ABSTRACT
Bradyrhizobium diazoefficiens, a soybean N2-fixing symbiont, possesses a dual flagellar system comprising a constitutive subpolar flagellum and inducible lateral flagella. Here, we analyzed the genomic organization and biosynthetic regulation of the lateral-flagellar genes. We found that these genes are located in a single genomic cluster, organized in two monocistronic transcriptional units and three operons, one possibly containing an internal transcription start site. Among the monocistronic units is blr6846, homologous to the class IB master regulators of flagellum synthesis in Brucella melitensis and Ensifer meliloti and required for the expression of all the lateral-flagellar genes except lafA2, whose locus encodes a single lateral flagellin. We therefore named blr6846 lafR (lateral-flagellar regulator). Despite its similarity to two-component response regulators and its possession of a phosphorylatable Asp residue, lafR behaved as an orphan response regulator by not requiring phosphorylation at this site. Among the genes induced by lafR is flbTL, a class III regulator. We observed different requirements for FlbTL in the synthesis of each flagellin subunit. Although the accumulation of lafA1, but not lafA2, transcripts required FlbTL, the production of both flagellin polypeptides required FlbTL. Moreover, the regulation cascade of this lateral-flagellar regulon appeared to be not as strictly ordered as those found in other bacterial species.
IMPORTANCE Bacterial motility seems essential for the free-living style in the environment, and therefore these microorganisms allocate a great deal of their energetic resources to the biosynthesis and functioning of flagella. Despite energetic costs, some bacterial species possess dual flagellar systems, one of which is a primary system normally polar or subpolar, and the other is a secondary, lateral system that is produced only under special circumstances. Bradyrhizobium diazoefficiens, an N2-fixing symbiont of soybean plants, possesses dual flagellar systems, including the lateral system that contributes to swimming in wet soil and competition for nodulation and is expressed under high energy availability, as well as under requirement for high torque by the flagella. The structural organization and transcriptional regulation of the 41 genes that comprise this secondary flagellar system seem adapted to adjust bacterial energy expenditures for motility to the soil's environmental dynamics.
KEYWORDS: Bradyrhizobium, flagella, expression, lafR, flbT, FlbT, LafR
INTRODUCTION
Flagellum-driven swimming motility—a characteristic trait of many bacterial species—is essential for the colonization of diverse niches in environments such as seas, freshwaters, sediments, soils, and the organs of plant or animal hosts. This form of bacterial locomotion requires the propulsion provided by flagella, as well as a guidance system mediated by chemotaxis (1).
Flagella are complex organelles formed by three main structures: a basal body that anchors the flagellum to the cell envelope, a filament that projects out from the cell often with a length greater than the cell body itself, and a hook that connects the basal body to the filament (see Fig. S1 in the supplemental material). The basal body, the most complex of these substructures, is responsible for two critical tasks: exporting the hook and filament proteins synthesized in the cytoplasm toward the extracellular space (2, 3) and providing the rotational motion of the flagella (4–6). For the first task, an export apparatus is embedded in the inner cell membrane ending in a rod that crosses the cell envelope and delivers the hook and filament polypeptides. Because the rod's internal channel is narrow, the polypeptides must pass through in a partially unfolded state, whose conformation is stabilized by specific chaperones through proton motive force and ATP hydrolysis as energy sources. For the second task, the basal body contains the flagellar motor, composed of a stator in the inner cell membrane and a rotor formed by a ring of several protein subunits that rotates inside the stator by the proton or sodium ion motive force. In turn, the hook and the filament are formed by the polymerization of thousands of monomers of structural proteins and are held together by specific hook-filament junctions (7, 8). The hook is a flexible connector that transmits motor rotation in the form of waves to the flagellar filament, whose extension in turn rotates while undulating like an Archimedean screw to drag or thrust the cell in an aqueous medium, depending on whether the flagellum is ahead or behind the cell, respectively (9). In Gram-negative bacteria, the whole structure passes through the inner membrane, the peptidoglycan layer, and the outer membrane, with each layer containing rings that behave like bushings. The >40 genes that encode the basal body, the hook, the filament, the rings, and the auxiliary and regulatory proteins lie in a limited number of operons that, together with the chemotaxis genes, comprise the flagellar regulon.
The synthesis and assemblage of the bacterial flagellum require a substantial organization in order to ensure that all the major structures are completed sequentially from the membrane-associated elements to the extracellular components (10, 11). Thus, depending on the species, the regulation of flagellum biosynthesis comprises three or four principal steps, whose stages have been carefully studied in model systems such as Caulobacter crescentus, Ensifer meliloti, Escherichia coli, Pseudomonas spp., Salmonella enterica, and Vibrio spp., among others (12–14). In general, a master regulator (class I) induces the expression of several flagellar operons (class II) that encode the basal body, the hook and hook-related proteins, and a regulator of the synthesis of the filament monomers or flagellins. Therefore, these flagellins (class III) are synthesized in the final step, with the basal body and the hook already in position. This strategy ensures that no energy is wasted on flagellin synthesis and export before these proteins are required (10, 13).
The rotation of the flagellar motor defines whether a bacterium swims in a given direction or erratically. In general, when the flagella rotate in a single direction, the bacteria swim in linear runs, whose stretches are interrupted when flagellar rotation switches direction or stops (15–18). The frequency at which these changes in direction occur are governed by the chemotaxis system in response to chemical stimuli (i.e., attractant or repellent gradients) present in the medium (18). As with the flagellar genes, the chemotaxis loci are often arranged in operons under control by the same regulators that govern the synthesis of flagellins and therefore are also grouped in class III of the flagellar regulon (13).
Bradyrhizobium diazoefficiens, the soybean N2-fixing symbiont, is a soil alphaproteobacterium that possesses two different flagellar systems with independent evolutionary origins (19, 20). One of these systems involves a subpolar flagellum closely related to the one in C. crescentus, while the other is characterized by lateral flagella similar to those in E. meliloti (20). The expression of each flagellar system is also different. The subpolar system seems constitutive within a range of conditions; by contrast, the expression of the lateral system requires arabinose as a carbon source or viscosity in the culture medium or the presence of obstacles in the swimming path (20–22). Hence, growth in liquid medium with mannitol as the carbon source does not permit the expression of the lateral flagella. Transcriptomic studies have also indicated that conditions of microoxia (23) or iron deficiency (24) prevent the expression of lateral-flagellar genes, whereas permanent exposure to moderate oxidative stress induces these loci (25). In the example of Ensifer meliloti and its close relative Brucella melitensis, flagellar expression is also under strict control by specific nutritional, physiological, and population-size requirements (26–29).
In contrast to other species having dual flagellar systems that use one exclusively for swimming in a liquid medium and the other only for swarming on surfaces, both flagellar systems of B. diazoefficiens may be utilized together in liquid medium and interact to produce an emergent swimming performance that allows the bacterium to continually swim alongside solid surfaces (20). Only the subpolar system, however, responds to chemotactic stimuli, whereas only the lateral system contributes to swimming in viscous-agar-containing medium (20). Although neither of these flagellar systems is required for the nodulation of soybean plants (30), bacterial motility might be essential for nodule occupation in competition against populations of compatible rhizobia in the soil (31). In earlier work, we obtained a derivative of B. diazoefficiens USDA 110 with higher motility by in vitro selection (21). The inoculation of this derivative on experimental soybean crops planted in soils with dense soybean-nodulating competitor populations resulted in an enhanced nodule occupation by the derivative and promoted a higher soybean grain yield (21, 31). Further studies indicated that the lateral-flagellar system of this derivative became derepressed upon culture in liquid medium with mannitol as the carbon and energy source (21, 22). Therefore, the control of lateral-flagellar synthesis in this species should take into account the cell's needs on the basis of the environmental conditions in order to coordinate the activity of both flagellar systems, which would appear to be essential for the symbiotic interaction with soybean plants. To better understand the regulation of these genetically controlled functions, in the study reported here, we investigated the organization and transcriptional control of the lateral-flagellar genes of B. diazoefficiens USDA 110.
RESULTS
Identification and characterization of lafR, the master regulator of lateral-flagellar synthesis in B. diazoefficiens USDA 110.
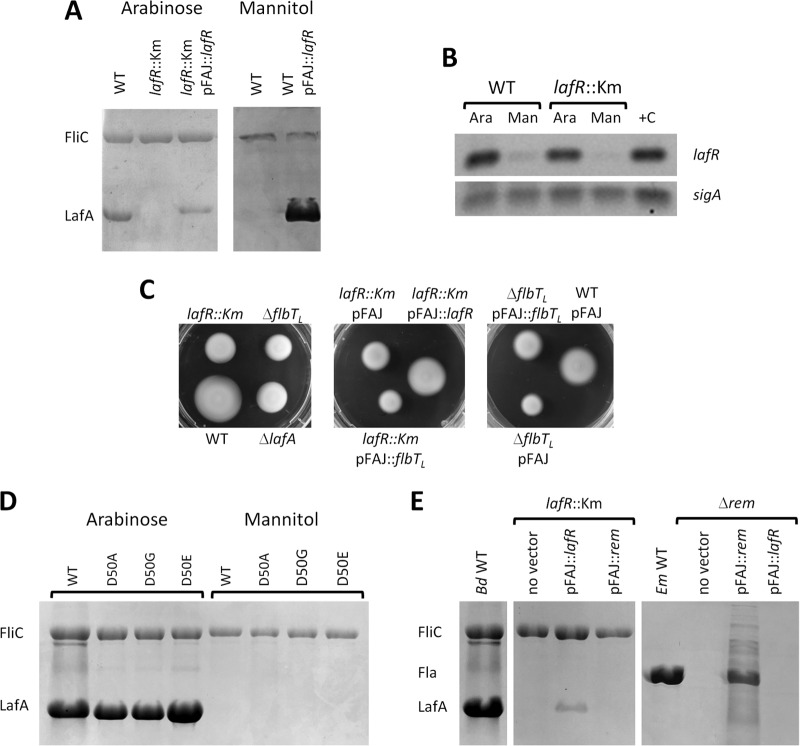
In E. meliloti, flagellar expression is controlled by a regulatory circuit composed of the LuxR-type master regulators VisNR and the OmpR-like transcriptional activator Rem (28). In a similar fashion, the LuxR-type VjbR and the OmpR-like FtcR are master regulators of flagellar expression in B. melitensis (32). Although the regulation circuit of neither VisNR nor VjbR is restricted to flagellar gene expression, the Rem and FtcR regulators seem to be more specific (28, 32). In addition, since the expression of rem and ftcR is regulated by VisNR and VjbR, respectively, the LuxR-type components were classified as class IA, whereas the OmpR-like components were considered class IB (28, 32). Within the complete genomic sequence of B. diazoefficiens USDA 110 (33), we could not find homologs to visNR or vjbR, but the locus tag blr6846 (Ga0076376_112362 in the reannotation at the IMG database)—located near the cluster of genes that encode the lateral flagella in B. diazoefficiens—encodes a predicted OmpR-like transcriptional-response regulator of 256 residues homologous to Rem and FtcR and harboring the typical receiver and helix-turn-helix DNA-binding domains (see Fig. S2A in the supplemental material). The high similarity among these three OmpR-like transcriptional-response regulators, as well as the position of blr6846 with respect to the lateral flagellar gene cluster, led us to suspect that blr6846 might be a master regulator of the synthesis of the lateral flagella in B. diazoefficiens. This suspicion was reinforced by the observation that the expression of blr6846 is dependent on the carbon source in a manner similar to that of the production of the lateral flagellins LafA1 and LafA2, since blr6846 is expressed in arabinose-grown cultures at substantially higher levels than in mannitol-containing cultures (Fig. 1A and B). In addition, a mutant harboring a kanamycin (Km)-resistant cassette inserted at base 7543291 in the middle of the coding sequence of blr6846, thus disrupting the connection between the receiver and helix-turn-helix DNA-binding domains (see Fig. S2B in the supplemental material), was found to lack lateral flagellins even when grown with arabinose as the carbon source (Fig. 1A). The production of the lateral flagellins was restored after introducing a wild-type (WT) copy of blr6846 in the pFAJ1708 replicative plasmid, indicating that the defective phenotype resulted from the disruption of the coding sequence of blr6846. In addition, the lateral flagellin production was restored in the wild-type strain grown with mannitol when blr6846 was expressed constitutively from the replicative plasmid (Fig. 1A), demonstrating that the expression of blr6846 was sufficient to produce the lateral flagellins under this condition. However, the polypeptide levels of LafA relative to those of FliC were variable in the complemented strains among the different experiments. Since pFAJ1708 is stable in B. diazoefficiens, we have no explanation for the observed instability in LafA recovery, which nevertheless does not rule out the conclusions that the lafR::Km mutation may be complemented in trans and that the presence of lafR in trans is sufficient for lateral flagellin synthesis in mannitol. The blr6846 mutant achieved a smaller swimming halo than the wild-type in soft agar, whose area was similar to that produced by the lateral flagellin-deficient ΔlafA mutant (Fig. 1C). This defect in swimming, observed in the mutants within the WT USDA 110 background, was also displayed by a blr6846 mutant in the LP 3004 background (the USDA 110 streptomycin [Sm]-resistant derivative) and by another blr6846 mutant in the LP 3008 background (the LP 3004 derivative with higher motility [data not shown]). In addition, motility of the lafR::Km mutant was restored by trans complementation with pFAJ::lafR (Fig. 1C), indicating that lafR expression was sufficient to produce functional lateral flagella. Taken together, these results corroborated that blr6846 might have roles similar to those of rem and ftcR, so that we refer to blr6846 here as lafR (lateral flagellum regulator) and have accordingly renamed the Km insertion mutation lafR::Km.
FIG 1.
Control of flagellin expression and motility by lafR in bacteria grown in liquid HMY with arabinose (Ara) or mannitol (Man) as a carbon source. (A) SDS-PAGE of extracellular B. diazoefficiens proteins of the subpolar flagellins (FliC, upper bands) and lateral flagellins (LafA, lower bands) in the wild-type (WT) and the lafR::Km extracts alone or in extracts from the WT and the lafR::Km strains complemented with a WT copy of lafR under the direction of the nptII promoter (pFAJ::lafR). (B) Agarose gel of RNA retrotranscripts amplified by RT-PCR of lafR in the WT or the lafR::Km mutant with the primers indicated in Fig. S2B and listed in Table S2 in the supplemental material compared to sigA as constitutive reference gene. A PCR from genomic DNA was performed as a positive control (+C). (C) Swimming motility in 0.3% (wt/vol) agar-containing AG medium. Left panel: wild type compared to lafR::Km, ΔflbTL, and ΔlafA mutants, the last one lacking lateral flagellins. Center panel: complementation of motility in the lafR::Km mutant with the pFAJ::lafR plasmid in comparison to the lafR::Km mutant carrying empty vector (pFAJ) or lafR::Km mutant carrying pFAJ::flbTL. Right panel: complementation of motility in the ΔflbTL mutant with the pFAJ::flbTL plasmid in comparison with ΔflbTL mutant carrying the empty vector (pFAJ). The results of all the complementations may be compared to the motility of the WT carrying the empty vector (WT pFAJ, right). (D) SDS-PAGE of B. diazoefficiens extracellular proteins—the subpolar flagellins (FliC, upper bands) and lateral flagellins (LafA, lower bands)—in the wild type and lafR point mutants D50A (with the Asp50 residue replaced by Ala), D50G (with Asp50 replaced by Gly), and D50E (with Asp50 replaced by Glu). (E) Composite SDS-PAGE of the subpolar (FliC, upper bands) and lateral (LafA, lower bands) flagellins of B. diazoefficiens or the FlaA-D flagellins of E. meliloti (Fla, middle bands). The flagellins are from B. diazoefficiens (Bd) wild-type and lafR::Km mutant either alone or complemented with pFAJ::lafR or with a wild-type copy of rem under the nptII promoter (pFAJ::rem), wild-type E. meliloti (Em WT), and the E. meliloti rem mutant (Δrem) either alone or complemented with pFAJ::rem or pFAJ::lafR. All the bacteria were grown on HMY with arabinose as the carbon source. The gels were run simultaneously in the same equipment.
LafR as an orphan response regulator.
A critical difference observed in the amino acid sequence of LafR with respect to its counterparts Rem and FtcR is the presence of an Asp residue at position 50 that is susceptible to phosphorylation as in the typical transcriptional response regulator OmpR (see Fig. S2A in the supplemental material). In contrast, Rem and FtcR possess a Glu at this position, the latter residue being larger than Asp by an additional methyl group (28, 32). Therefore, Rem and FtcR may function as though they were continually activated (32), whereas LafR might require phosphorylation for activation; this property would be consistent with the inducible nature of lateral flagella in B. diazoefficiens. We could not find, however, any putative histidine kinase associated with lafR. In order to elucidate this question, we constructed the single-substitution lafR(D50A), lafR(D50G), and lafR(D50E) mutants (see Table S1 in the supplemental material) in which the Asp50 residue was replaced by an Ala (without the carboxyl group of Asp), a Gly (without any residue), or a Glu (as in Rem and FtcR; see Fig. S2A in the supplemental material), respectively. Therefore, if phosphorylation of the Asp50 was required, neither lafR(D50A) nor lafR(D50G) would be activated, whereas lafR(D50E) would likely be constitutively active. Nevertheless, we observed the same profile of activation in all three mutants and in the wild type, i.e., the lateral flagellins were produced with arabinose but not with mannitol as the carbon source (Fig. 1D). Together, these results indicated that LafR behaves as an orphan response regulator whose activation seems not to require phosphorylation of Asp50 by a histidine kinase sensor.
The function of lafR cannot be replaced by rem.
To further characterize the possible similarity between lafR and rem, we looked for possible cross-complementation of these genes in B. diazoefficiens and E. meliloti mutants. Thus, we introduced a wild-type copy of rem carried by pFAJ1708 into the B. diazoefficiens lafR mutant and, reciprocally, introduced a wild-type copy of lafR into the E. meliloti rem mutant Rm2011mTn5STM.1.08.H02 (i.e., referred to as the Δrem mutant here [see Table S1 in the supplemental material]). The flagellins, however, were not observed in the cross-complemented strains (Fig. 1E). Since the controls complemented with the transcriptional regulator of the same species did produce flagellins, we concluded that either LafR or Rem are not stably expressed in E. meliloti or B. diazoefficiens, respectively, or the failure of cross-complementation in the experimental strains may be due to a lack of recognition in the interaction between the heterologous proteins and DNAs.
Operon organization in the lateral-flagellar gene cluster.
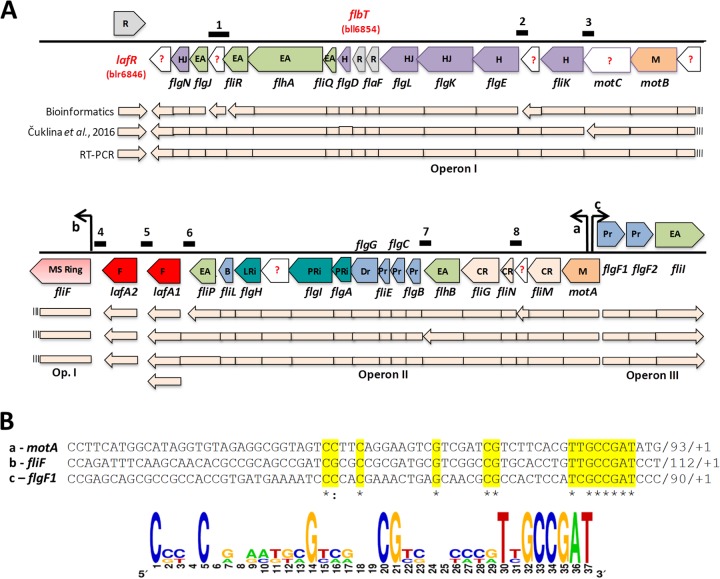
The 41 lateral-flagellar genes of B. diazoefficiens are grouped in a single cluster, although evidence for the origin of that acquisition through horizontal gene transfer could not be found (20). In addition, the lack of chemotaxis genes in the vicinity of this cluster—in agreement with the lack of chemotactic response exerted by the lateral flagellar system (20)—suggests that this cluster might constitute the complete lateral-flagellar regulon. Through the use of different bioinformatic tools (e.g., MicrobesOnline Database, ProOpDb, and DOOR2), we predicted the operon structure of the gene cluster that is schematized in Fig. 2A and found a new open reading frame between fliRL (bll6849) and flgJL (bll6850), whose sequence we named bll6849.5. According to this analysis, the lateral-flagellar gene cluster might be divided into at least five putative operons and three monocistronic transcriptional units: lafR, lafA1, and lafA2. All transcriptional units are conserved in E. meliloti and B. melitensis, but the synteny contains some differences (see Fig. S3 in the supplemental material). In a recent study, a genome-wide transcription start site map for B. diazoefficiens USDA 110 grown in peptone/salts/yeast extract/arabinose medium was established (34), indicating different operon structures for the lateral-flagellar region from those predicted by the bioinformatic analysis (Fig. 2A). To resolve this contradiction, we designed primers to amplify, by reverse transcription-PCR (RT-PCR), eight intergenic regions that should differ in the resulting transcripts according to whether the distribution of polycistronic mRNAs from this genomic cluster under our conditions is as predicted by bioinformatics or as reported in the experimental transcription start site mapping (34). The regions chosen were as follows: region 1 from 3′ flgJL (bll6849) to 5′ fliRL (bll6850; further encompassing the intergenic regions upstream and downstream from bll6849.5), region 2 from 3′ flgEL (bll6858) to 5′ bll6859, region 3 from 3′ fliKL (bll6860) to 5′ motC (bll6861), region 4 from 3′ fliFL (bll6864) to 5′ lafA2 (bll6865), region 5 from 3′ lafA2 (bll6865) to 5′ lafA1(bll6866), region 6 from 3′ lafA1 (bll6866) to 5′ fliPL (bll6867), region 7 from 3′ flgBL (bll6876) to 5′ flhBL (bll6877), and region 8 from 3′ fliNL (bll6879) to 5′ bll6880 (Fig. 2A). By this approach, we would be able to detect amplification only where the mRNA is polycistronic for the adjacent genes probed (35). We observed that no amplification occurred only between 3′ fliFL and 5′ lafA2, and between 3′ lafA2 and 5′ lafA1 (see Fig. S4 in the supplemental material), suggesting the existence of two operons encompassing bll6847-fliFL (operon I) and lafA1-motA (operon II), with lafA2 remaining as a monocistronic transcriptional unit situated between those two operons. In addition, lafR and flgF1L-fliIL (operon III) can be considered different transcriptional units since they are contained in the opposite strand (Fig. 2A).
FIG 2.
Operons of the lateral-flagellar gene cluster, indicating the transcription directions according to Rhizobase (http://genome.annotation.jp/rhizobase/Bradyrhizobium). (A) The genes identified in the cluster are classified by function as regulators (R, gray), unknown (?, white), hook and hook-filament junction (H and HJ, violet), export apparatus (EA, green), motor (M, orange), MS ring (pink), flagellins (F, red), basal body (B, blue), L-ring and P-ring (LRi and PRi, turquoise), distal and proximal rods (Dr and Pr, light blue), and C-ring (CR, light pink). Below this scheme, the operon structure is indicated according to: bioinformatics prediction (upper light-pink line), Čuklina et al. (34) (middle light-pink line), and our own results from RT-PCR (bottom light-pink line). Above the scheme, the positions of the deduced lafR-dependent promoters are shown as black arrows, and the positions of the intergenic amplicons predicted according to the RT-PCR strategy outlined in Fig. S4 in the supplemental material are shown as black segments numbered from 1 to 8. For the sake of simplicity, the L subscripts in the figure have been omitted from the name of each locus. (B) Sequence alignment of the conserved motifs found upstream from the transcription start sites (designated as +1) of the genes motA (row a), fliFL (row b), and flgFL1 (row c), the latter being located at the 5′ ends of operons I, II, and III, respectively (see panel A). The consensus sequence that may be deduced is indicated at the bottom of the panel.
LafR activation of three of the four transcriptional units.
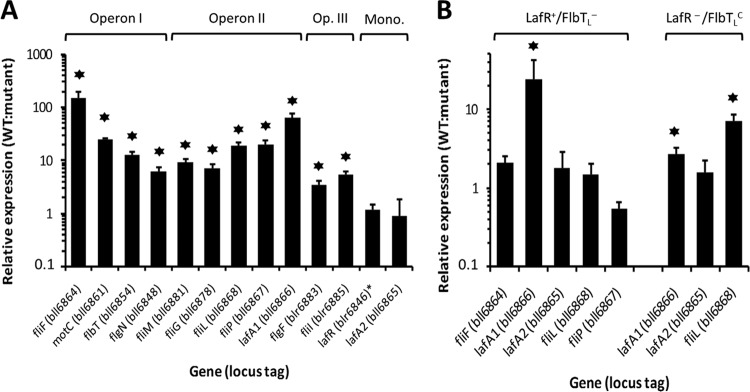
We analyzed the requirements for LafR with respect to the transcriptional profile of the lateral flagellar regulon by quantitative reverse transcription-PCR (qRT-PCR) with whole RNA from the wild type and the lafR::Km mutant. We chose as representative genes of each transcriptional unit—fliFL, motC, flbTL, and flgNL for operon I; fliML, fliGL, fliLL, fliPL, and lafA1 for operon II; and flgF1L and fliIL for operon III—along with the monocistronic transcriptional units lafR and lafA2 (Fig. 2A) for amplification. Figure 3A summarizes the results of the qRT-PCR assays. According to the relative expression levels obtained with the lafR::Km mutant in comparison to the wild type, we observed that lafR was not autoregulated but that, with respect to operons I to III, lafR was a positive regulator, a finding in agreement with the previous observation that the lafR::Km mutant was unable to produce lateral flagellins (Fig. 1). In contrast, mutation of lafR did not produce substantial changes in the abundance of the lafA2 transcript.
FIG 3.
Effects of mutations in lafR and flbTL on the mRNA accumulation of selected lateral-flagellar genes. (A) Transcription expression level in the wild-type strain relative to that of the lafR::Km mutant plus the standard deviations (SD), as determined by qRT-PCR from at least three independent biological replicas for the indicated genes (locus tags), the latter being representative of the different transcriptional units. Mono., monocistronic transcripts. The relative expression of lafR was evaluated with the primers indicated in Fig. S2B in the supplemental material, which amplify the 5′ end of lafR both in the wild-type and in the lafR::Km mutant. Stars, statistically significant differences (P < 0.05) from a threshold interval of 0.5 to 2.0 according to the Student t test. (B) Transcription expression level in the wild-type strain relative to that of the ΔflbTL strain (LafR+/FlbTL−, left) or that of lafR::Km carrying the plasmid pFAJ::flbTL (LafR−/FlbTLC, right) ± the SD, as determined by qRT-PCR from at least three independent biological replicas for the indicated genes, the latter having been selected to indicate the differential influence of flbTL on lafA1 expression. Stars, statistically significant differences (P < 0.05) from a threshold interval of 0.5 to 2.0 according to the Student t test. For the sake of simplicity, the L subscripts in the figure have been omitted from the name of each locus.
To search for possible common motifs in the 5′ untranslated regions (5′ UTRs) of the three operons, we performed sequence comparisons with the MEME Suite (36). A de novo search for these sequences located a possible common motif shared by the LafR-induced operons (Fig. 2B). The motif is located approximately at the same distance from the transcription start site base pair (+1) of each operon. Of relevance here is that a search with the MAST algorithm at the MEME server did not locate this motif either upstream from the E. meliloti rem sequence—a finding in agreement with the lack of cross-complementation between lafR and rem—or upstream from any other B. diazoefficiens gene, suggesting that this motif might be shared by the lafR-dependent promoters.
The lack of changes in lafA2 transcript accumulation between the wild type and the lafR::Km mutant indicates that another regulation is likely to be responsible for the inhibition of LafA2 polypeptide production in this mutant. We suspected that this role might be fulfilled by FlbT, a protein known as a translational regulator of flagellin synthesis in other bacteria (37, 38).
FlbTL as a positive regulator of flagellin synthesis.
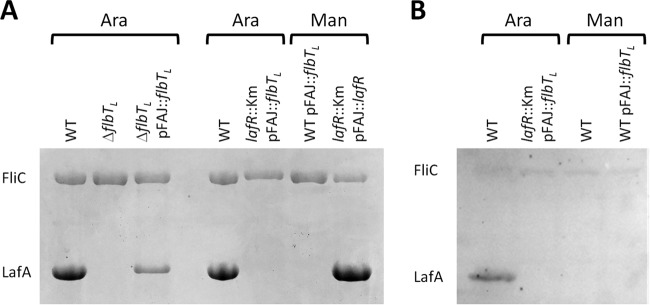
To investigate whether or not FlbTL regulates flagellin synthesis in B. diazoefficiens as in other bacteria, we constructed a flbTL deletion mutant by eliminating 195 bp from the middle of the coding region (between bases 7549514 and 7549709) without any alteration in the reading frame (see Fig. S5 in the supplemental material). This mutant (ΔflbTL) would thus be expected to produce an internally deleted gene product without any polar effects on genes downstream in the operon. We observed that the ΔflbTL (LafR+/FlbTL−) mutant was unable to produce LafA1 and LafA2 with arabinose as the carbon source and that this phenotype was reversed when the ΔflbTL mutant was complemented in trans with a wild-type copy of flbTL carried in the replicative plasmid pFAJ::flbTL (Fig. 4A). Likewise, the ΔflbTL mutant swimming motility in soft agar was compromised, similar to the analogous defect in ΔlafA and lafR::Km mutants, and this phenotype was partially complemented in trans (Fig. 1C). These results indicated that, as in B. melitensis (38), FlbTL is required for lateral flagellin synthesis.
FIG 4.
Control of flagellin expression by flbTL in bacteria grown in liquid HMY medium with arabinose (Ara) or mannitol (Man) as carbon source. (A) SDS-PAGE of the B. diazoefficiens extracellular subpolar (FliC) and lateral (LafA) flagellins in the wild-type, the ΔflbTL strain, the ΔflbTL strain complemented with the plasmid pFAJ:: flbTL that carries a wild-type copy of flbTL under the direction of the nptII promoter, or the lafR::Km strain complemented with the plasmid pFAJ::flbTL or pFAJ::lafR. (B) Western blots of the cellular B. diazoefficiens proteins FliC and LafA, as visualized by an anti-lafA polyclonal antiserum, from the wild-type strain either alone or complemented with the plasmid pFAJ::flbTL or from the lafR::Km strain complemented with the plasmid pFAJ::flbTL. The polyclonal anti-lafA serum exhibited some cross-reaction against FliC, whose activity in this experiment served as an internal standard.
Next, we extended the analysis of the transcriptional profile of the lateral-flagellar operons by incorporating an LafR−/FlbTLC strain (lafR::Km mutant complemented with flbTL in trans under the control of a constitutive promoter). Using the same approach as before (Fig. 3A), we compared the relative expression of motC and fliFL (operon I), fliML, fliLL, and lafA1(operon II), and lafA2 in the wild type to both LafR+/FlbTL− and the LafR−/FlbTLC genetic backgrounds. We observed that the deletion of flbTL had no effect on the expression of any of the genes tested, except for lafA1: in this gene, however, transcript accumulation was inhibited when flbTL bore a mutation similar to that of lafR (Fig. 3B). Because flbTL was itself induced by lafR, the observed effect of the lafR mutation on lafA1 expression could be indirect as a result of a downregulation of flbTL within the lafR::Km genetic background. Therefore, we evaluated the expression of lafA1 in the LafR−/FlbTLC genetic background and observed that the transcript accumulation of lafA1 was partially restored, although the relative expression of lafA1 in the wild type with respect to that of the mutant was still significant, indicating that flbTL had a partial influence on the control of lafA1 transcript accumulation. Conversely, the adjacent gene fliLL within the same operon responded only to lafR since that locus was downregulated within the LafR−/FlbTLC genetic background (similar to the result with the lafR::Km mutant) but was not affected in the LafR+/FlbTL− background (Fig. 3B).
Although a transcription start site of lafA1 could not be detected by RNA sequencing analysis (34), the pattern of differential regulation on the part of lafA1 with respect to the rest of operon II prompted us to investigate whether an internal promoter activity might be found within operon II upstream from lafA1. To this end, we cloned the DNA segments between the 3′ end of fliPL and the ATG of lafA1 (PlafA1) and between the 3′ end of lafA1 and the ATG of lafA2 (PlafA2) in the replicative plasmid pCB303 upstream from the promoterless lacZ site and measured the resultant β-galactosidase activity of the fusions. We observed more than twice the activity in the pCB::PlafA1 plasmid carrying the PlafA1::lacZ fusion than in the pCB::PlafA2 plasmid carrying the PlafA2::lacZ fusion, without differences among the wild-type, lafR::Km, and ΔflbTL genetic backgrounds (Fig. 5), thus indicating the existence of an active promoter upstream from lafA1, whose locus—as with lafA2—would not be under transcriptional control of LafR or FlbTL. To further investigate the difference in the responses of lafA1 and lafA2 to FlbTL, we compared the nucleotide sequences and RNA predicted structures of the 5′ UTRs of these transcripts. To this end, we used the sequence within the lafA2 5′ UTR beginning at the experimentally identified transcription start site located 118 nucleotides upstream from the ATG initiation codon (34) and extending through the first 17 codons of the coding sequence (39) in comparison to a putative 5′ UTR of lafA1 starting also 118 nucleotides upstream from the ATG and continuing through the first 17 codons of the coding sequence. Despite the high conservation among these sequences, we observed a gap near a sequence complementary to the ribosome binding site (RBS) in lafA2 (see Fig. S6A in the supplemental material). In four of seven predicted stable secondary structures of the lafA1 5′ UTR region, a small loop arose at the RBS complementary sequence, whose conformation might loosen RBS occlusion but would leave the ATG initiation codon in a double-helix stretch (see Fig. S6B in the supplemental material). In contrast, the other three structures, as well as the two predicted stable secondary structures of the lafA2 5′ UTR, had the RBS site in a double-helix stretch, but the ATG start codon was predicted in a single-strand region (see Fig. S6B and C in the supplemental material). Taken together, these results are consistent with the postulated presence of a constitutive internal active promoter in operon II for lafA1 transcription. The most plausible assumption to explain the above results may be that lafA1 mRNA stability might be controlled by FlbTL differently from that of lafA2 (Fig. 2A).
FIG 5.
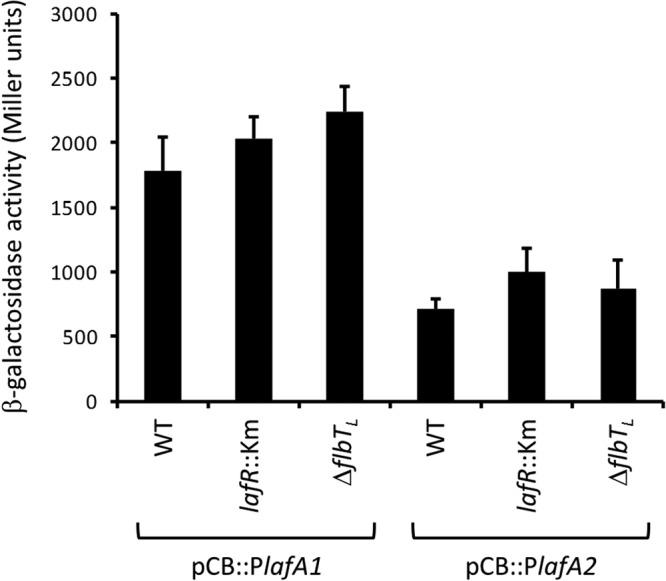
β-Galactosidase activities of pCB::PlafA1 and pCB::PlafA2 lacZ fusions within three genetic backgrounds. In the figure, the β-galactosidase activity in Miller units is plotted on the ordinate for each of the genetic backgrounds denoted on the abscissa. The two clonal fusions are indicated in brackets below the figure. Each mean value is from two independent clones measured in duplicate. Error bars indicate the SD.
Despite lafA1 and lafA2 transcript production, lateral flagellins were observed in neither B. diazoefficiens USDA 110 carrying pFAJ::flbTL cultured with mannitol nor the lafR::Km mutant carrying pFAJ::flbTL cultured with arabinose (Fig. 4A). We reasoned that these observations might be due to the lack of a filament export apparatus under conditions where LafR is not produced, thus leading to an accumulation of LafA1 and LafA2 inside the cell. To test for this possibility, we obtained total cellular proteins and performed a Western blot with an LafA-specific polyclonal antibody (22). As expected, the wild-type B. diazoefficiens cultured with mannitol did not accumulate LafA intracellularly, whereas the same strain cultured with arabinose contained a clearly visible band of binding by the anti-LafA antibody in the Western blot at the expected molecular mass. In contrast, complementation with flbTL in trans failed to restore LafA accumulation in either the wild-type cells grown with mannitol or the lafR::Km mutant grown with arabinose, where lafR was not expressed (Fig. 4B).
DISCUSSION
The lateral-flagellar genes of B. diazoefficiens lie in a single cluster encompassing 34,823 bp, organized in two monocistronic transcriptional units and three operons, with one of those three possibly having an independent internal promoter. In addition, no chemotaxis genes are present in this cluster. The expression of the three operons required the protein product of lafR; this gene constitutes one of the monocistronic transcriptional units identified. The regulation of LafR, together with the protein's sequence homology to known class IB master regulators, suggests that LafR is the class IB master regulator of lateral flagellum synthesis in B. diazoefficiens. In alphaproteobacteria there are different types of master regulators, including two-component systems such as the ctrA-cckA of Rhodobacter capsulatus (40) or OmpR-like transcriptional activators such as the rem of E. meliloti (28) or the ftcR of B. melitensis (32). These latter activators, in turn, are controlled by the respective LuxR-type systems, visNR and vjbR (32, 41, 42), which respond to cell cycle cues or environmental stimuli, indicating the need to activate or inactivate flagellar synthesis. In the particular example of B. diazoefficiens, the respiration rate might be just such a signal linked to the transcription of the lateral-flagellar regulon. Previous reports indicated that situations diminishing the respiration rate, such as microaerobiosis, the bacteroid state (23), or iron deficiency (24), downregulate the lateral-flagellar regulon transcription, whereas situations known to increase the oxygen consumption, such as permanent exposure to moderate oxidative stress (25) or the use of arabinose as the sole carbon source (43; C. Cogo, J. Pérez-Giménez, C. B. Rajeswari, M. F. Luna, and A. R. Lodeiro, unpublished data), promote that transcription. Moreover, these changes in transcription were not observed in the subpolar-flagellar regulon, indicating that these stimuli act specifically on the lateral flagella. Although we could not find visNR or vjbR homologs in B. diazoefficiens, the RegSR two-component system—which regulates the responses to microoxia—was reported to modify the expression specifically of lafR and the lateral-flagellar regulon after a switch from oxic to microoxic (O2 concentration < 0.5%) conditions (44). In agreement with these results, a TetR family transcriptional regulator was also found to repress the lateral-flagellar genes in a coordinated manner, along with genes encoding high-affinity cytochromes and oxidative-stress detoxification products, without affecting the subpolar-flagellar genes (45). Therefore, several stimuli related to the energy status of the cell are able to trigger lateral-flagellar expression in B. diazoefficiens without affecting subpolar-flagellar expression. Such stimuli seem transmitted to the class IB regulator lafR by different class IA regulators from those of E. meliloti or B. melitensis. In turn, lafR itself could be part of a two-component system, but two observations argue against this possibility. First, a putative histidine kinase could not be found for this system; second, the phosphorylatable Asp50 residue of LafR may be replaced by Ala, Gly, or Glu without alterations in the response of flagellin synthesis to the carbon source present in the culture medium, indicating that Asp50 is not phosphorylated in LafR.
The correlation between operon organization and flagellar substructures shown in Fig. S1 in the supplemental material indicates that all the genes induced by LafR are among the class II genes transcribed in the second step of the cascade, although the strict temporal order observed in other species (11) is not reflected by the distribution of the flagellar genes among the three operons. Most of the flagellar motor (4) is encoded in operon II, except the MS ring component FliFL and the stator protein MotB, both of whose loci are encoded in operon I. Moreover, the genes encoding the export apparatus (2, 3), whose component is also part of the basal body, are scattered among the three operons. The export of the hook and filament proteins from the cytoplasm to the extracellular space through the narrow space inside the rod may require (i) their recruitment at the export gate that is formed by FlhAL, FliQL, and FliRL (operon I) and FlhBL and FliPL (operon II), (ii) ATP hydrolysis catalyzed by FliIL (operon III), (iii) the chaperon activities of FlgNL (operon I) and FlgAL (operon II), and (iv) the control switch in the export sequence between hook and filament effected by FliKL (operon I). The structure of the rod apparatus and the rings that act as bushings in the membranes and the peptidoglycan layer are mostly encoded in operon II, except for flgFL, whose locus is in operon III. The gene flgJL that encodes the β-N-acetylglucosaminidase required for the hydrolysis of the peptidoglycan layer in order to allow passage of the P-ring and the rod (46), however, is in operon I (see Fig. S1 in the supplemental material). Therefore, a functional export apparatus in this flagellar system requires the expression of genes from the three operons. Some genes are also missing—such as fliD encoding the filament cap and the fliI-associated fliH and fliJ—whose loci might be encoded in the hypothetical open reading frames that we could not yet identify. In addition, a complementation by proteins from the subpolar-flagellar system cannot be discarded, although this possibility seems unlikely in view of the substantial difference in structure and function between the two flagellar systems.
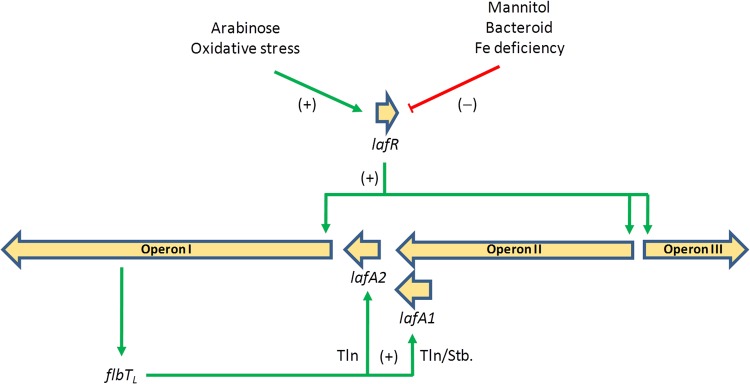
Moreover, operon I encodes FlbTL, a regulator whose homologs in C. crescentus and B. melitensis regulate the expression of class III genes (37, 38). Despite that homology, however, FlbT plays opposite roles in those systems: whereas in C. crescentus FlbT is an inhibitor of the translation of flagellin transcripts, in B. melitensis that protein is required for translation. The target site of FlbT-dependent regulation—it is reported to lie within the 5′ UTR of the mRNA—regulates translation and mRNA stability, but the existence of an as-yet-unknown intermediate for the formation of the FlbT/5′ UTR/mRNA complex might explain these opposite actions (39). Moreover, some effect of FlbT on the activity of the flagellin gene promoter has also been detected (38, 47). In the example of the B. diazoefficiens lateral flagella, the role of FlbTL in lafA1 and lafA2 expression was even more intriguing. As in B. melitensis, FlbTL was required for lateral-flagellin production (Fig. 4), including LafA1 and LafA2 in the low-molecular-mass band (20); however, as a striking exception to the known flagellar systems, the flagellin gene lafA1 lies at the 3′ end of operon II under transcriptional control of the class IB regulator LafR, instead of being a class III gene encoded in a monocistronic transcriptional unit. After an evaluation of the effects of LafR and FlbTL on fliLL and lafA1 transcript accumulation (Fig. 3B), LafR activity proved to be only in part required for lafA1 expression; this result is consistent with the existence of an internal promoter within operon II upstream of lafA1. The activity of this internal promoter would not be regulated by LafR or FlbTL, but the differences in nucleotide sequence and predicted secondary structure between the 5′ UTRs of lafA1 and lafA2, as well as the discrepancies in the responses of these genes to FlbTL with respect to transcript abundance, suggest that the lafA1 transcript would be more unstable than the lafA2 transcript in the absence of FlbTL. Therefore, both the expression and the subsequent mRNA stability of lafA1 seem to be under a mixture of class IB and class III regulation, whereas the lafA2 monocistronic transcript would require neither LafR nor FlbTL for accumulation, and in this instance the role of FlbTL might be restricted to some form of translational control. Neither LafA1 nor LafA2 seemed to accumulate in the cytoplasm of cells expressing flbTL from a multicopy plasmid when the export apparatus was not formed, indicating that an additional system coordinates LafA production and secretion. We identified flaFL as being located immediately upstream from flbTL in operon I. In C. crescentus and B. melitensis FlaF was described as a counterregulator of FlbT through an as-yet-unknown mechanism (38, 47). If FlaFL has a similar role in B. diazoefficiens, that protein might prevent FlbT-dependent translation of the lafA2 and lafA1 transcripts until the export apparatus becomes functional. Figure 6 presents the scheme of the regulatory circuit that may be deduced from our results.
FIG 6.
Scheme of the regulation of the lateral-flagellar genes that may be deduced from the present results. Cultivation with arabinose as the carbon source induces the expression of lafR, whereas cultivation with mannitol as the carbon source does not. LafR activates the transcription (Txn) of operons I, II, and III without any special hierarchical order among them, while the monocistronic lafA2 is transcribed independently of LafR. Operon I contains flbTL, whose locus, upon activation, acts as a translation (Tln) inducer of the monocistronic lafA2 and appears to stabilize (Stb) the lafA1 transcript from a promoter within operon II. In addition to the effects of arabinose and mannitol, evidence from the literature indicates that prolonged exposure to moderate oxidative stress also induces lafR and the lateral flagellar regulon (25), whereas situations of O2 limitation as the bacteroid state (23) or iron limitation (24) repress them. We also observed that viscosity and tortuousity of the medium induce lateral flagella (20) and that microoxia was reported as inhibiting lateral flagella genes expression (23). Therefore, the signal to which the expression of lateral-flagellar genes responds might be related to the energy status of the cell, apart from the specific carbon source available.
The present results indicate that the hierarchies of regulation at the level of transcription are not as strict as in model systems but instead seem more similar to the regulation in B. melitensis, where the flagella are synthesized during a short period in the bacterial culture (38). This lack of a hierarchy might constitute an adaptation to the use of these flagella only when required by the environmental conditions. In the example of the lateral flagella of B. diazoefficiens, the environmental conditions seem to be related either to energy availability and demand—in particular the availability of oxygen (23) and the carbon source (the present study)—or to a requirement for higher torque by the flagella, such as upon an alteration in the viscosity and/or the porosity of the medium (20). Monitoring of these situations might be essential in order for this bacterium to adapt its energy expenditures for motility to the soil's environmental dynamics.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Table S1 in the supplemental material summarizes the bacterial strains and plasmids used in this work. B. diazoefficiens was grown in HM salts (48) plus 0.1% yeast extract (HMY) with 0.5% mannitol or 0.5% arabinose as the carbon source. Total biomass was estimated by measurement of the optical density at 500 nm (OD500) and the number of viable bacteria by the number of CFU on yeast extract-mannitol (49) agar plates (YMA). For swimming-motility analysis, bacteria were inoculated with a sterile toothpick on semisolid AG medium (48) containing 0.3% (wt/vol) agar, and the motility halo was registered as described previously (21). For conjugation, a modified peptone salt-yeast extract medium (50) was used. Escherichia coli was grown in Luria-Bertani medium (51). Antibiotics were added to the media at the following concentrations (μg ml−1): streptomycin, 400 (B. diazoefficiens) or 100 (E. coli); spectinomycin, 200 (B. diazoefficiens) or 100 (E. coli); kanamycin, 150 (B. diazoefficiens) or 25 (E. coli); ampicillin, 200; gentamicin (Gm), 100 (B. diazoefficiens) or 10 (E. coli); chloramphenicol (Cm), 20 (B. diazoefficiens); and tetracycline (Tc), 20 or 50 (B. diazoefficiens in liquid or solid cultures) or 10 (E. coli).
Bioinformatic methods.
The multiple alignments were performed by means of CLUSTAL OMEGA (52) at the EMBL online server (53). The operon prediction was run in different online servers: the ProOpDB online server (54), the DOOR2 online server (55), and the MicrobesOnline server tools (56). Analysis of these results in comparison to the transcription start site map of B. diazoefficiens (34) was carried out using the Integrated Genome Browser (57). All the oligonucleotides were designed with Primer BLAST (58). The RNA secondary structure prediction was carried out with Mfold 2.3 at 30°C with the other parameters as the default (59). To find the common motif in the upstream DNA sequences of operons I, II, and III, the MEME Suite was used (36). The scheme of the motif was built with the WebLogo server (60).
Genetic techniques and DNA manipulation.
The cloning procedures—comprising DNA isolation, restriction digestion, ligation, and bacterial transformation—were performed as previously described (51). Bi- or triparental matings were performed with E. coli DH5α or S17-1, respectively, as previously described (61). Electroporation was performed with a GenePulser (Bio-Rad, Hercules, CA) at 1.5 V, 25 μF, and 200 Ω in a 0.1-cm-gap-width electroporation cuvette.
Oligonucleotide primers (see Table S1 in the supplemental material) were purchased from Life Technologies (Buenos Aires, Argentina). DNA amplification was performed by using PCR in a Bioer Life Express thermocycler (Hangzhou, China) with Taq DNA polymerase (Life Technologies, Buenos Aires, Argentina) for routine PCR or with KAPA HiFi hot start (HS) DNA polymerase (Kapabiosystems, Woburn, MA) for amplification of targets longer than 1,000 bp. The DNA sequencing was performed at Macrogen Corp. (Seoul, South Korea).
To construct the B. diazoefficiens lafR::Km mutant, specific primers for blr6846 were designed. The fragment between bp 7542881 and 7543632 was amplified from the B. diazoefficiens USDA 110 genomic DNA and cloned into the plasmid pG18mob2 (62) by means of an internal EcoRI site (at bp 7542999) and a HindIII site generated from the LafR_Rv primer to generate the plasmid pG::lafR. Next, the Km cassette from the plasmid pUC4K (63) was cloned into an internal BamHI site (at bp 7543291) of the lafR fragment to give plasmid pG::lafR::Km. The gene insertion of the Km cassette was performed by introducing the pG::lafR::Km into B. diazoefficiens wild-type strain USDA 110 by biparental mating, with recombinant selection by growth on Km/Cm YMA, with subsequent screening for Km resistance and Gm sensitivity in order to select for the double-crossover mutant. The resulting strain was designated lafR::Km; this strain carries the Km cassette insertion at the position 7543291 of the genomic DNA, thus disrupting the connection between the receiver and the helix-turn-helix domains of lafR (see Fig. S2B in the supplemental material).
The point mutations in the residue susceptible to phosphorylation (Asp50; see Fig. S2A in the supplemental material) were performed as described previously (51). In brief, PCR primers were designed complementary to the region spanning the mutation site of plasmid pG::lafR—that plasmid DNA being the template for the reaction—but with single complementary base changes for one residue in both strands of the site in order to generate the desired point mutation. The PCR under the direction of those primers then amplified with Pfu DNA polymerase (Thermo Fisher Scientific, Waltham, MA) the entire plasmid, including the mutation introduced in the primers, to give the double-stranded DNA for the new mutant plasmid. The PCR mix was then treated with DpnI, and the template was degraded. The reaction mix was desalted and transformed into E. coli DH5α. Because the position of the point mutation coincides with a SalI restriction site in the wild-type sequence, we screened the clones by looking for resistance to digestion with this endonuclease. The positive clones were then corroborated by DNA sequencing. The fragments with the point mutation were subcloned in the plasmid vector pK18mobsacB (64) to give the derivatives pKsacB::lafRD50A, pKsacB::lafRD50G, and pKsacB::lafRD50E. Each plasmid was transferred by mating to the wild-type strain, and simple crossovers were selected by Km resistance. Resolution of the plasmid was forced by plating the Km-resistant colonies in YMA supplemented with 10% (wt/vol) sucrose. The resulting clones were corroborated by PCR amplification and SalI digestion of the fragment.
To construct the B. diazoefficiens ΔflbTL mutant, the crossover PCR method described by Link et al. (65) was applied to generate an in-frame deletion of the coding sequence of bll6854 (flbTL). To this end, specific primers (FlbTUP_w and FlbTUP_Rv for PCR 1 and FlbTDW_Fw and FlbTDW_Rv for PCR 2) were designed for the amplification of upstream (118-bp) and downstream (99-bp) fragments of flbTL (PCR 1 and PCR 2 according to the methods in the references cited). Next, a PCR 3 reaction was run with primers FlbTUP_Fw and FlbTDW_Rv with an equal mixture of PCR 1 and PCR 2 products as the template. The product of this PCR contains the 5′ and 3′ portions of flbTL interrupted by a short synthetic sequence (21 bp) that replaces an internal 195-bp fragment of the 411-bp coding sequence of flbTL without affecting the reading frame (see Fig. S5 in the supplemental material). This construct was cloned into the pK18mobsacB vector to yield plasmid pKsacB::flbTL. This plasmid was transferred to the wild-type strain by biparental mating, and a resulting single crossover was selected by Km resistance. These simple recombinants were selected for further double crossovers by plating the bacteria in YMA supplemented with 10% (wt/vol) sucrose; thereafter, the resulting clones were subjected to PCR to distinguish the wild-type resolution of the plasmid from the mutant genotype. The correct in-frame deletion was verified by DNA sequencing.
Complementation experiments were performed by integrating the complete sequences of lafR or flbTL (amplified with the primer pair LafRextFw/LafRextRv or FlbTextFw/FlbTextRv, respectively) into a replicative vector. Stated in brief, the 1,009-bp lafR and the 592-bp flbTL target sequences were amplified from B. diazoefficiens USDA 110 chromosomal DNA and then cloned into the XbaI/KpnI sites of plasmid pFAJ1708 to create pFAJ::lafR and pFAJ::flbTL, respectively. The recombinant plasmids were cloned with the lafR or flbTL genes under the direction of the strong, constitutive nptII promoter (66). These constructions were all confirmed by sequencing. Finally, each plasmid harboring the complete lafR or flbTL gene was transferred into the desired B. diazoefficiens strain by biparental mating, selected by Tc resistance, and confirmed by PCR amplification and DNA sequencing.
To construct the lacZ fusions, the lafA1 and lafA2 promoter regions were amplified with the primer pairs promA1_Fw/promA1_Rv and promA2_Fw/promA2_Rv, respectively (see Table S1 in the supplemental material). The amplicons were digested with XbaI/PstI and then cloned into the same restriction sites of the promoterless plasmid vector pCB303 carrying the complete sequence of the β-galactosidase gene (67). These constructions were named pCB::PlafA1 and pCB::PlafA2, respectively. Each plasmid was transferred into the B. diazoefficiens USDA 110 strain by biparental mating, selected by Tc resistance, and confirmed by PCR amplification. The β-galactosidase activity was measured as described previously (51).
RNA extraction and RT-PCR.
Portions (13 ml) of B. diazoefficiens USDA 110 cells were harvested from liquid cultures, washed twice with 1 M NaCl, and disrupted with lysozyme in Tris-EDTA buffer (pH 8.0, 30 min, 37°C). Total RNA was extracted through the use of TRIzol (Life Technologies, Buenos Aires, Argentina), according to the manufacturer's instructions, and the quality and quantity of the extract determined with a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). Aliquots (0.125 μg) were treated with DNase I (30 min, 37°C), and the cDNA synthesized with Moloney murine leukemia virus reverse transcriptase (Life Technologies, Buenos Aires, Argentina) under the direction of random hexamer primers according to the manufacturer's instructions. To check the quality of the cDNA preparation, PCRs were performed with the primer pairs phaR_Fw/phaR_Rv and relA_Fw/relA_Rv (see Table S1 in the supplemental material) as described previously (61). The absence of contaminating DNA was demonstrated by the lack of PCR amplification in an RNA sample that was not subjected to reverse transcription. Primers for the housekeeping gene sigA were used as a positive control (68).
To determine the operon structure, three RT-PCRs were performed with the appropriate cDNAs for each fragment (see Fig. S4): two of the reactions amplified fragments of the coding sequence of each contiguous gene (positive control), while the third amplified a fragment encompassing the intergenic region between the target genes (35).
Quantitative real-time RT-PCR.
Amplification of the cDNAs obtained as described above was performed with the primers indicated in the Table S1 in the supplemental material for each gene in a Line-Gene instrument (Bioer, Hangzhou, China) and analyzed with Line-Gene K fluorescence quantitative detection system (v4.0.00 software). Ready-to-use iQ SYBR green Supermix (Bio-Rad, Hercules, CA) was used for all the reactions, according to the manufacturers' instructions. Normalized expression values were calculated based on the absolute quantities of the gene of interest relative to the value for sigA (68).
Flagellin separation and analysis.
The preparation of flagellins was carried out as described previously (30). Stated in brief, rhizobia grown in liquid medium to an OD500 of 1.0 were vortexed for 5 min and centrifuged at 10,000 × g for 30 min at 4°C. The supernatant was collected and incubated with 1.3% (vol/vol) polyethylene glycol 6000 and 166 mM NaCl for 2 h at 4°C. This suspension was centrifuged at 11,000 × g for 40 min at 4°C, and the pellet resuspended in phosphate-buffered saline. For analysis, the samples were boiled in Laemmli loading buffer for 10 min and then separated by SDS-PAGE (69). Polypeptide bands were revealed with Coomassie brilliant blue R250.
Total proteins were prepared after lysis by boiling. The cell pellet was washed with 1 M NaCl solution, resuspended in Laemmli loading buffer, and heated for 10 min at 100°C. The samples were then centrifuged at 14,000 × g for 10 min and analyzed by SDS-PAGE. After electrophoresis, the gels were stained with Coomassie brilliant blue R250. Western blots were prepared with specific anti-LafA polyclonal antibodies on the total proteins extracted from the cell pellets, as previously described (22, 70).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Donald Haggerty for English revision and Anke Becker (Marburg University, Germany) for kindly providing the E. meliloti strains.
This study was supported by the Agencia Nacional de Promoción de la Investigación Científica y Tecnológica (ANPCyT) and the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), both from Argentina. E.J.M., J.I.Q., M.J.A., and A.R.L. are members of the Scientific Career of CONICET. C.D. is a fellow of CONICET.
The funders had no role in study design or data collection and interpretation. We declare that we have no conflict of interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00253-17.
REFERENCES
- 1.Yuan J, Branch RW, Hosu BG, Berg HC. 2012. Adaptation at the output of the chemotaxis signaling pathway. Nature 484:233–236. doi: 10.1038/nature10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans LD, Hughes C, Fraser GM. 2014. Building a flagellum outside the bacterial cell. Trends Microbiol 22:566–572. doi: 10.1016/j.tim.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minamino T. 2014. Protein export through the bacterial flagellar type III export pathway. Biochim Biophys Acta 1843:1642–1648. doi: 10.1016/j.bbamcr.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Minamino T, Imada K. 2015. The bacterial flagellar motor and its structural diversity. Trends Microbiol 23:267–274. doi: 10.1016/j.tim.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Takekawa N, Nishiyama M, Kaneseki T, Kanai T, Atomi H, Kojima S, Homma M. 2015. Sodium-driven energy conversion for flagellar rotation of the earliest divergent hyperthermophilic bacterium. Sci Rep 5:12711. doi: 10.1038/srep12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg HC. 2016. The flagellar motor adapts, optimizing bacterial behavior. Prot Sci doi: 10.1002/pro.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujii T, Kato T, Namba K. 2009. Specific arrangement of alpha-helical coiled coils in the core domain of the bacterial flagellar hook for the universal joint function. Structure 17:1485–1493. doi: 10.1016/j.str.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Calladine CR, Luisi BF, Pratap JV. 2013. A “mechanistic” explanation of the multiple helical forms adopted by bacterial flagellar filaments. J Mol Biol 425:914–928. doi: 10.1016/j.jmb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magariyama Y, Ichiba M, Nakata K, Baba K, Ohtani T, Kudo S, Goto T. 2005. Difference in bacterial motion between forward and backward swimming caused by the wall effect. Biophys J 88:3648–3658. doi: 10.1529/biophysj.104.054049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarter LL. 2006. Regulation of flagella. Curr Opin Microbiol 9:180–186. doi: 10.1016/j.mib.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Altegoer F, Bange G. 2015. Undiscovered regions on the molecular landscape of flagellar assembly. Curr Opin Microbiol 28:98–105. doi: 10.1016/j.mib.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Aldridge P, Hughes KT. 2002. Regulation of flagellar assembly. Curr Opin Microbiol 5:160–165. doi: 10.1016/S1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- 13.Brown J, Faulds-Pain A, Aldridge P. 2009. The coordination of flagellar gene expression and the flagellar assembly pathway, p 99–120. In Jarrel KF. (ed), Pili and flagella: current research and future trends. Caister Academic Press, Trowbridge, Wiltshire, United Kingdom. [Google Scholar]
- 14.Smith TG, Hoover TR. 2009. Deciphering bacterial flagellar gene regulatory networks in the genomic era. Adv Appl Microbiol 67:257–295. doi: 10.1016/S0065-2164(08)01008-3. [DOI] [PubMed] [Google Scholar]
- 15.Gotz R, Schmitt R. 1987. Rhizobium meliloti swims by unidirectional, intermittent rotation of right-handed flagellar helices. J Bacteriol 169:3146–3150. doi: 10.1128/jb.169.7.3146-3150.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scharf B. 2002. Real-time imaging of fluorescent flagellar filaments of Rhizobium lupini H13-3: flagellar rotation and pH-induced polymorphic transitions. J Bacteriol 184:5979–5986. doi: 10.1128/JB.184.21.5979-5986.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei Y, Wang X, Liu J, Nememan I, Singh AH, Weiss H, Levin BR. 2011. The population dynamics of bacteria in physically structured habitats and the adaptive virtue of random motility. Proc Natl Acad Sci U S A 108:4047–4052. doi: 10.1073/pnas.1013499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sourjik V, Wingreen NS. 2012. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol 24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu R, Ochman H. 2007. Origins of flagellar gene operons and secondary flagellar systems. J Bacteriol 189:7098–7104. doi: 10.1128/JB.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quelas JI, Althabegoiti MJ, Jimenez-Sanchez C, Melgarejo AA, Marconi VI, Mongiardini EJ, Trejo SA, Mengucci F, Ortega-Calvo JJ, Lodeiro AR. 2016. Swimming performance of Bradyrhizobium diazoefficiens is an emergent property of its two flagellar systems. Sci Rep 6:23841. doi: 10.1038/srep23841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Althabegoiti MJ, López-García SL, Piccinetti C, Mongiardini EJ, Pérez-Giménez J, Quelas JI, Perticari A, Lodeiro AR. 2008. Strain selection for improvement of Bradyrhizobium japonicum competitiveness for nodulation of soybean. FEMS Microbiol Lett 282:115–123. doi: 10.1111/j.1574-6968.2008.01114.x. [DOI] [PubMed] [Google Scholar]
- 22.Covelli JM, Althabegoiti MJ, López MF, Lodeiro AR. 2013. Swarming motility in Bradyrhizobium japonicum. Res Microbiol 164:136–144. doi: 10.1016/j.resmic.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Pessi G, Ahrens CH, Rehrauer H, Lindemann A, Hauser F, Fischer HM, Hennecke H. 2007. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol Plant Microbe Interact 20:1353–1363. doi: 10.1094/MPMI-20-11-1353. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Sangwan I, Lindemann A, Hauser F, Hennecke H, Fischer HM, O'Brian MR. 2006. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol Microbiol 60:427–437. doi: 10.1111/j.1365-2958.2006.05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donati AJ, Jeon JM, Sangurdekar D, So JS, Chang WS. 2011. Genome-wide transcriptional and physiological responses of Bradyrhizobium japonicum to paraquat-mediated oxidative stress. Appl Environ Microbiol 77:3633–3643. doi: 10.1128/AEM.00047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei X, Bauer WD. 1998. Starvation-induced changes in motility, chemotaxis, and flagellation of Rhizobium meliloti. Appl Environ Microbiol 64:1708–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fretin D, Fauconnier A, Kohler S, Halling S, Leonard S, Nijskens C, Ferooz J, Lestrate P, Delrue RM, Danese I, Vandenhaute J, Tibor A, DeBolle X, Letesson JJ. 2005. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol 7:687–698. doi: 10.1111/j.1462-5822.2005.00502.x. [DOI] [PubMed] [Google Scholar]
- 28.Rotter C, Muhlbacher S, Salamon D, Schmitt R, Scharf B. 2006. Rem, a new transcriptional activator of motility and chemotaxis in Sinorhizobium meliloti. J Bacteriol 188:6932–6942. doi: 10.1128/JB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoang HH, Gurich N, González JE. 2008. Regulation of motility by the ExpR/Sin quorum-sensing system in Sinorhizobium meliloti. J Bacteriol 190:861–871. doi: 10.1128/JB.01310-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Althabegoiti MJ, Covelli JM, Pérez-Giménez J, Quelas JI, Mongiardini EJ, López MF, López-García SL, Lodeiro AR. 2011. Analysis of the role of the two flagella of Bradyrhizobium japonicum in competition for nodulation of soybean. FEMS Microbiol Lett 319:133–139. doi: 10.1111/j.1574-6968.2011.02280.x. [DOI] [PubMed] [Google Scholar]
- 31.López-García SL, Perticari A, Piccinetti C, Ventimiglia L, Arias N, De Battista J, Althabegoiti MJ, Mongiardini EJ, Pérez-Giménez J, Quelas JI, Lodeiro AR. 2009. In-furrow inoculation and selection for higher motility enhances the efficacy of Bradyrhizobium japonicum nodulation. Agron J 101:1–7. doi: 10.1016/S0065-2113(08)00801-8. [DOI] [Google Scholar]
- 32.Leonard S, Ferooz J, Haine V, Danese I, Fretin D, Tibor A, de Walque S, De Bolle X, Letesson JJ. 2007. FtcR is a new master regulator of the flagellar system of Brucella melitensis 16M with homologs in Rhizobiaceae. J Bacteriol 189:131–141. doi: 10.1128/JB.00712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, Kohara M, Matsumoto M, Shimpo S, Tsuruoka H, Wada T, Yamada M, Tabata S. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res 9:189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- 34.Čuklina J, Hahn J, Imakaev M, Omasits U, Förstner KU, Ljubimov N, Goebel M, Pessi G, Fischer HM, Ahrens CH, Gelfand MS, Evguenieva-Hackenberg E. 2016. Genome-wide transcription start site mapping of Bradyrhizobium japonicum grown free-living or in symbiosis: a rich resource to identify new transcripts, proteins and to study gene regulation. BMC Genomics 17:302. doi: 10.1186/s12864-016-2602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redondo-Nieto M, Lloret J, Larenas J, Barahona E, Navazo A, Martínez-Granero F, Capdevila S, Rivilla R, Martín M. 2008. Transcriptional organization of the region encoding the synthesis of the flagellar filament in Pseudomonas fluorescens. J Bacteriol 190:4106–4109. doi: 10.1128/JB.00178-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 37.Mangan EK, Malakooti J, Caballero A, Anderson P, Ely B, Gober JW. 1999. FlbT couples flagellum assembly to gene expression in Caulobacter crescentus. J Bacteriol 181:6160–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferooz J, Lemaire J, Letesson JJ. 2011. Role of FlbT in flagellin production in Brucella melitensis. Microbiology 157:1253–1262. doi: 10.1099/mic.0.044867-0. [DOI] [PubMed] [Google Scholar]
- 39.Anderson PE, Gober JW. 2000. FlbT, the posttranscriptional regulator of flagellin synthesis in Caulobacter crescentus, interacts with the 5′ untranslated region of flagellin mRNA. Mol Microbiol 38:41–52. doi: 10.1046/j.1365-2958.2000.02108.x. [DOI] [PubMed] [Google Scholar]
- 40.Lang AS, Beatty JT. 2002. A bacterial signal transduction system controls genetic exchange and motility. J Bacteriol 184:913–918. doi: 10.1128/jb.184.4.913-918.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sourjik V, Muschler P, Scharf B, Schmitt R. 2000. VisN and VisR are global regulators of chemotaxis, flagellar, and motility genes in Sinorhizobium (Rhizobium) meliloti. J Bacteriol 182:782–788. doi: 10.1128/JB.182.3.782-788.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tambalo DD, Del Bel KL, Bustard DE, Greenwood PR, Steedman AE, Hynes MF. 2010. Regulation of flagellar, motility and chemotaxis genes in Rhizobium leguminosarum by the VisN/R-Rem cascade. Microbiology 156:1673–1685. doi: 10.1099/mic.0.035386-0. [DOI] [PubMed] [Google Scholar]
- 43.Thorne DW, Burris RH. 1940. Respiratory enzyme systems in symbiotic nitrogen fixation: III. The respiration of Rhizobium from legume nodules and laboratory cultures. J Bacteriol 39:187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindemann A, Moser A, Pessi G, Hauser F, Friberg M, Hennecke H, Fischer HM. 2007. New target genes controlled by the Bradyrhizobium japonicum two-component regulatory system RegSR. J Bacteriol 189:8928–8943. doi: 10.1128/JB.01088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohkama-Ohtsu N, Honma H, Nakagome M, Nagata M, Yamaya-Ito H, Sano Y, Hiraoka N, Ikemi T, Suzuki A, Okazaki S, Minamisawa K, Yokoyama T. 2016. Growth rate of and gene expression in Bradyrhizobium diazoefficiens USDA110 due to a mutation in blr7984, a TetR family transcriptional regulator gene. Microbes Environ 31:249–259. doi: 10.1264/jsme2.ME16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herlihey FA, Moynihan PJ, Clarke AJ. 2014. The essential protein for bacterial flagella formation FlgJ functions as a β-N-acetylglucosaminidase. J Biol Chem 289:31029–31042. doi: 10.1074/jbc.M114.603944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Llewellyn M, Dutton RJ, Easter J, O'Donnol D, Gober JW. 2005. The conserved flaF gene has a critical role in coupling flagellin translation and assembly in Caulobacter crescentus. Mol Microbiol 57:1127–1142. doi: 10.1111/j.1365-2958.2005.04745.x. [DOI] [PubMed] [Google Scholar]
- 48.Sadowsky MJ, Tully RE, Cregan PB, Keyser HH. 1987. Genetic diversity in Bradyrhizobium japonicum serogroup 123 and its relation to genotype-specific nodulation of soybean. Appl Environ Microbiol 53:2624–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vincent JM. 1970. A manual for the practical study of the root nodule bacteria. IBP handbook 15. Blackwell Scientific Publications, Oxford, United Kingdom. [Google Scholar]
- 50.Regensburger B, Hennecke H. 1983. RNA polymerase from Rhizobium japonicum. Arch Microbiol 135:103–109. doi: 10.1007/BF00408017. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 52.Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41(Web Server issue):W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taboada B, Ciria R, Martínez-Guerrero CE, Merino E. 2012. ProOpDB: Prokaryotic Operon DataBase. Nucleic Acids Res 40(Database issue):D627–D631. doi: 10.1093/nar/gkr1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mao X, Ma Q, Zhou C, Chen X, Zhang H, Yang J, Mao F, Lai W, Xu Y. 2014. DOOR 2.0: presenting operons and their functions through dynamic and integrated views. Nucleic Acids Res 42(Database issue):D654–D659. doi: 10.1093/nar/gkt1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, Dubchak IL, Alm EJ, Arkin AP. 2010. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res 38(Database issue):D396–D400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freese NH, Norris DC, Loraine AE. 2016. Integrated genome browser: visual analytics platform for genomics. Bioinformatics 32:2089–2095. doi: 10.1093/bioinformatics/btw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quelas JI, Mongiardini EJ, Casabuono A, López-García SL, Althabegoiti MJ, Covelli JM, Pérez-Giménez J, Couto A, Lodeiro AR. 2010. Lack of galactose or galacturonic acid in Bradyrhizobium japonicum USDA 110 exopolysaccharide leads to different symbiotic responses in soybean. Mol Plant Microbe Interact 23:1592–1604. doi: 10.1094/MPMI-05-10-0122. [DOI] [PubMed] [Google Scholar]
- 62.Kirchner O, Tauch A. 2003. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J Biotechnol 104:287–299. doi: 10.1016/S0168-1656(03)00148-2. [DOI] [PubMed] [Google Scholar]
- 63.Vieira J, Messing J. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 64.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 65.Link AJ, Phillips D, Church GM. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dombrecht B, Vanderleyden J, Michiels J. 2001. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in gram-negative bacteria. Mol Plant Microbe Interact 14:426–430. doi: 10.1094/MPMI.2001.14.3.426. [DOI] [PubMed] [Google Scholar]
- 67.Schneider K, Beck CF. 1987. New expression vectors for identifying and testing signal structures for initiation and termination of transcription. Methods Enzymol 153:452–461. doi: 10.1016/0076-6879(87)53071-3. [DOI] [PubMed] [Google Scholar]
- 68.Hauser F, Lindemann A, Vuilleumier S, Patrignani A, Schlapbach R, Fischer H-M, Hennecke H. 2006. Design and validation of a partial-genome microarray for transcriptional profiling of the Bradyrhizobium japonicum symbiotic gene region. Mol Genet Genomics 275:55–67. doi: 10.1007/s00438-005-0059-7. [DOI] [PubMed] [Google Scholar]
- 69.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 70.Pérez-Giménez J, Covelli JM, López MF, Althabegoiti MJ, Ferrer-Navarro M, Mongiardini EJ, Lodeiro AR. 2012. Soybean seed lectin prevents the accumulation of S-adenosyl methionine synthetase and the S1 30S ribosomal protein in Bradyrhizobium japonicum under C and N starvation. Curr Microbiol 65:465–474. doi: 10.1007/s00284-012-0180-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.