ABSTRACT
The complex carbohydrates of terrestrial and marine biomass represent a rich nutrient source for free-living and mutualistic microbes alike. The enzymatic saccharification of these diverse substrates is of critical importance for fueling a variety of complex microbial communities, including marine, soil, ruminant, and monogastric microbiota. Consequently, highly specific carbohydrate-active enzymes, recognition proteins, and transporters are enriched in the genomes of certain species and are of critical importance in competitive environments. In Bacteroidetes bacteria, these systems are organized as polysaccharide utilization loci (PULs), which are strictly regulated, colocalized gene clusters that encode enzyme and protein ensembles required for the saccharification of complex carbohydrates. This review provides historical perspectives and summarizes key findings in the study of these systems, highlighting a critical shift from sequence-based PUL discovery to systems-based analyses combining reverse genetics, biochemistry, enzymology, and structural biology to precisely illuminate the molecular mechanisms underpinning PUL function. The ecological implications of dynamic PUL deployment by key species in the human gastrointestinal tract are explored, as well as the wider distribution of these systems in other gut, terrestrial, and marine environments.
KEYWORDS: Bacteroidetes, carbohydrate, carbohydrate-active enzymes (CAZymes), metabolism, microbiome, polysaccharide utilization loci (PULs), polysaccharides, symbiosis
INTRODUCTION
Complex carbohydrates, in the form of structural and storage polysaccharides, constitute the largest repository of metabolically accessible carbon in the biosphere (1, 2). The result of primary production, biomass carbohydrates thus present a ubiquitous energy source to fuel microbial life in both terrestrial and marine ecosystems (Fig. 1). Carbohydrate utilization is inextricably linked with the ability of microbes to persist in environments as diverse as freshwater, salt water, soil, and animal gastrointestinal tracts, particularly where competition for a common, and potentially temporally limited, pool of nutrients is fierce. The advent of improved culturing techniques and next-generation sequencing has granted us coveted access to a repository of genetic clues to the metabolic potential of key species across ecosystems (3–10). We are now in an era in which there is urgent need for post(meta)genomic functional analysis to elucidate the interplay between carbohydrate catabolism and microbial ecosystem dynamics.
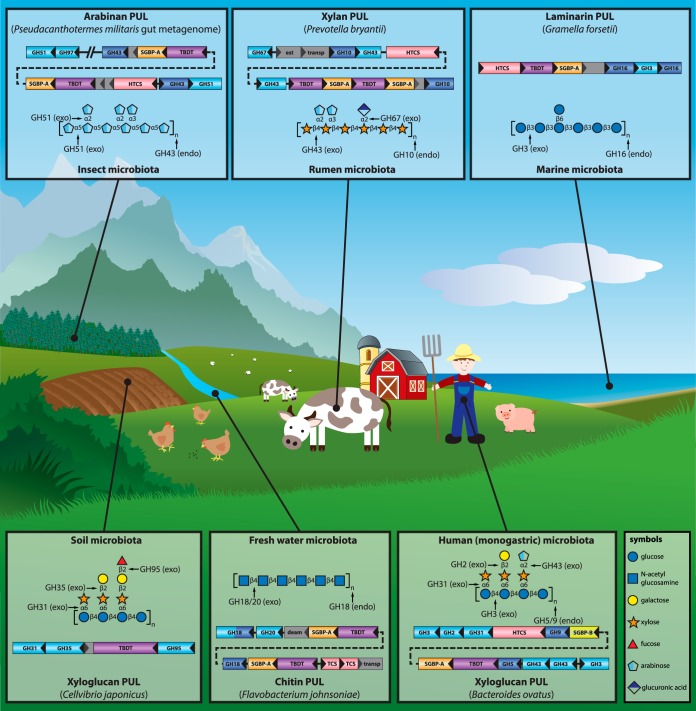
FIG 1.
The ecological distribution of PULs in nature. PULs are found in a variety of microbial communities, highlighting the global role of this polysaccharide utilization strategy. Each semitransparent box contains a representative bacterial PUL from a distinct microbial ecosystem, as well as the schematic structure of the target glycan. Clockwise from the top left are the arabinan PUL from an unidentified bacterium from the gut of Pseudocanthotermes militaris (93), the xylan PUL from Prevotella bryantii (97), the laminarin PUL from Gramella forsetii (101), the xyloglucan PUL from Bacteroides ovatus (78), the chitin PUL from Flavobacterium johnsoniae (112), and the xyloglucan PUL from Cellvibrio japonicus (126). Genes are colored according to protein function as follows: blue, endo-GH; cyan, exo-GH; orange, SusD-homologous SGBP; yellow, other SGBP; purple, TBDT; pink, (hybrid) two-component sensor (HTCS/TCS); gray, unknown or other function (est, esterase; transp, transporter; deam, deaminase). Monosaccharides are represented by Consortium for Functional Glycomics symbols (154).
THE UBIQUITY OF COMPLEX CARBOHYDRATES AND CAZymes
Complex carbohydrates—oligosaccharides and polysaccharides—are composed of a tremendous diversity of monosaccharide subunits and glycosidic linkages (11–13) (see Fig. 1 for examples). Therefore, a correspondingly large array of specific enzymes is required to effect complete saccharification and feed primary metabolism. Hence, there is demonstrable enrichment of genes encoding carbohydrate-active enzymes (CAZymes) in saprophytic and pathogenic microorganisms that attack plant and algal cell walls (14–17). Despite the complexity of dietary carbohydrates entering animal gastrointestinal tracts daily, most animal genomes are remarkably bereft of CAZyme-encoding genes (14, 18, 19). Humans, for example, are intrinsically able to digest only a small group of relatively simple dietary carbohydrates, namely, starch, lactose, and sucrose (14). These observations have spurred considerable interest, dating back decades, in the contribution of complex carbohydrate degradation by intestinal microbiota to the nutrition of monogastric and ruminant animals (14, 20–24).
The CAZyme classification initiated by Bernard Henrissat is a key foundation for genomic, biochemical, and structural studies of the proteins and enzymes involved in complex carbohydrate degradation (25, 26). The CAZy database presently groups, on the basis of amino acid sequence, 145 families of glycoside hydrolases (GHs), 103 families of glycosyltransferases, 26 families of polysaccharide lyases (PLs), 16 families of carbohydrate esterases (CEs), 13 families of redox auxiliary activities, and 81 families of associated noncatalytic carbohydrate-binding modules (27–31; see also www.cazy.org and www.cazypedia.org). A key feature of the CAZy classification is the dissection of open reading frames to reveal discrete, and sometimes complex, CAZyme modular organization (30), which significantly increases the accuracy of bioinformatic analyses.
The predictive power of the CAZy classification has proven to be remarkably robust. Within a given family, key active-site residues, the catalytic mechanism, and the overall three-dimensional fold are strictly conserved (with very few exceptions [32]), while some families are further grouped into clans on the basis of a conserved catalytic mechanism and tertiary structure (25). Substrate specificity is, however, less easily divined because many large families (some with tens of thousands of members) are “polyspecific,” i.e., encompassing members with distinct activity profiles, often on structurally related complex carbohydrates. Here, further division into subfamilies has been shown to be beneficial (28, 33–37). However, because of a comparative paucity of biochemically and structurally characterized members vis-à-vis the vast bulk of (meta)genomic sequence data (25-27, 38), bioinformatic predictions of CAZyme function in the context of microbial community ecology are still largely naive. Therefore, there is considerable scope to advance the field through a concerted, systems-based approach that incorporates microbial genetics, biochemistry, enzymology, and structural biology.
BACTEROIDETES AND THE PUL PARADIGM
Members of the Gram-negative phylum Bacteroidetes are widespread across diverse ecological niches, including marine, freshwater, and terrestrial habitats, and are notably abundant in microbiota of the alimentary canal. For example, Bacteroidetes bacteria, together with the members of the Gram-positive phylum Firmicutes, dominate the microbiota of the human colon (39, 40). Bacteroidetes bacteria are also profuse in the guts of plant biomass-consuming nonhuman animals (40–45). In animals, including humans, Bacteroidetes bacteria provide many symbiotic benefits, notably, the production of short-chain fatty acids by hydrolysis and fermentation of otherwise indigestible complex carbohydrates, which are absorbed and utilized by the epithelial cells of the gut (40, 46, 47).
A unique feature of Bacteroidetes genomes is the presence of polysaccharide utilization loci (PULs), a term first coined by Bjursell, Martens, and Gordon in 2006 (48) to describe clusters of colocalized, coregulated genes, the products of which orchestrate the detection, sequestration, enzymatic digestion, and transport of complex carbohydrates (49–51). PULs encode a complement of cell surface glycan-binding proteins (SGBPs), TonB-dependent transporters (TBDTs), CAZymes (most frequently GHs, but also PLs and CEs where substrate appropriate), and carbohydrate sensors/transcriptional regulators. The complexity of PULs often scales with that of their cognate substrates (Fig. 1) (52, 53) and may include ancillary enzymes such as proteases (54), sulfatases (55, 56), and phosphatases (57). These elegant systems constitute the major nutrient acquisition strategy deployed by Bacteroidetes bacteria and thus are intrinsically linked to the colonization of nutritional niches and the establishment of microbial ecosystems.
THE ARCHETYPAL PUL, THE Sus
The first evidence of a concerted molecular system for complex glycan degradation in Bacteroidetes bacteria was uncovered through pioneering studies of dietary starch utilization by the human gut symbiont Bacteroides thetaiotaomicron, which were initiated in the 1980s by Abigail Salyers and coworkers. Notably, initial cellular fractionation studies revealed that key enzymes and starch-binding proteins were individually localized to the cell surface, periplasmic space, and cytoplasm, thus suggesting the presence of a multiprotein carbohydrate-degrading system spanning both bacterial membranes (58). Indeed, subsequent studies precisely identified all of the components of this system, including proteins responsible for recognition and initial hydrolysis of starch at the outer membrane, translocation of glycans into the periplasm, further hydrolysis to monosaccharides, and transcriptional regulation (59–66). Together, eight genes were identified as part of a single gene cluster, collectively named the starch utilization system (Sus), which established a new paradigm of complex carbohydrate utilization (49, 51). Presently, the concerted operation of the Sus continues to be dissected through genetic, biochemical, and structural approaches (67–71) that, together with Salyers' seminal studies, outline a general cellular model for the study of other PULs (Fig. 2).
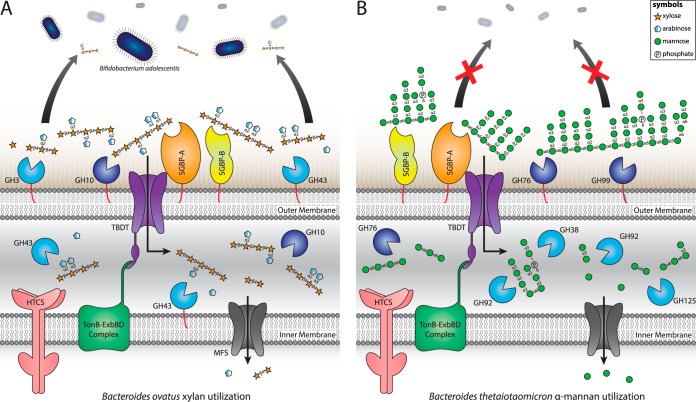
FIG 2.
Nutritional foraging strategies encoded by PULs and their roles in microbial ecological interactions. (A) In a distributive mechanism, utilization of wheat arabinoxylan by B. ovatus releases partial breakdown products (PBPs) that diffuse into the extracellular environment and support the growth of B. adolescentis (79). (B) In contrast, the selfish mechanism employed by B. thetaiotaomicron in the digestion of yeast α-mannans results in the rapid import of extracellular products into the periplasm, where saccharification is culminated (80). The concerted actions of these two PUL models drive syntrophic and cooperative networks in the context of the complex microbial environment of the gut microbiota. Proteins are colored as in Fig. 1. Monosaccharides are represented by Consortium for Functional Glycomics symbols (154).
MOLECULAR ARCHITECTURES OF PUL SUBSYSTEMS
A hallmark of canonical PULs is the presence of at least one sequential pair of susC and susD homologs (49) that encode an outer membrane TBDT (Fig. 1 and 2, purple) and an N-terminally lipidated SGBP (Fig. 1 and 2, orange), respectively. In light of the considerable structural diversity among SGBPs (see below), SusD homologs are referred to as SGBP-A proteins. Genetic studies have revealed that the TBDT and SGBP-A proteins are intimately associated; deletion of SGBP-A eliminates or reduces PUL function, yet growth can be rescued by complementation with SGBP-A variants in which substrate binding has been eliminated by site-directed mutagenesis (49, 67, 72). SGBP-A homologs show strong primary, secondary, and globular tertiary structural conservation, with topological variation in the extended substrate-binding surface accounting for carbohydrate specificity (72–76). Recent seminal crystal structures have revealed that SGBP-A homologs form flexible “lids” at the extracellular opening of their associated TBDTs and further highlight an integral role for SGBPs-A in selective nutrient transit (77).
Substrate binding is often assisted by one or more structurally distinct SGBPs (e.g., SGBP-B) (Fig. 1 and 2, yellow) that may have a specificity identical or complementary to that of SGBP-A. The lack of sequence conservation of the genes that encode these proteins often precludes definitive assignment as SusE (or SusF) homologs; hence, these auxiliary SGBPs are sometimes referred to as SusE positioned (or SusF positioned) with reference to their relative genetic organization. Despite this absence of sequence similarity, the crystal structures obtained to date have revealed that these N-terminal lipoproteins share extended multidomain tertiary structures that present substrate-binding faces in one or more distal C-terminal domains (70, 72).
Each PUL also contains a complement of CAZymes tasked with the dismantling of polysaccharides, beginning with the action of one or more cell surface-anchored endoglycanases (Fig. 1 and 2, blue) (78–80); in the Sus, this role is fulfilled by SusG, a GH13 endo-α1,4-glucanase (α-amylase) (65, 69). The resulting fragments are actively shuttled into the periplasmic space by the TBDT (e.g., SusC [61, 64]), where additional linkage-specific GHs (Fig. 1 and 2, cyan) act to release the component monosaccharides (or certain disaccharides) for metabolism in the cytosol. In the Sus, SusA (a GH13 α1,4-glucosidase) and SusB (a GH97 α1,6-glucosidase) are sufficient to hydrolyze all of the linkages in starch oligosaccharides (62, 71), while PULs directed toward more complex substrates typically have manifold exoglycosidases (reviewed in references 52 and 53; see Fig. 1 and 2 and below for specific examples).
PUL regulation is most commonly mediated by one of three mechanisms, the SusR sensor/regulator, extracytoplasmic function sigma (ECF-σ) factor–anti-σ-factor pairs, or hybrid two-component systems (HTCSs) (Fig. 1 and 2, pink). Other PUL-associated regulatory mechanisms include LacI, CRP, AraC (non-HTCS), SARP-OmpR, and classic TCSs (81). SusR is a predicted inner membrane-spanning receptor that binds starch-derived oligosaccharides (but not glucose) and triggers the upregulation of the remaining sus genes (63). Intriguingly, SusR appears to be the exception, rather than the rule, in the documented PUL catalogue (82). Rather, PUL regulation is most often orchestrated by ECF-σ–anti-σ pairs that are commonly associated with PULs targeting host-derived glycans (83) or HTCS proteins associated with PULs targeting a variety of plant cell wall carbohydrates (39, 84, 85). Regardless of the regulatory system utilized by a specific PUL, there generally appears to be a finely tuned interpretation of complex glycan signals that is necessary for a targeted, dynamic response; with some exceptions, monosaccharides are typically not inducers (39, 83, 86–89) and may, in fact, repress PUL expression (90).
PULOMICS
As introduced above, susC-susD pairs are hallmarks of PULs and have been used to enumerate PUL complements among the genomes of key human gut symbionts, including B. thetaiotaomicron (88 PULs), Bacteroides ovatus (126 PULs), and Bacteroides cellulosilyticus WH2 (113 PULs) (39, 48, 89). The abundance and diversity of PULs have been well documented as a result of these initiatives, which have enabled the comparative analysis of PULs from various gut Bacteroidetes bacteria and provided an essential foundation to understand nutrient niche colonization and community dynamics. For example, the genomes of B. thetaiotaomicron and B. ovatus both harbor ca. 100 PULs; however, strikingly few homologous PULs are shared between them, suggesting that these two symbionts have distinct glycan niches (39). Similarly, recent transcriptomic analyses indicated that B. xylanisolvens dynamically responds to discrete structures of pectins and xylans through several differentially regulated PULs (91, 92). Such studies reflect the exquisite nutrient adaptation of individual Bacteroidetes bacteria.
Beyond the human gut, PULs have increasingly been identified in a variety of Bacteroidetes species (Fig. 1) through (meta)genomic, transcriptomic, and proteomic approaches (93–96). PULs have been identified in species outside the Bacteroides genus in diverse environments, including the ruminal Prevotella bryantii (97), the freshwater dweller Flavobacterium johnsoniae (98), and the gut microbiota of the termite Pseudacanthotermes militaris (99). Distinctly, PULs from the gliding soil bacterium Cytophaga hutchinsonii lack individual susC-susD pairs, but instead, two such pairs are encoded elsewhere in the genome (100).
The discovery of specialized CAZymes tasked with dismantling unique polysaccharides is particularly well illustrated by the PULs of several marine species that utilize highly sulfated algal polysaccharides. For example, PULs from Zobellia galactanivorans (55), Formosa agariphila (56), and Gramella forsetii (101) contain CAZyme portfolios tuned to the unique monosaccharide residues of algal polysaccharides and, not surprisingly, are also enriched in sulfatases (see also SulfAtlas, a new sulfatase classification database from the Marine Glycobiology and ABiMS teams at the Station Biologique de Roscoff [102; http://abims.sb-roscoff.fr/sulfatlas/]).
Recently, the development of automatic prediction tools by the CAZy team has led to the genesis of the PUL database (PULDB; http://www.cazy.org/PULDB/index.php). The PULDB presently catalogs ca. 4,000 predicted PULs from >70 Bacteroidetes bacteria, including Alistipes, Bacteroides, Dysgonomonas, Odoribacter, Parabacteroides, Paraprevotella, Prevotella, and Tannerella species. A key feature of the PULDB is that it, like the CAZy database, is anchored by experimentally characterized PULs (81).
INSIGHT FROM INTEGRATED FUNCTIONAL CHARACTERIZATION
Large-scale (meta)genomic approaches have clearly been instrumental in PUL discovery, as well as predicting the metabolic potential of diverse Bacteroidetes bacteria. However, refined functional characterization at the molecular and cellular levels remains critical for a full understanding of the roles of PULs in microbial communities. In a few cases, genomic and transcriptomic studies have been coupled with biochemical analyses of individual CAZymes, e.g., β-xylanases (103–105), arabinofuranosidases (93), β-glucanases (106, 107), and alginate lyases (108). Recently, a series of high-impact studies have combined genetic, enzymological, biophysical, and structural techniques to comprehensively characterize the molecular functions of individual PULs.
The pioneering study deploying this approach described the differential utilization of the fructans levan [β(2,6) linked] and inulin [β(2,1) linked] by several human-gut symbiotic Bacteroides species, revealing that each harbors a set of linkage-specific enzymes that are instrumental in defining nutritional preferences (86). Notably, the heterologous expression of these enzymes in species that lacked homologous activities resulted in increased fitness of the recipient on the target polysaccharide, showcasing that differences in gene content between species can translate to increased fitness on inaccessible substrates. This work also represents the first structural and functional information obtained for the HTCS, highlighting the importance of the periplasmic sensor domain in the binding of small oligosaccharides. Subsequent insightful structural biology revealed that this binding event was accompanied by a unique “scissor blade” closing mechanism thought to aid in the transduction of the signal across the membrane to trigger the upregulation of associated PUL genes (88).
Starch and fructans, discussed above, are comparatively simple storage polysaccharides that are composed of a single monosaccharide repeating unit. One of the first examples of the comprehensive characterization of a PUL directed toward a more complex plant cell wall polysaccharide was that of the xyloglucan utilization locus (XyGUL) from the human gut symbiont B. ovatus (Fig. 1) (78). The saccharification pathway of this ubiquitous, highly branched dietary glycan was determined through the biochemical and structural characterization of all eight GHs and two SGBPs from the XyGUL, in harness with reverse genetics (72, 78, 109). These studies were instrumental in highlighting the adaptive evolution of GH cohorts among syntenic XyGULs. Furthermore, these syntenic XyGULs served as diagnostic markers of xyloglucan metabolic capacity among individual Bacteroides species and across human gut metagenomes (78).
Subsequently, comprehensive functional studies of PULs targeting other complex polysaccharides have been reported. A pair of xylan-targeting PULs from B. ovatus, PUL-XylS and PUL-XylL, was found to encode enzymes tailored for individual plant β-xylans that varied in their composition and branching (79). Recently, the detailed genetic, biochemical, and enzyme structural characterization of a galactomannan-specific PUL from B. ovatus revealed the interplay of two mannan-specific SGBPs, two GH26 endo-β-mannanases, and a GH36 exo-α-galactosidase in the deconstruction of this plant cell wall polysaccharide (110, 111). Among environmental bacteria, a complex chitin utilization locus from Flavobacterium johnsoniae has been extensively functionally characterized (112). Notable features of the system include two pairs of SusC/SusD homologs, a secreted chitinase composed of two GH18 modules separated by a chitin-binding module, and an intracellular glucosamine-6-phosphate deaminase. Taken together, these studies highlight the considerable insight systems-based analysis can bring to PUL structure-function studies in the context of the ecology of a variety of ecosystems.
In addition to common plant cell wall polysaccharides, specialized PULs devoted to the utilization of rare polysaccharides act to enhance the catabolic repertoire of selected gut species. The dynamic effects of the human diet on the adaptive evolution of the distal gut microbiota were recently highlighted in the seminal “sushi factor” study, which documented the presence of β-porphyranases (i.e., GH16 and GH86), algal polysaccharide-specific CAZymes, in the microbiota of Japanese populations (113). Notably, these CAZymes were found as part of PULs thought to be acquired by the gut bacterium Bacteroides plebeius via lateral gene transfer from porphyranolytic Z. galactanivorans associated with uncooked edible algae (i.e., nori) (114). Further evidence suggests that B. plebeius and other human gut Bacteroides spp. may have also acquired algal polysaccharide utilization genes from marine bacteria (108, 114).
The ability of gut bacteria to adapt to structurally complex dietary polysaccharides was highlighted by the discovery and detailed characterization of three PULs from B. thetaiotaomicron involved in the utilization of α-mannans from the yeast cell wall (80). Yeast residues in the intestine originate from either endogenous yeasts or the consumption of leavened foods and fermented beverages, products of technologies that have existed for only a few thousand years (57). Detailed biochemical and reverse genetic analyses of these B. thetaiotaomicron α-mannan and other PULs have been instrumental in enhancing our understanding of the roles of PUL acquisition in the evolving landscape of the gut. In this context, it is especially notable that one of the three B. thetaiotaomicron mannan PULs is located on a mobile element that is structurally similar and homologous to that harboring porphyran utilization genes in B. plebeius (53).
GIVE AND TAKE: THE ROLES OF GLYCAN UTILIZATION IN GUT ECOLOGY
The coordinated carbohydrate utilization systems of PULs represent an impressive evolutionary solution for capturing valuable carbon sources in competitive environments, which avoid the limitations of extracellular systems such as cellulosomes or freely diffusing enzymes employed by fungi and other bacteria. However, emerging research suggests that, in some cases, these apparently selfish PUL systems may be “leaky,” with particular benefit to the community; partial breakdown products (PBPs) released by the action of certain PULs can be shared with neighboring bacteria and support the dynamic response of microbial communities (115, 116).
This distributive mechanism has been observed in xylan utilization by B. ovatus, where simple oligosaccharides produced at the cell surface diffuse into the extracellular environment and are utilized by Bifidobacterium adolescentis, a species lacking the enzymatic machinery to catalyze this initial depolymerization step (Fig. 2A) (79). Interestingly, this form of syntrophy was observed only during the utilization of simple, linear xylans. These synergistic interactions therefore appear to be glycan and species specific and may reflect hierarchies in the selective metabolism of substrates. In this regard, the preferential degradation of some glycans over others is likely to play a central role in shaping the complex microbial relationships of the microbiota (117, 118).
The PUL-mediated liberation of PBPs contributes to the complex metabolic web of cross-feeding interactions that has been mapped between several Bacteroidales type strains (115), although in the context of the entire gut microbiota, these relationships are likely to be much more complex. For example, in some species, CAZymes are selectively packaged into outer membrane vesicles and released into the extracellular environment, where they are thought to mediate the production of free glycan fragments for use by the greater gut community (115, 119). Remarkably, certain species, such as B. ovatus, secrete enzymes that are not required for the utilization of glycans such as inulin by the bacterium itself or by its clonemates; rather, this effort appears to benefit other species in the gut community (116). This seemingly altruistic act results in significant fitness benefits for B. ovatus that are realized only in the context of a complex microbiota.
In contrast to the extensive and complex dynamic relationships that exist between cohorts of bacteria in the gut, certain species such as B. thetaiotaomicron exhibit relatively little collaboration during the digestion of complex glycans. This form of selfish metabolism is deployed by B. thetaiotaomicron during the utilization of yeast α-mannans, in which manno-oligosaccharides generated at the cell surface are rapidly imported into the periplasm for further breakdown, conferring no direct benefits on neighboring species (Fig. 2B) (80). To facilitate this process, surface mannanases appear to operate at a lower rate than homologous mannanases within the periplasm, ensuring that mass action effects do not impede transport.
As a result of the acquisition of rare carbohydrate-specific PULs and diverse glycan utilization strategies, glycan “generalists,” or microorganisms capable of metabolizing a range of glycans, within the gut are endowed with multiple foraging strategies to ensure their survival in this highly competitive ecosystem.
EXTENDING THE PUL PARADIGM
As discussed above, the concept of the PUL was originally defined in the context of Bacteroidetes systems containing the hallmark tandem susC-susD homologs encoding TBDT-SGBP pairs (48, 49, 81). TBDTs are not specific to Bacteroidetes bacteria but are broadly distributed across Gram-negative bacteria, including alpha- and gammaproteobacteria living in association with biomass debris. Inspection of the genomes of such organisms reveals that TBDT- and CAZyme-encoding genes may be colocalized—analogous to canonical Bacteroidetes PULs—although susD homologs and sensor/regulator systems are notably absent (108, 120–122). Despite their limitations, these TBDT/CAZyme-encoding clusters thus arguably comprise a type of “polysaccharide utilization locus.” Indeed, the coordinated action of such loci in the utilization of complex carbohydrates, including xylans and N-glycans, was first demonstrated in the plant pathogen Xanthomonas campestris pv. campestris (123–125). Arlat and coworkers thus advanced the term CUT (carbohydrate utilization locus-containing TBDT) to describe such systems (123).
Recently, an “abbreviated” XyGUL from the soil-dwelling, saprophytic gammaproteobacterium Cellvibrio japonicus (Fig. 1) has been the subject of reverse genetic, biochemical, and structural studies that highlight the concerted action of a TBDT and three periplasmic, side chain-specific exoglycosidases (126). Vis-à-vis the “complete” XyGUL of B. ovatus (Fig. 1) (78), the C. japonicus XyGUL lacks genes encoding a keystone extracellular endoxyloglucanase, which is provided elsewhere in the genome (127, 128). The lack of genes encoding a SusD (SGBP-A) homolog and a sensor/regulator system in the C. japonicus XyGUL, both of which are ubiquitous in Bacteroidetes PULs, mirrors observations in bacteria from other phyla (120). Furthermore, the TBDTs from C. japonicus and B. ovatus XyGULs have distinct amino acid sequences, despite being functionally homologous (50, 126).
Although necessarily distinct in structure from their Gram-negative counterparts, Gram-positive Firmicutes bacteria also deploy elaborate cell surface-associated systems for the utilization of soluble and insoluble polysaccharides, especially cellulose and resistant starch (40, 129–133). Recently, inducible, substrate-specific gene clusters targeting a variety of plant- and host-based glycans were identified in the genomes of the human gut symbionts Eubacterium rectale and Roseburia species (134). These “Gram-positive PULs” (gpPULs) contain a minimum of one CAZyme, a carbohydrate transport system (most commonly ATP-binding cassette transporters), and a LacI- or AraC-like transcriptional regulator (134).
In the broader perspective, the identification of colocalized genes encoding CAZymes and transporters presents a valuable tool for bioinformatic analyses of complex carbohydrate utilization in bacteria. Continued comprehensive molecular characterization of all flavors of PUL systems will be crucial for understanding the evolution of nutrient acquisition across phyla and ecosystems.
CONCLUSION AND FUTURE PERSPECTIVES
Recent studies have clearly demonstrated the importance of combining genetic, biochemical, and structural tools to fully dissect the function of individual PULs and reveal their specific roles in mediating the dynamics of carbohydrate utilization in diverse environments. In turn, well-characterized PULs serve as genetic markers, enabling the prediction of complex carbohydrate metabolism with greater reliability. A picture that has emerged is that individual Bacteroidetes bacteria in complex environments, such as the human gut, contain partially overlapping sets of PULs, which indicate both the ability to respond dynamically to nutrient availability and niche specialization within a web of species. Further, comparison of syntenic PULs across species highlights the ongoing evolution of PUL specificity through stepwise changes in CAZyme cohorts (78, 79, 86).
In light of the limited number of biochemically characterized PUL CAZymes, SGBPs, TBDTs, and sensor-regulators, molecular structure-function studies are only in their infancy. The generally small number of characterized CAZymes versus available sequence data and the limits this places on functional prediction have been discussed above. With respect to substrate binding and import, the interplay among SGBPs, TBDTs, and endoglycanases remains to be fully elucidated at the molecular level (68).
Likewise, the mechanisms by which regulatory systems within Bacteroidetes PULs respond to carbohydrates and how signals are transduced into gene expression are not fully understood. In particular, a complete structural and functional portrait of these complex membrane-spanning systems, as well as a detailed understanding of the genetic signatures targeted by these proteins (135), will help fill a significant gap in our understanding of how PULs are regulated and may help usher in an era of designer communities and personalized intestinal medicine (136–138). Recently, a series of cis-encoded small RNAs were discovered in association with a subset of PULs in Bacteroides fragilis (139). These molecules are postulated to play a role in the suppression of host glycan-specific PUL systems in Bacteroides species, potentially adding a new layer of regulation to the strictly controlled hierarchical expression of these PULs. The role of monosaccharides in specific feedback inhibition of PUL expression is likewise an emerging area (90).
Functional genomic studies of PULs (broadly defined here to include all aspects of molecular characterization from transcriptomics though structural biochemistry) have provided critical insights into nutrient acquisition strategies and microbial ecological interactions. These insights will continue to deepen our understanding of microbial enzyme systems in human and animal nutrition and health, as well as their involvement in driving fundamental environmental processes such as global carbon cycling. Successively, these basic research initiatives have lasting impacts on a variety of industries, serving to inform a wave of novel technologies with applications in industrial enzyme discovery (140, 141) and engineered microbial therapeutics (142–148).
Rapid advances toward next-generation solutions for animal agriculture that address losses in productivity, food safety, and/or food security that result from escalating restrictions on the use of antimicrobial growth promoters are paramount. In this regard, rigorous evaluation of prebiotic and probiotic outcomes (149) and establishment of realistic production benchmarks are mandatory before there will be further adoption by industry. Alternatively, the engineering of intestinal microorganisms, such as chimeric live vaccines (150, 151), CRISPR-based genome editing (152), and synthetic biology of secondary metabolism (153), holds vast potential and may ultimately transform how food is produced in the future.
Collectively, fundamental research on PUL function informs the development of a range of next-generation technologies aimed at the intentional manipulation of microbial communities, including bioengineered inducible synbiotic systems for metabolic selection and in vivo targeted delivery of therapeutics.
ACKNOWLEDGMENTS
Work in Lethbridge was supported by Alberta Innovates BioSolutions (grant BIOFS026) and Agriculture and Agri-Food Canada. Work in Vancouver was supported by operating grants from the Canadian Institutes of Health Research (MOP-137134 and MOP-142472) and infrastructure support from the Canadian Foundation for Innovation and the British Columbia Knowledge Development Fund.
Biographies

Julie M. Grondin obtained her B.Sc. (honors) in biochemistry from Queen's University in 2009, completing her undergraduate thesis project on protein-carbohydrate interactions under the supervision of Steven Smith. Fascinated by this project, she remained in Smith's group, where she deepened her study of these interactions, culminating in the completion of her Ph.D. in biochemistry in 2014. Specifically, her doctoral thesis focused on utilizing complementary structural biology methods, including nuclear magnetic resonance, X-ray crystallography, and small-angle X-ray scattering, to study the molecular determinants of carbohydrate recognition by large multimodular GHs from gut bacteria. In 2014, she joined D. Wade Abbott's group at Agriculture and Agri-Food Canada as a postdoctoral fellow, where she is currently engineering metabolic selection into a commensal gut bacterium with the purpose of applying these technologies toward antimicrobial therapeutic delivery in the gut.

Kazune Tamura obtained his B.Sc. degree from the University of British Columbia (UBC) in the Combined Honors in Biochemistry and Chemistry program, during which time he completed an undergraduate thesis project in the laboratory of Harry Brumer. His interest in enzymology and structural biology, combined with experiences in the laboratories of Lindsay Eltis (UBC) and Shinya Fushinobu (University of Tokyo), led to a fruitful year that motivated him to stay for graduate studies. He is now a second-year biochemistry and molecular biology Ph.D. student in the Brumer group working on elucidating the mechanism of microbial glycan utilization. Kazune considers himself fortunate to have the opportunity to collaborate with numerous world-class structural biologists and microbial geneticists, both locally and internationally.

Guillaume Déjean completed a 4-year degree program in bioengineering with a specialization in plant biotechnology at the University of Toulouse III in 2006 and obtained a Master 2 degree in plant biosciences from the same university in 2007. He then joined the group of Matthieu Arlat in the Laboratory of Plant-Microbe Interactions (Toulouse, France), where he earned a Ph.D. in microbiology in 2011 for investigating high-affinity nutrient-scavenging systems in phytopathogenic bacteria. In 2012, Guillaume joined the group of Joseph Mougous at the University of Washington to study type 6 secretion systems. Guillaume has been a member of the group of Harry Brumer since 2014, specializing in the genetics and biochemistry of the enzymes and binding proteins involved in complex carbohydrate utilization by the human gut microbiota.

D. Wade Abbott received his Ph.D. from the University of Victoria in 2005. He then studied the molecular basis of protein-carbohydrate interactions under Alisdair Boraston at the University of Victoria. In 2008, he joined Harry Gilbert's group at the Complex Carbohydrate Research Center, at the University of Georgia, where he investigated the functional genomics of carbohydrate utilization pathways from intestinal bacteria. Currently, he is a research scientist for Agriculture and Agri-Food Canada based at the Lethbridge Research and Development Centre and adjunct professor at the University of Lethbridge, in Lethbridge, Alberta, Canada. His research program investigates the mechanisms of complex carbohydrate modification by intestinal bacteria with a focus on enzyme discovery, bacterial bioengineering, and carbohydrate-based innovations for animal agriculture.

Harry Brumer was raised in northern lower Michigan and obtained his B.Sc. in biochemistry from Michigan State University in 1993. Keenly interested in the interface between chemistry and biology, he then obtained an M.Sc. in organic chemistry at the University of Illinois at Chicago in 1995, where he worked with R. M. Moriarty on the use of pentoses as chiral synthons for vitamin D derivatives. He then began an extended period of scientific globetrotting, first to the University of Manchester Institute of Science and Technology, where he obtained a Ph.D. in 1998 studying fungal carbohydrate enzymology under the supervision of M. L. Sinnott (comentored by Paul F. G. Sims). After postdoctoral research in glycochemistry with S. G. Withers at the University of British Columbia, he obtained his first faculty position in 1999 at the Royal Institute of Technology (Kungliga Tekniska Högskolan [KTH]) in Stockholm. There, he worked closely with molecular geneticist T. T. Teeri on fundamental and applied carbohydrate enzymology of relevance to plant and forest biotechnology. After rising to the rank of full professor at KTH, in 2011 he returned to Vancouver as a professor in the Michael Smith Laboratories and Department of Chemistry at the University of British Columbia. He currently also holds associate faculty positions in the Department of Botany and the Department of Biochemistry and Molecular Biology. His research group has diverse interests in the roles of CAZymes in biology and biotechnology, including complex glycan utilization by gut symbionts and environmental microbes.
REFERENCES
- 1.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 2.Malhi Y. 2002. Carbon in the atmosphere and terrestrial biosphere in the 21st century. Philos Trans A Math Phys Eng Sci 360:2925–2945. doi: 10.1098/rsta.2002.1098. [DOI] [PubMed] [Google Scholar]
- 3.Rossmassler K, Dietrich C, Thompson C, Mikaelyan A, Nonoh JO, Scheffrahn RH, Sillam-Dusses D, Brune A. 2015. Metagenomic analysis of the microbiota in the highly compartmented hindguts of six wood- or soil-feeding higher termites. Microbiome 3:56. doi: 10.1186/s40168-015-0118-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayorga OL, Kingston-Smith AH, Kim EJ, Allison GG, Wilkinson TJ, Hegarty MJ, Theodorou MK, Newbold CJ, Huws SA. 2016. Temporal metagenomic and metabolomic characterization of fresh perennial ryegrass degradation by rumen bacteria. Front Microbiol 7:1854. doi: 10.3389/fmicb.2016.01854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shinkai T, Mitsumori M, Sofyan A, Kanamori H, Sasaki H, Katayose Y, Takenaka A. 2016. Comprehensive detection of bacterial carbohydrate-active enzyme coding genes expressed in cow rumen. Anim Sci J 87:1363–1370. doi: 10.1111/asj.12585. [DOI] [PubMed] [Google Scholar]
- 6.Haney CH, Samuel BS, Bush J, Ausubel FM. 2015. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat Plants 1:15051. doi: 10.1038/nplants.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mhuantong W, Charoensawan V, Kanokratana P, Tangphatsornruang S, Champreda V. 2015. Comparative analysis of sugarcane bagasse metagenome reveals unique and conserved biomass-degrading enzymes among lignocellulolytic microbial communities. Biotechnol Biofuels 8:16. doi: 10.1186/s13068-015-0200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 9.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lasken RS, McLean JS. 2014. Recent advances in genomic DNA sequencing of microbial species from single cells. Nat Rev Genet 15:577–584. doi: 10.1038/nrg3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A. 2011. Plant cell walls: from chemistry to biology. Garland Science, New York, NY. [Google Scholar]
- 12.Synytsya A, Copikova J, Kim WJ, Park YI. 2015. Cell wall polysaccharides of marine algae, p 545–590. In Kim S-K. (ed), Springer handbook of marine biotechnology. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 13.Tailford LE, Crost EH, Kavanaugh D, Juge N. 2015. Mucin glycan foraging in the human gut microbiome. Front Genet 6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 15.Brouwer H, Coutinho PM, Henrissat B, de Vries RP. 2014. Carbohydrate-related enzymes of important Phytophthora plant pathogens. Fungal Genet Biol 72:192–200. doi: 10.1016/j.fgb.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Abot A, Arnal G, Auer L, Lazuka A, Labourdette D, Lamarre S, Trouilh L, Laville E, Lombard V, Potocki-Veronese G, Henrissat B, O'Donohue M, Hernandez-Raquet G, Dumon C, Leberre VA. 2016. CAZyChip: dynamic assessment of exploration of glycoside hydrolases in microbial ecosystems. BMC Genomics 17:671. doi: 10.1186/s12864-016-2988-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hehemann JH, Boraston AB, Czjzek M. 2014. A sweet new wave: structures and mechanisms of enzymes that digest polysaccharides from marine algae. Curr Opin Struct Biol 28:77–86. doi: 10.1016/j.sbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Zhang D, Lax AR, Henrissat B, Coutinho P, Katiya N, Nierman WC, Fedorova N. 2012. Carbohydrate-active enzymes revealed in Coptotermes formosanus (Isoptera: Rhinotermitidae) transcriptome. Insect Mol Biol 21:235–245. doi: 10.1111/j.1365-2583.2011.01130.x. [DOI] [PubMed] [Google Scholar]
- 19.Bärlocher F, Arsuffi TL, Newell SY. 1989. Digestive enzymes of the saltmarsh periwinkle Littorina irrorata (Mollusca: Gastropoda). Oecologia 80:39–43. doi: 10.1007/BF00789929. [DOI] [PubMed] [Google Scholar]
- 20.Cantarel BL, Lombard V, Henrissat B. 2012. Complex carbohydrate utilization by the healthy human microbiome. PLoS One 7:e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cummings JH, Macfarlane GT. 1997. Role of intestinal bacteria in nutrient metabolism. JPEN J Parenter Enteral Nutr 21:357–365. [DOI] [PubMed] [Google Scholar]
- 22.Savage DC. 1977. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 23.McNeil NI. 1984. The contribution of the large-intestine to energy supplies in man. Am J Clin Nutr 39:338–342. [DOI] [PubMed] [Google Scholar]
- 24.Denman SE, McSweeney CS. 2015. The early impact of genomics and metagenomics on ruminal microbiology. Annu Rev Anim Biosci 3:447–465. doi: 10.1146/annurev-animal-022114-110705. [DOI] [PubMed] [Google Scholar]
- 25.Davies GJ, Sinnott ML. 2008. Sorting the diverse: the sequence-based classifications of carbohydrate-active enzymes. Biochem J 416:26–32. http://www.biochemist.org/bio/03004/0026/030040026.pdf. [Google Scholar]
- 26.Davies GJ, Williams SJ. 2016. Carbohydrate-active enzymes: sequences, shapes, contortions and cells. Biochem Soc Trans 44:79–87. doi: 10.1042/BST20150186. [DOI] [PubMed] [Google Scholar]
- 27.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, Henrissat B. 2010. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J 432:437–444. doi: 10.1042/BJ20101185. [DOI] [PubMed] [Google Scholar]
- 29.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. 2013. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6:41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coutinho PM, Rancurel C, Stam M, Bernard T, Couto FM, Danchin EGJ, Henrissat B. 2009. Carbohydrate-active enzymes database: principles and classification of glycosyltransferases, p 89–118. In von der Lieth C-W, L̈utteke T, Frank M (ed), Bioinformatics for glycobiology and glycomics: an introduction. John Wiley & Sons, Ltd., London, United Kingdom. [Google Scholar]
- 31.Terrapon N, Lombard V, Drula E, Coutinho PM, Henrissat B. 2016. The CAZy database/the carbohydrate-active enzyme (CAZy) database: principles and usage guidelines, p 117–131. In Aoki-Kinoshita KF. (ed), A practical guide to using glycomics databases. Springer, Tokyo, Japan. [Google Scholar]
- 32.Gloster TM, Turkenburg JP, Potts JR, Henrissat B, Davies GJ. 2008. Divergence of catalytic mechanism within a glycosidase family provides insight into evolution of carbohydrate metabolism by human gut flora. Chem Biol 15:1058–1067. doi: 10.1016/j.chembiol.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aspeborg H, Coutinho PM, Wang Y, Brumer H III, Henrissat B. 2012. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol Biol 12:186. doi: 10.1186/1471-2148-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stam MR, Danchin EG, Rancurel C, Coutinho PM, Henrissat B. 2006. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of alpha-amylase-related proteins. Protein Eng Des Sel 19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- 35.Mewis K, Lenfant N, Lombard V, Henrissat B. 2016. Dividing the large glycoside hydrolase family 43 into subfamilies: a motivation for detailed enzyme characterization. Appl Environ Microbiol 82:1686–1692. doi: 10.1128/AEM.03453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLean R, Hobbs JK, Suits MD, Tuomivaara ST, Jones DR, Boraston AB, Abbott DW. 2015. Functional analyses of resurrected and contemporary enzymes illuminate an evolutionary path for the emergence of exolysis in polysaccharide lyase family 2. J Biol Chem 290:21231–21243. doi: 10.1074/jbc.M115.664847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathieu S, Henrissat B, Labre F, Skjak-Braek G, Helbert W. 2016. Functional exploration of the polysaccharide lyase family PL6. PLoS One 11:e0159415. doi: 10.1371/journal.pone.0159415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turnbaugh PJ, Henrissat B, Gordon JI. 2010. Viewing the human microbiome through three-dimensional glasses: integrating structural and functional studies to better define the properties of myriad carbohydrate-active enzymes. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:1261–1264. doi: 10.1107/S1744309110029088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White BA, Lamed R, Bayer EA, Flint HJ. 2014. Biomass utilization by gut microbiomes. Annu Rev Microbiol 68:279–296. doi: 10.1146/annurev-micro-092412-155618. [DOI] [PubMed] [Google Scholar]
- 41.Wong MT, Wang W, Lacourt M, Couturier M, Edwards EA, Master ER. 2016. Substrate-driven convergence of the microbial community in lignocellulose-amended enrichments of gut microflora from the Canadian beaver (Castor canadensis) and North American moose (Alces americanus). Front Microbiol 7:961. doi: 10.3389/fmicb.2016.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, Sorek R, Tringe SG, Podar M, Martin HG, Kunin V, Dalevi D, Madejska J, Kirton E, Platt D, Szeto E, Salamov A, Barry K, Mikhailova N, Kyrpides NC, Matson EG, Ottesen EA, Zhang X, Hernandez M, Murillo C, Acosta LG, Rigoutsos I, Tamayo G, Green BD, Chang C, Rubin EM, Mathur EJ, Robertson DE, Hugenholtz P, Leadbetter JR. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 43.Leser TD, Amenuvor JZ, Jensen TK, Lindecrona RH, Boye M, Moller K. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl Environ Microbiol 68:673–690. doi: 10.1128/AEM.68.2.673-690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu XY, Zhong T, Pandya Y, Joerger RD. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl Environ Microbiol 68:124–137. doi: 10.1128/AEM.68.1.124-137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knapp BA, Seeber J, Rief A, Meyer E, Insam H. 2010. Bacterial community composition of the gut microbiota of Cylindroiulus fulviceps (diplopoda) as revealed by molecular fingerprinting and cloning. Folia Microbiol (Praha) 55:489–496. doi: 10.1007/s12223-010-0081-y. [DOI] [PubMed] [Google Scholar]
- 46.Kristensen NB, Danfaer A, Agergaard N. 1998. Absorption and metabolism of short-chain fatty acids in ruminants. Arch Tierernahr 51:165–175. doi: 10.1080/17450399809381916. [DOI] [PubMed] [Google Scholar]
- 47.Campbell JM, Fahey GC Jr, Wolf BW. 1997. Selected indigestible oligosaccharides affect large bowel mass, cecal and fecal short-chain fatty acids, pH and microflora in rats. J Nutr 127:130–136. [DOI] [PubMed] [Google Scholar]
- 48.Bjursell MK, Martens EC, Gordon JI. 2006. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem 281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- 49.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem 284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hemsworth GR, Déjean G, Davies GJ, Brumer H. 2016. Learning from microbial strategies for polysaccharide degradation. Biochem Soc Trans 44:94–108. doi: 10.1042/BST20150180. [DOI] [PubMed] [Google Scholar]
- 51.Foley MH, Cockburn DW, Koropatkin NM. 2016. The Sus operon: a model system for starch uptake by the human gut Bacteroidetes. Cell Mol Life Sci 73:2603–2617. doi: 10.1007/s00018-016-2242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamaker BR, Tuncil YE. 2014. A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J Mol Biol 426:3838–3850. doi: 10.1016/j.jmb.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 53.Martens EC, Kelly AG, Tauzin AS, Brumer H. 2014. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol 426:3851–3865. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Renzi F, Manfredi P, Dol M, Fu J, Vincent S, Cornelis GR. 2015. Glycan-foraging systems reveal the adaptation of Capnocytophaga canimorsus to the dog mouth. mBio 6:e02507. doi: 10.1128/mBio.02507-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barbeyron T, Thomas F, Barbe V, Teeling H, Schenowitz C, Dossat C, Goesmann A, Leblanc C, Oliver Glockner F, Czjzek M, Amann R, Michel G. 2016. Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: example of the model algae-associated bacterium Zobellia galactanivorans DsijT. Environ Microbiol 18:4610–4627. doi: 10.1111/1462-2920.13584. [DOI] [PubMed] [Google Scholar]
- 56.Mann AJ, Hahnke RL, Huang S, Werner J, Xing P, Barbeyron T, Huettel B, Stuber K, Reinhardt R, Harder J, Glockner FO, Amann RI, Teeling H. 2013. The genome of the alga-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl Environ Microbiol 79:6813–6822. doi: 10.1128/AEM.01937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abbott DW, Martens EC, Gilbert HJ, Cuskin F, Lowe EC. 2015. Coevolution of yeast mannan digestion: convergence of the civilized human diet, distal gut microbiome, and host immunity. Gut Microbes 6:334–339. doi: 10.1080/19490976.2015.1091913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson KL, Salyers AA. 1989. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J Bacteriol 171:3192–3198. doi: 10.1128/jb.171.6.3192-3198.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson KL, Salyers AA. 1989. Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol 171:3199–3204. doi: 10.1128/jb.171.6.3199-3204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tancula E, Feldhaus MJ, Bedzyk LA, Salyers AA. 1992. Location and characterization of genes involved in binding of starch to the surface of Bacteroides thetaiotaomicron. J Bacteriol 174:5609–5616. doi: 10.1128/jb.174.17.5609-5616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reeves AR, D'Elia JN, Frias J, Salyers AA. 1996. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J Bacteriol 178:823–830. doi: 10.1128/jb.178.3.823-830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D'Elia JN, Salyers AA. 1996. Contribution of a neopullulanase, a pullulanase, and an alpha-glucosidase to growth of Bacteroides thetaiotaomicron on starch. J Bacteriol 178:7173–7179. doi: 10.1128/jb.178.24.7173-7179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D'Elia JN, Salyers AA. 1996. Effect of regulatory protein levels on utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol 178:7180–7186. doi: 10.1128/jb.178.24.7180-7186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reeves AR, Wang GR, Salyers AA. 1997. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol 179:643–649. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shipman JA, Cho KH, Siegel HA, Salyers AA. 1999. Physiological characterization of SusG, an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol 181:7206–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cho KH, Cho D, Wang GR, Salyers AA. 2001. New regulatory gene that contributes to control of Bacteroides thetaiotaomicron starch utilization genes. J Bacteriol 183:7198–7205. doi: 10.1128/JB.183.24.7198-7205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cameron EA, Kwiatkowski KJ, Lee BH, Hamaker BR, Koropatkin NM, Martens EC. 2014. Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis. mBio 5:e01441-14. doi: 10.1128/mBio.01441-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karunatilaka KS, Cameron EA, Martens EC, Koropatkin NM, Biteen JS. 2014. Superresolution imaging captures carbohydrate utilization dynamics in human gut symbionts. mBio 5:e02172. doi: 10.1128/mBio.02172-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koropatkin NM, Smith TJ. 2010. SusG: a unique cell-membrane-associated alpha-amylase from a prominent human gut symbiont targets complex starch molecules. Structure 18:200–215. doi: 10.1016/j.str.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 70.Cameron EA, Maynard MA, Smith CJ, Smith TJ, Koropatkin NM, Martens EC. 2012. Multidomain carbohydrate-binding proteins involved in Bacteroides thetaiotaomicron starch metabolism. J Biol Chem 287:34614–34625. doi: 10.1074/jbc.M112.397380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kitamura M, Okuyama M, Tanzawa F, Mori H, Kitago Y, Watanabe N, Kimura A, Tanaka I, Yao M. 2008. Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron. J Biol Chem 283:36328–36337. doi: 10.1074/jbc.M806115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tauzin AS, Kwiatkowski KJ, Orlovsky NI, Smith CJ, Creagh AL, Haynes CA, Wawrzak Z, Brumer H, Koropatkin NM. 2016. Molecular dissection of xyloglucan recognition in a prominent human gut symbiont. mBio 7:e02134–02115. doi: 10.1128/mBio.02134-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koropatkin N, Martens EC, Gordon JI, Smith TJ. 2009. Structure of a SusD homologue, BT1043, involved in mucin O-glycan utilization in a prominent human gut symbiont. Biochemistry 48:1532–1542. doi: 10.1021/bi801942a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. 2008. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bakolitsa C, Xu Q, Rife CL, Abdubek P, Astakhova T, Axelrod HL, Carlton D, Chen C, Chiu HJ, Clayton T, Das D, Deller MC, Duan L, Ellrott K, Farr CL, Feuerhelm J, Grant JC, Grzechnik A, Han GW, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kozbial P, Krishna SS, Kumar A, Lam WW, Marciano D, McMullan D, Miller MD, Morse AT, Nigoghossian E, Nopakun A, Okach L, Puckett C, Reyes R, Tien HJ, Trame CB, van den Bedem H, Weekes D, Hodgson KO, Wooley J, Elsliger MA, Deacon AM, Godzik A, Lesley SA, Wilson IA. 2010. Structure of BT_3984, a member of the SusD/RagB family of nutrient-binding molecules. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:1274–1280. doi: 10.1107/S1744309110032999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Phansopa C, Roy S, Rafferty JB, Douglas CW, Pandhal J, Wright PC, Kelly DJ, Stafford GP. 2014. Structural and functional characterization of NanU, a novel high-affinity sialic acid-inducible binding protein of oral and gut-dwelling Bacteroidetes species. Biochem J 458:499–511. doi: 10.1042/BJ20131415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glenwright AJ, Pothula KR, Bhamidimarri SP, Chorev DS, Baslé A, Firbank SJ, Zheng H, Robinson CV, Winterhalter M, Kleinekathöfer U, Bolam DN, van den Berg B. 2017. Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature 541:407–411. doi: 10.1038/nature20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, Creagh AL, Haynes CA, Kelly AG, Cederholm SN, Davies GJ, Martens EC, Brumer H. 2014. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Basle A, Morland C, Day AM, Zheng H, Rogers TE, Thompson P, Hawkins AR, Yadav MP, Henrissat B, Martens EC, Dupree P, Gilbert HJ, Bolam DN. 2015. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun 6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, Rogowski A, Hamilton BS, Chen R, Tolbert TJ, Piens K, Bracke D, Vervecken W, Hakki Z, Speciale G, Munoz-Munoz JL, Day A, Pena MJ, McLean R, Suits MD, Boraston AB, Atherly T, Ziemer CJ, Williams SJ, Davies GJ, Abbott DW, Martens EC, Gilbert HJ. 2015. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Terrapon N, Lombard V, Gilbert HJ, Henrissat B. 2015. Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics 31:647–655. doi: 10.1093/bioinformatics/btu716. [DOI] [PubMed] [Google Scholar]
- 82.Ravcheev DA, Godzik A, Osterman AL, Rodionov DA. 2013. Polysaccharides utilization in human gut bacterium Bacteroides thetaiotaomicron: comparative genomics reconstruction of metabolic and regulatory networks. BMC Genomics 14:873. doi: 10.1186/1471-2164-14-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sonnenburg ED, Sonnenburg JL, Manchester JK, Hansen EE, Chiang HC, Gordon JI. 2006. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc Natl Acad Sci U S A 103:8834–8839. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwalm ND, Townsend GE, Groisman EA. 2017. Prioritization of polysaccharide utilization and control of regulator activation in Bacteroides thetaiotaomicron. Mol Microbiol 104:32–45. doi: 10.1111/mmi.13609. [DOI] [PubMed] [Google Scholar]
- 86.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lynch JB, Sonnenburg JL. 2012. Prioritization of a plant polysaccharide over a mucus carbohydrate is enforced by a Bacteroides hybrid two-component system. Mol Microbiol 85:478–491. doi: 10.1111/j.1365-2958.2012.08123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lowe EC, Basle A, Czjzek M, Firbank SJ, Bolam DN. 2012. A scissor blade-like closing mechanism implicated in transmembrane signaling in a Bacteroides hybrid two-component system. Proc Natl Acad Sci U S A 109:7298–7303. doi: 10.1073/pnas.1200479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, Gordon JI. 2013. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol 11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schwalm ND, Townsend GE, Groisman EA. 2016. Multiple signals govern utilization of a polysaccharide in the gut bacterium Bacteroides thetaiotaomicron. mBio 7:e01342-16. doi: 10.1128/mBio.01342-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Despres J, Forano E, Lepercq P, Comtet-Marre S, Jubelin G, Chambon C, Yeoman CJ, Berg Miller ME, Fields CJ, Martens E, Terrapon N, Henrissat B, White BA, Mosoni P. 2016. Xylan degradation by the human gut Bacteroides xylanisolvens XB1A(T) involves two distinct gene clusters that are linked at the transcriptional level. BMC Genomics 17:326. doi: 10.1186/s12864-016-2680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Despres J, Forano E, Lepercq P, Comtet-Marre S, Jubelin G, Yeoman CJ, Miller ME, Fields CJ, Terrapon N, Le Bourvellec C, Renard CM, Henrissat B, White BA, Mosoni P. 2016. Unraveling the pectinolytic function of Bacteroides xylanisolvens using a RNA-seq approach and mutagenesis. BMC Genomics 17:147. doi: 10.1186/s12864-016-2472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arnal G, Bastien G, Monties N, Abot A, Anton Leberre V, Bozonnet S, O'Donohue M, Dumon C. 2015. Investigating the function of an arabinan utilization locus isolated from a termite gut community. Appl Environ Microbiol 81:31–39. doi: 10.1128/AEM.02257-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bennke CM, Kruger K, Kappelmann L, Huang S, Gobet A, Schuler M, Barbe V, Fuchs BM, Michel G, Teeling H, Amann RI. 2016. Polysaccharide utilisation loci of Bacteroidetes from two contrasting open ocean sites in the North Atlantic. Environ Microbiol 18:4456–4470. doi: 10.1111/1462-2920.13429. [DOI] [PubMed] [Google Scholar]
- 95.Pope PB, Mackenzie AK, Gregor I, Smith W, Sundset MA, McHardy AC, Morrison M, Eijsink VG. 2012. Metagenomics of the Svalbard reindeer rumen microbiome reveals abundance of polysaccharide utilization loci. PLoS One 7:e38571. doi: 10.1371/journal.pone.0038571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang L, Hatem A, Catalyurek UV, Morrison M, Yu Z. 2013. Metagenomic insights into the carbohydrate-active enzymes carried by the microorganisms adhering to solid digesta in the rumen of cows. PLoS One 8:e78507. doi: 10.1371/journal.pone.0078507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dodd D, Moon YH, Swaminathan K, Mackie RI, Cann IK. 2010. Transcriptomic analyses of xylan degradation by Prevotella bryantii and insights into energy acquisition by xylanolytic bacteroidetes. J Biol Chem 285:30261–30273. doi: 10.1074/jbc.M110.141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McBride MJ, Xie G, Martens EC, Lapidus A, Henrissat B, Rhodes RG, Goltsman E, Wang W, Xu J, Hunnicutt DW, Staroscik AM, Hoover TR, Cheng YQ, Stein JL. 2009. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol 75:6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bastien G, Arnal G, Bozonnet S, Laguerre S, Ferreira F, Faure R, Henrissat B, Lefevre F, Robe P, Bouchez O, Noirot C, Dumon C, O'Donohue M. 2013. Mining for hemicellulases in the fungus-growing termite Pseudacanthotermes militaris using functional metagenomics. Biotechnol Biofuels 6:78. doi: 10.1186/1754-6834-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xie G, Bruce DC, Challacombe JF, Chertkov O, Detter JC, Gilna P, Han CS, Lucas S, Misra M, Myers GL, Richardson P, Tapia R, Thayer N, Thompson LS, Brettin TS, Henrissat B, Wilson DB, McBride MJ. 2007. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl Environ Microbiol 73:3536–3546. doi: 10.1128/AEM.00225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kabisch A, Otto A, Konig S, Becher D, Albrecht D, Schuler M, Teeling H, Amann RI, Schweder T. 2014. Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes ‘Gramella forsetii’ KT0803. ISME J 8:1492–1502. doi: 10.1038/ismej.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barbeyron T, Brillet-Gueguen L, Carre W, Carriere C, Caron C, Czjzek M, Hoebeke M, Michel G. 2016. Matching the diversity of sulfated biomolecules: creation of a classification database for sulfatases reflecting their substrate specificity. PLoS One 11:e0164846. doi: 10.1371/journal.pone.0164846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hong PY, Iakiviak M, Dodd D, Zhang M, Mackie RI, Cann I. 2014. Two new xylanases with different substrate specificities from the human gut bacterium Bacteroides intestinalis DSM 17393. Appl Environ Microbiol 80:2084–2093. doi: 10.1128/AEM.03176-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang K, Pereira GV, Cavalcante JJ, Zhang M, Mackie R, Cann I. 2016. Bacteroides intestinalis DSM 17393, a member of the human colonic microbiome, upregulates multiple endoxylanases during growth on xylan. Sci Rep 6:34360. doi: 10.1038/srep34360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang M, Chekan JR, Dodd D, Hong PY, Radlinski L, Revindran V, Nair SK, Mackie RI, Cann I. 2014. Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes. Proc Natl Acad Sci U S A 111:E3708–E3717. doi: 10.1073/pnas.1406156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Naas AE, MacKenzie AK, Dalhus B, Eijsink VG, Pope PB. 2015. Structural features of a Bacteroidetes-affiliated cellulase linked with a polysaccharide utilization locus. Sci Rep 5:11666. doi: 10.1038/srep11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Naas AE, Mackenzie AK, Mravec J, Schuckel J, Willats WG, Eijsink VG, Pope PB. 2014. Do rumen Bacteroidetes utilize an alternative mechanism for cellulose degradation? mBio 5:e01401-14. doi: 10.1128/mBio.01401-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thomas F, Barbeyron T, Tonon T, Genicot S, Czjzek M, Michel G. 2012. Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteria to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ Microbiol 14:2379–2394. doi: 10.1111/j.1462-2920.2012.02751.x. [DOI] [PubMed] [Google Scholar]
- 109.Hemsworth GR, Thompson AJ, Stepper J, Sobala LF, Coyle T, Larsbrink J, Spadiut O, Goddard-Borger ED, Stubbs KA, Brumer H, Davies GJ. 2016. Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut. Open Biol 6:160142. doi: 10.1098/rsob.160142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bågenholm V, Reddy SK, Bouraoui H, Morrill J, Kulcinskaja E, Bahr CM, Aurelius O, Rogers T, Xiao Y, Logan DT, Martens EC, Koropatkin NM, Stålbrand H. 2017. Galactomannan catabolism conferred by a polysaccharide utilisation locus of Bacteroides ovatus: enzyme synergy and crystal structure of a beta-mannanase. J Biol Chem 292:229–243. doi: 10.1074/jbc.M116.746438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reddy SK, Bagenholm V, Pudlo NA, Bouraoui H, Koropatkin NM, Martens EC, Stalbrand H. 2016. A beta-mannan utilization locus in Bacteroides ovatus involves a GH36 alpha-galactosidase active on galactomannans. FEBS Lett 590:2106–2118. doi: 10.1002/1873-3468.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Larsbrink J, Zhu Y, Kharade SS, Kwiatkowski KJ, Eijsink VG, Koropatkin NM, McBride MJ, Pope PB. 2016. A polysaccharide utilization locus from Flavobacterium johnsoniae enables conversion of recalcitrant chitin. Biotechnol Biofuels 9:260. doi: 10.1186/s13068-016-0674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. 2010. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 114.Hehemann JH, Kelly AG, Pudlo NA, Martens EC, Boraston AB. 2012. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc Natl Acad Sci U S A 109:19786–19791. doi: 10.1073/pnas.1211002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rakoff-Nahoum S, Coyne MJ, Comstock LE. 2014. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol 24:40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rakoff-Nahoum S, Foster KR, Comstock LE. 2016. The evolution of cooperation within the gut microbiota. Nature 533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rogers TE, Pudlo NA, Koropatkin NM, Bell JS, Moya Balasch M, Jasker K, Martens EC. 2013. Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol Microbiol 88:876–890. doi: 10.1111/mmi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pudlo NA, Urs K, Kumar SS, German JB, Mills DA, Martens EC. 2015. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. mBio 6:e01282-15. doi: 10.1128/mBio.01282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Elhenawy W, Debelyy MO, Feldman MF. 2014. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 5:e00909-14. doi: 10.1128/mBio.00909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Neumann AM, Balmonte JP, Berger M, Giebel HA, Arnosti C, Voget S, Simon M, Brinkhoff T, Wietz M. 2015. Different utilization of alginate and other algal polysaccharides by marine Alteromonas macleodii ecotypes. Environ Microbiol 17:3857–3868. doi: 10.1111/1462-2920.12862. [DOI] [PubMed] [Google Scholar]
- 121.Eisenbeis S, Lohmiller S, Valdebenito M, Leicht S, Braun V. 2008. NagA-dependent uptake of N-acetyl-glucosamine and N-acetyl-chitin oligosaccharides across the outer membrane of Caulobacter crescentus. J Bacteriol 190:5230–5238. doi: 10.1128/JB.00194-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Neugebauer H, Herrmann C, Kammer W, Schwarz G, Nordheim A, Braun V. 2005. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J Bacteriol 187:8300–8311. doi: 10.1128/JB.187.24.8300-8311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denance N, Vasse J, Lauber E, Arlat M. 2007. Plant carbohydrate scavenging through tonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Déjean G, Blanvillain-Baufume S, Boulanger A, Darrasse A, Duge de Bernonville T, Girard AL, Carrere S, Jamet S, Zischek C, Lautier M, Sole M, Buttner D, Jacques MA, Lauber E, Arlat M. 2013. The xylan utilization system of the plant pathogen Xanthomonas campestris pv campestris controls epiphytic life and reveals common features with oligotrophic bacteria and animal gut symbionts. New Phytol 198:899–915. doi: 10.1111/nph.12187. [DOI] [PubMed] [Google Scholar]
- 125.Dupoiron S, Zischek C, Ligat L, Carbonne J, Boulanger A, Duge de Bernonville T, Lautier M, Rival P, Arlat M, Jamet E, Lauber E, Albenne C. 2015. The N-glycan cluster from Xanthomonas campestris pv. campestris: a toolbox for sequential plant N-glycan processing. J Biol Chem 290:6022–6036. doi: 10.1074/jbc.M114.624593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Larsbrink J, Thompson AJ, Lundqvist M, Gardner JG, Davies GJ, Brumer H. 2014. A complex gene locus enables xyloglucan utilization in the model saprophyte Cellvibrio japonicus. Mol Microbiol 94:418–433. doi: 10.1111/mmi.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Larsbrink J, Izumi A, Ibatullin FM, Nakhai A, Gilbert HJ, Davies GJ, Brumer H. 2011. Structural and enzymatic characterization of a glycoside hydrolase family 31 alpha-xylosidase from Cellvibrio japonicus involved in xyloglucan saccharification. Biochem J 436:567–580. doi: 10.1042/BJ20110299. [DOI] [PubMed] [Google Scholar]
- 128.Attia M, Stepper J, Davies GJ, Brumer H. 2016. Functional and structural characterization of a potent GH74 endo-xyloglucanase from the soil saprophyte Cellvibrio japonicus unravels the first step of xyloglucan degradation. FEBS J 283:1701–1719. doi: 10.1111/febs.13696. [DOI] [PubMed] [Google Scholar]
- 129.Ben David Y, Dassa B, Borovok I, Lamed R, Koropatkin NM, Martens EC, White BA, Bernalier-Donadille A, Duncan SH, Flint HJ, Bayer EA, Moraïs S. 2015. Ruminococcal cellulosome systems from rumen to human. Environ Microbiol 17:3407–3426. doi: 10.1111/1462-2920.12868. [DOI] [PubMed] [Google Scholar]
- 130.Moraïs S, Ben David Y, Bensoussan L, Duncan SH, Koropatkin NM, Martens EC, Flint HJ, Bayer EA. 2016. Enzymatic profiling of cellulosomal enzymes from the human gut bacterium, Ruminococcus champanellensis, reveals a fine-tuned system for cohesin-dockerin recognition. Environ Microbiol 18:542–556. doi: 10.1111/1462-2920.13047. [DOI] [PubMed] [Google Scholar]
- 131.Ze X, Ben David Y, Laverde-Gomez JA, Dassa B, Sheridan PO, Duncan SH, Louis P, Henrissat B, Juge N, Koropatkin NM, Bayer EA, Flint HJ. 2015. Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic Firmicutes bacterium Ruminococcus bromii. mBio 6:e01058-15. doi: 10.1128/mBio.01058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Smith SP, Bayer EA. 2013. Insights into cellulosome assembly and dynamics: from dissection to reconstruction of the supramolecular enzyme complex. Curr Opin Struct Biol 23:686–694. doi: 10.1016/j.sbi.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 133.Cockburn DW, Orlovsky NI, Foley MH, Kwiatkowski KJ, Bahr CM, Maynard M, Demeler B, Koropatkin NM. 2015. Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale. Mol Microbiol 95:209–230. doi: 10.1111/mmi.12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sheridan PO, Martin JC, Lawley TD, Browne HP, Harris HMB, Bernalier-Donadille A, Duncan SH, O'Toole PW, Scott KP, Flint HJ. 2016. Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes. Microb Genom 2:e000043. doi: 10.1099/mgen.0.000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wegmann U, Horn N, Carding SR. 2013. Defining the Bacteroides ribosomal binding site. Appl Environ Microbiol 79:1980–1989. doi: 10.1128/AEM.03086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, Brown EM, Schroeter K, Allen-Vercoe E. 2013. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome 1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Biddle AS, Black SJ, Blanchard JL. 2013. An in vitro model of the horse gut microbiome enables identification of lactate-utilizing bacteria that differentially respond to starch induction. PLoS One 8:e77599. doi: 10.1371/journal.pone.0077599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Woloszynek S, Pastor S, Mell JC, Nandi N, Sokhansanj B, Rosen GL. 2016. Engineering human microbiota: influencing cellular and community dynamics for therapeutic applications. Int Rev Cell Mol Biol 324:67–124. doi: 10.1016/bs.ircmb.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 139.Cao Y, Forstner KU, Vogel J, Smith CJ. 2016. cis-encoded small RNAs, a conserved mechanism for repression of polysaccharide utilization in Bacteroides. J Bacteriol 198:2410–2418. doi: 10.1128/JB.00381-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Raghavendra MP, Nayaka SC, Gupta VK. 2016. Microbial enzymes for conversion of biomass to bioenergy, p 1–26. In Gupta VK. (ed), Microbial enzymes in bioconversions of biomass. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 141.Goel G, Dagar SS, Raghav M, Bansal S. 2015. Rumen: an underutilised niche for industrially important enzymes, p 247–263. In Puniya AK, Singh R, Kamra DN (ed), Rumen microbiology: from evolution to revolution. Springer India, New Delhi, India. [Google Scholar]
- 142.Blanton LV, Barratt MJ, Charbonneau MR, Ahmed T, Gordon JI. 2016. Childhood undernutrition, the gut microbiota, and microbiota-directed therapeutics. Science 352:1533. doi: 10.1126/science.aad9359. [DOI] [PubMed] [Google Scholar]
- 143.Mimee M, Tucker AC, Voigt CA, Lu TK. 2016. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst 2:214. doi: 10.1016/j.cels.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 144.Farrar MD, Whitehead TR, Lan J, Dilger P, Thorpe R, Holland KT, Carding SR. 2005. Engineering of the gut commensal bacterium Bacteroides ovatus to produce and secrete biologically active murine interleukin-2 in response to xylan. J Appl Microbiol 98:1191–1197. doi: 10.1111/j.1365-2672.2005.02565.x. [DOI] [PubMed] [Google Scholar]
- 145.Mueller UG, Sachs JL. 2015. Engineering microbiomes to improve plant and animal health. Trends Microbiol 23:606–617. doi: 10.1016/j.tim.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 146.Horn N, Carvalho AL, Overweg K, Wegmann U, Carding SR, Stentz R. 2016. A novel tightly regulated gene expression system for the human intestinal symbiont Bacteroides thetaiotaomicron. Front Microbiol 7:1080. doi: 10.3389/fmicb.2016.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hamady ZZ, Scott N, Farrar MD, Wadhwa M, Dilger P, Whitehead TR, Thorpe R, Holland KT, Lodge JP, Carding SR. 2011. Treatment of colitis with a commensal gut bacterium engineered to secrete human TGF-beta1 under the control of dietary xylan 1. Inflamm Bowel Dis 17:1925–1935. doi: 10.1002/ibd.21565. [DOI] [PubMed] [Google Scholar]
- 148.Hamady ZZ, Scott N, Farrar MD, Lodge JP, Holland KT, Whitehead T, Carding SR. 2010. Xylan-regulated delivery of human keratinocyte growth factor-2 to the inflamed colon by the human anaerobic commensal bacterium Bacteroides ovatus. Gut 59:461–469. doi: 10.1136/gut.2008.176131. [DOI] [PubMed] [Google Scholar]
- 149.Angelakis E. 2017. Weight gain by gut microbiota manipulation in productive animals. Microb Pathog 106:162–170. doi: 10.1016/j.micpath.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 150.Nothaft H, Davis B, Lock YY, Perez-Munoz ME, Vinogradov E, Walter J, Coros C, Szymanski CM. 2016. Engineering the Campylobacter jejuni N-glycan to create an effective chicken vaccine. Sci Rep 6:26511. doi: 10.1038/srep26511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Price NL, Goyette-Desjardins G, Nothaft H, Valguarnera E, Szymanski CM, Segura M, Feldman MF. 2016. Glycoengineered outer membrane vesicles: a novel platform for bacterial vaccines. Sci Rep 6:24931. doi: 10.1038/srep24931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Selle K, Barrangou R. 2015. CRISPR-based technologies and the future of food science. J Food Sci 80:R2367–R2372. doi: 10.1111/1750-3841.13094. [DOI] [PubMed] [Google Scholar]
- 153.Nguyen QT, Merlo ME, Medema MH, Jankevics A, Breitling R, Takano E. 2012. Metabolomics methods for the synthetic biology of secondary metabolism. FEBS Lett 586:2177–2183. doi: 10.1016/j.febslet.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 154.Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T, Prestegard JJ, Schnaar RL, Freeze HH, Marth JD, Bertozzi CR, Etzler ME, Frank M, Vliegenthart JF, Lütteke T, Perez S, Bolton E, Rudd P, Paulson J, Kanehisa M, Toukach P, Aoki-Kinoshita KF, Dell A, Narimatsu H, York W, Taniguchi N, Kornfeld S. 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology 25:1323–1324. doi: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]