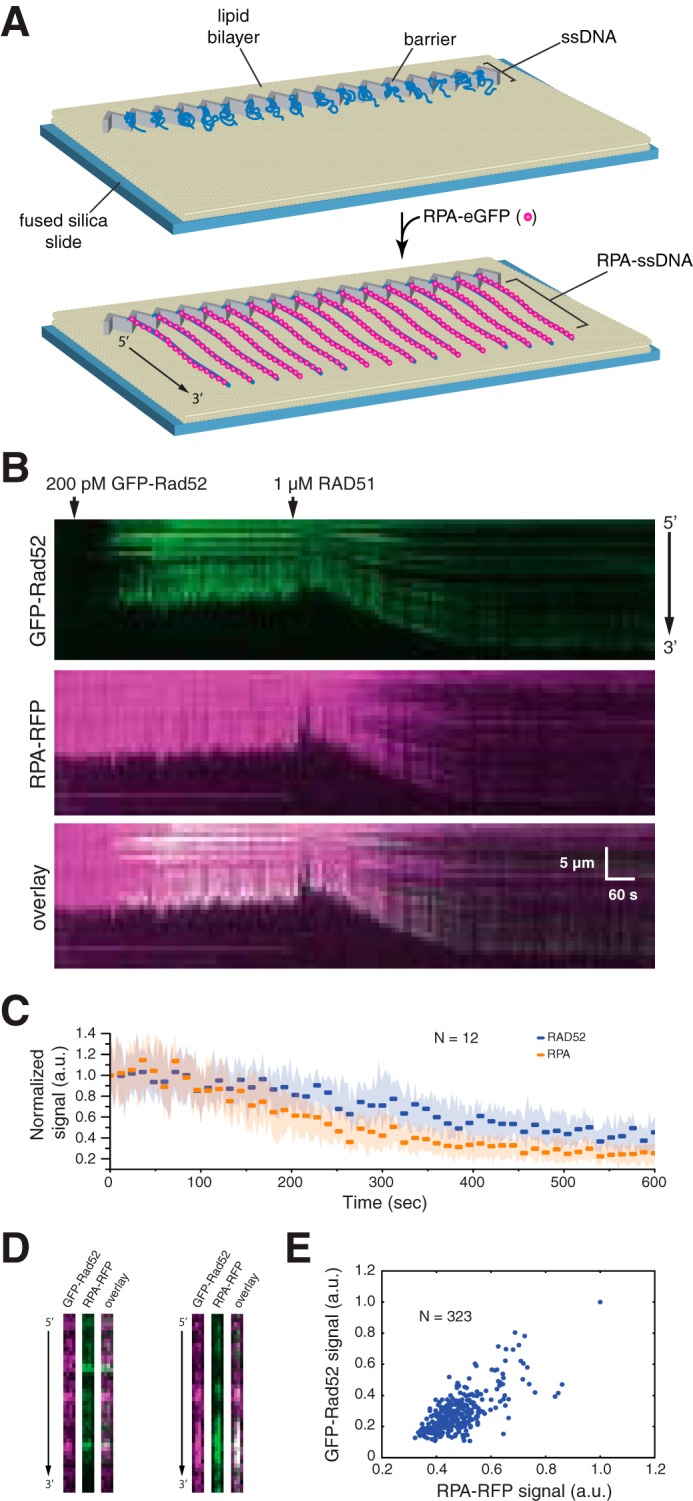
RAD52 and RPA behavior during presynaptic complex assembly. A, schematic illustration of the single-tethered ssDNA curtain assay. B, kymograms showing the binding of GFP-RAD52 (200 pm; green) to an RPA-RFP-ssDNA molecule (magenta), followed by the addition of 1 μm wild-type RAD51 (unlabeled). RAD51 binding coincides with a loss of GFP-RAD52 and RPA-RFP fluorescence signal that also coincides with extension of the ssDNA. C, graphic representation of the GFP-RAD52 and RPA-RFP fluorescence signal loss during presynaptic complex assembly. Each data point reflects the mean integrated signal intensity from entire ssDNA molecules (N = 12 molecules), and error bars represent standard deviation. D, side-by-side presentation of wide-field images of GFP-RAD52 (green) and RPA-RFP (magenta) showing co-localization of RAD52 and RPA clusters embedded within the presynaptic complex. E, scatter plot showing the correlation between GFP-RAD52- and RPA-RFP-binding distributions (based upon the normalized signal intensities) within the presynaptic complex.
Next, we injected 1 μm human RAD51 in buffer containing 30 mm Tris (pH 7.4), 1 mm MgCl2, 5 mm CaCl2, 100 mm KCl, 0.2 mg/ml BSA, 1 mm DTT, and 2 mm ATP. We then observed the reactions in real time with constant buffer flow; constant buffer flow is necessary to maintain the single-tethered ssDNA extended parallel to the sample chamber surface (65, 72). As expected, introduction of RAD51 into the sample chamber resulted in an increase in the apparent length of the ssDNA, which coincided with a loss of RPA-RFP fluorescence signal intensity (Fig. 6B). Interestingly, the GFP-RAD52 signal also decreased upon assembly of the presynaptic complex, indicating that RAD51 displaced some of the RAD52 from the ssDNA. Quantitation of the total fluorescence signal intensity integrated across the entire length of the ssDNA for GFP-RAD52 and RPA-RFP indicated that RAD51 binding under these conditions resulted in displacement of ∼75 and ∼60% of the RPA and RAD52, respectively (Fig. 6C).
Interestingly, some RAD52 remained bound to the ssDNA even after RAD51 binding (Fig. 6B). Side-by-side comparison of the GFP and RFP fluorescence signals demonstrated that RAD52 was co-localized with small clusters of RPA that also remained associated with the presynaptic complex after RAD51 binding (Pearson's r = 0.70; P < 1 × 10−5; N = 323) (Fig. 6, B and D). These findings show that small clusters of RAD52 and RPA can remain associated with the presynaptic complex, and these clusters are interspersed between the RAD51 filaments.
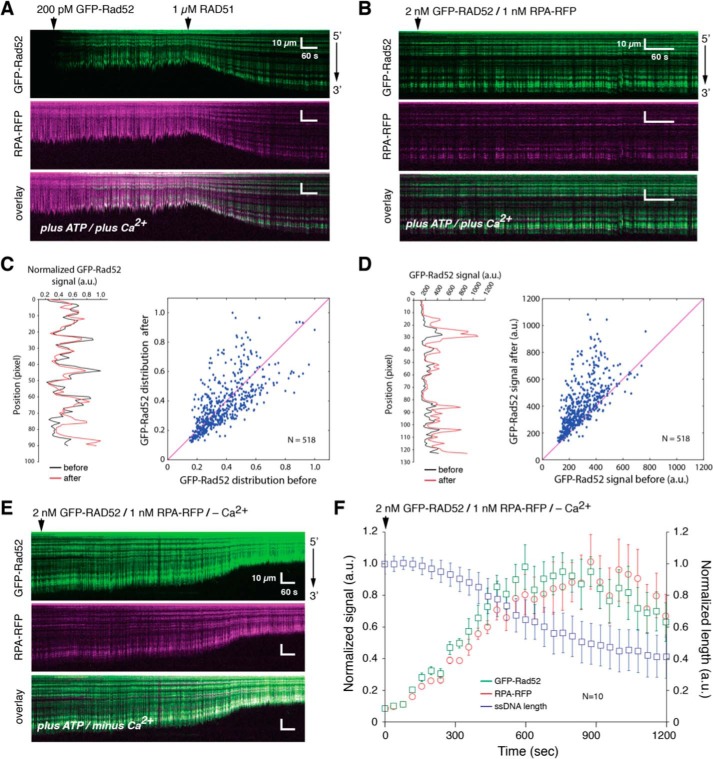
Binding of RAD52 on the presynaptic filament
We next sought to determine whether additional RAD52 could associate with the presynaptic complex after binding of RAD51. For this, we assembled RFP-tagged RPA-ssDNA and then added 200 pm GFP-RAD52, followed by 1 μm RAD51 in the presence of ATP and Ca2+, as indicted above. As expected, the addition of RAD51 caused the length of the ssDNA to increase, and some RPA-RFP and GFP-RAD52 remained bound to the ssDNA (Fig. 7A). Next, we injected an additional 2 nm GFP-RAD52 into the sample chamber while monitoring the length and fluorescence signal of the presynaptic complexes (Fig. 7B). Notably, the addition of just 600 pm GFP-RAD52 in the absence of RAD51 results in complete coverage of the ssDNA by RAD52 (Fig. 1E). However, we do not detect extensive new binding of GFP-RAD52 to the presynaptic complex, and we do not detect substantial new GFP-RAD52 binding to regions that had been previously devoid of RAD52 (Fig. 7B). This assertion was verified by comparing spatial distribution of the normalized GFP-RAD52 fluorescence signals along the presynaptic complex before and after the 2 nm GFP-RAD52 injection (Pearson's r = 0.67; P < 1 × 10−5; N = 518) (Fig. 7C). Even though GFP-RAD52 did not appear to bind to many new locations, there was still an overall increase in the fluorescence signal at locations corresponding to the pre-existing GFP-RAD52 clusters (Fig. 7D, right panel), which was confirmed by analysis of the fluorescence signal intensities before and after injection of 2 nm GFP-RAD52 (Fig. 7D, left panel). Together, these observations are most consistent with a model where ssDNA-bound RAD51 filaments exclude the binding of new RAD52, but additional RAD52 can interact with the presynaptic complex through association with pre-existing RAD52 clusters.
Figure 7.
RAD52 binding to the presynaptic complex. A, kymographs showing the presynaptic complex assembly on a RFP-RFP (magenta) bound ssDNA (single-tethered) in the presence of GFP-RAD52 (200 pm; green). B, kymographs showing the same presynaptic complex following injection of higher concentrations of GFP-RAD52 (2 nm) and RPA-RFP (1 nm). C, plot showing examples of the normalized spatial distribution (position) of GFP-RAD52 signal before and after the 2 nm GFP-RAD52 injection for one ssDNA (left panel) and scatter plot quantitating the correlation between the normalized GFP-RAD52 spatial distributions before and after the 2 nm GFP-RAD52 injection (right panel). D, plot showing examples of the raw signal intensity (not normalized) for GFP-RAD52 before and after the 2 nm GFP-RAD52 injection for one ssDNA (left panel) and scatter plot quantitating that the raw GFP-RAD52 signal increases after the 2 nm GFP-RAD52 injection (right panel). The magenta lines shown in the scatter plots for C and D represent the diagonal for reference. E, kymographs showing the dissociation of RAD51 from the ssDNA after removal of Ca2+ from the reaction buffer in the continued presence of 2 nm GFP-RAD52 (green) and 1 nm RPA-RFP (magenta); RAD51 dissociation is evidenced by the decrease in ssDNA. F, quantitation of the changes in total GFP-RAD52 and RPA-RFP signal intensities (integrated over entire ssDNA molecules) that coincide with RAD51 dissociation (N = 10 molecules), and error bars represent standard deviation. The images shown in A, B, and E show the same ssDNA molecule at each different stage of the experiment, allowing for direct visual comparisons.
Disassembly of the presynaptic filament
We next asked whether additional RAD52 could bind to the ssDNA after dissociation of RAD51. We have previously shown that Ca2+ promotes assembly of the human RAD51 presynaptic complex (72), consistent with prior reports from bulk biochemical assays (75, 76). Therefore, we initiated presynaptic complex disassembly by switching to buffer that contained 2 nm GFP-RAD52 and 1 nm RPA-RFP but lacked ATP and Ca2+. Under these conditions, the signal tethered ssDNA gradually shortened, indicating that RAD51 was undergoing dissociation, and at the same time the fluorescence signal increased for both GFP-RAD52 and RPA-RFP (Fig. 7, E and F). These results confirm that additional GFP-RAD52 and RPA-RFP can bind to the ssDNA substrate, but only after RAD51 has dissociated from the ssDNA. These results provide additional support for the conclusion that RAD52 cannot bind directly to human RAD51 filaments.
Discussion
Here we used ssDNA curtains to study the interactions of human RAD52 with the early intermediates of the homologous recombination pathway. We demonstrate that human RAD52 binds tightly to RPA-ssDNA. At low protein concentrations (50 pm), RAD52 binds as discrete complexes that are positioned randomly along the length of the RPA-ssDNA. When the concentration of RAD52 is increased to 600 pm, the protein appears to coat the entire length of the RPA-ssDNA. Interestingly, there is no evidence that RPA is displaced from the ssDNA even at these highest concentrations of RAD52, indicating that both proteins can extensively co-occupy the ssDNA prior to the arrival of RAD51. The RAD52 complexes are extremely stable, and we can see no evidence for RAD52 dissociation from the RPA-ssDNA complexes even when viewed over a 100-min observation period. In addition to these results with double-tethered ssDNA, we can also observe extensive RAD52-dependent compaction of single-tethered ssDNA molecules (not shown). This compaction can be reversed by displacement of RAD52 from the ssDNA by the addition of either 7 m urea (not shown). These observations are consistent with the ring-like structure of RAD52 and the fact that its ssDNA-binding domain follows the circular contour of this surface, which would necessitate a reduction in the apparent end-to-end distance for any bound ssDNA, as has been previously reported (51).
We find some striking similarities, as well as some notable differences, between the results we report here for human RAD52 and results we have previously reported for S. cerevisiae Rad52 (65). Both yeast and human RAD52 bind tightly to RPA-ssDNA, and both also appear to restrict the facilitated exchange of RPA when free RPA is present in solution. When RAD51 is added to the reactions, much of the RPA and RAD52 dissociate from the ssDNA, but small clusters of RPA and RAD52 remain interspersed between the resulting RAD51 filaments. Retention of these RPA-RAD52 clusters within the presynaptic complex is another trait shared between the yeast and human systems. We do not yet know whether these RPA-RAD52 clusters reflect biologically relevant breaks in the RAD51 filament, or whether they might arise because of the limited complement of HR proteins used in our in vitro assays. Future work will be necessary to explore these and other possibilities.
The most pronounced difference for the human and yeast RAD52 proteins occurs after addition of RAD51. For the S. cerevisiae proteins, we find that RPA and RAD52 can both bind extensively to the assembled presynaptic complex (65). These new binding events initiate at pre-existing clusters of RPA-Rad52 and can then appear to spread along the Rad51 filaments, but with no evidence that Rad51 is displaced from the ssDNA (65). Thus, the S. cerevisiae system appears to allow for assembly of more extensive Rad51-RPA-Rad52-ssDNA co-complexes. We have previously speculated that the presence of S. cerevisiae RPA and Rad52 in these co-complexes may facilitate some downstream step(s) in recombination (65), such as second strand capture, which is thought to require RAD52 (35). In contrast, we cannot detect extensive rebinding of either RPA or RAD52 to the human presynaptic complexes. We do detect some binding of additional RAD52 and RPA to pre-existing RPA-RAD52 clusters, but this rebinding is modest when compared with the yeast proteins, and we find no evidence that either human RPA or human RAD52 can spread into the RAD51 filaments. Therefore, if additional human RAD52 is required to participate in any downstream recombination steps, then it must enter the pathway at some stage downstream of RAD51 filament assembly and does not appear to interact extensively with the ssDNA-bound RAD51, or perhaps additional protein components of the presynaptic complex, such as the RAD51 paralogs, may be needed to allow further association of human RAD52. Future work will be necessary to explore these possibilities.
Interestingly, our results show that human RAD52 binds very tightly to RPA-ssDNA in vitro, with essentially no turnover on our experimental time scales. Fluorescently labeled RAD52 forms recombination foci that co-localize with DNA damage in vivo (66–68). Previous fluorescence recovery after photo-bleaching studies have shown that mammalian RAD52 undergoes rapid exchange between free and bound states within these repair foci, whereas RAD51 is largely immobile (69). Our in vitro studies do not necessarily contradict these findings because there are three key experimental differences that likely contribute to different outcomes for RAD52 turnover. First, we find that RAD52 binds extensively and tightly to RPA-ssDNA, but much of this bound protein is displaced upon addition of RAD51. There is a much more limited association of RAD52 with the presynaptic complex after RAD51 binding. The in vivo fluorescence recovery after photo-bleaching measurements were made under conditions where RAD51 was already present and so may not report on RAD52 association with RPA-ssDNA without RAD51 present (69). Second, our observations raise the question of what might be the relevant concentrations of RPA, RAD52 and RAD51 in vivo. Unfortunately, it is notoriously difficult to measure in vivo protein concentrations, although recent estimates suggest that human cells have on the order of 100 nm RAD51 and 1 μm RPA (77). We are unaware of any estimates for RAD52 concentrations in mammalian cells, although Rad52 concentrations in yeast have been reported from 1 to 20 nm (77, 78). However, we urge caution in interpreting these values with respect to either our data or the concentrations of these protein that are necessary for DNA repair in vivo, because the relevant in vivo concentrations must take into account the fact that these repair factors form subnuclear foci in response to DNA damage (79). Any interpretation of relevant in vivo protein concentrations requires an understanding of the local concentrations within repair foci, and to our knowledge such information is not yet available (79). Therefore, it remains a significant future challenge to define the concentrations of proteins within repair foci. Third, at this stage our experiments include only a very limited subset of the proteins known to associate with the presynaptic complex. It is possible, if not likely, that the presence of additional protein factors will have an impact upon the binding and turnover characteristics of RAD52, as well as any other proteins associated with the presynaptic complex. Similarly, RAD52 is regulated by both phosphorylation and sumoylation (80–85), and these post-translational modifications may have an impact upon its behavior in our assays. Future ssDNA curtain studies incorporating more recombination protein components and protein modifications may help continue to shed new light on the behaviors and properties of the human presynaptic complex.
Experimental procedures
Protein expression and purification
RPA was purified as previously described (72). His6-tagged human RAD52 and His6-tagged GFP-tagged human RAD52 were expressed in Escherichia coli Rosetta cells. For brevity, we refer to these His6-tagged proteins as RAD52 and GFP-RAD52 throughout. A single colony was inoculated into 1 liter of LB containing 50 μg/ml carbenicillin and induced at 0.8 A600 using 0.5 mm isopropyl β-d-thiogalactopyranoside. The cells were grown overnight at 16 °C and then harvested by centrifugation at 4000 × g for 25 min at 4 °C. The cell pellet was resuspended in lysis buffer (500 mm NaCl, 20 mm Tris, pH 7.5, 2 mm β-mercaptoethanol, 5 mm imidazole, 10% glycerol, and 0.5 mm PMSF) and then lysed by sonication. The lysate was clarified by centrifugation at 25,000 rpm for 30 min at 2 °C, and ammonium sulfate was added to achieve 30% saturation. A stir bar was placed in the beaker, and the mixture was stirred for 30 min at 4 °C. The solution was spun at 9000 rpm for 15 min at 4 °C, and the pellet was dissolved in NTA buffer (40 mm KPO4, pH 7.5, 600 mm NaCl, 20 mm imidazole, 4 mm β-mercaptoethanol) using a pipette. The resuspension was filtered through a Whatman filter to remove any undissolved precipitate. The protein solution is then bound to Ni-NTA resin (Qiagen) equilibrated in NTA buffer for 1 h in batch at 4 °C on a rotator. The Ni-NTA resin was then washed with NTA buffer with 30 mm imidazole and eluted using NTA buffer with 300 mm imidazole. The pooled fractions were dialyzed into R buffer (50 mm KCl, 20 mm Tris-HCl, pH 7.4, 1 mm DTT, 1 mm EDTA, 10% glycerol) and bound onto an ssDNA column (lyophilized powder, D8273; Sigma-Aldrich). The column was washed with R buffer with 110 mm KCl and eluted with a 0.05–3 m KCl gradient. The pooled fractions were dialyzed into R150 buffer (150 mm KCl, 20 mm Tris-HCl, pH 7.4, 1 mm DTT, 0.5 mm EDTA, 50% glycerol) overnight at 4 °C using 10,000 molecular weight cutoff SnakeSkinTM dialysis tubing (Thermo Fisher Scientific). The protein concentrations were measured using 280-nm absorbance.
Single-strand DNA annealing assays
These assays used two complementary oligonucleotides A (5′-AAA TAG ACA GAT CGC TGA GAT AGG TGC CTC ACT GAT TAA GCA TTG GTA ACT GTC AGA CCA AGT TTA CTC ATA TAT ACT TTA GAT TGA TTT-3′) and B (5′-AAA TCA ATC TAA AGT ATA TAT GAG TAA ACT TGG TCT GAC AGT TAC CAA TGC TTA ATC AGT GAG GCA CCT ATC TCA GCG ATC TGT CTA TTT-3′). Oligonucleotide A (90-mer, 3.6 μm nucleotide) and P32-labeled oligonucleotide B (90-mer, 3.6 μm nucleotide) were incubated in separate tubes with 0.6 μm RPA, as indicated, in 20 μl of reaction buffer (35 mm Tris-HCl, pH 7.5, 1 mm DTT, 2 mm MgCl2, 100 μg/ml BSA, and 50 mm KCl) at 30 °C for 5 min. RAD52 (0.7 μm) or GFP-RAD52 (0.7 μm) was added to the mixture containing oligonucleotide A, and the two reaction mixes were then combined and incubated at 30 °C. After the indicated incubation times, 9 μl of the reaction mixture was transferred to a tube containing 9 μl of 36 μm oligonucleotide B, 1% SDS, 1 μg/μl proteinase K, and then incubated for 5 min at 37 °C. The resulting DA species were resolved by 8% PAGE in 1× TAE buffer, and the percentage of product formation (dsDNA) was quantitated by phosphorimaging analysis.
Single-stranded DNA curtains
Flow cells were prepared as previously described (70, 71, 86). Biotinylated ssDNA was made by rolling circle replication using a circular M13 ssDNA template, a biotinylated primer, and phi29 DNA polymerase, as described (70). For the extension and visualization of the ssDNA curtain, hRPA-eGFP or hRPA-RFP (1 nm) was injected into the sample chamber at 1 ml/min in BSA buffer (40 mm Tris, pH 8.0, 2 mm MgCl2, 1 mm DTT, and 0.2 mg/ml BSA). After 2 min, 150 μl of 7 m urea was flushed through the sample chamber to help remove any remaining ssDNA secondary structure, and flow with 1 nm hRPA was continued for another 15 min to ensure that both ends of the ssDNA were anchored to the flow cell surface. This step is also performed for single-tethered ssDNA curtains.
TIRF microscopy and data analysis
Nikon Eclipse Ti-E with a Perfect Focus System microscope with Nikon CFI PLAN APO 60× objective was used for imaging. The signals were captured by two Andor iXon X3 EMCCD cameras. Temperatures for the experiments were maintained through an objective heater and a custom-made slide heater. The data from the cameras were acquired and saved on the computer with the NIS-Elements software and then converted to tiff stacks for analysis using Fiji (ImageJ 1.48b; Wayne Rasband, National Institutes of Health).
RAD52-binding experiments
All RAD52-binding experiments were performed at 37 °C using HR buffer (30 mm Tris-acetate, pH 7.5, 20 mm magnesium acetate, 50 mm KCl, 1 mm DTT, 0.2 mg/ml BSA). RAD52 binding measurements were made on preassembled RPA (wild-type or RFP)-ssDNA complexes in a double-tethered curtain unless otherwise indicated. Free RPA protein was washed away from the flow cell before binding of RAD52. RAD52-binding sites and intensity distributions were determined based on data from ssDNA-RPA-RFP curtains after the injection of 50 pm of RAD52-eGFP. The positions of RAD52 complexes were first determined by visual inspection. Intensity distribution is based on the raw pixel intensity measured at the positions of RAD52 complexes. RAD52 lifetime measurements were made with ssDNA-RPA-RFP curtains after the injection of 50 pm of RAD52-eGFP. After all free protein was washed away from the flow cell, buffer flow was stopped, and the complexes were monitored at 1- or 20-s intervals for a total of 12 min or 2 h, respectively, with 100-ms exposures. RPA chase experiments were conducted as previously described (65, 71, 72). After assembly of ssDNA-RPA-RFP, buffer flow without RPA was continued to wash out any residual fluorescent RPA molecules. RPA chase was done with an injection of 100 nm of wild-type RPA. In the case of RAD52, 50 pm of RAD52-eGFP was injected after the RPF-RFP was washed away from the flow cell. Free RAD52 was subsequently washed away before the wild-type RPA chase. RPA turnover was assessed through the quantitation of the RPA-RFP signal over time. Co-localization analysis was done by analyzing the normalized signal intensities for the green and the red channels individually (representing RAD52 or RPA signals) for each pixel along the ssDNA molecules. The co-localization of RAD52 and RPA after the wild-type RPA chase was evaluated using Pearson correlation analysis.
Presynaptic complex assembly
RAD51 filaments were formed at 37 °C using HRA buffer (30 mm Tris, pH 7.4, 1 mm MgCl2, 5 mm CaCl2, 100 mm KCl, 0.2 mg/ml BSA, 1 mm DTT, and 2 mm ATP). After forming ssDNA-RPA complexes, free RPA proteins were washed from the flow cell with HRA buffer flow for at least 2 min at 1 ml/min. RAD51 (1 μm) was injected through a 50-μl sample loop for double-tethered experiments (at 1 ml/min) or through a 1-ml loop for single tethered experiments (at 0.2 ml/min). The images were acquired at 2-s intervals. Co-localization analysis was done as described above by analyzing the normalized signal intensity for the green and red channels individually for each pixel along the ssDNA molecules. The correlation was evaluated using Pearson correlation analysis.
RAD52 and RPA binding to the presynaptic complex
Presynaptic complexes were formed as described above. After formation of the presynaptic complex, 2 nm GFP-RAD52 and 1 nm RPA-RFP were injected through the same 1-ml loop at 0.2 ml/min into the flow cell. Binding of RAD52 and or RPA were directly visualized through an increase in the signal intensity of the red and green channels. RPA and RAD52 co-localization analysis was done as described above. Where indicated, filament was disassembled through the removal of calcium and ATP from the HRA buffer, as previously described (72). The length of the single tethered DNA molecules during the disassembly of the presynaptic filament was determined by analyzing the smoothed kymograms and defining the local minimum in the first derivative of the signal intensities nearest to the bottom of the kymographs (where the 3′ ssDNA ends were located). The first derivative is expected to have a minimum peak when the signal decreases abruptly from the 3′ ends of DNA to the background; the pixel positions for these minima are used to approximate the lengths of the DNA molecules for each frame (time point) in the kymogram. These identified edges were then manually verified by eye.
Author contributions
C. J. M. and E. C. G. designed research; C. J. M. and Y. K. performed research; C. J. M., Y. K., P. S., and E. C. G. wrote the paper with input from all co-authors.
Acknowledgments
We thank Kyle Kaniecki, Luisina de Tullio, Brooks Crickard, and Justin Steinfeld for comments on the manuscript.
This work was supported by National Institutes of Health Grants R35GM118026 (to E. C. G.) and R01ES007061 and P01CA92584 (to P. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HR
- homologous recombination
- DSB
- double-stranded break
- ssDNA
- single-stranded DNA
- RPA
- replication protein A
- Ni-NTA
- nickel-nitrilotriacetic acid.
References
- 1. Cromie G. A., Connelly J. C., and Leach D. R. (2001) Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell 8, 1163–1174 [DOI] [PubMed] [Google Scholar]
- 2. Symington L. S., Rothstein R., and Lisby M. (2014) Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198, 795–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. San Filippo J., Sung P., and Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 4. Pâques F., and Haber J. E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jasin M., and Rothstein R. (2013) Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect Biol. 5, a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heyer W. D. (2015) Regulation of recombination and genomic maintenance. Cold Spring Harb. Perspect Biol. 7, a016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heyer W. D., Ehmsen K. T., and Liu J. (2010) Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mehta A., and Haber J. E. (2014) Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 6, a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cox M. M., Goodman M. F., Kreuzer K. N., Sherratt D. J., Sandler S. J., and Marians K. J. (2000) The importance of repairing stalled replication forks. Nature 404, 37–41 [DOI] [PubMed] [Google Scholar]
- 10. Carr A. M., and Lambert S. (2013) Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J. Mol. Biol. 425, 4733–4744 [DOI] [PubMed] [Google Scholar]
- 11. Brown M. S., and Bishop D. K. (2014) DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb. Perspect. Biol. 7, a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hunter N. (2015) Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lam I., and Keeney S. (2014) Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 7, a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Llorente B., Smith C. E., and Symington L. S. (2008) Break-induced replication: what is it and what is it for? Cell Cycle 7, 859–864 [DOI] [PubMed] [Google Scholar]
- 15. Anand R. P., Lovett S. T., and Haber J. E. (2013) Break-induced DNA replication. Cold Spring Harb. Perspect. Biol. 5, a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malkova A., and Ira G. (2013) Break-induced replication: functions and molecular mechanism. Curr. Opin. Genet. Dev. 23, 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aguilera A., and García-Muse T. (2013) Causes of genome instability. Annu. Rev. Genet. 47, 1–32 [DOI] [PubMed] [Google Scholar]
- 18. Prakash R., Zhang Y., Feng W., and Jasin M. (2015) Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 7, a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kass E. M., Moynahan M. E., and Jasin M. (2016) When genome maintenance goes badly awry. Mol. Cell 62, 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bianco P. R., Tracy R. B., and Kowalczykowski S. C. (1998) DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 3, D570–D603 [DOI] [PubMed] [Google Scholar]
- 21. Kowalczykowski S. C. (2015) An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7, a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morrical S. W. (2015) DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb. Perspect. Biol. 7, a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cejka P. (2015) DNA end resection: nucleases team up with the right partners to initiate homologous recombination. J. Biol. Chem. 290, 22931–22938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daley J. M., Niu H., Miller A. S., and Sung P. (2015) Biochemical mechanism of DSB end resection and its regulation. DNA Repair 32, 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Symington L. S. (2014) End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb. Perspect. Biol. 6, a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen R., and Wold M. S. (2014) Replication protein A: single-stranded DNA's first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. BioEssays 36, 1156–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wold M. S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 [DOI] [PubMed] [Google Scholar]
- 28. Sung P., and Klein H. (2006) Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7, 739–750 [DOI] [PubMed] [Google Scholar]
- 29. Bendixen C., Sunjevaric I., Bauchwitz R., and Rothstein R. (1994) Identification of a mouse homologue of the Saccharomyces cerevisiae recombination and repair gene, RAD52. Genomics 23, 300–303 [DOI] [PubMed] [Google Scholar]
- 30. Muris D. F., Bezzubova O., Buerstedde J. M., Vreeken K., Balajee A. S., Osgood C. J., Troelstra C., Hoeijmakers J. H., Ostermann K., and Schmidt H. (1994) Cloning of human and mouse genes homologous to RAD52, a yeast gene involved in DNA repair and recombination. Mutat. Res. 315, 295–305 [DOI] [PubMed] [Google Scholar]
- 31. Shinohara A., Shinohara M., Ohta T., Matsuda S., and Ogawa T. (1998) Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells 3, 145–156 [DOI] [PubMed] [Google Scholar]
- 32. New J. H., Sugiyama T., Zaitseva E., and Kowalczykowski S. C. (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391, 407–410 [DOI] [PubMed] [Google Scholar]
- 33. Sung P. (1997) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272, 28194–28197 [DOI] [PubMed] [Google Scholar]
- 34. Navadgi V. M., Dutta A., and Rao B. J. (2003) Human Rad52 facilitates a three-stranded pairing that follows no strand exchange: a novel pairing function of the protein. Biochemistry 42, 15237–15251 [DOI] [PubMed] [Google Scholar]
- 35. Sugiyama T., Kantake N., Wu Y., and Kowalczykowski S. C. (2006) Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 25, 5539–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mortensen U. H., Bendixen C., Sunjevaric I., and Rothstein R. (1996) DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. U.S.A. 93, 10729–10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sugiyama T., New J. H., and Kowalczykowski S. C. (1998) DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 95, 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lao J. P., Oh S. D., Shinohara M., Shinohara A., and Hunter N. (2008) Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol. Cell 29, 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McIlwraith M. J., and West S. C. (2008) DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol. Cell 29, 510–516 [DOI] [PubMed] [Google Scholar]
- 40. Game J. C., and Mortimer R. K. (1974) A genetic study of x-ray sensitive mutants in yeast. Mutat. Res. 24, 281–292 [DOI] [PubMed] [Google Scholar]
- 41. Symington L. S. (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66, 630–670, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hanamshet K., Mazina O. M., and Mazin A. V. (2016) Reappearance from obscurity: mammalian Rad52 in homologous recombination. Genes (Basel) 7, E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu J., and Heyer W. D. (2011) Who's who in human recombination: BRCA2 and RAD52. Proc. Natl. Acad. Sci. U.S.A. 108, 441–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singleton M. R., Wentzell L. M., Liu Y., West S. C., and Wigley D. B. (2002) Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl. Acad. Sci. U.S.A. 99, 13492–13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kagawa W., Kurumizaka H., Ishitani R., Fukai S., Nureki O., Shibata T., and Yokoyama S. (2002) Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol. Cell 10, 359–371 [DOI] [PubMed] [Google Scholar]
- 46. Stasiak A. Z., Larquet E., Stasiak A., Müller S., Engel A., Van Dyck E., West S. C., and Egelman E. H. (2000) The human Rad52 protein exists as a heptameric ring. Curr. Biol. 10, 337–340 [DOI] [PubMed] [Google Scholar]
- 47. Van Dyck E., Hajibagheri N. M., Stasiak A., and West S. C. (1998) Visualisation of human rad52 protein and its complexes with hRad51 and DNA. J. Mol. Biol. 284, 1027–1038 [DOI] [PubMed] [Google Scholar]
- 48. Reddy G., Golub E. I., and Radding C. M. (1997) Human Rad52 protein promotes single-strand DNA annealing followed by branch migration. Mutat. Res. 377, 53–59 [DOI] [PubMed] [Google Scholar]
- 49. Rothenberg E., Grimme J. M., Spies M., and Ha T. (2008) Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc. Natl. Acad. Sci. U.S.A. 105, 20274–20279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grimme J. M., Honda M., Wright R., Okuno Y., Rothenberg E., Mazin A. V., Ha T., and Spies M. (2010) Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 38, 2917–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brouwer I., Zhang H., Candelli A., Normanno D., Peterman E. J., Wuite G. J., and Modesti M. (2017) Human RAD52 captures and holds DNA strands, increases DNA flexibility, and prevents melting of duplex DNA: implications for DNA recombination. Cell Reports 18, 2845–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rijkers T., Van Den Ouweland J., Morolli B., Rolink A. G., Baarends W. M., Van Sloun P. P., Lohman P. H., and Pastink A. (1998) Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 18, 6423–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamaguchi-Iwai Y., Sonoda E., Buerstedde J. M., Bezzubova O., Morrison C., Takata M., Shinohara A., and Takeda S. (1998) Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol. 18, 6430–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Feng Z., Scott S. P., Bussen W., Sharma G. G., Guo G., Pandita T. K., and Powell S. N. (2011) Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl. Acad. Sci. U.S.A. 108, 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chun J., Buechelmaier E. S., and Powell S. N. (2013) Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol. Cell. Biol. 33, 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lok B. H., Carley A. C., Tchang B., and Powell S. N. (2013) RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene 32, 3552–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ciccia A., and Symington L. S. (2016) Stressing out about RAD52. Mol. Cell 64, 1017–1019 [DOI] [PubMed] [Google Scholar]
- 58. Bhowmick R., Minocherhomji S., and Hickson I. D. (2016) RAD52 facilitates mitotic DNA synthesis following replication stress. Mol. Cell 64, 1117–1126 [DOI] [PubMed] [Google Scholar]
- 59. Sotiriou S. K., Kamileri I., Lugli N., Evangelou K., Da-Ré C., Huber F., Padayachy L., Tardy S., Nicati N. L., Barriot S., Ochs F., Lukas C., Lukas J., Gorgoulis V. G., Scapozza L., and Halazonetis T. D. (2016) Mammalian RAD52 functions in break-induced replication repair of collapsed DNA replication forks. Mol. Cell 64, 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lok B. H., and Powell S. N. (2012) Molecular pathways: understanding the role of Rad52 in homologous recombination for therapeutic advancement. Clin. Cancer Res. 18, 6400–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huang F., Goyal N., Sullivan K., Hanamshet K., Patel M., Mazina O. M., Wang C. X., An W. F., Spoonamore J., Metkar S., Emmitte K. A., Cocklin S., Skorski T., and Mazin A. V. (2016) Targeting BRCA1- and BRCA2-deficient cells with RAD52 small molecule inhibitors. Nucleic Acids Res. 44, 4189–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chandramouly G., McDevitt S., Sullivan K., Kent T., Luz A., Glickman J. F., Andrake M., Skorski T., and Pomerantz R. T. (2015) Small-molecule disruption of RAD52 rings as a mechanism for precision medicine in BRCA-deficient cancers. Chem. Biol. 22, 1491–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sullivan K., Cramer-Morales K., McElroy D. L., Ostrov D. A., Haas K., Childers W., Hromas R., and Skorski T. (2016) Identification of a small molecule inhibitor of RAD52 by structure-based selection. PLoS One 11, e0147230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cramer-Morales K., Nieborowska-Skorska M., Scheibner K., Padget M., Irvine D. A., Sliwinski T., Haas K., Lee J., Geng H., Roy D., Slupianek A., Rassool F. V., Wasik M. A., Childers W., Copland M., Müschen M., Civin C. I., and Skorski T. (2013) Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood 122, 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gibb B., Ye L. F., Kwon Y., Niu H., Sung P., and Greene E. C. (2014) Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nat. Struct. Mol. Biol 21, 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lisby M., Rothstein R., and Mortensen U. H. (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. U.S.A. 98, 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu Y., Li M., Lee E. Y., and Maizels N. (1999) Localization and dynamic relocalization of mammalian Rad52 during the cell cycle and in response to DNA damage. Curr. Biol. 9, 975–978 [DOI] [PubMed] [Google Scholar]
- 68. Liu Y., and Maizels N. (2000) Coordinated response of mammalian Rad51 and Rad52 to DNA damage. EMBO Reports 1, 85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Essers J., Houtsmuller A. B., van Veelen L., Paulusma C., Nigg A. L., Pastink A., Vermeulen W., Hoeijmakers J. H., and Kanaar R. (2002) Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21, 2030–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gibb B., Silverstein T. D., Finkelstein I. J., and Greene E. C. (2012) Single-stranded DNA curtains for real-time single-molecule visualization of protein-nucleic acid interactions. Anal. Chem. 84, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gibb B., Ye L. F., Gergoudis S. C., Kwon Y., Niu H., Sung P., and Greene E. C. (2014) Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One 9, e87922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ma C. J., Gibb B., Kwon Y., Sung P., and Greene E. C. (2017) Protein dynamics of human RPA and RAD51 on ssDNA during assembly and disassembly of the RAD51 filament. Nucleic Acids Res. 45, 749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ma C. J., Steinfeld J. B., and Greene E. C. (2017) Single-stranded DNA curtains for studying homologous recombination. Methods Enzymol. 582, 193–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Graham J. S., Johnson R. C., and Marko J. F. (2011) Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA. Nucleic Acids Res. 39, 2249–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bugreev D. V., and Mazin A. V. (2004) Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. U.S.A. 101, 9988–9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mazina O. M., and Mazin A. V. (2004) Human Rad54 protein stimulates DNA strand exchange activity of hRad51 protein in the presence of Ca2+. J. Biol. Chem. 279, 52042–52051 [DOI] [PubMed] [Google Scholar]
- 77. Kulak N. A., Pichler G., Paron I., Nagaraj N., and Mann M. (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 [DOI] [PubMed] [Google Scholar]
- 78. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., and Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 79. Lisby M., and Rothstein R. (2015) Cell biology of mitotic recombination. Cold Spring Harb. Perspect. Biol. 7, a016535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kitao H., and Yuan Z. M. (2002) Regulation of ionizing radiation-induced Rad52 nuclear foci formation by c-Abl-mediated phosphorylation. J. Biol. Chem. 277, 48944–48948 [DOI] [PubMed] [Google Scholar]
- 81. Honda M., Okuno Y., Yoo J., Ha T., and Spies M. (2011) Tyrosine phosphorylation enhances RAD52-mediated annealing by modulating its DNA binding. EMBO J. 30, 3368–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sacher M., Pfander B., Hoege C., and Jentsch S. (2006) Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 8, 1284–1290 [DOI] [PubMed] [Google Scholar]
- 83. Ohuchi T., Seki M., Branzei D., Maeda D., Ui A., Ogiwara H., Tada S., and Enomoto T. (2008) Rad52 sumoylation and its involvement in the efficient induction of homologous recombination. DNA Repair 7, 879–889 [DOI] [PubMed] [Google Scholar]
- 84. Bergink S., Ammon T., Kern M., Schermelleh L., Leonhardt H., and Jentsch S. (2013) Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51-Rad52 interaction. Nat. Cell Biol. 15, 526–532 [DOI] [PubMed] [Google Scholar]
- 85. Torres-Rosell J., Sunjevaric I., De Piccoli G., Sacher M., Eckert-Boulet N., Reid R., Jentsch S., Rothstein R., Aragón L., and Lisby M. (2007) The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 9, 923–931 [DOI] [PubMed] [Google Scholar]
- 86. Krejci L., Altmannova V., Spirek M., and Zhao X. (2012) Homologous recombination and its regulation. Nucleic Acids Res. 40, 5795–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]