ABSTRACT
Rta, an Epstein-Barr virus (EBV) immediate-early protein, reactivates viral lytic replication that is closely associated with tumorigenesis. In previous studies, we demonstrated that in epithelial cells Rta efficiently induced cellular senescence, which is an irreversible G1 arrest likely to provide a favorable environment for productive replications of EBV and Kaposi's sarcoma-associated herpesvirus (KSHV). To restrict progression of the cell cycle, Rta simultaneously upregulates CDK inhibitors and downregulates MYC, CCND1, and JUN, among others. Rta has long been known as a potent transcriptional activator, thus its role in gene repression is unexpected. In silico analysis revealed that the promoter regions of MYC, CCND1, and JUN are common in (i) the presence of CpG islands, (ii) strong chromatin immunoprecipitation (ChIP) signals of CCCTC-binding factor (CTCF), and (iii) having at least one Rta binding site. By combining ChIP assays and DNA methylation analysis, here we provide evidence showing that Rta binding accumulated CpG methylation and decreased CTCF occupancy in the regulatory regions of MYC, CCND1, and JUN, which were associated with downregulated gene expression. Stable residence of CTCF in the viral latency and reactivation control regions is a hallmark of viral latency. Here, we observed that Rta-mediated decreased binding of CTCF in the viral genome is concurrent with virus reactivation. Via interfering with CTCF binding, in the host genome Rta can function as a transcriptional repressor for gene silencing, while in the viral genome Rta acts as an activator for lytic gene loci by removing a topological constraint established by CTCF.
IMPORTANCE CTCF is a multifunctional protein that variously participates in gene expression and higher-order chromatin structure of the cellular and viral genomes. In certain loci of the genome, CTCF occupancy and DNA methylation are mutually exclusive. Here, we demonstrate that the Epstein-Barr virus (EBV) immediate-early protein, Rta, known to be a transcriptional activator, can also function as a transcriptional repressor. Via enriching CpG methylation and decreasing CTCF reloading, Rta binding efficiently shut down the expression of MYC, CCND1, and JUN, thus impeding cell cycle progression. Rta-mediated disruption of CTCF binding was also detected in the latency/reactivation control regions of the EBV genome, and this in turn led to viral lytic cycle progression. As emerging evidence indicates that a methylated EBV genome is a preferable substrate for EBV Zta, the other immediate-early protein, our results suggest a mechanistic link in understanding the molecular processes of viral latent-lytic switch.
KEYWORDS: Epstein-Barr virus, immediate-early protein Rta, lytic cycle reactivation, CTCF, DNA methylation, cell cycle arrest, genome topology, immediate-early protein Rta
INTRODUCTION
Epstein-Barr virus (EBV) is an oncogenic virus that is associated with a variety of epithelial malignancies, including nonkeratinizing nasopharyngeal carcinoma (NPC), gastric adenocarcinoma, and salivary gland carcinoma (1). The life cycle of EBV is divided into two stages: latent and lytic cycles. Accumulating evidence indicates that lytic cycle replication of EBV is closely associated with NPC development. Serological screening of NPC patients indicated that the titers of antibodies to EBV lytic proteins were elevated prior to the occurrence of NPC (2–4). In addition, higher antibody titers are correlated with advanced pathological staging of NPC. NPC patients in remission had undetectable or low levels of EBV-specific IgA; conversely, patients with recurrent disease had higher titers of EBV-specific IgA (5). These results raise the possibility that reactivation of EBV plays a role in NPC pathogenesis.
EBV Rta is an immediate-early protein encoded by the EBV genome that is essential and sufficient to reactivate a latent EBV genome in both epithelial and B cells (6, 7). Rta is a transcriptional activator that recognizes Rta binding sites in the promoter regions of its target genes. Rta can activate gene expression directly by binding to Rta binding sites or indirectly by associating with other DNA-binding factors, including SP1 and ATF7IP/MCAF1 (8–12). Moreover, in 293TetER and TW01TetER cells, which were established as doxycycline-inducible systems for Rta expression in HEK293 cells and NPC-TW01 cells, respectively, Rta expression caused cell cycle arrest in the G1 phase, followed by cellular senescence (13). In addition, in latently EBV-infected 293TetER cells, Rta expression turned on the lytic cycle program and resulted in the production of encapsidated virions in the culture medium. Surprisingly, Rta was also capable of reactivating a latent Kaposi's sarcoma-associated herpesvirus (KSHV) genome in the same cell background (14). Furthermore, results from microarray analysis indicated that Rta expression not only upregulated but also downregulated the expression of a number of cell cycle regulators (14). For example, MYC, which is known to be a suppressor of cellular senescence (15, 16) and an inhibitor of KSHV reactivation (17), is downregulated by Rta.
In a mitotic cell, the MYC gene is embedded in a gene-poor environment whose expression is heavily determined by CCCTC-binding factor (CTCF), an 11-zinc finger DNA-binding protein recognizing GC-rich elements (reviewed in reference 18). Among the many functions of CTCF that are exerted to control MYC expression, one is to protect the P2 promoter region from DNA methylation, an important step that is required for high-level MYC mRNA synthesis in proliferating cells (19, 20). In addition, through recognizing ∼20,000 sites in the human genome, CTCF plays multiple roles in genome organization, including transcriptional regulator, enhancer-blocking insulator, chromatin barrier insulator, alternative mRNA splicing regulator, and higher-order chromatin structure coordinator (21, 22).
Besides the human genome, CTCF also participates in delineating the latent EBV and KSHV genomes through occupying ∼20 sites on each of the viral episomes (23, 24). In the KSHV genome, CTCF-cohesin loops the latency control region (LCR), located between the divergent promoters for LANA and K14 and the lytic reactivation control region (referred to as RCR in the present study) to maintain viral latency (24, 25). When CTCF was removed from these loci, either by depletion of CTCF, treatment with sodium butyrate, or deletion of CTCF binding sites in the LCR, the conformation of the KSHV genome was loosened (26) and viral lytic reactivation was initiated (24, 25). In the EBV genome, binding of CTCF not only loops oriP with Cp or Qp, which determines the promoter usage in different latency types (27), but also loops oriP with the 3′-untranslated region of LMP1 to regulate latent gene expression (23, 28). Loss of CTCF protection at these sites led to elevated DNA methylation, altering the expression patterns of latent genes. Importantly, lytic cycle reactivation was slightly increased under such conditions (28, 29).
It is known that DNA methylation is established by DNA methyltransferases (DNMTs). Three active DNMTs have been identified in mammalian cells, namely, DNMT1, DNMT3A, and DNMT3B (30). Reactive oxygen species (ROS) is one of the modulators critical in coordinating the epigenetic modifications, including DNA methylation and histone acetylation (31). For example, ROS production might result in gene silencing via enhancement of DNA binding activity of DNMT1 (32, 33) or increasing the protein levels of DNMT1 and DNMT3B (34, 35).
Here, we provided evidence linking the following molecular events in Rta-inducible 293 cells harboring a latent EBV genome: (i) Rta expression is accompanied by ROS production that might be a trigger for increased DNA methylation in host CpG islands, (ii) increased CpG DNA methylation in the regulatory regions of MYC, CCND1, and JUN interferes with CTCF reloading, leading to gene silencing, and (iii) increased CpG DNA methylation also interferes with CTCF binding in the latent-lytic control regions of the EBV genome, which results in EBV latent-lytic switch. Taken together, these results reveal a previously undocumented mechanism of action of EBV Rta. Via interference with CTCF binding to both the host and viral genomes, Rta simultaneously and efficiently arrests cell cycle progression and expedites viral lytic cycle replication.
RESULTS
EBV Rta binding in the promoter regions is associated with downregulation of MYC, CCND1, and JUN.
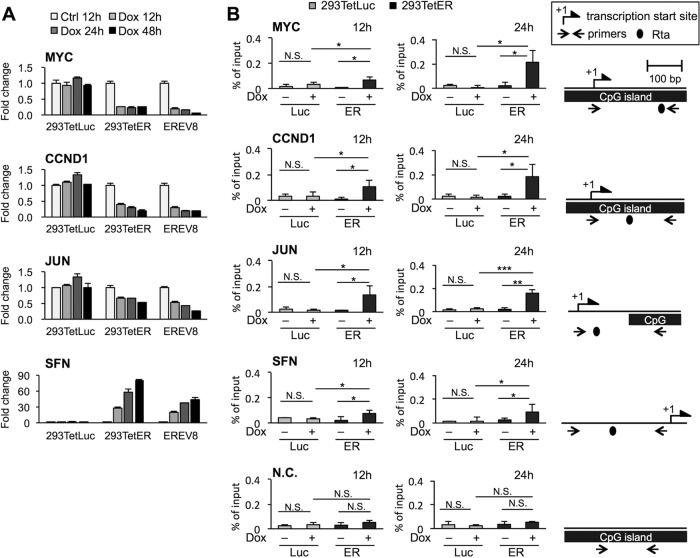
EBV Rta is known to be a potent transcriptional activator. However, we observed that the expression of Rta efficiently downregulated transcription of a battery of positive cell cycle regulators, including MYC, CCND1, and JUN, effectively leading to cell cycle arrest (14). This phenomenon is universal in all doxycycline (Dox)-inducible Rta-expressing cell lines, including TW01TetER, 293TetER, and Rta-inducible EBV replication systems in HEK293 cells, dubbed EREV8 (Fig. 1A and reference 14). Previous studies demonstrated that the consensus Rta binding sites are 16 to 18 nucleotides in length with high GC content, including the two flanking core elements (5′-GNCC-3′ and 5′-GGNG-3′) (8–10). Sequence analysis of the promoter regions (upstream, 2,000 bp; downstream, 200 bp) of MYC, CCND1, and JUN genes revealed that all of them contain potential Rta binding sites, and some of them are located in CpG islands (Fig. 1B). In order to validate that EBV Rta does bind to these cellular promoters, chromatin immunoprecipitation (ChIP) assay was performed using specific antibodies against EBV Rta and normal rabbit IgG, which served as a control to estimate nonspecific binding signals. In addition, the promoter region of SFN, which is known to be positively regulated upon Rta binding (12), served as a positive control. The results indicated that EBV Rta bound to the promoter regions of MYC, CCND1, JUN, and SFN at an early time point (12 h) after Dox treatment (Fig. 1B). The bindings persisted until 24 h and were specific, since rabbit IgG control yielded no signal (see Fig. S1A in the supplemental material). Importantly, there is no Rta binding signal in the MYC gene body, a negative-control region without an Rta binding site (Fig. 1B). In parallel, ChIP assay was performed in 293TetLuc cells with or without Dox induction, and no binding signal of Rta was observed (Fig. 1B). These results suggested that Rta binding in the promoter regions of MYC, CCND1, and JUN is associated with a downregulation of gene expression.
FIG 1.
EBV Rta-mediated gene repression is associated with Rta binding in promoters of target genes. (A) The levels of transcripts of MYC, CCND1, JUN, and SFN at the indicated time points in untreated (Ctrl) and doxycycline (Dox)-treated 293TetLuc, 293TetER, and EREV8 cells were measured by real-time RT-PCR assays. Data are presented as the means ± standard deviations (SD) from triplicate PCR results. Two independent experiments were performed; one representative data set is shown. (B) ChIP assays of EBV Rta in the cellular promoters of 293TetLuc (Luc) and 293TetER (ER) cells. Untreated (−) and Dox-treated (+) cells were harvested at the indicated time points and subjected to ChIP assays using specific antibodies against EBV Rta or normal rabbit IgG, which served as a control to exclude nonspecific binding (Fig. S1A). The eluted DNA fragments were quantified by real-time PCR analysis as a percentage of the input using the ΔCT method. The MYC gene body without Rta binding site served as a negative control (N.C.). Error bars depict the means ± SD from four independent experiments. Student's t test was used to evaluate the significant difference between the indicated data sets. ***, P < 0.005; **, P < 0.01; *, P < 0.05; N.S., not significant. Schematic diagrams of transcription start sites, CpG islands, and potential Rta binding sites in the target promoter region are denoted on the right. Lengths of promoters are illustrated to scale.
EBV Rta expression increases CpG methylation and decreases CTCF binding in the cellular promoters.
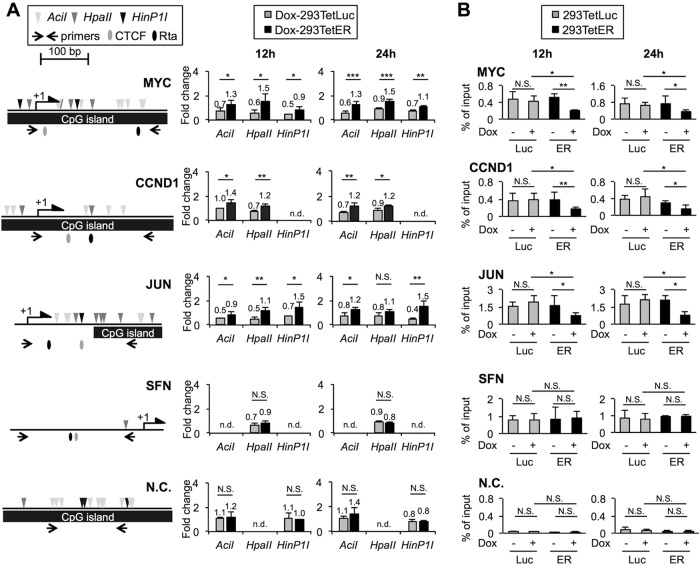
In addition to Rta binding sites, in the promoter regions of MYC, CCND1, and JUN, canonical CTCF binding sites (36–39) were also identified (Fig. 2A, left). Importantly, Gombert and Krumm previously established a positive regulatory role of CTCF for MYC expression in which CTCF prevents local DNA methylation by binding to the P2 promoter region of the MYC gene (20). Furthermore, a recent report indicated that up to 41% of CTCF bindings are sensitive to CpG methylation, suggesting a tight linkage between DNA methylation and CTCF occupancy genome-wide (40). Therefore, we hypothesized that Rta binding to these promoter regions affects their CpG DNA methylation state, which in turn influences CTCF occupancy. To assess this possibility, genomic DNAs of untreated or Dox-treated 293TetLuc and 293TetER cells prepared at 12 h and 24 h were digested with CpG methylation-sensitive restriction enzymes, including AciI, HpaII, and HinP1I, followed by real-time PCR analysis. As increased CpG methylation protects DNA from methylation-sensitive enzyme digestion, larger amounts of PCR product would indicate higher levels of methylation in the specified region. Of note, because DNA methylation is a dynamic process and cellular context dependent, the CpG methylation level in each indicated DNA fragment of Dox-treated cells was normalized to that in the same region of untreated cell counterparts. As shown in Fig. 2A, in Dox-treated 293TetLuc cells, decreased levels of CpG methylation (0.4× to 0.9×) were observed in the promoter regions of MYC, CCND1, and JUN. The decreased CpG methylation levels in these regulatory regions were associated with increased mRNA levels in the same time frame (Fig. 1A), suggesting that an active transcription process was ongoing in the Dox-treated 293TetLuc cells. In contrast, in Dox-treated 293TetER cells, increased levels of CpG methylation (1.1× to 1.5×) were observed in the comparable regions of MYC, CCND1, and JUN, which is consistent with downregulation of mRNA levels (Fig. 1A), reinforcing the notion that accumulated CpG methylation is linked to gene silencing.
FIG 2.
EBV Rta expression increases DNA methylation and decreases CTCF binding in the promoter regions of MYC, CCND1, and JUN. (A, left) Schematic diagrams of methylation-sensitive restriction enzyme sites, CTCF binding sites, and Rta binding sites in each target promoter region. These regions contain no EcoRI site, thus EcoRI served as an input control for AciI, HpaII, and HinP1I. The MYC gene body without Rta and CTCF binding sites served as a negative control (N.C.). Lengths of promoters are illustrated to scale. (Right) CpG methylation levels in the cellular promoters of 293TetLuc and 293TetER cells. Cellular DNAs of paired untreated and doxycycline (Dox)-treated (12 and 24 h) cells were extracted and subjected to restriction enzyme digestions. DNA fragments protected by each methylation-sensitive enzyme were quantified by real-time PCR. Fold changes of each restriction enzyme assessment denote the relative CpG methylation levels in the Dox-treated cells compared to their untreated counterparts. Error bars depict the means ± SD from four independent experiments. Student's t test was used to evaluate the significant difference between the indicated data set. ***, P < 0.005; **, P < 0.01; *, P < 0.05; N.S., not significant; n.d., not determined. (B) ChIP assays of CTCF in the cellular promoters of 293TetLuc and 293TetER cells. Untreated (−) and Dox-treated (+) cells harvested at 12 h and 24 h were subjected to ChIP assays using specific antibodies against CTCF or normal rabbit IgG, which served as a control to exclude nonspecific binding (Fig. S1A). The eluted DNA fragments were quantified by real-time PCR analysis as a percentage of input using the ΔCT method. Error bars represent the means ± SD from four independent experiments. Student's t test was used to evaluate the significant difference between the indicated data sets. **, P < 0.01; *, P < 0.05; N.S., not significant.
To assess each CTCF's occupancy in these regulatory regions upon Dox treatment, formaldehyde-fixed chromatins of the aforementioned cells were subjected to ChIP assays by using a CTCF-specific antibody and a negative-control antibody, normal rabbit IgG. The eluted DNAs were evaluated by real-time PCR assays. As shown in Fig. 2B, the binding signals of CTCF were not significantly altered in the Dox-treated 293TetLuc cells; however, the bindings of CTCF in the promoter regions of MYC, CCND1, and JUN were evidently decreased in the Dox-treated 293TetER cells. Compared to the protein kinetics of CTCF in Dox-treated 293TetER cells, detachment of CTCF binding occurred earlier (12 h) than a slight decrement of CTCF protein level (24 h) in Rta-expressing cells (Fig. S2A), suggesting that disengagement of CTCF from DNA is due to triggers other than protein downregulation. As a control, CTCF occupancy in SFN did not fluctuate in both types of Dox-treated cells, which implies that methylation of CpG islands contributes to the alteration of CTCF occupancies. In addition, by using in vitro DNA affinity precipitation assay (DAPA), we observed that Rta even enhanced the binding ability of CTCF on a naive, unmethylated DNA probe (Fig. S2B). Thus, methylation of DNA could be the main blockade for CTCF accessibility. Taken together, these results support our hypothesis that Rta binding is associated with elevated CpG methylation levels and CTCF detachment in the promoter regions of MYC, CCND1, and JUN.
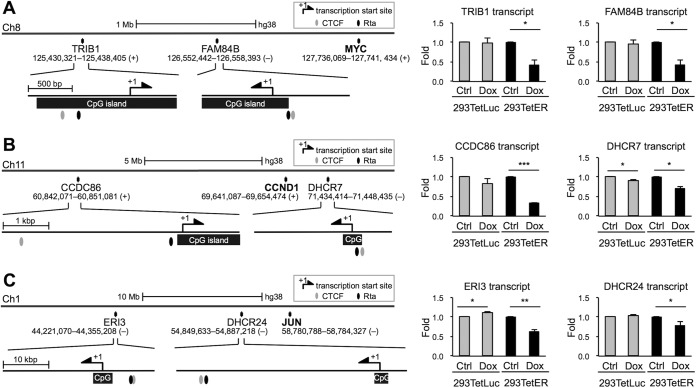
Common features of gene loci downregulated by Rta.
Previously, in our microarray data sets (GSE24587) (14), it was noticeable that a number of genes located near MYC, CCND1, and JUN were also repressed by Rta expression. We inspected the regulatory DNA sequences of these genes, including TRIB1 and FAM84B, located in chromosome (chr) 8 (near MYC), CCDC86 and DHCR7, located in chr 11 (near CCND1), and ERI3 and DHCR24, located in chr 1 (near JUN), and we found that most of these promoters (upstream, 2,000 bp; downstream, 200 bp) harbor CpG islands, putative Rta and CTCF binding sites (Fig. 3). These phenomena were reminiscent of those elements located in MYC, CCND1, and JUN. Thus, we measured the RNA expression levels of these genes in Rta-expressed cells by using real-time reverse transcription-PCR (RT-PCR) assays. As expected, most of the tested transcripts were decreased in Dox-treated 293TetER cells but not altered in 293TetLuc cells (Fig. 3). Therefore, it is likely that Rta exerts a unified function to downregulate the expression of cellular genes clustered in certain loci of the host genome.
FIG 3.
Promoter regions of Rta-downregulated genes share similar molecular features. Shown are schematic diagrams of the promoter features in Rta-repressed genes, including TRIB1 and FAM84B near MYC in chromosome 8 (A), CCDC86 and DHCR7 near CCND1 in chromosome 11 (B), and ERI3 and DHCR24 near JUN in chromosome 1 (C). The transcript levels of the indicated genes at 12 h in untreated (Ctrl) and doxycycline (Dox)-treated 293TetLuc and 293TetER cells were measured by real-time RT-PCR assays. Data are presented as the means ± SD from triplicate PCR results. Two independent experiments were performed; one representative data set is shown. Student's t test was used to assess the statistical significance. ***, P < 0.005; **, P < 0.01; *, P < 0.05.
EBV Rta expression is accompanied by ROS production and increased DNMT1/DNMT3A binding activity in MYC, JUN, and CCND1 promoters.
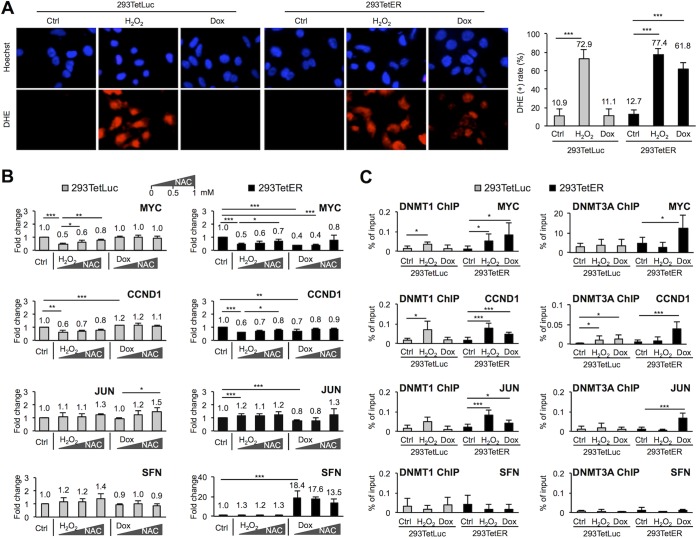
DNA methylation is established by the DNMT family, which includes DNMT1, DNMT3A, and DNMT3B. The methylation activities are highly associated with protein levels or binding abilities of DNMTs. Thus, we first assessed the transcript levels of each DNMT in the mRNA microarray data set (GSE24587) (14). We found that the transcript levels of each DNMT all were stably maintained from 24 h to 48 h after Dox treatment in 293TetER cells (not shown). To address whether Rta recruits DNMTs to the target promoters, thereby enhancing the local methylation process, immunoprecipitation assays were performed in a set of 293TetER cells with or without Dox treatments. The results showed no convincing interactions can be detected between endogenous DNMTs and Rta (not shown). Recently, emerging evidence established a crucial role of reactive oxygen species (ROS) in epigenomic regulations, particularly in enhancing the enzyme activity of DNMTs (reviewed in reference 41). In addition, the ROS/MAPK pathway was linked to EBV and KSHV lytic cycle reactivation (42–44). These results are consistent with our microarray data in that several antioxidant-related genes were significantly modulated upon Rta expression in Dox-inducible 293 and TW01 cells, including the upregulation of the glutathione pathway (NQO1, GPX3, and GSS) and downregulation of the thioredoxin pathway (GCLM, SLC7A11/XCT, and CD44) (GSE24587) (14). While the underlying mechanism is still elusive, we assessed whether Rta expression alters the cellular redox state. To this end, intracellular oxidative stress was determined in Dox-treated 293TetLuc and 293TetER cells using dihydroethidium (DHE) staining. Exposure of cell aliquots to 500 μM hydrogen peroxide (H2O2) served as a positive control for ROS production. As shown in Fig. 4A, ∼73% 293TetLuc and ∼77% 293TetER cells were stained with DHE upon H2O2 stimulation. Interestingly, compared to the untreated controls, a significant increase of ROS production was detectable in Dox-treated 293TetER (∼62%) but not in 293TetLuc cells, suggesting that Rta expression is accompanied by ROS production.
FIG 4.
EBV Rta expression is accompanied by ROS production and increased binding activity of DNMT1/DNMT3A in target cellular promoters. (A) Cellular levels of reactive oxygen species (ROS) were measured by using dihydroethidium (DHE) staining (200× magnifications). 293TetLuc and 293TetER cells were incubated with culture medium (Ctrl), 500 μM H2O2, or 50 ng/ml doxycycline (Dox) for 6 h, followed by Hoechst 33258 or DHE staining. Percentages of DHE-positive cells, determined by counting ≥300 cells (200× magnifications) for each treatment, are summarized on the right. Error bars indicate the means ± SD from four independent experiments. (B) Rta-mediated MYC, CCND1, and JUN downregulation was blocked by N-acetyl-l-cysteine (NAC). 293TetLuc and 293TetER cells were pretreated with 0.5 or 1 mM NAC for 1 h followed by 500 μM H2O2 or 50 ng/ml Dox treatment for 6 h. Cellular RNAs were extracted and analyzed by real-time RT-PCR. Data are presented as the means ± SD from four independent experiments. The statistical significances of untreated and drug-treated cells were evaluated by Student's t test: ***, P < 0.005; **, P < 0.01; *, P < 0.05. (C) Binding activities of DNMT1 and DNMT3A were increased in Rta-expressed cells. Untreated (Ctrl), H2O2-treated, and Dox-treated (6 h) 293TetLuc and 293TetER cells were subjected to ChIP assays using specific antibodies against DNMT1 and DNMT3A. Normal rabbit IgG was used as a control to estimate nonspecific binding (Fig. S1B). In the real-time PCR assays, data are shown as percentages of input using the ΔCT method. Error bars represent the means ± SD from four independent experiments. Student's t test was used to assess statistical significance: ***, P < 0.005; **, P < 0.01; *, P < 0.05.
To monitor whether ROS production is associated with gene silencing, the transcript levels of MYC, CCND1, and JUN were determined in untreated, H2O2-, or Dox-treated 293TetER and 293TetLuc cells. In parallel, SFN was included as a control. We observed that in both 293TetLuc and 293TetER, H2O2 treatments decreased the RNA expression levels of MYC and CCND1, which can be partially reversed by the ROS scavenger N-acetyl-l-cysteine (NAC) (Fig. 4B, first two rows). However, these phenomena were only detectable in Dox-treated 293TetER and not 293TetLuc cells, indicating that ROS production was present only in Rta expression cells. These results are consistent with the DHE staining shown in Fig. 4A. To our surprise, downregulation of JUN was not evident in H2O2-treated 293TetLuc or 293TetER cells, suggesting that different dosages of H2O2 or different temporal regulation schemes are required for silencing of JUN expression. Interestingly, downregulation of JUN- and NAC-mediated reversion was still observed in Dox-treated 293TetER cells (Fig. 4B, third row). Importantly, expression of SFN was not affected by H2O2 in either cell (Fig. 4B, fourth row), reinforcing that Rta-mediated gene silencing is ROS specific. Taken together, these data suggested that an abrupt production of ROS, either through H2O2 or EBV Rta expression, contributes to downregulation of certain host genes, including MYC, CCND1, and JUN.
To assess the binding ability of DNMTs in the target promoters, ChIP assays were performed using antibodies against DNMT1, DNMT3A, and DNMT3B. In four independent experiments, elevated binding of DNMT1 and DNMT3A was consistently observed in the promoters of MYC, CCND1, and JUN in Dox-treated 293TetER cells but not in the untreated cell counterparts (Fig. 4C). Unfortunately, ChIP assay for DNMT3B was unsuccessful, which might be attributable to the low affinity of antibody-chromatin binding during ChIP assay. In addition, in the ROS production time frame (6 h), no obvious alterations in protein levels of DNMT1, DNMT3A, and DNMT3B were detected (Fig. S2C). Taken together, these results provided a potential mechanism by which Rta-mediated ROS production provokes DNMT1 and DNMT3A, accumulated in a subset of cellular promoters, to inhibit gene expression.
Rta-mediated MYC, CCND1, and JUN downregulation can be recapitulated in NP cells.
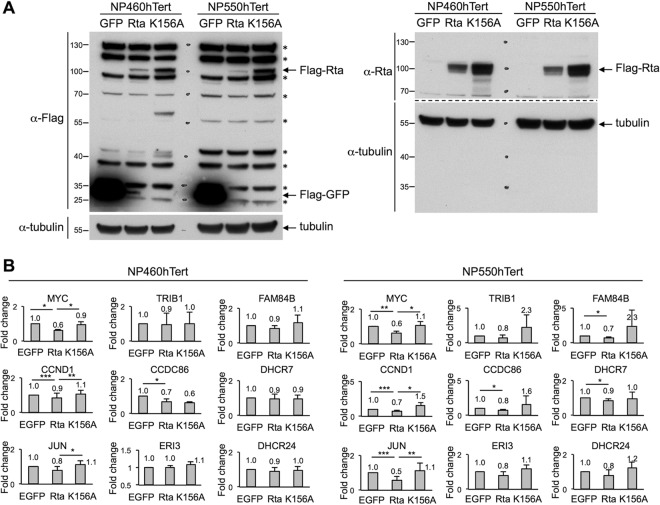
In order to confirm the gene repression function of EBV Rta in a more physiologically relevant cell type, two telomerase-immortalized nasopharyngeal (NP) cells, NP460hTert and NP550hTert (45, 46), were transiently expressed with EBV Rta by using a lentiviral transduction protocol, followed by Western blot analysis and real-time RT-PCR assays. In parallel, lentivirions of green fluorescent protein (GFP) and an Rta DNA binding-defective mutant, K156A (47), were included as controls. As shown in Fig. 5A, all of the ectopically expressed proteins were expressed in both NP460hTert and NP550hTert cells; however, the expression level of the wild-type Rta was much less than that of GFP and Rta K156A (Fig. 5A, Flag-Rta versus Flag-GFP). The instability of full-length Rta polypeptide was recently linked to conjugation of cellular E3 ubiquitin ligases, including RNF4 and TRIM5α, by which Rta was efficiently ubiquitinated and subjected to degradation (48, 49).
FIG 5.
Rta decreased the RNA expression levels of cellular genes in NP cells. Lentivirions of GFP, Rta, and K156A (an Rta DNA binding-defective mutant) were transduced into NP460hTert and NP550hTert cells by using a lentiviral transduction system. (A) At 24 h postransduction, the protein extracts were harvested and subjected to Western blot analysis by using specific antibodies against FLAG tag (left) and Rta (right). α-Tubulin served as a loading control. An asterisk denotes nonspecific cellular bands detected by FLAG-tag antibody in NP cell lines. (B) At 48 h postransduction, total RNAs were harvested and the expression levels of each indicated cellular genes were measured by real-time RT-PCR analysis. Data are presented as the means ± SD from four independent experiments. Statistical evaluation was performed with Student's t test: ***, P < 0.005; **, P < 0.01; *, P < 0.05.
Although at lower quantity, the patterns of Rta-mediated gene suppression were still evident in these two NP cells. For MYC, CCND1, and JUN, Rta, but not Rta K156A, decreased the expression of these genes in both cells (Fig. 5B). For genes that are located near MYC, CCND1, and JUN, although not statistically significant, the expression levels of most of them were decreased by Rta but not Rta K156A (Fig. 5B). Taken together, these results indicate that, similar to the situation in Dox-inducible Rta cell lines, EBV Rta-mediated gene silencing, for which the DNA binding motif is crucial, is conserved in certain genetic loci across different cell backgrounds.
CTCF represses the lytic replications of EBV in EREV8 cells.
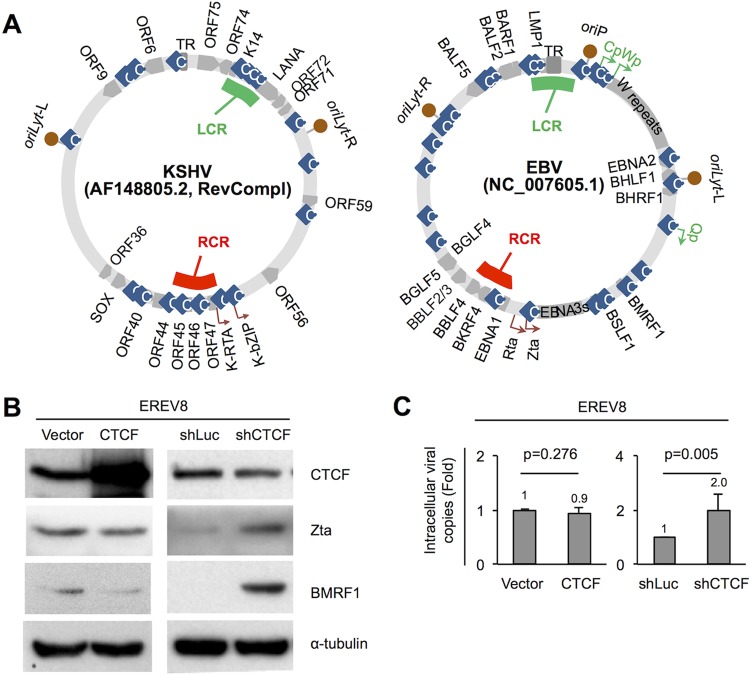
CTCF has been documented as a critical molecule in maintaining the EBV and KSHV genomes in their latent conformations (50). Previous studies from Lieberman's laboratory have established that CTCF loops the latency control region (LCR) and lytic reactivation control region (RCR) of KSHV episomes to repress lytic cycle replications (24–26). The KSHV LCR is located upstream of LANA (Fig. 6A). LANA is expressed in every latently KSHV-infected cell and is essential for sustaining viral episomes (24). The RCR of KSHV is located at the promoter region of ORF45 (Fig. 6A), which is ∼3 kb upstream of the transcriptional start site of the immediate-early gene K-RTA (25). To define similar regions in the EBV genome, the annotated genomic maps of EBV and KSHV were laid out side by side, with the genome of KSHV in a reverse-complement direction so that most homologous proteins were comparable. By this analysis, the binding sites of CTCF in the LMP1/LMP2 locus and the BKRF4 promoter region were designated EBV LCR and RCR, respectively (Fig. 6A). We next investigated the role of CTCF in maintaining EBV latency by using an Rta-inducible EBV replication system, EREV8 (14). Previously, we noticed that the viral genomes in ∼5% of EREV8 populations spontaneously enter the lytic cycle during regular cell culture maintenance. To answer whether the level of this spontaneous lytic replication would be altered by the CTCF concentration, ectopic expression of CTCF or short hairpin RNA (shRNA)-mediated CTCF depletion was conducted in EREV8 cells for 3 days, followed by Western blot analysis and intracellular viral DNA determination. As expected, in CTCF-overexpressing cells, decreases in viral lytic protein expression (Zta and BMRF1) and intracellular viral DNAs (∼0.9×) were observed; in CTCF-depleted cells, increases in viral lytic protein expression and intracellular viral DNAs (∼2×) were detected (Fig. 6B and C). Together, these results supported the notion that CTCF plays a suppressive role in EBV lytic cycle replication.
FIG 6.
CTCF represses viral lytic cycle replication in EREV8 cells. (A, left) Sequences of KSHV strain AF148805.2 (137,969 bp) and EBV strain NC_007605.1 (171,823 bp) were retrieved from NCBI, followed by manual alignment and annotation using SnapGene Viewer (GSL Biotech LLC, IL). Functional orthologs of KSHV and EBV were found to be located at similar positions of the maps, including (clockwise) TR|TR, oriLyt-R|oriLyt-L, ORF59|BMRF1, ORF56|BSLF1, K-bZIP|Zta, K-RTA|Rta, BKRF4|ORF45, ORF44|BBLF4, ORF40|BBLF2_3, SOX|BGLF5, ORF36|BGLF4, oriLyt-L|oriLyt-R, ORF9|BALF5, and ORF6|BALF2. The latency control region (LCR; green strip) is located at the KSHV LANA promoter and EBV LMP1/LMP2 region, respectively. The lytic reactivation control region (RCR; red strip) is located upstream of KSHV K-RTA and EBV BKRF4, respectively. Information on CTCF binding sites (light-blue rhombi) was adapted from references 23–26, 29, and 83. (B and C) Expression plasmids of vector control (vector), CTCF, shRNA of luciferase (shLuc), and shRNA of CTCF (shCTCF) were transfected into EREV8 cells for 3 days. (B) Protein extracts from cells with CTCF overexpression (48 h) or shRNA of CTCF (72 h) were harvested and subjected to Western blot analysis for CTCF, Zta, and BMRF1 (lytic protein) expression; α-tubulin served as a loading control. (C) Intracellular EBV DNAs collected at 72 h postransduction were quantitated by real-time PCR analysis using the EBV DNA polymerase fragment as the amplification target. Data are presented as the means ± SD from three independent experiments. Statistical evaluations were performed with Student's t test.
EBV Rta expression disrupts the binding of CTCF in the EBV genomes.
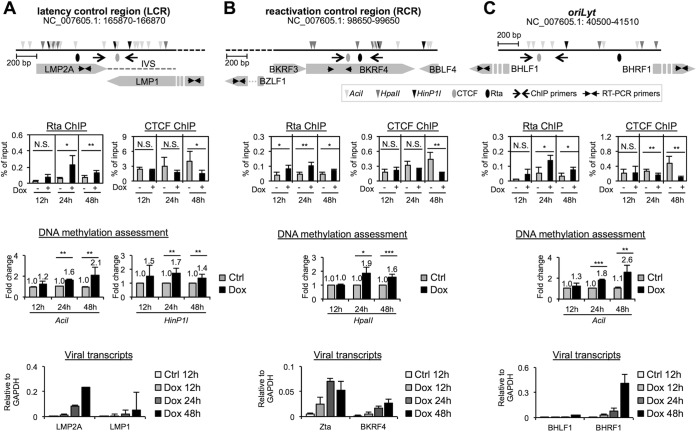
In addition to LCR and RCR, the third region in the viral genome that attracted our attention was the replication origin of the lytic cycle, oriLyt. Evidence to date indicates that the immediate-early protein Zta, also known as the oriLyt-binding protein, preferentially binds to methylated viral DNA and is central to efficient lytic replication (51–54). In addition, El-Guindy et al. demonstrated that binding of Rta onto EBV oriLyt is essential for viral lytic DNA replication (55). As a step to explore whether Rta expression perturbs the CTCF occupancy in the LCR, RCR, and oriLyt regions of the EBV genome, which in turn lead to latency disruption, Rta and CTCF ChIP assays were performed in paired untreated and Dox-treated EREV8 cells for 12 h, 24 h, and 48 h. Of note, in this set of experiments, 100 μg/ml phosphonoacetic acid (PAA) was added to the culture medium of the untreated and Dox-treated cells. This step was to block viral genomes that undergo lytic cycle replication, since a high copy number of lytic replicating DNAs will interfere with data interpretation. As depicted in Fig. 7A, in the LCR locus, Rta binding was detectable at 24 h after Dox treatment, followed by CTCF disassociation at 48 h (Fig. 7A, 2nd row). Increases of DNA methylation (3rd row) and viral transcripts (LMP2A and LMP1) (4th row) were evident at 24 h and 48 h, respectively. In the RCR locus, Rta binding and Zta expression were noticeable as early as 12 h after treatment (Fig. 7B, 2nd and 4th rows, respectively), followed by increased DNA methylation at 24 h (3rd row) and CTCF detachment at 48 h (2nd row, right). In the oriLyt region, Rta binding, CTCF disassociation, and DNA methylation were similarly noticeable at 24 h (Fig. 7C, 2nd and 3rd rows). Interestingly, there was an abrupt induction of BHRF1 transcription at 48 h after Dox treatment, suggesting a relief of transcription repression at this time point. It is noteworthy that some of the increased CpG methylation detected here is located in the gene body (Fig. 7A, LMP2A and LMP1, and B, BKRF4), which is one of the signatures correlated with high RNA expression levels (56).
FIG 7.
EBV Rta expression decreases CTCF occupancy in the viral genome. (First row) Schematic diagrams of methylation-sensitive restriction enzyme sites (triangles), CTCF binding sites (gray ellipses), and Rta binding sites (black ellipses) in the latency control region (LCR) (A), lytic reactivation control region (RCR) (B), and oriLyt (C) of the EBV genome (NC_007605.1). (Second row) ChIP assays of EBV Rta and CTCF bindings in the LCR (A), RCR (B), and oriLyt (C) regions. Untreated (−) and Dox-treated (+) EREV8 cells were subjected to ChIP assays. Normal rabbit IgG served as a control to exclude nonspecific binding (Fig. S1C). The eluted DNA was evaluated by real-time PCR as a percentage of the input by using the ΔCT method. Data are presented as means ± SD from four independent experiments. The statistical significances between the indicated data sets were evaluated by Student's t test. ***, P < 0.005; **, P < 0.01; *, P < 0.05; N.S., not significant. (Third row) Methylation state assessment of EBV LCR (A), RCR (B), and oriLyt (C). Total DNAs of untreated (Ctrl) and Dox-treated (Dox) EREV8 cells were prepared and subjected to methylation-sensitive restriction enzyme digestion, followed by real-time PCR assays. Fold changes denote the relative CpG methylation levels in the Dox-treated EREV8 cells compared to their untreated controls at specified time points. Error bars depict the means ± SD from four independent experiments. (Fourth row) Expression kinetics of viral transcripts located in LCR (A), RCR (B), and oriLyt (C). Viral transcripts of untreated (Ctrl) and Dox-treated (Dox) EREV8 cells at the indicated time points were determined by using real-time RT-PCR analysis. Data are presented as the means ± SD from two independent experiments.
Taking the time frame into consideration, Rta binding in the LCR and RCR was accompanied by increasing DNA methylation at earlier time points (12 to 24 h), followed by detachment of CTCF (48 h). This phenomenon is delayed from what has been observed in the host MYC, CCND1, and JUN promoter regions, in which gene silencing (Fig. 1A), Rta binding (Fig. 1B), CpG methylation (Fig. 2A), and CTCF detachment (Fig. 2B) all occurred as early as 12 h after Dox treatment. It was reported that MYC is a positive regulator for CTCF mRNA expression (57). Thus, we suggested that the mechanism of Rta-mediated disruption of CTCF binding in the viral genomes is partially due to MYC reduction.
Based on these results, we hypothesized that Rta-mediated DNA methylation not only reduces the binding of CTCF to alter the configuration of viral genomes but also provides a proper methylated environment for the transactivation activity of Zta.
DISCUSSION
EBV Rta is known to be a transcriptional activator. Here, we demonstrate that Rta also acts as a transcriptional repressor by turning off important positive cell cycle-related genes, including MYC, CCND1, and JUN. In the promoters that are negatively regulated by Rta, Rta-mediated ROS production enhances the binding ability of DNMT1 and DNMT3A, causing increased methylation of local CpGs. Increased methylation of these genomic loci prevents the binding of CTCF to its binding sites, which leads to a blockage in cell cycle progression. Similar phenomena, namely, the binding of Rta, increased DNA methylation, and decreased CTCF occupancy was also observed in a coexisting EBV genome. However, the detachment of CTCF from the viral genome facilitated lytic cycle gene expression and, thus, viral replication. The key determinant that differentiated the two opposing outcomes of CTCF detachment was that CTCF acts as a methylation blocker in the host cellular promoters but functions as an insulator in the viral genome. The binding of CTCF in the viral latency/reactivation control loci of viral genomes partitions active (latent) from inactive (lytic) gene expression.
Successful latent infections of EBV and KSHV have been frequently documented in host cells whose proliferative potentials are abnormally high. For example, CCND1 was stabilized by EBV EBNA3C, which facilitated the G1-to-S transition during EBV infection of primary B cells and in EBV-positive cancer cell lines (58). Similarly, in nasopharyngeal epithelial cells, a preceding overexpression of CCND1 was required for stable EBV infection (45). In KSHV-infected BCBL1 cells, LANA associated with JUN to enhance its transcriptional activity for interleukin-6 expression, which is required for BCBL1 growth (59). In the case of MYC, an important positive cell cycle regulator, EBV EBNA3C and KSHV vIRF-3 are known to upregulate and stabilize MYC expression during EBV and KSHV infections, respectively, to promote host cell proliferation (60–62). In addition, by inhibiting the transactivation function of EBV Zta and repressing the expression of KSHV K-RTA, MYC acts as a suppressor of EBV and KSHV lytic cycle replication (17, 63).
In contrast, lytic replication of herpesvirus occurs preferentially in the G1 phase of the cell cycle (64), a less competitive environment for resources required for viral DNA replication. For example, when EBV undergoes lytic replication, host cells are protected from apoptosis and the DNA synthetic machinery is blocked; however, the activities of certain S-phase regulators are increased (65, 66). In this regard, previously we demonstrated that EBV Rta potently transactivates a number of CDK inhibitors, including p21, p27, and SFN, in various epithelial cells (12, 13). Here, we revealed another function of Rta in cell cycle arrest, namely, silencing gene expression of MYC, CCND1, and JUN. Taking these findings together, upon its expression, Rta acts to efficiently decrease the proliferative potential of host cells.
Two possible effects by which ROS exerts increases in DNA methylation have been proposed: one is to increase protein levels of DNMTs, and the other is to increase DNA binding affinity of DNMTs (32, 67, 68). Here, we provide evidence that Rta expression is accompanied by ROS production that increases the DNA binding ability of DNMT1 and DNMT3A in the CpG islands of MYC, CCND1, and JUN (Fig. 4C). In addition, it is reported that ROS is one of the physiological modulators in reactivating viral lytic cycle replications. Oxidative stress efficiently induces lytic gene expression in EBV-infected NPC and gastric carcinoma cells (42, 69) and in KSHV-infected primary effusion lymphoma cells (43). Thus, our results provide a link between EBV Rta-induced ROS production and lytic cycle progression. Alternatively, DNA methylation is catalyzed by DNMTs and removed by the DNA demethylases, namely, TET families (70). Whether Rta also influences TET protein detachment in the host and viral genomes is worthy of further investigation.
The mechanisms underlying CTCF-mediated viral lytic gene repression are not completely understood. Chen et al. reported that deletion of a CTCF binding site in the LMP1/LMP2 control region of the EBV genome increased the levels of both circular and linear viral genomes in the lymphoid cell lines, although the expression of the immediate-early protein Zta was unaltered (28). Similarly, deletion of the CTCF binding site in KSHV LCR led to increased expression of the lytic loci K14 and ORF74, suggesting that CTCF plays a role in restricting lytic gene expression (24). On the other hand, the results of CTCF depletion in cells latently infected with KSHV are controversial (25, 26, 71). In BCBL1 cells latently infected with KSHV, CTCF depletion led to the disruption of viral genome looping and decreased transcription of lytic genes, including K-RTA, K14, and ORF74 (26). However, in the same cell background, decreased CTCF binding to the KSHV lytic reactivation control region resulted in increased expression of K-RTA, ORF49, and ORF45 and increased synthesis of linear genomes (25, 71). Importantly, chemical induction of KSHV lytic cycle replication by sodium butyrate, alone or in combination with tetradecanoyl phorbol acetate, disturbed the CTCF occupancy in the viral genome and enhanced viral lytic gene expression (24–26). Here, in our EREV8 cell systems, overexpression of CTCF repressed and depletion of CTCF enhanced spontaneous lytic replication (Fig. 6B and C), supporting that CTCF has a suppressive role in EBV lytic cycle progression.
It has been demonstrated that a methylated EBV genome is essential for viral lytic reproduction (51, 53, 72), as the immediate-early protein Zta preferentially binds to methylated viral promoters (54, 73–75). In 293T cells stably infected with an EBV genome, high occupancies of Zta were observed in methylated promoters of BALF2, BMRF1, Rta, and BHLF1, whereas these bindings were disrupted once the EBV genome is hypomethylated (54). In addition, during the early stages of the lytic cycle in IgG-induced EBV-positive Akata cells, Zta associated with the promoters of BKRF4, BLLF2, and BRRF1. These promoters contain CpG-containing Zta response elements (CpG-ZREs). Highly methylated ZREs were associated with high Zta occupancies (73). Our data suggest that Rta expression provides methylation marks in the viral promoters for Zta binding during latent-lytic cycle transition. More experiments are required to validate this hypothesis.
MATERIALS AND METHODS
Cell culture.
293TetLuc and 293TetER are doxycycline (Dox)-inducible cell lines as a result of transferring pLenti4-Flag-Luc and pLenti4-Flag-Rta, respectively, into T-REx 293 cells (Invitrogen, CA) as described elsewhere (13). EREV8 is a 293TetER derivative line that harbors a latent EBV genome, essentially as described in reference 14. These cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% Tet system approved fetal bovine serum (FBS; Clontech Laboratories, CA), 5 μg/ml blasticidin-S-HCl (Invitrogen, CA), and 200 μg/ml zeocin (Invitrogen). To maintain the latently infected viral genomes, EREV8 cultures were further supplemented with 400 μg/ml G418 (Amresco, OH) (14). HEK293T and 293FT (ViraPower, Invitrogen) cells were maintained in DMEM containing 10% FBS. Human telomerase reverse transcriptase immortalized primary nasopharyngeal epithelial cells, NP460hTert and NP550hTert (45, 46), were maintained in a 1:1 mixture of K-sfm and DF-K medium, which was developed to maintain human keratinocytes by J. G. Rheinwald's laboratory (76).
Plasmids and DNA transfection.
pCMV6-AC-CTCF was purchased from OriGene Technologies, Inc. (Rockville, MD). A vector control plasmid was generated by deleting the coding region of CTCF in pCMV6-AC-CTCF using SacII (New England BioLabs, MA), followed by self-ligation. Subconfluent EREV8 cells (6 × 105 cells/well) were seeded in a 6-well plate 1 day before transfection. Two hundred nanograms of the indicated plasmids was transfected into EREV8 cells using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, cells were replaced with fresh culture medium and cultured for an additional 2 days before cell harvest.
Lentivirus-based shRNA knockdown of CTCF.
Five short hairpin RNA (shRNA) clones for each target protein (CTCF or luciferase) were purchased from the National RNAi Core (Academia Sinica, Taiwan). High-titer lentivirus was prepared according to the Core's instruction manual by using 293FT (ViraPower, Invitrogen) as a packaging cell line and Lipofectamine 2000 as a plasmid DNA transfection reagent. A pilot study using GFP-lentivirions was conducted to optimize the target cell plating and growth, viral dose, and assay times. EREV8 cells were seeded in 6-well plates at a density of 6 × 105 cells/well and transduced with the indicated lentivirions at a multiplicity of infection of 4 for 24 h in the presence of 8 μg/ml Polybrene (Sigma-Aldrich, St. Louis, MO). After replacing medium with fresh medium, cells were cultured for an additional 48 h before cell harvest.
Lentivirus transduction of EBV Rta in nasopharyngeal epithelial cells.
GFP, Rta, and Rta mutant (K156A) lentivirus were prepared according to the instruction manual of the ViraPower lentiviral expression system (Invitrogen) by using HEK293T as a packaging cell line and Lipofectamine 3000 (Thermo Fisher, Waltham, MA) as a plasmid DNA transfection reagent. NP460hTert and NP550hTert cells were seeded in a 6-well plate at a density of 5 × 105 cells/well and transduced with the indicated lentivirions for 24 h in the presence of 8 μg/ml Polybrene. After replacing with fresh medium, cells were cultured for an additional 24 h before cell harvest.
Determination of ROS production.
Cells with 6-h treatments of 500 μM hydrogen peroxide (H2O2) or 50 ng/ml doxycycline were incubated with 10 μM dihydroethidium (DHE; Sigma-Aldrich) and 10 μg/ml Hoechst 33258 (Sigma-Aldrich) at 37°C for 1 h in the dark. Intracellular superoxide (O2−) was monitored by changes in fluorescence intensity resulting from intracellular probe oxidation. After being washed with phosphate-buffered saline (PBS), the cells were fixed and inspected under a fluorescence microscope. For each treatment, the percent positive cell count was quantified by counting ≥300 cells.
Western blot analysis.
Cell lysates extracted by radioimmunoprecipitation assay (RIPA) buffer were subjected to SDS-PAGE separation and transferred onto nitrocellulose membranes. The membranes were blocked for 1 h in 1× Tris-buffered saline–Tween 20 (TBST) containing 5% nonfat milk and incubated with the indicated primary antibodies at 4°C overnight. The blots were washed three times with 1× TBST for 5 min each. The blots were incubated with peroxidase-conjugated secondary antibody in blocking buffer for 1 h at room temperature. Blots were washed three times with 1× TBST for 5 min each and developed by SuperSignal West Pico chemiluminescent substrate kit (Pierce Biotechnology, IL).
Antibodies.
Anti-BMRF1 (88A9) and anti-Zta (4F10) are mouse monoclonal antibodies developed in-house and purified from hybridoma supernatants as described previously (77, 78). Rabbit polyclonal antibodies for Flag tag and EBV Rta were prepared by LTK BioLaboratories (Taoyuan, Taiwan). The following antibodies were purchased from commercial suppliers: α-tubulin (05-829; EMD Millipore Corporation, MA), CTCF (2899; Cell Signaling Technology, MA), DNMT1 (GTX116011; GeneTex, Inc., Hsinchu, Taiwan), DNMT3A (sc-20703; Santa Cruz Biotechnology, Inc., TX), EBV Rta (11-008; Argene, Verniolle, France), and normal rabbit IgG (sc-2027; Santa Cruz Biotechnology, Inc.).
Semiquantitative real-time PCR.
Each real-time PCR is composed of 4 μl diluted DNA templates in H2O, 5 μl Power SYBR green master mix (Applied Biosystems, CA), and 1 μl primer mix (2 μM). The reaction was conducted and detected by a StepOnePlus real-time PCR system (Applied Biosystems). PCRs were performed in triplicate to obtain a mean value for each sample.
RT-PCR.
One microgram of RNA was reverse transcribed to cDNAs by using SuperScript III reverse transcriptase (Invitrogen) in a 20-μl reaction mixture. One hundredth of the resulting cDNAs was used as the DNA template in each real-time PCR. The expression level of each gene was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the ΔCT method. Primer sequences used for each experiment are listed in Table S1.
Chromatin immunoprecipitation assay.
For ChIP assay, cells were trypsinized and fixed by freshly prepared 1% formaldehyde at room temperature for 10 min. Glycine solution (125 mM) was added for 5 min to stop the fixation, followed by washing the cells with ice-cold PBS two times. Cell pellets were resuspended in cell lysis buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1× protease inhibitor, 1× phosphatase inhibitor) and incubated on ice for 15 min with brief vortexing every 5 min. After centrifugation at 1,500 rpm at 4°C for 5 min, the cell pellets were resuspended in nuclear lysis buffer (50 mM Tris [pH 8.0], 10 mM EDTA, 1% SDS) at a density of 4 × 107 cells/ml. A Diagenode Bioruptor (Diagenode Inc., NJ) was used to sonicate genomic DNA into 500- to ∼1,000-bp DNA fragments according to the manufacturer's protocol, followed by centrifugation at 10,000 rpm for 10 min to remove insoluble materials. Per IP reaction, sheared chromatins from 106 cells were immunoprecipitated by using appropriate amounts of the indicated antibodies at 4°C overnight. After incubation with 50 μl protein G Mag Sepharose Xtra (GE Healthcare Bio-Sciences, Uppsala, Sweden) for 2 h, protein G Sepharose-chromatin complexes were sequentially washed once in low-salt wash buffer (20 mM Tris-HCl [pH 8.0], 2 mM EDTA, 150 mM NaCl, 0.1% SDS, and 1% Triton X-100), once in high-salt buffer (20 mM Tris-HCl [pH 8.0], 2 mM EDTA, 500 mM NaCl, 0.1% SDS, 1% Triton X-100), once in LiCl buffer (10 mM Tris-HCl [pH 8.1], 0.25 M LiCl, 1 mM EDTA, 1% NP-40, 1% deoxycholic acid), and once in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The DNA-protein complex was eluted by freshly prepared elution buffer (1% SDS and 0.1 M NaHCO3) with gentle rotation at room temperature for 30 min. DNA-protein complex was reversed by using 10 μl 5 M NaCl at 65°C for 4 h, followed by proteinase K treatment. The DNAs were recovered by a QIAquick PCR purification kit (Qiagen). One-hundredth of the eluted DNA was subjected to real-time PCR analysis to determine the occupancies of target protein. PCRs were performed in triplicate to obtain a mean value for each sample in each experiment. Primers were designed by using NCBI Primer-Blast. The sequences are listed in Table S1.
DNA methylation analysis by using methylation-sensitive restriction enzymes.
Genomic DNAs were extracted by a DNeasy blood and tissue kit (Qiagen, Hilden, Germany). For each sample, 5 μg DNA was digested by AciI, HpaII, and HinP1I (NEB Inc.) at 37°C for 30 min, followed by heat inactivation to eliminate enzyme activity. EcoRI (NEB Inc.), which is insensitive to CpG methylation, was run in parallel to serve as a DNA input control. One-hundredth of the digested DNA was subjected to real-time PCR analysis using the same primer sets as those described for the ChIP experiments. For each restriction enzyme assessment, fold change was calculated by comparing the EcoRI input control-normalized values in the treated and untreated groups using the 2ΔΔCT method.
Quantification of intracellular EBV genomic DNA.
Intracellular EBV genomic DNA was extracted by a DNeasy blood and tissue kit (Qiagen) and eluted in 150 μl H2O. One hundredth of the eluted DNA was subjected to real-time PCR analysis. Serial dilutions with known copy numbers of EBV genome from Raji cellular DNA (50 copies/cell) were used as standards in titrating EBV genome copies. The primer sequences are listed in Table S1.
Computational analysis.
CTCF binding signals in the cellular promoters were identified by tracing the ENCODE Transcription Factor Binding Tracks in the UCSC Genome Browser (79, 80), followed by filtrating CTCF binding sites specific to HEK293 cells curated in database CTCFBSDB 2.0 (81, 82).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Taiwan NHRI CA-106-PP-05 and MOST 104-2314-B-400-010.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00736-17.
REFERENCES
- 1.Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat Rev Cancer 4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 2.Ji MF, Wang DK, Yu YL, Guo YQ, Liang JS, Cheng WM, Zong YS, Chan KH, Ng SP, Wei WI, Chua DT, Sham JS, Ng MH. 2007. Sustained elevation of Epstein-Barr virus antibody levels preceding clinical onset of nasopharyngeal carcinoma. Br J Cancer 96:623–630. doi: 10.1038/sj.bjc.6603609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chien YC, Chen JY, Liu MY, Yang HI, Hsu MM, Chen CJ, Yang CS. 2001. Serologic markers of Epstein-Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. N Engl J Med 345:1877–1882. doi: 10.1056/NEJMoa011610. [DOI] [PubMed] [Google Scholar]
- 4.Cao SM, Liu Z, Jia WH, Huang QH, Liu Q, Guo X, Huang TB, Ye W, Hong MH. 2011. Fluctuations of Epstein-Barr virus serological antibodies and risk for nasopharyngeal carcinoma: a prospective screening study with a 20-year follow-up. PLoS One 6:e19100. doi: 10.1371/journal.pone.0019100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henle G, Henle W. 1976. Epstein-Barr virus-specific IgA serum antibodies as an outstanding feature of nasopharyngeal carcinoma. Int J Cancer 17:1–7. doi: 10.1002/ijc.2910170102. [DOI] [PubMed] [Google Scholar]
- 6.Zalani S, Holley-Guthrie E, Kenney S. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci U S A 93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ragoczy T, Heston L, Miller G. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol 72:7978–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruffat H, Sergeant A. 1994. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res 22:1172–1178. doi: 10.1093/nar/22.7.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen LW, Chang PJ, Delecluse HJ, Miller G. 2005. Marked variation in response of consensus binding elements for the Rta protein of Epstein-Barr virus. J Virol 79:9635–9650. doi: 10.1128/JVI.79.15.9635-9650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoebe EK, Wille C, Hopmans ES, Robinson AR, Middeldorp JM, Kenney SC, Greijer AE. 2012. Epstein-Barr virus transcription activator R upregulates BARF1 expression by direct binding to its promoter, independent of methylation. J Virol 86:11322–11332. doi: 10.1128/JVI.01161-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang LK, Chung JY, Hong YR, Ichimura T, Nakao M, Liu ST. 2005. Activation of Sp1-mediated transcription by Rta of Epstein-Barr virus via an interaction with MCAF1. Nucleic Acids Res 33:6528–6539. doi: 10.1093/nar/gki956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang SY, Hsieh MJ, Chen CY, Chen YJ, Chen JY, Chen MR, Tsai CH, Lin SF, Hsu TY. 2012. Epstein-Barr virus Rta-mediated transactivation of p21 and 14-3-3sigma arrests cells at the G1/S transition by reducing cyclin E/CDK2 activity. J Gen Virol 93:139–149. doi: 10.1099/vir.0.034405-0. [DOI] [PubMed] [Google Scholar]
- 13.Chen YL, Chen YJ, Tsai WH, Ko YC, Chen JY, Lin SF. 2009. The Epstein-Barr virus replication and transcription activator, Rta/BRLF1, induces cellular senescence in epithelial cells. Cell Cycle 8:58–65. doi: 10.4161/cc.8.1.7411. [DOI] [PubMed] [Google Scholar]
- 14.Chen YJ, Tsai WH, Chen YL, Ko YC, Chou SP, Chen JY, Lin SF. 2011. Epstein-Barr virus (EBV) Rta-mediated EBV and Kaposi's sarcoma-associated herpesvirus lytic reactivations in 293 cells. PLoS One 6:e17809. doi: 10.1371/journal.pone.0017809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. 2007. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci U S A 104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang D, Mannava S, Grachtchouk V, Tang WH, Patil S, Wawrzyniak JA, Berman AE, Giordano TJ, Prochownik EV, Soengas MS, Nikiforov MA. 2008. C-MYC overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene 27:6623–6634. doi: 10.1038/onc.2008.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Chen S, Feng J, Deng H, Sun R. 2010. Myc is required for the maintenance of Kaposi's sarcoma-associated herpesvirus latency. J Virol 84:8945–8948. doi: 10.1128/JVI.00244-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips JE, Corces VG. 2009. CTCF: master weaver of the genome. Cell 137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiorentino FP, Giordano A. 2012. The tumor suppressor role of CTCF. J Cell Physiol 227:479–492. doi: 10.1002/jcp.22780. [DOI] [PubMed] [Google Scholar]
- 20.Gombert WM, Krumm A. 2009. Targeted deletion of multiple CTCF-binding elements in the human C-MYC gene reveals a requirement for CTCF in C-MYC expression. PLoS One 4:e6109. doi: 10.1371/journal.pone.0006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ong CT, Corces VG. 2014. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vietri Rudan M, Hadjur S. 2015. Genetic tailors: CTCF and cohesin shape the genome during evolution. Trends Genet 31:651–660. doi: 10.1016/j.tig.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Arvey A, Tempera I, Tsai K, Chen HS, Tikhmyanova N, Klichinsky M, Leslie C, Lieberman PM. 2012. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe 12:233–245. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen HS, Wikramasinghe P, Showe L, Lieberman PM. 2012. Cohesins repress Kaposi's sarcoma-associated herpesvirus immediate early gene transcription during latency. J Virol 86:9454–9464. doi: 10.1128/JVI.00787-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang H, Wiedmer A, Yuan Y, Robertson E, Lieberman PM. 2011. Coordination of KSHV latent and lytic gene control by CTCF-cohesin mediated chromosome conformation. PLoS Pathog 7:e1002140. doi: 10.1371/journal.ppat.1002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tempera I, Klichinsky M, Lieberman PM. 2011. EBV latency types adopt alternative chromatin conformations. PLoS Pathog 7:e1002180. doi: 10.1371/journal.ppat.1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen HS, Martin KA, Lu F, Lupey LN, Mueller JM, Lieberman PM, Tempera I. 2014. Epigenetic deregulation of the LMP1/LMP2 locus of Epstein-Barr virus by mutation of a single CTCF-cohesin binding site. J Virol 88:1703–1713. doi: 10.1128/JVI.02209-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tempera I, Wiedmer A, Dheekollu J, Lieberman PM. 2010. CTCF prevents the epigenetic drift of EBV latency promoter Qp. PLoS Pathog 6:e1001048. doi: 10.1371/journal.ppat.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson KD. 2001. DNA methylation, methyltransferases, and cancer. Oncogene 20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 31.Afanas'ev I. 2014. New nucleophilic mechanisms of ros-dependent epigenetic modifications: comparison of aging and cancer. Aging Dis 5:52–62. doi: 10.14336/AD.2014.050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. 2005. Recruitment of DNA methyltransferase I to DNA repair sites. Proc Natl Acad Sci U S A 102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, Casero RA, Sears CL, Baylin SB. 2011. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campos AC, Molognoni F, Melo FH, Galdieri LC, Carneiro CR, D'Almeida V, Correa M, Jasiulionis MG. 2007. Oxidative stress modulates DNA methylation during melanocyte anchorage blockade associated with malignant transformation. Neoplasia 9:1111–1121. doi: 10.1593/neo.07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He J, Xu Q, Jing Y, Agani F, Qian X, Carpenter R, Li Q, Wang XR, Peiper SS, Lu Z, Liu LZ, Jiang BH. 2012. Reactive oxygen species regulate ERBB2 and ERBB3 expression via miR-199a/125b and DNA methylation. EMBO Rep 13:1116–1122. doi: 10.1038/embor.2012.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. 2007. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci U S A 104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, Thurman RE, Kaul R, Myers RM, Stamatoyannopoulos JA. 2012. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res 22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Q, Ni X. 2015. ROS-mediated DNA methylation pattern alterations in carcinogenesis. Curr Drug Targets 16:13–19. doi: 10.2174/1389450116666150113121054. [DOI] [PubMed] [Google Scholar]
- 42.Huang SY, Fang CY, Wu CC, Tsai CH, Lin SF, Chen JY. 2013. Reactive oxygen species mediate Epstein-Barr virus reactivation by N-methyl-N′-nitro-N-nitrosoguanidine. PLoS One 8:e84919. doi: 10.1371/journal.pone.0084919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Feng J, Sun R. 2011. Oxidative stress induces reactivation of Kaposi's sarcoma-associated herpesvirus and death of primary effusion lymphoma cells. J Virol 85:715–724. doi: 10.1128/JVI.01742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye F, Zhou F, Bedolla RG, Jones T, Lei X, Kang T, Guadalupe M, Gao SJ. 2011. Reactive oxygen species hydrogen peroxide mediates Kaposi's sarcoma-associated herpesvirus reactivation from latency. PLoS Pathog 7:e1002054. doi: 10.1371/journal.ppat.1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsang CM, Yip YL, Lo KW, Deng W, To KF, Hau PM, Lau VM, Takada K, Lui VW, Lung ML, Chen H, Zeng M, Middeldorp JM, Cheung AL, Tsao SW. 2012. Cyclin D1 overexpression supports stable EBV infection in nasopharyngeal epithelial cells. Proc Natl Acad Sci U S A 109:E3473–E3482. doi: 10.1073/pnas.1202637109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li HM, Man C, Jin Y, Deng W, Yip YL, Feng HC, Cheung YC, Lo KW, Meltzer PS, Wu ZG, Kwong YL, Yuen AP, Tsao SW. 2006. Molecular and cytogenetic changes involved in the immortalization of nasopharyngeal epithelial cells by telomerase. Int J Cancer 119:1567–1576. doi: 10.1002/ijc.22032. [DOI] [PubMed] [Google Scholar]
- 47.Heilmann AM, Calderwood MA, Portal D, Lu Y, Johannsen E. 2012. Genome-wide analysis of Epstein-Barr virus Rta DNA binding. J Virol 86:5151–5164. doi: 10.1128/JVI.06760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang HH, Chen CS, Wang WH, Hsu SW, Tsai HH, Liu ST, Chang LK. 2016. TRIM5alpha promotes ubiquitination of Rta from Epstein-Barr virus to attenuate lytic progression. Front Microbiol 7:2129. doi: 10.3389/fmicb.2016.02129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang YC, Yoshikai Y, Hsu SW, Saitoh H, Chang LK. 2013. Role of RNF4 in the ubiquitination of Rta of Epstein-Barr virus. J Biol Chem 288:12866–12879. doi: 10.1074/jbc.M112.413393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen HS, Lu F, Lieberman PM. 2013. Epigenetic regulation of EBV and KSHV latency. Curr Opin Virol 3:251–259. doi: 10.1016/j.coviro.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bergbauer M, Kalla M, Schmeinck A, Gobel C, Rothbauer U, Eck S, Benet-Pages A, Strom TM, Hammerschmidt W. 2010. CpG-methylation regulates a class of Epstein-Barr virus promoters. PLoS Pathog 6:e1001114. doi: 10.1371/journal.ppat.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woellmer A, Arteaga-Salas JM, Hammerschmidt W. 2012. BZLF1 governs CpG-methylated chromatin of Epstein-Barr Virus reversing epigenetic repression. PLoS Pathog 8:e1002902. doi: 10.1371/journal.ppat.1002902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalla M, Gobel C, Hammerschmidt W. 2012. The lytic phase of Epstein-Barr virus requires a viral genome with 5-methylcytosine residues in CpG sites. J Virol 86:447–458. doi: 10.1128/JVI.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wille CK, Nawandar DM, Henning AN, Ma S, Oetting KM, Lee D, Lambert P, Johannsen EC, Kenney SC. 2015. 5-Hydroxymethylation of the EBV genome regulates the latent to lytic switch. Proc Natl Acad Sci U S A 112:E7257–E7265. doi: 10.1073/pnas.1513432112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.El-Guindy A, Ghiassi-Nejad M, Golden S, Delecluse HJ, Miller G. 2013. Essential role of Rta in lytic DNA replication of Epstein-Barr virus. J Virol 87:208–223. doi: 10.1128/JVI.01995-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 57.Chau CM, Zhang XY, McMahon SB, Lieberman PM. 2006. Regulation of Epstein-Barr virus latency type by the chromatin boundary factor CTCF. J Virol 80:5723–5732. doi: 10.1128/JVI.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saha A, Halder S, Upadhyay SK, Lu J, Kumar P, Murakami M, Cai Q, Robertson ES. 2011. Epstein-Barr virus nuclear antigen 3C facilitates G1-S transition by stabilizing and enhancing the function of cyclin D1. PLoS Pathog 7:e1001275. doi: 10.1371/journal.ppat.1001275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.An J, Sun Y, Rettig MB. 2004. Transcriptional coactivation of c-Jun by the KSHV-encoded LANA. Blood 103:222–228. doi: 10.1182/blood-2003-05-1538. [DOI] [PubMed] [Google Scholar]
- 60.Lo AK, Lo KW, Tsao SW, Wong HL, Hui JW, To KF, Hayward DS, Chui YL, Lau YL, Takada K, Huang DP. 2006. Epstein-Barr virus infection alters cellular signal cascades in human nasopharyngeal epithelial cells. Neoplasia 8:173–180. doi: 10.1593/neo.05625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banerjee S, Lu J, Cai Q, Saha A, Jha HC, Dzeng RK, Robertson ES. 2013. The EBV latent antigen 3C inhibits apoptosis through targeted regulation of interferon regulatory factors 4 and 8. PLoS Pathog 9:e1003314. doi: 10.1371/journal.ppat.1003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baresova P, Pitha PM, Lubyova B. 2012. Kaposi sarcoma-associated herpesvirus vIRF-3 protein binds to F-box of Skp2 protein and acts as a regulator of c-Myc protein function and stability. J Biol Chem 287:16199–16208. doi: 10.1074/jbc.M111.335216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin Z, Yin Q, Flemington E. 2004. Identification of a negative regulatory element in the Epstein-Barr virus Zta transactivation domain that is regulated by the cell cycle control factors c-Myc and E2F1. J Virol 78:11962–11971. doi: 10.1128/JVI.78.21.11962-11971.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flemington EK. 2001. Herpesvirus lytic replication and the cell cycle: arresting new developments. J Virol 75:4475–4481. doi: 10.1128/JVI.75.10.4475-4481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inman GJ, Binne UK, Parker GA, Farrell PJ, Allday MJ. 2001. Activators of the Epstein-Barr virus lytic program concomitantly induce apoptosis, but lytic gene expression protects from cell death. J Virol 75:2400–2410. doi: 10.1128/JVI.75.5.2400-2410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kudoh A, Fujita M, Kiyono T, Kuzushima K, Sugaya Y, Izuta S, Nishiyama Y, Tsurumi T. 2003. Reactivation of lytic replication from B cells latently infected with Epstein-Barr virus occurs with high S-phase cyclin-dependent kinase activity while inhibiting cellular DNA replication. J Virol 77:851–861. doi: 10.1128/JVI.77.2.851-861.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kang KA, Zhang R, Kim GY, Bae SC, Hyun JW. 2012. Epigenetic changes induced by oxidative stress in colorectal cancer cells: methylation of tumor suppressor RUNX3. Tumour Biol 33:403–412. doi: 10.1007/s13277-012-0322-6. [DOI] [PubMed] [Google Scholar]
- 68.Zhang R, Kang KA, Kim KC, Na SY, Chang WY, Kim GY, Kim HS, Hyun JW. 2013. Oxidative stress causes epigenetic alteration of CDX1 expression in colorectal cancer cells. Gene 524:214–219. doi: 10.1016/j.gene.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 69.Hagemeier SR, Barlow EA, Meng Q, Kenney SC. 2012. The cellular ataxia telangiectasia-mutated kinase promotes Epstein-Barr virus lytic reactivation in response to multiple different types of lytic reactivation-inducing stimuli. J Virol 86:13360–13370. doi: 10.1128/JVI.01850-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu H, Zhang Y. 2014. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li DJ, Verma D, Mosbruger T, Swaminathan S. 2014. CTCF and Rad21 act as host cell restriction factors for Kaposi's sarcoma-associated herpesvirus (KSHV) lytic replication by modulating viral gene transcription. PLoS Pathog 10:e1003880. doi: 10.1371/journal.ppat.1003880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kalla M, Schmeinck A, Bergbauer M, Pich D, Hammerschmidt W. 2010. AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc Natl Acad Sci U S A 107:850–855. doi: 10.1073/pnas.0911948107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramasubramanyan S, Kanhere A, Osborn K, Flower K, Jenner RG, Sinclair AJ. 2012. Genome-wide analyses of Zta binding to the Epstein-Barr virus genome reveals interactions in both early and late lytic cycles and an epigenetic switch leading to an altered binding profile. J Virol 86:12494–12502. doi: 10.1128/JVI.01705-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wille CK, Nawandar DM, Panfil AR, Ko MM, Hagemeier SR, Kenney SC. 2013. Viral genome methylation differentially affects the ability of BZLF1 versus BRLF1 to activate Epstein-Barr virus lytic gene expression and viral replication. J Virol 87:935–950. doi: 10.1128/JVI.01790-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu KP, Heston L, Park R, Ding Z, Wang'ondu R, Delecluse HJ, Miller G. 2013. Latency of Epstein-Barr virus is disrupted by gain-of-function mutant cellular AP-1 proteins that preferentially bind methylated DNA. Proc Natl Acad Sci U S A 110:8176–8181. doi: 10.1073/pnas.1301577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rheinwald JG, Hahn WC, Ramsey MR, Wu JY, Guo Z, Tsao H, De Luca M, Catricala C, O'Toole KM. 2002. A two-stage, p16(INK4A)- and p53-dependent keratinocyte senescence mechanism that limits replicative potential independent of telomere status. Mol Cell Biol 22:5157–5172. doi: 10.1128/MCB.22.14.5157-5172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsai CH, Liu MT, Chen MR, Lu J, Yang HL, Chen JY, Yang CS. 1997. Characterization of monoclonal antibodies to the Zta and DNase proteins of Epstein-Barr virus. J Biomed Sci 4:69–77. doi: 10.1007/BF02255596. [DOI] [PubMed] [Google Scholar]
- 78.Tsai CH, Williams MV, Glaser R. 1991. Characterization of two monoclonal antibodies to Epstein-Barr virus diffuse early antigen which react to two different epitopes and have different biological function. J Virol Methods 33:47–52. [DOI] [PubMed] [Google Scholar]
- 79.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, Lee BT, Barber GP, Harte RA, Diekhans M, Long JC, Wilder SP, Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. 2013. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res 41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bao L, Zhou M, Cui Y. 2008. CTCFBSDB: a CTCF-binding site database for characterization of vertebrate genomic insulators. Nucleic Acids Res 36:D83–D87. doi: 10.1093/nar/gkm875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ziebarth JD, Bhattacharya A, Cui Y. 2013. CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res 41:D188–D194. doi: 10.1093/nar/gks1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holdorf MM, Cooper SB, Yamamoto KR, Miranda JJ. 2011. Occupancy of chromatin organizers in the Epstein-Barr virus genome. Virology 415:1–5. doi: 10.1016/j.virol.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.