ABSTRACT
Oritavancin is a lipoglycopeptide antibiotic that exhibits potent activities against vancomycin-resistant Gram-positive pathogens. Oritavancin differs from vancomycin by a hydrophobic side chain attached to the drug disaccharide, which forms a secondary binding site to enable oritavancin binding to the cross-linked peptidoglycan in the cell wall. The mode of action of secondary binding site was investigated by measuring the changes in the peptidoglycan composition of Staphylococcus aureus grown in the presence of desleucyl-oritavancin at subinhibitory concentration using liquid chromatography-mass spectrometry (LC-MS). Desleucyl-oritavancin is an Edman degradation product of oritavancin that exhibits potent antibacterial activities despite the damaged d-Ala–d-Ala binding site due to its functional secondary binding site. Accurate quantitative peptidoglycan composition analysis based on 83 muropeptide ions determined that cell walls of S. aureus grown in the presence of desleucyl-oritavancin showed a reduction of peptidoglycan cross-linking, increased muropeptides with a tetrapeptide-stem structure, decreased O-acetylation of MurNAc, and increased N-deacetylation of GlcNAc. The changes in peptidoglycan composition suggest that desleucyl-oritavancin targets the peptidoglycan template to induce cell wall disorder and interferes with cell wall maturation.
IMPORTANCE Oritavancin is a lipoglycopeptide antibiotic with a secondary binding site that targets the cross-linked peptidoglycan bridge structure in the cell wall. Even after the loss of its primary d-Ala–d-Ala binding site through Edman degradation, desleucyl-oritavancin exhibits potent antimicrobial activities through its still-functioning secondary binding site. In this study, we characterized the mode of action for desleucyl-oritavancin's secondary binding site using LC-MS. Peptidoglycan composition analysis of desleucyl-oritavancin-treated S. aureus was performed by determining the relative abundances of 83 muropeptide ions matched from a precalculated library through integrating extracted ion chromatograms. Our work highlights the use of quantitative peptidoglycan composition analysis by LC-MS to provide insights into the mode of action of glycopeptide antibiotics.
KEYWORDS: Staphylococcus aureus, peptidoglycan, oritavancin, LC-MS, cell wall, cross-link, glycopeptide antibiotics, vancomycin
INTRODUCTION
Oritavancin is a potent lipoglycopeptide antibiotic active against multidrug-resistant Gram-positive pathogens, including vancomycin-resistant enterococci (VRE), methicillin-resistant Staphylococcus aureus, vancomycin-intermediate S. aureus, and vancomycin-resistant S. aureus (VRSA) (1–3). Like vancomycin, oritavancin binds to the d-Ala–d-Ala of the peptidoglycan (PG) stem (Fig. 1a) in lipid II to inhibit the transglycosylation step of PG biosynthesis (Fig. 1b). By binding to lipid II, oritavancin prevents the transfer of PG repeat units from lipid II to the growing nascent PG and results in cytoplasmic accumulation of Park's nucleotide (4, 5). In VRE and VRSA, the d-Ala–d-Ala terminus of the PG stem is replaced by a depsipeptide, d-Ala–d-Lac. This depsipeptide substitution reduces the vancomycin binding affinity by replacing one of the five hydrogen bonds with an electrostatic repulsion (6). While vancomycin is ineffective against VRE and VRSA (4, 7), oritavancin exhibits potent bactericidal activities against these vancomycin-resistant pathogens (1–3).
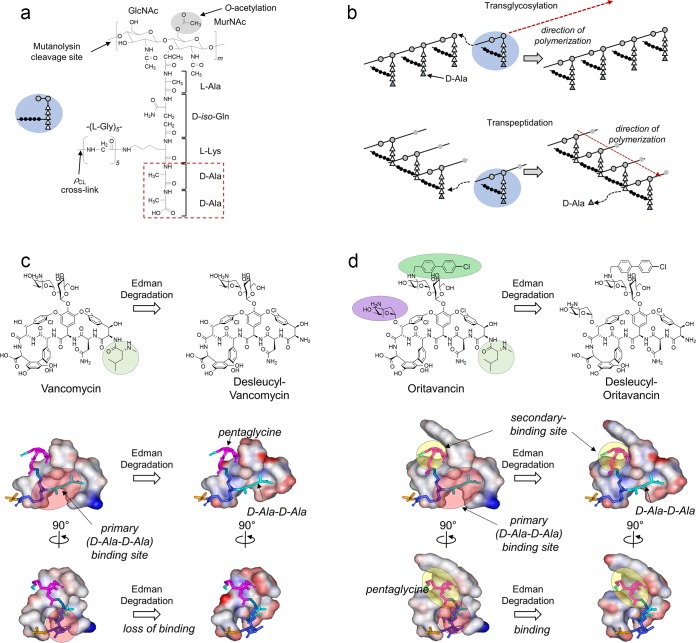
FIG 1.
Peptidoglycan structure and biosynthesis. (a) Chemical structure of an S. aureus peptidoglycan (PG) repeat unit. A single repeat unit consists of a disaccharide (GlcNAc and MurNAc), a pentapeptide stem (l-Ala–d-iso-Gln–l-Lys–d-Ala–d-Ala), and a pentaglycine bridge structure. Mutanolysin cleaves the β(1-4) glycosidic linkage between MurNAc and GlcNAc. The d-Ala–d-Ala terminus of the PG repeat unit (red box) is the primary binding site for vancomycin. The inset shows a schematic representation of a PG repeat unit, with the disaccharide as gray-filled circles, amino acids in the pentapeptide stem as open triangles, and the pentaglycine bridge as filled circles. (b) PG polymerization through two distinct steps with different enzymes. First, transglycosylase elongates the disaccharide glycan chain. Transpeptidase then interconnects the resulting glycan chains by forming cross-links between the N terminus of the pentaglycine bridge from one PG unit and the penultimate d-Ala of the PG stem from a neighboring glycan chain. The terminal d-Ala (blue triangle) is removed by the transpeptidase during cross-linking. (c) Chemical structures and space-filling models of vancomycin (left) and desleucyl-vancomycin (right) bound to PG. In the space-filling models, the d-Ala–d-Ala dipeptide is shown as a stick in light blue, d-iso-Gln and l-Lys are in dark blue, and pentaglycine is in magenta. Edman degradation of vancomycin cleaves the N-methylleucine (highlighted circle) and results in desleucyl-vancomycin with a damaged d-Ala–d-Ala binding site, which abolishes the antibacterial activity. (d) Chemical structures and space-filling models of oritavancin (left) and desleucyl-oritavancin (des-Ori) (right) bound to PG. Edman degradation of oritavancin results in des-Ori, which still exhibits potent antibacterial activity despite the damaged d-Ala–d-Ala binding site (red circle) due to its secondary binding site (yellow circle). The secondary binding site targeting the PG pentaglycine bridge structure is formed by the hydrophobic side chain of oritavancin and the aglycon structure.
The chemical structure of oritavancin differs from that of vancomycin (Fig. 1c and d) by an alkylation of a chlorobenzylphenyl side chain (green) at the 4-epi-vancomsamine on the fourth amino acid and an additional 4-epi-vancomsamine (purple) at the sixth amino acid of the aglycon. Since oritavancin readily forms dimers in solution and binds to membrane vesicles (8), drug dimerization and membrane anchoring mediated by the hydrophobic side chain of oritavancin are thought to increase its binding to the d-Ala–d-Lac-terminated PG stem structure to overcome the vancomycin resistance in VRE (9). However, in situ characterizations of oritavancin (10) and oritavancin-like glycopeptide (4, 11, 12) binding sites in intact whole cells of S. aureus by solid-state nuclear magnetic resonance (NMR) have shown that lipoglycopeptides do not bind to the cell wall as dimers or partition to the bacterial membrane. The only instance of oritavancin's hydrophobic side chain being found inserted into the lipid bilayer was when it was complexed to an isolated protoplast membrane of S. aureus. In isolated protoplast membrane, nascent PG strands that surround the cytoplasmic membrane are disrupted and extend away from the membrane. The perturbed PG organization exposes the lipid bilayer and permits oritavancin binding to the membrane (13).
A model structure of oritavancin bound to a PG repeat unit based on solid-state NMR distance constraints (4, 14) is shown in Fig. 1d. The model provides structural insights into the role of oritavancin's hydrophobic side chain. Two distinct binding sites are visible in the model: (i) the primary binding site, which is the aglycon structure identical to that of vancomycin for the binding of d-Ala–d-Ala (15), and (ii) the secondary binding site, which is a cleft formed between the hydrophobic side chain and the aglycon structure that targets the PG bridge structure (4). To investigate the mode of action of the secondary binding site, the primary-binding site in oritavancin was damaged through Edman degradation by cleaving N-methlylleucine from the aglycon structure. Unlike the Edman degradation product of vancomycin, des-N-methylleucyl-vancomycin (Fig. 1c), which is devoid of any antimicrobial activities, des-N-methylleucyl-oritavancin (referred to as des-Ori here) retains potent antimicrobial activities against both vancomycin-susceptible and -resistant pathogens (4). Thus, the secondary binding site in des-Ori is responsible for enhanced activities against vancomycin-resistant pathogens by compensating for the damaged d-Ala–d-Ala binding site, enabling it to bind to PG in intact whole cells of S. aureus (4, 7). In this study, we investigated the mode of action of the secondary binding site by analyzing changes to the PG composition of S. aureus grown in the presence of des-Ori at a sub-MIC using liquid chromatography-mass spectrometry (LC-MS).
RESULTS
Inhibition of S. aureus growth by desleucyl-oritavancin.
des-Ori was added to S. aureus (ATCC 6538P) during mid-exponential growth (optical density at 600 nm [OD600] of 0.6) to final drug concentrations of 0, 1, 5, 10, 15, and 20 μg/ml (Fig. 2a, dashed lines). Despite the damaged d-Ala–d-Ala binding site, des-Ori inhibited the growth of S. aureus. Concentration-dependent inhibition of S. aureus growth measured by plotting the OD600 of cultures at 100 min after des-Ori addition as a function of des-Ori concentration (Fig. 2b). Growth recovery was determined by plotting the time required for the resumption of growth by measuring the interval between des-Ori addition and the inflection point of the growth curve as a function of des-Ori concentration (Fig. 2c).
FIG 2.
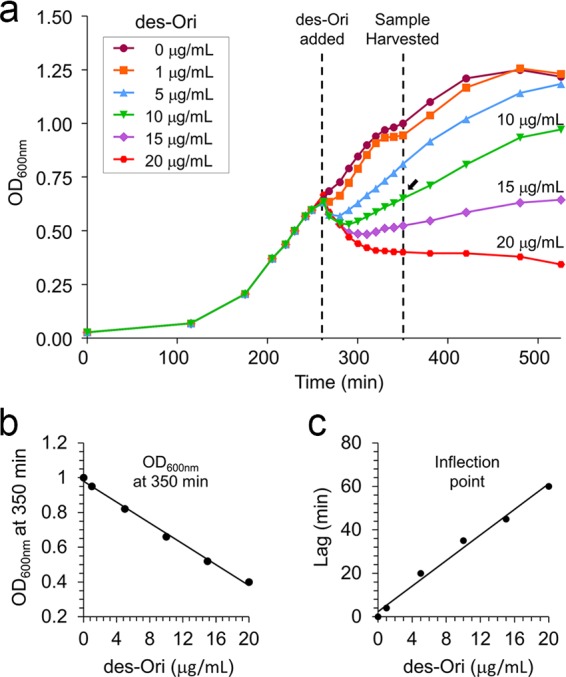
S. aureus growth in the presence of desleucyl-oritavancin. (a) Growth curve of S. aureus (ATCC 6538P) in the presence of des-Ori, monitored by measuring the optical density at 600 nm (OD600). des-Ori was added to S. aureus at an OD600 of 0.6, and the cells were harvested after 100 min of growth in the presence of des-Ori. Cell walls of S. aureus were synthesized in the presence of des-Ori at concentrations of 0, 1, 5, 10, 15, and 20 μg/ml during mid-exponential growth (dashed lines). (b and c) Following the des-Ori addition, S. aureus exhibited concentration-dependent growth inhibition as determined by (i) OD600 measurements after 100 min of growth in the presence of des-Ori (b) and (ii) the time delay to the point of inflection, which indicates the recovery of growth (Fig. 2c).
Identification and quantification of muropeptide species.
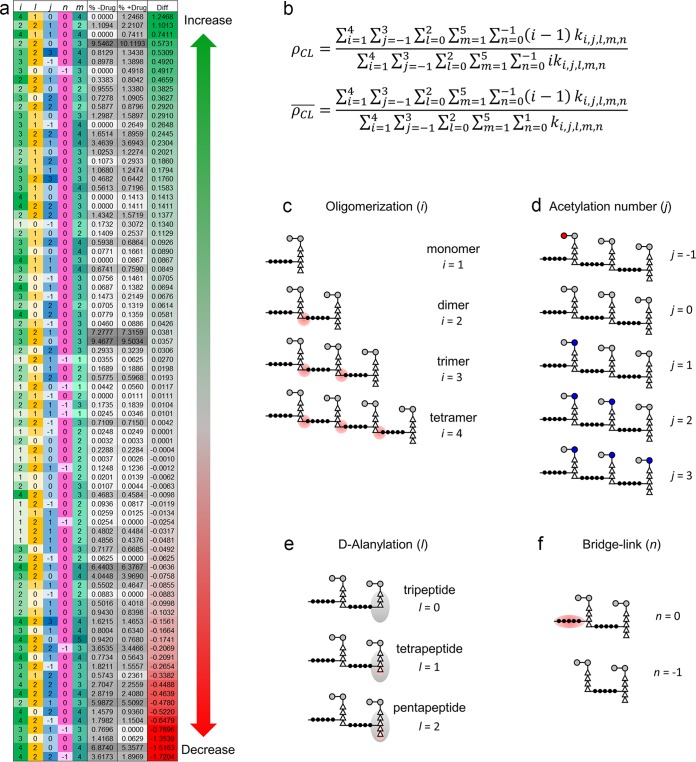
LC-MS analysis was performed on mutanolysin-digested muropeptide fragments from isolated cell walls of S. aureus grown in the presence of des-Ori at 10 μg/ml for 100 min (Fig. 2, arrow). Mutanolysin is a N-acetylmuramidase that cleaves the β(1-4) glycosidic bond between MurNAc and GlcNAc of PG with the PG modifications intact. The muropeptide fragments then were identified by matching the observed m/z values to the library of calculated m/z values that were generated in silico for muropeptide fragments from known chemical modifications (16). The abundance of each muropeptide species was measured by integrating the extracted ion chromatogram (XIC) for the selected ion. The relative abundance for each muropeptide was then calculated by dividing the integrated XIC of the selected ion by the total integral sum for all ion species. This approach differs from earlier approaches that relied on the integration of the UV absorption of chromatographically resolved peaks, where each peak was assumed to consist of a single muropeptide type. However, in our LC-MS, we found that every elution peak during the chromatographic separation contained multiple muropeptide species due to the extraordinary chemical diversity that resulted from various PG modifications. The use of XIC overcomes this limitation by integration of the selected ion from the total ion chromatogram to provide accurate quantification of muropeptide species despite the overlapping elution times. Of 960 unique muropeptide-derived ions generated for the in silico library, 83 muropeptide ions with masses ranging from 937 Da (monomer) to 4,870 Da (tetramer) were identified. Figure 3a shows all identified muropeptide species with the associated PG modifications and their relative abundances as measured by percent composition in isolated cell walls of S. aureus grown in the absence or presence of des-Ori. The PG modifications that were considered for the analysis were numbers of cross-links (Fig. 3c), O-acetylation and N-deacetylation (Fig. 3d), PG stem alanylation (Fig. 3e), and PG bridge attachment (Fig. 3f).
FIG 3.
The S. aureus peptidoglycan fragment ions examined and their relative abundances. (a) Each row shows a unique ion and its properties by column: i (green), number of PG repeat units in the muropeptide; l (yellow), number of d-Alas in the terminal peptide stem; j (blue), acetylation state of the fragment; n (pink), presence of a cross-linking pentaglycine bridge at the terminal subunit; and m (teal), charge state of the ion. The columns “% −Drug” and “% +Drug” list the average relative abundances of each ion with respect to all the ions being examined for S. aureus in the absence and presence of des-Ori (n = 3). The column “Diff” shows the difference between average relative abundances with and without the antibiotic. (b) Calculated PG cross-linking efficiency (ρCL) and average cross-linking per muropeptide (ρCL). The summation element ki,j,l,m,n, represents the integrals of the ion's abundance of a muropeptide as listed in panel a. (c to f) Schematic representations of muropeptide fragments based on the number of PG repeat units (c), acetylation (d), d-alanylation (e), and bridge link attachment (f). The index of summation i is the number of PG repeat units found in a muropeptide (d), j is the PG acetylation state (d), l is the d-alanylation of the PG stem (e), m is the charge state of muropeptide ions, and n is the pentaglycine bridge attachment (f).
Effects of des-Ori on PG cross-links.
The PG cross-linking efficiency (ρCL) and average cross-linking per muropeptide (ρCL) were calculated for mutanolysin-digested isolated cell walls of S. aureus based on the equations shown in Fig. 3b. The cross-linking efficiency is the percentage of cross-links per PG subunit as defined by Driehuis et al. (17), while the average cross-linking per muropeptide is the average number of cross-links found per muropeptide fragment. The summation element ki,j,l,m,n in the equations represents the abundance integral of the muropeptides listed in Fig. 3a. The first index of summation, i, is the number of PG repeat units found per muropeptide fragment (Fig. 3c). The summation was truncated after i = 4 due to the low abundance of oligomeric species greater than tetramers. The index of summation j represents the PG acetylation, ranging from −1 to 3 (Fig. 3d). The index of summation l is the number of d-Alas in the PG stem of the terminal PG subunit, which was from 0 (tripeptide stem), 1 (tetrapeptide stem), or 2 (pentapeptide stem) (Fig. 3e). The index of summation m is the charge state of muropeptide ions from +1 to +5. The index of summation n represents muropeptides with (0) or without (−1) a pentaglycine bridge structure on the terminal PG subunit (Fig. 3f).
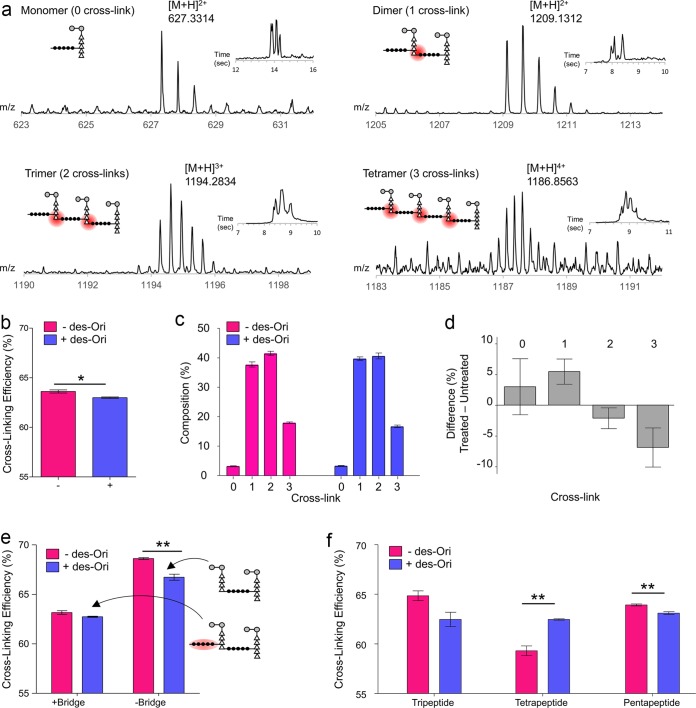
Figure 4a shows the mass spectra of the unmodified PG monomer, dimer, trimer, and tetramer and their associated extracted ion chromatograms (insets) from the mutanolysin-digested cell walls of untreated S. aureus. The calculated cross-linking efficiency (ρCL) for S. aureus grown in the absence of antibiotic was 63.32% ± 0.51%, which was reduced to 62.99% ± 0.23% when grown in the presence of des-Ori at a sub-MIC (Fig. 4b). The calculated average cross-linking per muropeptide (ρCL) for S. aureus grown in the absence of antibiotic was 1.987 ± 0.031 and was reduced to 1.938 ± 0.005 (mean ± 95% confidence interval; P = 0.078) when grown in the presence of des-Ori. The overall change in the cross-linking efficiency of des-Ori-treated S. aureus is small, because the effect corresponds to the mode of action of des-Ori at a sub-MIC. As shown in Fig. 2, addition of des-Ori (10 μg/ml) to S. aureus during mid-exponential growth (OD660 of 0.6) is not lethal, as the growth recovers within 40 min following the addition of antibiotic. The full extent of des-Ori inhibition of transpeptidase activity at a sub-MIC is apparent when the PG compositions of the observed muropeptides are plotted as a function of PG oligomerization (Fig. 4c). S. aureus treated with des-Ori show increases in peptidoglycan monomers and dimers accompanied by simultaneous decreases in trimers and tetramers (Fig. 4d).
FIG 4.
Inhibition of the transpeptidation step of peptidoglycan biosynthesis in S. aureus from desleucyl-oritavancin. (a) Mass spectra of PG monomers, dimers, trimers, and tetramers from mutanolysin-digested cell walls of untreated S. aureus. Corresponding extracted ion chromatograms and schematic representations of the muropeptide structure are shown as insets. Cross-links are highlighted by red circles. The extracted ion chromatogram (XIC) for the selected ion is shown as insets. (b) Calculated PG cross-linking efficiencies (ρCL) for S. aureus grown in the absence (−) and presence (+) of desleucyl-oritavancin at 10 μg/ml (*, P = 0.0238). (c) Normalized integral sum of ion chromatograms of the muropeptide species listed in Fig. 3a classified according to the number of cross-links. (d) Percent difference in normalized integral sums of ion chromatograms of cross-linked muropeptide species, obtained by subtracting “−des-Ori” from “+des-Ori” in panel c. S. aureus grown in the presence of desleucyl-oritavancin shows an increase in PG monomers and dimers and a decrease in trimers and tetramers. (e) Cross-linking efficiencies and absence of pentaglycine cross-linking bridge. (f) Terminal PG stem lengths and cross-linking efficiency. All error bars represent standard errors of the mean (n = 3). An unpaired Student t test was used to calculate P values (*, P < 0.05; **, P < 0.01).
Effects of des-Ori on PG bridge link.
More pronounced transpeptidase inhibition activity by des-Ori is observed when cross-linking efficiencies are calculated for the muropeptide fragments based on the bridge link attachment at the donor PG stem. Figure 4e shows the calculated cross-linking efficiencies for muropeptides with or without the terminal pentaglycine bridge structure from isolated cell walls of S. aureus grown in the absence (red) or in the presence (blue) of des-Ori. Schematic representations of PG dimers are shown as insets. For S. aureus grown in the absence of des-Ori, the cross-linking efficiency for muropeptides without the terminal bridge structure (69%) is higher than that of muropeptides with the bridge (63%). Higher degrees of cross-linking for the muropeptides without the bridge indicate that these muropeptides are associated with highly cross-linked regions of the cell wall. With the addition of des-Ori, cross-linking efficiencies are reduced for both muropeptides with and without the terminal pentaglycine bridge structure. However, a greater degree of reduction in cross-linking is observed for the muropeptides without the bridge, which is reduced from 69% to 67% for S. aureus grown in the presence of des-Ori (see Table S1 in the supplemental material).
Effects of des-Ori on PG stem length.
The change in cross-linking efficiency was further characterized in relation to the PG stem length, which is modified by sequential cleavage of d-Ala–d-Ala by dd-, and ld-carboxypeptidases (Table S1). For untreated S. aureus, the cross-linking efficiency for muropeptides with a tripeptide stem was 65%, which is higher than that of muropeptides with a tetrapeptide stem (59%) (Fig. 4f). This indicates high ld-carboxypeptidase activity associated with the cross-linked PG in mature cell wall (18). However, when S. aureus is grown in the presence of des-Ori, the cross-linking efficiency for the muropeptides with a tripeptide stem decreases, while the efficiency for the muropeptides with a tetrapeptide PG stem increases. The reduction in dd-, and ld-carboxypeptidase activities suggest accumulation of immature cell wall in des-Ori-treated S. aureus.
Effects of des-Ori on PG acetylation.
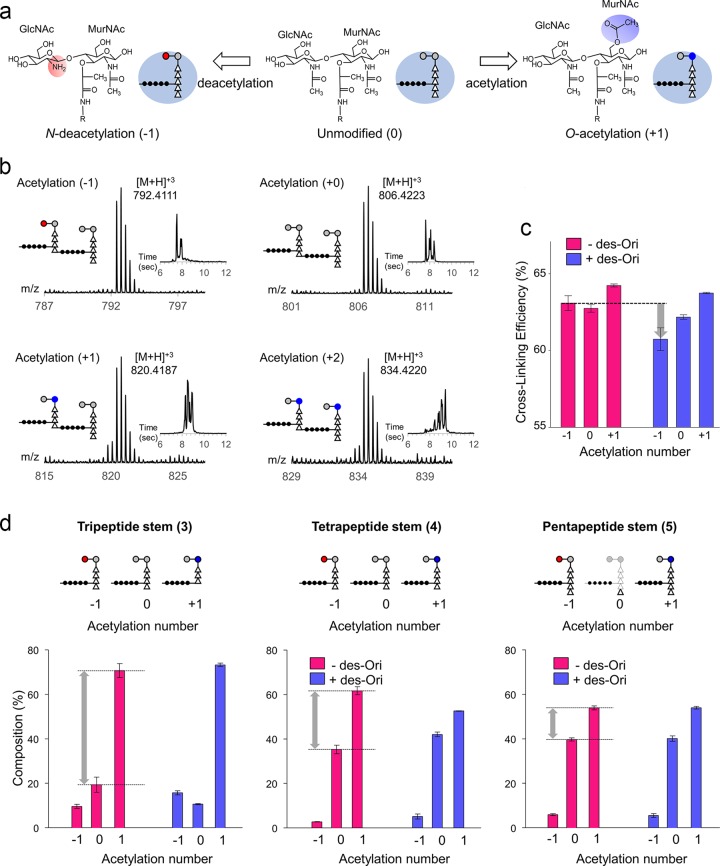
O-Acetylation of MurNAc and N-deacetylation of GlcNAc are two major PG modifications that confer lysozyme resistance to facilitate the evasion of host innate immunity (19) (Fig. 5a). Figure 5b shows the mass spectra and their corresponding extracted ion chromatograms (insets) of PG dimers for the net acetylation states of −1, 0, 1, and 2 from mutanolysin-digested cell walls of untreated S. aureus. An acetylation state of −1 represents N-deacetylation of a GlcNAc (Fig. 5b, red circle), 0 represents unmodified PG dimer, +1 represents O-acetylation of one MurNAc, and +2 represents O-acetylation of two MurNAcs (blue circles). Since each O-acetylation and N-deacetylation changes the muropeptide mass by 42 Da, the effects of des-Ori on the PG acetylation state were determined by characterizing the mass shifts in muropeptide fragments.
FIG 5.
Acetylation of S. aureus peptidoglycan. (a) The disaccharide of the PG repeat unit, MurNAc-GlcNAc, is chemically modified by O-acetylation of MurNAc (+1 Ac) or N-deacetylation of GlcNAc (−1). (b) Mass spectra and extracted ion chromatograms of PG dimers terminating with a pentapeptide stem with acetylation states of −1, 0, 1, and 2 from mutanolysin-digested cell walls of untreated S. aureus. A schematic representation of the proposed muropeptide structure for each acetylation state is shown as an inset. The extracted ion chromatograms (XIC) for the selected ions are shown as insets. (c) Acetylation number and cross-linking efficiency. (d) Normalized percent composition by acetylation number based on terminal PG stem lengths. All error bars represent standard errors of mean (n = 3).
The calculated cross-linking efficiencies for muropeptides with the acetylation states of −1, 0, and +1 are provided in Table S2 in the supplemental material and shown as a bar graph in Fig. 5c. The calculated ρCL values for untreated S. aureus are comparable for all acetylation states, except for slightly higher cross-linking observed for the muropeptides with an acetylation state of +1 (64%). With the addition of des-Ori, the cross-linking efficiencies for all acetylated muropeptide species in S. aureus are reduced. In particular, we found that the cross-linking efficiency of the N-deacetylated muropeptides was disproportionally affected by des-Ori (gray arrow). The specific role of PG N-deacetylation in S. aureus remains unclear, although N-deacetylation has been shown to be associated with increased pathogenicity and evasion of the host innate immune response by Enterococcus faecalis (20), Streptococcus pneumoniae (21), and Listeria monocytogenes (22). As O-acetylation has been shown to increase in cultures that enter the stationary growth phase (23), decreased cross-linking efficiencies for O-acetylated and N-deacetylated muropeptides suggest that des-Ori interferes with the cell wall maturation process.
PG acetylation and stem modifications.
The muropeptide composition was broken down into PG stem lengths and glycan acetylation state, and the normalized percentile compositions are provided in Table S3 in the supplemental material. Figure 5d shows bar graphs of percentile muropeptide composition by the PG stem structures, with tripeptide (left), tetrapeptide (middle), and pentapeptide (right) colored according to presence (blue) and absence (red) of des-Ori. The muropeptides with the tripeptide PG stem are heavily O-acetylated (+1), at approximately four times the abundance of unmodified muropeptides (0) and seven times that of the N-deacetylated muropeptides (−1). The difference in abundance between O-acetylated and unmodified muropeptides, shown as gray arrows (Fig. 5d), lessens as the PG stem length increases from tripeptide to pentapeptide. This result is consistent with preferential O-acetylation of the muropeptides with a tripeptide PG stem that are highly cross-linked (Fig. 4f). Hence, the cell walls of S. aureus grown in the absence of des-Ori have PG that is highly cross-linked with O-acetylated disaccharides with a tripeptide stem structure. In contrast, the cell walls of S. aureus grown in the presence of des-Ori resulted in PG with reduced cross-linking, reduced O-acetylation and N-deacetylation of disaccharides, and an increase in muropeptides with a tetrapeptide stem structure (Fig. 4f).
DISCUSSION
The interaction of glycopeptide antibiotics with the cell wall in vivo is complex and involves more than d-Ala–d-Ala binding (24). For disaccharide-modified lipoglycopeptides, the drug's hydrophobic side chain forms a secondary binding site that enables targeting of the cross-linked bridge structure of PG (Fig. 1d) (25–27). Unlike the Edman degradation product of vancomycin, which is devoid of any activity, des-Ori retains potent antimicrobial activities against both vancomycin-susceptible and -resistant pathogens despite the damaged d-Ala–d-Ala binding site (28, 29). This activity is mediated by the secondary binding site, which allows des-Ori binding to cross-linked PG in the cell wall (4, 7), as well as increased binding affinity to lipid II that has a pentaglycine bridge attached (30).
The PG composition analysis of S. aureus grown in the presence of des-Ori by LC-MS has determined that des-Ori inhibits the transpeptidation step of PG biosynthesis, which is in agreement with the reduced PG cross-linking in intact whole cells of S. aureus determined by solid-state NMR (31). In addition, LC-MS analysis has revealed that des-Ori preferentially reduces the cross-linking efficiencies of (i) muropeptides without a PG bridge (Fig. 4e), (ii) muropeptides with a tripeptide stem structure (Fig. 4f), and (iii) muropeptides that are N-deacetylated (Fig. 5c). Interestingly, des-Ori-treated S. aureus showed an overall increase in N-deacetylated muropeptides (Fig. 5d), but the cross-linking efficiency for these muropeptides was significantly reduced (see Table S2 in the supplemental material). We attribute these changes to des-Ori binding to the PG template at the inner layer of mature cell wall close to the cell membrane where the nascent PG is incorporated. This interaction with the PG template interferes with both the alignment and orientation of nascent PG stem and bridge structures, which are necessary for efficient incorporation of these new PGs into mature cell wall as mediated by the transpeptidase (26, 32–34). Thus, subsequent PG biosynthesis based on these defective templates results in the ensuing disorder of the cell wall, with reduced PG cross-linking, altered PG acetylation, and immature PG (improper PG stem modifications), which leaves bacteria highly susceptible to lysis.
MATERIALS AND METHODS
S. aureus cell wall isolation.
The procedure for the cell wall isolation is described elsewhere (10). des-Ori was added to the final concentration of 10 μg/ml to S. aureus (ATCC 6538P) grown in 200 ml of tryptic soy broth (TSB) at an OD600 of 0.6. After 100 min of further growth, the cells were harvested, resuspended in 2 ml of phosphate-buffered saline, and disrupted using a bead beater (Disruptor Genie; Scientific Industries) with 0.5-mm-diameter glass beads at 24°C. Eight cycles of 1 min of disruption separated by 1 min of rest to prevent overheating of the sample were used. Disrupted cells were separated from glass beads using the Steriflip 20-μm nylon net vacuum filtration system (EMD Millipore), followed by centrifugation at 25,000 × g for 3 min at 24°C. The remains of the cells were boiled in 2% sodium dodecyl sulfate solution for 30 min and then washed five times with deionized water through centrifugation at 25,000 × g for 3 min. The pellet was resuspended in 2 ml of 20 mM Tris buffer (pH 8.0), 200 μg of DNase was added to the suspension, and the sample was incubated for 24 h at 37°C. Three hundred micrograms of trypsin was added to the mixture, and it was incubated for additional 24 h at 37°C. The isolated cell walls were pelleted by centrifugation at 100,000 × g for 5 min at 24°C and washed with 20 mM Tris buffer, pH 8.0.
Mutanolysin digestion of S. aureus cell walls.
The cell wall isolate of S. aureus suspended in 2 ml of 20 mM Tris buffer (pH 8.0) was first digested using 500 U of mutanolysin (Sigma-Aldrich) at room temperature for 24 h. Five hundred units of mutanolysin was then added to the mixture and left for additional 24 h of incubation to ensure thorough digestion of the cell walls. Following the mutanolysin digestion, insoluble cell wall fragments were removed by use of a 0.45-μm centrifuge filter (Amicon), and the solubilized PG fraction was collected through a 30,000-molecular-weight-cutoff (MWCO) filter (Amicon) by centrifugation at 14,800 rpm (Thermo Scientific) for 5 min. Glycans in mutanolysin-digested muropeptides were reduced by adding sodium borohydride stock solution to final concentration of 10 mg/ml in 1 ml of borate buffer at pH 9.0. The reduction was carried out at 24°C for 30 min and quenched by addition of 120 μl of 85% phosphoric acid. The cell wall digest was frozen and lyophilized. Lyophilized peptidoglycan samples were cleaned for mass spectrometry analysis using 100-μl Pierce C18 tips (Thermo Scientific).
UPLC.
Waters C18 nanoACQUITY ultraperformance liquid chromatography (UPLC) and a Waters Synapt G2 high-definition mass spectrometer (HDMS) time-of-flight (TOF) mass analyzer were used to analyze the mutanolysin-digested peptidoglycan of S. aureus. For the UPLC separation, 1 μl of the sample in 50% acetonitrile–0.1% formic acid was injected into a 5-μl sample loop and loaded onto the column under isocratic condition of 99% buffer A (0.1% formic acid in methanol) and 1% buffer B (100% acetonitrile) for 1 min. The separation was carried out on a Waters nanoACQUITY reverse-phase C18 BEH column (75 μm by 100 mm; packed with 1.7-μm beads with a 130-Å pore size) with a nanoACQUITY Symmetry C18 trap column (180 μm by 20 mm; 5-μm beads with a 100-Å pore size) by applying a linear gradient to 50% buffer B for 60 min. After each separation, the column was regenerated under isocratic conditions with 85% buffer B for 5 min, followed by a linear gradient to 98% buffer A for 1 min, and then equilibrated at 98% buffer A for 23 min. The flow rate was constant at 0.450 μl/min throughout the analysis.
MS.
Eluents were analyzed with a Waters Synapt G2 HDMS-TOF mass analyzer operating under positive-ion mode. Electrospray ionization (ESI) was performed on the sample with a spray voltage of 35 V and a capillary voltage of 3.5 kV. The mass analyzer was optimized for an m/z range of 100 to 2,000. Fibrinopeptide B (Glu-Fib) was used as the calibration lock mass for the mass analyzer. Tandem mass spectrometry (MS/MS) fragmentation was accomplished through collision-induced dissociation with nitrogen gas at 120°C and 2.00 × 105 Pa. The transfer collision and the trap collision energies were 5 V and 30 V, respectively.
LC-MS data analysis.
Data analysis was carried out using MassLynx (Waters). MATLAB (MathWorks) was used for the calculation of exact masses and isotopic distributions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Adel Rafai Far from The Medicines Company for providing oritavancin and desleucyl-oritavancin for the study.
This paper is based on work supported by the National Institutes of Health Grant GM116130.
S.J.K. conceived and designed the experiments. J.D.C., E.E.F., A.N.T., and A.J.R. performed the experiments. J.D.C., E.E.F., A.N.T., and S.J.K. analyzed the data. All authors contributed to the writing of the manuscript.
We declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00278-17.
REFERENCES
- 1.Belley A, Arhin FF, Sarmiento I, Deng H, Rose W, Moeck G. 2013. Pharmacodynamics of a simulated single 1,200-milligram dose of oritavancin in an in vitro pharmacokinetic/pharmacodynamic model of methicillin-resistant Staphylococcus aureus infection. Antimicrob Agents Chemother 57:205–211. doi: 10.1128/AAC.01428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arhin FF, Sarmiento I, Moeck G. 2013. Oritavancin retains bactericidal activity in vitro against standard and high inocula of heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA). Int J Antimicrob Agents 41:397–398. doi: 10.1016/j.ijantimicag.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Arhin FF, Sarmiento I, Parr TR Jr, Moeck G. 2012. Activity of oritavancin and comparators in vitro against standard and high inocula of Staphylococcus aureus. Int J Antimicrob Agents 39:159–162. doi: 10.1016/j.ijantimicag.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 4.Kim SJ, Cegelski L, Stueber D, Singh M, Dietrich E, Tanaka KS, Parr TR, Far AR, Schaefer J. 2008. Oritavancin exhibits dual mode of action to inhibit cell-wall biosynthesis in Staphylococcus aureus. J Mol Biol 377:281–293. doi: 10.1016/j.jmb.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cegelski L, Kim SJ, Hing AW, Studelska DR, O' Connor RD, Mehta AK, Schaefer J. 2002. Rotational-echo double resonance characterization of the effects of vancomycin on cell wall synthesis in Staphylococcus aureus. Biochemistry 41:13053–13058. doi: 10.1021/bi0202326. [DOI] [PubMed] [Google Scholar]
- 6.Walsh CT, Fisher SL, Park IS, Prahalad M, Wu Z. 1996. Bacterial resistance to vancomycin: five genes and one missing hydrogen bond tell the story. Chem Biol 3:21–28. doi: 10.1016/S1074-5521(96)90079-4. [DOI] [PubMed] [Google Scholar]
- 7.Kim SJ, Matsuoka S, Patti GJ, Schaefer J. 2008. Vancomycin derivative with damaged d-Ala-d-Ala binding cleft binds to cross-linked peptidoglycan in the cell wall of Staphylococcus aureus. Biochemistry 47:3822–3831. doi: 10.1021/bi702232a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westwell MS, Bardsley B, Dancer RJ, Try AC, Williams DH. 1996. Cooperativity in ligand binding expressed at a model cell membrane by the vancomycin group antibiotics. Chem Commun (Camb) 3:580–590. [Google Scholar]
- 9.Williams DH. 1996. The glycopeptide story—how to kill the deadly ‘superbugs’. Nat Prod Rep 13:469–477. doi: 10.1039/NP9961300469. [DOI] [PubMed] [Google Scholar]
- 10.Kim SJ, Cegelski L, Studelska DR, O' Connor RD, Mehta AK, Schaefer J. 2002. Rotational-echo double resonance characterization of vancomycin binding sites in Staphylococcus aureus. Biochemistry 41:6967–6977. doi: 10.1021/bi0121407. [DOI] [PubMed] [Google Scholar]
- 11.Kim SJ, Tanaka KS, Dietrich E, Rafai Far A, Schaefer J. 2013. Locations of the hydrophobic side chains of lipoglycopeptides bound to the peptidoglycan of Staphylococcus aureus. Biochemistry 52:3405–3414. doi: 10.1021/bi400054p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SJ, Cegelski L, Preobrazhenskaya M, Schaefer J. 2006. Structures of Staphylococcus aureus cell-wall complexes with vancomycin, eremomycin, and chloroeremomycin derivatives by 13C{19F} and 15N{19F} rotational-echo double resonance. Biochemistry 45:5235–5250. doi: 10.1021/bi052660s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SJ, Singh M, Schaefer J. 2009. Oritavancin binds to isolated protoplast membranes but not intact protoplasts of Staphylococcus aureus. J Mol Biol 391:414–425. doi: 10.1016/j.jmb.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehta AK, Cegelski L, O'Connor RD, Schaefer J. 2003. REDOR with a relative full-echo reference. J Magn Reson 163:182–187. doi: 10.1016/S1090-7807(03)00078-8. [DOI] [PubMed] [Google Scholar]
- 15.Nitanai Y, Kikuchi T, Kakoi K, Hanamaki S, Fujisawa I, Aoki K. 2009. Crystal structures of the complexes between vancomycin and cell-wall precursor analogs. J Mol Biol 385:1422–1432. doi: 10.1016/j.jmb.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Chang JD, Foster EE, Yang H, Kim SJ. 2017. Quantification of the d-Ala-d-Lac-terminated peptidoglycan structure in vancomycin-resistant Enterococcus faecalis using a combined solid-state nuclear magnetic resonance and mass spectrometry analysis. Biochemistry 56:612–622. doi: 10.1021/acs.biochem.6b00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Driehuis F, de Jonge B, Nanninga N. 1992. Cross-linkage and cross-linking of peptidoglycan in Escherichia coli: definition, determination, and implications. J Bacteriol 174:2028–2031. doi: 10.1128/jb.174.6.2028-2031.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atrih A, Bacher G, Allmaier G, Williamson MP, Foster SJ. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J Bacteriol 181:3956–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke AJ, Dupont C. 1992. O-acetylated peptidoglycan: its occurrence, pathobiological significance, and biosynthesis. Can J Microbiol 38:85–91. doi: 10.1139/m92-014. [DOI] [PubMed] [Google Scholar]
- 20.Benachour A, Ladjouzi R, Le Jeune A, Hebert L, Thorpe S, Courtin P, Chapot-Chartier MP, Prajsnar TK, Foster SJ, Mesnage S. 2012. The lysozyme-induced peptidoglycan N-acetylglucosamine deacetylase PgdA (EF1843) is required for Enterococcus faecalis virulence. J Bacteriol 194:6066–6073. doi: 10.1128/JB.00981-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vollmer W, Tomasz A. 2002. Peptidoglycan N-acetylglucosamine deacetylase, a putative virulence factor in Streptococcus pneumoniae. Infect Immun 70:7176–7178. doi: 10.1128/IAI.70.12.7176-7178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, Psylinakis E, Bouriotis V, Hugot JP, Giovannini M, Coyle A, Bertin J, Namane A, Rousselle JC, Cayet N, Prevost MC, Balloy V, Chignard M, Philpott DJ, Cossart P, Girardin SE. 2007. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci U S A 104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfeffer JM, Strating H, Weadge JT, Clarke AJ. 2006. Peptidoglycan O acetylation and autolysin profile of Enterococcus faecalis in the viable but nonculturable state. J Bacteriol 188:902–908. doi: 10.1128/JB.188.3.902-908.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang J, Zhou H, Preobrazhenskaya M, Tao P, Kim SJ. 2016. The carboxyl terminus of eremomycin facilitates binding to the non-d-Ala-d-Ala segment of the peptidoglycan pentapeptide stem. Biochemistry 55:3383–3391. doi: 10.1021/acs.biochem.6b00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SJ, Schaefer J. 2008. Hydrophobic side-chain length determines activity and conformational heterogeneity of a vancomycin derivative bound to the cell wall of Staphylococcus aureus. Biochemistry 47:10155–10161. doi: 10.1021/bi800838c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh M, Kim SJ, Sharif S, Preobrazhenskaya M, Schaefer J. 2015. REDOR constraints on the peptidoglycan lattice architecture of Staphylococcus aureus and its FemA mutant. Biochim Biophys Acta 1848:363–368. doi: 10.1016/j.bbamem.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SJ, Singh M, Preobrazhenskaya M, Schaefer J. 2013. Staphylococcus aureus peptidoglycan stem packing by rotational-echo double resonance NMR spectroscopy. Biochemistry 52:3651–3659. doi: 10.1021/bi4005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge M, Chen Z, Onishi HR, Kohler J, Silver LL, Kerns R, Fukuzawa S, Thompson C, Kahne D. 1999. Vancomycin derivatives that inhibit peptidoglycan biosynthesis without binding d-Ala-d-Ala. Science 284:507–511. doi: 10.1126/science.284.5413.507. [DOI] [PubMed] [Google Scholar]
- 29.Goldman RC, Baizman ER, Longley CB, Branstrom AA. 2000. Chlorobiphenyl-desleucyl-vancomycin inhibits the transglycosylation process required for peptidoglycan synthesis in bacteria in the absence of dipeptide binding. FEMS Microbiol Lett 183:209–214. doi: 10.1111/j.1574-6968.2000.tb08959.x. [DOI] [PubMed] [Google Scholar]
- 30.Munch D, Sahl HG. 2015. Structural variations of the cell wall precursor lipid II in Gram-positive bacteria—impact on binding and efficacy of antimicrobial peptides. Biochim Biophys Acta 1848:3062–3071. doi: 10.1016/j.bbamem.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Kim SJ, Singh M, Sharif S, Schaefer J. 2017. Desleucyl-oritavancin with a damaged d-Ala-d-Ala binding site inhibits the transpeptidation step of cell-wall biosynthesis in whole cells of Staphylococcus aureus. Biochemistry 56:1529–1535. doi: 10.1021/acs.biochem.6b01125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharif S, Kim SJ, Labischinski H, Chen J, Schaefer J. 2013. Uniformity of glycyl bridge lengths in the mature cell walls of fem mutants of methicillin-resistant Staphylococcus aureus. J Bacteriol 195:1421–1427. doi: 10.1128/JB.01471-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SJ, Singh M, Sharif S, Schaefer J. 2014. Cross-link formation and peptidoglycan lattice assembly in the FemA mutant of Staphylococcus aureus. Biochemistry 53:1420–1427. doi: 10.1021/bi4016742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SJ, Chang J, Singh M. 2015. Peptidoglycan architecture of Gram-positive bacteria by solid-state NMR. Biochim Biophys Acta 1848:350–362. doi: 10.1016/j.bbamem.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.