Abstract
RNA polymerase II (pol II) is required for the transcription of all protein-coding genes and as such represents a major enzyme whose activity is tightly regulated. Transcriptional initiation therefore requires numerous general transcriptional factors and cofactors that associate with pol II at the core promoter to form a pre-initiation complex. Transcription factor IIA (TFIIA) is a general cofactor that binds TFIID and stabilizes the TFIID–DNA complex during transcription initiation. Previous studies showed that TFIIA can make contact with the DNA sequence upstream or downstream of the TATA box, and that the region bound by TFIIA could overlap with the elements recognized by another factor, TFIIB, at adenovirus major late core promoter. Whether core promoters contain a DNA motif recognized by TFIIA remains unknown. Here we have identified a core promoter element upstream of the TATA box that is recognized by TFIIA. A search of the human promoter database revealed that many natural promoters contain a TFIIA recognition element (IIARE). We show that the IIARE enhances TFIIA-promoter binding and enhances the activity of TATA-containing promoters, but represses or activates promoters that lack a TATA box. Chromatin immunoprecipitation assays revealed that the IIARE activates transcription by increasing the recruitment of pol II, TFIIA, TAF4, and P300 at TATA-dependent promoters. These findings extend our understanding of the role of TFIIA in transcription, and provide new insights into the regulatory mechanism of core promoter elements in gene transcription by pol II.
Keywords: DNA-protein interaction, gene regulation, gene transcription, general transcription factor (GTF), promoter, SELEX, TFIIA, core promoter element
Introduction
Eukaryotic transcription initiation by RNA polymerase II requires numerous transcription factors and cofactors to nucleate at the core promoter to form a pre-initiation complex (1, 2). The core promoter plays a critical role during transcriptional initiation and contains a number of DNA sequence elements such as the TATA box, the initiator, the downstream promoter element, the transcription factor IIB (TFIIB)4 recognition elements (BREs) and others (1–4). These elements are recognized by general transcription factors and cofactors (5–9), and assist to direct and orientate pre-initiation complex formation at the promoter. Core promoter elements not only regulate the activity of transcription but also determine transcription start site selection (4, 10, 11). Genome-wide studies have revealed that many genes lack so-called “canonical” core promoter elements, suggesting that other core promoter elements remain to be discovered. Indeed, a recent study showed that the core promoter element, DTIE, directs transcription start site selection of genes with TATA-less promoters (12). Nevertheless, it has been proposed that core promoter elements may not be essential for transcription of some genes in vivo (13), suggesting that canonical core promoter elements fine-tune physiological responses for specific genes (4). It has been shown that many “noncanonical” promoters instead contain epigenetic marks including histone modifications (H3K4me3 and H3K27me3) and DNA marks such as enhancers, CpG islands, and ATG deserts (14–16). The mechanisms by which canonical or noncanonical core promoters regulate transcriptional initiation are not fully understood.
Transcription factor TFIIA comprises three subunits, α, β, and γ; TFIIAα/β and TFIIAγ are encoded by different genes (17–19). The precursor of TFIIAα/β can be digested by taspase 1, but uncleaved TFIIAα/β remains active in transcriptional regulation (20). Recent studies showed that the cleavage of TFIIA by taspase 1 is involved in a number of molecular and biological processes (21–24). Although TFIIA was originally characterized as a general transcription factor, TFIIA is dispensable in transcription in vitro (25–26); perhaps, TFIIA is better to be described as a general cofactor because it acts as an anti-repressor or co-activator in transcriptional regulation (27–31). TFIIA can counteract the inhibitory roles of TAF1 and BTAF1 during TBP binding to the TATA box as well as the repressive effects of NC2 and HMGB1 on transcription (27, 28). TFIIA has also been shown to stabilize TFIID binding to DNA by interacting with transcriptional activators, TBP-associated factors (29, 30, 32–34), and TBP-related factors (35–37). It has been proposed that TFIIA induces the disassociation of TBP dimers and promotes the association between TBP and the TATA box promoter (38). TFIIA stabilizes the TBP–TATA box complex through direct contact on the face of TBP opposite to the TFIIB-binding side (39, 40). Studies using cryoelectron microscopy revealed that TFIIA and the transcription activator Rap1 cooperatively commit TFIID in transcription initiation (41).
Previous studies showed that human TFIIA can make specific contacts with the DNA immediately upstream of the TATA box (4, 39, 42–44). In our previous work, mutations of BRE consensus bases within the adenovirus major late (AdML) promoter inhibit the formation of TFIIA–TBP–DNA complexes, suggesting that a sequence-specific TFIIA DNA-binding region might overlap with the sequence of the BRE (45). In this study, we confirm that TFIIA makes direct contact with the sequence immediately upstream of the TATA box at the AdML promoter using combined molecular approaches. Using this information, we identified a core promoter element upstream of the TATA box that is recognized by TFIIA. We show that the TFIIA recognition element regulates transcription activity in a promoter context-dependent manner and determine the mechanism by which the TFIIA recognition element regulates transcription at the AdML promoter.
Results
TFIIA makes direct contact with the DNA sequence upstream of the TATA box at the AdML promoter
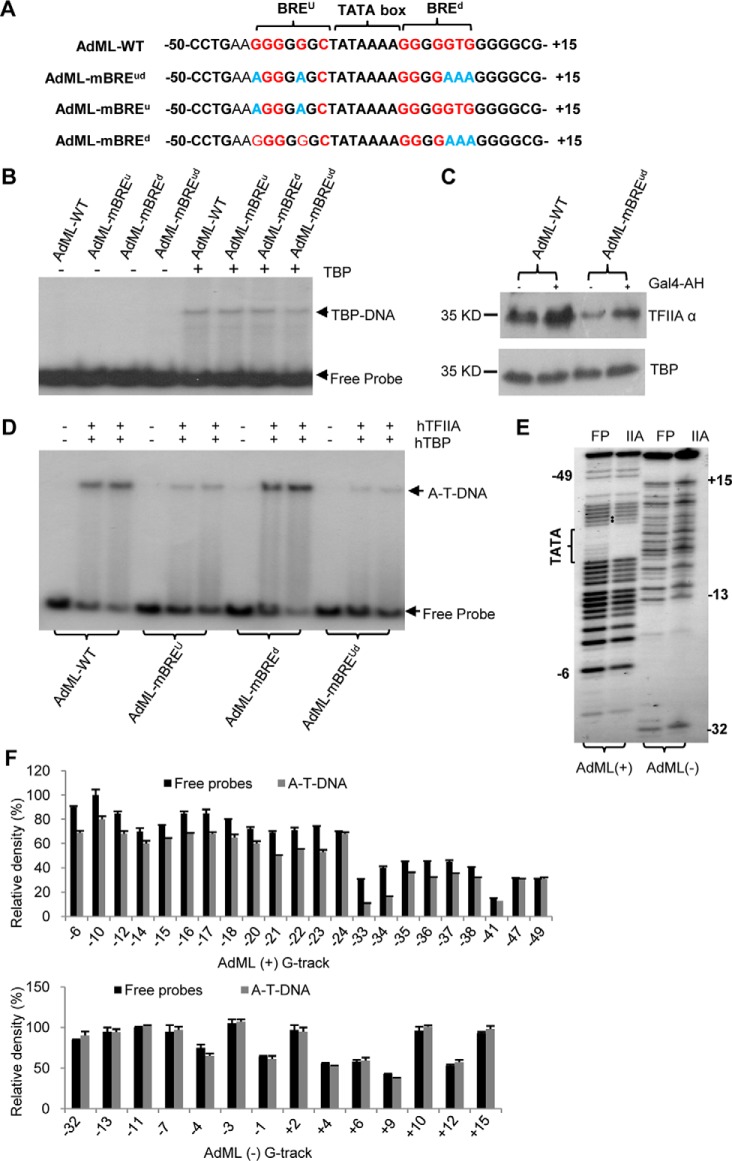
Our previous work showed that mutations of BREu and BREd consensus bases within the AdML promoter (AdML-mBREud, Fig. 1A) inhibit formation of a TFIIA–TBP–DNA complex when compared with wild type AdML promoter, suggesting that a TFIIA DNA-binding region could overlap with the BREu or BREd of the AdML promoter (45). To exclude the possibility that mutations of BREud consensus bases might affect TBP binding to the promoter DNA, bandshift assays were performed using TBP and radiolabeled promoter DNA fragments. As shown in Fig. 1B, the formation of TBP–DNA complexes showed minimal difference between the wild type AdML and AdML derivatives, indicating that mutations of the BREu or BREd consensus bases within the AdML promoter do not affect the formation of a TBP–DNA complex. To confirm this observation, protein–DNA binding assays were performed using HEK293T nuclear extract and promoter DNA immobilized on magnetic beads. TFIIA showed decreased binding to the AdML–mBREud promoter when compared with the wild type AdML promoter. However, TBP binding to DNA showed little difference between these two promoters (Fig. 1C), confirming that mutations of BREud consensus bases do not affect TBP binding to DNA, but significantly reduce TFIIA recruitment. To determine the specific contribution of the BREu and BREd elements to TFIIA recruitment, bandshift assays were performed using native hTFIIA, recombinant hTBP, and radiolabeled DNA fragments containing either a mutated BREu or a mutated BREd (Fig. 1A). The result shows that mutations in the BREu but not the BREd reduced the formation of a TFIIA–TBP–DNA complex compared with the wild type AdML promoter (Fig. 1D). This suggests that the TFIIA DNA-binding region overlaps with the BREu at the AdML promoter. To determine whether TFIIA interacts with specific bases within the BREu, DNA methylation interference assays were performed using native hTFIIA, recombinant hTBP, and radiolabeled AdML promoter DNA fragments. The data show that two G bases immediately upstream of the TATA box displayed weak signals at the positive strand of AdML (the black solid dots in lane IIA, Fig. 1E). This observation was verified by density quantification for the G tracks (−33 and −34 in G tracks, Fig. 1F); indicating that TFIIA can interact with at least two bases upstream of the TATA box. Taken together, these data confirm that TFIIA makes direct contact with the sequence immediately upstream of the TATA box at the AdML promoter.
Figure 1.
TFIIA makes direct contacts with the sequence upstream of the TATA box at the AdML promoter. A, the DNA sequences for wild type AdML and AdML derivatives showing the TATA box (bold), BRE consensus bases (red), and BRE mutated bases (light blue). B, mutations of the BREu (or BREd) consensus bases in AdML promoter did not affect the formation of TBP–DNA complex. Bandshift assays were performed with 200 ng of TBP and 1 μCi of radiolabeled wild type AdML or AdML derivatives and detected by autoradiography. C, mutations of the BREud consensus bases in the AdML promoter reduced TFIIA recruitment to promoter DNA but did not reduce TBP binding. Protein–DNA binding assays were performed using DNA-immobilized streptavidin magnetic beads and HEK293T nuclear extract, followed by incubation, washing, and elution. The eluted samples were detected by Western blotting with the antibodies against TFIIA and TBP, respectively. Equivalent volumes of elution (10 μl) was loaded in each lane for Western blotting. GAL4–AH, an activator comprised of a GAL4 DNA-binding domain and an acidic peptide (α helix). D, mutations of the BREu consensus bases in the AdML promoter decreased the formation of the TFIIA–TBP–DNA complex. EMSA was performed using recombinant hTBP, native hTFIIA, and radioactive wild type AdML or AdML derivatives and detected by autoradiography. A-T-DNA, TFIIA–TBP–DNA complex. The sequence for AdML wild type and its derivatives are indicated as in A. E, methylation interference assays showing TFIIA contacts with the bases immediately upstream of the TATA box. The TATA box (left bracket) and TFIIA contacting sites (solid black dots) are indicated in AdML positive strand. FP, free probe; IIA, TFIIA. F, relative density of G tracks for both AdML promoter positive and negative strands in the methylation interference assays performed in E. The relative density for the G-tracks of both free probe and the DNA from TFIIA–TBP–DNA complex was analyzed by ImageJ.
Derivation of TFIIA recognition element upstream of the TATA box and its prevalence in natural promoters
Previous studies using photocross-linking showed that TFIIA subunits, α and β, interact with a number of bases immediately upstream of the TATA box (on both positive and negative strands) at the AdML promoter (42). Our methylation interference analysis revealed the contacts between TFIIA and the two G bases of the positive strand upstream of the AdML TATA box (the black solid dots in Fig. 2A). Using this information, we generated a core promoter DNA library that contains randomized bases between positions −41 and −32 within AdML promoter derivatives (Fig. 2A). The randomized DNA was labeled with [α-32P]ATP and subjected to protein–DNA binding selection through bandshift assays (SELEX). After seven rounds of SELEX, the efficiency of selection was examined by a bandshift assay. Fig. 2B shows that formation of a TFIIA–TBP–DNA complex was significantly enhanced, indicating that the affinity of TFIIA to DNA was increased by SELEX. The DNA fragments were then cloned into pGEM3 and 50 clones sequenced across the promoter region. A TFIIA-binding consensus sequence was determined by the frequency of occurrence for each base. As shown in Fig. 2C, the consensus sequence exhibited strong preference for G or T, with exclusion of A and C at the region between −41 and −39 and the region between −35 and −32. Based on this information, we derived a consensus sequence: 5′-G-T/G-G-G/C/A-G/C-A/G-T-G/T-G/T-T-3′. Here we refer to the consensus sequence upstream of the TATA box as TFIIA recognition element (this element is abbreviated as IIARE, intentionally distinguished from the ARE (androgen response element)). The IIARE displayed four-base identity to the BREu (−38 to −32), and six-base identity to wild type AdML (−41 and −32). To gain insight into the prevalence of the IIARE in natural promoters, we searched the promoters from the human promoter database (epd.vital-it.ch/) (52, 56). There were 1957 TATA-containing promoters and 2058 TATA-less promoters used to analyze the presence of IIARE consensus bases in the region between −41 and −32 at the natural promoters. The result shows that 50.3% of the TATA-containing promoters contain over 5 consensus bases of the IIARE, 26.5% of promoters over 6 consensus bases, and 9.96% of promoters over 7 consensus bases. However, the IIARE showed a higher prevalence at the TATA-less promoters, particularly where the number of IIARE consensus bases is ≥7 or 8 (Table 1). The prevalence of the IIARE in TATA-less natural promoters is similar to the 5–8% of natural promoters that contain a TATA box (1). These results suggest that the IIARE is potentially present in a large proportion of natural promoters. Whether the IIARE in these promoters is functional requires further study.
Figure 2.
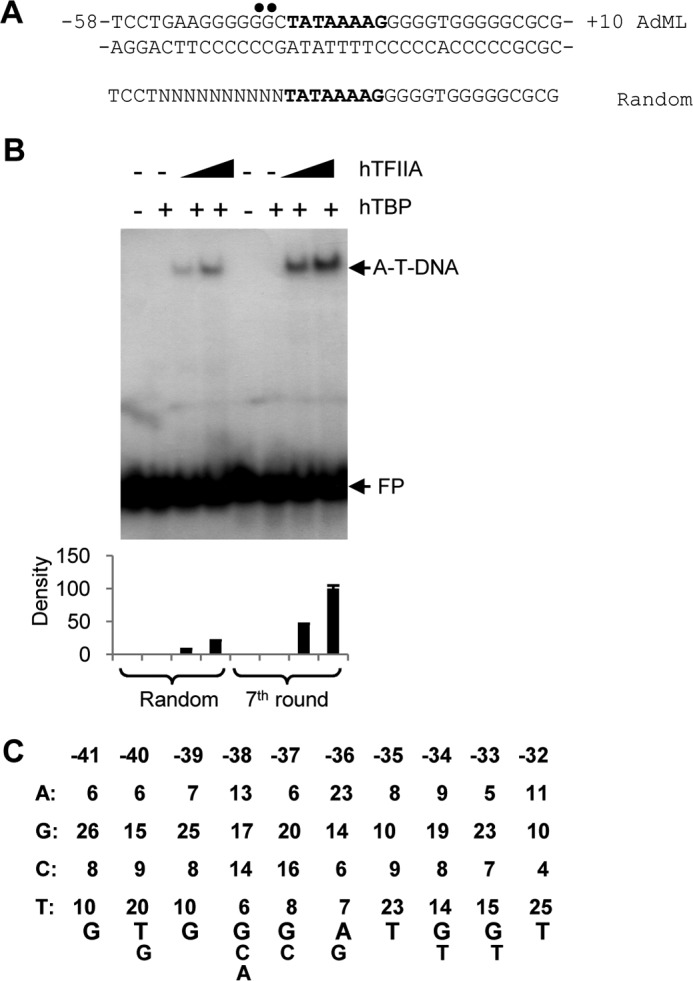
A TFIIA recognition element upstream of the TATA box derived from random selection and DNA sequencing. A, the sequences for wild type AdML and the randomized AdML positive strand. TFIIA contacting bases in the AdML promoter are marked with solid black dots and the TATA box is shown in bold character on the positive strand. The randomized bases (10 consecutive Ns) are located upstream of the TATA box (bold). B, bandshift assay showing increased enrichment of the TFIIA–TBP–DNA complex after seven rounds of selection compared with the original randomized DNA pool (Random). EMSA was performed with 100 ng of recombinant hTBP and native hTFIIA (50 or 100 ng) and detected by autoradiography. Relative density for EMSA is presented (bottom panel). FP, free probe; A-T-DNA, TFIIA–TBP–DNA complex. C, the frequency of base occurrence at each position over the randomized region (−41 to −32 nucleotides) and the consensus bases of the TFIIA recognition region (below). 50 clones in total were screened and sequenced after seven rounds of selection. The consensus bases are determined by the frequency of occurrence (>25%) at each position. The size of letter reflects the frequency of occurrence for each base.
Table 1.
The prevalence of IIARE in human natural promoters
| Number of IIARE consensus bases | ≥5 | ≥6 | ≥7 | ≥8 |
|---|---|---|---|---|
| Percentage ± S.D. in TATA-containing natural promoters | 50.3 ± 0.095 | 26.5 ± 0.062 | 9.96 ± 0.023 | 2.4 ± 0.014 |
| Percentage ± S.D. in TATA-less natural promoters | 58.4 ± 0.134 | 28.7 ± 0.095 | 15.5 ± 0.041 | 7.9 ± 0.021 |
The IIARE mutation in TATA-dependent promoters inhibits TFIIA recruitment and promoter activity
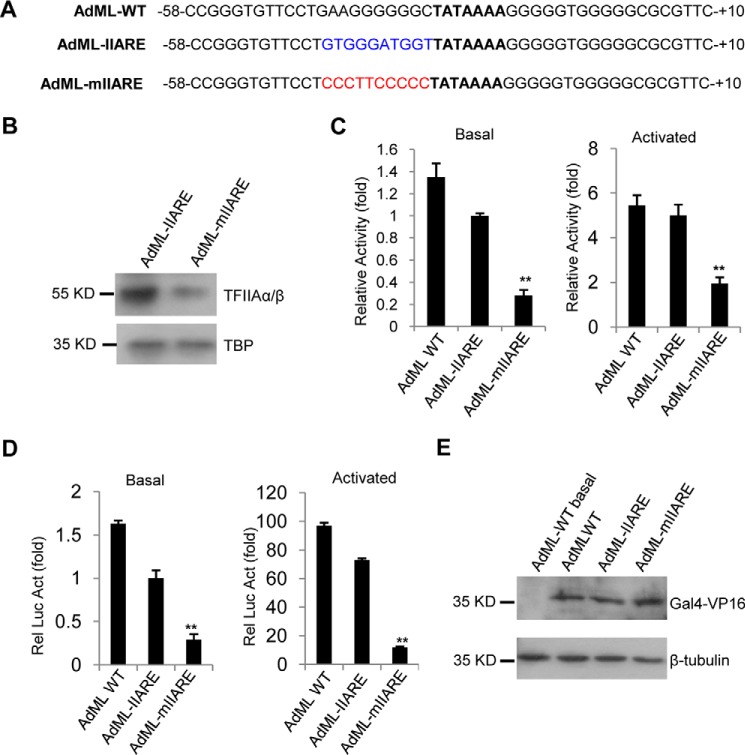
To determine whether the IIARE consensus that we derived affects the affinity of TFIIA to DNA, we generated AdML promoter derivatives that contain an optimal IIARE(AdML-IIARE) or a defective IIARE (AdML–mIIARE) (Fig. 3A). Promoter DNA was immobilized with streptavidin magnetic beads and binding assays were performed using reconstituted hTFIIA and recombinant hTBP. Stably bound proteins were detected by Western blot analysis. As shown in Fig. 3B, TFIIA exhibited decreased binding to the promoter containing a defective IIARE (AdML-mIIARE) when compared with the promoter containing an optimal IIARE (AdML-IIARE). Thus, the IIARE within AdML promoter derivatives enhances TFIIA binding to DNA, which is consistent with the observation from the random selection (Fig. 2B). To determine the effect of the IIARE on the activities of these promoters, in vitro transcription assays were performed using HEK293T nuclear extract and reporter vectors driven by either the AdML-IIARE or AdML-mIIARE promoters. Because the vector does not contain any eukaryotic promoter except the tested promoter, transcripts can be detected using RT-qPCR as described previously (46). The AdML–mIIARE promoter showed significant reduction in transcriptional activity in either the absence or presence of GAL4–VP16 when compared with the AdML–IIARE promoter or the wild type AdML promoter (Fig. 3C, left and right panels). To verify these observations, transient transfection of the promoter-driven reporters was performed with HEK293T cells in the absence or presence of GAL4–VP16 and the promoter activity was determined by luciferase assay. The IIARE defective promoter, AdML–mIIARE, showed reduced activity regardless of the co-expression of GAL4–VP16 when compared with the AdML–IIARE promoter or the AdML–WT promoter (Fig. 3D). Taken together, these data indicate that the IIARE within the AdML promoter is required for efficient TFIIA recruitment and positively regulates promoter activity.
Figure 3.
Mutation of the IIARE in the ADML promoter reduces TFIIA binding to DNA and promoter activity. A, the DNA sequences for the wild type AdML core promoter and its derivatives showing the TATA box (bold), IIARE consensus bases (blue), and mutated IIARE consensus bases (red). B, mutation of the IIARE upstream of the TATA box reduces TFIIA binding to the promoter DNA. Protein–DNA binding assays were performed with recombinant hTFIIA, hTBP, and promoter DNA-immobilized streptavidin magnetic beads. The eluted samples were analyzed by Western blotting with the indicated antibodies. Equivalent volumes of elution (10 μl) was loaded in each lane for Western blotting. C, in vitro transcription assays showing the relative activity for the promoters of AdML–WT, AdML–IIARE, and AdML–mIIARE in the absence (left) or presence (right) of GAL4–VP16. Relative activity was obtained by comparing the basal activity of the AdML–IIARE promoter with that of the AdML–WT promoter or that of AdML derivative promoters in the absence or presence of Gal4–VP16, where the basal activity for the AdML–IIARE promoter was arbitrarily set as 1. D, luciferase assays showing the effect of the IIARE mutation on reporter activity. Relative luciferase activity was obtained by comparing the basal activity of the AdML–IIARE promoter with that of the AdML–WT promoter or AdML derivative promoters with or without the co-expression of Gal4–VP16. The basal activity for the AdML–IIARE promoter was arbitrarily set as 1. E, immunoblotting of the samples used in D. Western blot analysis was performed using antibodies against Gal4 or β-tubulin. Equivalent amounts of protein (10 μg) were loaded in each lane for Western blotting. Each column in C and D represents the mean ± S.E. of three independent experiments. Significant difference was analyzed by comparing the activity of the IIARE-containing promoter with that of the IIARE-defect promoter. *, p < 0.05; **, p < 0.01. p value was obtained by Student's t test.
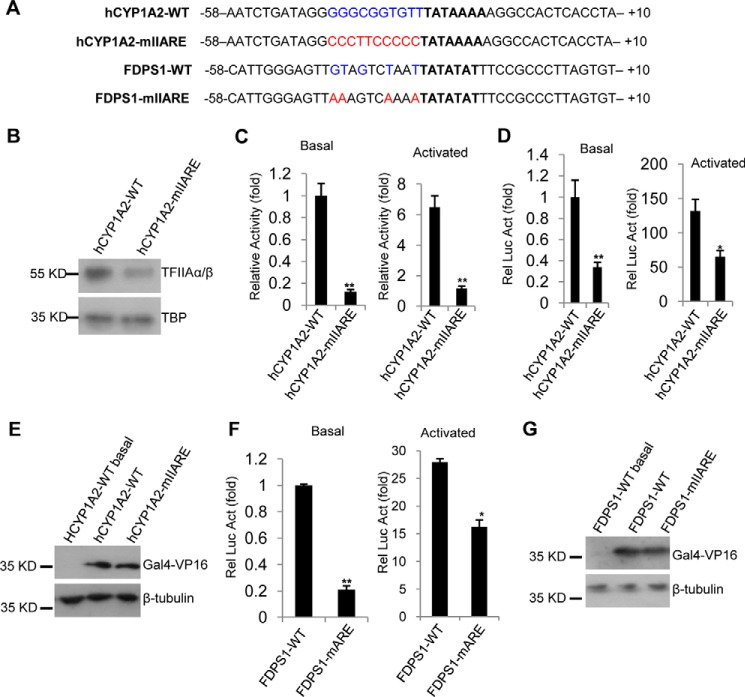
We next determined if our observations can be extended to natural promoters that contain a IIARE consensus sequence. A search of the human promoter database revealed that the cytochrome P450 1A2 (hCYP1A2) promoter contains a typical IIARE and a canonical TATA box (Fig. 4A). DNA fragments containing either the wild type hCYP1A2 core promoter or a mutant IIARE derivative were synthesized and cloned into the reporter vector pGL3-basic. Using these DNA fragments and vector-based sequences, protein–DNA binding assays, in vitro transcription assays, and luciferase assays were performed as described in the legend to Fig. 3. The results from these experiments showed that an intact IIARE within the hCYP1A2 promoter is required for TFIIA recruitment (Fig. 4B) and efficient promoter activity (Fig. 4, C–E), confirming a positive role for the IIARE in TATA-containing promoters.
Figure 4.
Mutation of the IIARE in natural promoters reduces TFIIA binding to DNA and promoter activity. A, the DNA sequences for the natural core promoters and their derivatives showing the TATA box (bold), the IIARE consensus bases (blue), and the mutated IIARE consensus bases (red). B, mutation of the IIARE reduces TFIIA binding to the hCYP1A2 promoter. C, in vitro transcription assays showing the relative activity for the promoters of hCYP1A2–WT and hCYPEA2–mIIARE in the presence or absence of GAL4–VP16. D, luciferase assays showing the effect of mutation of the IIARE on the expression of a reporter gene driven by the human CYP1A2 gene promoter or its derivatives. E, immuoblotting of the samples to confirm the co-expression of Gal4–VP16 used in C. F, luciferase assays showing the effect of IIARE mutation on the expression of a reporter gene driven by the human FDPS1 gene promoter. G, immuoblotting of the samples with the co-expression of Gal4–VP16 used in E. Protein–DNA binding assays, in vitro transcription assays, luciferase assays, and Western blotting were performed as described in the legend to Fig. 3. Each column in C, D, and F represents the mean ± S.E. of three independent experiments. Significant difference was analyzed by comparing the activity of the IIARE-containing promoter with that of the IIARE-defect promoter. *, p < 0.05; **, p < 0.01. p value was obtained by Student's t test.
The promoters of AdML–WT and hCYP1A2–WT also contain a BREu consensus sequence, raising the possibility that the IIARE-defective promoters also affect the BREu consensus bases. To exclude the possibility of interference by mutation of BREu, the natural promoter that directs expression of farnesyldiphosphate synthase (FDPS1) was tested. The FDPS1 promoter contains a TATA box and five IIARE consensus bases, but only one BREu consensus base (−38, Fig. 4A). An FDPS1 promoter mutant derivative was generated through altering four of the IIARE consensus bases (Fig. 4A). The wild type FDPS1 promoter and IIARE-defective derivative were then cloned into pGL3-basic, followed by a reporter assay. The results show that mutations of the IIARE consensus sequence within the FDPS1 promoter significantly reduced expression of the reporter gene (Fig. 4F). However, we note that inhibition of promoter activity in the mIIARE derivative can be partially alleviated by co-expression of GAL4–VP16 (Fig. 4F). This effect is likely due to TFIIA interaction with activator (29) providing an alternative mechanism for TFIIA recruitment. Taken together, these data suggest that the IIARE plays a positive regulatory role in both basal and activated transcription at promoters containing a TATA box.
The IIARE is a negative element in AdML–IIARE promoter derivatives that lack a TATA box
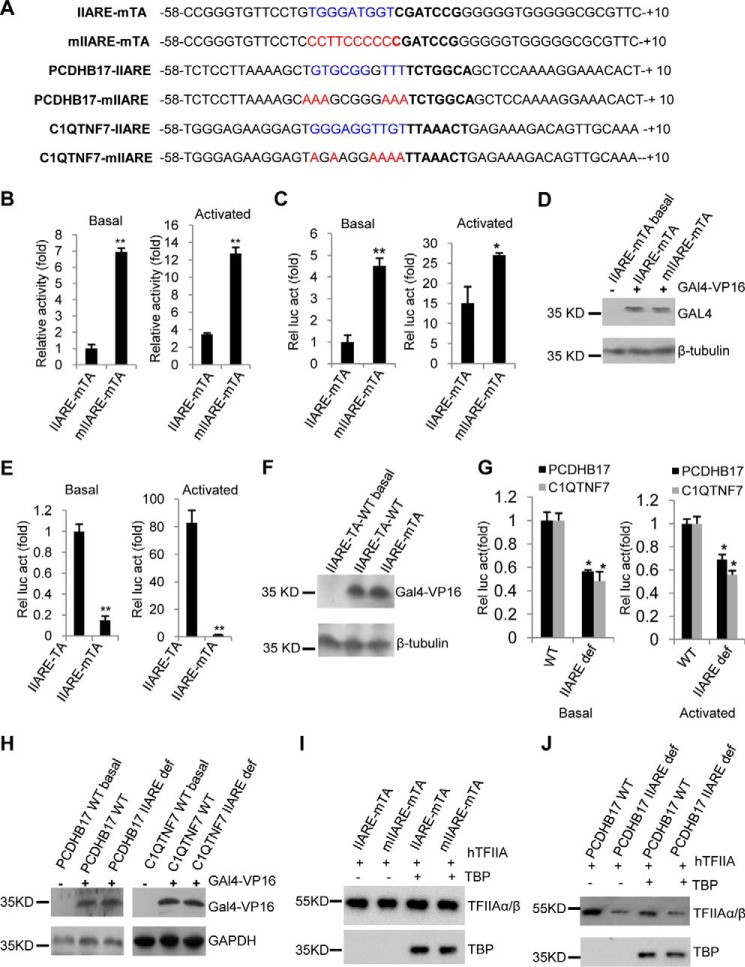
Several previous studies have showed that core promoter elements can work alone or in combination with other elements in transcription regulation (4), and that the function of core promoter elements depends on the context of the promoter (9). Indeed, TFIIA can stabilize TBP binding to the TATA box (39, 40). We therefore asked whether the role of the IIARE in gene transcription is dependent on the TATA box. To answer the question, AdML derivatives that lack a TATA box, but contain an optimal IIARE (AdML–IIARE–mTATA) or a defective IIARE (AdML–mIIARE–mTATA), were synthesized (Fig. 5A) and cloned into pGL3-basic. In vitro transcription assays were performed with these vectors. Intriguingly, the promoter of AdML–mIIARE–mTATA showed a significant increase in transcription activity in the absence or presence of GAL4–VP16 when compared with the AdML–IIARE–mTATA promoter (Fig. 5B). This suggests that the IIARE plays an inhibitory role in the absence of a TATA box. To confirm this observation, transient transfection assays were performed with HEK293T cells with the promoter derivatives. The data show that the AdML–mIIARE–mTATA promoter was significantly more active than the AdML–IIARE–mTATA promoter in both the absence and presence of GAL4–VP16 (Fig. 5, C and D). To exclude the possibility that mutation of the TATA box might solely account for this observation, luciferase assays were performed using the reporter vector driven by either the AdML–IIARE–TATA or AdML–IIARE–mTATA promoters. The data confirm that mutation of the TATA box significantly reduced the activity of the reporter gene (Fig. 5, E and F), suggesting that the TATA box mutation does not contribute to the increased activity caused by mutation of IIARE.
Figure 5.
The effect of IIARE mutation on the transcriptional activity of the AdML promoters that lack a TATA box. A, the DNA sequences for core promoters lacking a TATA box and their derivatives showing the IIARE consensus bases (blue), the mutated IIARE consensus bases (red), and the sequences around the TATA region (−31 to −25; bold). B, mutation of the IIARE significantly increased transcription activity in vitro of the AdML promoter derivatives lacking a TATA box. C, mutation of the IIARE within the AdML–IIARE–mTATA promoter significantly increased the expression of the reporter gene. D, immunoblotting analyses to conform the co-expression of GAL4–VP16 in the samples used in C. E, luciferase assays showing the effect of mutations in the TATA box on the expression of a reporter gene driven by the IIARE-containing AdML promoter. F, Western blotting of the samples to confirm co-expression of GAL4–VP16 in the samples used in E. G, luciferase assays showing the effect of mutation of the IIARE on the activity of natural promoters that lack a the TATA box. H, Western blotting to confirm the co-expression of GAL4–VP16 in the samples used in G. I and J, Western blotting showing the effect of the IIARE mutation on TFIIA binding to the promoters that lack the TATA box. In vitro transcription assays, luciferase assays, protein–DNA binding assays, and Western blotting were performed as described in the legend to Fig. 3. Abbreviations of the tested promoters is as follows: IIARE-mTA, AdML–IIARE–mTATA; mIIARE-mTA, AdML–mIIARE–mTATA; IIARE-TA, AdML–IIARE–mTATA. Each column in B, C, F, and G represents the mean ± S.E. of three independent experiments. Significant difference was analyzed by comparing the activity of the IIARE-containing promoter with that of the IIARE-defect promoter. *, p < 0.05; **, p < 0.01. p value was obtained by Student's t test.
We next determined whether the IIARE functions as a negative element in a natural promoter that lacks a TATA box. To this end, two natural TATA-less promoters, from the PCDHB17 and C1QTNF7 genes, were identified by searching the human promoter database. Core promoter DNA fragments of the PCDHB17 and C1QTNF7 genes and their derivatives containing a mutant IIARE were synthesized (Fig. 5A). They were cloned into pGL3-basic vector and then transfected into 293T cells. Unexpectedly, mutation of the IIARE significantly reduced expression of the reporter genes driven by the PCDHB17 or C1QTNF7 promoters (Fig. 5, G and H), suggesting that the IIARE positively regulates transcription of the PCDHB17 and C1QTNF7 promoters. Unlike at the AdML derivatives that lack a functional TATA element, these results are consistent with those from the promoters containing the TATA box. To understand how a defective TATA box affects TFIIA binding to the promoter, protein–DNA assays were performed using promoter DNA fragments and recombinant proteins TFIIA and TBP. The data show that promoters lacking the TATA box were still bound by TFIIA regardless of the presence of TBP (Fig. 5, I and J). However, mutation of the IIARE did not affect TFIIA recruitment to the AdML derivative promoters that lack a functional TATA box (Fig. 5I). In contrast, mutation of the IIARE in the PCDHB17 promoter (which is naturally TATA-less) reduced TFIIA recruitment (Fig. 5J). Taken together, these data imply that the IIARE regulates transcription activity in a promoter context-dependent manner.
The IIARE positively regulates transcription by increasing recruitment of pol II, TFIIA, TAF4, and p300 at the AdML–IIARE promoter
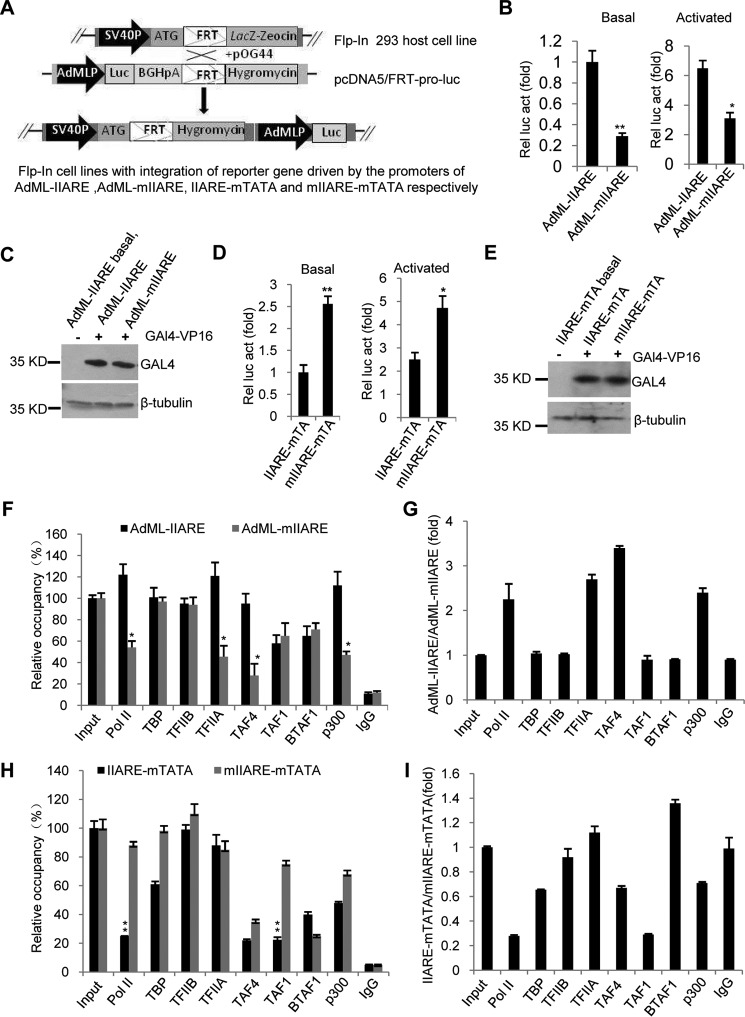
We have determined that the IIARE can play a positive and negative role in transcription at the AdML–IIARE and AdML–IIARE–mTATA promoters, respectively (Figs. 3, C and D, and 5, B and C). Whether this observation can be reproduced in vivo remains unclear. We therefore generated stable 293 cell line derivatives using the Flp-In system as shown in Fig. 6A. The Flp-In 293 stable cell lines were used for luciferase assays in both the absence and presence of GAL4–VP16. The results show that mutation of the IIARE significantly reduced the activity of the reporter gene driven by the AdML–mIIARE promoter compared with the AdML–IIARE promoter (Fig. 6, B and C). In contrast, compared with the AdML–IIARE–mTATA derivative, we observed an increase in activity of the reporter gene driven by the AdML–mIIARE–mTATA promoter (Fig. 6, D and E). These results are consistent with those from the assays in vitro (Figs. 3 and 5, B and C).
Figure 6.
The IIARE positively modulates transcription by increasing the recruitment of pol II, TFIIA, TAF4, and P300 at the promoter of AdML-IIARE. A, the scheme showing the generation of Flp-In 293 stable cell lines that integrate promoter-directed luciferase gene derivatives. B, mutation of the IIARE within the promoter of AdML–IIARE significantly reduced reporter gene expression in vivo. Flp-In 293 stable cell lines were co-transfected with pcDNA–β-Gal and pcDNA–Gal4–VP16 or control pcDNA3.1, the cell lysate was used to detect luciferase activity. Relative luciferase activity was obtained as described in the legend to Fig. 3D. C, immunoblotting analyses of the samples to confirm expression of Gal4–VP16 in samples used in B. Immunoblotting was performed as described in the legend to Fig. 3E. D, luciferase assays showing the effect of IIARE mutation on reporter gene expression for the AdML derivatives that lack a TATA box. Luciferase assays were performed as in B. E, immunoblotting analyses for the samples to confirm expression of Gal4–VP16 used in D. Immunoblotting was performed as described in the legend to Fig. 3E. F, ChIP-qPCR assays showing the relative occupancy for the indicated factors at the promoters of AdML–IIARE or AdML–mIIARE. G, comparison of the relative occupancy for indicated factors between the promoters of AdML–IIARE and AdML–mIIARE. H, ChIP-qPCR assays showing the relative occupancy for the indicated factors at the promoters of AdML–IIARE–mTATA or AdML–mIIARE–mTATA. I, comparison of the relative occupancy for the indicated factors at the promoters of AdML–IIARE–mTATA and AdML–mIIAR–mTATA. The relative occupancy was obtained by comparing the enrichment of promoter DNA from the ChIP sample for individual factors with that from input. 1 ng of genomic DNA was used to perform qPCR for input, which is equivalent to 0.01% of sample used for ChIP assay of individual factor. Each column in B, D, F, and H represents the mean ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01. p value was obtained by Student's t test.
To understand how the IIARE regulates transcription of the AdML–IIARE or AdML–mIIARE promoters, ChIP assays were performed by using the Flp-In 293 stable cell lines and the antibodies indicated in Fig. 6F. As shown in Fig. 6, F and G, the occupancy of TFIIA significantly decreased at the promoter of AdML–mIIARE compared with that at the promoter of AdML–IIARE. This is consistent with the results from protein-DNA binding assays in vitro (Fig. 3B). Strikingly, the occupancies for pol II, TAF4, and P300 at the promoter of AdML–mIIARE decreased 2–3-fold, respectively, compared with recruitment at the AdML–IIARE promoter, suggesting that the IIARE enhances recruitment of these factors to the promoter of AdML–IIARE. However, the occupancies for TBP, TFIIB, TAF1, and BTAF showed little difference between the AdML–IIARE and AdML–mIIARE promoters.
To understand how the IIARE affects transcription of the ADML promoter derivative that lack a TATA box, ChIP assays were performed with the stable cell lines integrated with these promoters. Fig. 6H shows that the occupancy of TFIIA showed little change between the promoters of AdML–IIARE–mTATA and AdML–mIIARE–mTATA, which is consistent with the observation for protein–DNA binding assay (Fig. 5I). However, the occupancies of pol II or TAF1 at the AdML–IIARE–mTATA promoter were significantly lower than that at the AdML–mIIARE–mTATA promoter (Fig. 6, H and I), suggesting that the IIARE inhibits the transcription activity by reducing the recruitment of pol II and TAF1 at the AdML–IIARE–mTATA promoter. The occupancies of TFIIB, TBP, TAF4, and P300 did not show a significant difference between the promoters except BTAF1 recruitment was slightly reduced at the mutant IIARE derivative. The data show that TBP was recruited at both of the promoters in vivo even though they contained a mutant TATA box. TBP can bind other general factors and cofactors such as TFIIB and TAFs, which likely provides an alternative means of recruitment independent on the TATA element (1). Taken together, the data in Fig. 6 suggest that the IIARE enhances transcription of the AdML promoter by increasing the recruitment of pol II, TFIIA, TAF4, and P300 at the promoter, but inhibits transcription of the AdML promoter that lacks a TATA element by reducing the recruitment of pol II and TAF1 at the promoter.
Discussion
Previous studies revealed that TFIIA makes direct contacts with up to 9 base pairs upstream of the TATA box (42, 47) and 2 base pairs downstream of the TATA box (42). Crystal structure analyses of TFIIA–TBP–DNA using yeast TBP and TFIIA core domains showed that the β-barrel of TOA 1 interacts with 3–5 bases upstream of the TATA box, but not with the bases downstream of the TATA box (39, 44). In this study, we have confirmed that TFIIA makes direct contacts with the DNA sequence upstream of the TATA box (Fig. 1, D and E), in particular with the two G bases immediately upstream of the TATA box on the positive strand of the AdML promoter (Fig. 1, E and F). This result is in agreement with the previous findings (43) and suggests the presence of a TFIIA recognition region upstream of the TATA box at the AdML promoter. Using this information, we then defined a TFIIA recognition element (IIARE) upstream of the AdML TATA box through SELEX and DNA sequencing. This element displays strong preference for G and T, and against A and C at its two ends (Fig. 2C). In this regard the IIARE consensus sequence is similar to the BREd where TFIIB also favors G or T, although the specific sequence of consensus bases is distinct (9). The IIARE is distinct from the BREu (8), in which TFIIB prefers bases G and C. Another feature of the IIARE is that the central region (−38 to −36) displays less preference to G or T than the flanking bases (−41 to −39 and −35 to −32) (Fig. 2C). Previous photocross-linking studies showed that TFIIAβ mainly cross-linked to the sequence immediately upstream of the TATA box, whereas TFIIAα was mostly cross-linked with the sequence further upstream of TFIIAβ–cross-linked region. This spacing between the TFIIAα and TFIIAβ contact regions (42) could potentially contribute to the difference in base preference within the IIARE (Fig. 2C).
We found mutation of the IIARE severely affected TFIIA binding to promoter DNA and reduced promoter activity regardless in both the absence and presence of the transcriptional activator GAL4–VP16 (Figs. 3, 4, and 5, G and J). This coincided with reduced binding of TFIIA to the promoter DNA. However, when artificial AdML promoter derivatives that lack a TATA box were tested for the same experiments, we observed a negative function for the IIARE (Fig. 5, B–D). Thus, our results suggest that IIARE modulates transcription in promoter context-dependent manner. Previous studies showed that both BREu and BREd can also positively or negatively regulate activity of the core promoter (9, 48). The BREd regulates transcription activity in a promoter context-dependent manner (9); whereas the BREu plays different roles in basal and activator-dependent transcription (48). These observations suggest that the role of core promoter elements in transcription relies on specific conditions, either in the context of the core promoter or the interplay of gene-specific transcriptional regulators. Indeed, previous in vivo studies suggest that the function of a core promoter element depends on the specific physiological environment (13).
ChIP assays showed that mutation of the IIARE significantly reduced the recruitment of pol II, TFIIA, TAF4, and P300 at the AdML promoter (Fig. 6, F and G). Others have reported that the transcriptional coactivator P300 promotes the formation of a TFIIA–TBP–DNA complex through acetylation of TFIIA (49). In addition, TFIIA binds to TAF4 to assist pre-initiation complex (PIC) assembly at the core promoter (33). Thus, a IIARE-dependent increase in the occupancy of TFIIA could subsequently enhance the recruitment of TAF4 and P300 at the promoter with an optimal IIARE. TAF1 and BTAF1 have been shown to compete with TFIIA to bind the TBP–DNA complex and inhibit transcription (50, 51). However, the recruitment of TAF1 and BTAF1 was not affected by mutation of the IIARE within the AdML promoter that contains a functional TATA box (Fig. 6, F and G). Surprisingly, mutation of the IIARE within the ADML promoter containing a defective TATA element increased the recruitment of both pol II and TAF1 (Fig. 6, H and I). It is possible that mutation of the IIARE combined with defective TBP–TATA interactions increases the potential of TAF1 to bind the Initiator element. In addition, TAF1 can play dual roles in transcriptional regulation through phosphorylation of histones to activate transcription (1). It is therefore possible that TFIIA–IIARE contacts modulate the recruitment of TAF1 in the absence of TBP–TATA interactions.
Our ChIP assays revealed that the subunits of TFIID differentially associated with either the ARE-containing promoter or ARE-defective promoter. Previous studies suggest that TAFs can form multiple complexes, some of which do not contain TBP (1). Moreover, protein-coding genes show distinct requirements for individual TAFs (53), and TAFs are differentially recruited at the promoters of many active genes as stem cell differentiation progresses (54). Therefore, it is plausible that TFIIA–IIARE interactions can influence the TAF dependence of the core promoter and thus their requirement at individual genes.
In this study we identified a core promoter element upstream of the TATA box that is recognized by TFIIA. The IIARE modulates transcription in a promoter context-dependent manner. The IIARE enhances transcription by increasing the recruitment of pol II, TFIIA, TAF4, and p300 at the AdML promoter. The IIARE can acts as a negative element at the AdML promoter when the TATA element is rendered inactive. In this context the IIARE inhibits the association of TAF1 and pol II with the AdML-IIARE. These findings not only extend our understanding of the role of TFIIA in gene transcription, but also provide new insights into the regulatory mechanism of core promoter elements in transcription by pol II.
Experimental procedures
Plasmids, proteins, and site-directed mutagenesis
Human cDNAs for the TFIIA subunits (α/β and γ) and TBP were cloned into the plasmid pET30a. The cDNA for HA-tagged TFIIAα/β was cloned into pcDNA3.1(+). The promoter DNA fragments for AdML, hCYP1A2, FDPS1, PCDHB17, C1QTNF7, and their derivatives were initially cloned into the pGEM3 plasmid. A DNA fragment comprised of nine GAL4–DNA recognition sites was inserted immediately upstream of the promoter derivatives; the nine GAL4–DNA-binding sites along with the promoter DNA fragment were then transferred to the reporter vector pGL3-basic. The cDNA encoding GAL4–VP16 was cloned into pET30a and pcDNA3.1(+). Recombinant proteins for TFIIAα/β, TFIIAγ, TBP, and GAL4–VP16 were expressed with Escherichia coli BL21 (DE3; Agilent) and purified with nickel-agarose as described in the manufacturer's manual (Qiagen). Reconstituted TFIIA was obtained through denaturing the recombinant proteins TFIIAα/β and TFIIAγ with protein denature buffer (50 mm Tris-Cl, pH 7.4,, 1 mm EDTA, 100 mm NaCl, 0.1 mm PMSF, and 7 m urea) for 4 h, and then renaturing the proteins by dialysis (20 mm HEPES, pH 7.6, 1 mm EDTA, 100 mm NaCl, 0.1 mm PMSF, 10% glycerol, and 0–7 m urea); the concentration of urea in dialyzing buffer was decreased at 1 m per 5 h from 7 to 0 m during the dialysis. Native TFIIA was purified from HEK293T nuclear extract as described previously (55). All chemicals were purchased from Sigma.
Bandshift assays and methylation interference
One microgram of core promoter DNA for wild type AdML-BRE promoter and its derivatives (AdML-mBREud, AdML-mBREd and AdML-mBREu) was labeled with 2.5 μCi of [γ-32P]ATP (GE Healthcare) and 2.5 units of T4 polynucleotide kinase (Promega). Bandshift assays were performed with 0.1 pmol of radiolabeled AdML core promoter DNA, 5 or 10 μg of native human TFIIA (hTFIIA) protein, and 10 μg of recombinant human TBP (hTBP) protein as described previously (45). Methylation interference was performed using 1 pmol of radiolabeled AdML core promoter DNA, 50 μg of native human TFIIA protein, and 50 μg of recombinant hTBP protein as described previously (9).
Immobilized protein-DNA binding assays and Western blot analysis
Overlapping oligonucleotides for wild type AdML core promoter (AdML-WT) and its derivatives or wild type hCYP1A2p, PCDHB17, and their derivatives were synthesized and modified with biotin at the 5′ end of the positive strands (TianyiHuiyuan Co.). One μl of 25 m forward and reverse oligonucleotides were used for overlapping PCR in 50 μl of reaction mixture. The PCR product was purified by the PCR clean kit (Axygen) and immobilized with streptavidin magnetic beads (Thermo) in 100 μl of 1× PBS buffer at 4 °C overnight. The beads were then washed with 1× PBS buffer 3 times and collected with a magnetic stand (Promega). The DNA-immobilized beads were incubated with HEK293T nuclear extract or reconstituted hTFIIA and recombinant hTBP at 4 °C for 4 h. The beads were washed with buffer D (20 mm HEPES, 100 mm KCl, 0.2 mm EDTA, 1 mm DTT, 0.2 mm PMSF, 20% glycerol) 3 times, DNA-bound proteins were eluted with 1× SDS loading buffer and the samples analyzed by Western blotting. The antibodies for Western blot analysis were purchased from Abcam (TFIIA, ab50821; TBP, ab51841; and β-Tubulin, ab6046) and Santa Cruz Biotechnology (GAL4, SC-577).
Random selection
The oligonucleotides for the AdML derivative used in random selection was synthesized as follows: 5′-CGTGACCGGGTGTTCCTNNNNNNNNNNTATAAAAGGGGGTGGGGGCGCGTTCGTCCTCACTCTCTTCCGCATCGCTGT-3′. This DNA fragment contains 10 consecutive randomized nucleotides immediately upstream of the TATA box, the randomized region was synthesized with equimolar nucleotides (Taiyihuiyuan Co.). The primers used for PCR and cloning were as follows: RSF, 5′-CGCGGATCCCGTGACCGGGTGTTCCT-3′; RSR, CGCGAATTCACAGCGCATGCGAATTCCCATG-3′. PCR primers, RSF and RSR, contain restriction sites recognized by BamHI and EcoRI, respectively, to assist DNA fragment cloning. Random Selection (SELEX) was performed as described previously (9). Briefly, in the first cycle, 0.1 μl of 25 μm AdML template and 1 μl of 25 μm primers (RSF and RSR, respectively) were used for PCR in 50 μl of reaction mixture containing 200 μm dNTP, 1 μl of [α-32P]dATP (2.5 μCi), and 5 units of Taq DNA polymerase (Roche Applied Science). The radioactive PCR product was purified on a 5% native polyacrylamide gel. Bandshift assays were performed using 0.2 pmol of purified DNA fragment, 10 μg of native hTFIIA, and 10 μg of recombinant hTBP as described previously (45). The DNA was recovered from the shifted band and amplified by PCR within 50 μl of reaction mixture as described above. The amplified radioactive DNA was used for the second round of selection. After seven rounds of selection in total, the recovered DNA fragments were amplified by PCR without the addition of radioactive isotope, followed by digestion with BamHI and EcoRI and cloned into pGEM3. Fifty clones in total were screened and used for sequencing of the promoter region. The frequency of occurrence for each base over the randomized region (from −41 to −32) was counted and the consensus bases were determined by the frequency of occurrence.
In vitro transcription assays
In vitro transcription assays were performed using the non-radioactive method as described in our recent work (46). In this method, pGL3-basic reporter gene vector driven by promoter or promoter derivative was used for DNA template. One hundred ng of DNA template were used for RNA synthesis in the reaction mixture (25 μl of HeLa nuclear extract, 3 μl of 100 mm MgCl2, 3 μl of 10 mm NTP) in the absence or presence of 1 μg of GAL4–VP16. When the reaction finished, the samples were subject to DNA template depletion, hybridization, and primer extension, the transcript was detected by using RT-qPCR and our uniquely designed primers (46). Relative transcriptional activity was analyzed by Bio-Rad CFX Manager 3.0 software.
Cell culture, transfection, and luciferase assay
HEK293T cells were cultured in 12-well plates with high-glucose DMEM Complete medium. Transient transfection for HEK293T cells or Flp-In 293 stable cell lines was performed with Turbofect reagent (Thermo). Luciferase assays were performed with the dual-light detection system (Thermo). Briefly, HEK293T cells were co-transfected with 1.5 μg of reporter vector driven by wild type promoter or its IIARE derivatives, 1 μg of vector expressing β-galactosidase and 0.5 μg of vector expressing GAL4–VP16 or empty vector pcDNA3.1(+). 36 h post-transfection, the cells were harvested and disrupted with the lysis buffer provided by the manufacturer (Thermo). After centrifugation at 1200 rpm for 5 min, 3 μl of the cell lysate were used to detect luciferase activity with the dual-light detection system. Luciferase activity was normalized by the activity of β-galactosidase within the same sample. Relative luciferase activity was obtained by comparing luciferase activity between wild type promoter and its IIARE-defect promoter, in which the basal activity of the wild type promoter was arbitrarily set as 1.
Generation of Flp-In 293 stable cell lines and ChIP assay
Flp-In 293 stable cell lines were generated with the Flp-In system (Invitrogen). A Flp-In 293 host cell line that has been integrated with the fragment of FRT–LacZ–Zeocin was provided by Taylor lab (The University of Manchester). The DNA fragments for AdML–IIARE–Luc, AdML–mIIARE–Luc, AdML–IIARE–mTATA–Luc, and AdML–mIIARE–mTATA–Luc were cloned into the vector pcDNA5/FRT, respectively. The vector pcDNA5/FRT bearing a promoter-driven reporter gene (pcDNA5/FRT-pro-luc) and the vector expressing a recombinase (pOG44) were co-transfected into the 293 host cell line with Lipofectamine 2000 (Thermo). 48 h post-transfection, selection was initiated by adding hygromycin to the cells at the final concentration of 200 μg/ml. The cell line from a single colony was obtained by diluting the cells into 96-well plates. The positive cell lines were determined by detecting the expression of luciferase gene.
ChIP assays for pol II and transcription factors or cofactors were performed using Flp-In 293 stable cell line that was integrated with a promoter-driven luciferase gene as described previously (45). The ChIP samples were detected by qPCR using Bio-Rad Real Time Detection System and analyzed by Bio-Rad CFX manager 3.0. The relative occupancy was obtained by comparing the enrichment of promoter DNA from the ChIP sample of each factor with that from the Input sample, 1 ng of genomic DNA was used to perform qPCR for input, which is equivalent to 0.01% of the sample used for the ChIP assay of individual factor. Fold-change was obtained by comparing the relative occupancy of a factor between the promoters with or without ARE. The antibodies for ChIP assays were purchased from Santa Cruz Biotechnology (pol II, SC-21751; TBP, SC-204; TFIIB, SC-225; TAF1, SC-735; TAF4, SC-136093; BTAF1, SC-8139; and P300, SC-584) and Abcam (TFIIA, Ab50821).
Author contributions
J. W. and S. Z. performed in vitro transcription assays and reporter assays, Western blot analysis, ChIP assays, and data analysis. W. H. cloned natural promoters and derivatives. Y. W. expressed and purified TFIIA and derivatives. H. P. assisted in the protein–DNA binding assay. P. S. provided facilities and consumables for performing some of experiments such as bandshift assays. Y. Z. cloned the AdML–IIARE promoter and its derivatives. S. G. E. R. designed and supported a part of experiments and revised the manuscript. W. D. designed most of the experiments, performed bandshift assays, random selection, protein–DNA binding assays, and data analysis, gained the funding, and wrote the manuscript.
Acknowledgment
We thank Professor Steve Taylor at University of Manchester for providing the Flp-In 293 host cell line.
This work was supported by National Natural Science Foundation of China Project Grants 31271395 and 31673157 (to W. D.). The authors declare that they have no conflicts of interest with the contents of this article.
- TFIIB
- transcription factor IIB
- BRE
- TFIIB recognition elements
- qPCR
- quantitative PCR
- TFIIA
- transcription factor IIA
- NC2
- negative cofactor 2
- TFIID
- transcription factor IID
- GAL4–VP16
- an recombinant activator consist of GAL4 DNA-binding domain and herpes virus protein 16 activation domain
- TBP
- TATA-binding protein
- TAF
- TBP-associated factor
- BTAF1
- B-TFIID TBP–associated protein 1
- HMGB1
- high mobility group box 1 protein
- IIARE
- TFIIA recognition element
- BREu
- TFIIB recognition element upstream of the TATA box
- BREd
- TFIIB recognition element downstream of the TATA box
- AdML
- adenovirus major late promoter
- AdML-BREud
- adenovirus major late promoter containing TFIIB recognition elements at upstream and downstream of the TATA box
- AdML-BREu
- adenovirus major late promoter containing a TFIIB recognition elements at upstream of the TATA box
- AdML–IIARE
- adenovirus major late promoter containing a TFIIA recognition element
- ADML–mIIARE
- adenovirus major late promoter containing a mutated TFIIA recognition element
- AdML–IIARE–mTATA
- adenovirus major late promoter containing a TFIIA recognition element and a mutated TATA box
- AdML–mIIARE–mTATA
- adenovirus major late promoter containing a mutated TFIIA recognition element and a mutated TATA box
- hCYP1A2
- human cytochrome p450 1A2 gene
- FDPS1
- farnesyldiphosphate synthase 1 gene
- PCDHB17
- protocadherin β17 gene
- C1QTNF7
- C1q and tumor necrosis factor-related protein 7 gene
- pol II
- polymerase II
- SELEX
- systematic evolution of ligands by exponential enrichment.
References
- 1. Thomas M. C., and Chiang C. M. (2006) The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 [DOI] [PubMed] [Google Scholar]
- 2. Venters B. J., and Pugh B. F. (2009) How eukaryotic genes are transcribed. Crit. Rev. Biochem. Mol. Biol. 44, 117–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Butler J. E., and Kadonaga J. T. (2002) The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16, 2583–2592 [DOI] [PubMed] [Google Scholar]
- 4. Roy A. L., and Singer D. S. (2015) Core promoters in transcription: old problem, new insights. Trends Biochem. Sci. 40, 165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smale S. T., and Kadonaga J. T. (2003) The RNA polymerase II core promoter. Annu. Rev. Biochem. 72, 449–479 [DOI] [PubMed] [Google Scholar]
- 6. Shao H., Revach M., Moshonov S., Tzuman Y., Gazit K., Albeck S., Unger T., and Dikstein R. (2005) Core promoter binding by histone-like TAF complexes. Mol. Cell. Biol. 25, 206–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chalkley G. E., and Verrijzer C. P. (1999) DNA binding site selection by RNA polymerase II TAFs: a TAFII250-TAFII150 complex recognizes the initiator. EMBO J. 18, 4835–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lagrange T., Kapanidis A. N., Tang H., Reinberg D., and Ebright R. H. (1998) New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12, 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deng W., and Roberts S. G. (2005) A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 19, 2418–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Danino Y. M., Even D., Ideses D., and Juven-Gershon T. (2015) The core promoter: at the heart of gene expression. Biochim. Biophys. Acta 1849, 1116–1131 [DOI] [PubMed] [Google Scholar]
- 11. Reiter W. D., Hüdepohl U., and Zillig W. (1990) Mutational analysis of an archae bacterial promoter: essential role of a TATA box for transcription efficiency and start-site selection in vitro. Proc. Natl. Acad. Sci. U.S.A. 87, 9509–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marbach-Bar N., Bahat A., Ashkenazi S., Golan-Mashiach M., Haimov O., Wu S. Y., Chiang C. M., Puzio-Kuter A., Hirshfield K. M., Levine A. J., and Dikstein R. (2016) DTIE, a novel core promoter element that directs start site selection in TATA-less genes. Nucleic Acids Res. 44, 1080–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barbash Z. S., Weissman J. D., Campbell J. A. Jr., Mu J., and Singer D. S. (2013) Major histocompatibility complex class I core promoter elements are not essential for transcription in vivo. Mol. Cell. Biol. 33, 4395–4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Voigt P., Tee W. W., and Reinberg D. (2013) A double take on bivalent promoters. Genes Dev. 27, 1318–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee M. P., Howcroft K., Kotekar A., Yang H. H., Buetow K. H., and Singer D. S. (2005) ATG deserts define a novel core promoter subclass. Genome Res. 15, 1189–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ramirez-Carrozzi V. R., Braas D., Bhatt D. M., Cheng C. S., Hong C., Doty K. R., Black J. C., Hoffmann A., Carey M., and Smale S. T. (2009) A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138, 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeJong J., Bernstein R., and Roeder R. G. (1995) Human general transcription factor TFIIA: characterization of a cDNA encoding the small subunit and requirement for basal and activated transcription. Proc. Natl. Acad. Sci. U.S.A. 92, 3313–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma D., Watanabe H., Mermelstein F., Admon A., Oguri K., Sun X., Wada T., Imai T., Shiroya T., and Reinberg D. (1993) Isolation of a cDNA encoding the largest subunit of TFIIA reveals functions important for activated transcription. Genes Dev. 7, 2246–2257 [DOI] [PubMed] [Google Scholar]
- 19. Ozer J., Moore P. A., Bolden A. H., Lee A., and Rosen C. A., and Lieberman P. M. (1994) Molecular cloning of the small (γ) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 8, 2324–2335 [DOI] [PubMed] [Google Scholar]
- 20. Zhou H., Spicuglia S., Hsieh J. J., Mitsiou D. J., Høiby T., Veenstra G. J., Korsmeyer S. J., and Stunnenberg H. G. (2006) Uncleaved TFIIA is a substrate for taspase 1 and active in transcription. Mol. Cell. Biol. 26, 2728–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Malecová B., Caputo V. S., Lee D. F., Hsieh J. J., and Oelgeschläger T. (2015) Taspase1 processing alters TFIIA cofactor properties in the regulation of TFIID. Transcription 6, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takeda S., Sasagawa S., Oyama T., Searleman A. C., Westergard T. D., Cheng E. H., and Hsieh J. J. (2015) Taspase1-dependent TFIIA cleavage coordinates head morphogenesis by limiting Cdkn2a locus transcription. J. Clin. Invest. 125, 1203–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suzuki H., Isogai M., Maeda R., Ura K., and Tamura T. A. (2015) TBP-like protein (TLP) interferes with Taspase1-mediated processing of TFIIA and represses TATA box gene expression. Nucleic Acids Res. 43, 6285–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oyama T., Sasagawa S., Takeda S., Hess R. A., Lieberman P. M., Cheng E. H., and Hsieh J. J. (2013) Cleavage of TFIIA by Taspase1 activates TRF2-specified mammalian male germ cell programs. Dev. Cell 27, 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Van Dyke M. W., Roeder R. G., and Sawadogo M. (1988) Physical analysis of transcriptional pre-initiation complex assembly on a class II gene promoter. Science 241, 1335–1338 [DOI] [PubMed] [Google Scholar]
- 26. Wu S. Y., and Chiang C. M. (1998) Properties of PC4 and an RNA polymerase II complex in directing activated and basal transcription in vitro. J. Biol. Chem. 273, 12492–12498 [DOI] [PubMed] [Google Scholar]
- 27. Xie J., Collart M., Lemaire M., Stelzer G., and Meisterernst M. (2000) A single point mutation in TFIIA suppresses NC2 requirement in vivo. EMBO J. 19, 672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ge H., and Roeder R. G. (1994) The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J. Biol. Chem. 269, 17136–17140 [PubMed] [Google Scholar]
- 29. Clemens K. E., Piras G., Radonovich M. F., Choi K. S., Duvall J. F., DeJong J., Roeder R., and Brady J. N. (1996) Interaction of the human T-cell lymphotropic virus type 1 Tax transactivator with transcription factor IIA. Mol. Cell. Biol. 16, 4656–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dion V., and Coulombe B. (2003) Interaction of a DNA-bound transcriptional activator with TBP-TFIIA-TFIIB-Promoter quaternary complex. J. Biol. Chem. 278, 11495–11501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kobayashi N., Boyer T. G., and Berk A. J. (1995) A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol. Cell. Biol. 15, 6465–6473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Solow S., Salunek M., Ryan R., and Lieberman P. M. (2001) TAFII250 phosphorylates human transcription factor IIA on serine residues important for TBP binding and transcription activity. J. Biol. Chem. 276, 15886–15892 [DOI] [PubMed] [Google Scholar]
- 33. Yokomori K., Admon A., Goodrich J. A., Chen J. L., and Tjian R. (1993) Drosophila TFIIA-L is processed into two subunits that are associatedwith the TBP/TAF complex. Genes Dev. 7, 2235–2245 [DOI] [PubMed] [Google Scholar]
- 34. Robinson M. M., Yatherajam G., Ranallo R. T., Bric A., Paule M. R., and Stargell L. A. (2005) Mapping and functional characterization of the TAF11 interaction with TFIIA. Mol. Cell. Biol. 25, 945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hansen S. K., Takada S., Jacobson R. H., Lis J. T., and Tjian R. (1997) Transcription properties of a cell type-specific TATA-binding protein TRF. Cell 91, 71–83 [DOI] [PubMed] [Google Scholar]
- 36. Rabenstein M. D., Zhou S., Lis J. T., and Tjian R. (1999) TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc. Natl. Acad. Sci. U.S.A. 96, 4791–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schimanski B., Nguyen T. N., and Günzl A. (2005) Characterization of a multisubunit transcription factor complex essential for spliced leader RNA gene transcription in Trypanosoma brucei. Mol. Cell. Biol. 25, 7303–7313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coleman R. A., Taggart A. K., Burma S., Chicca J. J. 2nd, and Pugh B. F. (1999) TFIIA regulates TBP and TFIID dimers. Mol. Cell 4, 451–457 [DOI] [PubMed] [Google Scholar]
- 39. Tan S., Hunziker Y., Sargent D. F., and Richmond T. J. (1996) Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 381, 127–151 [DOI] [PubMed] [Google Scholar]
- 40. Oelgeschläger T., Chiang C. M., and Roeder R. G. (1996) Topology and reorganization of a human TFIID-promoter complex. Nature 382, 735–738 [DOI] [PubMed] [Google Scholar]
- 41. Papai G., Tripathi M. K., Ruhlmann C., Layer J. H., Weil P. A., and Schultz P. (2010) TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature 465, 956–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lagrange T., Kim T. K, Orphanides G., Ebright Y. W., Ebright R. H., and Reinberg D. (1996) High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc. Natl. Acad. Sci. U.S.A. 93, 10620–10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bleichenbacher M., and Tan S., and Richmond T. J. (2003) Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes. J. Mol. Biol. 332, 783–793 [DOI] [PubMed] [Google Scholar]
- 44. Geiger J. H., Hahn S., Lee S., and Sigler P. B. (1996) Crystal structure of the yeast TFIIA/TBP/DNA complex. Science 272, 830–836 [DOI] [PubMed] [Google Scholar]
- 45. Deng W., Malecová B., Oelgeschläger T., and Roberts S. G. (2009) TFIIB recognition elements control the TFIIA-NC2 axis in transcriptional regulation. Mol. Cell. Biol. 29, 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang J., Zhao S., Zhou Y., Wei Y., and Deng W. (2015) Establishment and validation of a non-radioactive method for in vitro transcription assay using primer extension and quantitative real time PCR. PLOS ONE 10, e0135317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Coulombe B., Li J., and Greenblatt J. (1994)Topological localization of the human transcription factors IIA, IIB, TATA box-binding protein, and RNA polymerase II-associated protein 30 on a class II promoter. J. Biol. Chem. 269, 19962–19967 [PubMed] [Google Scholar]
- 48. Evans R., Fairley J. A., and Roberts S. G. (2001) Activator-mediated disruption of sequence-specific DNA contacts by the general transcription factor TFIIB. Genes Dev. 15, 2945–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mitsiou D. J., and Stunnenberg H. G. (2003) p300 is involved in formation of the TBP–TFIIA-containing basal transcription complex, TAC. EMBO J. 22, 4501–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kokubo T., Gong D. W., Yamashita S., Horikoshi M., Roeder R. G., and Nakatani Y. (1993) Drosophila 230-kD TFIID subunit, a functional homolog of the human cell cycle gene product, negatively regulates DNA binding of the TATA box-binding subunit of TFIID. Genes Dev. 7, 1033–1046 [DOI] [PubMed] [Google Scholar]
- 51. Auble D. T., and Hahn S. (1993) An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 7, 844–856 [DOI] [PubMed] [Google Scholar]
- 52. Dreos R., Ambrosini G., Périer R. C., Bucher P. (2015) The eukaryotic promoter database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 43, D92–D96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zaborowska J., Taylor A., Roeder R. G., and Murphy S. (2012) A novel TBP-TAF complex on RNA polymerase II-transcribed snRNA genes. Transcription 3, 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maston G. A., Zhu L. J., Chamberlain L., Lin L., Fang M., and Green M. R. (2012) Non-canonical TAF complexes regulate active promoters in human embryonic stem cells. Elife 1, e00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ge H., Martinez E., Chiang C. M., and Roeder R. G. (1996) Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 274, 57–71 [DOI] [PubMed] [Google Scholar]
- 56. Dreos R., Ambrosini G., Groux R., Cavin Périer R., Bucher P. (2017) The eukaryotic promoter database in its 30th year: focus on non-vertebrate organisms. Nucleic Acids Res. 45, D51–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]