Abstract
Reactive Oxygen Species (ROS) and Reactive Nitrogen Species (RNS) were initially regarded mainly as metabolic by-products with damaging properties. Over the last decade, our understanding of their role in metabolism was drastically changed and they were recognized as essential mediators in cellular signaling cascades, as well as modulators of biochemical pathways. Proteostasis is highly affected by the various levels of intracellular and extracellular free radicals with either mild or severe outcomes. As part of the proteostatic network, the proteasome system is equally affected by redox alterations. This short review summarizes the effects of oxidative stress on proteasome status while it also recapitulates conditions and processes where redox alterations signal changes to proteasome expression, assembly and function.
Abbreviations: Abcd1, ATP binding cassette subfamily D member 1; AD, Alzheimer's disease; ALD, adrenoleukodystrophy; ALS, amyotrophic lateral sclerosis; AP-1, activator protein-1; APP, amyloid precursor protein; CNS, central nervous system; DDI, DNA-damage inducible 1; D-gal, D-galactose; DS, down syndrome; E1, ubiquitin-activating enzyme; E2, ubiquitin-conjugating enzyme; E3, ubiquitin-ligase; E4, ubiquitin-prolongation enzyme; ER, endoplasmic reticulum; ΕRK, extracellular signal-regulated kinase; ESCs, embryonic stem cells; γ-GCS, γ-glutamylcysteinesynthetase; GPX1, glutathione peroxidase-1; GST, glutathione s-transferase; HD, Huntington's disease; HFL-1, human fetal lung fibroblasts; HIF-1, hypoxia inducible factor 1; HO-1, heme oxygenase 1; HSF, heat shock factor; IFN-γ, interferon-γ; JNKs, c-Jun N-terminal κinases; Keap1, kelch-like ECH associated protein 1; Maf, musculoaponeurotic fibrosarcoma protein; MAPKs, mitogen- activated protein kinases; MEFs, mouse embryonic fibroblasts; NF-kB, nuclear factor-kappa B; Nrf2, nuclear factor (erythroid-derived-2)-like 2; NOX, NADPH oxidase; NQO-1, NAD(P)H quinoneoxidoreductase- 1; PARP, poly ADP-ribose polymerase; PKC, protein kinase C; ROS, reactive oxygen species; RNS, reactive nitrogen species; SKN-1, skinhead-1; SESN, sestrin; SINGs, stress-induced nuclear granules; SIRT-1, sirtuin-1; SOD1, superoxide dismutase 1; TNF, tumor necrosis factor; UBC1, ubiquitin-conjugating enzyme 1; UCHL1, C-terminal hydrolase L1; UPS, ubiquitin proteasome system; 18α GA, 18α-glycyrrhetinic acid
Keywords: Oxidative stress, Proteasome, Free radicals, Redox status
Graphical abstract
1. Oxidative stress
Organisms are obliged to live in an environment that contains 78% nitrogen and 21% oxygen. Therefore, they have adapted to that environment and this is reflected by the mechanisms they possess to maintain a balance between oxidants and antioxidants [1]. Oxidative stress is a term that is used to describe the imbalance between oxidants and antioxidants that occur in favor of oxidants [2].
1.1. Free radicals
Free radicals are in general defined as chemical species that possess unpaired electrons. There are two major species; a) Reactive oxygen species (ROS) that are molecules derived from O2 and, b) Reactive nitrogen species (RNS) that are molecules derived from nitrogen and oxygen (nitric oxide – NO) [1]. Increase in cellular ROS and RNS levels is caused by both endogenous and exogenous sources. Exogenous sources include smoke, air pollutants, radiation (UV and ionizing) and several drugs. The majority of endogenous ROS are produced through respiration in the mitochondria while the endoplasmic reticulum (ER) and several enzymes are additional ROS sources. In general most of the enzymes that metabolize oxygen may produce ROS i.e. NADPH oxidase (NOX), cytochrome P450 enzymes, lipooxygenases and cyclooxygenase. Superoxide anions (O2-), hydrogen peroxide (H2O2) and hydroxyl radicals (OH∙) are the most important cellular ROS [1]. Hydroxyl radicals are released by metal storage proteins and heme groups while free copper or iron ions are released from iron-sulphur clusters [3]. Intracellular ROS levels are important. Low levels may lead to cell proliferation, differentiation and in general they can act as signaling messengers, whereas high levels lead to cell death, apoptosis and senescence as they affect all biological macromolecules such as lipids, proteins and nucleic acids. Consequently, the regulation of cellular ROS levels is highly important [2].
1.2. Redox signaling: major pathways involved
ROS are considered as important signaling messengers in the cell, playing a key role in numerous pathways [4]. Nevertheless, when oxidative stress occurs, cellular systems become dysfunctional and cells are led to death. Upon mild or severe oxidative stress, many transcription factors and signaling pathways are triggered, indicating that ROS can promote alterations not only in the gene but also in the protein expression [5].
ROS are involved in the activation of several serine/threonine kinases. There are two major examples; mitogen- activated protein kinases (MAPKs) and protein kinase C (PKC). MAPKs include the extracellular signal-regulated kinases (ERK), c-Jun N-terminal kinases (JNKs) and p38 kinases that are involved in cell proliferation, differentiation and apoptosis. It has been shown that mild oxidative stress has a mitogenic effect through the activation of the growth factor receptor-related ERK pathways. Under mild oxidative stress, p38 promotes mitotic arrest while upon severe oxidative stress JNK and p38 regulate cellular apoptotis [6]. PKC is the other important serine/threonine kinase that alters the expression of various genes during oxidative stress. PKC is highly responsive to oxidative stress due to its numerous cysteines that can be modified by oxidants. It is involved in various pathways such as cell death, growth and stress response [7].
Several transcription factors such as Nrf2 (Nuclear factor (erythroid-derived-2) like-2), NF-kB (Nuclear factor-kappa B), tumor protein p53, HIF-1 (Hypoxia inducible factor 1), FOXO (Forkhead box class O) and AP-1 (Activator protein-1) are also affected by ROS. There are three nuclear factor (erythroid-derived-2) related factors (Nrfs) namely Nrf1, Nrf2 and Nrf3. Nrf1 and Nrf2 have been shown to play a key role in oxidative response. Nrf2 is a transcription factor that under normal conditions is bound to Keap1 (Kelch-like ECH Associated Protein 1) in the cytoplasm, it is ubiquitinated by the E3 ligase complex, Keap1-Cul3-Rbx-1 and degraded by the proteasome [8]. Under oxidative stress, Keap1 is modified and the E3 ligase complex is inactivated allowing Nrf2 to accumulate and translocate to the nucleus where it heterodimerizes with small Maf proteins (Musculoaponeuroticfibrosarcoma proteins) to bind antioxidant-response elements (ARE), thus driving the transcription of several protective genes including NQO-1 (NAD(P)H quinoneoxidoreductase- 1), HO-1 (Heme Oxygenase 1), GSTs (Glutathione S-Transferase), γ-GCS (γ-Glutamylcysteinesynthetase) and several proteasome subunits. In that sense, Nrf2 and its inhibitor Keap1 are sensors of oxidative stress and Nrf2 promotes cell survival. Impairment of Keap1 and Nrf2 leads cells to reduced response or to Nrf2overactivation [6]. Upon proteasome inhibition, Nrf1 forms heterodimers with small Mafs on the ARE of proteasome subunits to promote their gene expression [9]. Under normal conditions, Nrf1 is located in the ER and degraded by the proteasome. In response to proteasome inhibition, Nrf1 is cleaved and thus gets activated and translocated to the nucleus promoting the expression of proteasome subunits [10]. In support, Nrf1 knock out MEFs (Mouse embryonic fibroblasts) fail to produce proteasomes upon treatment with the proteasome inhibitor YU101 [11]. DDI2 (DNA-damage inducible 1 homolog 2) has been found to be necessary for the activation of Nrf1. Upon deletion of DDI2, an increase in the cytosolic form of Nrf1 occurs and production of proteasomes is reduced [12]. Surprisingly, proteasomes are needed for Nrf1 activation since Nrf1 produces proteasomes less effectively upon exposure to high concentrations of proteasome inhibitors [13]. In Caenorhabditis elegans, SKN-1 (Skinhead-1; the ortholog of Nrf1/2/3) has a crucial role in response to oxidative stress by promoting the expression of detoxifying genes and the expression of proteasome genes upon proteasome inhibition [14]. It has been shown that for SKN-1 activation following proteasome impairment a peptide-N- glycanase and DDI1, a conserved aspartic protease, are required [15]. Similar observations have been made in Saccharomyces cerevisiae with the stress-regulated transcription factor Rpn4 governing the expression of proteasome genes in response to stress and proteasome inhibition [16].
NF-kB is a transcription factor that activates genes responsible for cell growth, differentiation, inflammation and cell death [17]. NF-kB can be activated by a variety of signals including ROS that are considered as second messengers involved in its activation via tumor necrosis factor (TNF) and interleukin-1 [18].
P53 has an important impact on the cellular redox state. Under normal conditions, p53 activates the expression of several antioxidant proteins, such as SESN1(mammalian sestrin homolog), SESN2, and GPX1(glutathione peroxidase-1) while reduced p53 levels enhance ROS production [19].
AP-1 is another transcription factor that is responsive to oxidative stress driving the expression of genes involved in cell proliferation, differentiation and apoptosis [20].
HIF-1 consists of HIF-1α and HIF-1β subunits. HIF-1β expression is independent of O2whereas HIF-1α subunit is modulated by the relative O2levels. During hypoxia, HIF-1α translocates into the nucleus and dimerizes with HIF-1β subunit and p300/CBP or other coactivators inducing the expression of genes that assist the cells to overcome the hypoxic stress [21].
FOXO transcription factor can be activated by oxidative signals through various mechanisms. For example, upon oxidative stress FOXO is phosphorylated by JNK and Mst1 thus being translocated in the nucleus and activated. FOXO may also get deacetylated by SIRT-1 (sirtuin-1) upon oxidative stress thus acquiring enhanced activity and exhibiting increased DNA binding activity [22]. Monoubiquitination enhances the activity of FOXO proteins whereas poly-ubiquitination drives them to proteasome degradation [5].
The intensity of oxidative stress is crucial not only for the final outcome and the cellular fate but also for the signaling pathway that will be eventually modulated. For example during mild stress, Nrf2 is activated transcribing antioxidant enzymes but when the intensity of the stress is high, NF-kB, AP-1, MAPKs and HSF (heat shock factor) are activated in order to transcribe antioxidant enzymes, inflammatory proteins and heat shock proteins. Finally, under high oxidative stress the cell is driven to death by necrosis and apoptosis; there is not specific evidence regarding the pathway that mediates this response [2].
2. Proteasome system
Oxidative stress affects the correct function of cellular and molecular mechanisms that assures cellular integrity, leading to homeostasis deregulation. Proteostasis is highly disturbed upon oxidative stress with the proteasome system playing a pivotal role.
2.1. Structure
Proteasomes are large protein complexes responsible for the proper regulation of the cellular protein load. They constitute one of the main cellular proteolytic mechanisms. The proteasome consists of catalytic and regulatory subunits. The basic particle of the proteasome is the 20S core, a barrel-shaped complex formed through the assembly of four protein rings. The outer rings are identical and consist of seven different α (alpha) regulatory subunits (α1–7). The α-rings embrace two identical inner rings that consist of seven different β (beta) catalytic subunits (β1–7) [23]. Three β subunits possess catalytic properties; β1 with caspase-like activity (peptidyl-glutamyl-peptide-hydrolase), β2 with trypsin-like activity and β5 with chymotrypsin-like activity [24].
The barrel-shaped 20S proteasome forms a gated channel through which a limited number of peptides and proteins enter [25]. To alter the gate conformation and allow the degradation of a wider range of proteins (ubiquitinated, damaged and misfolded proteins), 19S regulatory complexes bind onto the 20S proteasome. The assembly of one or two 19S regulatory complexes on either end of the core proteasome leads to the formation of 26S or 30S complexes, respectively. The 19S regulatory complex consists of a “base” and a “lid” [26]. The base is the component that is adjusted to the core and through its hexameric ring of six AAA-ATPases (Rpt1-6) can modify the conformation of the α-gated channel. Four non-ATPases are also part of the 19S “base”. The horseshoe-shaped “lid” is attached to the base and consists of nine non-ATPases.
2.2. Function
The proteasome is able to recognize and degrade peptides and proteins either to maintain the equilibrium between normal protein production and degradation, or to eliminate the damaged, misfolded/unfolded or pathogenic proteins. The 20S proteasome is able to recognize and promote the degradation of already unfolded, damaged (misfolded, oxidized) proteins in an ATP-independent manner [27], [28]. In contrast, the 26S/30S complexes need ATP to complete the degradation process. Proteins targeted to be degraded by the 26S/30S complexes are tagged with a poly-ubiquitin tail that is recognized by the 19S regulatory complex. Upon substrate recognition, 19S ATP-dependent subunits are activated to unfold the substrate and facilitate its entrance through the gate and, finally, its route to the catalytic subunits [29]. The conjugation of the poly-ubiquitin chain on the protein target is effectuated through the consecutive recruitment of four types of enzymes; the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), the ubiquitin-ligase (E3) [30] and the ubiquitin-prolongation enzyme (E4) [31]. The classification of the latter is still under discussion.
The immunoproteasome is a specific proteasome type that is induced by IFN-γ (interferon-γ), TNF-α, or lipopolysaccharides [32]. It consists of the inducible 20S proteasome (i20S with the inducible forms iβ1, iβ2 and iβ5) and mainly the 11S regulatory particle. Finally, in mammals, hybrid proteasomes are also found, containing both 19S and 11S regulatory complexes [33]. This hybrid proteasome can degrade substrates both ATP- dependently and -independently in a more efficient way than the immunoproteasome [34], but its role is not yet fully elucidated.
2.3. Proteasome manipulation
Proteasome-mediated degradation has been shown to be manipulated both genetically and/or through the use of compounds [26], [35], [36]. Under specific conditions (such as aging, age-related conditions and diseases), the activity of the proteasome has been shown to be compromised. Several studies have focused on the activation of the proteasome (either in cellular models or in model organisms) to alleviate the negative effects of the above-mentioned processes [37], [38], [39], [40], [41], [42], [43], [44]. The proteasome activity has been shown to be increased through the induced expression of several subunits (core and/or regulatory ones) that ultimately lead to enhanced proteasome assembly and/or function. Compound-mediated proteasome activation has been achieved either through transcriptional up-regulation of proteasome subunits or through direct binding on the proteasomes that confers conformational changes and therefore results in the establishment of an opened (and therefore active) proteasome form [45].
2.4. Effects of oxidative stress on proteasome status
The proteasomes, and more specifically the 20S complex, possess a major role in the maintenance of redox balance by recognizing and removing the oxidatively modified or damaged proteins. Since protein oxidation promotes protein unfolding, the 20S complex is recruited to cleave those substrates in an ATP-independent manner [46], [47]. Proteasome-mediated degradation has been shown to be enhanced by more than 10 times upon exposure to H2O2 or O2- [48]. Following oxidative stress, the 26S proteasome was shown to disassemble in order to retrieve intact 20S and 19S particles [49]. Moreover, 20S proteasomes have been shown to be more resistant to oxidative stress in contrast to the proper function of 26S proteasomes that is highly affected [50]. Additionally, the immunoproteasome has been shown to degrade oxidatively modified proteins in a more efficient way when the 11S regulatory particle is attached on it rather than working alone [51], [52]. Nevertheless, constitutive exposure to reactive species does not only affect cellular proteins but also the proteasome (and specifically the 26S proteasome) itself [53]. Moreover, several post-translational modifications of specific subunits like glutathionylation, carbonylation, glycoxidation and lipoxidation have been reported to occur upon oxidative stress [54]. Specifically, S-glutathionylation has been shown to be a post-translational modification that is involved in the redox regulation of the proteasome [55]. As a result, proteasome activity is impaired leading to a vicious cycle between heavily oxidized cellular protein load (that results to cross-linked protein aggregates) and their inhibitory action on the proteasome function [56]. The interplay between the proteasome and oxidative stress is further suggested by the observation that cells treated with proteasome inhibitors exhibit increased ROS levels [57]. There is data indicating that NADH/NAD+ has a key role in the proteasome function during redox regulation. NADH has been shown to maintain the proteasome amounts in normal levels [58]. Additionally, the proteasome inhibitor bortezomib becomes more effective upon depletion of NAD+ in myeloma cells thus suggesting the active interplay between NAD+ and the proteasome [59].
As mentioned above, Nrf2 that is activated upon oxidative stress has been shown to regulate the expression of proteasome genes [60]. Recently, this adaptive response has been revealed to be sex- and age-dependent in Drosophila melanogaster [61]. Nrf2/ Keap-1 axis was also shown to play a key role in stem cell differentiation where ROS levels are constantly altered. More specifically, upon treatment with sulphoraphane, Nrf2 becomes activated and controls the pluripotency of human embryonic stem cells (hESCs) through the regulation of proteasome expression and elevation of their proteasome activity [62]. During oxidative stress after As (III) exposure of Ub-deficient N2a neuroblastoma cells, UPS is impaired. This impairment is related to the increase of p65-Nrf1 in the nucleus, a form that competes with Nrf2 and p95-Nrf1, leading to decreased proteasome activity and expression [14].
3. Conditions and processes where redox alterations signal changes to proteasome function
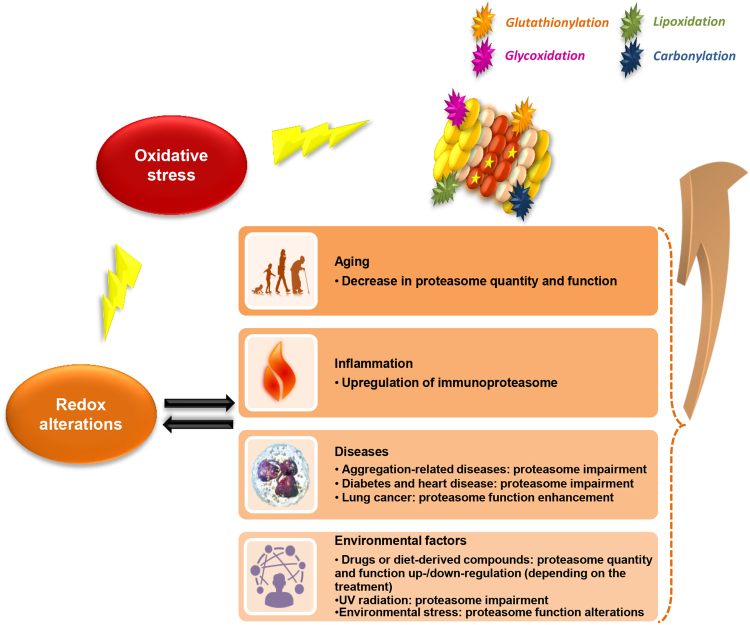
Oxidative stress seems to play a major role in various conditions and diseases, such as aging, inflammation, diabetes, cancer and neurodegenerative diseases. In many of them, the observed redox alterations have been also shown to induce changes on the proteasome function. Fig. 1 summarizes conditions and processes that affect the proteasome status through redox alterations.
Fig. 1.
Effects of redox alterations on proteasome status. Oxidative stress promotes redox alterations on cellular level that affect proteasome structure, assembly and function. The proteasome is also subjected to post-translational modifications itself (glutathionylation, carbonylation, lipoxidation and glycoxidation-placed randomly in the figure-) due to oxidative stress [54]. Various conditions and processes (i.e. aging, diseases, inflammation and environmental factors) promote redox alterations that affect the proteasome status. The active sites of the proteasome are depicted with stars in the β-subunits.
3.1. Aging
Aging is a process that occurs in all organisms leading to dysfunction of cells and tissues and ultimately to organismal deterioration. Upon aging progression and cellular senescence, the proteasome and its activities have been shown to be reduced [63], [64]. Moreover, ROS levels are elevated (mainly due to the downstream mitochondrial dysfunction combined with a dysregulation of repair mechanisms; [65]) and a disruption of the equilibrium between oxidants and antioxidants occurs leading to an accumulation of damaged and oxidized proteins. The accumulation of inhibitory cross-linked proteins and ROS products in the cell may explain at least partially the observed decreased proteasome activity upon aging. Finally, the observed reduced Nrf2 activity during aging may also affect the proteasome gene expression [66].
3.2. Inflammation
Increased ROS levels are observed during inflammation and infectious diseases. Specifically ROS produced by macrophages and neutrophils assist the immune system's response. During inflammation, pathways such as JNK, MAPK and the transcription factors AP-1 and NF-kB that are normally involved in redox regulation are also induced [57]. Upon inflammation, the immunoproteasome is also upregulated [67]. On top of its role to respond to oxidative stress, many studies suggest that the immunoproteasome has an important role during viral infection by regulating CD8 T cell responses, the production of proinflammatory cytokines and the activation of the NF-κB pathway [68].
3.3. Diseases
In pathological conditions, redox alterations are usually more intense resulting in complete homeostasis and proteostasis imbalance as observed in neurodegenerative and cardiovascular diseases, diabetes and cancer. Proteasome function can be either negatively or positively regulated during disease-related redox modifications.
Aggregation-prone proteins play a key role in the development of neurodegenerative diseases. Under such conditions, redox imbalance leads to highly oxidatively-modified proteins that tend to accumulate and create aggregates resulting in proteasome impairment. In turn, proteasomes that are not functional, cannot degrade the damaged proteins and aggregates that are formed and, consequently, a feedback loop is established towards elevated levels of oxidative stress and aggregated proteins.
Adrenoleukodystrophy (ALD) is a disease induced by the loss of function of Abcd1 (ATP binding cassette subfamily D member 1) peroxisomal transporter, where high levels of oxidative stress impair proteasomal and mitochondrial function. As an adaptive mechanism to limit the damage, induction of the immunoproteasome occurs [69]. Treatment with antioxidants restores the proteasome function and prevents the immunoproteasome induction [69].
Alzheimer's disease (AD) is characterized by the aggregation-prone Amyloid beta peptide (Aβ); its accumulation in the brain of AD patients induces high levels of oxidative stress resulting in proteasome inhibition [70], [71]. Furthermost, patients with Down Syndrome (DS) tend to exhibit an early onset AD phenotype because DS-associated oxidative stress impairs the ubiquitin-mediated proteasome degradation leading to a three-fold increase in the amount of the Amyloid Precursor Protein (APP; its cleavage leads to the production of Aβ) as compared to the levels in AD patients without DS [72].
Similarly, during the progression of Huntington's disease (HD; characterized by the aggregation of huntingtin protein) and Amyotrophic Lateral Sclerosis (ALS; characterized by superoxide dismutase 1, SOD1, aggregation) deleterious redox modifications occur due to oxidatively-modified protein aggregates that negatively affect the proteasomal degradation mechanism and the mitochondrial function in the synapses of the Central Nervous System (CNS) [65].
Parkinson's disease (PD) is an α-synuclein-associated disease where iron, calcium and ROS levels are elevated and levels of glutathione remain low, thus resulting in increased oxidative damage and mitochondrial malfunction. This cascade of events leads to the degeneration of the dopaminergic neurons as oxidatively-modified and aggregated α-synuclein impairs the proteasome and inhibits the mitochondrial complex I creating a feedback loop towards additional proteasome oxidation and inactivation [73]. Recently, under early onset PD conditions, it has been shown that the proteasome adapts to oxidative stress in a TCF11/Nrf1-dependent manner [74].
Diabetes is a metabolic disease with severe complications. One of the factors that contribute to the impairment of blood glucose regulation is the systemic oxidative stress and the inability of pancreatic β-cells to produce anti-oxidant enzymes, among others [75], thus leading to proteasomal impairment and aggregation of polyubiquitinated proteins [76]. Nrf2 upregulation restores the proteasome activities and mediates adaptation to oxidative stress [77], in contrast to bortezomib-mediated proteasome inhibition that further negatively affects the diabetes-mediated disrupted cellular redox status [78]. On the other hand, atherosclerotic plaques of type 2 diabetes patients have been found to exhibit high levels of nitrotyrosine and O2- (oxidative stress markers) together with high levels of 20S proteasome activities and ubiquitin derived from CD68+ macrophages that assist the maintenance of a healthier environment [79]. Palmitate treatment induces insulin resistance and promotes UPS and autophagy activation. Treatment with curcumin alleviates the palmitate-induced ER stress with a parallel suppression of the 20S peptidase activity and induction of autophagy [80].
Increased ROS levels in patients with ischemia and reperfusion of the heart lead to decreased levels of proteasome function, whereas elevated levels of oxidized proteins induced by sympathetic signaling result in acute increase of the 26S proteasome activities in patients with hypertrophia [81].
Cancer cells tend to adapt to oxidative stress via Nrf2 upregulation [82]. A positive feedback loop between gankyrin (19S subunit RPN-10) and Nrf2 rescues cancer cells from oxidative stress and limits ROS levels in hepatocarcinoma. More specifically, gankyrin binds Keap1 leading to the inhibition of Nrf2polyubiquitination and degradation [83]. Similarly, Nrf2 is upregulated through the inhibition of its ubiquitin-mediated degradation in multiple myeloma resulting in low ROS levels [84]. On the other hand, in chronic conditions of lung cancer, cachexia is a symptom that is related to the increased levels of protein oxidation, PARP (poly ADP-ribose polymerase) activity and ubiquitin-mediated degradation leading to fiber atrophy [85].
3.4. Environmental factors, supplements and drugs
Dietary habits and supplementation, drug treatment and cellular/organismal environment can affect the cellular redox status.
Several natural or chemical compounds have been shown to affect the crosstalk between the proteasome and redox regulation. Quercetin and oleuropein exhibit both anti-aging and anti-oxidant properties through the stimulation of proteasome function [86], [87]. Treatment with 18α-glycyrrhetinic acid (18α GA) promotes the proteasome assembly and activity in HFL-1 human primary cells in an Nrf2-dependent manner leading to parallel decreased ROS levels [38]. The positive effects of 18αGA on proteasome status have been shown to be evolutionary conserved; verified at the level of a multi-cellular organism (C. elegans) [42]. The flavonoid quercetin, a known Nrf2 activator [88], was shown to enhance proteasome activities, to promote increased oxidative stress resistance and to confer enhanced longevity in HFL-1 cells [86].
The anti-alcoholism drug disulfiram can also inhibit the ubiquitin-mediated proteasome degradation [89] and this inhibition was suggested to be related to the redox-associated apoptosis in melanoma cells [90].
Deficiency in vitamin D promotes redox alterations that ultimately lead to 20S proteasome hyperactivation resulting in muscular atrophy [91]. Hypoxia has been suggested to promote similar effects on muscles. More specifically, hypoxic environment increases the oxidative stress levels resulting in elevated levels of proteasome- and autophagy-mediated degradation [92]. Administration of ethanol promotes redox alterations that lead to decreased proteasome activities [93].
Radiation can also trigger redox alterations. UV radiation promotes protein oxidation leading to increased cellular oxidative stress levels and decreased levels of proteasome activities without however affecting the 20S or 26S proteasome conformation [94]. D-galactose (D-gal) is a constituent of human diet and has been found to induce oxidative stress and to promote senescence in primary auditory cortex cells by affecting the proteasome activity [95]. Overexpression of the ubiquitin C-Terminal Hydrolase L1 (UCHL1) leads to increased proteasome activity and alters the aging process in the auditory cortex.
Environmental stress like oxidative stress, salt stress and starvation not only induce protein misfolding but also the acute formation of distinct subnuclear structures that are called stress induced nuclear granules (SINGs) in the nematode C. elegans [96]. SINGs contain ubiquitin molecules and assembled proteasomes ready to respond to stress.
Even in plants, overexpression of the ubiquitin-conjugating enzyme UBC1 rescues tobacco from cadmium toxicity and oxidative stress by activating the 20/26S proteasome [97].
4. Conclusions
The interplay between the proteasome function and regulation and the cellular redox status is constant and bidirectional. Depending on the level of redox alterations, the effects on proteasome status may vary from beneficial to devastating. Identification of the mechanisms that are involved in this interplay will enable us to limit the catastrophic effects and to multiply the positive outcomes.
Acknowledgments
Cited work from our lab was supported by the following Research Funding Programs: Thales “GenAge” (QΑLΗS ΑP:10479/3.7.12 MIS380228) and “MAESTRO” co-financed by the European Union (European Social Fund) and Greek National Funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) and a Scientific Project funded by Empirikion Foundation to NC.
References
- 1.Ye Z.W., Zhang J., Townsend D.M., Tew K.D. Oxidative stress, redox regulation and diseases of cellular differentiation. Biochim. Biophys. Acta. 2015;1850(8):1607–1621. doi: 10.1016/j.bbagen.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lushchak V.I. Free radicals, reactive oxygen species, oxidative stress and its classification. Chem.-Biol. Interact. 2014;224:164–175. doi: 10.1016/j.cbi.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 3.Nathan C., Cunningham-Bussel A. Beyond oxidative stress: an immunologist's guide to reactive oxygen species. Nat. Rev. Immunol. 2013;13(5):349–361. doi: 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egea J., Fabregat I., Frapart Y.M., Ghezzi P., Gorlach A., Kietzmann T., Kubaichuk K., Knaus U.G., Lopez M.G., Olaso-Gonzalez G., Petry A., Schulz R., Vina J., Winyard P., Abbas K., Ademowo O.S., Afonso C.B., Andreadou I., Antelmann H., Antunes F., Aslan M., Bachschmid M.M., Barbosa R.M., Belousov V., Berndt C., Bernlohr D., Bertran E., Bindoli A., Bottari S.P., Brito P.M., Carrara G., Casas A.I., Chatzi A., Chondrogianni N., Conrad M., Cooke M.S., Costa J.G., Cuadrado A., Dang P. My-Chan, Smet B. De, Debelec-Butuner B., Dias I.H.K., Dunn J.D., Edson A.J., Assar M. El, El-Benna J., Ferdinandy P., Fernandes A.S., Fladmark K.E., Forstermann U., Giniatullin R., Giricz Z., Gorbe A., Griffiths H., Hampl V., Hanf A., Herget J., Hernansanz-Agustin P., Hillion M., Huang J., Ilikay S., Jansen-Durr P., Jaquet V., Joles J.A., Kalyanaraman B., Kaminskyy D., Karbaschi M., Kleanthous M., Klotz L.O., Korac B., Korkmaz K.S., Koziel R., Kracun D., Krause K.H., Kren V., Krieg T., Laranjinha J., Lazou A., Li H., Martinez-Ruiz A., Matsui R., McBean G.J., Meredith S.P., Messens J., Miguel V., Mikhed Y., Milisav I., Milkovic L., Miranda-Vizuete A., Mojovic M., Monsalve M., Mouthuy P.A., Mulvey J., Munzel T., Muzykantov V., Nguyen I.T.N., Oelze M., Oliveira N.G., Palmeira C.M., Papaevgeniou N., Pavicevic A., Pedre B., Peyrot F., Phylactides M., Pircalabioru G.G., Pitt A.R., Poulsen H.E., Prieto I., Rigobello M.P., Robledinos-Anton N., Rodriguez-Manas L., Rolo A.P., Rousset F., Ruskovska T., Saraiva N., Sasson S., Schroder K., Semen K., Seredenina T., Shakirzyanova A., Smith G.L., Soldati C.M., Sousa B.C., Spickett C.M., Stancic A., Stasia M.J., Steinbrenner H., Stepanic V., Steven S., Tokatlidis K., Tuncay E., Turan B., Ursini F., Vacek S.P., Vajnerova O., Valentova K., Breusegem F. Van, Varisli L., Veal E.A., Yalcin A.S., Yelisyeyeva O., Zarkovic N., Zatloukalova M., Zielonka J., Touyz R.M., Papapetropoulos A., Grune T., Lamas S., Schmidt H., Lisa F. Di, Daiber A. European contribution to the study of ROS: a summary of the findings and prospects for the future from the COST action BM1203 (EU-ROS) Redox Biol. 2017;13:94–162. doi: 10.1016/j.redox.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Q. Transcriptional responses to oxidative stress: pathological and toxicological implications. Pharmacol. Ther. 2010;125(3):376–393. doi: 10.1016/j.pharmthera.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y., Du Y., Le W., Wang K., Kieffer N., Zhang J. Redox control of the survival of healthy and diseased cells. Antioxid. Redox Signal. 2011;15(11):2867–2908. doi: 10.1089/ars.2010.3685. [DOI] [PubMed] [Google Scholar]
- 7.Gopalakrishna R., Jaken S. Protein kinase C signaling and oxidative stress. Free Radic. Biol. Med. 2000;28(9):1349–1361. doi: 10.1016/s0891-5849(00)00221-5. [DOI] [PubMed] [Google Scholar]
- 8.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y., Xiang Y. Molecular and cellular basis for the unique functioning of Nrf1, an indispensable transcription factor for maintaining cell homoeostasis and organ integrity. Biochem. J. 2016;473(8):961–1000. doi: 10.1042/BJ20151182. [DOI] [PubMed] [Google Scholar]
- 10.Steffen J., Seeger M., Koch A., Kruger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell. 2010;40(1):147–158. doi: 10.1016/j.molcel.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Radhakrishnan S.K., Lee C.S., Young P., Beskow A., Chan J.Y., Deshaies R.J. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell. 2010;38(1):17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koizumi S., Irie T., Hirayama S., Sakurai Y., Yashiroda H., Naguro I., Ichijo H., Hamazaki J., Murata S. The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction. eLife. 2016;5 doi: 10.7554/eLife.18357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sha Z., Goldberg A.L. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol.: CB. 2014;24(14):1573–1583. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee D., Ryu K.Y. Effect of cellular ubiquitin levels on the regulation of oxidative stress response and proteasome function via Nrf1. Biochem. Biophys. Res. Commun. 2017;485(2):234–240. doi: 10.1016/j.bbrc.2017.02.105. [DOI] [PubMed] [Google Scholar]
- 15.Lehrbach N.J., Ruvkun G. Proteasome dysfunction triggers activation of SKN-1A/Nrf1 by the aspartic protease DDI-1. eLife. 2016;5 doi: 10.7554/eLife.17721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dohmen R.J., Willers I., Marques A.J. Biting the hand that feeds: Rpn4-dependent feedback regulation of proteasome function. Biochim. Biophys. Acta. 2007;1773(11):1599–1604. doi: 10.1016/j.bbamcr.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Pande V., Ramos M.J. Molecular recognition of 15-deoxy-delta(12,14)-prostaglandin J2 by nuclear factor-kappa B and other cellular proteins. Bioorg. Med. Chem. Lett. 2005;15(18):4057–4063. doi: 10.1016/j.bmcl.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 18.Baud V., Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001;11(9):372–377. doi: 10.1016/s0962-8924(01)02064-5. [DOI] [PubMed] [Google Scholar]
- 19.Liu B., Chen Y., St Clair D.K. ROS and p53: a versatile partnership. Free Radic. Biol. Med. 2008;44(8):1529–1535. doi: 10.1016/j.freeradbiomed.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess J., Angel P., Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 2004;117(Pt 25):5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 21.Hagg M., Wennstrom S. Activation of hypoxia-induced transcription in normoxia. Exp. Cell Res. 2005;306(1):180–191. doi: 10.1016/j.yexcr.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 22.Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Hu L.S., Cheng H.L., Jedrychowski M.P., Gygi S.P., Sinclair D.A., Alt F.W., Greenberg M.E. Vol. 303. Science; New York, N.Y.: 2004. pp. 2011–2015. (Stress-Dependent Regulation of FOXO Transcription Factors by the SIRT1 Deacetylase). [DOI] [PubMed] [Google Scholar]
- 23.Coux O., Tanaka K., Goldberg A.L. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 24.Groll M., Ditzel L., Lowe J., Stock D., Bochtler M., Bartunik H.D., Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386(6624):463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 25.Groll M., Bajorek M., Kohler A., Moroder L., Rubin D.M., Huber R., Glickman M.H., Finley D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000;7(11):1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 26.Papaevgeniou N., Chondrogianni N. The ubiquitin proteasome system in Caenorhabditis elegans and its regulation. Redox Biol. 2014;2:333–347. doi: 10.1016/j.redox.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breusing N., Grune T. Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol. Chem. 2008;389(3):203–209. doi: 10.1515/BC.2008.029. [DOI] [PubMed] [Google Scholar]
- 28.Davies K.J. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83(3–4):301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 29.Ciechanover A., Stanhill A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim. Biophys. Acta. 2014;1843(1):86–96. doi: 10.1016/j.bbamcr.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 31.Koegl M., Hoppe T., Schlenker S., Ulrich H.D., Mayer T.U., Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96(5):635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 32.Stratford F.L., Chondrogianni N., Trougakos I.P., Gonos E.S., Rivett A.J. Proteasome response to interferon-gamma is altered in senescent human fibroblasts. FEBS Lett. 2006;580(16):3989–3994. doi: 10.1016/j.febslet.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 33.Tanahashi N., Murakami Y., Minami Y., Shimbara N., Hendil K.B., Tanaka K. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J. Biol. Chem. 2000;275(19):14336–14345. doi: 10.1074/jbc.275.19.14336. [DOI] [PubMed] [Google Scholar]
- 34.Hendil K.B., Khan S., Tanaka K. Simultaneous binding of PA28 and PA700 activators to 20 S proteasomes. Biochem. J. 1998;332(Pt 3):749–754. doi: 10.1042/bj3320749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chondrogianni N., Gonos E.S. Structure and function of the ubiquitin-proteasome system: modulation of components. Prog. Mol. Biol. Transl. Sci. 2012;109:41–74. doi: 10.1016/B978-0-12-397863-9.00002-X. [DOI] [PubMed] [Google Scholar]
- 36.Kornitzer D., Ciechanover A. Modes of regulation of ubiquitin-mediated protein degradation. J. Cell. Physiol. 2000;182(1):1–11. doi: 10.1002/(SICI)1097-4652(200001)182:1<1::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 37.Chondrogianni N., Georgila K., Kourtis N., Tavernarakis N., Gonos E.S. 20S proteasome activation promotes life span extension and resistance to proteotoxicity in Caenorhabditis elegans. FASEB J.: Off. Publ. Fed. Am. Soc. Exp. Biol. 2015;29(2):611–622. doi: 10.1096/fj.14-252189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapeta S., Chondrogianni N., Gonos E.S. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J. Biol. Chem. 2010;285(11):8171–8184. doi: 10.1074/jbc.M109.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapetanou M., Chondrogianni N., Petrakis S., Koliakos G., Gonos E.S. Proteasome activation enhances stemness and lifespan of human mesenchymal stem cells. Free Radic. Biol. Med. 2017;103:226–235. doi: 10.1016/j.freeradbiomed.2016.12.035. [DOI] [PubMed] [Google Scholar]
- 40.Liu G., Rogers J., Murphy C.T., Rongo C. EGF signalling activates the ubiquitin proteasome system to modulate C. elegans lifespan. EMBO J. 2011;30(15):2990–3003. doi: 10.1038/emboj.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papaevgeniou N., Chondrogianni N. UPS activation in the battle against aging and aggregation-related diseases: an extended review. Methods Mol. Biol. 2016;1449:1–70. doi: 10.1007/978-1-4939-3756-1_1. [DOI] [PubMed] [Google Scholar]
- 42.Papaevgeniou N., Sakellari M., Jha S., Tavernarakis N., Holmberg C.I., Gonos E.S., Chondrogianni N. 18alpha-glycyrrhetinic acid proteasome activator decelerates aging and Alzheimer's disease progression in caenorhabditis elegans and neuronal cultures. Antioxid. Redox Signal. 2016;25(16):855–869. doi: 10.1089/ars.2015.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tonoki A., Kuranaga E., Tomioka T., Hamazaki J., Murata S., Tanaka K., Miura M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell. Biol. 2009;29(4):1095–1106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vilchez D., Morantte I., Liu Z., Douglas P.M., Merkwirth C., Rodrigues A.P., Manning G., Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489(7415):263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 45.Chondrogianni N., Voutetakis K., Kapetanou M., Delitsikou V., Papaevgeniou N., Sakellari M., Lefaki M., Filippopoulou K., Gonos E.S. Proteasome activation: an innovative promising approach for delaying aging and retarding age-related diseases. Ageing Res. Rev. 2015;23(Pt A):37–55. doi: 10.1016/j.arr.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Aiken C.T., Kaake R.M., Wang X., Huang L. Oxidative stress-mediated regulation of proteasome complexes. Mol. Cell. Proteom.: MCP. 2011;10(5) doi: 10.1074/mcp.M110.006924. (R110 006924) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldberg A.L. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426(6968):895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 48.Davies K.J., Goldberg A.L. Oxygen radicals stimulate intracellular proteolysis and lipid peroxidation by independent mechanisms in erythrocytes. J. Biol. Chem. 1987;262(17):8220–8226. [PubMed] [Google Scholar]
- 49.Livnat-Levanon N., Kevei E., Kleifeld O., Krutauz D., Segref A., Rinaldi T., Erpapazoglou Z., Cohen M., Reis N., Hoppe T., Glickman M.H. Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 2014;7(5):1371–1380. doi: 10.1016/j.celrep.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 50.Hohn A., Konig J., Grune T. Protein oxidation in aging and the removal of oxidized proteins. J. Proteom. 2013;92:132–159. doi: 10.1016/j.jprot.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Ferrington D.A., Gregerson D.S. Immunoproteasomes: structure, function, and antigen presentation. Prog. Mol. Biol. Transl. Sci. 2012;109:75–112. doi: 10.1016/B978-0-12-397863-9.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pickering A.M., Koop A.L., Teoh C.Y., Ermak G., Grune T., Davies K.J. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 2010;432(3):585–594. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reinheckel T., Ullrich O., Sitte N., Grune T. Differential impairment of 20S and 26S proteasome activities in human hematopoietic K562 cells during oxidative stress. Arch. Biochem. Biophys. 2000;377(1):65–68. doi: 10.1006/abbi.2000.1717. [DOI] [PubMed] [Google Scholar]
- 54.Hohn T.J., Grune T. The proteasome and the degradation of oxidized proteins: part III-Redox regulation of the proteasomal system. Redox Biol. 2014;2:388–394. doi: 10.1016/j.redox.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silva G.M., Netto L.E., Simoes V., Santos L.F., Gozzo F.C., Demasi M.A., Oliveira C.L., Bicev R.N., Klitzke C.F., Sogayar M.C., Demasi M. Redox control of 20S proteasome gating. Antioxid. Redox Signal. 2012;16(11):1183–1194. doi: 10.1089/ars.2011.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friguet B., Szweda L.I. Inhibition of the multicatalytic proteinase (proteasome) by 4-hydroxy-2-nonenal cross-linked protein. FEBS Lett. 1997;405(1):21–25. doi: 10.1016/s0014-5793(97)00148-8. [DOI] [PubMed] [Google Scholar]
- 57.Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82(1):47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 58.Tsvetkov P., Myers N., Eliav R., Adamovich Y., Hagai T., Adler J., Navon A., Shaul Y. NADH binds and stabilizes the 26S proteasomes independent of ATP. J. Biol. Chem. 2014;289(16):11272–11281. doi: 10.1074/jbc.M113.537175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cagnetta A., Cea M., Calimeri T., Acharya C., Fulciniti M., Tai Y.T., Hideshima T., Chauhan D., Zhong M.Y., Patrone F., Nencioni A., Gobbi M., Richardson P., Munshi N., Anderson K.C. Intracellular NAD(+) depletion enhances bortezomib-induced anti-myeloma activity. Blood. 2013;122(7):1243–1255. doi: 10.1182/blood-2013-02-483511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vriend J., Reiter R.J. The Keap1-Nrf2-antioxidant response element pathway: a review of its regulation by melatonin and the proteasome. Mol. Cell. Endocrinol. 2015;401:213–220. doi: 10.1016/j.mce.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 61.Pomatto L.C.D., Wong S., Carney C., Shen B., Tower J., Davies K.J.A. The age- and sex-specific decline of the 20s proteasome and the Nrf2/CncC signal transduction pathway in adaption and resistance to oxidative stress in Drosophila melanogaster. Aging. 2017;9(4):1153–1185. doi: 10.18632/aging.101218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jang J., Wang Y., Kim H.S., Lalli M.A., Kosik K.S. Nrf2, a regulator of the proteasome, controls self-renewal and pluripotency in human embryonic stem cells. Stem Cells (Dayt. Ohio) 2014;32(10):2616–2625. doi: 10.1002/stem.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chondrogianni N., Petropoulos I., Franceschi C., Friguet B., Gonos E.S. Fibroblast cultures from healthy centenarians have an active proteasome. Exp. Gerontol. 35(6-7) 2000:721–728. doi: 10.1016/s0531-5565(00)00137-6. [DOI] [PubMed] [Google Scholar]
- 64.Chondrogianni N., Stratford F.L., Trougakos I.P., Friguet B., Rivett A.J., Gonos E.S. Central role of the proteasome in senescence and survival of human fibroblasts: induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J. Biol. Chem. 2003;278(30):28026–28037. doi: 10.1074/jbc.M301048200. [DOI] [PubMed] [Google Scholar]
- 65.Riederer B.M., Leuba G., Elhajj Z. Oxidation and ubiquitination in neurodegeneration. Exp. Biol. Med. 2013;238(5):519–524. doi: 10.1177/1535370213488484. [DOI] [PubMed] [Google Scholar]
- 66.Zhang H., Davies K.J., Forman H.J. Oxidative stress response and Nrf2 signaling in aging. Free Radic. Biol. Med. 2015;88(Pt B):314–336. doi: 10.1016/j.freeradbiomed.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnston-Carey H.K., Pomatto L.C., Davies K.J. The immunoproteasome in oxidative stress, aging, and disease. Crit. Rev. Biochem. Mol. Biol. 2015;51(4):268–281. doi: 10.3109/10409238.2016.1172554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCarthy M.K., Weinberg J.B. The immunoproteasome and viral infection: a complex regulator of inflammation. Front. Microbiol. 2015;6:21. doi: 10.3389/fmicb.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pujol A. Novel therapeutic targets and drug candidates for modifying disease progression in adrenoleukodystrophy. Endocr. Dev. 2016;30:147–160. doi: 10.1159/000439340. [DOI] [PubMed] [Google Scholar]
- 70.Bonet-Costa V., Pomatto L.C., Davies K.J. The proteasome and oxidative stress in Alzheimer's disease. Antioxid. Redox Signal. 2016;25(16):886–901. doi: 10.1089/ars.2016.6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X., Petranovic D. Amyloid-beta peptide-induced cytotoxicity and mitochondrial dysfunction in yeast. FEMS Yeast Res. 2015;15(6) doi: 10.1093/femsyr/fov061. [DOI] [PubMed] [Google Scholar]
- 72.Tramutola A., Di Domenico F., Barone E., Arena A., Giorgi A., di Francesco L., Schinina M.E., Coccia R., Head E., Butterfield D.A., Perluigi M. Polyubiquitinylation profile in down syndrome brain before and after the development of Alzheimer neuropathology. Antioxid. Redox Signal. 2017;26(7):280–298. doi: 10.1089/ars.2016.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dias V., Junn E., Mouradian M.M. The role of oxidative stress in Parkinson's disease. J. Park. Dis. 2013;3(4):461–491. doi: 10.3233/JPD-130230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sotzny F., Schormann E., Kuhlewindt I., Koch A., Brehm A., Goldbach-Mansky R., Gilling K.E., Kruger E. TCF11/Nrf1-mediated induction of proteasome expression prevents cytotoxicity by rotenone. Antioxid. Redox Signal. 2016;25(16):870–885. doi: 10.1089/ars.2015.6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lenzen S., Drinkgern J., Tiedge M. Low antioxidant enzyme gene expression in pancreatic islets compared with various other mouse tissues. Free Radic. Biol. Med. 1996;20(3):463–466. doi: 10.1016/0891-5849(96)02051-5. [DOI] [PubMed] [Google Scholar]
- 76.Costes S., Huang C.J., Gurlo T., Daval M., Matveyenko A.V., Rizza R.A., Butler A.E., Butler P.C. Beta-cell dysfunctional ERAD/ubiquitin/proteasome system in type 2 diabetes mediated by islet amyloid polypeptide-induced UCH-L1deficiency. Diabetes. 2011;60(1):227–238. doi: 10.2337/db10-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uruno A., Yagishita Y., Yamamoto M. The Keap1-Nrf2 system and diabetes mellitus. Arch. Biochem. Biophys. 2015;566:76–84. doi: 10.1016/j.abb.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 78.Al-Khalili L., de Castro Barbosa T., Ostling J., Massart J., Cuesta P.G., Osler M.E., Katayama M., Nystrom A.C., Oscarsson J., Zierath J.R. Proteasome inhibition in skeletal muscle cells unmasks metabolic derangements in type 2 diabetes. Am. J. Physiol. Cell Physiol. 2014;307(9):C774–C787. doi: 10.1152/ajpcell.00110.2014. [DOI] [PubMed] [Google Scholar]
- 79.Marfella R., D'Amico M., Esposito K., Baldi A., Di Filippo C., Siniscalchi M., Sasso F.C., Portoghese M., Cirillo F., Cacciapuoti F., Carbonara O., Crescenzi B., Baldi F., Ceriello A., Nicoletti G.F., D'Andrea F., Verza M., Coppola L., Rossi F., Giugliano D. The ubiquitin-proteasome system and inflammatory activity in diabetic atherosclerotic plaques: effects of rosiglitazone treatment. Diabetes. 2006;55(3):622–632. doi: 10.2337/diabetes.55.03.06.db05-0832. [DOI] [PubMed] [Google Scholar]
- 80.Ye M., Qiu H., Cao Y., Zhang M., Mi Y., Yu J., Wang C. Curcumin improves palmitate-induced insulin resistance in human umbilical vein endothelial cells by maintaining proteostasis in endoplasmic reticulum. Front. Pharmacol. 2017;8:148. doi: 10.3389/fphar.2017.00148. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 81.Drews O., Taegtmeyer H. Targeting the ubiquitin-proteasome system in heart disease: the basis for new therapeutic strategies. Antioxid. Redox Signal. 2014;21(17):2322–2343. doi: 10.1089/ars.2013.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., Scrimieri F., Winter J.M., Hruban R.H., Iacobuzio-Donahue C., Kern S.E., Blair I.A., Tuveson D.A. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475(7354):106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang C., Tan Y.X., Yang G.Z., Zhang J., Pan Y.F., Liu C., Fu J., Chen Y., Ding Z.W., Dong L.W., Wang H.Y. Gankyrin has an antioxidative role through the feedback regulation of Nrf2 in hepatocellular carcinoma. J. Exp. Med. 2016;213(5):859–875. doi: 10.1084/jem.20151208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Riz I., Hawley T.S., Marsal J.W., Hawley R.G. Noncanonical SQSTM1/p62-Nrf2 pathway activation mediates proteasome inhibitor resistance in multiple myeloma cells via redox, metabolic and translational reprogramming. Oncotarget. 2016;7(41):66360–66385. doi: 10.18632/oncotarget.11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chacon-Cabrera A., Mateu-Jimenez M., Langohr K., Fermoselle C., Garcia-Arumi E., Andreu A.L., Yelamos J., Barreiro E. Role of PARP activity in lung cancer-induced cachexia: effects on muscle oxidative stress, proteolysis, anabolic markers, and phenotype. J. Cell. Physiol. 2017 doi: 10.1002/jcp.25851. [DOI] [PubMed] [Google Scholar]
- 86.Chondrogianni N., Kapeta S., Chinou I., Vassilatou K., Papassideri I., Gonos E.S. Anti-ageing and rejuvenating effects of quercetin. Exp. Gerontol. 2010;45(10):763–771. doi: 10.1016/j.exger.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 87.Katsiki M., Chondrogianni N., Chinou I., Rivett A.J., Gonos E.S. The olive constituent oleuropein exhibits proteasome stimulatory properties in vitro and confers life span extension of human embryonic fibroblasts. Rejuvenation Res. 2007;10(2):157–172. doi: 10.1089/rej.2006.0513. [DOI] [PubMed] [Google Scholar]
- 88.Tanigawa S., Fujii M., Hou D.X. Action of Nrf2 and Keap1 in ARE-mediated NQO1 expression by quercetin. Free Radic. Biol. Med. 2007;42(11):1690–1703. doi: 10.1016/j.freeradbiomed.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 89.Lovborg H., Oberg F., Rickardson L., Gullbo J., Nygren P., Larsson R. Inhibition of proteasome activity, nuclear factor-KappaB translocation and cell survival by the antialcoholism drug disulfiram. Int. J. Cancer. 2006;118(6):1577–1580. doi: 10.1002/ijc.21534. [DOI] [PubMed] [Google Scholar]
- 90.Cen D., Gonzalez R.I., Buckmeier J.A., Kahlon R.S., Tohidian N.B., Meyskens F.L., Jr. Disulfiram induces apoptosis in human melanoma cells: a redox-related process. Mol. Cancer Ther. 2002;1(3):197–204. [PubMed] [Google Scholar]
- 91.Bhat M., Ismail A. Vitamin D treatment protects against and reverses oxidative stress induced muscle proteolysis. J. Steroid Biochem. Mol. Biol. 2015;152:171–179. doi: 10.1016/j.jsbmb.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 92.de Theije C.C., Langen R.C., Lamers W.H., Gosker H.R., Schols A.M., Kohler S.E. Differential sensitivity of oxidative and glycolytic muscles to hypoxia-induced muscle atrophy. J. Appl. Physiol. 2015;118(2):200–211. doi: 10.1152/japplphysiol.00624.2014. [DOI] [PubMed] [Google Scholar]
- 93.Donohue T.M., Jr., Thomes P.G. Ethanol-induced oxidant stress modulates hepatic autophagy and proteasome activity. Redox Biol. 2014;3:29–39. doi: 10.1016/j.redox.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bulteau A.L., Moreau M., Nizard C., Friguet B. Impairment of proteasome function upon UVA- and UVB-irradiation of human keratinocytes. Free Radic. Biol. Med. 2002;32(11):1157–1170. doi: 10.1016/s0891-5849(02)00816-x. [DOI] [PubMed] [Google Scholar]
- 95.Zhang Y., Huang X., Zhao X.Y., Hu Y.J., Sun H.Y., Kong W.J. Role of the ubiquitin C-terminal hydrolase L1-modulated ubiquitin proteasome system in auditory cortex senescence. ORL J. Oto-Rhino-Laryngol. Relat. Spec. 2017;79(3):153–163. doi: 10.1159/000468944. [DOI] [PubMed] [Google Scholar]
- 96.Sampuda K.M., Riley M., Boyd L. Stress induced nuclear granules form in response to accumulation of misfolded proteins in Caenorhabditis elegans. BMC Cell Biol. 2017;18(1):18. doi: 10.1186/s12860-017-0136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bahmani R., Kim D., Lee B.D., Hwang S. Over-expression of tobacco UBC1 encoding a ubiquitin-conjugating enzyme increases cadmium tolerance by activating the 20S/26S proteasome and by decreasing Cd accumulation and oxidative stress in tobacco (Nicotiana tabacum) Plant Mol. Biol. 2017 doi: 10.1007/s11103-017-0616-6. [DOI] [PubMed] [Google Scholar]