Abstract
There is significant clinical need for new antifungal agents to manage infections with pathogenic species such as Cryptococcus neoformans. Because the purine biosynthesis pathway is essential for many metabolic processes, such as synthesis of DNA and RNA and energy generation, it may represent a potential target for developing new antifungals. Within this pathway, the bifunctional enzyme adenylosuccinate (ADS) lyase plays a role in the formation of the key intermediates inosine monophosphate and AMP involved in the synthesis of ATP and GTP, prompting us to investigate ADS lyase in C. neoformans. Here, we report that ADE13 encodes ADS lyase in C. neoformans. We found that an ade13Δ mutant is an adenine auxotroph and is unable to successfully cause infections in a murine model of virulence. Plate assays revealed that production of a number of virulence factors essential for dissemination and survival of C. neoformans in a host environment was compromised even with the addition of exogenous adenine. Purified recombinant C. neoformans ADS lyase shows catalytic activity similar to its human counterpart, and its crystal structure, the first fungal ADS lyase structure determined, shows a high degree of structural similarity to that of human ADS lyase. Two potentially important amino acid differences are identified in the C. neoformans crystal structure, in particular a threonine residue that may serve as an additional point of binding for a fungal enzyme-specific inhibitor. Besides serving as an antimicrobial target, C. neoformans ADS lyase inhibitors may also serve as potential therapeutics for metabolic disease; rather than disrupt ADS lyase, compounds that improve the stability the enzyme may be used to treat ADS lyase deficiency disease.
Keywords: crystal structure, enzyme kinetics, fungi, nucleoside/nucleotide biosynthesis, pathogenesis, adenylosuccinate lyase, virulence
Introduction
The link between defects in primary metabolism and disease has been recognized for over a century. The physician Alfred Garrod (1) first associated abnormal metabolism with gout in 1848, and his son Archibald Garrod (2) later combined these observations with the concept of Mendelian inheritance to produce his seminal work, Inborn Errors of Metabolism, in 1909. Since that time, numerous other medical conditions caused by abnormalities in primary metabolism have been identified (3–6). These include defects in the production of the nitrogenous bases that are essential for DNA and RNA synthesis, energy metabolism, and signal transduction. At present, over 35 enzyme defects in humans related to purine or pyrimidine biosynthesis have been characterized, and of these at least 17 are associated with serious clinical consequences (7, 8).
One of the first such conditions to be identified was the rare autosomal recessive disease adenylosuccinate lyase deficiency (OMIM 103050) associated with the corresponding purine biosynthetic enzyme (EC 4.3.2.2) (9). Jaeken and van den Berghe (9) identified three patients with psychomotor delay and autism that had high levels of the purine biosynthesis intermediates adenylosuccinate (ADS)3 and succinylaminoimidazole carboxamide riboside (SAICAR) in their urine, plasma, and cerebrospinal fluid. These purines were identified in 1956 from Saccharomyces cerevisiae as the substrates of ADS lyase, which converts them into AMP and fumarate (10), and AICAR (aminoimidazole carboxamide riboside) and fumarate, respectively (Fig. 1, A and B) (11).
Figure 1.
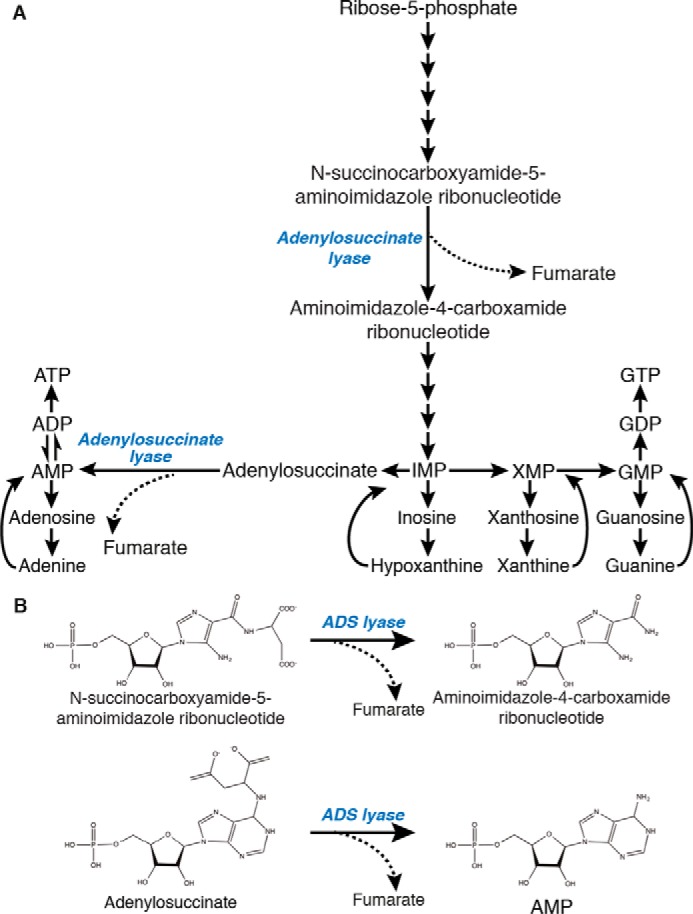
ADS lyase in the purine biosynthesis pathway. A, ADS lyase is responsible for the β-elimination of fumarate from the substrates SAICAR and ADS. B, β-elimination of fumarate from substrates SAICAR and adenylosuccinate by ADS lyase for the production of aminoimidazole-4-carboxamine ribonucleotide and AMP, respectively.
ADS lyase belongs to the fumarase C superfamily, a group of enzymes that perform the β-elimination of fumarate from their substrates (12–14). Structures of ADS lyase from nine species of bacteria, one species of archaea, and six species of eukaryotes (Homo sapiens, Schistosoma mansoni, Plasmodium vivax, Leishmania donovani, Trypanosoma brucei, and Caenorhabditis elegans (15, 16)) have been determined. All form the homotetramer arrangement characteristic of members of the fumarase C superfamily.
The four protomers in the ADS lyase tetramer are oriented such that three contribute to each of the four active sites. In the best-studied ADS lyase structure from humans (PDB code 2J91), 12 residues are associated with substrate binding (His-86, Arg-85, Thr-111, Gln-241, Arg-329, Leu-331, Ser-334, and Arg-338 from the fist protomer, Arg-20, Arg-303, and Lys-295 from the second, and Thr-158 from the third) (15). Once the substrate is docked, the flexible C3 loop of the second protomer is hypothesized to close on the active site, allowing the catalytic Ser-289 to come in contact with the substrate and remove its Cβ proton. The catalytic His-159 residue from the third protomer then protonates N-6 of the substrate, causing the C-N bond to break and release fumarate with the corresponding purine (15, 17).
To date, investigation of ADS lyase as a therapeutic target has been limited to Plasmodium falciparum, where it has been proposed as a potential focus for antimalarial agent development (18). Many parasitic protozoa such as P. falciparum lack key enzymes required for the de novo synthesis of the purine intermediate IMP and are dependent on salvaging purine precursors from the host to synthesize ATP and GTP. As ADS lyase is a post-IMP enzyme in the pathway, it has been retained in these species, and by extension, so has its pre-IMP ability to produce AICAR from SAICAR, although this does not play a role in P. falciparum metabolism, as this substrate is not naturally present in the parasite. SAICAR analogs could therefore serve as the basis for the development of novel antimalarial agents (18).
In contrast, the possible role of this enzyme as an antifungal target has not yet been investigated, nor has the structure in any member of the kingdom Fungi. Recent studies in the major fungal pathogens have highlighted the potential of purine biosynthetic enzymes as drug targets (19–23). For example, the encapsulated yeast Cryptococcus neoformans has been shown to require IMP dehydrogenase, GMP synthase, ADS synthase, and AIR carboxylase to successfully infect a mammalian model (19, 20, 23–25).
There is significant unmet clinical need for new antifungal agents that target species such as C. neoformans. Although globally distributed, meningoencephalitis caused by this basidiomycete is particularly prevalent in sub-Saharan Africa where incidence of HIV is high and mortality rates can reach 75% (26); a major contributor to this mortality is the toxicity, high cost, poor availability, and limited efficacy of antifungal agents employed in its treatment (27, 28). Even in developed countries, the typical combination therapy composed of amphotericin B, flucytosine, and fluconazole has not altered significantly in over two decades despite unacceptably high mortality, patient relapse, and antifungal resistance being observed (29–31).
Here we present an investigation of ADS lyase in C. neoformans. Deletion of the ADS lyase gene ADE13 shows it is essential for C. neoformans survival in a purine poor environment, such as in a murine model of infection. Analysis of enzyme kinetics and crystal structure determination of recombinantly purified ADS lyase protein has highlighted the high degree of similarity between this enzyme and its human ortholog, suggesting that unlike other purine biosynthesis enzymes studied thus far, C. neoformans ADS lyase may facilitate important insights into the function of the human enzyme in addition to serving as an antimicrobial target.
Results
Identification of the ADS lyase-encoding gene in C. neoformans
To characterize ADS lyase from C. neoformans, the corresponding gene was identified in the genome of type strain H99 using the S. cerevisiae ortholog Ade13 in a reciprocal best-hit BLAST analysis (32). A single hit was observed, indicating that as with other purine biosynthetic genes identified so far, the gene is present in single copy in this clinically important pathogen. Located on chromosome 8, the locus was designated as CNAG_03270 in the published H99 genome (33); subsequently employing CNAG_03270 as the query sequence in a BLAST search of the S. cerevisiae genome identified ADE13 as the only statistically significant hit in that species. As is standard practice in C. neoformans, the gene CNAG_03270 has therefore been named ADE13 after the S. cerevisiae ortholog, whose predicted product is 71.4% identical at the amino acid level. In comparison, C. neoformans Ade13 is 68.4% identical to ADS lyase from humans.
Ade13 is essential for adenine prototrophy in C. neoformans
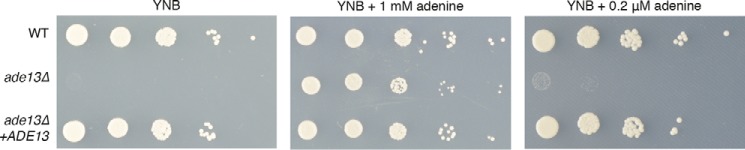
To verify the role of the ADE13 product in purine metabolism, we performed targeted gene deletion via biolistic transformation of C. neoformans type strain H99. As predicted for a mutant lacking ADS lyase activity, the ade13Δ mutant was only able to grow on yeast nitrogen base (YNB) when supplemented with sufficient exogenous adenine; as with S. cerevisiae, the ade13Δ mutant was an adenine auxotroph (Fig. 2).
Figure 2.
C. neoformans ADE13 role in ATP biosynthesis. Growth of 10-fold serial dilutions of wild-type (WT), ade13Δ and ade13Δ+ADE13 strains of C. neoformans on YNB medium (A); YNB medium supplemented with 1 mm adenine (B); and YNB medium supplemented with 0.2 μm adenine (C).
However, in contrast to in vitro growth assays supplemented with 1 mm adenine, the human central nervous system is extremely purine poor. To verify that the CNS would not provide sufficient salvageable adenine to support growth of C. neoformans cells lacking the biochemical activity encoded by ADE13, we repeated our phenotypic tests using 0.2 μm adenine, the concentration found in human cerebrospinal fluid (34, 35). Growth was not restored, supporting the hypothesis that loss of Ade13 function would abrogate growth during the infection process (Fig. 2). In keeping with the observed adenine auxotrophy originating from loss of the ADE13 gene, reintroduction of the wild-type allele into the well characterized Safe Haven on chromosome 1 (24) to generate the strain ade13Δ+ADE13 restored growth on all media tested (Fig. 2). Overall, these data indicate that ADE13 encodes ADS lyase.
Loss of Ade13 affects production of C. neoformans virulence factors
The ability of C. neoformans to infect a host requires a range of virulence factors that protect against host defenses, scavenge nutrients, facilitate dissemination, and enable infiltration of a variety of tissue types. We investigated the in vitro production of several of these virulence factors using test media supplemented with 1 mm adenine to observe the effects of the ade13Δ mutant independently from adenine auxotrophy-associated growth defects.
The ability to produce proteases is key to enabling C. neoformans to disseminate into host tissues during infection and to cross the blood-brain barrier (36). When grown on adenine-supplemented BSA media, the production of proteases by the ade13Δ mutant was compromised at both 30 and 37 °C (Fig. 3A). The production of the pigment melanin in C. neoformans protects from the oxidants produced by host effector cells (37). When grown on adenine-supplemented l-3,4-dihydroxyphenylalanine (l-DOPA) media, the ade13Δ mutant exhibited reduced melanin production at 37 °C (Fig. 3B). A differentiating feature of Cryptococcus from other fungal pathogens is its polysaccharide capsule that serves as a protective barrier against the immune system. Following growth in adenine-supplemented RPMI 1640 media, India ink staining revealed diminished capsule production by the ade13Δ mutant at both 30 and 37 °C (Figs. 3C and supplemental Fig. S1).
Figure 3.
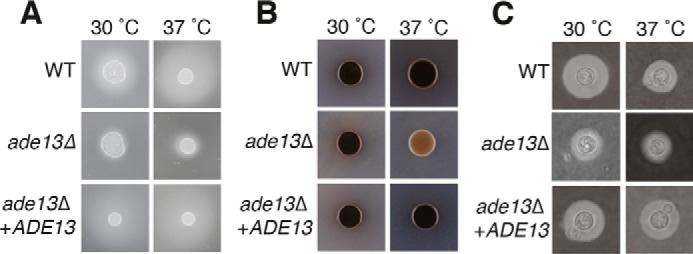
Loss of ADE13 influences the production of C. neoformans virulence factors. A, protease production was determined on YNB medium with amino acids and 0.1% bovine serum albumin plates, with strains at 30 and 37 °C for 48 h. The presence of proteases is observed by the halo ring around the colony. B, melanin production was determined on l-DOPA medium with strains incubated at 30 and 37 °C for 48 h. Pigmentation is observed by darkening of the colony. C, C. neoformans strains were incubated in RPMI 1640 medium, 10% fetal bovine serum, and 1 mm adenine at 30 and 37 °C. At 30 h, cells were stained with India ink. The capsule is observed by the exclusion of ink particles.
Overall, even with the addition of exogenous adenine, key adaptive mechanisms of C. neoformans required for successful infection were severely compromised in the ade13Δ mutant, and these were restored upon reintroduction of the wild-type allele by inserting it at the Safe Haven site (24).
Ade13 is crucial for virulence in a murine inhalation model
Given the defect in virulence-associated phenotypes and requirement for adenine concentrations that far exceed those present in the CNS, it was anticipated that the ade13Δ mutant would be unable to establish a wild-type infection in a mammal, such as in the well established C. neoformans murine inhalation infection model (38). Although all mice infected with wild-type C. neoformans or complemented ade13Δ+ADE13 strains succumbed to infection within 25 days, the mice infected with the ade13Δ mutant survived and continued to gain weight. The lack of symptoms associated with a C. neoformans infection continued until the end point of the experiment (45 days; Fig. 4A). In contrast to mice infected with the wild-type and complemented strains, the organ fungal burden from sacrificed animals infected with the ade13Δ mutant showed that the infection had been cleared (Fig. 4, B and C).
Figure 4.
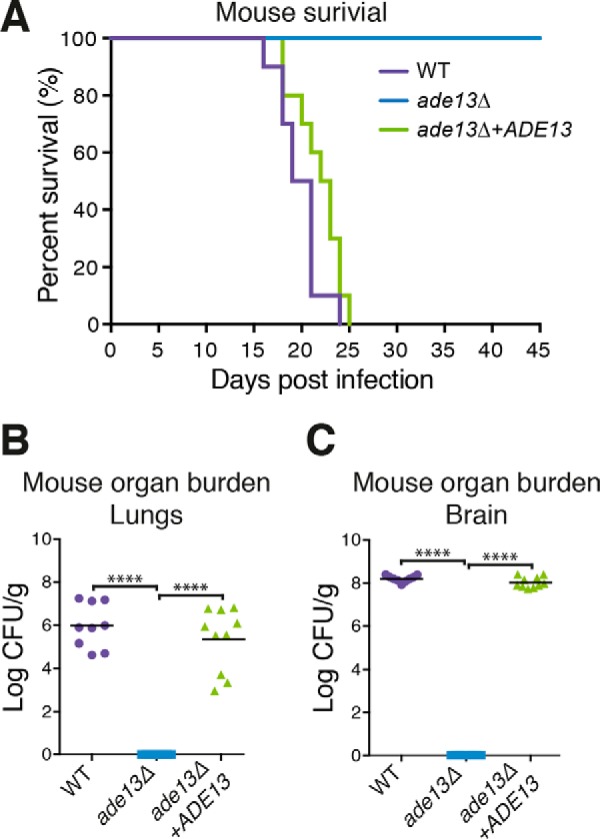
Virulence of the ade13Δ mutant in mice. A, virulence of the ade13Δ mutant in a murine model of infection. 6-Week-old female BALB/c mice were infected with WT, ade13Δ, or ade13Δ+ADE13 C. neoformans strains (n = 10) and survival was monitored over 45 days. Posthumous organ burden was calculated in colony forming units (CFU)/g of brain (B) and lungs (C). ****, p < 0.0001.
Comparison of C. neoformans ADS lyase catalytic activity to the enzyme from other species
Given that our mutant experiments revealed that the product of the ADE13 gene is essential for successful infection in a mammalian model, and by extension a potential drug target, we next investigated its enzymatic activity. His-tagged C. neoformans Ade13 was expressed in Escherichia coli and purified via immobilized metal affinity chromatography and size exclusion chromatography (SEC) for use in steady-state kinetic assays (supplemental Fig. S2). ADS lyase activity assays were optimized as part of the investigation, and performed at the optimal temperature of 37 °C and pH 8.
The β-elimination of fumarate from the substrate adenylosuccinate was measured and exhibited Michaelis-Menten kinetics (Table 1) with a Km of 22.2 ± 2.9 μm and a kcat of 25.2 s−1. Kinetic data are also available for ADS lyase from a number of other species including humans (39). Here, the elimination of fumarate from adenylosuccinate had a Km of 1.8 ± 0.3 μm and kcat of 97.0 s−1. These data are also consistent with the kinetic parameters of recombinant ADS lyase from the parasites L. donovani and P. falciparum, which have been reported to have Km values of 24.0 and 32.0 ± 1.7 μm, and kcat values of 28.0 and 7.5 s−1, respectively (18, 40). Although assays for different species used different temperatures, all show remarkably similar catalytic profiles (18, 40).
Table 1.
Comparison of kinetic parameters of ADS lyase from different organisms
Assay conditions were: C. neoformans (40 mm PPB, pH 8, 37 °C), S. cerevisiae (50 mm sodium phosphate buffer, pH 7.0, 35 °C), E. coli (50 mm HEPES buffer, pH 8.5, 25 °C), M. tuberculosis and M. smegmatis (succinic acid, sodium dihydrogen phosphate and glycine in ratio 2:7:7, pH 7.6, 37 °C), P. falciparum (50 mm PPB, pH 7.4, 25 °C), L. donovani (20 mm HEPES-KOH, pH 7, 25 °C), B. subtilis (50 mm HEPES, pH 7, 25 °C), H. sapiens (150 mm NaCl, pH 7, 25 °C). Values are shown ± S.E.
| Species | Kcat | Km | Kcat/Km | References |
|---|---|---|---|---|
| s−1 | μm | μm−1s−1 | ||
| C. neoformans | 25.2 | 22.2 ± 2.9 | 1.1 | This work |
| S. cerevisiae | NDa | 12 | ND | 10 |
| E. coli | ND | ND | 16.5 | 17 |
| M. tuberculosis | 0.1 ± 0.0 | 204.2 ± 48.2 | 0.0005 | 76 |
| M. smegmatis | 0.7 ± 0.0 | 43.7 ± 2.6 | 0.02 | 76 |
| P. falciparum | 7.5 ± 0.7 | 32.0 ± 1.7 | 0.23 | 18 |
| L. donovani | 28.0 | 24.0 | ND | 40 |
| B. subtilis | 1.3 ± 0.2 | 3.5 ± 0.4 | 3.8 × 105 | 77 |
| H. sapiens | 97.0 ± 5.2 | 1.8 ± 0.3 | 53.9 | 39 |
a ND denotes no data.
Crystal structure of C. neoformans ADS lyase
Based on SEC coupled with MALLS, C. neoformans ADS lyase is a tetramer (220 kDa) in solution (supplemental Fig. S2); this result is consistent with the oligomeric state of the human and E. coli enzymes, which were also reported to be tetramers (15, 17, 41).
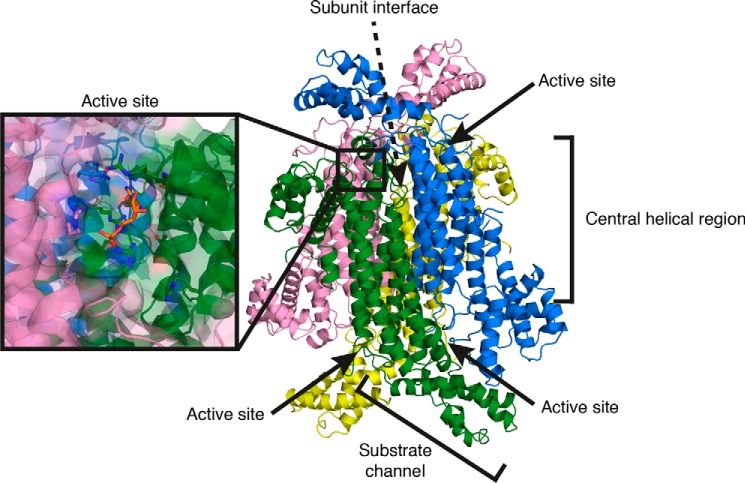
To compare the C. neoformans enzyme with its human counterpart, the crystal structure of unliganded C. neoformans ADS lyase was determined (PDB code 5V4L). Consistent with the MALLS result and the structures from other species, C. neoformans ADS lyase is a tetramer with four active sites, each containing residues contributed by three of the protomers. Each protomer is 479 residues in length, forming 13 α-helices and two β-strands, resulting in an S-shape overall (Fig. 5). The tetramer assembly consists of adjacent subunits arranged antiparallel to each other, with two active sites on one side and two on the other side of the complex.
Figure 5.
Crystal structure of C. neoformans ADS lyase. Crystal structure at 2.1-Å resolution, showing the tetramer arrangement of ADS lyase. The inset shows the active site location at protomer interfaces.
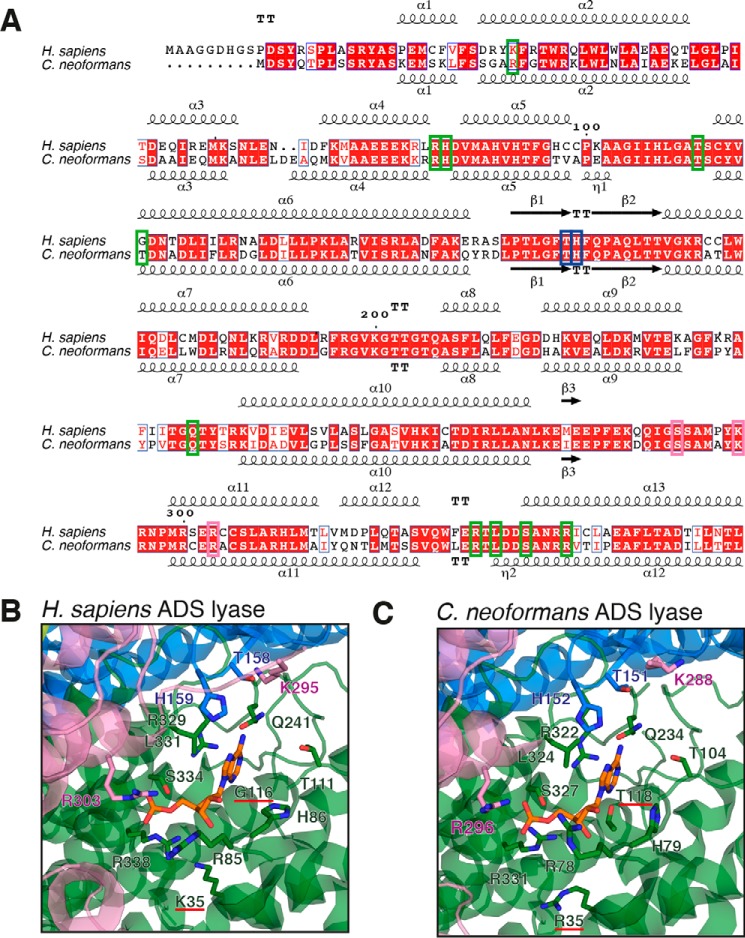
Pairwise structural alignment of the C. neoformans and human ADS lyase structures returns a close match, with an overall root mean square deviation value of 1.90 Å, similar to that of the Mycobacterium smegmatis enzyme (PDB 4NLE), which has an root mean square deviation value of 1.84 Å compared with the human ADS lyase structure (PDB 4FFX). The 12 residues associated with substrate binding in human ADS lyase are identical in C. neoformans. Further analysis of the active site cavity reveals two changes: human Gly-116 and Lys-35 correspond to Thr-118 and Arg-35, respectively, in C. neoformans (Fig. 6, B and C). Although lysine to arginine is a conservative change, the orientation of the arginine in C. neoformans expands the binding pocket by an additional 2 Å, which may allow for a compound to favor C. neoformans ADS lyase over the human enzyme. The difference between a glycine and a threonine in the active site results in surface differences and can be exploited through the design of specific interactions of the inhibitory compound with the threonine side chain in the fungal enzyme. A number of antifungal compounds, including fluconazole, exploit tighter binding to the active site of the fungal protein, despite limited differences to the human counterpart (42, 43). These differences could potentially be exploited in the rational design of fungal-specific ADS lyase inhibitors. Several substitutions that lead to ADS lyase deficiency disease are present outside the substrate pocket, suggesting other differences between the fungal and human enzymes may affect the active site indirectly; studies with substrate analogs will enable a deeper understanding of the mode of action of ADS lyase and consequently identify further strategies to achieve antifungal specificity.
Figure 6.
Active site comparison of human and C. neoformans ADS lyases. A, sequence alignment of ADS lyases from human and C. neoformans. Residues corresponding to the active site are boxed represented by colors corresponding to subunit A (green), B (blue), and C (pink). B and C, active-site residues from human (His-86, Arg-85, Thr-111, Gln-241, Arg-329, Leu-331, Arg-338, His-159, Arg-303, Lys-295, Thr-158) and Gly-116 and Lys-35 underlined in red (B) and C. neoformans (His-79, Arg-78, Thr-104, Gln-234, Arg-322, Leu-324, Arg-331, His-152, Arg-3296, Lys-288, Thr-151) and Thr-118 and Arg-35 underlined in red (C) ADS lyases, with side chains of active-site residues shown in stick representation. The colors correspond to the subunit in which they belong. The bound AMP (shown in “orange”) corresponds to the structure of human ADS lyase-ANP complex (PDB code 2J91) and was modeled into the C. neoformans ADS lyase active site for visualization purposes.
The tetramer interface contains a number of residues that are not conserved between the fungal and the human enzymes. Although protein–protein interfaces are more difficult to target with small-molecule compounds, compared with enzyme active sites, the differences in the interfaces may serve as an alternative foundation for antifungal drug design.
As well as serve as a potential antifungal drug target, C. neoformans may be a useful model organism in which to study the ADS lyase deficiency disease in humans; it is still unclear if the symptoms of this disease arise from the accumulation of SAICAR and/or ADS, or from the perturbation of ATP and GTP biosynthesis. Studies of the human enzyme have identified 36 mutations associated with ADS lyase deficiency disease in humans that can be broadly assigned to four regions (supplemental Table S2). C. neoformans ADS lyase could therefore be useful in the design of novel antifungals, or in finding new therapeutics that could assist in treatment of the human disease. By finding compounds that bind and stabilize ADS lyase in a genetically tractable model organism such as C. neoformans, a treatment for ADS lyase deficiency disease could be found that increases the activity of the defective enzyme, alleviating symptoms. This method of therapeutically stabilizing a protein has been previously shown in the development of treatments for amyotrophic lateral sclerosis (44).
In contrast to other purine biosynthetic enzymes studied in this fungal pathogen, which differ more from their human counterparts in activity and structure, C. neoformans ADS lyase shows a high degree of similarity. Although this perhaps makes ADS lyase a less attractive target for antifungal design, differences discussed above suggest strategies that can be exploited to find ligands specific to the fungal enzyme. Furthermore, the similarity makes the fungal enzyme an alternative model to understand the molecular basis of the effects of mutations leading to human ADS lyase deficiency disease.
Discussion
Although antifungal agents that target purine biosynthesis have not yet been developed, there is increasing data that suggests this aspect of primary metabolism may be a druggable target for broad spectrum, non-toxic and affordable therapies to treat life-threatening fungal infections. Two GMP biosynthetic enzymes (IMP dehydrogenase and GMP synthase) and one ATP biosynthetic enzyme (ADS synthetase) have already been characterized at the genetic, enzymatic, and structural levels in C. neoformans as potential antifungal drug targets, and each have key active site differences compared with their human orthologs that could potentially enable the development of fungus-specific inhibitors (19, 20, 23). In contrast to these, the bifunctional nature of ADS lyase influences not only ATP biosynthesis, but also production of the key purine intermediate IMP (11); by disrupting IMP biosynthesis, the de novo production of both ATP and GTP is lost, suggesting that the inhibition of this enzyme may represent an even more powerful antifungal strategy.
As with the other purine de novo biosynthesis enzymes that have been characterized in C. neoformans, loss of ADS lyase results in auxotrophy and an inability to establish a life-threatening infection in a mouse model. Organ burden analysis revealed that the mutant was cleared from the infected animals, a result consistent with our observations that concentrations of adenine equivalent to that found in the human CNS were insufficient to restore growth of the mutant in vitro.
To gauge the suitability of this enzyme as a drug target, we investigated the biochemical activity and structure and compared them to the ortholog from the human host. In stark contrast to IMP dehydrogenase, GMP synthase, and ADS synthetase, C. neoformans ADS lyase exhibits a remarkably similar kinetic profile to its human counterpart, a similarity also shared with the equivalent enzyme from P. falciparum that has been proposed as a antimalarial target (18, 45). However, the protozoan enzyme exhibits only 15% identity to the human enzyme, whereas the C. neoformans enzyme exhibits 69% identity.
Crystal structure determination revealed that the similarity between the human and fungal enzymes is not limited to their kinetic profiles; the structure of the active site is also highly conserved between C. neoformans and human ADS lyase. Two amino acid changes are found in the active site pocket, and although the corresponding residues are not predicted to be involved in substrate binding, these glycine-to-threonine and lysine-to-arginine differences lead to surface changes in the active site pocket and can be exploited to design fungal enzyme-specific ligands. Substrate analogs have served as useful inhibitors of the purine biosynthesis pathway in a range of species (46–50) and taking advantage of this difference could enable the development of fungal ADS lyase inhibitors; this change is conserved in other fungi.
Another fungal-specific mode of disruption may be to target the tetramer assembly. Shown to be essential for activity, these regions are also a greater point of difference between C. neoformans and human ADS lyase. Of the three protomer interface residues mutated in ADS lyase deficiency only one is identical in C. neoformans. Designing a compound that disrupts tetramer formation based on these differences would likely be more challenging than developing a substrate structural analog, but given the lack of successful novel antifungals in the last 20 years, such an alternative strategy could still be attractive.
Beyond serving as a target in antifungal development, further studies of C. neoformans ADS lyase may facilitate the study of ADS lyase deficiency at a genetic level. Structural studies of human ADS lyase have enabled mapping of the mutations that cause ADS lyase deficiency in dozens of patients, as well as their biochemical consequences (15, 51). More than two-thirds of these residues are identical in C. neoformans. The R246H mutation is the most common cause of disease, present in one-third of all patients; this arginine residue is present in C. neoformans. The clinical manifestation resulting from the R426H mutation varies significantly; however, in vitro enzymatic assays of the mutant revealed it to be thermally unstable. The wild-type arginine residue is on the surface of the substrate channel; when mutated, an arginine-mediated interaction with Gln-409 and Asp-422 residues is disrupted (51–56). These two residues are also present in C. neoformans. Although these key mutations associated with this human disease are known, it is still unclear which of their physiological consequences are responsible for disease phenotypes, the impact on purine biosynthesis, or the accumulation of pathway intermediates. A close homolog of the ADS lyase that has similar structural features, such as ADS lyase from C. neoformans in vivo studies and genetic manipulation may determine these underlying causes for the disease.
In addition to its role in ADS lyase deficiency, the enzyme has also been shown to have up to 3-fold higher activity in tumors compared with healthy cells (57, 58), and increased ADS lyase activity was found to be a reliable indicator of hepatic, prostate, and breast tumors (59, 60). Further investigation of this enzyme may therefore not only serve to improve survival rates of ADS lyase deficiency patients where activity of the enzyme is low, but also assist in the development of agents to treat cancers where activity of the enzyme is high. A close eukaryotic homolog of the human enzyme with a solved structure and known biochemistry, such as from C. neoformans, could be employed to better understand these roles of ADS lyase.
Experimental procedures
Bioinformatic analyses
The C. neoformans type strain H99 genome sequence used in this study was reported by Janbon and colleagues (33). The gene encoding ADS lyase was identified in the C. neoformans genome by reciprocal best-hit BLAST analysis querying with the S. cerevisiae Ade13 protein.
Strains and media
Strains were stored in 15% glycerol at −80 °C until needed, and once grown were used for no longer than 2 weeks. Non-auxotrophic C. neoformans strains were cultured in liquid yeast peptone dextrose (YPD) media (1% yeast extract, 2% bacto-peptone, 2% glucose) at 30 °C and maintained on solidified YPD (additional 2% agar) at 4 °C unless stated otherwise. The adenine auxotrophic ade13Δ mutant was cultured in liquid YNB media (BD Bioscience) supplemented with 2% glucose, 10 mm ammonium sulfate, and 1 mm adenine at 30 °C and maintained at 4 °C on solidified YNB (additional 2% agar) supplemented as before unless otherwise stated. E. coli was cultured at 37 °C in either lysogeny broth (LB) (1% tryptone, 0.5% yeast extract, 1% sodium chloride) or terrific broth (TB) (1.2% tryptone, 2.4% yeast extract, 0.4% glycerol) supplemented with 100 μg/ml of ampicillin and 50 μg/ml of kanamycin and maintained on solidified LB (additional 2% agar) supplemented with antibiotics as described above.
Molecular techniques
The sequence of oligonucleotides used are given in supplemental Table S1. The ADE13 deletion construct was generated using overlap PCR, by using primers UQ1746 and UQ1749 to join the ADE13 5′ region (UQ1746 and UQ1747) to the G418 resistance marker NEO (UQ1832 and UQ1833) and the ADE13 3′ region (UQ1748 and UQ1749). H99 genomic DNA was used as the ADE13 template and plasmid pJAF1 as the template for the NEO cassette (61). The overlap construct was transformed into type strain H99 via biolistic transformation using a Bio-Rad He-1000 Biolistic device (Bio-Rad) and media containing 100 μg/ml of G418 and 1 mm adenine to select for the ade13Δ mutant strain. For complementation, the ADE13 gene was PCR-amplified (primers UQ1746 and UQ1749) from H99 genomic DNA, digested with NheI and XhoI, and cloned into the Safe Haven nourseothricin resistance vector pSDMA25 (24) cut with SpeI and XhoI to generate pKLB02. pKLB02 was subsequently linearized with BaeI and transformed into the ade13Δ mutant strain, selecting for nourseothricin resistance (100 μg/ml). Gene deletion and complemented strain validation were carried out on genomic DNA prepared by the CTAB protocol (62), digested, electrophoresed on 1% TAE-agarose gels, and Southern-blotted onto Hybond-XL membrane (GE Healthcare) using standard procedures (62, 63). Probes were generated from H99 using primers UQ1746 and UQ1749 with the Rediprime II kit and [α-32P]dCTP (PerkinElmer Life Sciences). Blots were hybridized at 65 °C and membranes were exposed onto Fuji Super RX medical X-ray film (Fujifilm, Japan).
Phenotypic assays
Production of melanin was assayed on l-DOPA medium containing 1 mm adenine (64). Urease assays were performed on Christensen's agar plus 1 mm adenine (65), and protease assays were performed on YNB with amino acids and ammonium sulfate supplemented with 2% glucose, 0.1% bovine serum albumin (BSA), and 1 mm adenine (Sigma). Strains were spotted at least 3 cm apart to prevent phenotypic cross-talk. Images were collected after 24–92 h incubation at 30 or 37 °C. Assays were performed in biological triplicate.
For capsule assays, strains were incubated in RPMI 1640 media (Life Technologies) supplemented with 2% glucose, 10% fetal bovine serum (Life Technologies), and 1 mm adenine with shaking at 30 or 37 °C. At 30 and 96 h, cells were stained with India ink (BD Diagnostics) and imaged with a Leica DM2500 microscope and DFC425C camera (Leica, Germany). At least 10 independent images were taken and the relative capsule diameter of 50 cells from each culture was determined as described by Zaragoza and colleagues (66). Assays were performed in biological triplicate and one-way analysis of variance tests with Sidak's post-test were employed in GraphPad Prism version 7.0 (GraphPad Software) to compare replicates to identify significant differences.
Murine inhalation model of cryptococcosis
For murine infection assays, 6-week-old female BALB/c mice (Animal Resources Centre, Australia) were infected by nasal inhalation (38). For each strain, 10 mice were inoculated with a 50-μl drop containing 5 × 105 C. neoformans cells. A maximum of five mice were housed per individually ventilated cage (Tecniplast, USA) with Bed-o'Cobs 1/8 inch bedding (The Andersons, USA), Crink-l'Nest nesting material (The Andersons, USA), and cardboard as environmental enrichment. Mice were provided Rat and Mouse Cubes (Specialty Feeds, Australia) and water ad libitum. Each mouse was examined and weighed twice daily for the duration of the experiment, with affected mice euthanized via CO2 inhalation once body weight had decreased to 80% of pre-infection weight or they exhibited symptoms consistent with infection. Death after CO2 inhalation was confirmed by pedal reflex prior to dissection. Brain, lungs, liver, spleen, and kidneys were collected, homogenized in 1 ml of PBS using a TissueLyser II (Qiagen, Germany), serially diluted, and plated on YNB supplemented with 100 μg/ml of ampicillin, 50 μg/ml of kanamycin, and 25 μg/ml of chloramphenicol. Plates were incubated at 30 °C, and after 48 h colonies were counted and used to calculate colony-forming units per g of organ. Kaplan-Meier survival curves were plotted using GraphPad Prism 7.0 (GraphPad Software). Significance was analyzed using the log-rank test, whereas organ burden significance was determined using a one-way analysis of variance with Tukey's multiple comparisons test. p values of <0.05 were considered significant.
Ethics statement
This study was carried out in strict accordance with the recommendations in the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes by the National Health and Medical Research Council. The protocol was approved by the Molecular Biosciences Animal Ethics Committee of The University of Queensland (AEC approval number: SCMB/008/11/UQ/NHMRC). Infection was performed under methoxyflurane anesthesia, and all efforts were made to minimize suffering through adherence to the Guidelines to Promote the Wellbeing of Animals Used for Scientific Purposes as put forward by the National Health and Medical Research Council.
Expression and purification of C. neoformans ADSL
Total RNA from YPD-grown C. neoformans strain H99 was extracted using TRIzol (Invitrogen). Intron-free cDNA was synthesized using the SuperScript III First Strand Synthesis System (Invitrogen). The ADE13 ORF was PCR-amplified using primers designed to introduce SspI and XhoI restriction sites (UQ2085 and UQ2086) and the amplicon cloned into the pCRII-TOPO vector (Invitrogen) to give pCAM123. The SspI/XhoI fragment from pCAM123 was then subcloned into the E. coli expression vector pQE-30 (Qiagen, Germany) cut with SspI/XhoI to introduce an N-terminal His6 tag (MRGSHHHHHHGS) to give pCAM126. pCAM126 was subsequently transformed into E. coli strain BL21(DE3) pLysS (Novagen, Japan) and grown in LB supplemented with 100 mg/ml of ampicillin and 35 mg/ml of kanamycin at 37 °C to an A600 of ∼1. Cultures were then induced with 1 mm isopropyl β-d-1-thiogalactopyranoside and grown for a further 5 h at 37 °C. Cells were harvested and resuspended in lysis buffer (50 mm HEPES, pH 8.0, 300 mm NaCl, 30 mm imidazole, 1 mm DTT, and 1 mm PMSF) before disruption with a Sonifier W-450 Digital Ultrasonic Cell Disruptor sonicator (Branson). Following centrifugation, the supernatant was loaded onto a 5-ml HisTrap Fast Flow column (GE Healthcare) to purify the His-tagged protein by immobilized nickel-affinity chromatography. The protein was eluted in a linear gradient of 30–500 mm imidazole, showing a single elution peak. Peak fractions were pooled, concentrated, and further purified using a Superdex 200 SEC column (GE Healthcare). Protein was eluted at a rate of 2.5 ml/min with SEC buffer (10 mm HEPES, pH 7.5, 150 mm NaCl, and 1 mm DTT) using an ÄKTApurifier FPLC system. Peak fractions were combined and concentrated to ∼11 mg/ml and flash-frozen in liquid nitrogen for storage at −80 °C.
Steady-state kinetics
ADS lyase activity was monitored spectrophotometrically using a Cary60 UV-visible spectrophotometer (Agilent). The ADS-to-AMP reaction was monitored by the decrease in absorbance at 280 nm as described for the P. falciparum enzyme (18). Following optimization for the C. neoformans enzyme (pH range tested, 6–10; temperature range tested, 20–45 °C), 40 mm potassium phosphate buffer (pH 8.0) was used. Assays were performed in triplicate at 37 °C with purified, recombinant C. neoformans ADS lyase used at 2 nm final concentration. A differential extinction coefficient (Δϵ), 100 mm−1 cm−1, was used to calculate the specific activity of the enzyme in units (U) per milligram of ADS lyase (15). As previous studies of ADS lyase from Bacillus subtilis and human revealed that correct quaternary structure formation of the protein required incubation at 25 °C for 2 h prior to assays, to ensure restoration of enzymatic activity following freezing, the same protocol was followed (15, 67). Data for the ADS-to-AMP reaction were fitted to the Michaelis-Menten equation using GraphPad Prism 7.0 (GraphPad Software).
SEC-coupled MALLS
MALLS was coupled with SEC using a Superdex 75 5/150 size exclusion column (GE Healthcare) and performed using a Dawn Heleos II 18-angle light-scattering detector coupled with an Optilab rEX refractive index detector (Wyatt Technology). Measurements were performed at room temperature with a flow rate of 0.5 ml/min in 10 mm HEPES (pH 7.5), 150 mm NaCl, and 1 mm DTT. The sample volume was 30 μl with a protein concentration of 5 mg/ml. Molecular mass calculations were performed using the Astra 5.3 software (Wyatt Technology). Input of the refractive increment (dn/dc values) was set at 0.186 in molecular mass calculations (68).
Crystallization
Initial crystal hits were obtained by sparse matrix screening using Index and PEG/Ion screens (Hampton Research) and optimized by Additive Screen (Hampton Research). The crystals used for diffraction experiments were obtained via hanging-drop vapor diffusion after 2 days by mixing 1 μl of protein solution at 11 mg/ml with 1 μl of well solution containing 0.1 m BisTris/citric acid (pH 6.5), 17% PEG 3350, and 0.12 m sodium chloride at 20 °C. X-ray diffraction data were collected on the MX1 beamline of the Australian Synchrotron, Melbourne, Australia, using Blu-Ice (69) and processed and scaled using XDS (70) and Scala (71). The C. neoformans ADS lyase structure was solved by molecular replacement using Phaser (72) in the PHENIX suite version 1.1.4 (73), with the human ADS lyase structure (PDB code 4FFX) as a template. The resulting model was refined with data between 20 and 2.1-Å resolution and model building between rounds of refinement was performed with Coot version 0.8.1 (74). Structure validation was performed using MolProbity version 4.3 (75).
Author contributions
J. L. C., K. L. B., U. K., S. J. W., B. K., and J. A. F. conceived and designed the experiments; J. C., K. L. B., R. D. B., A. E. K., and M. T. performed the experiments; J. L. C., K. L. B., U. K., S. J. W., B. K., and J. A. F. analyzed the data; J. C. and J. A. F. wrote the paper; S. J. W., A. A. B. R., M. S. B., M. A. C., and B. K. contributed to editing and writing.
Supplementary Material
Acknowledgments
X-ray diffraction data collection was undertaken on the MX1 beamline at the Australian Synchrotron, Victoria, Australia. We also acknowledge the use of the University of Queensland Remote Operation Crystallization and X-ray Diffraction Facility (UQ ROCX).
This work was supported in part by National Health and Medical Research Council (NHMRC) Grant APP1049716 (to J. A. F. and M. S. B.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1 and S2 and Figs. S1 and S2.
The atomic coordinates and structure factors (code 5V4L) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ADS
- adenylosuccinate
- SAICAR
- succinylaminoimidazole carboxamide riboside
- AICAR
- aminoimidazole carboxamide riboside
- SEC
- size exclusion chromatography
- MALLS
- multi-angle laser light scattering
- YNB
- yeast nitrogen base
- l-DOPA
- l-3,4-dihydroxyphenylalanine
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PDB
- Protein Data Bank.
References
- 1. Garrod A. B. (1848) Observations on certain pathological conditions of the blood and urine, in gout, rheumatism, and Bright's disease. Med. Chir. Trans. 31, 83–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garrod A. E. S. (1909) Inborn errors of metabolism, H. Frowde and Hodder & Stoughton, London [Google Scholar]
- 3. Lesch M., and Nyhan W. L. (1964) A familial disorder of uric acid metabolism and central nervous system function. Am. J. Med. 36, 561–570 [DOI] [PubMed] [Google Scholar]
- 4. Mackenzie D. Y., and Woolf L. I. (1959) Maple syrup urine disease; an inborn error of the metabolism of valine, leucine, and isoleucine associated with gross mental deficiency. Br. Med. J. 1, 90–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carlyll H. B., and Mott F. W. (1911) Seven cases of amaurotic idiocy (Tay-Sachs disease). Proc. R. Soc. Med. 4, 147–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lanpher B., Brunetti-Pierri N., and Lee B. (2006) Inborn errors of metabolism: the flux from Mendelian to complex diseases. Nat. Rev. Genet. 7, 449–460 [DOI] [PubMed] [Google Scholar]
- 7. Kelley R. E., and Andersson H. C. (2014) Disorders of purines and pyrimidines. Handb. Clin. Neurol. 120, 827–838 [DOI] [PubMed] [Google Scholar]
- 8. Simmonds H. A., Duley J. A., Fairbanks L. D., and McBride M. B. (1997) When to investigate for purine and pyrimidine disorders. Introduction and review of clinical and laboratory indications. J. Inherit. Metab. Dis. 20, 214–226 [DOI] [PubMed] [Google Scholar]
- 9. Jaeken J., and Van den Berghe G. (1984) An infantile autistic syndrome characterised by the presence of succinylpurines in body fluids. Lancet 2, 1058–1061 [PubMed] [Google Scholar]
- 10. Carter C. E., and Cohen L. H. (1956) The preparation and properties of adenylosuccinase and adenylosuccinic acid. J. Biol. Chem. 222, 17–30 [PubMed] [Google Scholar]
- 11. Miller R. W., Lukens L. N., and Buchanan J. M. (1959) Biosynthesis of the purines: XXV: the enzymatic cleavage of N-(5-amino-1-ribosyl-4-imidazolylcarbonyl)-l-aspartic acid 5′-phosphate. J. Biol. Chem. 234, 1806–1811 [PubMed] [Google Scholar]
- 12. Weaver T., and Banaszak L. (1996) Crystallographic studies of the catalytic and a second site in fumarase C from Escherichia coli. Biochemistry 35, 13955–13965 [DOI] [PubMed] [Google Scholar]
- 13. Simpson A., Bateman O., Driessen H., Lindley P., Moss D., Mylvaganam S., Narebor E., and Slingsby C. (1994) The structure of avian eye lens δ-crystallin reveals a new fold for a superfamily of oligomeric enzymes. Nat. Struct. Biol. 1, 724–734 [DOI] [PubMed] [Google Scholar]
- 14. Shi W., Dunbar J., Jayasekera M. M., Viola R. E., and Farber G. K. (1997) The structure of l-aspartate ammonia-lyase from Escherichia coli. Biochemistry 36, 9136–9144 [DOI] [PubMed] [Google Scholar]
- 15. Ray S. P., Deaton M. K., Capodagli G. C., Calkins L. A., Sawle L., Ghosh K., Patterson D., and Pegan S. D. (2012) Structural and biochemical characterization of human adenylosuccinate lyase (ADSL) and the R303C ADSL deficiency-associated mutation. Biochemistry 51, 6701–6713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vedadi M., Lew J., Artz J., Amani M., Zhao Y., Dong A., Wasney G. A., Gao M., Hills T., Brokx S., Qiu W., Sharma S., Diassiti A., Alam Z., Melone M., et al. (2007) Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol. Biochem. Parasitol. 151, 100–110 [DOI] [PubMed] [Google Scholar]
- 17. Tsai M., Koo J., Yip P., Colman R. F., Segall M. L., and Howell P. L. (2007) Substrate and product complexes of Escherichia coli adenylosuccinate lyase provide new insights into the enzymatic mechanism. J. Mol. Biol. 370, 541–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bulusu V., Srinivasan B., Bopanna M. P., and Balaram H. (2009) Elucidation of the substrate specificity, kinetic and catalytic mechanism of adenylosuccinate lyase from Plasmodium falciparum. Biochim. Biophys. Acta 1794, 642–654 [DOI] [PubMed] [Google Scholar]
- 19. Morrow C. A., Valkov E., Stamp A., Chow E. W., Lee I. R., Wronski A., Williams S. J., Hill J. M., Djordjevic J. T., Kappler U., Kobe B., and Fraser J. A. (2012) De novo GTP biosynthesis is critical for virulence of the fungal pathogen Cryptococcus neoformans. PLoS Pathog. 8, e1002957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blundell R. D., Williams S. J., Arras S. D., Chitty J. L., Blake K. L., Ericsson D. J., Tibrewal N., Rohr J., Koh Y. Q., Kappler U., Robertson A. A., Butler M. S., Cooper M. A., Kobe B., and Fraser J. A. (2016) Disruption of de novo adenosine triphosphate (ATP) biosynthesis abolishes virulence in Cryptococcus neoformans. ACS Infect. Dis. 2, 651–663 [DOI] [PubMed] [Google Scholar]
- 21. Jiang L., Zhao J., Guo R., Li J., Yu L., and Xu D. (2010) Functional characterization and virulence study of ADE8 and GUA1 genes involved in the de novo purine biosynthesis in Candida albicans. FEMS Yeast Res. 10, 199–208 [DOI] [PubMed] [Google Scholar]
- 22. Rodriguez-Suarez R., Xu D., Veillette K., Davison J., Sillaots S., Kauffman S., Hu W., Bowman J., Martel N., Trosok S., Wang H., Zhang L., Huang L. Y., Li Y., Rahkhoodaee F., et al. (2007) Mechanism-of-action determination of GMP synthase inhibitors and target validation in Candida albicans and Aspergillus fumigatus. Chem. Biol. 14, 1163–1175 [DOI] [PubMed] [Google Scholar]
- 23. Chitty J. L., Tatzenko T. L., Williams S. J., Koh Y. Q., Corfield E. C., Butler M. S., Robertson A. A., Cooper M. A., Kappler U., Kobe B., and Fraser J. A. (2017) GMP synthase is required for virulence factor production and infection by Cryptococcus neoformans. J. Biol. Chem. 292, 3049–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arras S. D., Chitty J. L., Blake K. L., Schulz B. L., and Fraser J. A. (2015) A genomic safe haven for mutant complementation in Cryptococcus neoformans. PLoS ONE 10, e0122916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perfect J. R., Toffaletti D. L., and Rude T. H. (1993) The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect. Immun. 61, 4446–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park J., Rajasingham R., Smith R., and Boulware D. (2014) Update on the global burden of cryptococcosis. Mycoses, Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- 27. Loyse A., Dromer F., Day J., Lortholary O., and Harrison T. S. (2013) Flucytosine and cryptococcosis: time to urgently address the worldwide accessibility of a 50-year-old antifungal. J. Antimicrob. Chemother. 68, 2435–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loyse A., Thangaraj H., Easterbrook P., Ford N., Roy M., Chiller T., Govender N., Harrison T. S., and Bicanic T. (2013) Cryptococcal meningitis: improving access to essential antifungal medicines in resource-poor countries. Lancet Infect. Dis. 13, 629–637 [DOI] [PubMed] [Google Scholar]
- 29. Friese G., Discher T., Füssle R., Schmalreck A., and Lohmeyer J. (2001) Development of azole resistance during fluconazole maintenance therapy for AIDS-associated cryptococcal disease. AIDS 15, 2344–2345 [DOI] [PubMed] [Google Scholar]
- 30. Perlin D. S. (2007) Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bicanic T., Harrison T., Niepieklo A., Dyakopu N., and Meintjes G. (2006) Symptomatic relapse of HIV-associated cryptococcal meningitis after initial fluconazole monotherapy: the role of fluconazole resistance and immune reconstitution. Clin. Infect Dis. 43, 1069–1073 [DOI] [PubMed] [Google Scholar]
- 32. Dorfman B. Z. (1969) The isolation of adenylosuccinate synthetase mutants in yeast by selection for constitutive behavior in pigmented strains. Genetics 61, 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Janbon G., Ormerod K. L., Paulet D., Byrnes E. J. 3rd, Yadav V., Chatterjee G., Mullapudi N., Hon C. C., Billmyre R. B., Brunel F., Bahn Y. S., Chen W., Chen Y., Chow E. W., et al. (2014) Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 10, e1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eells J. T., and Spector R. (1983) Purine and pyrimidine base and nucleoside concentrations in human cerebrospinal fluid and plasma. Neurochem. Res. 8, 1451–1457 [DOI] [PubMed] [Google Scholar]
- 35. Rodríguez-Núñez A., Camiña F., Lojo S., Rodríguez-Segade S., and Castro-Gago M. (1993) Concentrations of nucleotides, nucleosides, purine bases and urate in cerebrospinal fluid of children with meningitis. Acta Paediatr. 82, 849–852 [DOI] [PubMed] [Google Scholar]
- 36. Xu C. Y., Zhu H. M., Wu J. H., Wen H., and Liu C. J. (2014) Increased permeability of blood-brain barrier is mediated by serine protease during Cryptococcus meningitis. J. Int. Med. Res. 42, 85–92 [DOI] [PubMed] [Google Scholar]
- 37. Salas S. D., Bennett J. E., Kwon-Chung K. J., Perfect J. R., and Williamson P. R. (1996) Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. J. Exp. Med. 184, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cox G. M., Mukherjee J., Cole G. T., Casadevall A., and Perfect J. R. (2000) Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stone R. L., Zalkin H., and Dixon J. E. (1993) Expression, purification, and kinetic characterization of recombinant human adenylosuccinate lyase. J. Biol. Chem. 268, 19710–19716 [PubMed] [Google Scholar]
- 40. Boitz J. M., Strasser R., Yates P. A., Jardim A., and Ullman B. (2013) Adenylosuccinate synthetase and adenylosuccinate lyase deficiencies trigger growth and infectivity deficits in Leishmania donovani. J. Biol. Chem. 288, 8977–8990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kmoch S., Hartmannová H., Stibůrková B., Krijt J., Zikánová M., and Sebesta I. (2000) Human adenylosuccinate lyase (ADSL), cloning and characterization of full-length cDNA and its isoform, gene structure and molecular basis for ADSL deficiency in six patients. Hum. Mol. Genet. 9, 1501–1513 [DOI] [PubMed] [Google Scholar]
- 42. Ghannoum M. A., and Rice L. B. (1999) Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12, 501–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hitchcock C. A., Dickinson K., Brown S. B., Evans E. G., and Adams D. J. (1990) Interaction of azole antifungal antibiotics with cytochrome P-450-dependent 14α-sterol demethylase purified from Candida albicans. Biochem. J. 266, 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Auclair J. R., Boggio K. J., Petsko G. A., Ringe D., and Agar J. N. (2010) Strategies for stabilizing superoxide dismutase (SOD1), the protein destabilized in the most common form of familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 107, 21394–21399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marshall V. M., and Coppel R. L. (1997) Characterisation of the gene encoding adenylosuccinate lyase of Plasmodium falciparum. Mol. Biochem. Parasitol. 88, 237–241 [DOI] [PubMed] [Google Scholar]
- 46. Christopherson R. I., Lyons S. D., and Wilson P. K. (2002) Inhibitors of de novo nucleotide biosynthesis as drugs. Acc. Chem. Res. 35, 961–971 [DOI] [PubMed] [Google Scholar]
- 47. Skipper H. E., Thomson J. R., Elion G. B., and Hitchings G. H. (1954) Observations on the anticancer activity of 6-mercaptopurine. Cancer Res. 14, 294–298 [PubMed] [Google Scholar]
- 48. Mendelsohn L. G., Shih C., Schultz R. M., and Worzalla J. F. (1996) Biochemistry and pharmacology of glycinamide ribonucleotide formyltransferase inhibitors: LY309887 and lometrexol. Invest. New Drugs 14, 287–294 [DOI] [PubMed] [Google Scholar]
- 49. Franklin T. J., and Cook J. M. (1969) The inhibition of nucleic acid synthesis by mycophenolic acid. Biochem. J. 113, 515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sweeney M. J., Hoffman D. H., and Esterman M. A. (1972) Metabolism and biochemistry of mycophenolic acid. Cancer Res. 32, 1803–1809 [PubMed] [Google Scholar]
- 51. Zikanova M., Skopova V., Hnizda A., Krijt J., and Kmoch S. (2010) Biochemical and structural analysis of 14 mutant adsl enzyme complexes and correlation to phenotypic heterogeneity of adenylosuccinate lyase deficiency. Hum. Mutat. 31, 445–455 [DOI] [PubMed] [Google Scholar]
- 52. Marie S., Cuppens H., Heuterspreute M., Jaspers M., Tola E. Z., Gu X. X., Legius E., Vincent M. F., Jaeken J., Cassiman J. J., and Van den Berghe G. (1999) Mutation analysis in adenylosuccinate lyase deficiency: eight novel mutations in the re-evaluated full ADSL coding sequence. Hum. Mutat. 13, 197–202 [DOI] [PubMed] [Google Scholar]
- 53. Edery P., Chabrier S., Ceballos-Picot I., Marie S., Vincent M. F., and Tardieu M. (2003) Intrafamilial variability in the phenotypic expression of adenylosuccinate lyase deficiency: a report on three patients. Am. J. Med. Genet. A 120A, 185–190 [DOI] [PubMed] [Google Scholar]
- 54. Jurecka A., Zikanova M., Tylki-Szymanska A., Krijt J., Bogdanska A., Gradowska W., Mullerova K., Sykut-Cegielska J., Kmoch S., and Pronicka E. (2008) Clinical, biochemical and molecular findings in seven Polish patients with adenylosuccinate lyase deficiency. Mol. Genet. Metab. 94, 435–442 [DOI] [PubMed] [Google Scholar]
- 55. Henneke M., Dreha-Kulaczewski S., Brockmann K., van der Graaf M., Willemsen M. A., Engelke U., Dechent P., Heerschap A., Helms G., Wevers R. A., and Gärtner J. (2010) In vivo proton MR spectroscopy findings specific for adenylosuccinate lyase deficiency. NMR Biomed 23, 441–445 [DOI] [PubMed] [Google Scholar]
- 56. Donti T. R., Cappuccio G., Hubert L., Neira J., Atwal P. S., Miller M. J., Cardon A. L., Sutton V. R., Porter B. E., Baumer F. M., Wangler M. F., Sun Q., Emrick L. T., and Elsea S. H. (2016) Diagnosis of adenylosuccinate lyase deficiency by metabolomic profiling in plasma reveals a phenotypic spectrum. Mol. Genet. Metab. Rep. 8, 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jackson R. C., Morris H. P., and Weber G. (1976) Increased adenylosuccinase activity in hepatomas and kidney tumors. Life Sci. 18, 1043–1048 [DOI] [PubMed] [Google Scholar]
- 58. Jackson R. C., Morris H. P., and Weber G. (1977) Enzymes of the purine ribonucleotide cycle in rat hepatomas and kidney tumors. Cancer Res. 37, 3057–3065 [PubMed] [Google Scholar]
- 59. Mack D. O., Lewis E. M., Butler E. M., Archer W. H., and Smith L. D. (1985) A comparison of succinyladenylate lyase activity and serum sialic acid as markers of malignancy. Biochem. Med. 34, 327–334 [DOI] [PubMed] [Google Scholar]
- 60. Reed V. L., Mack D. O., and Smith L. D. (1987) Adenylosuccinate lyase as an indicator of breast and prostate malignancies: a preliminary report. Clin. Biochem. 20, 349–351 [DOI] [PubMed] [Google Scholar]
- 61. Fraser J. A., Subaran R. L., Nichols C. B., and Heitman J. (2003) Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2, 1036–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pitkin J. W., Panaccione D. G., and Walton J. D. (1996) A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142, 1557–1565 [DOI] [PubMed] [Google Scholar]
- 63. Sambrook J., F. E. and Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 64. Chaskes S., and Tyndall R. L. (1975) Pigment production by Cryptococcus neoformans from para- and ortho-Diphenols: effect of the nitrogen source. J. Clin. Microbiol. 1, 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Christensen W. B. (1946) Urea decomposition as a means of differentiating Proteus and Paracolon cultures from each other and from Salmonella and Shigella types. J. Bacteriol. 52, 461–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zaragoza O., Fries B. C., and Casadevall A. (2003) Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO2. Infect. Immun. 71, 6155–6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. De Zoysa Ariyananda L., and Colman R. F. (2008) Evaluation of types of interactions in subunit association in Bacillus subtilis adenylosuccinate lyase. Biochemistry 47, 2923–2934 [DOI] [PubMed] [Google Scholar]
- 68. Wen J., Arakawa T., and Philo J. S. (1996) Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal. Biochem. 240, 155–166 [DOI] [PubMed] [Google Scholar]
- 69. McPhillips T. M., McPhillips S. E., Chiu H. J., Cohen A. E., Deacon A. M., Ellis P. J., Garman E., Gonzalez A., Sauter N. K., Phizackerley R. P., Soltis S. M., and Kuhn P. (2002) Blu-Ice and the distributed control system: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 9, 401–406 [DOI] [PubMed] [Google Scholar]
- 70. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Winn M. D. (2003) An overview of the CCP4 project in protein crystallography: an example of a collaborative project. J. Synchrotron Radiat. 10, 23–25 [DOI] [PubMed] [Google Scholar]
- 72. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 75. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Banerjee S., Agrawal M. J., Mishra D., Sharan S., Balaram H., Savithri H. S., and Murthy M. R. (2014) Structural and kinetic studies on adenylosuccinate lyase from Mycobacterium smegmatis and Mycobacterium tuberculosis provide new insights on the catalytic residues of the enzyme. FEBS J. 281, 1642–1658 [DOI] [PubMed] [Google Scholar]
- 77. Brosius J. L., and Colman R. F. (2000) A key role in catalysis for His89 of adenylosuccinate lyase of Bacillus subtilis. Biochemistry 39, 13336–13343 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.