Abstract
Ubiquinone (UQ), also referred to as coenzyme Q, is a widespread lipophilic molecule in both prokaryotes and eukaryotes in which it primarily acts as an electron carrier. Eleven proteins are known to participate in UQ biosynthesis in Escherichia coli, and we recently demonstrated that UQ biosynthesis requires additional, nonenzymatic factors, some of which are still unknown. Here, we report on the identification of a bacterial gene, yqiC, which is required for efficient UQ biosynthesis, and which we have renamed ubiK. Using several methods, we demonstrated that the UbiK protein forms a complex with the C-terminal part of UbiJ, another UQ biogenesis factor we previously identified. We found that both proteins are likely to contribute to global UQ biosynthesis rather than to a specific biosynthetic step, because both ubiK and ubiJ mutants accumulated octaprenylphenol, an early intermediate of the UQ biosynthetic pathway. Interestingly, we found that both proteins are dispensable for UQ biosynthesis under anaerobiosis, even though they were expressed in the absence of oxygen. We also provide evidence that the UbiK–UbiJ complex interacts with palmitoleic acid, a major lipid in E. coli. Last, in Salmonella enterica, ubiK was required for proliferation in macrophages and virulence in mice. We conclude that although the role of the UbiK–UbiJ complex remains unknown, our results support the hypothesis that UbiK is an accessory factor of Ubi enzymes and facilitates UQ biosynthesis by acting as an assembly factor, a targeting factor, or both.
Keywords: bioenergetics, coenzyme Q10 (CoQ10), electron transfer, Escherichia coli (E. coli), microbiology, ubiquinone
Introduction
Isoprenoid quinones are conserved in almost all living organisms and function primarily as electron and proton transporters in photosynthetic and respiratory chains (1). Quinones are composed of a polar redox-active head group coupled to a lipid side chain, which varies in both length and degree of saturation (1). Menaquinone (MK)4 and ubiquinone (UQ) are the main isoprenoid quinones and differ with respect to the structure of the head group, a naphthalene ring (in MK) and a benzene ring (in UQ). MK is present in both bacteria and archaea, whereas UQ is restricted to α-, β-, and γ-proteobacteria but is also found in eukaryotes (1–3). It is now commonly accepted that MK arose before UQ (4, 5). MK and demethylmenaquinone (DMK) function predominantly in anaerobic respiratory chains, whereas UQ is the major electron carrier used for reduction of oxygen by cytochrome oxidases (1). Quinone biosynthesis has been extensively studied in Escherichia coli but is still incompletely characterized. The octaprenyl side chain is common to UQ and (D)MK, and chorismate is the precursor of the benzoquinone and napthoquinone rings.
The enzymes catalyzing the different steps of UQ biosynthesis have been described (6). They catalyze the functionalization (prenylation, decarboxylation, hydroxylation, and methylation) of the phenyl ring of the 4-hydroxybenzoate precursor. In total 11 proteins, named from UbiA to -J and UbiX are required for UQ biosynthesis. The hydroxylases UbiI, UbiF, and UbiH use dioxygen as a co-substrate and are therefore active only in aerobic conditions (7, 8). The hydroxylases that participate in anaerobic UQ biosynthesis have yet to be identified (6). It is noticeable that only UbiE, a methylase, participates in both UQ and MK biosynthesis. Our recent investigations demonstrated that UQ biosynthesis requires additional factors that seem not to carry out enzymatic function. For example, we have recently discovered that UbiJ is required for UQ biosynthesis in aerobic conditions in both E. coli and Salmonella enterica (9). However, the role of UbiJ in UQ biosynthesis remains enigmatic.
In the present study, we discovered a novel gene, namely yqiC, important for UQ biosynthesis under aerobiosis in E. coli and S. enterica. Contrary to a recent report (10), MK biosynthesis was not impaired in the yqiC mutant. We therefore propose to change the name of yqiC to ubiK. The protein from E. coli was characterized biochemically and structurally. Interestingly, we demonstrate that UbiK physically interacts with UbiJ and forms an UbiK2–UbiJ1 complex. A Salmonella ubiK mutant is growth-deficient under aerobic conditions and the defect is exacerbated at higher temperatures. Consistently, Salmonella ubiK mutant is defective for proliferation in macrophages and mice infection. Overall, our results demonstrate the implication of UbiK in aerobic UQ biosynthesis and provide an additional illustration of the importance of UQ for pathogenic bacteria proliferation.
Results
ubiK is a new gene involved in aerobic UQ8 biosynthesis
A high-throughput study previously analyzed the growth of E. coli strains from the Keio single gene deletion library in more than 300 conditions and established a phenotypic signature for each mutant strain (11). Functional connection between genes with correlated phenotypic signatures could be predicted and the phenomic profiles of most ubi mutants were indeed highly correlated (http://ecoliwiki.net/tools/chemgen/)5 (11). Interestingly, the ubiI mutant had the highest correlation coefficient with the yqiC mutant (hereafter called ubiK), prompting us to investigate whether ubiK might play a role in UQ8 biosynthesis.
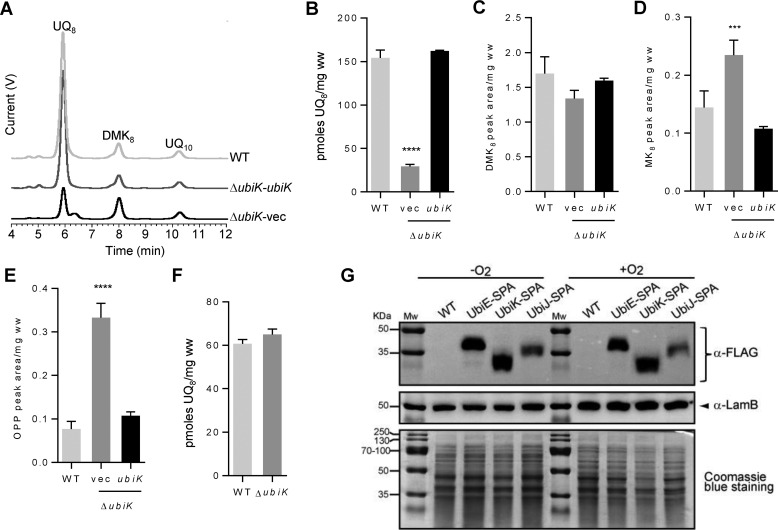
The ubiK mutant had no growth defect in LB medium under aerobic conditions (supplemental Fig. S1A). Its UQ8 content, as determined by HPLC analysis coupled to electrochemical detection (ECD), decreased down to 18% compared with the wild-type strain (Fig. 1, A and B). Complementation of the ubiK mutant with a plasmid carrying ubiK restored the wild-type UQ8 level (Fig. 1, A and B). DMK8 and MK8 were optimally detected at 247 nm (supplemental Fig. S1B) and we found that the levels of DMK8 were unchanged in the ubiK mutant (Fig. 1C), whereas those of MK8 increased slightly (Fig. 1D). HPLC chromatograms recorded at 275 nm showed a compound that eluted at 6.4 min, right after UQ8 in the ubiK mutant (supplemental Fig. S1C). Mass spectrometry analysis revealed a main peak at m/z 656.5 (M + NH4+) consistent with octaprenylphenol (OPP, C46H70O) (supplemental Fig. S1D). The accumulation of OPP, an intermediate in the biosynthesis of UQ8, has been observed previously in several ubi mutants deficient in UQ8 (12, 13). The increased level of OPP in the ΔubiK mutant was diminished to the WT level upon transformation by the plasmid carrying ubiK (Fig. 1E), showing that OPP accumulation correlated with UQ8 deficiency. The UQ8 content of the ubiK mutant strain after anaerobic growth in LB was found to be similar to WT (Fig. 1F). Finally, via insertion of a sequential affinity purification tag (SPA tag) at the 3′ end of the ubiK gene in the E. coli chromosome, UbiK levels were quantified by Western blot analysis after growth under aerobic and anaerobic conditions. We also analyzed the levels of SPA-tagged UbiE and UbiJ. Levels of the three proteins were not influenced by the growth conditions (Fig. 1G). This result was consistent with a recent study, which demonstrated that ubiJ and ubiK genes were regulated neither by Fnr nor ArcA, two major transcription factors that respond to oxygen levels (14). Altogether, our results showed that deletion of the ubiK gene impacts UQ8 biosynthesis in aerobic conditions but does not decrease (D)MK biosynthesis.
Figure 1.
A, HPLC ECD analyses of lipid extracts from 1 mg of E. coli WT or ΔubiK cells transformed with either the empty pK vector (vec) or the pK vector carrying the ubiK gene grown in LB medium in aerobic conditions. The chromatograms are representative of three independent experiments. The peaks corresponding to UQ8, DMK8, OPP, and the coenzyme Q10 (UQ10) standard are indicated. B, quantification of cellular UQ8 content of the E. coli strains described in A based on ECD signal (peak area). C and D, quantification of cellular DMK8 and MK8 from chromatograms at 247 nm (see supplemental Fig. S1B). E, quantification (peak area) of OPP from chromatograms at 275 nm (see supplemental Fig. S1D). B–E, mean ± S.E.; n = 3–5; ***, p < 0.001; ****, p < 0.0001, one-way analysis of variance. F, ECD quantification of cellular UQ8 content of the E. coli WT and ΔubiK cells grown in anaerobic conditions in LB medium; n = 3. G, immunodetection of SPA-tagged proteins UbiE, UbiK, and UbiJ after growth in LB medium under aerobic (+O2) or anaerobic (−O2) conditions. Equal loading was verified with immunodetection of the protein LamB and staining of the SDS-PAGE with Coomassie Blue dye. Mw, molecular mass.
UbiK is almost exclusively present in Proteobacteria
E. coli UbiK (UniProtKB entry Q46868) is a 96-residue protein that belongs to the BMFP (Brucella membrane fusogenic protein) superfamily (Pfam PF04380). Bioinformatics analysis revealed that the 596 members of the BMFP family known to date (Pfam 30.0, March 2017, http://xfam.org/)5 are almost exclusively found in α-, β-, and γ-proteobacteria, where UQ8 is present (Ref. 40 and supplemental Fig. S2). Interestingly, ubiK sequences are not found in the archaebacterial kingdom where MK is the major quinone (1). This exclusive presence of ubiK in the proteobacteria phylum is consistent with the involvement of ubiK in UQ8 biosynthesis and not in MK biosynthesis. An alignment of a subset of UbiK sequences from diverse proteobacteria species is shown in supplemental Fig. S3. The average sequence identity of all proteins from this family is 34%. One should note that the C-terminal end of UbiK sequences, corresponding to the last 16 amino acid residues in the E. coli protein, is poorly conserved among all different species. Consistent with this, we found the deletion of the C terminus (residues 81–96) had no effect on UbiK function because this construct yielded wild-type levels of UQ8 in the ΔubiK strain (not shown).
UbiK forms a trimer
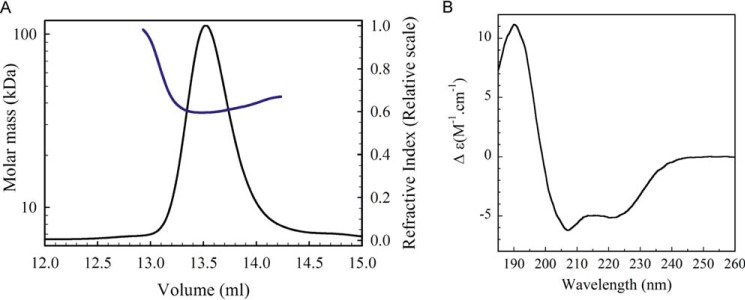
The ubiK gene from E. coli was cloned without any tag into a pACYCDuet-1 plasmid and the protein was over-expressed in the BL21(DE3) E. coli strain. High yield of pure and soluble UbiK could be obtained (10 mg/liter of culture). To determine its absolute molar mass, multi-angle light scattering coupled to size exclusion chromatography (SEC-MALS) experiments were performed. UbiK showed a single homogeneous peak with a molecular mass of 32 kDa (Rh = 2.9 ± 0.08 nm), thus corresponding to a trimer (molecular mass for monomer = 11.3 kDa), with an elongated hydrodynamic shape (f/f0 = 1.5) (Fig. 2A). Its secondary structure was analyzed by circular dichroism (CD) spectroscopy. The CD spectrum of UbiK exhibited two minima at 208 and 222 nm, indicating the prevalence of helices (Fig. 2B). Analysis of the spectrum using K2D3 and CDSSTR softwares allowed to estimate the percentage of helical structure of 52–55%, random coils of 42–45%, and β-sheets of 3–4%. Using analytical ultracentrifugation with fluorescence detection, a trimeric oligomer was observed at a concentration down to 250 nm, at which point the protein aggregates suggested it is unstable as a monomer. A thermal unfolding experiment was performed by monitoring the CD spectrum of UbiK as a function of temperature (supplemental Fig. S4). The midpoint of the unfolding transition (TM) was 66.5 ± 0.1 °C and the observed conformational transitions were fully reversible, which allowed us to determine the Van't Hoff enthalpy of unfolding, ΔH = 250.6 ± 2 kJ/mol. These values are indicative of a very stable protein, in accordance with a coiled-coil protein structure (15).
Figure 2.
A, SEC-MALS analysis of UbiK from E. coli (injection of 100 μl of UbiK at 2 mg/ml on a Superdex 200 10/300 GL increase column, 50 mm Tris-HCl, 150 mm NaCl buffer, pH 7.5. Solid and blue lines correspond to refraction index signal and molar mass, respectively. B, far-UV circular dichroism spectrum (185–260 nm) of E. coli UbiK (50 mm Tris-HCl, 150 mm NaCl buffer, pH 7.5).
Structural model of UbiK
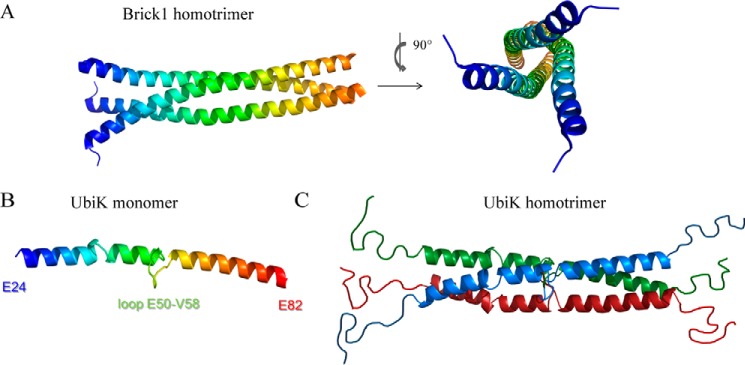
UbiK and human Brick1 share 25% sequence identity. Human Brick1 (UniProtKB entry Q8WUW1) forms the core subunit of the Wave (Wiskott-Aldrich VErprolin proteins) complex involved in actin polymerization. Brick1 contributes to the architecture of the complex by serving as a platform on which the four other proteins assemble. To gain insights into the structural organization of the UbiK trimer, a structural model was generated using Swiss-Model (http://swissmodel.expasy.org/). The PDB codes for the main templates used were: 3PP5 (Brick1 from Dictyostelium discoideum), 3P8C chain E (Brick1 from Homo sapiens), and 4LL7 (She3p from Saccharomyces cerevisiae) (16). When purified alone, Brick1 assembles as an α-helical triple coiled-coil along a well conserved hydrophobic patch (17) (Fig. 3A). The hydrophobic core of the Brick1 trimer bundle is also seen within the UbiK sequence (supplemental Fig. S3). The best model of UbiK monomer calculated with Swiss-Model thus consisted of two long aligned α helices connected by a loop in the central core of the protein and disordered domains at the N- and C-terminal ends (Fig. 3B). The secondary structures contents of this model matched the ones determined by circular dichroism (Fig. 2B): ∼53% of α-helices and 47% of random coils. On this basis we propose a model for the UbiK homotrimer, displaying the same fold as the triple coiled-coil structure in Brick1 (Fig. 3C).
Figure 3.
A, crystal structure of the α-helical coiled-coil Brick1 homotrimer from D. discoideum (PDB code 3PP5); B, predicted structural model of E. coli UbiK (Swiss model); C, hypothesized model of the α-helical coiled-coil UbiK homotrimer, based on sequence identity, CD, and SEC-MALS. N and C termini are predicted as disordered regions.
UbiK interacts with other Ubi proteins
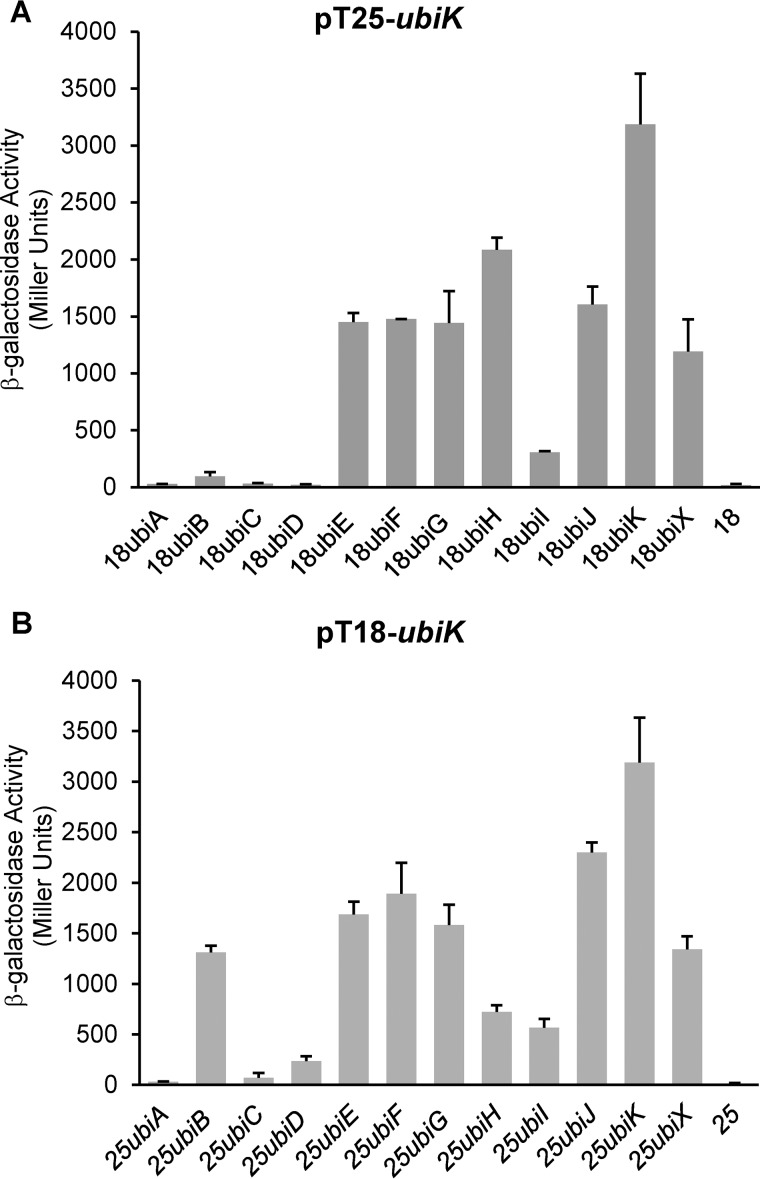
To answer whether UbiK interacts with other Ubi proteins in vivo, we used the bacterial BACTH two-hybrid reporter system that monitors both direct and indirect interactions. This method is based on functional complementation between adenylate cyclase fragments T18 and T25 expressed from two compatible plasmids (18). Adenylate cyclase activity is restored only when proteins fused to T18 and T25 interact. In line with our results about the oligomerization of UbiK, it was found to interact with itself (Fig. 4, A and B). In addition, UbiK interacted with the following Ubi proteins: UbiB, -E, -F, -G, -H, -I, -J, and -X (Fig. 4, A and B). More specifically, UbiK interacted with UbiE, -F, -G, -H, -I, -J, -K, and -X proteins regardless of whether it was fused to the T18 or T25 proteins. UbiK also interacted with UbiB but only when it was fused to the T18 moiety. Last, UbiK failed to interact with UbiA, UbiC, and UbiD (Fig. 4, A and B). Altogether, these results suggested that UbiK plays a pivotal role in E. coli UQ8 biosynthesis.
Figure 4.
Interactions between UbiK and the other Ubi proteins. A bacterial two-hybrid system was used to detect in vivo protein-protein interactions. Values of β-galactosidase activities, calculated in Miller units, with standard errors are shown: pT25-ubiK (A) or pT18-ubiK (B) were analyzed with ubiJ, -E, -I, -K, -B, -D, -G, -A, -X, -H, or -F genes, respectively.
UbiK interacts directly with UbiJ
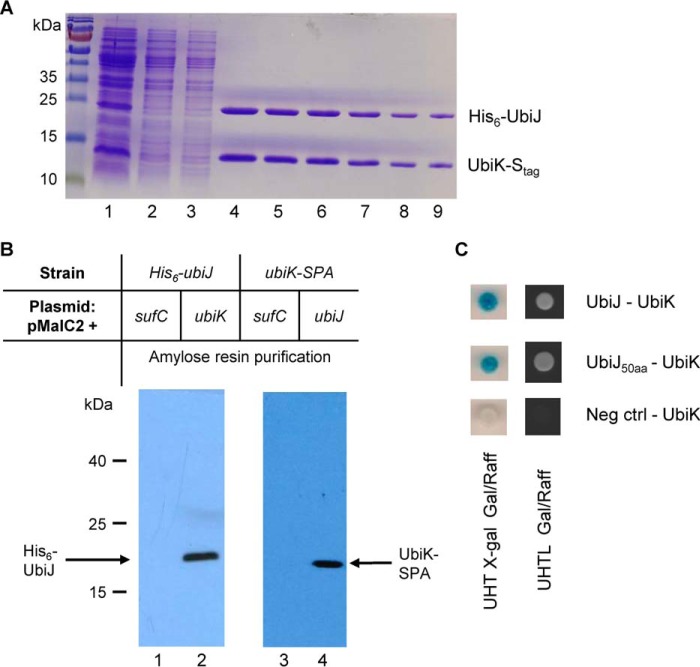
To further substantiate the idea that UbiK had a pleiotropic role in UQ8 biosynthesis, we used co-expression and co-purification assays to test the connection between UbiK and other Ubi proteins. For this we choose a subset of Ubi candidates among those suggested to partner with UbiK by the bacterial two-hybrid assay above. Likewise, UbiG, -J, -X, -E, and -J were fused to a His6 tag and UbiK to an S tag. Soluble fractions containing the His6-tagged proteins were purified on nickel-nitrilotriacetic acid resin, and UbiK was detected exclusively in extracts containing UbiJ–His6 (Fig. 5A). Interactions between the UbiK and UbiJ proteins were further investigated using pulldown assays. ubiK, ubiJ, and sufC (used as a negative control) genes were fused at their 5′-end with the malE gene. The chimeric proteins, MBP–UbiK and MBP–SufC, were purified in strains carrying a chromosomal copy of the his6–ubiJ gene, whereas on the other hand MBP–UbiJ and MBP–SufC were purified from a strain containing an ubiK–SPA gene. Soluble extracts of each strain were loaded on an amylose column. The column was then extensively washed and finally a solution of maltose (10 mm) was applied. MBP–UbiK and His6–UbiJ were co-eluted (Fig. 5B, lane 2) as MBP–UbiJ and UbiK–SPA (Fig. 5B, lane 4). MBP–SufC was always eluted alone (Fig. 5B, lanes 1 and 3). His6–UbiJ and UbiK–SPA were identified by immunoblotting using anti-His (Gene Tex) and anti-FLAG (Sigma) antibodies, respectively. Thus, biochemical analyses allowed us to confirm that UbiK interacts with UbiJ. Last, yeast two-hybrid-based tests were run as they report on direct interactions and might eventually allow to pinpoint zones of interaction. Evidence of an UbiK–UbiJ direct interaction was found to depend on the C terminus 50 amino acids of UbiJ only (UbiJ(151–201)) (Fig. 5C). No interaction was observed using the N terminus part of UbiJ (data not shown). Altogether, these results established unambiguously that UbiK and UbiJ interact specifically and form a complex in vivo.
Figure 5.
A, UbiJ-His and UbiK-S tag were co-expressed from the pETDuet-6his-ubiJ/ubiK-S tag plasmid. Cell extracts were passed over a nickel-nitrilotriacetic acid column. Imidazole gradient was applied and the two proteins were co-eluted. Lane 1, soluble cell extract; lane 2, flow through; lane 3, wash; lanes 4–9, imidazole eluted peak fractions. B, pulldown assays were carried out to investigate the interactions between the UbiK and UbiJ proteins. ubiK, ubiJ, and sufC (negative control) genes were fused at their 5′ end with the malE gene and the chimeric proteins were purified in His6–ubiJ (lanes 1 and 2) or ubiK–SPA (lanes 3 and 4) backgrounds. Soluble extracts of each strain were loaded onto an amylose column, the column was washed and a solution of maltose was applied. MBP–UbiK and His6–UbiJ were co-eluted (lane 2) as MBP–UbiJ and UbiK–SPA (lane 4). C, interactions between UbiK and UbiJ (top), UbiK and the 50 C-terminal amino acids of UbiJ (middle), and UbiK and an empty plasmid (bottom) were analyzed by a yeast two-hybrid reporter system. Diploid strains producing pairs of Ubi proteins were tested on solid medium for activation of the lacZ gene and the intensity of interaction was monitored by a gradient from dark blue (strong interaction) to white (no interaction) (left). The ability to grow on selective medium lacking leucine was also monitored during 16 h at 30 °C (right).
Biochemical characterization of the E. coli UbiK–UbiJ complex
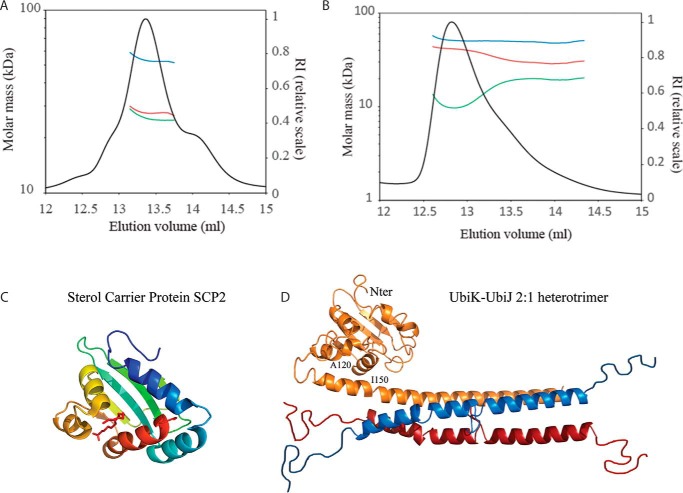
E. coli UbiK and UbiJ were co-expressed in E. coli BL21 strain, using the T7 promoter pETDuet-1 expression vector. A two-step purification consisting of an affinity column (His Trap HP) followed by size exclusion chromatography (HiLoad 16/600 Superdex 200 pg) resulted in three well separated protein peaks (supplemental Fig. S5). The low-molecular mass peak, called P3, with a molar mass of ∼18–20 kDa based on SDS-PAGE, was analyzed by ESI-MS and peptide mass fingerprint (supplemental Fig. S6). P3 did not contain any UbiK protein but only two main fragments of UbiJ: one major species lacking the last 56 C-terminal residues (UbiJ(Δ146–201)) and one minor species, lacking the last 63 C-terminal residues (UbiJ(Δ138–201)) (supplemental Figs. S5 and S6). These truncated forms are most likely a result of proteolysis during purification. In contrast, the first (minor, P1) and the second (major, P2) peaks contained both UbiK and UbiJ in seemingly different ratios. They were both subjected to SEC-MALS and analytical ultracentrifugation. SEC-MALS analysis of P2, eluting at 13.4 ml, led to a single molar mass (49 kDa) with a hydrodynamic radius of 3.6 ± 0.4 nm and a frictional ratio f/f0 of 1.7 that matches an elongated heterocomplex of two monomers of UbiK (28 kDa) and one monomer of UbiJ (21 kDa) (Fig. 6A). Sedimentation velocity experiments revealed three different species, with the sedimentation coefficients: 2.8 s20,w (∼60%), corresponding to a 2:1 UbiK–UbiJ complex; 3.8 s20,w (∼20%); 5.6 s20,w (∼10%) corresponding to larger oligomeric species (supplemental Fig. S7A). The stoichiometry of the dominant species remained unchanged with increased protein concentrations (supplemental Fig. S7B), indicating a stable UbiK–UbiJ (2:1) heterocomplex. P1 eluting at 12.8 ml (Fig. 6B) consists mainly of two species: the minor component is UbiK homotrimer and the major component is a heterotrimer of UbiK–UbiJ (2:1). Evidence for the presence of the UbiK homotrimer in P1 is given by sedimentation velocity experiments. The 2.2 s20,w species in P1 is only seen by interference (supplemental Fig. S7C), and not in absorbance (supplemental Fig. S7D), thus indicating the presence of UbiK only, and has furthermore, a sedimentation coefficient similar to that of the homotrimeric UbiK (2.1 s20,w) (supplemental Fig. S7E). The heterotrimer of UbiK–UbiJ in P1 has a different sedimentation coefficient (3.2 s20,w) than the main species in P2 (2.8 s20,w) probably due to having a different 3D conformation. This is further suggested by the varying frictional ratio seen between P2 and P1, 1.80 f/f0 and 1.55 f/f0, respectively. This higher frictional ratio seen for P2 highlights its more elongated form and explains the higher sedimentation coefficient.
Figure 6.
SEC-MALS analysis of the two complexes P1 (A) and P2 (B) of UbiK–UbiJ. Samples were run on a Superdex 200 Increase 10/300 GL. Blue, red, and green curves correspond to the molar masses of the UbiK–UbiJ complex, UbiK and UbiJ, respectively (protein-conjugate method). C, crystal structure of the sterol carrier protein SCP2 from Yarrowia lipolytica, with palmitic acid ligand. D, hypothesized model of the E. coli UbiK–UbiJ 2:1 heterotrimer complex, made by Phyre 2. UbiJ monomer is colored orange, UbiK dimer is colored blue and red. The template for the N-terminal domain of UbiJ(1–120) was PDB 4JGX (SCP2 from Y. lipolytica); the template for the C-terminal domain (151–195) was PDB 3QH9 (human coiled-coil liprin) and the template for the middle domain (132–150) was PDB 1M1J (fibrinogen α subunit).
To further confirm the UbiJ C-terminal interaction with UbiK, we produced three plasmids encoding UbiK without an S tag alongside three truncated versions of UbiJ (His tag), UbiJ(Δ121–201), UbiJ(Δ1–120), and UbiJ(Δ1–145) (supplemental Fig. S8). Upon coexpression and purification of UbiK and UbiJ truncations by nickel affinity column, we observed that the C terminus of UbiJ (UbiJ(Δ1–120) and UbiJ(Δ1–145)) is essential for binding UbiK and that the N terminus (UbiJ(Δ121–201)) is not able to bind UbiK. These in vitro results confirm the in vivo double hybrid assays (Fig. 5C) that show that only the C terminus is responsible for UbiK binding.
In the absence of a three-dimensional structure, we also built a model for the heterotrimer UbiK–UbiJ complex. Sequence homologies between UbiK and Brick1 and between UbiJ and Sterol Carrier Protein (SCP-2) proteins (supplemental Fig. S9 and 6C), which are intracellular lipid transfer proteins, as well as in silico structural predictions allowed us to propose the model shown in Fig. 6D. In this model the UbiK–UbiJ 2:1 heterocomplex results from the formation of a three-helix bundle between two monomers of UbiK and the α-helical C terminus of one monomer of UbiJ, in full agreement with the latter being responsible for UbiJ binding to UbiK.
UbiK–UbiJ bind lipids
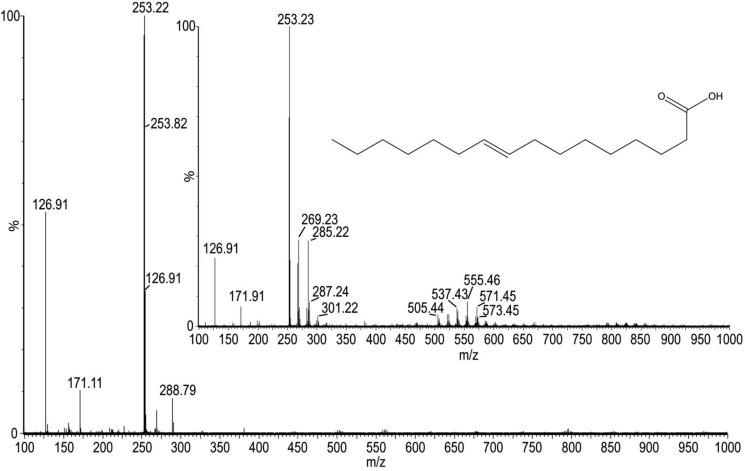
E. coli UbiJ is made of an N-terminal sterol carrier protein (SCP-2) domain (1–120) and a putative C-terminal coiled-coil domain (121–201) (supplemental Fig. S9). SCP-2, also known as nonspecific lipid transfer protein, is a small ubiquitous protein of unknown function. Several existing X-ray or NMR structures of SCP-2 proteins revealed the protein accommodates an internal and hydrophobic cavity suitable for lipid binding (19–23). As a matter of fact, fatty acid molecules, such as palmitic and stearic acids, were observed in the various SCP-2 members. Therefore, we tested if a lipid molecule might be bound to the purified UbiK–UbiJ complex. The purified UbiK–UbiJ P2 complex was treated with methanol, heated at 95 °C, and centrifuged. The supernatant was then injected onto an ESI/MS apparatus equipped with a triple TOF detector in a negative mode (Fig. 7). Interestingly, in this supernatant a mass at m/z = 253.22 was identified as palmitoleic acid (commercial standard m/z = 253.23). Palmitoleic acid, (9Z)-hexadec-9-enoic acid, is a C16 monounsaturated lipid that is largely found in E. coli, along with other C14 and C18 fatty acids: it represents 35% of the lipids within the membrane of the bacterium, whereas palmitic acid represents 45%, vaccenic acid 18%, and myristic acid 2%. Similar experiments using UbiK alone revealed no lipid binding (data not shown).
Figure 7.
ESI-TOF-MS of UbiK–UbiJ P2 supernatant. The m/z 263.22 seen in the UbiK–UbiJ sample corresponds to the exact mass of palmitoleic acid, m/z 263.23 (commercial standard inset).
The Salmonella ubiK gene is required for UQ8 biosynthesis, intracellular proliferation, and virulence in mice
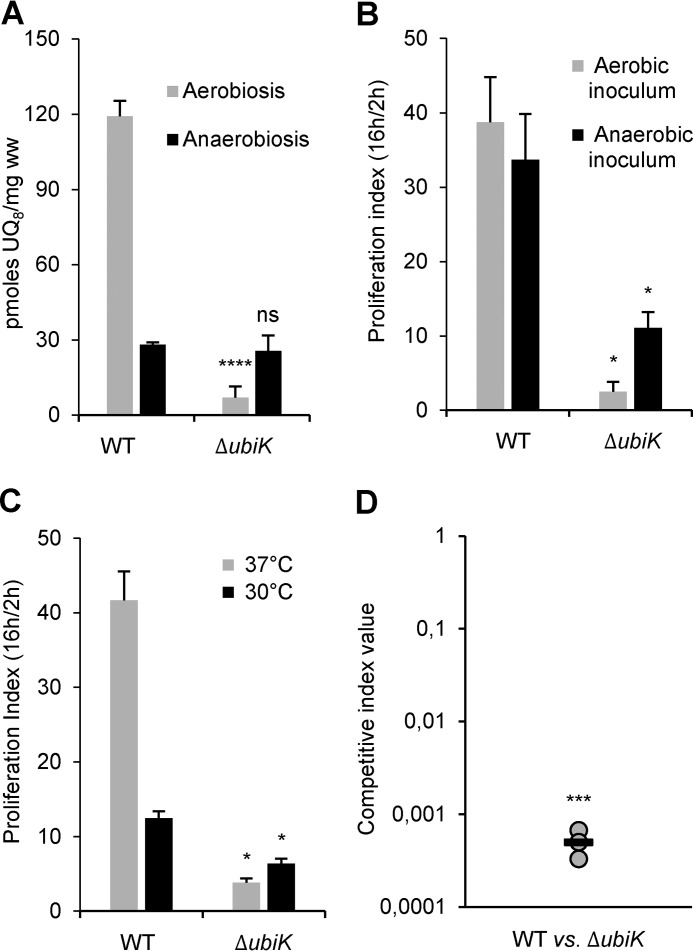
Previously, we found that ubiJ was required for UQ8 biosynthesis and intracellular proliferation in S. enterica (9). Given the functional interaction between UbiK and UbiJ in E. coli reported above, we constructed an ubiK mutant in S. enterica and measured its UQ8 content. Compared with the wild-type strain, we found a 16-fold decrease in UQ8 when the ubiK mutant was grown in LB under aerobic conditions (Fig. 8A). Complementation of this mutant led to an increase of the UQ8 level to ∼70% of the WT (supplemental Fig. S10A). The levels of DMK8 and MK8 were not significantly different between ΔubiK and WT strains (supplemental Fig. S10B). As already observed in E. coli, the UQ8 content of the ubiK mutant was not different from that of the WT after growth in anaerobic conditions (Fig. 8A). Next, RAW 264.7 mouse macrophages were infected with the wild-type strain and the ubiK mutant. Bacterial proliferation was assayed by calculating the proliferation index as the ratio of the number of intracellular bacteria at 16 h post-infection to that at 2 h post-infection. Because important differences were found in the function of UQ8 content in the presence or absence of oxygen, bacterial inoculum were grown either aerobically or anaerobically. The ubiK mutant exhibited a proliferation index 16-fold lower compared with its wild-type parent with an aerobic inoculum and 3-fold lower with an anaerobic inoculum (Fig. 8B). Interestingly, the intracellular proliferation of the ubiK mutant was found to be higher when the infection was performed with an anaerobic inoculum (11) compared with an aerobic inoculum (2.5) (Fig. 8B). Because an ubiK mutant was previously shown to exhibit a thermosensitive phenotype in Salmonella (24), we monitored the growth of wild-type and an ubiK mutant at different temperatures and indeed found that growth impairment was exacerbated at higher temperatures (supplemental Fig. S10C). Thus, to investigate the impact of the temperature on the intracellular proliferation capacity of the ubiK mutant, infection of RAW 264.7 mouse macrophages was carried out at 30 and 37 °C. The proliferation index of the ubiK mutant was severely affected compared with the wild-type strain at 37 °C (12-fold), whereas the difference was much more reduced at 30 °C (2-fold) (Fig. 8C). The UQ8 deficiency of the ubiK mutant compared with the wild-type strain was less severe at 30 °C than at 37 °C (9- and 16-fold, respectively) (supplemental Figs. S8A and S10D), which suggest that the increased proliferation index of the ubiK mutant at 30 °C may be due to the higher UQ8 content. Last, mixed infections of mice were carried out using the ubiK mutant and wild-type strain to determine the competitive index (CI) and compare the virulence of the two strains (25). Mice were infected intraperitoneally and the ubiK mutant was found to be highly affected for its virulence in the animal (CI = 0.0005) (Fig. 8D). Collectively, these results confer to ubiK a key role in UQ8 biosynthesis and in Salmonella virulence.
Figure 8.
A, UQ8 content of S. enterica WT and ΔubiK cells after growth at 37 °C in LB medium in aerobic (O2) or anaerobic conditions. Mean ± S.E.; n = 4; ns, nonsignificant; ****, p < 0.0001, unpaired two-tailed t test. B and C, Salmonella strains (WT and ΔubiK) were grown overnight under aerobic (gray bars, B) or anaerobic conditions (black bars, B) in LB at 37 °C. Opsonized bacteria were phagocytosed by RAW 264.7 cells. The experiments were carried out at 37 °C (gray bars, C) or 30 °C (black bars, C). Two and 16 h post-infection, mouse macrophages were lysed for enumeration of intracellular bacteria (gentamicin-protected) determined by colony-forming unit (cfu) counts. The values shown represent the proliferation index calculated as a ratio of the intracellular bacteria between 16 and 2 h postinfection. Results are the mean ± S.D. of at least three independent experiments each in triplicate. Asterisks indicate a statistically significant difference between the ubiK mutant and the WT. *, p ≤ 0.05 (Mann-Whitney U test). D, BALB/c mice were inoculated intraperitoneally with a 1:1 mixture of ubiK mutant and wild-type Salmonella strains. Forty-eight hours post-inoculation, spleens were harvested for bacterial counts. Competitive indexes of wild-type versus ubiK mutant strains in mice were determined. Each circle represents one mouse and the horizontal bar corresponds to the mean. A one-sample t test was used to determine whether the CI was significantly different from 1. ***, p ≤ 0.001.
Discussion
UQ8 acts as a membrane-embedded electron shuttle and is a key molecule for bacteria to use respiratory metabolism for growth. Biosynthesis of UQ8 includes a series of enzymatic steps in which a benzene ring undergoes a series of modifications (prenylation, decarboxylation, methylation, and hydroxylation). Recently, however, we obtained evidence that non-enzymatic biogenesis factors might also be required. Indeed, we identified UbiJ as a new factor involved in UQ8 biosynthesis in E. coli and Salmonella under aerobiosis (9). In the present study, we identified an additional new biogenesis factor, called UbiK, which is also required for UQ8 biosynthesis and is unlikely to carry out any enzymatic function. Phenotypic and biochemical characterization revealed that UbiK and UbiJ function as a complex that contributes to as yet unclear step(s) in UQ8 biosynthesis. We collected evidence that allow us to discuss their possible role in helping other enzymatic Ubi proteins to carry out their function.
Our results and interpretation differ from a recent study that concluded that UbiK was involved in MK biosynthesis in Salmonella (10). This conclusion was based on an analysis of the quinone content of an ubiK mutant. We did not confirm such results as, in our analysis, the levels of (D)MK8 were not decreased in the ubiK mutant compared with the wild-type strain in both E. coli and S. enterica Typhimurium. We believe that this discrepancy is due to the erroneous identification of Salmonella endogenous MK8 in the HPLC chromatograms by Wang et al. (10). Indeed, these authors used commercial vitamin K2 (MK4) as a standard to identify the retention time of MK8 in chromatograms of Salmonella lipid extracts, whereas MK4 and MK8 have an entirely different retention time on C18 columns.
The occurrence of an UbiK–UbiJ complex was demonstrated by using multiple molecular, biochemical, and physical methods. Besides all evidences for the occurrence of an UbiK–UbiJ complex, we noticed marked changes in the behavior of UbiJ and UbiK depending on whether the proteins were overexpressed individually or together. Indeed, whereas UbiK alone is a trimer, it is converted to a dimer upon interaction with one UbiJ monomer, and whereas UbiJ is insoluble on its own it is perfectly soluble when interacting with UbiK. In the absence of a three-dimensional crystal structure, we are able to propose a model for that complex. In this model a UbiJ monomer and UbiK dimer make a heterotrimeric complex allowed by the formation of a three-helix bundle from two monomers of UbiK and the helical C terminus of UbiJ.
Human Brick1 (UniProtKB entry Q8WUW1) forms the core subunit of the Wave (Wiskott-Aldrich VErprolin proteins) complex involved in actin polymerization. Interestingly, the 400-kDa Wave complex consists of five proteins and Brick1 (HSPC300) contributes to the architecture of this mega complex by serving as a platform to the other proteins (16). On this basis, one can speculate that UbiK, and by extension the UbiK–UbiJ complex, might participate to some sort of platform. Such a role would be consistent with the fact that none of them are predicted to carry out an enzymatic function, and that inactivation of both drastically reduces the level of UQ8 but lead only to the accumulation of the early intermediate OPP, which is not diagnostic of a defect in a specific biosynthetic step. The hypothesis that all Ubi enzymes are part of, and function as a mega complex has received some support in bacteria and is well established in eukaryotes (26–28). That the UbiK–UbiJ is instrumental in the formation of such a complex is supported by data of the bacterial two-hybrid analysis. Indeed, UbiK was found to be connected, directly or not, with most Ubi proteins. Hence it is tempting to think of the UbiK–UbiJ complex as a facilitator of the formation (folding, assembly, and nucleation) of a Ubi mega complex. Another possibility, which is not exclusive from the one just discussed, is that UbiK–UbiJ helps location or interaction of the Ubi enzymes with the membrane. Indeed, a conundrum is that whereas UQ8 biosynthesis must take place within the membrane environment, only one of the Ubi proteins, UbiA, is actually membrane-bound. The obvious need of locating the Ubi enzymes or complex to the membrane could be fulfilled by the UbiK–UbiJ complex. This putative function is consistent with our demonstration that UbiK–UbiJ is able to bind lipids.
During the course of the infection, Salmonella can be exposed to different stressful conditions and must negotiate anaerobic and aerobic environments (29). In this work, we provided evidence that UbiK is required for full multiplication in macrophages and full virulence in mice. This is in agreement with a previous study by Carrica et al. (24) who reported that ubiK was important for Salmonella virulence in a mouse model. However, because the ubiK mutant exhibits a thermosensitive phenotype, it is difficult to sort out the global defect from the specific contribution to virulence. Interestingly, we found the ubiK mutant proliferation to be partially recovered when the bacterial inoculum was grown anaerobically or when the infection was carried out at 30 °C, two conditions during which UQ8 levels increased in the ubiK background. This is both consistent with our previous proposal for a requirement of UQ8 for an efficient intracellular proliferation and in support of the hypothesis that Salmonella intracellular lifestyle relies on aerobic respiration and oxygen availability to survive and proliferate within host cells.
Experimental procedures
Bacterial strains and growth conditions
E. coli strains were grown in Luria-Bertani (LB) medium at 37 °C in Erlenmeyer flasks filled to 1/10 of the maximal volume and shaken at 150 rpm (aerobic conditions). For anaerobic growth, Hungate tubes were used and the LB medium was extensively degassed with argon before the tubes were closed and autoclaved. Cultures were inoculated through the septum of the Hungate tube with a syringe fitted with a sterile needle. Ampicillin (50 μg/ml), kanamycin (25 μg/ml), and chloramphenicol (25 μg/ml) were added when needed. Deletion of various genes and concomitant insertion of an antibiotic resistance cassette was carried out using λ-Red-mediated recombination (30). Mutations were moved to the wild-type E. coli strain MG1655 by P1 transductions and to the wild-type S. enterica strain 12023 by P22 transductions.
Plasmid constructions
The ubiK and ubiJ inserts were obtained by PCR amplification using MG1655 as template and the oligonucleotides 5-EcoRI-ubiK/3-XhoI-ubiK and 5-EcoRI-ubiJ/3-XhoI-ubiJ, respectively. ubiK and ubiJ inserts were EcoRI-XhoI digested and inserted into EcoRI-SalI-digested pTrc99A or pBAD24 plasmids, yielding the pTrc-ubiK and pTrc-ubiJ plasmids or pA-ubiK and pA-ubiJ plasmids, respectively.
To overproduce MBP–UbiK and MBP–UbiJ, the ubiK and ubiJ genes were cloned into the pMal-c2 vector downstream of the Ptac promoter. The ubiK and ubiJ inserts were obtained by EcoRI-HindIII digestion of the pTrc-ubiK and pTrc-ubiJ plasmid and the restricted fragments were inserted into EcoRI-HindIII-digested pMal-c2, yielding pMal-ubiK and pMal-ubiJ.
The pEG202 and pJG4–5 vectors were used to express UbiK, UbiJ, and UbiJ50aa proteins fused to the DNA-binding protein LexA and to the transcriptional activation motif B42, respectively. The inserts were obtained by PCR using MG1655 chromosomal DNA as a template and 5-EcoRI-ubiK/3-XhoI-ubiK, 5-EcoRI-ubiJ/3-XhoI-ubiJ, and 5-EcoRI-ubiJ50aa/3-XhoI-ubiJ primers. The inserts were digested by EcoRI-XhoI. The restricted ubi genes containing inserts were cloned into the EcoRI-XhoI-digested pEG202 and pJG4-5, yielding the pB42 and pLexA plasmid series, respectively.
To construct pETDuet-ubiK-S-tag. The coding sequence of the E. coli ubiK gene was PCR amplified from an MG1655 strain by using the 5-NdeI-ubiK and 3-XhoI-del-stop-ubiK primers, digested using NdeI and XhoI, and cloned into pETDuet-1, yielding pETDuet-ubiK-S-tag. To construct pETDuet-ubiK-no-tag, the ubiK gene was PCR amplified by using the 5-NdeI-ubiK and 3-XhoI-ubiK primers, digested using NdeI and XhoI, and cloned into pETDuet-1, yielding pETDuet-ubiK-no-tag.
ubiJ insert was obtained by EcoRI-HindIII digestion of the pTrc99A-6his-ubiJ vector. The restricted fragment was inserted into pETDuet-ubiK-S-tag EcoRI-HindIII-digested, yielding pETDuet-6his-ubiJ/ubiK-S-tag.
To modify pETDuet-6his-ubiJ/ubiK-S-tag and remove the S tag, a TGA stop codon was inserted using 5-ubiKstop and 3-ubiKstop primers. The pETDuet-6his-ubiJ/ubiK (without S tag) was used to generate the three pETDuet-ubiJ truncated versions: UbiJ(Δ121–201), UbiJ(Δ1–120), and UbiJ(Δ1–145). All pETDuet plasmids versions were verified by DNA sequencing and then transformed into E. coli BL21(DE3).
Strain construction
The yqiC::Kan mutation from the BW25113 strain (Keio library) was transduced into a MG1655 strain by P1 transduction. To build the His6-ubiJ strain, an insert carrying the lacZ-aadA7(SpcR)-ptac-His6-ubiJ-lacZ gene was obtained after PvuII-ScaI digestion of the pGEMT-aadA7(SpcR)-ptac-His6-ubiJ vector. In the resulting fragment, the His6-ubiJ gene is under control of the Ptac promoter. Replacement of the lacZ allele by the LacZ-aadA7(SpcR)-ptac-His6-ubiJ-LacZ gene was carried out in the ubiJ strain as described by Datsenko and Wanner (30).
The DY330 ubiC-SPA strain encompassing the SPA tag DNA sequence and the kanamycin antibiotic resistance marker cassette (KanR) was used as a template in polymerase chain reaction (PCR) amplification. A 5-ubiK-SPA gene-specific forward primer, located immediately upstream of the ubiK gene stop codon, and a 3-ubiK-Kan gene-specific reverse primer, located immediately downstream of the target gene stop codon, were used to amplify the SPA tag and KanR cassette. The purified PCR product is subsequently targeted to integrate at the 3′ end (immediately upstream of the native stop codon) of an ubiK gene in the BW25113 strain in which the λ-Red recombination machinery is expressed. The ubiK–SPA–Kan was transduced through P1 phage from BW25113 to MG1655.
Analyses of the quinone content
Quinone extraction and quantification by HPLC-ECD analyses were performed as previously described (31). UQ10 was used as a standard and a precolumn guard cell set at +650 mV allowed the quinones to be detected in their oxidized form. ECD was performed with a Coulochem III (Thermo Fisher) equipped with a 5011A analytical cell (E1, −650 mV; E2, +650 mV), and UV detection at multiple wavelengths was performed with a diode array detector. The peaks for DMK8 and MK8 were integrated from the 247-nm chromatograms and for OPP from the 275-nm chromatograms. Similarly to the UQ8 signal, the peak areas were corrected for sample loss during the lipid extraction based on the recovery of the UQ10 standard and values were expressed per mg of cell wet weight. When mass spectrometry (MS) detection was needed, the flow was divided after the diode array detector with an adjustable split valve (Analytical Scientific Instruments) to allow simultaneous EC (60% of the flow) and MS (40% of the flow) detections. MS detection was performed in positive mode with electrospray ionization on a MSQ Plus spectrometer (Thermo Fisher). The probe temperature was 450 °C, the cone voltage was 80 V, and MS spectra were recorded between m/z 600 and 880 with a scan time of 0.4 s.
Western blot analysis
E. coli cells were harvested in exponential phase, washed with PBS buffer, and heated at 95 °C for 10 min in Laemmli buffer. Equal amounts of E. coli protein extracts were analyzed on a 15% acrylamide SDS-PAGE gel. Immunoblot analyses were performed with polyclonal antibodies raised against LamB (courtesy of A. Meinke, Intercell AG), monoclonal antibodies were raised against the FLAG epitope (Sigma) included in the SPA tag (32) and monoclonal antibodies raised against the His epitope (Gene Tex). Immunodetection was performed using goat anti-mouse HRP-conjugated (Bethyl Laboratories) or goat anti-rabbit HRP-conjugated (Covalab) and the Clarity Western ECL substrate (Bio-Rad). Primary antibodies were used at the following dilutions: anti-LamB, 1/2000, and anti-FLAG epitope, 1/5000.
Bacterial two-hybrid reporter system
The bacterial two-hybrid reporter system is based on functional complementation between Bordetella pertussis adenylate cyclase fragments, T18 and T25, expressed separately from two compatible plasmid replicons (18). Adenylate cyclase activity is restored only when proteins fused to T18 and T25 interact. Functional reconstitution of B. pertussis adenylate cyclase in an E. coli Δcya lac+ strain was monitored by assaying the activity of a cAMP-CRP-dependent lac promoter of a chromosomally encoded lac operon. Fusions were constructed with T18 and T25 present at the N-terminal of Ubi proteins.
Yeast two-hybrid system
The yeast two-hybrid system assay was performed as described by Golemis et al. (33). The β-galactosidase activity from diploid cells, obtained by mating of strains EGY48 and RF206 carrying the appropriate plasmid, was detected on plates containing X-gal (33) and quantified from liquid culture. The β-galactosidase activity was expressed as nanomoles of o-nitrophenyl-β-d-galactoside hydrolyzed per min/mg of protein. The diploid cells were also assayed for the expression of the Leu reporter gene. Diploid strains were grown in liquid selective CM medium lacking leucine.
Pulldown of MBP hybrid proteins
MG1655 ubiK–SPA cells transformed with pMal–sufC and pMal–ubiJ and MG1655 His6–ubiJ cells transformed with pMal–sufC and pMal–ubiK were grown at 37 °C in 200 ml of LB medium to an A600 of 1.0. Induction was performed with 0.1 mm IPTG, which was added for 3.5 h at 37 °C. The bacterial pellet was resuspended in 8 ml of buffer A (100 mm Tris-HCl, pH 7.5, 50 mm NaCl) and disrupted twice by French pressure treatment. The cell lysate was centrifuged at high speed for 15 min at 4 °C. Supernatants were added to 300 μl of amylose resin (New England Biolabs). The mixture was stored for 3.5 h at 4 °C. The resin was then washed with 30 resin volumes of buffer A. The recombinant proteins, MBP–UbiK, MBP–UbiJ, and MBP–SufC, were eluted with buffer A added with 10 mm maltose. For detecting His6–UbiJ, 0.5-ml elution fractions concentrated ×10 on Microcon (Millipore) were analyzed by immunoblotting using anti-His antibodies (Gene Tex), whereas for detecting UbiK-SPA protein, aliquots of the 0.5-ml fractions were analyzed using anti-FLAG (Sigma) antibodies.
Expression and purification of UbiK
Overexpression was performed in BL21(DE3) E. coli strain transformed with pETDuet-ubiK-no-tag. Cells were grown with shaking (200 rpm) at 37 °C, in LB medium containing ampicillin (100 μg/ml), until they reached A600 nm = 0.5, when overexpression was induced by addition of IPTG (0.4 mm). After induction, cells were grown for 4 h at 37 °C, and then harvested by centrifugation at 5000 × g for 10 min. All subsequent operations were carried out at 4 °C. Cells were resuspended in 5 volumes of buffer B (50 mm Tris-HCl, pH 7.5, 20 mm NaCl, 1 mm Pefabloc) and lysed by sonication (Branson Digital Sonifier, Amplitude 40% for 10 min). Nucleic acids were precipitated by streptomycin sulfate (2% w/v) and the crude extracts were then submitted to ultracentrifugation at 180,000 × g for 90 min (Optima XPN-80, rotor 50.2 Ti, Beckman Coulter). The resulting supernatant was loaded onto a Hiprep 26/10 Desalting column (GE Healthcare). The resulting desalted proteins were loaded onto a Hitrap Q FF (5 ml, GE Healthcare) equilibrated with buffer A. Bound proteins were eluted with a gradient of NaCl (0–500 mm) in 50 mm Tris-HCl, pH 7.5, buffer, and UbiK was eluted at 120 mm NaCl. The most pure UbiK fractions were pooled and loaded onto a HiLoad 26/600 Superdex 200 pg equilibrated with 50 mm Tris-HCl, pH 7.5, 150 mm NaCl. The elution volume of UbiK was 185 ml. UbiK fractions were pooled, frozen in liquid N2, then stored at −80 °C.
Expression and purification of UbiK–UbiJ complexes
Overexpression was performed in BL21(DE3) E. coli strains transformed with pETDuet-6his-ubiJ/ubiK-S-tag. Cells were grown with shaking (200 rpm) at 37 °C, in LB medium containing ampicillin (100 μg/ml), until they reached A600 nm = 0.4, when overexpression was induced by addition of IPTG (0.2 mm). After induction, cells were grown for 4 h at 37 °C, and then harvested by centrifugation at 5000 × g for 10 min. All subsequent operations were carried out at 4 °C. Cells were resuspended in 5 volumes of buffer B and lysed by sonication (Branson Digital Sonifier, Amplitude 40% for 10 min). Crude extracts were then submitted to ultracentrifugation at 180,000 × g for 90 min (Optima XPN-80, rotor 50.2 Ti, Beckman Coulter). The resulting supernatant was loaded onto a Hitrap Chelating HP column (5 ml, GE Healthcare) equilibrated with buffer A. Bound proteins were eluted with a gradient of imidazole (0–500 mm) in buffer, 50 mm Tris-HCl, pH 7.5, 150 mm NaCl. The most pure UbiK–UbiJ complex fractions were pooled and loaded onto a HiLoad 26/600 Superdex 200 pg equilibrated with 50 mm Tris-HCl, pH 7.5, 150 mm NaCl. The fractions corresponding to peak 1 and peak 2 fractions were pooled, frozen in liquid N2, stored at −80 °C, and then submitted to SEC-MALS analysis.
Circular dichroism
Circular dichroism in the far-UV region was performed at 20 °C using a spectropolarimeter (Jasco J-810) equipped with a water-cooled Peltier unit. UbiK spectra were recorded in a 0.01-mm path length cell (121.QS, Hellma) from 185 to 260 nm with 11 mg/ml of UbiK sample in 50 mm Tris-HCl, 150 mm NaCl, pH 7.5, buffer. For each sample, an averaged spectrum was produced by merging three consecutive scans; the spectra were corrected using buffer baseline measured under the same conditions. Data were recorded in mdeg then converted to Δϵ (m−1 cm−1). Estimations of secondary structure were predicted by using CDSSTR, SELCON3, and CONTIN methods included in the DICHROWEB server (34), or using the K2D3 method (35).
SEC-MALS analyses
Purified UbiK and copurified UbiK–UbiJ P1 and P2 were analyzed using an HPLC-MALS system (Shimadzu) equipped with three detectors, the light scattering detector (mini DAWN TREOS system, Wyatt Technology), the refractive index detector (Optilab T-rEX, Wyatt Technology), and the UV detector from HPLC (SPD-20A, Shimadzu). 200-μg samples of UbiK and UbiK–UbiJ P1 and P2 were injected in a Superdex 200 10/300 GL Increase column (GE Healthcare Life Sciences) equilibrated in 50 mm Tris-Cl, pH 7.5, 150 mm NaCl buffer at a flow rate of 0.5 ml min−1. Molar masses of proteins were calculated using ASTRA 6.1 software (Wyatt Technology) using a refractive index increment (dn/dc) value of 0.183 ml g−1.
Molecular weights of UbiK and UbiJ within the UbiK–UbiJ complexes were determined by using the protein-conjugate method. This method is based on the use of the three signals (light scattering, absorbance, and differential refractive index) to determine the molecular weight of the protein and of the conjugate (DNA, RNA, glycosylation, detergent etc.) in complex. Formalism of the protein-conjugate method was described in Veesler et al. (36) with a protein–detergent complex. By analogy, the system can be applied to a protein A–protein B complex if mass extinction coefficient are different (ϵA is 2 or 3 times higher than ϵB). As dn/dc of protein is relatively constant (37), dn/dc were fixed to 0.183 ml g−1. α is the weight proportion of protein A, 1-α corresponds to the weight proportion of protein B. CA and CB are the respective weight concentrations of proteins A and B.
| (Eq. 1) |
| (Eq. 2) |
Using Equation 3, the concentration of CAB can be expressed as the following.
| (Eq. 3) |
Where Δn is the variation of refractive index. Mass extinction coefficient of AB complex can be expressed as shown.
| (Eq. 4) |
By using Beer-Lambert equation and Equation 4, CAB can be expressed as shown.
| (Eq. 5) |
By using Equations 3 and 5, we obtain,
| (Eq. 6) |
For which α can be inferred.
| (Eq. 7) |
As dn/dc are identical, the term OD280nm((dn/dc)A − (dn/dc)B) is negligible. Therefore, the terms α, 1-α, CA, and CB can be calculated by the protein-conjugate method and can be used to determine the molecular weights of protein A and protein B in a AB complex if ϵA and ϵB are significatively different. Concerning UbiK and UbiJ complexes, the UbiJ extinction coefficient (0.951 ml/(mg cm)) was predicted from the protein amino acid sequences with the ProtParam tool (expasy.org/protparam/) and UbiK extinction coefficient was determined experimentally (0.059 ml/(mg cm)) using the “UV extension from RI peak” method included in the ASTRA software.
Analytical ultracentrifuge (AUC)
Sedimentation velocity experiments were conducted with a Beckman XL-I (Beckman-Coulter, Palo Alto, CA) AUC using an An-50Ti rotor. Increasing concentrations of UbiK and UbiK–UbiJ complexes P1 and P2 were loaded into Epon charcoal-filled two-sector 12-mm path length cells. 400 μl of samples and 410 μl of buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl) were centrifuged at 42,000 rpm (128,297 × g) and 20 °C. Absorbance at 280 nm and interference scans were collected every 5 min and sedimentation velocity data were analyzed using SEDFIT software (38). Viscosity and density parameters were measured by using, respectively, a Anton Paar microviscosimeter and density meter (DMA 4500). Partial specific volumes of proteins were predicted by SEDNTERP software.
Contrary to Multi-Signal Sedimentation Velocity experiments (39), which require 3 signals (2 different wavelengths in absorbance and interferometry) to determine protein stoichiometry, the present case is easier because the extinction coefficient of UbiK is close to zero (see SEC-MALS under “Results”). So the interferometric signal (ΔJ) is due to both complex components, whereas the absorbance signal is only due to UbiJ. ΔJUbiJ/UbiK, ΔJUbiJ, and ΔJUbiK correspond to fringes displaced, respectively, due to the UbiJ–UbiK complex, UbiJ and UbiK. ϵIF UbiJ, ϵIF UbiK correspond to molar increments of UbiJ and UbiK in Rayleigh interferometry and ϵUbiJ corresponds to a molar extinction coefficient at 280 nm (all three last parameters are predicted thanks to calculator module of SEDFIT software by using protein sequences). l is the path length of analytical ultracentrifugation centerpiece (1.2 cm).
| (Eq. 8) |
Thanks to Equation 8 of Padrick (39), we can write the following.
| (Eq. 9) |
As only UbiJ absorbs we can apply the Beer-Lambert equation.
| (Eq. 10) |
OD280 nm is the optical density at 280 nm. Finally, by combining Equations 8–10, we obtain the following.
| (Eq. 11) |
| (Eq. 12) |
After integration of each peak obtained in c(s) distribution by SEDFIT, ΔJUbiJ/UbiK, OD280 nm are determined, CUbiJ and CUbiK are calculated to obtain the molar ratio of UbiK and UbiJ in UbiK–UbiJ complex.
Fluorescence sedimentation velocity experiments were performed thanks to the Aviv fluorescence detection system (AU-FDS) and UbiK was labeled by monolith NT-115 Blue NHS (Nanotemper Technology) according to the manufacturer's instructions. Labeled protein was diluted from 60 nm to 3 pm with the buffer cited previously; to prevent protein adsorption, BSA was added at a final concentration of 0.1 mg/ml for the concentration of labeled UbiK below 1 nm. To study the oligomeric state of UbiK at concentrations higher than 60 nm, unlabeled UbiK was added to 60 nm fluorescent protein to reach the desired concentration. Samples were sedimented in the same conditions used for absorbance/interference experiments except no reference is required during fluorescence experiments.
Infection of macrophages with S. enterica strains
RAW 264.7 macrophages were seeded at a density of 4 × 105 cells per well in 6-well tissue culture plates containing DMEM with 10% fetal bovine serum (FBS) (HyClone). S. enterica strains were grown overnight with or without oxygen and at 30 or 37 °C as indicated in the figure legends. Afterward, these cultures were opsonized in DMEM containing FBS and normal mouse serum (10%, Perbio) for 30 min. Bacteria were added to the monolayers at a multiplicity of infection 10:1, centrifuged at 400 × g for 5 min at 4 °C, and incubated for 30 min at 37 °C in 5% CO2. The macrophages were washed three times, and incubated with DMEM containing FBS and 100 μg/ml of gentamicin for 60 min, after which the gentamicin concentration was decreased to 10 μg/ml for the remainder of the experiment. For enumeration of intracellular bacteria, macrophages were washed two times with PBS, lysed with 0.1% Triton X-100, and a dilution series was plated on LB agar.
Virulence assays in mice
Eight-week-old female BALB/c mice (Charles River Laboratories) were inoculated intraperitoneally with equal amounts of S. enterica wild-type and ubiK mutant strains for a total of 105 bacteria per mouse. The spleens were harvested 48 h after inoculation, then homogenized. Bacteria were recovered and enumerated after plating a dilution series onto LB agar and LB agar with kanamycin. CI were determined for each mouse. The CI is defined as the ratio between the ubiK mutant and wild-type strains within the output (bacteria recovered from the mouse after infection) divided by their ratios within the input (initial inoculum).
Statistical methods
Results are presented as mean ± S.E. Data sets were compared by unpaired two-tailed t test or an analysis of variance using GraphPad Prism 7 software.
Author contributions
F. B., F. P., M. F., M. L., and L. A. conceived and designed the experiments. L. L., C. F., L. A., M. H. C., S. B. H., B. F., D. H., C. M. D., B. R., L. P., and C. V. performed the experiments. D. C. preformed the mass spectrometry analyses. J. C. contributed reagents/materials/analysis tools. F. B., F. P., M. F., M. L., and L. A. wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank the FB group for discussions and suggestions. This work has benefited from the facilities and expertise of the Macromolecular Interaction Platform of I2BC (UMR 9198) supported by FRISBI.
This work was supported by the Agence Nationale de la Recherche (ANR), ANR Blanc (An)aéroUbi ANR-15-CE11-0001-02, Aix-Marseille Université (AMU), Centre National de la Recherche Scientifique (CNRS) Grant PICS07279, and French State Program ”Investissements d'Avenir“ Grants “LABEX DYNAMO” and ANR-11-LABX-0011. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S10.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- MK
- menaquinone
- UQ
- ubiquinone
- DMK
- demethylmenaquinone
- OPP
- 3-octaprenylphenol
- SEC-MALS
- size exclusion chromatography-multi-angle light scattering
- SCP
- sterol carrier protein
- AUC
- analytical ultracentrifuge
- CI
- competitive index
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- ECD
- electrochemical detection
- SPA
- sequential peptide affinity
- MBP
- maltose-binding protein
- PDB
- Protein Data Bank.
References
- 1. Nowicka B., and Kruk J. (2010) Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta 1797, 1587–1605 [DOI] [PubMed] [Google Scholar]
- 2. Collins M. D., and Jones D. (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 45, 316–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhi X.-Y., Yao J.-C., Tang S.-K., Huang Y., Li H.-W., and Li W.-J. (2014) The futalosine pathway played an important role in menaquinone biosynthesis during early prokaryote evolution. Genome Biol. Evol. 6, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schoepp-Cothenet B., Lieutaud C., Baymann F., Verméglio A., Friedrich T., Kramer D. M., and Nitschke W. (2009) Menaquinone as pool quinone in a purple bacterium. Proc. Natl. Acad. Sci. U.S.A. 106, 8549–8554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ravcheev D. A., and Thiele I. (2016) Genomic analysis of the human gut microbiome suggests novel enzymes involved in quinone biosynthesis. Front. Microbiol. 7, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aussel L., Pierrel F., Loiseau L., Lombard M., Fontecave M., and Barras F. (2014) Biosynthesis and physiology of coenzyme Q in bacteria. Biochim. Biophys. Acta 1837, 1004–1011 [DOI] [PubMed] [Google Scholar]
- 7. Hajj Chehade M., Loiseau L., Lombard M., Pecqueur L., Ismail A., Smadja M., Golinelli-Pimpaneau B., Mellot-Draznieks C., Hamelin O., Aussel L., Kieffer-Jaquinod S., Labessan N., Barras F., Fontecave M., and Pierrel F. (2013) ubiI, a new gene in Escherichia coli coenzyme Q biosynthesis, is involved in aerobic C5-hydroxylation. J. Biol. Chem. 288, 20085–20092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alexander K., and Young I. G. (1978) Alternative hydroxylases for the aerobic and anaerobic biosynthesis of ubiquinone in Escherichia coli. Biochemistry 17, 4750–4755, [DOI] [PubMed] [Google Scholar]
- 9. Aussel L., Loiseau L., Hajj Chehade M., Pocachard B., Fontecave M., Pierrel F., and Barras F. (2014) ubiJ, a new gene required for aerobic growth and proliferation in macrophage, is involved in coenzyme Q biosynthesis in Escherichia coli and Salmonella enterica serovar Typhimurium. J. Bacteriol. 196, 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang K.-C., Huang C.-H., Ding S.-M., Chen C.-K., Fang H.-W., Huang M.-T., and Fang S.-B. (2016) Role of yqiC in the pathogenicity of Salmonella and innate immune responses of human intestinal epithelium. Front. Microbiol. 7, 1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nichols R. J., Sen S., Choo Y. J., Beltrao P., Zietek M., Chaba R., Lee S., Kazmierczak K. M., Lee K. J., Wong A., Shales M., Lovett S., Winkler M. E., Krogan N. J., Typas A., and Gross C. A. (2011) Phenotypic landscape of a bacterial cell. Cell 144, 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poon W. W., Davis D. E., Ha H. T., Jonassen T., Rather P. N., and Clarke C. F. (2000) Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J. Bacteriol. 182, 5139–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hsu A. Y., Poon W. W., Shepherd J. A., Myles D. C., and Clarke C. F. (1996) Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry 35, 9797–9806 [DOI] [PubMed] [Google Scholar]
- 14. Federowicz S., Kim D., Ebrahim A., Lerman J., Nagarajan H., Cho B. K., Zengler K., and Palsson B. (2014) Determining the control circuitry of redox metabolism at the genome-scale. PLoS Genet. 10, e1004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mason J. M., and Arndt K. M. (2004) Coiled coil domains: stability, specificity, and biological implications. Chembiochem 5, 170–176 [DOI] [PubMed] [Google Scholar]
- 16. Chen Z., Borek D., Padrick S. B., Gomez T. S., Metlagel Z., Ismail A. M., Umetani J., Billadeau D. D., Otwinowski Z., and Rosen M. K. (2010) Structure and control of the actin regulatory WAVE complex. Nature 468, 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Derivery E., Fink J., Martin D., Houdusse A., Piel M., Stradal T. E., Louvard D., and Gautreau A. (2008) Free Brick1 is a trimeric precursor in the assembly of a functional wave complex. PloS One 3, e2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karimova G., Pidoux J., Ullmann A., and Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stolowich N. J., Petrescu A. D., Huang H., Martin G. G., Scott A. I., and Schroeder F. (2002) Sterol carrier protein-2: structure reveals function. Cell Mol. Life Sci. CMLS. 59, 193–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Berti F. P., Capaldi S., Ferreyra R., Burgardt N., Acierno J. P., Klinke S., Monaco H. L., and Ermácora M. R. (2013) The crystal structure of sterol carrier protein 2 from Yarrowia lipolytica and the evolutionary conservation of a large, non-specific lipid-binding cavity. J. Struct. Funct. Genomics 14, 145–153 [DOI] [PubMed] [Google Scholar]
- 21. Dyer D. H., Lovell S., Thoden J. B., Holden H. M., Rayment I., and Lan Q. (2003) The structural determination of an insect sterol carrier protein-2 with a ligand-bound C16 fatty acid at 1.35-Å resolution. J. Biol. Chem. 278, 39085–39091 [DOI] [PubMed] [Google Scholar]
- 22. García F. L., Szyperski T., Dyer J. H., Choinowski T., Seedorf U., Hauser H., and Wüthrich K. (2000) NMR structure of the sterol carrier protein-2: implications for the biological role. J. Mol. Biol. 295, 595–603 [DOI] [PubMed] [Google Scholar]
- 23. Haapalainen A. M., van Aalten D. M., Meriläinen G., Jalonen J. E., Pirilä P., Wierenga R. K., Hiltunen J. K., and Glumoff T. (2001) Crystal structure of the liganded SCP-2-like domain of human peroxisomal multifunctional enzyme type 2 at 1.75-Å resolution. J. Mol. Biol. 313, 1127–1138 [DOI] [PubMed] [Google Scholar]
- 24. Carrica M. C., Craig P. O., García-Angulo V. A., Aguirre A., García-Véscovi E., Goldbaum F. A., and Cravero S. L. (2011) YqiC of Salmonella enterica serovar Typhimurium is a membrane fusogenic protein required for mice colonization. BMC Microbiol. 11, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beuzón C. R., and Holden D. W. (2001) Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3, 1345–1352 [DOI] [PubMed] [Google Scholar]
- 26. González-Mariscal I., García-Testón E., Padilla S., Martín-Montalvo A., Pomares-Viciana T., Vazquez-Fonseca L., Gandolfo-Domínguez P., and Santos-Ocaña C. (2014) Regulation of coenzyme Q biosynthesis in yeast: a new complex in the block. IUBMB Life 66, 63–70 [DOI] [PubMed] [Google Scholar]
- 27. Marbois B., Gin P., Gulmezian M., and Clarke C. F. (2009) The yeast Coq4 polypeptide organizes a mitochondrial protein complex essential for coenzyme Q biosynthesis. Biochim. Biophys. Acta 1791, 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Floyd B. J., Wilkerson E. M., Veling M. T., Minogue C. E., Xia C., Beebe E. T., Wrobel R. L., Cho H., Kremer L. S., Alston C. L., Gromek K. A., Dolan B. K., Ulbrich A., Stefely J. A., Bohl S. L., et al. (2016) Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol. Cell 63, 621–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marteyn B., Scorza F. B., Sansonetti P. J., and Tang C. (2011) Breathing life into pathogens: the influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell Microbiol. 13, 171–176 [DOI] [PubMed] [Google Scholar]
- 30. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pelosi L., Ducluzeau A.-L., Loiseau L., Barras F., Schneider D., Junier I., and Pierrel F. (2016) Evolution of ubiquinone biosynthesis: multiple proteobacterial enzymes with various regioselectivities to catalyze three contiguous aromatic hydroxylation reactions. mSystems 1, e00091–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bouveret E., and Brun C. (2012) Bacterial interactomes: from interactions to networks. Methods Mol. Biol. 804, 15–33 [DOI] [PubMed] [Google Scholar]
- 33. Golemis E. A., Giurys J., and Brent R. (1994) Two hybrid systems/interaction traps. in The bacteriophages (Calendar R., ed) John Wiley & Sons, New York [Google Scholar]
- 34. Whitmore L., and Wallace B. A. (2008) Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 35. Louis-Jeune C., Andrade-Navarro M. A., and Perez-Iratxeta C. (2012) Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins 80, 374–381 [DOI] [PubMed] [Google Scholar]
- 36. Veesler D., Blangy S., Siponen M., Vincentelli R., Cambillau C., and Sciara G. (2009) Production and biophysical characterization of the CorA transporter from Methanosarcina mazei. Anal. Biochem. 388, 115–121 [DOI] [PubMed] [Google Scholar]
- 37. Zhao H., Brown P. H., and Schuck P. (2011) On the distribution of protein refractive index increments. Biophys. J. 100, 2309–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Padrick S. B., Deka R. K., Chuang J. L., Wynn R. M., Chuang D. T., Norgard M. V., Rosen M. K., and Brautigam C. A. (2010) Determination of protein complex stoichiometry through multisignal sedimentation velocity experiments. Anal. Biochem. 407, 89–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., Potter S. C., Punta M., Qureshi M., Sangrador-Vegas A., Salazar G. A., Tate J., and Bateman A. (2016) The Pfam protein families database: towards a more sustainable future Nucleic Acids Res. 44, D279–D285 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.