Abstract
Cyclic electron flow around photosystem I (CEF) is critical for balancing the photosynthetic energy budget of the chloroplast by generating ATP without net production of NADPH. We demonstrate that the chloroplast NADPH dehydrogenase complex, a homolog to respiratory Complex I, pumps approximately two protons from the chloroplast stroma to the lumen per electron transferred from ferredoxin to plastoquinone, effectively increasing the efficiency of ATP production via CEF by 2-fold compared with CEF pathways involving non-proton-pumping plastoquinone reductases. By virtue of this proton-pumping stoichiometry, we hypothesize that NADPH dehydrogenase not only efficiently contributes to ATP production but operates near thermodynamic reversibility, with potentially important consequences for remediating mismatches in the thylakoid energy budget.
Keywords: electron transfer complex, photosynthesis, plant biochemistry, proton motive force, proton pump, cyclic electron flow
Introduction
The canonical “Z-scheme” model of photosynthetic linear electron flow (LEF,3 supplemental Fig. S1A) uses a series of two photochemical reaction centers to extract electrons from water at photosystem II (PSII) and transfer them to NADPH through photosystem I (PSI) (1). The electron transfer reactions are coupled to proton transfer, generating an electrochemical gradient of protons, the proton motive force (Δp), that drives the synthesis of ATP. Because of the strong coupling of electron and proton transfer reactions (2, 3), LEF should produce a fixed (or rigid) ratio of ATP/NADPH (for review, see Ref. 4). In contrast, downstream metabolic reactions impose variable demands for ATP and NADPH, requiring dynamic adjustments of the photosynthetic energy budget to avoid “metabolic congestion” that can lead to buildup of products or depletion of substrates that can result in photodamage (2, 3). Indeed, LEF alone should not be able to power the Calvin-Benson cycle. With one proton deposited in the lumen during water oxidation and two protons translocated by the cytochrome bf complex catalyzing through the Q-cycle, LEF should result in the translocation of three protons for each electron transferred to NADPH (5–7); with a H+/ATP ratio of 4.67 at the ATP synthase (8), LEF should produce 2.6 ATPs for every 2 NADPHs, a deficit of 0.4 ATP/2 NADPH compared with the ratio required to sustain the Calvin-Benson cycle.
There is substantial evidence that cyclic electron flow around photosystem I (CEF) plays an important role in balancing the ATP/NADPH energy budget (3, 4, 9, 10). The generally accepted model for CEF involves the transfer of highly reducing electrons from photoexcited PSI centers to plastoquinone (PQ) through a PQ reductase, resulting in the formation of plastoquinol (PQH2) with the uptake of protons from the chloroplast stroma. The PQH2 is then oxidized by the cytochrome bf complex and returned to the oxidizing site of PSI via plastocyanin or cytochrome c6.
There are several proposed CEF pathways in higher plant chloroplasts that differ at the level of the plastoquinone reductase (for review, see Ref. 11; see also supplemental Fig. S1, B and C). One of these involves the thylakoid ferredoxin:plastoquinone oxidoreductase complex, for historical reasons called the NADPH dehydrogenase complex (see “Discussion”), which is homologous to respiratory Complex I (12, 13). Another pathway, termed the ferredoxin:quinone reductase (FQR), is sensitive to antimycin A (AA) and proposed to involve a complex of the PGR5/PGRL1 proteins that is able to transfer electrons from ferredoxin (Fd) to PQ (11, 14–17).
Both of these CEF routes are conserved across the flowering plants even though they may appear redundant, i.e. both function in CEF by transferring electrons from ferredoxin to PQ (15, 17, 18). However, the PQ reductases in these two pathways are structurally distinct, possibly giving clues about their relative functions. Of particular interest is the homology of NDH to the bacterial or respiratory type I NADH:quinone reductases (Complex I) (12, 19, 20) that are known to couple reduction of quinones to the pumping of up to two protons for each electron transferred to quinone, efficiently generating Δp to drive ATP synthesis (21, 22). In contrast, the FQR is likely to catalyze a simpler, more direct mode of PQ reduction (14, 17) that is very unlikely to be coupled to proton translocation, much like the type II (non-proton-pumping) NAD(P)H:quinone oxidoreductases such as NDA2 and NDH2, as found in the mitochondrial respiratory chains of plants, certain fungi, and protozoa. Non-proton motive NDA/NDH2-type enzymes are also widespread in bacteria and the plastids of certain green algae (23, 24).
If the NDH is functionally similar to Complex I, it could have the capability to use the energy liberated during PQ reduction to translocate protons across the thylakoid against the electrochemical gradient and thus increase the H+/e− stoichiometry of CEF. Although proton-pumping activity for NDH has been proposed in previous work (25, 26), it has not been experimentally demonstrated. Such activity would have a large impact on the energy balance of the chloroplast by allowing for more efficient balancing of the ATP/NADPH budget with relatively low turnover rates. On the other hand, as discussed below, a high coupling ratio for proton-pumping to electron transfer could impose significant thermodynamic and kinetic limitations or even allow plastoquinol to reduce NADP+. In this work we used a series of complementary approaches to assess the possibility that NDH acts as a proton pump both in vitro and in vivo.
Results
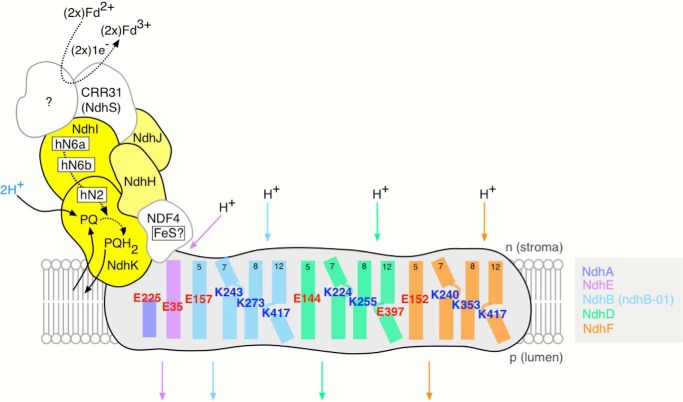
Sequence conservation in NDH of residues essential for proton-pumping
Fig. 1 shows a structural schematic of Arabidopsis thaliana NDH indicating the likely organization and conservation of charged residues within the membrane domain, which are considered to be essential for proton translocation in respiratory Complex I. Complete sequence alignments for the membrane subunits depicted in Fig. 1 for the respective NDH subunits from A. thaliana, Spinacia oleracea (spinach), and Nicotiana tabacum (tobacco) and Complex I from Escherichia coli, Thermus thermophilus, Yarrowia lipolytica (yeast), and Bos taurus are presented in supplemental Fig. S2, A–E).
Figure 1.
Structural schematic of NDH showing conserved residues essential for proton-pumping within the membrane domain (gray) and likely organization of the electron donor domain (yellow). Subunits equivalent to the NADH-binding domain of respiratory Complex I (the N-module in the terminology of Brandt; 67) have not been identified in NDH. The (plasto)quinone-binding Q-module is extant in NDH and formed from NdhH, -I, -J, and -K. Structural motifs for the Complex I iron–sulfur clusters N6a, N6b (NdhI), and N2 (NdhK) are conserved in NDH, although their presence has yet to be confirmed spectroscopically. As such, the h- prefix for these putative clusters in indicates their homology to Complex I. The subunits NDF4 and CRR31/NdhS are unique to NDH. NDF4 is likely to contain and iron–sulfur (FeS) cluster, and CRR31/NdhS has been proposed to be a constituent of the Fd binding site (although it may not be a redox-active participant) (68, 69). Other stromal subunits that have been identified as NDH components but are unlikely to directly participate in the electron transfer pathway are not shown here (70, 71). Highlighted charged residues within NdhB, -D, -E, and -F in the membrane domain are conserved between NDH and Complex I and are considered essential for proton translocation. Residues are numbered according to the A. thaliana sequence data. Helices are numbered according to their Complex I homologues. Predicted discontinuities in helices 7 and 12 of NdhB, -D, and -F as observed in the atomic structure of T. thermophilus Complex I are shown. Note that the NdhB gene is duplicated in Arabidopsis (the sequences of B-01 and B-02 are identical). Complete predicted sequence alignments for the membrane domain subunits shown here are presented in supplemental Fig. S2.
Four discrete proton channels exhibiting highly characteristic sequence conservation have been identified in the atomic structures of the membrane domains of T. thermophilus and Y. lipolytica Complex I (21, 27, 28). Three of these channels are contained within the “antiporter-like” subunits equivalent to NdhB, -D, and -F, and the fourth channel was from an association of subunits equivalent to NdhA, -C, -E, and -G. This latter group also forms part of the quinone-binding site within the enzyme. The lysine and glutamate residues located within the center of the lipid bilayer are key mechanistic features of these proton channels and are conserved among Complex I and NDH (displayed in Fig. 1). Mutagenesis of these residues has been shown to impair the proton-pumping capacity of E. coli Complex I (for review, see Ref. 22). The conservation of these intramembrane lysine and glutamate residues between Complex I and NDH provides circumstantial evidence for the protonmotive activity of the latter enzyme, which was then investigated experimentally as described below.
Proton-pumping activity of chloroplast NDH probed in vitro
In vitro ATP generation in the dark via protonmotive NDH activity
The low abundance of NDH in plant thylakoids generally precludes the use of proteoliposome reconstitution experiments to directly assess protonmotive activity, as has been used for respiratory Complex I (19, 29). As such, we developed an in vitro assay (illustrated in Fig. 2A) for use with coupled thylakoid preparations designed to report ATP generation only in the presence of a proton-pumping, NDH-type plastoquinone reductase. ATP synthesis was monitored using firefly luciferase luminescence in spinach thylakoids. Spinach was chosen, as it provides a reliable (and abundant) source for the isolation of coupled thylakoids. To avoid interference from light-driven ATP production through photophosphorylation, experiments were conducted in strict darkness in the presence of dichloromethyl urea (DCMU), an inhibitor of PSII, and/or tridecyl stigmatellin (an inhibitor of the cytochrome bf complex; see below). The NDH reaction was initiated by the addition of exogenous (oxidized) Fd, NADPH, and the PQ analogue decylplastoquinone (dPQ). Oxidized Fd (the electron donor to NDH) was reduced by NADPH through endogenous ferredoxin:NADP oxidoreductase (FNR) activity. The suspension medium was well buffered so that the uptake of protons onto dPQ should not have affected the pH of the extra-lumenal space, and thus, Δp should only be produced by the transfer of protons in the thylakoid lumen. As shown in Fig. 2B, the addition of Fd and NADPH alone did not induce luminescence, but further addition of dPQ resulted in a sustained, reproducible increase in luminescence, reflecting the production of ∼10 nmol of ATP/mg chlorophyll−1 min−1. This rate was ∼1% of the steady-state (LEF) ATP production rate reported for spinach thylakoid suspensions illuminated at 250 μmol of photons m−2 s−1 using ferricyanide as the Hill oxidant (30). The (comparatively) low rate of ATP production supported by NDH activity in the in vitro assay described in this section should be understood in terms of (i) the energetically unfavorable reduction of ferredoxin by NADPH and, thus, low electron donor concentration and (ii) the low abundance of NDH and the predominance of the (non-protonmotive) FQR CEF pathway in spinach (see “Discussion”).
Figure 2.
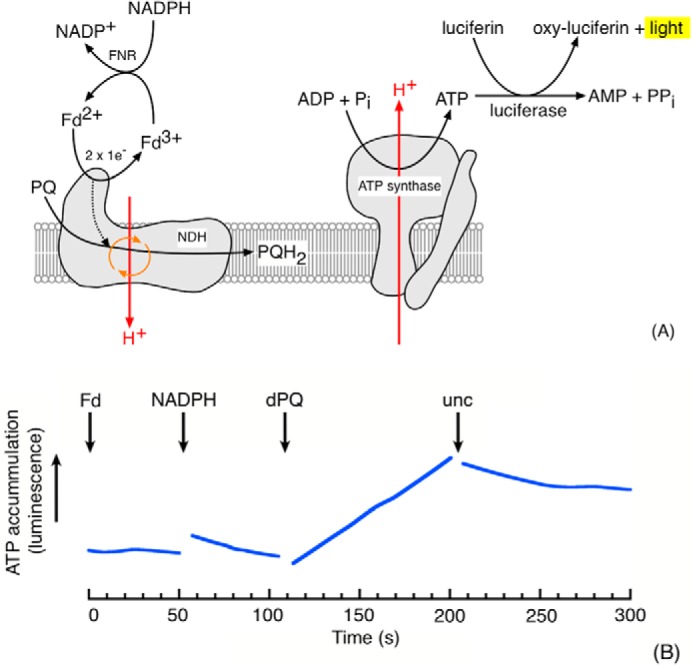
Evidence for involvement of a proton pump in CEF. A, schematic of experimental rationale showing the coupling of proton-pumping NDH activity with ATP synthesis and subsequent detection by luciferase-mediated luminescence. A non-protonmotive NDH would be incapable of generating the proton gradient required for ATP synthesis. B, ATP synthesis, monitored by luciferase luminescence, in DCMU-treated S. oleracea thylakoids (50 μg of Chl/ml) in the dark after the addition of 5 μm Fd, 100 μm NADPH, and 50 μm dPQ. The assay buffer consisted of 10 mm HEPES (pH 7.5), 10 mm KCl, 5 mm MglCl2, 2 mm ADP, 2 mm potassium phosphate, 2 mm DTT, 100 μm diadenosine pentaphosphate, and 10 μm DCMU. Premixed Enliten recombinant luciferase/luciferin reagent (used as supplied by Promega) was added to a final concentration of 8% (v/v). The addition of valinomycin and nigericin (10 μm each) is indicated by unc. Representative data (fitted in Kaleidagraph (Synergy Software) by the locally weighted least square error method after application of a five-point smooth) are shown; discontinuities in the data are due to the removal of mixing artifacts on substrate addition. The rate of ATP synthesis on dPQ addition was ∼10 nmol of ATP/mg of Chl/min.
For the in vitro proton pumping assay described here the order of substrate addition was not critical but that NADPH, Fd, and dPQ were all required, implying that the oxidation of NADPH through Fd to dPQ was coupled to the synthesis of ATP (supplemental Fig. S3). This result is consistent with the previously reported requirements of Fd for NDH activity (18).
The observed ATP synthesis was dependent on the generation of Δp as demonstrated by its abolition upon the addition of uncouplers (valinomycin with nigericin, see the arrow marked unc in Fig. 2B). A number of additional controls were performed to verify the identities of the processes participating to the ATP generation observed in this luciferase-based assay. FQR activity was not expected to contribute to the Δp generation observed here, and as expected ATP generation was insensitive to the addition of 10 μm antimycin A (considered an inhibitor of FQR activity; Ref. 17) (supplemental Fig. S4). The ATP synthesis was insensitive to tridecyl stigmatellin (supplemental Fig. S5), a potent inhibitor of the bf complex indicating that the observed ATP synthesis also did not involve electron or proton translocation by the bf complex. Likewise, the activity was insensitive to oligomycin (supplemental Fig. S5), a specific inhibitor of mitochondrial ATP synthase, indicating that the reaction was not catalyzed by contaminant mitochondrial respiration.
Linkage of NDH activity to generation of Δp probed by post illumination fluorescence kinetics
As a second, independent approach to determining the proton coupling of NDH, we assessed the effects of Δp on the reduction of the PQ pool using the “post illumination fluorescence” signal, a transient rise in chlorophyll fluorescence observed in the dark after a sustained period of actinic illumination in leaves and chloroplast preparations. This fluorescence signal is generally considered to be related to the activity of NDH and results from the transfer of electrons from stromal donors to the PQ pool, which equilibrates with the QA quinone molecule in photosystem II, resulting in elevated chlorophyll a fluorescence yield (Fig. 3A; also Ref. 12).
Figure 3.
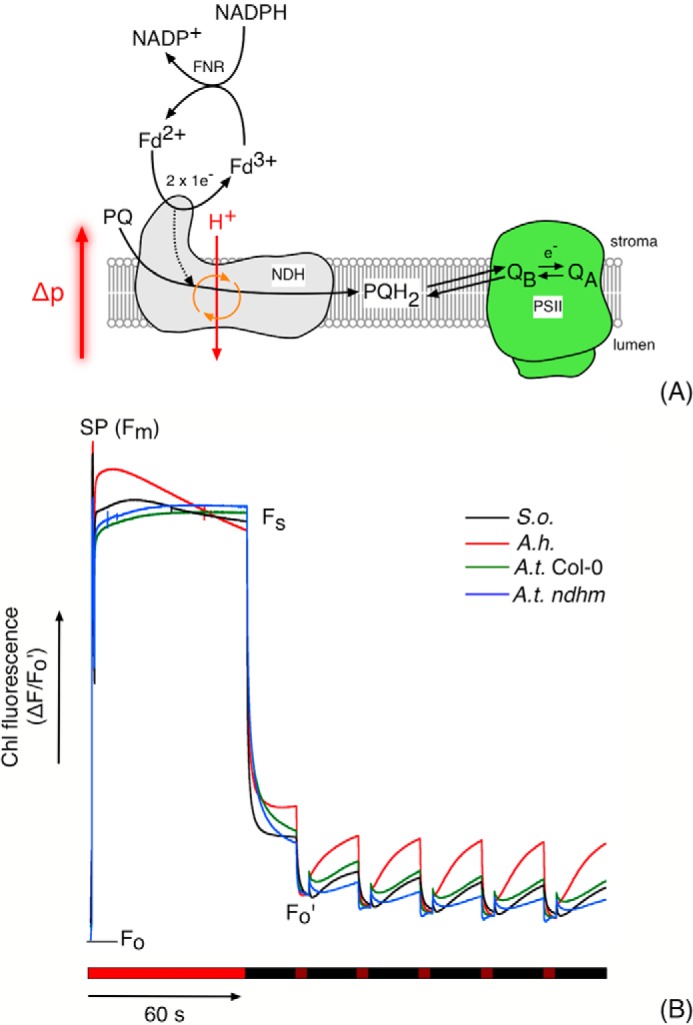
A, mechanism of association of NDH-mediated PQ pool reduction and the post illumination fluorescence rise. PSII, QA, and QB represent photosystem II and the primary and second quinone electron acceptors in PSII, respectively. As a protonmotive enzyme, NDH activity is subject to thermodynamic back pressure from the transthylakoid protonmotive force (Δp). B, the post illumination fluorescence rise in S. oleracea (S.o.), A. hybridus (A.h.), A. thaliana-Columbia (A.t. Col-0), and A.t. ndhM thylakoid preparations. Assay conditions consisted of thylakoids suspended at 50 μg Chl/ml in 10 mm HEPES (pH 7.5), 10 mm KCl, 5 mm MgCl2 supplemented with 5 μm Fd, and 100 μm NADPH. The periods of actinic (620 nm, 250 μmol of photons m−2 s−1) and far-red (720 nm, 50 μmol of photons m−2 s−1) illumination are indicated by bright and dark red bars under the fluorescence data. Periods of darkness are indicated by black bars. SP refers to a saturating actinic flash (5000 μmol of photons m−2 s−1). Fo, Fm, Fs, and Fo′ indicate the fluorescence levels in the dark, during the saturating flash, during the steady state under actinic illumination, and under far-red illumination respectively. The position of Fo is indicated by the gray bar. Representative unsmoothed data (normalized to Fo′) from three biological replicates are presented.
Fig. 3B compares the kinetics of fluorescence yield changes in thylakoid preparations from S. oleracea, Amaranthus hybridus, and A. thaliana (wild-type Col-0 and the NDH knock-out ndhm; Ref. 31) using 100 μm NADPH as the electron donor in the presence of exogenous (spinach) Fd.
The experiments were initiated by measurements of the fluorescence level in the dark, F0, followed by a brief pulse saturating actinic light to estimate for the maximal fluorescence yield, FM, allowing for normalization of results from preparation to preparation. This treatment was followed by illumination with ∼250 μmol of photons m−2 s−1 red light for 60 s to establish steady-state electron transfer. After illumination, the actinic light was switched off, and chlorophyll fluorescence yield was recorded. Upon switching off the actinic light, the fluorescence yield values decayed initially to a level well above the F0 value, likely indicating QA was not completely reoxidized in the dark. To fully oxidize PQ, we periodically illuminated with far-red light (50 μmol photons m−2 s−1), which preferentially excites PSI, resulting in quenching of the fluorescence signal. After the far-red light, fluorescence yield increased again, reflecting re-reduction of the PQ pool and QA by NADPH/Fd. These re-reduction responses, which we term post illumination fluorescence rise (PIFR), were maintained over multiple cycles of far-red illumination and dark recovery, indicating that the pool of reductant was not depleted during the experiments.
The largest PIFR response was observed in A. hybridus, a NAD-ME C4 species with an elevated level of NDH in mesophyll chloroplasts compared with bundle-sheath (32). The amplitude of the PIFR in S. oleracea and A. thaliana (Col-0) was ∼65 and 50% that observed in A. hybridus, respectively. The PIFR in Arabidopsis was diminished in the ndhm line, which lacks a functional NDH complex (31), confirming that the PIFR was related to NDH activity. The residual (20%) PIFR in ndhm presumably reflects the activity of the (non-protonmotive) FQR pathway or the recently demonstrated direct reduction of PSII electron acceptors (33). To test these assignments, we repeated the experiments in the presence of 1 mm hydroxylamine and 10 μm DCMU, abolishing variable fluorescence from PSII. The PIFR was inhibited under these conditions (supplemental Fig. S6), confirming that the PIFR observed in the absence of DCMU and hydroxylamine was related to PQ pool reduction (mediated by NDH) and subsequent redox equilibration with QA (34). An anaerobic control for plastid terminal (plastoquinol) oxidase (PTOX) activity on the kinetics of the PIFR was performed in spinach and A. hybridus thylakoid preparations (supplemental Fig. S7). When normalizing the fluorescence data to F0′, the extent of the A. hybridus PIFR was increased by 15–20% during the 20-s transient under anaerobic conditions compared with aerobic. The initial rate of the PIFR was also observed to increase ∼1.5-fold under anaerobic conditions in this species. The PIFR extents were unaffected by anaerobiosis in S. oleracea thylakoid preparations. These data presumably reflect higher PTOX activity in A. hybridus thylakoid preparations compared with S. oleracea.
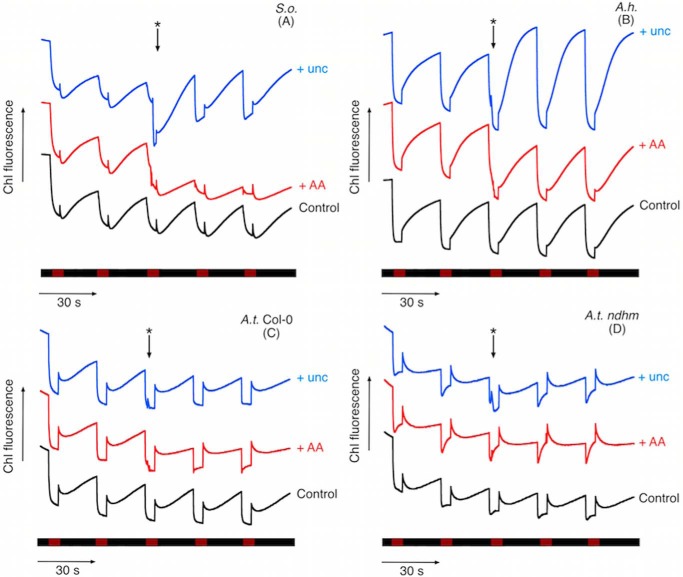
It is generally considered that the FQR-dependent pathway of CEF is sensitive to AA, whereas the NDH-dependent pathway is insensitive. The addition of 10 μm AA reduced the amplitude of the PIFR by ∼60% in S. oleracea and A. thaliana (Col-0) thylakoids and almost completely in A. thaliana (ndhm) thylakoid preparations (Fig. 4, A–D). In contrast, the amplitude of the rise in A. hybridus thylakoid preparations was largely unaffected by AA, consistent with an NDH-dominated CEF pathway in this species (32). These data provide a useful example for a point that is often overlooked in the literature; that is, that both NDH and FQR activity contribute to the PIFR (which is to be expected, because both activities result in quinone reduction) (35), and the contribution of these activities varies between species. Accordingly, the PIFR is reported to be diminished (but not abolished) in the A. thaliana pgr5 mutant (36), which is considered to be affected in CEF.
Figure 4.
The effect of uncoupling (unc, 10 μm nigericin + 10 μm valinomycin) and 10 μm AA on the post illumination fluorescence rise in S. oleracea (S.o.) (A), A. hybridus (A.h.) (B), A. thaliana-Columbia (A.t. Col-0) (C), and A.t. ndhM thylakoid (D) preparations. Effectors were added during the course of the experiments as indicated by the asterisked arrow above the fluorescence data. Other conditions are as in Fig. 3.
Collapsing the transthylakoid Δp through the use of valinomycin and nigericin in the presence of 10 mm K+, which should uncouple the thylakoid membranes, resulted in a rapid, 2-fold increase in the amplitude and rate of the PIFR observed in S. oleracea and A. hybridus chloroplast preparations (Fig. 4, A–D). Dissipation of the Δp allows the protonmotive NDH complex to operate unimpeded by thermodynamic “back pressure” created by the light-induced proton gradient across the thylakoid membrane. The PIFR was less sensitive to proton uncoupling in A. thaliana (Col-0) and ndhm mutant (Fig. 4, C and D), likely reflecting the larger contribution from the AA-sensitive, and non proton-pumping FQR pathways, which should not be hindered by Δp backpressure.
Proton-pumping activity of chloroplast NDH probed in vivo
As discussed in Sacksteder et al. (6) and Avenson et al. (37), the initial slope of the dark interval relaxation kinetics (DIRK) of the electrochromic shift (ECS) signal during brief dark intervals can be analyzed to obtain vH+, an estimate of the relative fluxes of protons generated by the light reactions. Previous work (38) showed the rates of CEF can be estimated by comparing vH+, which is driven by both LEF and CEF, with electron transfer through PSII, which only contributes to LEF. Here, we call this method the “proton-LEF” method.
Here we introduce a method, modified from the proton-LEF approach, to estimate the ratio of proton translocation to electron transfer through LEF and CEF. In the new approach we compared vH+ with light-driven proton fluxes to electron transfer through PSI (vP700), measured using the method modified by that of Fan et al. (65). Each turnover of LEF or CEF should result in one electron transferred to P700, so that
| (Eq. 1) |
By contrast, vH+ should depend on both the rates and proton stoichiometries of CEF and LEF; therefore,
| (Eq. 2) |
where nCEF and nLEF represent the H+/e− ratios for CEF and LEF. The slope of the relationship between vH+ and vP700 will be,
| (Eq. 3) |
where m = vH+/vP700. Our previous work showed that, in wild type A. thaliana, under non-stressed conditions, CEF is <10% that of LEF (37). By contrast, in the high cyclic electron flow 1 (hcef1) mutant, CEF is elevated, whereas LEF is decreased so that CEF contributes ∼90% of electron and proton fluxes (38). Thus, to the first approximation, Col-0 and hcef1 have essentially all LEF and CEF, respectively, and one can easily derive a rough estimate the relative proton to electron stoichiometries for CEF and LEF,
| (Eq. 4) |
where nCEF and nLEF are the proton to electron stoichiometries for CEF and LEF, and mCol-0 and mhcef1 are the ratios of vH+ to vP700 (for Col-0 and hcef1, respectively (we also derived a more complex equation that considers the residual levels of LEF and CEF in Col-0 and hcef1, but the results were within 20% that of those using the simplified equation above). The value of nLEF is likely to be 3 (6), i.e. for each electron through LEF one proton is released from water oxidation and two were from the Q-cycle. In the absence of a proton pumping PQ reductase, nCEF = 2, i.e. 2 protons transported by the Q-cycle, and thus, we would expect mhcef1 < mCol-0. If CEF occurs through an NDH that pumps on proton per electron, we would expect mhcef1 ≈ mCol-0. Finally, if NDH pumps 2 protons per e− (i.e. 4 protons per NAD(P)H) we would expect mhcef1 > mCol-0, as we clearly observes. If one assumes that the contributions to CEF were constant throughout the experiments, then the ratio of slopes for the relationship between vH+ and vP700 should approximate the ratio of n (H+/e−) values. Assuming n = 3 for LEF, fitting the data in Fig. 5 yields a wide range of possible values for H+/e− ratio of slopes, from ∼4.5 to 8.8. Given this broad uncertainty, it is probably not possible to obtain an accurate estimate of n from these data. However, because the points for hcef1 (predominantly CEF) all fall above the curve for the wild type, which is predominantly LEF, we can make the quantitative conclusion that H+/e− for CEF is larger than that for LEF in this mutant.
Figure 5.
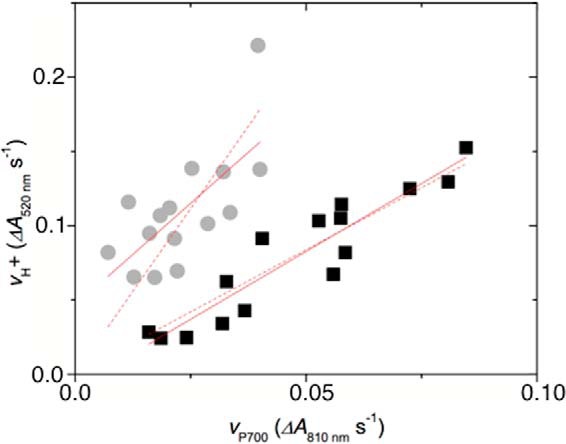
Evidence for involvement of a proton pump in CEF in vivo. Relative rates of light-induced thylakoid proton flux (vH+) were estimated by the initial slope of the decay of the electrochromic shift. The rates of turnover PSI (vP700) were estimated by the rate of decay of the P700+ absorbance changes upon rapid transition from light to dark under conditions where P700+ accumulates (700-nm illumination). Data are from intact leaves of Col-0 (black) and hcef1 (gray) under increasing light intensities. Lines represent best free fit of the data (solid lines, slopes of 1.8 and 2.75 for Col-0 and hcef1) or forced through the origin (dashed lines, slopes of 1.6 ± 0.3 and 4.7 ± 0.8 for Col-0 and hcef1).
Discussion
The function of plastid NDH complex has been the subject of considerable controversy (39). NDH is known to catalyze the reduction of PQ by reduced Fd and thus has often been suggested to play a role in CEF (18, 35, 38, 40). However, other CEF pathways, particularly that involving the FQR, have been proposed to constitute the major route for CEF (14–16, 41), although it is clear that CEF can proceed rapidly even in the absence of FQR, through the NDH complex (35, 38, 40, 42).
The fact that NDH is homologous to proton-pumping Complex I (NADH:UQ oxidoreductase) of mitochondria and related prokaryotic systems suggests proton-pumping capacity may also be conserved in plastids, with substantial consequences for the energetics of CEF. In support of this possibility, key residues required for proton-pumping (22, 28, 43) are also conserved in the plastid complex (Figs. 1 and supplemental Fig. S2).
In this work we used several complementary approaches to provide the first direct evidence for the predicted proton-pumping capacity of NDH. In one approach we developed an in vitro assay that reported dark ATP synthesis in coupled, thylakoid preparations under conditions strictly dependent upon NDH activity (Fig. 2 and supplemental Figs. S3–S5). Secondly, the extent of the PIFR, a chlorophyll fluorescence phenomenon arising from the NDH-mediated reduction of the PQ pool (Fig. 3), increased on the addition of uncouplers to thylakoid preparations from S. oleracea, A. hybridus, and A. thaliana (Col-0) (Fig. 4). This latter observation is consistent with a protonmotive NDH complex with a pumping (H+/e−) stoichiometry of two or more so that its enzymatic activity is controlled by the thermodynamic back pressure from transmembrane Δp (as discussed below, proton-pumping capacities of <2H+/e− should not be thermodynamically limited by physiologically accessible Δp extents.) Finally, in vivo data obtained on the high CEF hcef1 mutant indicated a H+/e− stoichiometry for CEF of about four (Fig. 5), requiring an NDH that pumps about two protons per electron transferred to PQ, consistent with the published ratios for respiratory Complex I of ∼2H+/e− (22, 43, 44).
A proton-pumping NDH should have several important consequences. First, it effectively doubles the amount of ATP produced for a given rate of CEF, making it a more efficient mechanism to balance the ATP/NADPH budget, i.e. compared with non-proton-pumping FQR, fewer turnovers would be required to sustain the required ATP production. The actual contribution of NDH-related CEF to chloroplast bioenergetics has been in dispute. Estimates suggest a chloroplast NDH content of ∼0.05 per PSI in N. tabacum (12). To meet the ATP demands imposed by the Calvin-Benson cycle, NDH-mediated CEF would need to translocate protons at a rate of ∼13% the LEF rate (37). With a fixed ratio of 3 H+/e− for LEF, a 1H+/e− NDH would need to turnover 13% the rate of LEF, whereas a 2H+/e− NDH would need to turnover 9% of the rate of LEF. Assuming NDH has an activity similar to that of respiratory Complex I, which has maximal sustained turnover numbers of ∼200 e− s−1 (45, 46), we estimate that wild-type levels of NDH could support a minimum rate of CEF of ∼4 e− s−1 per PSI complex or ∼4% of maximal LEF. This rate, in the absence of environmental stresses, is reasonable, as NDH likely works in parallel with the FQR complex and other processes like the malate shunt or the water-water cycle (for review, see Ref. 4) to balance the chloroplast energy budget. Under stressed conditions leading to ROS formation (47, 48), in chloroplasts of certain C4 plants (32), and in the high CEF mutant hcef1 (38, 40), where high rates of CEF are observed, the content of NDH subunits increase substantially, likely allowing the bulk transfer of electrons through NDH-mediated CEF to be much higher. For example, ∼15-fold higher levels of NDH were observed in the high CEF mutant hcef1 (38).
Furthermore, a proton-pumping NDH is expected to be required for non-photosynthetic energy transduction in higher plant plastids, as in the proposed chlororespiratory pathway (49–51) to involve NDH and the plastid terminal oxidase. Our finding that NDH is a proton pump adds critical support for this possibility because without coupling PQ reduction to proton translocation the proposed pathway would not be able to conserve energy in Δp and ATP synthesis (see also Ref. 26).
Although a proton-pumping NDH provides a higher ATP output for CEF, the coupling of the forward reaction to the generation of Δp should also thermodynamically constrain the extent of the overall reaction. At equilibrium, the relationship between protons translocated into the lumen against Δp per electron traversing a redox span of ΔEh mV is given by Equation 5,
| (Eq. 5) |
where n represents the H+/e− ratio. With Fd as the electron donor to NDH, ΔEh equals 520 mV assuming 90% reduction of both the PQ and Fd pools (conditions that may be met during photosynthetic induction) at a stromal pH of 7.5. Assuming a Δp of 180 mV across the thylakoid membrane in the light, NDH would be capable of acting as a 2H+/e− pump. This assertion also holds for a predominantly oxidized (90%) Fd pool. However, if the redox state of Fd comes into equilibrium with that of NADPH, as would be expected under many conditions (e.g. when LEF is limited by turnover of the Calvin-Benson cycle, diminishing kinetic controlling factors on NDH activity), then the NDH energetics become significantly more constrained. As illustrated in supplemental Fig. S8, certain physiologically relevant combinations of redox poise and Δp would limit the forward (plastoquinone reductase) NDH reaction and may even allow for reverse reactions as discussed further below. One may expect these conditions to be produced during rapid light transients where large Δp can be generated (1, 9).
The predicted thermodynamic linkage between PQ reduction and proton translocation is clearly demonstrated in our results showing that PIFR associated with CEF is constrained by Δp (Fig. 4). It is noteworthy that we would not expect to observe significant thermodynamic backpressure if H+/e− was lower than two (44). For example, if H+/e− for NDH was 1, the equilibrium constant for reduction of PQ by NADPH with Δp = 180 mV should be 5 × 106, allowing for essentially full reduction of PQ and QA. This analysis thus strongly supports the involvement of a NDH with a coupling stoichiometry of 2H+/e−. In addition, the quinone reductase reaction catalyzed by a 1H+/e− NDH (or non-protonmotive enzyme such as NDA/NDH2; Ref. 23) would be insensitive to the presence of uncouplers under physiological conditions and would not manifest the transient post illumination fluorescence rise response observed in Fig. 4 upon valinomycin and nigericin addition. Similarly, CEF catalyzed by the PGR5/FQR-pathway as observed via the PIFR would be expected to be uncoupler-insensitive.
This thermodynamic backpressure effect on NDH also likely explains the unusual kinetics of the transient PIFR seen in vivo, which appears to indicate a rate of PQ reduction that is much slower than could sustain expected rates of CEF (see Discussion in Ref. 52). More likely, the slow kinetics represents a combination of NDH activity and the relaxation of Δp in the dark that allows the reaction to proceed over a slow time course dictated by dissipation of Δp in the dark. As such, the previous conclusion (52) that the PIFR represents a very slow rate of PQ pool reduction should be reassessed, as these kinetics probably reflect, not a slow NDH, but the shifting of equilibrium between stromal reductants and the PQ pool as Δp is dissipated in the dark. PTOX activity, acting as a slow drain of electrons from the PQ pool under aerobic conditions (49, 50), may also be expected to modify the kinetics of the fluorescence transients observed in Fig. 3, although the effect of anaerobiosis on the PIFR transients was observed to be relatively minor in S. oleracea and A. hybridus (supplemental Fig. S7).
The thermodynamic sensitivity of the reaction catalyzed by NDH would provide a simple “self-control” mechanism for regulation of the chloroplast energy budget under conditions of low ATP demand (i.e. high ΔGATP) and favors the partitioning of electrons into downstream metabolism. For instance, a deficit of ATP would favor a low Δp and a more reduced NADPH pool, allowing the NDH reaction to proceed. An excess of ATP might result in high Δp, constraining the quinone reductase activity of NDH. The fact that the extent of the NDH reaction is limited by Δp implies that the system reaches quasi-equilibrium; the enzyme can be considered to be approaching the “static head” condition at normal operating Δp (≈180 mV). As such, increasing Δp should favor the operation of NDH in the “reverse” direction (assuming no other limiting kinetic factors), consuming Δp, and functioning as a plastoquinol:NADP+ oxidoreductase under conditions of a moderately high Δp (>180 mV) when the PQ pool is predominantly (90%) reduced (supplemental Fig. S8, A–C). This condition would fall within the expected ranges of redox and Δp conditions expected in vivo and might be expected to occur under fluctuating environmental conditions. Although in the current study we do not directly demonstrate that the reverse reaction occurs under physiological conditions, we note that functional reversal of NDH-like complexes has been observed in respiratory homologs of this enzyme (53–55).
A 1H+/e−-coupled (or non-protonmotive) NDH would be energetically incapable of significant reverse reaction under physiological conditions. The reverse reaction catalyzed by the 2H+/e− thylakoid NDH becomes increasingly energetically favorable as the pool of oxidized acceptor (NADP+) and magnitude of Δp increases. With Fd as the electron acceptor, the reversibility of NDH is expected to be strongly thermodynamically limited below a Δp of ≈ 200 mV (supplemental Fig. S8C). However, rapid oxidation of Fd molecules (by ferredoxin:NADP oxidoreductase and other downstream processes) reduced by PSI and NDH activity would facilitate this reverse reaction. Thus, under conditions of high Δp, we propose that chloroplasts may conduct electron flow from water, via PSII, to NADPH without the direct participation of PSI, effectively a “pseudo-linear” pathway for photosynthetic electron transfer that could operate under special conditions of high Δp and reduced PQH2. NDH may be considered to be acting as a “metabolic buffer” enzyme in this hypothesis, smoothing oscillations in ATP/NADPH supply and demand. For example, activation of the pseudo-linear pathway may have large consequences for regulation of metabolic processes that are modulated by the redox state of NADPH through the thioredoxin system, possibly explaining why NDH knockouts exhibit strong phenotypes during the induction of photosynthesis when high Δp and imbalances of redox state are expected or when subjected to environmental stress, such as decreased CO2, fluctuating light intensity, or chilling (56, 57). A similar pseudo-linear electron transfer pathway for photoreduction of NADP+ in the absence of photosystem I activity has been postulated to exist in the alga Chlamydomonas reinhardtii, although this organism was later found to lack a protonmotive plastid NDH complex (50, 58).
The trade-offs between efficiency and thermodynamic constraints may also explain why there are multiple routes of differentially regulated CEF pathways in chloroplasts. Additionally, as noted by Allen (59), the redox poise of the electron donor and acceptor pool will also affect CEF activity, with FQR and NDH potentially in competition for substrate. Under some conditions, highly efficient, but slower, synthesis of ATP via the NDH complex would be favored. However, NDH activation may be slow, taking ∼20 min (40), and a more rapidly activated pathway may be needed for rapid responses. In addition, there may also be conditions where higher Δp than that afforded by the NDH pathway may be beneficial, perhaps to acidify the lumen and down-regulate photosynthesis (9). We thus propose that the NDH and FQR modes of CEF are activated stepwise when the downstream metabolism decreases ATP/NADPH. First, a reducing environment is generated as NADPH accumulates, and CEF is activated through the non-proton-pumping, FQR-dependent (antimycin A-sensitive) pathway. Under conditions in which this route of CEF is not able to augment the ATP deficit and restore redox homeostasis to the chloroplast, ROS is generated and activates the proton-pumping NDH complex. This would increase the yield of ATP formation per e− transfer, increasing the efficiency of ATP production via CEF. This route of CEF is not as rapidly activated as the FQR, and long-term ROS generation leads to not only activation of already assembled complexes but an increase in total NDH content (40, 47, 48).
Experimental procedures
Reagents, plant materials, and growth conditions
Reagents were purchased from Sigma unless otherwise noted.
A. thaliana and A. hybridus (the latter supplied by Swallowtail Garden Seeds, Santa Rosa, CA) plants were grown in a 16:8 light:dark photoperiod (white fluorescent illumination at 100 μmol of photons m−2 s−1) at 21 °C. Experiments using Col-0 and A. hybridus and were performed at 3–4 weeks of age, whereas slow-growing hcef1 was used at the same developmental stage as Col-0 (around 6–7 weeks of age). Chloroplasts were prepared from A. hybridus, Col-0 and ndhm (Salk_087707) plants 3 weeks after germination. Spinach was obtained from a local market, with chloroplasts prepared on the morning of purchase.
Chloroplasts were prepared from S. oleracea, A. thaliana, and A. hybridus leaves as described in Seigneurin Berny et al. (60), and chlorophyll was determined as in Porra et al. (61). Chloroplast intactness with regard to proton coupling was routinely monitored using the Δp-dependent acridine dye fluorescence quenching assay described (62).
Spectroscopic measurements
All spectroscopic measurements were made using an integrated diode emitter array (IDEA) spectrophotometer/fluorimeter (63). Plants were poised by illuminating with 700-nm actinic light to favor oxidation of PSI and enable measurement of electron flux through PS I by analysis of dark interval relaxation kinetics.
Transthylakoid proton flux (vH+, ΔA520 nm m−2 s−1) was calculated using the ECS of the carotenoids at 520 nm as described in Strand et al. (40) and Avenson et al. 64 and references within supplemental Fig. S9. To correct for variability in leaf pigmentation between Col-0 and hcef1, the total extent of ECS in hcef1 was normalized to the fraction of Col-0 per area of leaf chlorophyll content as described previously for hcef1 (38, 42).
The redox state of PSI was monitored using absorbance changes at 820 nm in a protocol modified from Fan et al. (65). The initial rate of P700+ re-reduction kinetics were used as the relative rate of electron transfer through PSI (vP700, ΔA820 nm m−2 s−1) (supplemental Fig. S9).
Post illumination chlorophyll a fluorescence transients were measured as described in Burrows et al. (66) and Shikanai et al. (66) with modifications. Thylakoids were preilluminated with 250 μmol of photons m−2 s−1 (620 nm) for 1 min. After the light to dark transition, the measuring beam (505 nm, 0.5 μmol of photons m−2 s−1) was pulsed at a 10-Hz interval. Chlorophyll fluorescence changes during the dark interval were monitored for 145 s, with 5-s pulses of far-red illumination (720 nm, 50 μmol of photons m−2 s−1) every 20 s.
In vitro ATP synthesis assays
Proton-pumping in vitro was measured as ATP production in the dark using the Promega (Enliten) luciferase/luciferin reagent kit with a laboratory-constructed photomultiplier-based phosphoroscope. Osmotically ruptured S. oleracea chloroplasts (prepared in the presence of 2 mm reduced DTT) were present at a chlorophyll concentration of 50 μg/ml. DCMU was present at 10 μm. Fd and NADPH were present at 5 μm and 100 μm, respectively. The assay buffer (pH 7.5) consisted of 10 mm HEPES, 2 mm potassium phosphate, 10 mm KCl, 5 mm MgCl2, and 2 mm DTT and was supplemented with 2 mm ADP and 100 μm diadenosine pentaphosphate (an adenylate kinase inhibitor). Premixed Enliten recombinant luciferase/luciferin reagent (used as supplied by Promega) was added to a final concentration of 8% (v/v). Proton-pumping was initiated by the addition of 50 μm dPQ, and the proton gradient was collapsed by the addition of 10 μm nigericin and 10 μm valinomycin. The plastid ATP synthase was activated by red actinic illumination (50 μmol of photons m−2 m−1, 625 nm) of the chloroplast suspension for 2 min immediately before the addition of DCMU, ADP, Enliten reagent, and the start of data collection.
Author contributions
D. D. S and N. F. designed the experiments and collected, analyzed, and interpreted the data. D. M. K. designed the experiments, constructed the instrumentation, and analyzed and interpreted the data. All authors contributed to the drafting of the manuscript, and all authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgment
We thank Dr. Jeffrey Cruz for helpful discussions.
This work was supported by Grant DE-FG02-11ER16220 from the Photosynthetic Systems program from Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy (to D. M. K.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S9 and Equation 2.
We dedicate this paper to the memory of Dr. Derek Bendall (1930–2014), pioneer of cyclic electron flow research and mentor to N. F.
- LEF
- linear electron flow
- PSI
- Photosystem I
- PSII
- Photosystem II
- Δp
- protonmotive force
- vH+
- transthylakoid proton flux
- vP700
- rate of PSI turnover
- AA
- antimycin A
- CEF
- cyclic electron flow
- DCMU
- dichloromethyl urea
- (d)PQ
- (decyl)plastoquinone
- ECS
- electrochromic shift
- Fd
- ferredoxin
- FQR
- ferredoxin:(plasto)quinone reductase
- NDA/NDH2
- non-protonmotive NAD(P)H dehydrogenases
- NDH
- (protonmotive) NAD(P)H dehydrogenase complex
- PIFR
- post illumination fluorescence rise
- PTOX
- plastid terminal (plastoquinol) oxidase.
References
- 1. Eberhard S., Finazzi G., and Wollman F.-A. (2008) The dynamics of photosynthesis. Annu. Rev. Genet. 42, 463–515 [DOI] [PubMed] [Google Scholar]
- 2. Kramer D. M., Avenson T. J., Kanazawa A., Cruz J. A., Ivanov B., and Edwards G. (2004), The relationship between photosynthetic electron transfer and its regulation. In Chlorophyll a fluorescence: a signature of photosynthesis (Papageorgiou G. C., and Govindjee, ed.) pp 251–278, Springer, Dordrecht, The Netherlands [Google Scholar]
- 3. Cruz J. A., Avenson T. J., Kanazawa A., Takizawa K., Edwards G. E., and Kramer D. M. (2005) Plasticity in light reactions of photosynthesis for energy production and photoprotection. J. Exp. Bot. 56, 395–406 [DOI] [PubMed] [Google Scholar]
- 4. Kramer D. M., and Evans J. R. (2011) The importance of energy balance in improving photosynthetic productivity. Plant Physiol. 155, 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mitchell P. (1975) Protonmotive Q-cycle-general formulation. FEBS Lett. 59, 137–139 [DOI] [PubMed] [Google Scholar]
- 6. Sacksteder C. A., Kanazawa A., Jacoby M. E., and Kramer D. M. (2000) The proton to electron stoichiometry of steady-state photosynthesis in living plants: a proton-pumping Q cycle is continuously engaged. Proc. Natl. Acad. Sci. U.S.A. 97, 14283–14288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cramer W. A., Hasan S. S., and Yamashita E. (2011) The Q cycle of cytochrome bc complexes: a structure perspective. Biochim. Biophys. Acta 1807, 788–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poetsch A., Rexroth S., Heberle J., Link T. A., Dencher N. A., and Seelert H. (2003) Characterisation of subunit III and its oligomer from spinach chloroplast ATP synthase. Biochim. Biophys. Acta 1618, 59–66 [DOI] [PubMed] [Google Scholar]
- 9. Joliot P., and Johnson G. N. (2011) Regulation of cyclic and linear electron flow in higher plants. Proc. Natl. Acad. Sci. U.S.A. 108, 13317–13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iwai M., Takizawa K., Tokutsu R., Okamuro A., Takahashi Y., and Minagawa J. (2010) Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464, 1210–1213 [DOI] [PubMed] [Google Scholar]
- 11. Strand D. D., and Kramer D. M. (2014) Control of non-photochemical exciton quenching by the proton circuit of photosynthesis. In Non-Photochemical Fluorescence Quenching and Energy Dissipation in Plants, Algae, and Cyanobacteria in Advances in Photosynthesis and Respiration (Demmig-Adams B., Garab G., Adams W. W. III, and Govindjee, eds.) pp 387–408, Springer, Dordrecht, The Netherlands [Google Scholar]
- 12. Burrows P. A., Sazanov L. A., Svab Z., Maliga P., and Nixon P. J. (1998) Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17, 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peng L., Yamamoto H., and Shikanai T. (2011) Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim. Biophys. Acta 1807, 945–953 [DOI] [PubMed] [Google Scholar]
- 14. Bendall D., and Manasse R. (1995) Cyclic photophosphorylation and electron-transport. Biochim. Biophys. Acta 1229, 23–38 [Google Scholar]
- 15. Munekage Y., Hojo M., Meurer J., Endo T., Tasaka M., and Shikanai T. (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110, 361–371 [DOI] [PubMed] [Google Scholar]
- 16. DalCorso G., Pesaresi P., Masiero S., Aseeva E., Schünemann D., Finazzi G., Joliot P., Barbato R., and Leister D. (2008) A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132, 273–285 [DOI] [PubMed] [Google Scholar]
- 17. Hertle A. P., Blunder T., Wunder T., Pesaresi P., Pribil M., Armbruster U., and Leister D. (2013) PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol. Cell 49, 511–523 [DOI] [PubMed] [Google Scholar]
- 18. Yamamoto H., Peng L., Fukao Y., and Shikanai T. (2011) An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23, 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sazanov L. A., Burrows P. A., and Nixon P. J. (1998) The plastid ndh genes code for an NADH-specific dehydrogenase: isolation of a complex I analogue from pea thylakoid membranes. Proc. Natl. Acad. Sci. U.S.A. 95, 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Friedrich T., and Scheide D. (2000) The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 479, 1–5 [DOI] [PubMed] [Google Scholar]
- 21. Efremov R. G., Baradaran R., and Sazanov L. A. (2010) The architecture of respiratory complex I. Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 22. Friedrich T. (2014) On the mechanism of respiratory complex I. J. Bioenerg. Biomembr. 46, 255–268 [DOI] [PubMed] [Google Scholar]
- 23. Melo A. M., Bandeiras T. M., and Teixeira M. (2004) New insights into type II NAD(P)H:quinone oxidoreductases. Microbiol. Mol. Biol. Rev. 68, 603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Desplats C., Mus F., Cuiné S., Billon E., Cournac L., and Peltier G. (2009) Characterization of Nda2, a plastoquinone-reducing type II NAD(P)H dehydrogenase in Chlamydomonas chloroplasts. J. Biol. Chem. 284, 4148–4157 [DOI] [PubMed] [Google Scholar]
- 25. Wang C., Yamamoto H., and Shikanai T. (2015) Role of cyclic electron transport around photosystem I in regulating proton motive force. Biochim. Biophys. Acta 1847, 931–938 [DOI] [PubMed] [Google Scholar]
- 26. Nawrocki W. J., Tourasse N. J., Taly A., Rappaport F., and Wollman F.-A. (2015) The plastid terminal oxidase: its elusive function points to multiple contributions to plastid physiology. Annu. Rev. Plant Biol. 66, 49–74 [DOI] [PubMed] [Google Scholar]
- 27. Baradaran R., Berrisford J. M., Minhas G. S., and Sazanov L. A. (2013) Crystal structure of the entire respiratory complex I. Nature 494, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hunte C., Zickermann V., and Brandt U. (2010) Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329, 448–451 [DOI] [PubMed] [Google Scholar]
- 29. Steimle S., Bajzath C., Dörner K., Schulte M., Bothe V., and Friedrich T. (2011) Role of subunit NuoL for proton translocation by respiratory complex I. Biochemistry 50, 3386–3393 [DOI] [PubMed] [Google Scholar]
- 30. Davenport J. Q., and McCarty R. E. (1986) Relationships between rates of steady-state ATP synthesis and the magnitude of the proton-activity gradient across thylakoid membranes. Biochim. Biophys. Acta 851, 136–145 [Google Scholar]
- 31. Rumeau D., Bécuwe-Linka N., Beyly A., Louwagie M., Garin J., and Peltier G. (2005) New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell 17, 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takabayashi A., Kishine M., Asada K., Endo T., and Sato F. (2005) Differential use of two cyclic electron flows around photosystem I for driving CO2-concentration mechanism in C4 photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 102, 16898–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fisher N., and Kramer D. M. (2014) Non-photochemical reduction of thylakoid photosynthetic redox carriers in vitro: Relevance to cyclic electron flow around photosystem I? Biochim. Biophys. Acta 1837, 1944–1954 [DOI] [PubMed] [Google Scholar]
- 34. Diner B. A. (1977) Dependence of the deactivation reactions of photosystem II on the redox state of plastoquinone pool A varied under anaerobic conditions; equilibria on the acceptor side of photosystem II. Biochim. Biophys. Acta 460, 247–258 [DOI] [PubMed] [Google Scholar]
- 35. Sazanov L. A., Burrows P. A., and Nixon P. J. (1998) The chloroplast Ndh complex mediates the dark reduction of the plastoquinone pool in response to heat stress in tobacco leaves. FEBS Lett. 429, 115–118 [DOI] [PubMed] [Google Scholar]
- 36. Gotoh E., Matsumoto M., Ogawa K., Kobayashi Y., and Tsuyama M. (2010) A qualitative analysis of the regulation of cyclic electron flow around photosystem I from the post-illumination chlorophyll fluorescence transient in Arabidopsis: a new platform for the in vivo investigation of the chloroplast redox state. Photosynth. Res. 103, 111–123 [DOI] [PubMed] [Google Scholar]
- 37. Avenson T. J., Cruz J. A., Kanazawa A., and Kramer D. M. (2005) Regulating the proton budget of higher plant photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 102, 9709–9713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Livingston A. K., Cruz J. A., Kohzuma K., Dhingra A., and Kramer D. M. (2010) An Arabidopsis mutant with high cyclic electron flow around photosystem I (hcef) involving the NADPH dehydrogenase complex. Plant Cell 22, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shikanai T. (2007) Cyclic electron transport around photosystem I: genetic approaches. Annu. Rev. Plant Biol. 58, 199–217 [DOI] [PubMed] [Google Scholar]
- 40. Strand D. D., Livingston A. K., Satoh-Cruz M., Froehlich J. E., Maurino V. G., and Kramer D. M. (2015) Activation of cyclic electron flow by hydrogen peroxide in vivo. Proc. Natl. Acad. Sci. U.S.A. 112, 5539–5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Munekage Y., Hashimoto M., Miyake C., Tomizawa K.-I., Endo T., Tasaka M., and Shikanai T. (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429, 579–582 [DOI] [PubMed] [Google Scholar]
- 42. Livingston A. K., Kanazawa A., Cruz J. A., and Kramer D. M. (2010) Regulation of cyclic electron flow in C3 plants: differential effects of limiting photosynthesis at ribulose-1,5-bisphosphate carboxylase/oxygenase and glyceraldehyde-3-phosphate dehydrogenase. Plant Cell Environ. 33, 1779–1788 [DOI] [PubMed] [Google Scholar]
- 43. Sazanov L. A. (2015) A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 16, 375–388 [DOI] [PubMed] [Google Scholar]
- 44. Wikström M., and Hummer G. (2012) Stoichiometry of proton translocation by respiratory complex I and its mechanistic implications. Proc. Natl. Acad. Sci. U.S.A. 109, 4431–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hirst J. (2013) Mitochondrial complex I. Annu. Rev. Biochem. 82, 551–575 [DOI] [PubMed] [Google Scholar]
- 46. Fato R., Estornell E., Di Bernardo S., Pallotti F., Parenti Castelli G., and Lenaz G. (1996) Steady-state kinetics of the reduction of coenzyme Q analogs by complex I (NADH:ubiquinone oxidoreductase) in bovine heart mitochondria and submitochondrial particles. Biochemistry 35, 2705–2716 [DOI] [PubMed] [Google Scholar]
- 47. Casano L. M., Martín M., and Sabater B. (2001) Hydrogen peroxide mediates the induction of chloroplastic Ndh complex under photooxidative stress in barley. Plant Physiol. 125, 1450–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lascano H. R., Casano L. M., Martín M., and Sabater B. (2003) The activity of the chloroplastic Ndh complex is regulated by phosphorylation of the NDH-F subunit. Plant Physiol. 132, 256–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bennoun P. (2002) The present model for chlororespiration. Photosynth. Res. 73, 273–277 [DOI] [PubMed] [Google Scholar]
- 50. Peltier G., and Cournac L. (2002) Chlororespiration. Annu. Rev. Plant Biol. 53, 523–550 [DOI] [PubMed] [Google Scholar]
- 51. Rumeau D., Peltier G., and Cournac L. (2007) Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 30, 1041–1051 [DOI] [PubMed] [Google Scholar]
- 52. Groom Q. J., Kramer D. M., Crofts A. R., and Ort D. R. (1993) The non-photochemical reduction of plastoquinone in leaves. Photosynth. Res. 36, 205–215 [DOI] [PubMed] [Google Scholar]
- 53. Jackson J. B., and Crofts A. R. (1968) Energy-linked reduction of nicotinamide adenine dinucleotides in cells of Rhodospirillum rubrum. Biochem. Biophys. Res. Commun. 32, 908–915 [DOI] [PubMed] [Google Scholar]
- 54. Cobley J. G. (1976) Energy-conserving reactions in phosphorylating electron-transport particles from Nitrobacter winogradskyi: activation of nitrite oxidation by the electrical component of the protonmotive force. Biochem. J. 156, 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hirst J., King M. S., and Pryde K. R. (2008) The production of reactive oxygen species by complex I. Biochem. Soc. Trans. 36, 976–980 [DOI] [PubMed] [Google Scholar]
- 56. Horváth E. M., Peter S. O., Joët T., Rumeau D., Cournac L., Horváth G. V., Kavanagh T. A., Schäfer C., Peltier G., and Medgyesy P. (2000) Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 123, 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li X. G., Duan W., Meng Q. W., Zou Q., and Zhao S. J. (2004) The function of chloroplastic NAD(P)H dehydrogenase in tobacco during chilling stress under low irradiance. Plant Cell Physiol 45, 103–108 [DOI] [PubMed] [Google Scholar]
- 58. Peltier G., and Thibault P. (1988) Oxygen exchange studies in Chlamydomonas mutants deficient in photosynthetic electron transport: evidence for photosystem II-dependent oxygen uptake in vivo. Biochim. Biophys. Acta 936, 319–324 [Google Scholar]
- 59. Allen J. F. (2003) Cyclic, pseudocyclic and noncyclic photophosphorylation: new links in the chain. Trends Plant Sci. 8, 15–19 [DOI] [PubMed] [Google Scholar]
- 60. Seigneurin Berny D., Salvi D., Joyard J., and Rolland N. (2008) Purification of intact chloroplasts from Arabidopsis and spinach leaves by isopycnic centrifugation. Curr. Protoc. Cell Biol. 40, 3.30.1–3.30.14 ( 10.1002/0471143030.cb0330s40) [DOI] [PubMed] [Google Scholar]
- 61. Porra R. J., Thompson W., and Kriedemann P. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 [Google Scholar]
- 62. Strand D. D., Fisher N., Davis G. A., and Kramer D. M. (2016) Redox regulation of the antimycin A sensitive pathway of cyclic electron flow around photosystem I in higher plant thylakoids. Biochim. Biophys. Acta 1857, 1–6 [DOI] [PubMed] [Google Scholar]
- 63. Hall C. C., Cruz J., Wood M., Zegerac R., De Mars D., Carpenter J. F., Kanazawa A., and Kramer D. M. (2013) Photosynthetic measurements with the Idea Spec: an integrated diode emitter array spectrophotometer/fluorometer. In Photosynthesis Research for Food, Fuel, and Future (Kuang T., Lu C., and Zhang L., eds.) pp 184–188, Springer, Heidelberg, Germany [Google Scholar]
- 64. Avenson T. J., Cruz J. A., and Kramer D. M. (2004) Modulation of energy-dependent quenching of excitons in antennae of higher plants. Proc. Natl. Acad. Sci. U.S.A. 101, 5530–5535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fan D. Y., Nie Q., Hope A. B., Hillier W., Pogson B. J., and Chow W. S. (2007) Quantification of cyclic electron flow around photosystem I in spinach leaves during photosynthetic induction. Photosynth. Res. 94, 347–357 [DOI] [PubMed] [Google Scholar]
- 66. Shikanai T., Endo T., Hashimoto T., Yamada Y., Asada K., and Yokota A. (1998) Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. U.S.A. 95, 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brandt U. (2006) Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75, 69–92 [DOI] [PubMed] [Google Scholar]
- 68. Hu P., Lv J., Fu P., and Hualing M. (2013) Enzymatic characterization of an active NDH complex from Thermosynechococcus elongatus. FEBS Lett. 587, 2340–2345 [DOI] [PubMed] [Google Scholar]
- 69. Yamamoto H., and Shikanai T. (2013) In planta mutagenesis of Src homology 3 domain-like fold of NdhS, a ferredoxin-binding subunit of the chloroplast NADH dehydrogenase-like complex in Arabidopsis: a conserved Arg-193 plays a critical role in ferredoxin binding. J. Biol. Chem. 288, 36328–36337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ifuku K., Endo T., Shikanai T., and Aro E.-M. (2011) Structure of the chloroplast NADH dehydrogenase-like complex: nomenclature for nuclear-encoded subunits. Plant Cell Physiol. 52, 1560–1568 [DOI] [PubMed] [Google Scholar]
- 71. Suorsa M., Sirpiö S., and Aro E.-M. (2009) Towards characterization of the chloroplast NAD(P)H dehydrogenase complex. Mol. Plant 2, 1127–1140 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.