Abstract
Post-translational modifications of histone proteins regulate numerous cellular processes. Among these modifications, trimethylation of lysine 36 in histone H3 (H3K36me3) and acetylation of lysine 16 in histone H4 (H4K16ac) have important roles in transcriptional regulation and DNA damage response signaling. However, whether these two epigenetic histone marks are mechanistically linked remains unclear. Here we discovered a new pathway through which H3K36me3 stimulates H4K16ac upon DNA double-strand break (DSB) induction in human cells. In particular, we examined, using Western blot analysis, the levels of H3K36me3 and H4K16ac in cells after exposure to various DSB-inducing agents, including neocarzinostatin, γ rays, and etoposide, and found that H3K36me3 and H4K16ac were both elevated in cells upon these treatments. We also observed that DSB-induced H4K16 acetylation was abolished in cells upon depletion of the histone methyltransferase gene SET-domain containing 2 (SETD2) and the ensuing loss of H3K36me3. Furthermore, the H3K36me3-mediated increase in H4K16ac necessitated lens epithelium–derived growth factor p75 splicing variant (LEDGF), which is a reader protein of H3K36me3, and the KAT5 (TIP60) histone acetyltransferase. Mechanistically, the chromatin-bound LEDGF, through its interaction with KAT5, promoted chromatin localization of KAT5, thereby stimulating H4K16 acetylation. In this study, we unveiled cross-talk between two important histone epigenetic marks and defined the function of this cross-talk in DNA DSB repair.
Keywords: DNA repair, epigenetics, histone acetylation, histone methylation, posttranslational modification (PTM)
Introduction
In eukaryotes, packing of genomic DNA into chromatin creates a physical barrier limiting the access of DNA by protein machineries involved in various activities of DNA metabolism, including DNA repair (1, 2). Thus, repair of DNA lesions in chromatin necessitates chromatin remodeling, histone exchange, and/or post-translational modifications of histones (3).
Among the plethora of post-translational modifications of histone proteins, methylation of lysine 36 in histone H3 (H3K36)2 is a key and widespread modification on chromatin, and it has been implicated in RNA splicing as well as DNA replication, transcription, methylation, and repair (4–6). In mammalian cells, mono-, di-, and trimethylation of H3K36 (H3K36me, H3K36me2, and H3K36me3, respectively) are catalyzed by distinct methyltransferases; nuclear SET domain (NSD)–containing methyltransferases, including NSD1, NSD2 and NSD3, induce H3K36me and H3K36me2, whereas SET domain-containing 2 (SETD2) is the major enzyme for catalyzing H3K36me3 (5). Previous studies showed that methylation of H3K36 plays important roles in DNA double-strand break (DSB) repair (7–9) and mismatch repair (6). In mammalian cells, Metnase-mediated H3K36me2 was found to be DNA damage–inducible, and this methylation mark enables the recruitment and stabilization of NBS1 and Ku70 near DNA DSB sites, thereby promoting DSB repair via the non-homologous end-joining pathway (10). In addition, the SETD2-catalyzed H3K36me3 enables the recruitment of CtIP to sites of DNA DSBs and promotes resection at DSB ends (7). Reducing the H3K36me3 level through knockdown of SETD2 or overexpression of an H3K36me2/3 demethylase gene, KDM4A/JMJD2A, in cells led to defective repair of DNA DSBs via the homologous recombination (HR) pathway (7, 8). Moreover, H3K36me3 is required for the recruitment of the mismatch recognition protein hMutSα onto chromatin through interaction with the PWWP domain of hMSH6, thereby enabling mismatch repair (6).
The H3K36me3-binding proteins LEDGF and MRG15 also promote DNA DSB repair (7, 11–13). LEDGF specifically recognizes H3K36me3 via its PWWP domain, which stimulates CtIP-mediated DNA end resection during HR-mediated repair of DNA DSBs (7, 13). MRG15 is a conserved chromodomain protein that binds specifically to H3K36me3 (14); after DNA damage, MRG15 interacts with PALB2 and activates the HR pathway (11).
Acetylation of lysine 16 in histone H4 (H4K16ac) is another important histone modification that functions in DNA damage repair (15, 16). hMOF and KAT5 are the two major acetyltransferases catalyzing the formation of H4K16ac (17–19). Incorporation of synthetic H4K16ac into nucleosomes prevents their compaction into 30-nm chromatin fibers in vitro (20). Additionally, the H4K16ac level is up-regulated upon exposure to ionizing radiation (17), and this acetylation is thought to modulate DNA DSB repair pathway choice by favoring HR via promoting the chromatin localization of BRCA1 while restricting the binding of 53BP1 to chromatin (21, 22).
Despite the importance of H3K36me3 and H4K16ac in DNA repair in mammalian cells, it remains elusive whether there exists a mechanistic link between these two histone epigenetic marks. In this context, Arabidopsis MRG domain proteins are known to bridge H3K36me3 and histone H4 acetylation to regulate flowering time (23). Therefore, we decided to assess the potential link between these two important histone epigenetic marks in mammalian cells. In this study, we generated, by employing the CRISPR/Cas9-based genome-editing method, isogenic HEK293T cells depleted of the SETD2, LEDGF, MRG15, or KAT5 gene and analyzed the temporal responses of H3K36me3 and H4K16ac in these cells following exposure to neocarzinostatin (NCS), a DNA DSB-inducing agent. We found that DNA DSB induction stimulated a transient increase in H3K36me3 that enhanced the binding of LEDGF to chromatin, thereby promoting the chromatin localization of KAT5 and the acetylation of H4K16. Thus, our results uncovered a novel cross-talk between H3K36me3 and H4K16ac in the context of DNA DSB repair.
Results
DNA DSB formation stimulated transient increases in H3K36me3 and H4K16ac, and the elevation in H4K16ac required SETD2
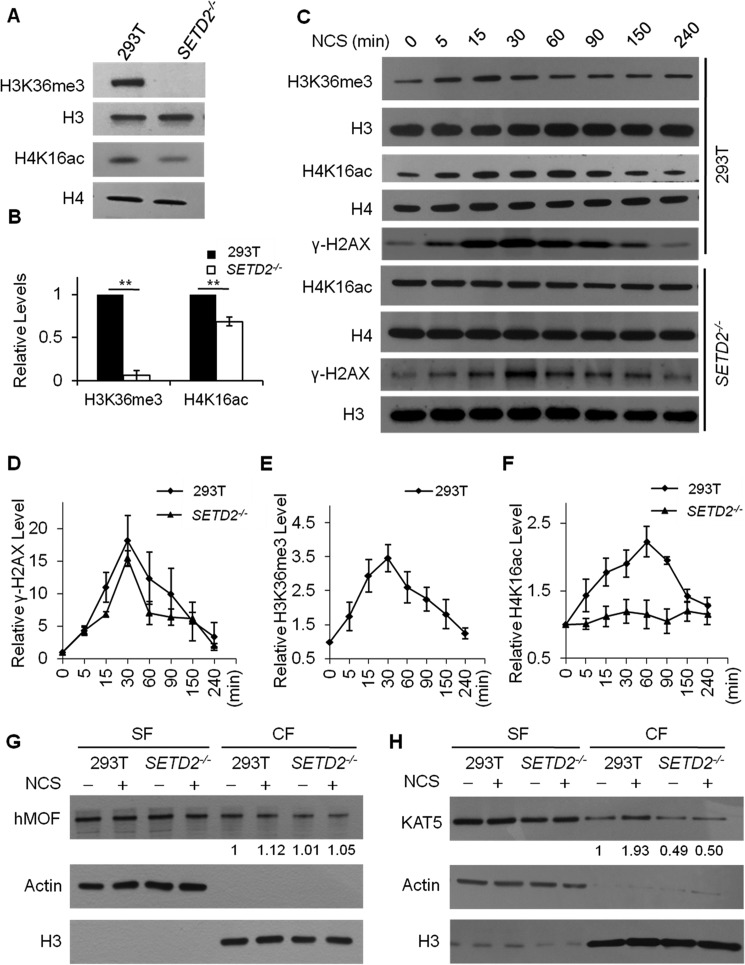
To examine whether there is cross-talk between H3K36me3 and H4K16ac, we generated H3K36me3-deficient cells by knocking out the major H3K36 trimethyltransferase SETD2 using the CRISPR/Cas9 genome-editing method (supplemental Fig. S1A) and examined the levels of these two histone epigenetic marks by Western blot analysis (Fig. 1). As expected, H3K36me3 was almost entirely abolished in SETD2−/− cells (Fig. 1), which is accompanied with a significant drop (by ∼30%) in the basal level of H4K16ac (Fig. 1, A and B), supporting that H3K36me3 promotes H4K16ac.
Figure 1.
Exposure to DNA DSB–inducing agents led to elevated levels of H3K36me3 and H4K16ac, and SETD2−/− cells exhibited a decreased level of H4K16ac. A, the levels of H3K36me3 and H4K16ac were decreased in SETD2−/− cells, where histones H3 and H4 were used as references, respectively. B, quantification of the Western blot results showed diminished H3K36me3 and H4K16ac levels in SETD2−/− cells. **, p < 0.01. The p values were calculated using two-tailed, unpaired Student's t test. C–F, time course experiments showed a transient elevation in the levels of γH2AX, H3K36me3, and H4K16ac in HEK293T (293T) cells after treatment with 100 ng/ml NCS, where histones H4, H3, and H4 were used as references, respectively. The NCS-induced increase in H4K16ac was abolished in SETD2−/− cells. The quantification results displayed in D–F represent the mean and S.E. of results obtained from three independent biological replicates conducted on 3 separate days for all experiments, except that the time course experiment for H3K36me3 was conducted in five independent biological replicates performed on 5 separate days (see supplemental Fig. S2 for Western blot images obtained from other biological replicates). G and H, the chromatin localization of KAT5, but not hMOF, was increased in HEK293T cells after NCS treatment, and this increase in chromatin binding of KAT5 was lost in SETD2−/− cells. Actin and histone H3 were employed as loading controls for the soluble fraction (SF) and chromatin fraction (CF) fractions, respectively. The relative values below the lanes were calculated from the ratios of band intensities of CF over SF, which were normalized against histone H3 and actin, respectively.
Because both H3K36me3 and H4K16ac function in DNA DSB repair, we next examined the temporal responses of these two histone epigenetic marks in HEK293T cells following exposure to NCS, a radiomimetic agent capable of inducing DNA DSBs in cells. We found that NCS exposure led to transient increases in the levels of H3K36me3 and H4K16ac in HEK293T cells that peak at ∼30 and 60 min, respectively, and return to nearly normal levels at ∼4 h (Fig. 1, C–F and supplemental Fig. S2). In addition, the time-dependent alteration in the level of H3K36me3 following NCS treatment mirrors that of γ-H2AX (Fig. 1, D–E), which reflects the level of DNA DSBs (24). Similar transient increases in γ-H2AX, H3K36me3, and H4K16ac were also observed in HEK293T cells upon exposure to other DNA DSB-inducing agents, including γ rays and etoposide (supplemental Fig. S3). Furthermore, exposure of HeLa and U2OS cells to NCS also stimulated an elevation in H3K36me3 and H4K16ac (supplemental Fig. S4). We next asked whether this NCS-stimulated increase in H4K16ac requires SETD2-mediated H3K36me3. Indeed, our results showed that genetic depletion of SETD2 in HEK293T cells abolished the NCS-induced elevation in H4K16ac, although the NCS-triggered transient increase in γ-H2AX was not affected by the loss of SETD2 (Fig. 1, C–F and supplemental Fig. S2). This result underscored that H3K36me3 is indispensable for DNA damage-induced H4K16ac.
KAT5 is recruited to chromatin and induces H4K16ac upon DNA DSB induction, which is abolished in cells deficient in SETD2
Having demonstrated the dependence of H4K16 acetylation on H3K36me3 after DNA DSB induction, we next explored the histone acetyltransferases involved in this process by assessing the chromatin localizations of the two known H4K16 acetyltransferases, hMOF and KAT5, following NCS exposure (Fig. 1, G and H). Our results revealed that NCS treatment did not elicit chromatin localization of hMOF in HEK293T cells or isogenic cells depleted of SETD2 (Fig. 1G). By contrast, chromatin-bound KAT5 was substantially increased, by 1.6–1.9-fold, in HEK293T cells after NCS exposure, and this DNA damage-triggered chromatin localization of KAT5 was abrogated in HEK293T cells depleted of SETD2, although the expression level of KAT5 was not altered upon SETD2 depletion (Fig. 1H and supplemental Fig. S6). Hence, these results support that the DNA damage-stimulated recruitment of KAT5 to chromatin necessitates SETD2-mediated trimethylation of H3K36.
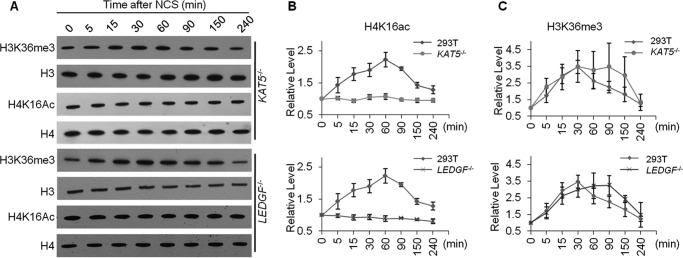
To further substantiate the above finding, we knocked out the KAT5 gene in HEK293T cells (supplemental Fig. S1A) and assessed its effect on the temporal responses of H3K36me3 and H4K16ac after NCS exposure. Our results showed that, despite the increase in H3K36me3, the level of H4K16ac in KAT5−/− cells did not change after NCS treatment, underscoring the crucial role of KAT5 in NCS-triggered H4K16ac (Fig. 2, A–C and supplemental Fig. S5).
Figure 2.
Temporal responses of H3K36me3 and H4K16ac levels in different cell lines following DNA DSB induction. A–C, time course experiments revealed the dynamic changes in H3K36me3 and H4K16ac levels following NCS treatment. Shown are the Western blot results (A) and the quantification data for the relative levels of H3K36me3 and H4K16ac (B and C). The relative levels of these two histone modifications were calculated using histones H3 and H4 as references, respectively. The quantification data represent the mean and S.E. of results obtained from three independent biological replicates for all experiments, with the exception that the time course experiment for H3K36me3 was conducted in five independent biological replicates; each biological replicate was conducted on a separate day. Western blot images for data obtained from other biological replicates for LEDGF−/− and KAT5−/− cells are shown in supplemental Fig. S5.
LEDGF functions in the DNA damage-stimulated increase in H4K16ac
We next explored the reader proteins of H3K36me3 that transduce the signal from H3K36me3 to H4K16ac. In this respect, mortality factor 4-like protein 1 (MRG15) and lens epithelium–derived growth factor p75 splicing variant (LEDGF) were shown previously to bind to H3K36me3 through their chromodomain and PWWP domain, respectively (13, 14), and both proteins function in DNA DSB repair (12, 13). Hence, we also depleted MRG15 and LEDGF genes using CRISPR/Cas9 (supplemental Fig. S1) and examined the impact of their depletion on the NCS-stimulated increase in H4K16ac. It turned out that genetic ablation of LEDGF, but not MRG15, abolished the NCS-induced elevation in H4K16ac (Fig. 2, A–C, and supplemental Figs. S5 and S7). Similar to what we observed for KAT5−/− cells, the NCS-triggered elevation in H3K36me3 was still observed in LEDGF−/− cells (Fig. 2, A–C, and supplemental Fig. S5). In this vein, it is worth noting that the basal levels of H4K16ac were not changed upon depletion of the LEDGF or KAT5 gene (supplemental Fig. S7C). Therefore, our results support a crucial role of LEDGF in facilitating DNA damage–induced H4K16ac.
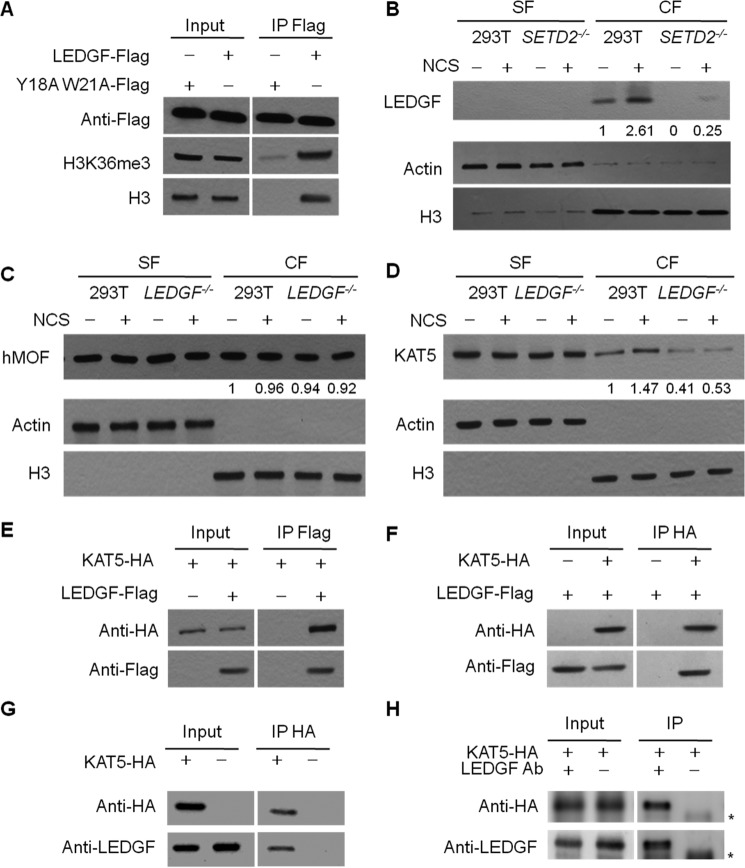
LEDGF interacts with H3K36me3 and KAT5 in cells and promotes DNA damage-induced recruitment of KAT5 to chromatin
LEDGF possesses a conserved PWWP domain that is characterized by its strong binding affinity toward methylated lysine in histone proteins (7, 13, 25). Thus, we examined the interaction between LEDGF and H3K36me3 by immunoprecipitation, where we employed FLAG-tagged LEDGF and its variant form, with two aromatic amino acid residues in the binding pocket of the PWWP domain being mutated to alanines to pull down H3K36me3 and histone H3. Our results showed that, although wild-type LEDGF could pull down histone H3 and H3K36me3, the PWWP domain mutant displayed markedly reduced binding affinities toward H3K36me3 and histone H3 (Fig. 3A; results from another biological replicate are shown in supplemental Fig. S8A). We further assessed the chromatin localization of LEDGF in SETD2−/− cells. In HEK293T cells, substantial recruitment of LEDGF to chromatin could be observed after NCS treatment; genetic depletion of SETD2, however, led to a pronounced drop in the amount of LEDGF in the chromatin fraction (Fig. 3B and supplemental Fig. S8, B and C). This result indicates a pivotal role of H3K36me3 in tethering LEDGF to chromatin.
Figure 3.
LEDGF facilitates the recruitment of KAT5 to chromatin following DNA DSB induction. A, FLAG-tagged LEDGF, but not LEDGF-Y18AW21A, could pull down histone H3 and H3K36me3. IP, immunoprecipitation. B, chromatin localization of LEDGF protein in HEK293T and SETD2−/− cells. Knockout of SETD2 confers diminished chromatin localization of LEDGF. C and D, the chromatin localization of C-terminally FLAG-tagged KAT5 (KAT5-FLAG), but not hMOF, was increased in HEK293T cells upon NCS treatment, and this increase in chromatin binding of KAT5-FLAG was abolished in LEDGF−/− cells. Actin and histone H3 were employed as loading controls for the SF and chromatin CF, respectively. The values below the lanes were calculated from the band intensities normalized against histone H3. E and F, reciprocal pulldown experiments revealed the interaction between LEDGF and KAT5. KAT5-HA and LEDGF-FLAG were co-transfected into cells, and pulldown experiments were conducted using anti-HA beads or anti-FLAG beads. Results from another biological replicate is shown in supplemental Fig. S8, F and G. G and H, reciprocal pulldown experiments revealed the interaction between endogenous LEDGF and ectopically expressed HA-KAT5. Results from another biological replicate are shown in supplemental Fig. S8, H and I. The nonspecific band in H was labeled with an asterisk. LEDGF Ab, LEDGF antibody used for immunoprecipitation.
We also assessed the role of LEDGF in the recruitment of KAT5 to chromatin (Fig. 3, C and D). We found that the depletion of LEDGF, while not affecting the expression level of KAT5 protein (supplemental Fig. S6C), nearly abolished the DNA damage-stimulated localization of KAT5 to chromatin, which is similar to the observations made for SETD2−/− cells (Fig. 3D and supplemental Fig. S8, D and E). The chromatin localization of hMOF, however, was not perturbed by the absence of LEDGF, and this observation is not dependent on whether the cells are exposed to NCS (Fig. 3C).
We also investigated the interaction between LEDGF and KAT5 in cells. Plasmids for expressing HA-tagged KAT5 and FLAG-tagged LEDGF were co-transfected into HEK293T cells, and reciprocal pulldown experiments revealed the interaction between the two ectopically expressed proteins (Fig. 3, E and F and supplemental Fig. S8, F and G). Similar immunoprecipitation experiments using lysates from cells transfected with HA-tagged KAT5 alone showed the interaction between the ectopically expressed KAT5 and endogenous LEDGF (Fig. 3, G and H and supplemental Fig. S8, H and I). It is worth noting that we did not conduct pulldown experiments for monitoring the interaction between endogenous KAT5 and LEDGF because of the lack of a suitable antibody for endogenous KAT5. However, we were not able to detect the interaction between recombinant LEDGF and KAT5 purified from Escherichia coli, suggesting that the interaction between the two proteins may be indirect (i.e. involving other proteins in human cells) or modulated by post-translational modifications of LEDGF and/or KAT5. Cumulatively, our results support the notion that DNA DSB induction leads to increased H3K36me3, which stimulates the recruitment of LEDGF to chromatin through interaction with its PWWP domain, and promotes the chromatin localization of KAT5.
SETD2 and LEDGF play important roles in stimulating H3K36me3 and H4K16ac at a specifically generated DNA DSB site
Having demonstrated the responses in global levels of H3K36me3 and H4K16ac following DNA damage induction by NCS, we next examined the levels of H3K36me3 and H4K16ac in DNA regions surrounding an I-SceI–generated DNA DSB site using ChIP followed by real-time quantitative PCR analysis. Real-time PCR targets were located ∼500 and 2500 bp from the I-SceI–induced DNA DSB site in U2OS-DRGFP cells (Fig. 4A) (26). It turned out that the levels of H3K36me3 and H4K16ac at the proximal site (i.e. 500 bp from the DNA DSB locus) were pronouncedly elevated, whereas no substantial increases were found at the distal site (i.e. 2500 bp away from the DNA DSB site; Fig. 4, B and C). After knockdown of SETD2 or LEDGF by siRNA, the -fold changes in H4K16ac enrichment at the DNA DSB site were markedly diminished, again supporting that H3K36me3 and its reader protein LEDGF assume crucial roles in the DNA DSB-induced H4K16ac (Fig. 4, B and C).
Figure 4.
Changes in H3K36me3 and H4K16ac levels around a site-specifically generated DNA DSB site, and the modulations of the levels of these two histone modifications by SETD2 or LEDGF. A, schematic showing the locations of two sets of target fragments used in the ChIP assay. The numbers below indicate the numbers of base pairs between the target fragments and the I-SceI cleavage site. B and C, the relative occupancies (enrichment) of H3K36me3 (B) and H4K16ac (C) as revealed by ChIP assay. U2OS cells were transfected with or without siRNA targeting SETD2 or LEDGF gene for 48 h. The I-SceI plasmid was subsequently transfected into cells, and the cells were harvested 4 h following transfection. Cells without I-SceI transfection were used as negative control. For ChIP experiments, the relative enrichment was calculated by comparing the antibody binding results with the amounts of input DNA, and each value was normalized against the negative control without I-SceI transfection. The data represent the mean and S.E. of results from three biological replicates. D, a model showing the H3K36me3-H4K16ac cross-talk in response to DNA DSB induction and the involvement of LEDGF and KAT5 in this cross-talk.
Discussion
Both H3K36me3 and H4K16ac were shown previously to be important in HR-mediated repair of DNA DSBs, where H3K36me3 stimulates the binding of CtIP to chromatin through LEDGF (7–9), and H4K16ac promotes chromatin localization of BRCA1 by limiting the binding of 53BP1 to H4K20me2-marked chromatin (22). In this study, we demonstrated for the first time that H3K36me3 can be stimulated by DNA DSB-inducing agents and that these two DNA damage signaling pathways are mechanistically coupled.
H3K36me3 could be stimulated by DNA DSB–inducing agents
We provided multiple lines of evidence to support that H3K36me3 could be increased upon DNA DSB formation. In particular, we found that the global level of H3K36me3 increased gradually in HEK293T, HeLa, and U2OS cells after NCS treatment, which peaks at ∼30 min and returns to an almost normal level at ∼4 h (Fig. 1, C and E, and supplemental Fig. S2 and S4). This transient increase in H3K36me3 was also manifested in HEK293T cells upon treatment with other agents that induce DNA DSBs, including γ rays and etoposide (supplemental Fig. S3). In addition, the temporal response in H3K36me3 parallels that of γ-H2AX (Fig. 1 and supplemental Figs. S2 and S3), an indicator of DNA DSB levels (24). Moreover, our ChIP-PCR analysis results showed elevated levels of H3K36me3 at a locus proximal to the I-SceI–generated DNA DSB site for chromatin samples isolated 4 h following I-SceI transfection (Fig. 4). At first glance, these findings appear to be incongruent with previous ChIP analysis results revealing the lack of increase in H3K36me3 following DNA DSB induction (7, 27). However, the previous ChIP experiments were conducted for chromatin samples isolated from cells at a much longer time interval following I-SceI transfection (18–24 h), and our time-course study clearly demonstrated the transient nature of the increase in H3K36me3 (Fig. 1 and supplemental Figs. S3 and S4).
DNA damage–induced H3K36me3 enhances the chromatin binding of LEDGF and KAT5
Others have shown that SETD2-mediated H3K36me3 is required for DSB repair pathway choice by promoting HR through the recruitment of LEDGF and CtIP to chromatin (7–9). We further substantiated these previous findings by demonstrating that the CRISPR/Cas9-mediated genetic ablation of SETD2 in HEK293T cells substantially perturbed the chromatin localization of LEDGF and that the interaction between histone H3 and LEDGF requires an intact PWWP domain of the latter protein (Fig. 3).
In human cells, H4K16 acetylation is mainly catalyzed by hMOF and KAT5 (28, 29). In MOF-depleted cells, non-homologous end-joining– and HR-mediated repair of DNA DSBs was inhibited (15). KAT5 participates in DNA DSB repair through the NuA4-KAT5 complex, which acetylates histones H2AX and H4 near DSB sites (17, 30, 31). Furthermore, KAT5 can acetylate lysine residues on non-histone proteins (32). For instance, KAT5 catalyzes the acetylation of ataxia telangiectasia-mutated (ATM), and this acetylation activates ATM and confers heightened cellular resistance toward ionizing radiation (33, 34). We found that DNA DSB induction enhances the localization of KAT5, but not hMOF, to chromatin (Fig. 1, G and H), and this DNA damage–stimulated chromatin localization of KAT5 depends on H3K36me3 and LEDGF (Figs. 1H and 3D). Thus, we uncovered a novel regulatory mechanism of KAT5 in DNA damage response and repair.
H3K36me3-H4K16ac cross-talk in DNA DSB repair
Our results revealed a novel histone cross-talk where, upon DNA DSB induction, H3K36me3 stimulates H4K16ac. We found that the DNA damage–induced transient increase in H4K16ac is abolished in human cells depleted of the H3K36 trimethyltransferase SETD2 (Fig. 1). Our notion that H3K36me3 promotes H4K16ac also finds support in the observation that the increase in H3K36me3 precedes that of H4K16ac following DNA DSB induction (Figs. 1 and 2). H4K16ac is a very important signal in DNA DSB repair, where it modulates DSB repair pathway choice (35, 36). Introduction of synthetic H4K16ac into nucleosomes was shown to suppress their compaction into 30-nm chromatin fibers (20); thus, H4K16ac elicits an open chromatin environment that is permissive for DNA DSB repair. In addition, elevated acetylation of H4K16 diminishes the binding of 53BP1 to H4K20me2-decorated chromatin, which enhances the binding of BRCA1 to chromatin and HR repair (21, 22). Thus, the discovery that H3K36me3 and H4K16ac are mechanistically coupled in HR-mediated repair of DNA DSBs provides important new knowledge about the modulation of DNA repair by histone epigenetic marks. In this respect, it is worth noting that SETD2 is among the most frequently mutated genes in human cancers (37), and SETD2 is known to function in maintaining genomic integrity (38, 39). Our study suggests that the function of SETD2 function in tumor suppression and genomic maintenance may arise not only from its direct role in inducing H3K36me3 but also its indirect impact on promoting H4K16ac.
Taken together, our results support a mechanistic model where DNA DSB induction stimulates the SETD2-dependent H3K36me3 (Fig. 4D). Trimethylation of H3K36 promotes the chromatin localization of its reader protein, LEDGF, which binds to KAT5 and facilitates recruitment of KAT5 to chromatin, thereby elevating H4K16ac. Thus, we revealed a novel cross-talk between H3K36me3 and H4K16ac in the context of DNA DSB repair in human cells and defined the molecular mechanism for this cross-talk.
Experimental procedures
Cell culture
HEK293T, HeLa, and U2OS cells were purchased from the ATCC (Manassas, VA). U2OS cells harboring a chromosomally integrated copy of the DRGFP reporter were kindly provided by Prof. Jeremy M. Stark (40). Cells were cultured in Dulbecco's modified Eagle's medium (ATCC) supplemented with 10% FBS (Invitrogen) and 100 unit/ml penicillin/streptomycin. All cells were cultured at 37 °C in a humidified incubator with 5% CO2.
CRISPR/Cas9-mediated genome editing in HEK293T cells
CRISPR/Cas9-mediated genome editing was performed as described previously (41, 42). An online single guide RNA design tool (http://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design)3 was employed for designing the single guide RNA. The guide sequences used in this study were as follows (the protospacer adjacent motif (PAM) sequence is underlined): human SETD2 gene, AAATATAAATGGATCTCCTGGGG; human MRG15 gene, ACCTTTGCTTCATAAAGAAGAGG; human LEDGF gene, TACAAATCACTTACTCGAGCTGG; human KAT5 gene, CAGTGGCAGCCCAGCCAGGACGG.
Oligodeoxyribonucleotides corresponding to target sequences were obtained from IDT and inserted into the hSpCas9 plasmid pX330 (Addgene, Cambridge, MA). The constructed plasmids were then transfected into HEK293T cells using Lipofectamine 2000 (Invitrogen), and individual cells were cultured for further analysis. Successful knockout of the corresponding genes was confirmed by genomic DNA sequencing and/or Western blot analyses.
Histone extraction, chromatin fractionation, and Western blot
Core histones were extracted from cultured human cells following procedures published previously (43). Briefly, cell pellets were washed with 0.5 ml of lysis buffer containing 0.25 m sucrose, 10 mm MgCl2, 0.5 mm PMSF, 50 mm Tris (pH 7.4), and 0.5% Triton X-100. The pellets were then resuspended in 0.5 ml of the same buffer and kept at 4 °C overnight. The histones were extracted from the cell lysis mixture with 0.4 m sulfuric acid by incubation at 4 °C for at least 4 h with continuous vortexing. Histones were then precipitated with cold acetone and redissolved in water. For detection of histone modifications, different cell lines were treated with or without NCS (100 ng/ml), etoposide (50 μm), or γ rays (5 Gy), and the cells were harvested at different time points for histone extraction. The NCS-induced temporal alterations in H3K36me3 and H4K16ac in HEK293T cells and isogenic cells depleted of SETD2, KAT5, LEDGF, or MRG15 were conducted in at least three biological replicates, each of which was performed on a separate day.
Chromatin fractionation was performed following the protocol described by Aygun et al. (44). Briefly, the cells were lysed using cytoplasmic lysis buffer (10 mm Tris-HCl (pH 8.0), 0.34 m sucrose, 3 mm CaCl2, 2 mm MgCl2, 0.1 mm EDTA, 1 mm DTT, 0.5% Nonidet P-40, and protease inhibitor mixture) for 30 min on ice, and the intact nuclei were subsequently pelleted by centrifugation at 5000 rpm for 2 min. Nuclei were lysed with nuclear lysis buffer (20 mm HEPES (pH 7.9), 1.5 mm MgCl2, 1 mm EDTA, 150 mm KCl, 0.1% Nonidet P-40, 1 mm DTT, 10% glycerol, and protease inhibitor mixture) by homogenization. After centrifugation at 14,000 rpm for 30 min, the chromatin-enriched pellet fraction was incubated in a chromatin isolation buffer (20 mm HEPES (pH 7.9), 1.5 mm MgCl2, 150 mm KCl, 10% glycerol, protease inhibitor mixture, and 0.15 unit/μl benzonase (Sigma)) on ice for 2 h. Debris was then removed by centrifugation at 5000 rpm for 2 min and the supernatant collected as the chromatin fraction. Protein concentrations in the soluble and chromatin fractions were determined by Bradford assay (Bio-Rad). For detection of KAT5, a plasmid encoding C-terminally FLAG-tagged KAT5 (kindly provided by Dr. Yingli Sun) was transfected into different cell lines prior to chromatin fractionation and Western blot analysis.
For Western blot analysis, protein samples were separated by SDS-PAGE and transferred to a nitrocellulose membrane (Bio-Rad). The membrane was then incubated in a solution containing the appropriate primary antibodies and 5% BSA for 2 h. After washing, the membrane was incubated in a 5% blotting grade blocker (Bio-Rad) containing suitable secondary antibodies for 2 h. The immunoblots were detected using ECL Western blotting detection reagent (Amersham Biosciences). Primary antibodies used in this study included γ-H2AX (Cell Signaling Technology, 9718S, 1:3000), H3K36me3 (Abcam, ab9050, 1:5000), histone H3 (Cell Signaling Technology, 9715S, 1:10,000), H4K16ac (Millipore, 07-329, 1:10,000), histone H4 (Cell Signaling Technology, 13919S, 1:5000), actin (Cell Signaling Technology, 4967S, 1:5000), MRG15 (Cell Signaling Technology, 14098S, 1:3000), LEDGF (Abcam, ab177159, 1:3000), hMOF (Abcam, ab72056, 1:5000), anti-FLAG (Cell Signaling Technology, 2368S, 1:8000), and anti-HA (Sigma, H6908, 1:5000).
Pulldown and immunoprecipitation assay
For pulldown assay, HEK293T cells were transfected with plasmids encoding KAT5-HA and LEDGF-FLAG. Cells were collected and lysed in CelLytic M cell lysis reagent (Sigma) with 1× protease inhibitor mixture (Sigma) for 30 min on ice. After centrifugation, the lysate (supernatant) was incubated with pre-equilibrated anti-FLAG M2 affinity gel (Sigma) or anti-HA magnetic beads (Thermo Fisher Scientific) at 4 °C for 4 h. Beads were washed with 20 mm Tris (pH 8.0), 100 mm KCl, and 0.1% IGEPAL CA-630 (Sigma) five times and boiled in SDS-PAGE loading dye for SDS-PAGE and Western blot analysis, following the procedures described above. For immunoprecipitation, HEK293T cells were transfected with plasmids encoding KAT5-HA. The cells were then collected and lysed in CelLytic M cell lysis reagent (Sigma) with 1× protease inhibitor mixture (Sigma) for 30 min on ice. After centrifugation, the lysate (supernatant) was incubated with or without LEDGF antibody at 4 °C for 1 h. The pre-equilibrated protein A/G Plus beads (Thermo Fisher Scientific) were subsequently added and incubated at 4 °C for 4 h. Beads were washed with 20 mm Tris (pH 8.0), 100 mm KCl, and 0.1% IGEPAL CA-630 (Sigma) five times and boiled in SDS-PAGE loading dye for SDS-PAGE and Western blot analyses following the aforementioned procedures.
ChIP and real-time PCR
ChIP experiments were conducted as described previously (45). Approximately 2 × 106 cells were used for chromatin isolation. Generally, washed cells were resuspended in Douncing buffer (10 mm Tris-HCl (pH 7.5), 4 mm MgCl2, 1 mm CaCl2, and 1× protease inhibitor mixture). After homogenization, micrococcal nuclease (Worthington Biochemicals) was used to fragment the chromatin, and the reaction was terminated with 0.5 m EDTA. A hypotonic buffer (0.2 mm EDTA (pH 8.0), 0.1 mm benzamidine, 0.1 mm phenylmethylsulfonyl fluoride, 1.5 mm dithiothreitol, and 1× protease inhibitor mixture) was then added to the mixture and incubated on ice for 1 h. ChIP assays were performed with 1 μg of anti-H3K36me3 or anti-H4K16ac antibody. Immunoprecipitated chromatin was eluted with a buffer containing 100 mm NaHCO3 and 1% SDS at 68 °C for 2 h. DNA was purified using the QIAquick PCR purification kit (Qiagen) and used for real-time PCR analysis. Real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad) on the CFX96 real-time PCR detection system (Bio-Rad). Primers used in real-time PCR were as follows (F1 and F2 refer to DNA fragments that are ∼500 and 2500 bp away from the I-SceI–induced DSB site, respectively): ChIP-F1-F, TCTTCTTCAAGGACGACGGCAACT; ChIP-F1-R, TTGTAGTTGTACTCCAGCTTGTGC; ChIP-F2-F, CCGCGACGTCTGTCGAGAAG; ChIP-F2-R, GCCGATGCAAAGTGCCGATA. The siRNA target sequences for SETD2 and LEDGF were 5′-GAAACCGUCUCCAGUCUGU-3′ and 5′-GCAAUGAGGAUGUGACUAA-3′, respectively.
Author contributions
L. L. and Y. W. conceived the project, designed the experiments, analyzed the data, and wrote the manuscript. L. L. conducted all the experiments.
Supplementary Material
Acknowledgments
We thank Profs. Jeremy M. Stark and Yingli Sun for providing cell lines and plasmids.
This work was supported by National Institutes of Health Grant R21 ES025392. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S8.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- H3K36
- lysine 36 in histone H3
- H3K36me
- H3K36 monomethylation
- H3K36me2
- H3K36 dimethylation
- H3K36me3
- H3K36 trimethylation
- NSD
- nuclear SET domain
- DSB
- double-strand break
- HR
- homologous recombination
- LEDGF
- lens epithelium–derived growth factor p75 splicing variant
- H4K16ac
- acetylation of lysine 16 in histone H4
- NCS
- neocarzinostatin
- ATM
- ataxia telangiectasia-mutated
- CF
- chromatin fraction
- SF
- soluble fraction.
References
- 1. Zhou V. W., Goren A., and Bernstein B. E. (2011) Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 12, 7–18 [DOI] [PubMed] [Google Scholar]
- 2. Suganuma T., and Workman J. L. (2011) Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 80, 473–499 [DOI] [PubMed] [Google Scholar]
- 3. Smeenk G., and van Attikum H. (2013) The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu. Rev. Biochem. 82, 55–80 [DOI] [PubMed] [Google Scholar]
- 4. Varier R. A., and Timmers H. T. (2011) Histone lysine methylation and demethylation pathways in cancer. Biochim. Biophys. Acta 1815, 75–89 [DOI] [PubMed] [Google Scholar]
- 5. Wagner E. J., and Carpenter P. B. (2012) Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li F., Mao G., Tong D., Huang J., Gu L., Yang W., and Li G. M. (2013) The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell 153, 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pfister S. X., Ahrabi S., Zalmas L. P., Sarkar S., Aymard F., Bachrati C. Z., Helleday T., Legube G., La Thangue N. B., Porter A. C., and Humphrey T. C. (2014) SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 7, 2006–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carvalho S., Vítor A. C., Sridhara S. C., Martins F. B., Raposo A. C., Desterro J. M., Ferreira J., and de Almeida S. F. (2014) SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife 3, e02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aymard F., Bugler B., Schmidt C. K., Guillou E., Caron P., Briois S., Iacovoni J. S., Daburon V., Miller K. M., Jackson S. P., and Legube G. (2014) Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fnu S., Williamson E. A., De Haro L. P., Brenneman M., Wray J., Shaheen M., Radhakrishnan K., Lee S. H., Nickoloff J. A., and Hromas R. (2011) Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. U.S.A. 108, 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hayakawa T., Zhang F., Hayakawa N., Ohtani Y., Shinmyozu K., Nakayama J., and Andreassen P. R. (2010) MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. J. Cell Sci. 123, 1124–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sy S. M., Huen M. S., and Chen J. (2009) MRG15 is a novel PALB2-interacting factor involved in homologous recombination. J. Biol. Chem. 284, 21127–21131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daugaard M., Baude A., Fugger K., Povlsen L. K., Beck H., Sørensen C. S., Petersen N. H., Sorensen P. H., Lukas C., Bartek J., Lukas J., Rohde M., and Jäättelä M. (2012) LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat. Struct. Mol. Biol. 19, 803–810 [DOI] [PubMed] [Google Scholar]
- 14. Zhang P., Du J., Sun B., Dong X., Xu G., Zhou J., Huang Q., Liu Q., Hao Q., and Ding J. (2006) Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 34, 6621–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharma G. G., So S., Gupta A., Kumar R., Cayrou C., Avvakumov N., Bhadra U., Pandita R. K., Porteus M. H., Chen D. J., Cote J., and Pandita T. K. (2010) MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol. Cell Biol. 30, 3582–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krishnan V., Chow M. Z., Wang Z., Zhang L., Liu B., Liu X., and Zhou Z. (2011) Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 108, 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu J., Chen Y., Lu L. Y., Wu Y., Paulsen M. T., Ljungman M., Ferguson D. O., and Yu X. (2011) Chfr and RNF8 synergistically regulate ATM activation. Nat. Struct. Mol. Biol. 18, 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Renaud E., Barascu A., and Rosselli F. (2016) Impaired TIP60-mediated H4K16 acetylation accounts for the aberrant chromatin accumulation of 53BP1 and RAP80 in Fanconi anemia pathway-deficient cells. Nucleic Acids Res. 44, 648–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun Y., Jiang X., Xu Y., Ayrapetov M. K., Moreau L. A., Whetstine J. R., and Price B. D. (2009) Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol. 11, 1376–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., and Peterson C. L. (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 21. Hsiao K. Y., and Mizzen C. A. (2013) Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J. Mol. Cell Biol. 5, 157–165 [DOI] [PubMed] [Google Scholar]
- 22. Tang J., Cho N. W., Cui G., Manion E. M., Shanbhag N. M., Botuyan M. V., Mer G., and Greenberg R. A. (2013) Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20, 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu Y., Gan E. S., Zhou J., Wee W. Y., Zhang X., and Ito T. (2014) Arabidopsis MRG domain proteins bridge two histone modifications to elevate expression of flowering genes. Nucleic Acids Res. 42, 10960–10974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iacovoni J. S., Caron P., Lassadi I., Nicolas E., Massip L., Trouche D., and Legube G. (2010) High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 29, 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eidahl J. O., Crowe B. L., North J. A., McKee C. J., Shkriabai N., Feng L., Plumb M., Graham R. L., Gorelick R. J., Hess S., Poirier M. G., Foster M. P., and Kvaratskhelia M. (2013) Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 41, 3924–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu Y., Scully R., Sobhian B., Xie A., Shestakova E., and Livingston D. M. (2011) RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 25, 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pei H., Zhang L., Luo K., Qin Y., Chesi M., Fei F., Bergsagel P. L., Wang L., You Z., and Lou Z. (2011) MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470, 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Akhtar A., and Becker P. B. (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5, 367–375 [DOI] [PubMed] [Google Scholar]
- 29. Ikura T., Ogryzko V. V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., and Nakatani Y. (2000) Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463–473 [DOI] [PubMed] [Google Scholar]
- 30. Murr R., Loizou J. I., Yang Y. G., Cuenin C., Li H., Wang Z. Q., and Herceg Z. (2006) Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 8, 91–99 [DOI] [PubMed] [Google Scholar]
- 31. Jha S., Shibata E., and Dutta A. (2008) Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol. Cell Biol. 28, 2690–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carrozza M. J., Utley R. T., Workman J. L., and Côté J. (2003) The diverse functions of histone acetyltransferase complexes. Trends Genet. 19, 321–329 [DOI] [PubMed] [Google Scholar]
- 33. Sun Y., Jiang X., Chen S., Fernandes N., and Price B. D. (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U.S.A. 102, 13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun Y., Xu Y., Roy K., and Price B. D. (2007) DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol. Cell Biol. 27, 8502–8509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chapman J. R., Taylor M. R., and Boulton S. J. (2012) Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 [DOI] [PubMed] [Google Scholar]
- 36. Panier S., and Boulton S. J. (2014) Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 15, 7–18 [DOI] [PubMed] [Google Scholar]
- 37. Lawrence M. S., Stojanov P., Mermel C. H., Robinson J. T., Garraway L. A., Golub T. R., Meyerson M., Gabriel S. B., Lander E. S., and Getz G. (2014) Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T., Shimamura T., Sato-Otsubo A., Nagae G., Suzuki H., Nagata Y., Yoshida K., Kon A., Suzuki Y., Chiba K., et al. (2013) Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 45, 860–867 [DOI] [PubMed] [Google Scholar]
- 39. Zhu X., He F., Zeng H., Ling S., Chen A., Wang Y., Yan X., Wei W., Pang Y., Cheng H., Hua C., Zhang Y., Yang X., Lu X., Cao L., et al. (2014) Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat. Genet. 46, 287–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gunn A., and Stark J. M. (2012) I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 920, 379–391 [DOI] [PubMed] [Google Scholar]
- 41. Sakuma T., Nishikawa A., Kume S., Chayama K., and Yamamoto T. (2014) Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 4, 5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu J., Li L., Wang P., You C., Williams N. L., and Wang Y. (2016) Translesion synthesis of O4-alkylthymidine lesions in human cells. Nucleic Acids Res. 44, 9256–9265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiong L., Ping L., Yuan B., and Wang Y. (2009) Methyl group migration during the fragmentation of singly charged ions of trimethyllysine-containing peptides: precaution of using MS/MS of singly charged ions for interrogating peptide methylation. J. Am. Soc. Mass Spectrom. 20, 1172–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aygün O., Svejstrup J., and Liu Y. (2008) A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl. Acad. Sci. U.S.A. 105, 8580–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maunakea A. K., Nagarajan R. P., Bilenky M., Ballinger T. J., D'Souza C., Fouse S. D., Johnson B. E., Hong C., Nielsen C., Zhao Y., Turecki G., Delaney A., Varhol R., Thiessen N., Shchors K., et al. (2010) Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.