ABSTRACT
Bats serve as a reservoir for various, often zoonotic viruses, including significant human pathogens such as Ebola and influenza viruses. However, for unknown reasons, viral infections rarely cause clinical symptoms in bats. A tight control of viral replication by the host innate immune defense might contribute to this phenomenon. Transcriptomic studies revealed the presence of the interferon-induced antiviral myxovirus resistance (Mx) proteins in bats, but detailed functional aspects have not been assessed. To provide evidence that bat Mx proteins might act as key factors to control viral replication we cloned Mx1 cDNAs from three bat families, Pteropodidae, Phyllostomidae, and Vespertilionidae. Phylogenetically these bat Mx1 genes cluster closely with their human ortholog MxA. Using transfected cell cultures, minireplicon systems, virus-like particles, and virus infections, we determined the antiviral potential of the bat Mx1 proteins. Bat Mx1 significantly reduced the polymerase activity of viruses circulating in bats, including Ebola and influenza A-like viruses. The related Thogoto virus, however, which is not known to infect bats, was not inhibited by bat Mx1. Further, we provide evidence for positive selection in bat Mx1 genes that might explain species-specific antiviral activities of these proteins. Together, our data suggest a role for Mx1 in controlling these viruses in their bat hosts.
IMPORTANCE Bats are a natural reservoir for various viruses that rarely cause clinical symptoms in bats but are dangerous zoonotic pathogens, like Ebola or rabies virus. It has been hypothesized that the interferon system might play a key role in controlling viral replication in bats. We speculate that the interferon-induced Mx proteins might be key antiviral factors of bats and have coevolved with bat-borne viruses. This study evaluated for the first time a large set of bat Mx1 proteins spanning three major bat families for their antiviral potential, including activity against Ebola virus and bat influenza A-like virus, and we describe here their phylogenetic relationship, revealing patterns of positive selection that suggest a coevolution with viral pathogens. By understanding the molecular mechanisms of the innate resistance of bats against viral diseases, we might gain important insights into how to prevent and fight human zoonotic infections caused by bat-borne viruses.
KEYWORDS: Mx protein, bat, bunyavirus, Ebola virus, influenza, interferons, orthomyxovirus, vesicular stomatitis virus
INTRODUCTION
Bats are hosts to a broad range of different viruses, therein also comprising a reservoir of potential zoonotic pathogens (1–3). For example, rhabdoviruses, including rabies virus and other lyssaviruses, are commonly detected in various bat species (4–6). Highly pathogenic filoviruses of the Marburgvirus and Ebolavirus genera have been detected in African fruit bats of the Pteropodidae family (7, 8). There is serological evidence that bats of the Phyllostomidae family found in Guatemala and Peru are frequently infected with previously unknown influenza A-like viruses (9, 10). Moreover, serological and viral nucleic acid sequencing data in African and Asian bats of the Rhinolophidae and Vespertilionidae families suggest infections with bunyaviruses of the Nairovirus and Hantavirus genera (11–13). However, most of these pathogens, with the exception of rabies virus and bat lyssaviruses (6, 14), seem to be under strong host control, as they do not cause obvious disease in bats (15). Therefore, the innate immune response may contribute to early control of these viruses (16), preventing disease but eventually allowing viral persistence and shedding. Unveiling the potential mechanisms leading to this phenomenon is of utmost importance to understand the ecology of zoonotic viruses, to explain the pathogenicity of these viruses for humans and other dead-end hosts, and potentially to identify new ways to control these viruses once they have crossed the species barrier.
The interferon (IFN) system navigates the innate antiviral defense of mammalian hosts through induction of IFN-stimulated genes (ISGs). Invading viral pathogens are sensed by cellular pattern recognition receptors, leading to the secretion of type I (IFN-α/β) and type III (IFN-λ) interferons. These cytokines activate their respective receptors and induce an intracellular antiviral state, thereby suppressing virus replication. Critical components of this innate antiviral defense system have also been described for bats (17–22). Moreover, constant elevated expression of type I IFNs in Pteropus alecto was recently described, and thus constitutive expression of ISGs might contribute to an early stringent control of invading viral pathogens in bats (23).
Myxovirus resistance (Mx) genes are prominent antiviral ISGs that are exclusively induced by type I and type III IFNs (24, 25). Mx proteins are large GTPases that were initially described as inhibitors of influenza virus replication (26). Most mammals encode two Mx paralogs called Mx1 and Mx2, or MxA and MxB for the human gene products. Their structures resemble that of dynamin-like large GTPases, with an N-terminal globular GTP-binding (G) domain and a C-terminal stalk that are both connected by a bundle-signaling element (BSE) (27). More recent studies broadened the antiviral spectrum of Mx proteins from tick-transmitted orthomyxoviruses, rhabdoviruses, and bunyaviruses to HIV and large DNA viruses (26). The antiviral action of Mx proteins is based on recognition of viral target structures like viral ribonucleoprotein complexes, leading to mislocalization or even disruption of the viral structures (28–32). Accordingly, escape from Mx restriction has been described for human influenza A viruses (FLUAV) and HIV-1 through mutations in the viral nucleocapsid proteins, the main structural component of the ribonucleoprotein complexes (33, 34).
To better understand how bats are capable of controlling viral infections and coexisting with potentially damaging pathogens, we analyzed the antiviral function of bat Mx1 proteins and compared their activity with that of the well-characterized human MxA ortholog. Mx1 cDNAs from seven different bat species that were previously described as hosts of zoonotic viral pathogens were tested for inhibition of a range of viruses that are related to viral agents commonly found in bats. We found interesting differences in their antiviral specificity that are discussed with respect to viral persistence in the bat hosts.
(This work was conducted by J. Fuchs in partial fulfillment of the requirements for a PhD from the Faculty of Biology of the University of Freiburg, Freiburg, Germany.)
RESULTS
Cloning of bat Mx1 cDNAs.
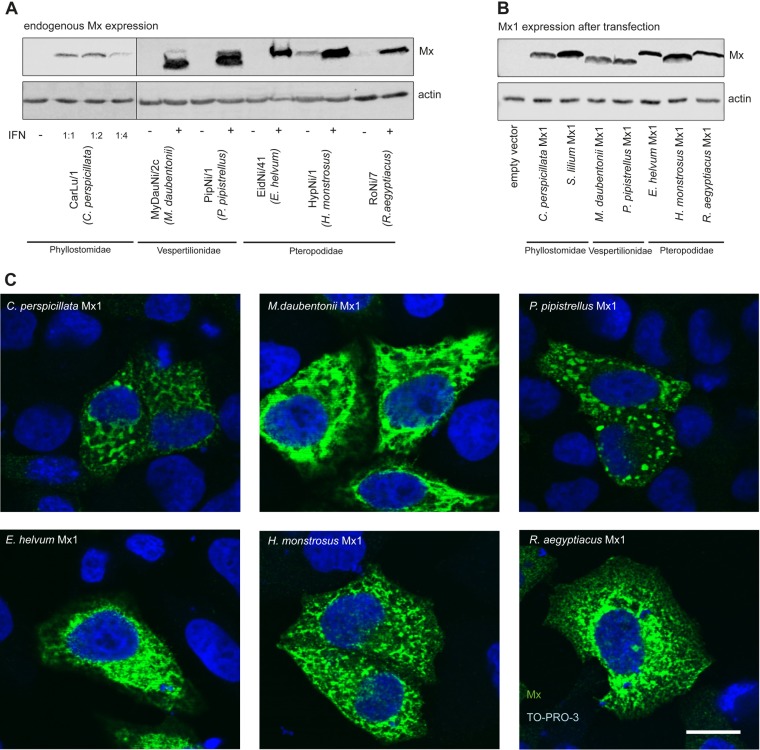
We chose cell cultures from six species out of three bat families to clone the Mx1 cDNAs. Immortalized bat cells of Carollia perspicillata (Phyllostomidae), Myotis daubentonii and Pipistrellus pipistrellus (Vespertilionidae), and Eidolon helvum, Hypsignathus monstrosus, and Rousettus aegyptiacus (Pteropodidae) were treated with 1000 U/ml of IFN-α to induce Mx gene expression, and 24 h later total RNA was extracted. To investigate Mx gene expression, lysates of untreated and IFN-stimulated cells were analyzed by Western blotting using a pan-specific monoclonal anti-Mx antibody (M143) recognizing the highly conserved G domain of Mx (35). All IFN-treated cell cultures showed strong bands at the expected molecular mass, around 78 kDa (Fig. 1A), with the exception of CarLu/1 cells from C. perspicillata. The appearance of an Mx doublet for the Verspertilionidae species might result from molecular mass differences between the simultaneously expressed Mx1 and Mx2 paralogs. CarLu/1 cells did not respond to human IFN-α (data not shown). To produce endogenous type I IFN, CarLu/1 cells were infected with Rift Valley fever virus (RVFV) clone 13, a known inducer of type I IFN (36). The virus-inactivated supernatant of the infected cells but not that of mock-treated cells induced an Mx-specific band in CarLu/1 cell culture (Fig. 1A).
FIG 1.
Expression and localization of bat Mx1. (A) Immortalized bat cell lines were treated with (+) or without (−) 1000 U/ml of human IFN-α B/D variant for 24 h, except CarLu/1. CarLu/1 cells were treated with an autologous IFN preparation (see Materials and Methods) in three different dilutions. Cells were lysed and the endogenous Mx proteins were detected by Western blotting using the Mx-specific monoclonal antibody M143. Detection of β-actin was used as a loading control. (B) 293T cells were transfected with 300 ng of Mx1 plasmid DNA per well of a 12-well plate for 24 h, and cell lysates were subjected to Western blot analysis as described for panel A. (C) HeLa cells were transfected with 1 μg of Mx1 plasmid DNA per well of a 6-well plate for 24 h. The cells were fixed and stained with the Mx-specific M143 antibody (green). Genomic DNA was detected using TO-PRO-3 (blue). Images were recorded by confocal fluorescence microscopy. The scale bar represents 15 μm.
cDNAs were synthesized by reverse transcription of the extracted poly(A)+ RNAs. Subsequent sequencing of PCR and 5′ and 3′ rapid amplification of cDNA ends (RACE) products yielded the complete cDNA sequences. RNA isolated from Sturnira lilium cells was provided by M. Schwemmle, Freiburg, Germany. The sequence information was used to amplify and clone the full-length Mx1 open reading frames (ORFs). To examine the molecular masses and intracellular localization of the recombinant bat Mx1 proteins encoded by the cloned Mx1 cDNA expression constructs, 293T and HeLa cells were transfected with the expression plasmids and the cells were analyzed by Western blotting and immunofluorescence using the Mx-specific monoclonal antibody M143 (35). The recombinant bat Mx1 showed distinct, family-specific migration patterns of the protein bands that overall correspond to the calculated molecular masses of the deduced amino acid sequences of about 75.4 kDa for Phyllostomidae Mx1, 75 kDa for Vespertilionidae Mx1, and 75.2 kDa for Pteropodidae Mx1 (Fig. 1B). All recombinant bat Mx1 accumulated in the cytoplasm of the transfected cells in a dotted or diffuse reticular pattern (Fig. 1C), very similar to the published distribution of human MxA (37).
The nucleotide sequences showed relatively high identities to other members of the same bat family in a phylogenetic tree analysis (Fig. 2A). Moreover, the deduced Mx1 amino acid sequences showed about 70% identity to Mx1 of other bat families (Fig. 2C). An alignment of the amino acid sequences of all available bat Mx1 sequences (see Fig. S1 in the supplemental material) reveals an overall high sequence similarity except for the highly variable N-terminal part before the first BSE and the L4 loop in the stalk domain. The phylogenetic analysis showed that bat Mx1 genes form a separate branch within the mammalian Mx1 tree (Fig. 2B). The close similarity to other mammalian Mx1 proteins is reflected by the sequence identities of the bat Mx1 sequences to the human MxA sequence (Fig. 2C). Interestingly, the cDNA clones of all bat Mx1 genes showed allelic variations that were mostly silent but led in some species to amino acid changes (Fig. 2C). According to our sequence alignment analyses, we defined the cDNA clones with the most common nucleotide or amino acid variation at particular positions as allele 1, which was used for further characterization.
FIG 2.
Phylogenetic analysis of bat Mx1. (A) Phylogenetic tree of bat Mx1 using a nucleotide sequence alignment with human MxA as an outgroup. (B) Phylogenetic tree of mammalian Mx1 nucleotide sequences. Scale bars represent genetic distances (nucleotide substitutions per site). (C) Allelic variants of bat Mx1 and their sequence identity to E. helvum Mx1 and human MxA as determined via multiple-protein alignment.
Antiviral activity of bat Mx1 against orthomyxoviruses.
Mx proteins are best known for their inhibitory effect on influenza A virus (FLUAV) replication (38, 39). The antiviral activity of Mx proteins is dependent on a functional GTPase activity (40). Therefore, we used GTPase-inactive Mx mutants as negative controls. We inactivated the highly conserved phosphate-binding loop by exchanging lysines at position 83 in MxA and in the bat Mx1 to alanine, resulting in a block of GTP hydrolysis (40). To assess the antiviral potential of bat Mx1, we used a polymerase reconstitution or minireplicon assay of an avian influenza A virus (H5N1) that is known for its high sensitivity to the action of human MxA (41). Coexpression of the viral polymerase subunits and the viral nucleoprotein (NP), together with an artificial RNA minigenome encoding the firefly luciferase reporter gene, reconstitutes a transcription and replication competent viral ribonucleoprotein complex (41). Additional transfection of wild-type MxA suppressed 89% of the viral polymerase activity compared to the activity in the presence of the inactive MxA(K83A) mutant (Fig. 3A). Cotransfection of the bat Mx1 expression plasmids also reduced viral polymerase activity, whereas the inactive mutants had no or in some cases even a slight enhancing effect on reporter gene expression (Fig. 3A). Western blot analysis showed comparable accumulations of the recombinant proteins in the cell lysates. Interestingly, the two bat Mx1 proteins from S. lilium and C. perspicillata, belonging to the Phyllostomidae family, reduced viral gene expression comparable to that with human MxA. Mx1 of M. daubentonii and E. helvum showed roughly 75% inhibition of reporter gene expression, whereas Mx1 of P. pipistrellus, R. aegyptiacus, and H. monstrosus reduced the activity of the viral polymerase only by about 40% (Fig. 3A).
FIG 3.
Antiviral activity of bat Mx1 against orthomyxoviruses. 293T cells were cotransfected with plasmids encoding the components of different orthomyxoviral polymerase reconstitution systems, including the polymerase subunits PB1, PB2, and PA, the NP, and a viral minigenome coding for firefly luciferase reporter under the control of a viral promoter (pPol-I FF-Luc). Expression plasmids for the indicated Mx1 constructs and for Renilla luciferase under the control of a constitutive promoter (RLuc) were cotransfected. At 24 h posttransfection, cells were lysed and firefly and Renilla luciferase activities were determined. (A and B) FLUAV H5N1 (A/Vietnam/1203/04) minireplicon system using 10 ng of PB2, 10 ng of PB1, 10 ng of PA, 100 ng of NP, 50 ng of Pol-I FF-Luc, 10 ng of RLuc, and 300 ng of Mx1 expression plasmids or the corresponding inactive mutants with a K-to-A (A) or T-to-A (B) change. Mx1 protein and NP expression was monitored by Western blotting using Mx-specific M143 (A) or FLAG-specific (B) antibody. (C) To measure the bat Mx1 effect on viral growth, 293T cells (6-well format) were transfected with 800 ng of Mx1 expression plasmids 16 h prior to infection with SC35M (H7N7, Kan1 NP) with an MOI of 0.001. At 48 h after infection, the supernatants were harvested and titrated. Viral titer of the empty vector control was set to 100% (corresponding to 105 to 106 PFU/ml in four independent experiments). (D) THOV minireplicon system using 10 ng of PB2, 10 ng of PB1, 10 ng of PA, 25 ng of NP, 50 ng of Pol-I FF-Luc, 10 ng of RLuc, and 300 ng of Mx1 expression plasmids. At 24 h posttransfection, the cell lysates were analyzed for luciferase activity and for Mx1 protein and NP expression by Western blotting. Firefly luciferase activity was normalized to Renilla luciferase activity (FF/RL). The empty vector control was set to 100%. Significance was calculated with a one-sided Student t test (n = 3). ns, nonsignificant. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). The numbers above the columns indicate fold changes in viral polymerase activity (A, B, and D) or viral titer (C) in the presence of wild-type Mx1 compared to the inactive control.
For the following studies, we limited our analysis to one representative bat Mx1 candidate from each family. Furthermore, we used FLAG-tagged bat Mx1 for a more comparable detection of the recombinant proteins in the Western blot analysis. To improve comparability of the expression of the inactive Mx1 mutants that were used as negative controls with that of the wild-type constructs, we switched to a threonine-to-alanine change in the conserved switch I region of the GTP binding domain, threonine-103 in MxA, resulting in GTPase-inactive mutants with more robust expression levels (40). Accordingly, repetition of the FLUAV minireplicon system under these optimized conditions (Fig. 3B) reproduced the results achieved in the initial analysis whose results are shown in Fig. 3A.
Next we tested the inhibitory activity of the bat Mx1 proteins against viral replication in infected cells. 293T cells were transfected with the Mx expression constructs and then infected with a low dose of a recombinant avian FLUAV that shows enhanced sensitivity to the expression of human MxA (42). Accordingly, the titers of virus progeny in the supernatants of human MxA-expressing cells were reduced by almost 2 logs in comparison to cells expressing the antiviral inactive MxA(T103A) mutant (Fig. 3C). Likewise, bat Mx1-expressing cells showed reduction of virus titers in the supernatants, with E. helvum Mx1 showing the highest and P. pipistrellus Mx1 showing a somewhat reduced antiviral effect (Fig. 3C). These results were in line with those of the minireplicon assays whose results are shown in Fig. 3A and B. Notably, viral NP was not detectable by Western blotting in the Mx-expressing cells at 48 h postinfection (p.i.), indicating an efficient block of viral gene expression by bat Mx1 (Fig. 3C).
To investigate the antiviral potential of bat Mx1 proteins against orthomyxoviruses in more detail, we tested whether they had an inhibitory effect on the polymerase complex of Thogoto virus (THOV), a tick-transmitted orthomyxovirus with high sensitivity to the antiviral effect of human MxA (43). Interestingly, recombinant bat Mx1 showed almost no activity against the viral polymerase complex of THOV (Fig. 3D), whereas the expression of human MxA led to a 99% reduction of the viral polymerase activity, as published previously (44). This indicates an intriguing antiviral specificity of bat Mx1 among different orthomyxoviruses.
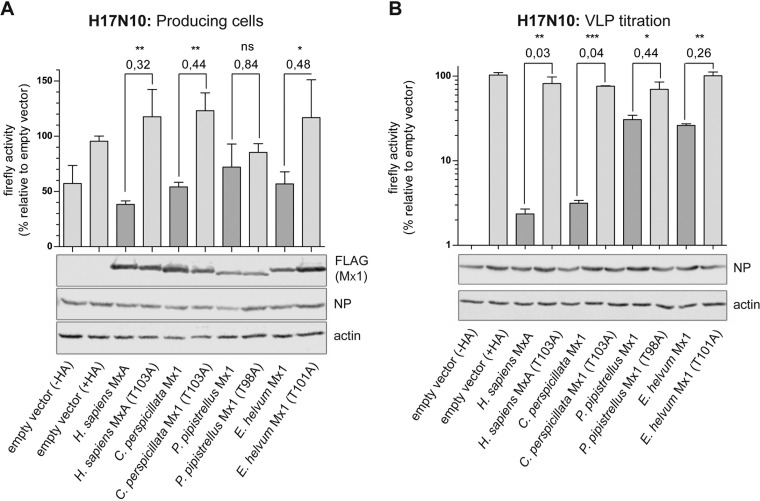
Recently, influenza A-like virus infections were identified in bats of the Phyllostomidae family by detection of viral nucleic acids and positive serology (9, 10). Since the influenza A-like bat virus itself has not been isolated so far, we used artificial, replication-deficient but transcriptionally active virus-like particles (VLPs) to analyze the effect of bat Mx1 proteins. The VLPs carried a firefly luciferase minigenome, the polymerase subunits and NP of the bat (H17N10) virus, and all other structural elements as well as the surface glycoproteins of an H7N7 influenza virus with broad tissue specificity as described before (45).
The H17N10 VLPs were produced in 293T cells (producing cells) in the presence of cotransfected Mx1. At 48 h posttransfection, the supernatants containing newly produced VLPs were collected and the firefly luciferase activity in the lysates of the producing cells was determined as a measure of viral polymerase activity (Fig. 4A). Coexpression of human MxA reduced reporter gene expression to 32%, Mx1 of C. perspicillata and E. helvum reduced reporter gene expression to about 46%, and Mx1 of P. pipistrellus resulted in no significant reduction compared to the empty vector (with hemagglutinin [HA]) control lacking Mx (Fig. 4A). In addition, we determined release of progeny VLPs in the supernatants of the transfected producer cells by incubating naive indicator cells (MDCK II) with the supernatants. To support transcription of the viral reporter minigenome carried by the VLPs, the indicator cells were additionally infected with an SC35M (H7N7) helper virus. The firefly luciferase activity detected in the indicator cells is a measure of the amount of VLPs (VLP titration [Fig. 4B]). Interestingly, human MxA and C. perspicillata Mx1 reduced VLP formation by 96%, whereas Mx1 of E. helvum and P. pipistrellus showed only reduced inhibitory capacity (Fig. 4B). As a control, VLPs were produced in the absence of Mx1 and the viral hemagglutinin protein. Consequently, the supernatant did not contain functional VLPs and reporter gene expression was not detectable in the lysates of the indicator cells (Fig. 4B). Western blot analysis showed similar expression levels of Mx proteins and of transfected bat NP in the producing cells (Fig. 4A). Comparable infection of the indicator cells with the SC35M helper virus was monitored by equal levels of expression of viral NP (Fig. 4B).
FIG 4.
Influence of bat Mx1 on the polymerase activity of bat influenza (H17N10). (A) A FLUAV (H17N10) minireplicon system using 10 ng of PB2, 10 ng of PB1, 10 ng of PA, 10 ng of NP, and 50 ng of Pol-I FF-Luc was cotransfected with the helper plasmids encoding the additional structural proteins from H7N7 virus as well as 300 ng of Mx1 expression plasmids (producing cells). At 48 h posttransfection, supernatants were collected and the cell lysates were analyzed for firefly luciferase activity and for Mx1 protein and NP expression by Western blotting. (B) FLUAV (H17N10) VLP titration. Indicator cells (MDCK II) were coinfected with 50 μl of the VLP-producing cell supernatants and with the SC35M helper virus for 24 h. As a control, indicator cells were treated with a comparable amount of donor cell supernatant produced in the absence of the viral hemagglutinin (−HA). Firefly luciferase activity was measured in cell lysates, and infection of indicator cells with SC35M helper virus was monitored by Western blotting detecting NP. The empty vector control was set to 100%. Significance was calculated with a one-sided Student t test (n = 3). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). The numbers above the columns indicate fold changes in viral polymerase activity in the presence of wild-type Mx1 compared to the inactive control.
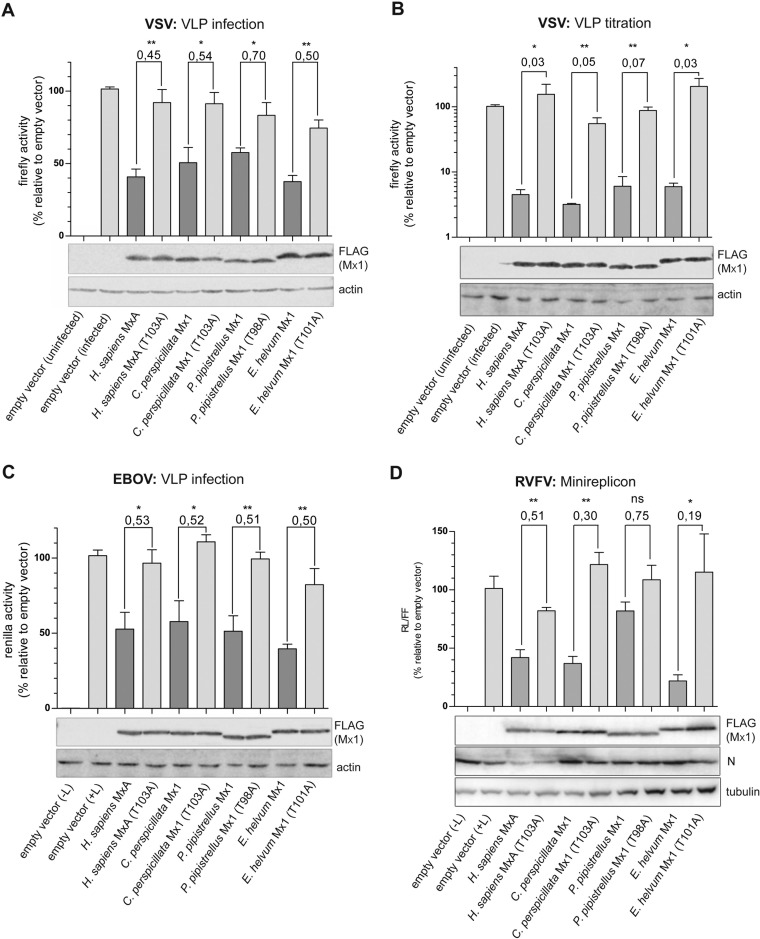
Antiviral activity of bat Mx1 against VSV.
Human MxA efficiently blocks replication of vesicular stomatitis virus (VSV) (38). To test the antiviral activity of bat Mx1 proteins against rhabdoviruses, we used a VSV glycoprotein (VSV-G)-pseudotyped, propagation-incompetent VSV replicon expressing firefly luciferase, VSV*ΔG(FLuc) (46). VLP infection of the Mx1-transfected cells resulted in the expression of luciferase correlating to the viral polymerase activity. Transfection of human MxA and of the bat Mx1 proteins blocked viral polymerase activity by 50% compared to that of the inactive controls (Fig. 5A). However, in this experimental setup, Mx1 expression plasmids might not have been present in all infected cells, thereby diluting a possible inhibitory effect. Therefore, we repeated this experiment with cells cotransfected with a VSV-G expression plasmid together with the different Mx1 constructs. Because VSV*ΔG(FLuc) replicon lacks the glycoprotein gene, we reconstituted viral particle formation only in the transfected cells coexpressing VSV-G and Mx1. Formation of the newly formed pseudotyped viral particles in the supernatants of the cotransfected cells was determined by transferring them onto naive indicator cells (VLP titration [Fig. 5B]). In this setting, particle formation was reduced by about 95% due to the action of human MxA or the bat Mx1 constructs compared to their inactive controls (Fig. 5B). Taken together, these results indicate that bat Mx1 proteins inhibit VSV gene expression as well as replication of the viral genome.
FIG 5.
Block of VSV, EBOV, and RVFV replication by bat Mx1 and human MxA. (A) 293T cells were transfected with 300 ng of the indicated Mx1 constructs (12-well format). At 24 h posttransfection, the cells were infected with VSV*ΔG(FLuc) for 24 h. Firefly luciferase activity was determined in the cell lysates. Mx1 expression in cell lysates was controlled by Western blotting. (B) 293T cells were cotransfected with 300 ng of VSV-G and 300 ng of the indicated Mx1 constructs (12-well format) and were infected with VSV*ΔG(FLuc) 24 h later. At 24 h postinfection, 50-μl volumes of the supernatants of the transfected and infected cells were used to infect naive 293T indicator cells (24-well format) to analyze the amount of newly produced particles. Firefly luciferase activity was measured in the indicator cell lysates. Luciferase activity carried by VLPs produced in cells transfected with empty vector was set to 100%. Significance was calculated with a one-sided Student t test (n = 3 [A] and n = 5 [B]). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). (C) 293T cells (12-well format) were cotransfected with 50 ng of NP, 375 ng of L, 50 ng of VP35, 30 ng of VP30, 90 ng of TIM-1, and 300 ng of the indicated Mx1 expression plasmids 24 h prior to infection with EBOV VLPs. As a control, cells were treated with comparable amounts of VLP preparation produced in the absence of the L construct (−L). At 24 h postinfection, the activity of the Renilla luciferase encoded by the viral genome was determined. The empty vector control (without Mx expression) was set to 100%. Mx1 expression was controlled by Western blotting. Significance was calculated with a one-sided Student t test (n = 3). *, P ≤ 0.05; **, P ≤ 0.01). (D) 293T cells were transfected with 250 ng of the indicated Mx1 constructs together with plasmids encoding RVFV N, L, and M and the Renilla luciferase-encoding minigenome (250 ng each). At 48 h posttransfection, cells were lysed and the Renilla luciferase activity measured. As controls either RVFV L was omitted or the Mx1 plasmid was replaced with an empty vector. The activity of the empty vector control was set to 100%. Statistical significance was calculated with a one-sided Student t test (n = 3). *, P ≤ 0.05; **, P ≤ 0.01). Cell lysates were subjected to Western blot analysis using FLAG-, RVFV N-, and β-tubulin-specific antibodies.
Antiviral effect against EBOV.
Old World fruit bats were identified as possible reservoirs of Ebola virus (EBOV) (7, 8). Therefore, the antiviral activity of bat Mx1 against EBOV was determined using a newly established VLP assay (47). An artificial tetracistronic minigenome encoding the viral VP24, VP40, and GP structural proteins and a Renilla luciferase reporter was cotransfected with expression plasmids encoding the viral RNA polymerase L, the phosphoprotein VP35, the transcription initiation factor VP30, and the nucleoprotein. These helper plasmids support transcription and replication of the viral minigenome as well as packaging and formation of VLPs. To test the effect of Mx1 on EBOV replication, Mx1 expression plasmids were cotransfected with the viral helper plasmids encoding L polymerase, VP35, VP30, NP, and the cellular TIM-1 adhesion factor. The last was included to enhance susceptibility of the cells to infection with EBOV VLPs at 24 h posttransfection. Using this approach, a robust Renilla luciferase expression was detected in the cell lysates (Fig. 5C). Omission of the L helper plasmid abolished expression of the luciferase encoded by the minigenome, indicating the dependency of reporter gene expression on viral polymerase activity. Coexpression of human MxA or the three bat Mx1 proteins reduced luciferase activity to about 50% compared to the activity measured in the presence of the respective inactive mutants, indicating that bat Mx1 proteins are able to control EBOV polymerase activity.
Antiviral effect against bunyaviruses.
Infections with bunyaviruses, including hantavirus and nairovirus, have been detected in bats from Africa and Asia (11–13). To test a possible antiviral function of bat Mx1 against bunyaviruses, we used a polymerase reconstitution-minireplicon system for Rift Valley fever virus (RVFV) (48). The cells were cotransfected with plasmids encoding Mx1 and expression constructs coding for viral L, M, and N proteins together with a Renilla luciferase-encoding minigenome construct. In this setting, MxA reduced the activity of the viral polymerase by 50% compared to that of its inactive control (Fig. 5D). Interestingly, coexpression of Mx1 of E. helvum and C. perspicillata showed an even stronger effect, with 80% and 60% inhibition, respectively, whereas Mx1 of P. pipistrellus did not reduce the viral polymerase activity significantly (Fig. 5D), indicating differences in antibunyavirus activities of the Mx1 proteins from different bat species.
Evolutionary analysis of bat Mx1.
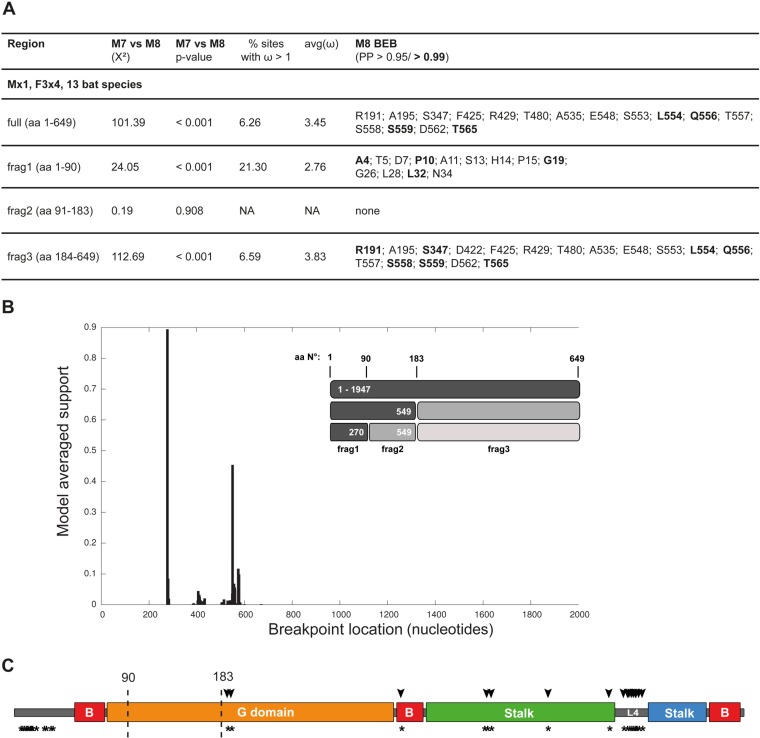
After confirmation of the antiviral capacity of the bat Mx1 proteins against a diverse range of RNA viruses, we used our Mx1 cDNA sequences and in addition other publicly available bat Mx1 sequences to analyze the evolution of the bat Mx1 genes with respect to their phylogenetic relationship. We tested whether rates of nonsynonymous changes (dN) exceeded synonymous changes (dS) using the PAML software suite (49), with a dN/dS (ω) ratio greater 1 indicating positive selection. Analysis of the 13 bat Mx1 sequences (Fig. 2A) revealed 16 positively selected sites, mostly in the stalk, with 9 of these positions concentrated in the flexible loop L4 (Fig. 6A and C, arrowheads). The calculated likelihood values and posterior probabilities were robust under various codon frequency models (F3x4, F1x4, and F61) and the initial ω values used.
FIG 6.
Positive selection of bat Mx1. (A) Results of the evolutionary analysis for positively selected sites in full-length bat Mx1 (amino acids [aa] 1 to 649) of 13 bat Mx1 coding sequences as presented in Fig. 2A. In addition, positive selection was analyzed for the fragments of the GARD analysis—frag1 (aa 1 to 90), frag2 (aa 91 to 183), and frag3 (aa 184 to 649)—with respect to the sequence of M. daubentonii. P values were achieved by performing chi-squared tests on twice the difference of the computed log likelihood values of the models disallowing (M7) or allowing (M8) dN/dS (ω) values of >1. The BEB column lists rapidly evolving sites with a dN/dS value of >1 and a posterior probability of >0.95, determined by the Bayes empirical Bayes implemented in CODEML. Indels were removed from the alignment prior to evolutionary analyses. (B) Evidence of evolutionary breakpoints in the alignment of the 13 bat Mx1 sequences (as shown in Fig. S1 in the supplemental material) identified with GARD. The best-fitting nucleotide substitution model (HKY85) was applied. The GARD fragments and the supported breakpoints are indicated. (C) Illustration of the primary structure of bat Mx1 adapted to the crystal structure of MxA (27): the unstructured regions (gray), the bundle signaling elements (B, red), the G domain (orange), the stalk (green, blue), and loop 4 (L4). Amino acid positions correspond to the full-length Mx1 sequence of M. daubentonii. The rapidly evolving sites are indicated as arrowheads or asterisks (for the GARD fragments in panel B). Breakpoints of recombination identified by GARD are indicated by vertical dashed lines.
A recent phylogenetic analysis of mammalian Mx1 genes detected multiple recombination events between Mx paralogs that might influence the evolutionary history of different Mx fragments and therefore the comparative analysis of Mx orthologs (50). We analyzed the bat Mx1 sequences for recombination events using GARD, a genetic algorithm for recombination detection (51), and identified two recombination breakpoints (Fig. 6B). Both breakpoints are located near exon/intron boundaries, when referring to the genomic sequence of M. lucifugus as a reference (ENSMLUG00000011447). Breakpoint 1 is located at the 5′ end of exon 3 and breakpoint 2 at the 3′ end of exon 4. Therefore, fragment 1 comprises exons 1 and 2 and the first 5 nucleotides (nt) of exon 3. Fragment 2 comprises the rest of exon 3 and exon 4, except the last 15 nt at the 3′ end of exon 4. Fragment 3 comprises those 15 nt of exon 4 and exons 5 to 13. Breakpoints in similar regions of mammalian Mx genes have been recently described by Mitchell et al. for the analyses of rodent and primate Mx homologs (50). We independently repeated this recombination analysis of the bat Mx1 sequences using the RDP tool (52) and confirmed the most significant breakpoints, with slightly shifted positions compared to the GARD analysis: nucleotide position 246 instead of 270 and 554 instead of 549. The identified cDNA fragments code for important functional regions in the Mx structure. Fragment 1 encodes the first helix of the BSE, fragment 2 highly conserved elements of the GTP-binding domain and fragment 3 the C-terminal part of the G domain and the stalk (Fig. 6C). However, the detected breakpoints in our bat Mx1 cDNA analysis did not show a significant topological incongruence based on a Kishino Hasegawa (KH) test (53) applied after the GARD analysis. Most likely, such KH-insignificant breakpoints arise due to variation in branch lengths between fragments. This could indicate forms of recombination or other processes, like heterotachy.
An extended analysis for positions under positive selection was separately performed for the fragments obtained by the GARD analysis. Interestingly, fragment 1 now showed several residues under positive selection within the N-terminal region in addition to the positions identified in the C-terminal fragment 3 (Fig. 6A and C, asterisks). We performed the same calculations for the fragments resulting from the RDP analysis with no impact on the significant positively selected sites identified above. Again, fragment 2 showed a strong purifying selection of all residues, attesting to its evolutionary conservation. The two hot spots of positive selection in the overall conserved structure of the protein might be of specific importance for evolutionary adaptation processes in the host-pathogen arms race between bats and their viral infectious agents.
DISCUSSION
Bats are recognized as an important reservoir of potential zoonotic viruses (54). However, it is unclear how bats deal with the infection, replication, and possible persistence of these viruses, which often induce severe to fatal infections in humans upon zoonotic transmission. In this study, we evaluated the role of the IFN-induced bat Mx1 proteins in the control of diverse RNA viruses and found a broad antiviral spectrum similar to that of the human MxA ortholog against orthomyxo-, rhabdo-, filo-, and bunyaviruses.
Our phylogenetic analysis of Mx1 proteins grouped the bat Mx1 sequences according to their affiliation in the three different bat families (55). Within the individual families, bat Mx1 showed around 80% sequence identity. Between the families, the bat Mx1 sequences displayed a reduced (around 70%) identity, which is comparable with the sequence identity to other mammalian Mx1 proteins, e.g., human MxA. This may reflect the long, about 40 to 50 million years, independent evolutionary history and diversification of Mx1 genes in the individual Chiroptera families (56).
A detailed examination of the mammalian Mx1 sequences revealed a high proportion of invariant amino acid residues under purifying selection, supporting the view of Mx proteins as dynamin-like molecular machines with a sophisticated, highly conserved structure and mechanism (27) that allow evolution-driven variations only in a few flexible regions (44, 50). Accordingly, when analyzing the bat Mx1 sequences, we identified residues under positive selection in two variable and surface-exposed regions. Most residues under positive selection were identified in the N terminus ahead of the first BSE helix and in the C-terminal loop L4 (Fig. 6). The accumulation of residues under positive selection in these two surface-exposed, variable regions of Mx1 indicates a likely structural flexibility of these two regions compared to the overall high structural conservation of the majority of the bat Mx1 molecules. Interestingly, positively selected positions in the N terminus were identified only when individual gene segments were analyzed. Using GARD analysis, we detected two breakpoints resulting in three gene segments, suggesting exchanges between ancient bat Mx paralogs by recombination events (Fig. 6B and C). A comparable analysis using the orthologous primate MxA genes identified clusters of positively selected residues in the L4 loop, which has been previously identified as a determinant of Mx antiviral specificity (44). Positively selected positions in the N-terminal region were identified in the paralogous primate MxB genes (50). The various positively selected residues in these two regions of bat Mx1 molecules argue for a long-standing conflict of bats with diverse viral agents (57), which circulated or are still present in the animals.
The selection of diversifying mutations in the bat Mx1 sequences suggests a critical role of bat Mx1 in the innate immune response to various viral pathogens. A multitude of diverse viral agents has been identified in various bat species that possibly executed a significant pressure driving the evolution of bat Mx1 orthologs (54). We first tested the activity of bat Mx1 against three different orthomyxoviruses because influenza A-like virus infections have been detected in bats of the Phyllostomidae family (9, 10). Using a polymerase reconstitution assay reflecting the activity of the polymerase complex of an avian H5N1 and infection experiments with an H7N7 influenza virus, we detected a quite remarkable suppression activity by most of the bat Mx1 proteins, comparable to the activity of human MxA (Fig. 3) (41, 58). Remarkably, however, the tick-transmitted THOV polymerase complex was not significantly affected by bat Mx1. Although bats are known to be infested by ticks and could come in contact with arboviruses (59, 60), infections of bats with Thogoto viruses have not been reported. THOV is extremely sensitive to human MxA (Fig. 3D) (43, 61) and murine Mx1 (62) in cell culture as well as in vivo. Therefore, the insensitivity of THOV to three different bat Mx1 proteins is astonishing and suggests that bats never had contact with this virus and did not have the chance to evolve their Mx1 proteins to powerful anti-THOV weapons. In contrast, replication of the recently described bat influenza A-like (H17N10) virus (9) was efficiently controlled by bat Mx1. The production of new VLPs (Fig. 4B) was much more strongly inhibited than the transcriptional activity of the viral polymerase complex (Fig. 4A), in line with previous results indicating that human MxA preferentially targets viral genome replication (39). In this setting, especially Mx1 of C. perspicillata showed a strong inhibitory effect comparable to that of human MxA (Fig. 4B). These results point to a family-specific control of influenza A-like viruses in bats in which H17N10 virus infections have been detected (10), suggesting an arms race between host innate immunity and bat influenza viruses that runs in favor of the bat host.
In addition, we tested three additional representatives of negative-strand RNA viruses of the Rhabdoviridae, Filoviridae, and Bunyaviridae. Bat Mx1 suppressed the activity of the viral polymerases with an efficacy that was comparable to that of human MxA (Fig. 5). Using a recently established EBOV-like particle infection system (47), we showed for the first time an antiviral effect of Mx proteins against filoviruses (Fig. 5C). Particularly Mx1 of E. helvum, a member of the African fruit bats, which are assumed to serve as reservoirs for EBOV and other filoviruses (7, 8, 63), showed a significant suppression of the viral polymerase activity, suggesting a role in the control of EBOV replication in this species.
The inhibitory activity of E. helvum Mx1 was the most effective one in all three virus systems mentioned above (Fig. 5). This contrasts with the moderate inhibitory effect of E. helvum Mx1 on the H17N10 influenza A-like viral replication (Fig. 4B) and suggests formation of individual antiviral specificities during bat Mx1 evolution. It is worth mentioning that we tested the activity of bat Mx1 proteins only in the context of human 293T cells, and additional factors from the bat cell environment might further modulate their activity.
The three RNA viruses VSV, EBOV, and RVFV have a cytoplasmic replication phase, whereas influenza viruses replicate within the cell nucleus. Bat Mx1 proteins accumulated in the cytoplasm of transfected cells (Fig. 1C), very similar to the localization of the orthologous human MxA (37). The molecular action of human MxA against RNA viruses has been intensely studied and reveals a spectrum of strategies to block viral replication (26). However, the fundamental basis of its action is the recognition of viral nucleocapsids or ribonucleoprotein complexes, in which the viral RNA genome is tightly associated with the viral nucleoprotein and the viral polymerase complex. For example, the replication of VSV is inhibited at an early step of the viral life cycle. It was shown that already primary transcription of incoming viral nucleocapsids is blocked by MxA (38). Accordingly, isolated ribonucleoprotein complexes of VSV are directly hampered by purified, recombinant MxA (31). Bunyaviruses are efficiently controlled by MxA as well (61, 64, 65). MxA binds to the viral nucleoprotein, sequesters newly synthesized nucleoprotein or viral ribonucleoprotein complexes in endoplasmic reticulum (ER)-associated aggregates, and efficiently blocks transcription and amplification of the viral genome (30, 48, 66). Orthomyxoviruses might be affected by a similar mechanism. Human MxA binds the nucleoprotein of influenza A virus and THOV and prevents the translocation of viral nucleocapsids into the nucleus, the site of orthomyxovirus transcription and genome replication (29, 67, 68). However, incoming nucleocapsids of influenza viruses may escape the early recognition by MxA and may reach the cell nucleus to start primary transcription. In this situation secondary transcription is blocked, possibly by MxA-mediated sequestration of newly synthesized viral nucleoprotein (39, 67). In our future research, we will focus on detailed molecular analyses of the antiviral action of the orthologous bat Mx1 proteins, especially their action against filoviruses. This can possibly improve strategies to fight emerging viral pathogens.
Zhou et al. recently reported an elevated, constitutive expression of IFN-α in unstimulated cells of Pteropus alecto (Pteropodidae), which was not further enhanced by viral infection (23). In our present analysis, we used Mx1 expression as a marker of type I IFN activity as shown previously in cell culture and in vivo (24, 25). Analyzing cells from three different bat families, including Pteropodidae, we detected only minor expression of Mx proteins in untreated cells, which was strongly upregulated following treatment of the cells with IFN-α (Fig. 1A). The Mx1 induction in the bat cells was comparable to the induction of MxA in human alveolar epithelial A549 cells that were treated in parallel (data not shown). However, in our experimental setup we did not analyze accumulation of Mx1 transcripts in untreated bat cell cultures and did not consider the role of IFN-λ in bat Mx1 induction, which was shown to be important for Mx1 expression in human, mouse, and bat systems (21, 24, 25). Astonishingly, bat cells from the Pteropodidae and Vespertilionidae families responded to treatment with the human IFN-α B/D variant, whereas CarLu cells (Phyllostomidae) were unresponsive. They responded only to authentic conditioned medium of virus-infected CarLu cells, indicating interesting family-specific differences in the IFN-α receptor specificity. Infection with RVFV clone 13 induced comparable levels of Mx1 in all bat cells used in our study, including CarLu cells (data not shown). Accordingly, the accumulation of Mx1 argues for a virus-induced type I or type III IFN production and upregulation of Mx1 as described previously (17, 21). Therefore, bat Mx1 expression can be used as a reliable marker for the detection of type I and type III IFN activity during virus infection and IFN treatment.
The overall efficient inhibition of various viral pathogens by bat Mx1 indicates that this ISG product exhibits a central role in the control of viral replication in bats. Of note, we cannot speculate about the identity of the pathogens that drove the ancient arms race with bat Mx1, resulting in positive selected positions in today's bat species. Since the bat Mx1 cDNAs showed rather comparable antiviral activities against the limited spectrum of RNA viruses employed in the present study, our results cannot fully explain the evolution of bat Mx1 proteins. Follow-up studies should extend the spectrum of viruses tested for bat Mx1 sensitivity and will refine the connection of Mx1 antiviral capacity with the genetic evolution of these important components of the innate antiviral defense.
MATERIALS AND METHODS
Cells, IFN treatment, synthesis of CarLu IFN, and RNA extraction.
Bat cell cultures were obtained from the cell culture collection of the Institute of Virology, University of Bonn. All bat cell cultures were immortalized by lentiviral transduction with the simian virus 40 (SV40) large T antigen and tested negative for mycoplasma, SV5, lyssavirus, and filovirus contaminations as described previously (17). The species type of the cell cultures was confirmed by amplification of the COX1 gene, encoding cytochrome c, as previously described (69). Cultures of human embryonic kidney (HEK) 293T cells, human cervical carcinoma HeLa cells, Madin-Darby canine kidney epithelial cells (MDCK II), Carollia perspicillata CarLu/1 (70), Myotis daubentonii MyDauNi/2c (17), Pipistrellus pipistrellus PipNi/1 (17), Eidolon helvum EidNi/41 (17), Hypsignathus monstrosus HypNi/1 (71), and Rousettus aegyptiacus RoNi/7 (17) cells were cultivated in Dulbecco's modified Eagle's medium (DMEM) with 5 to 15% fetal calf serum (FCS) at 37°C.
To induce Mx expression, cells were treated for 24 h with 1,000 U/ml of human IFN-α B/D (72). CarLu/1 cells were unresponsive to this treatment. Therefore, they were infected with RVFV clone 13 (36) at a multiplicity of infection (MOI) of 1 for 48 h. The supernatant with endogenous IFN was harvested, dialyzed against glycine buffer (pH 2), and subsequently used for IFN treatment of naive CarLu/1 cells. Total RNA of IFN-stimulated cells was isolated using the TRIzol RNA isolation reagent (Life Technologies). Total RNA extracted from Sturnira lilium cells was kindly provided by M. Schwemmle, Freiburg, Germany.
cDNA cloning.
Total RNA (1 μg) was reverse transcribed using the RevertAid H Minus First Strand cDNA synthesis kit (Thermo Fisher Scientific). In order to clone cDNA from the coding region of bat Mx, PCRs were performed with primers specific for conserved regions of Mx sequences. Mx1 specific primer sequences for the different bat species are listed in Table S1 in the supplemental material. To determine the remaining 5′ and 3′ terminal sequences, rapid amplification of cDNA ends (RACE) was performed using a 5′/3′ RACE 2nd Generation kit (Roche). RACE primers were designed based on the sequence information of the previously obtained bat Mx cDNA fragments. All amplification products were sequenced and assembled in silico. The open reading frames (ORFs) were determined and cloned into the expression vector pCAGGS under the control of the chicken actin promoter (73) using specific primers which harbored an N-terminal FLAG tag. The GTPase-inactive mutants corresponding to the change in human MxA(T103A) at position 103 were cloned by a mutagenesis PCR with a primer set that harbored the mutation.
Transfection, Western blotting, and immunofluorescence analysis.
Transfections of 50 to 80% confluent 293T cells were performed with JetPEI (Polyplus). For Western blotting, cells were lysed in 1× passive lysis buffer (Promega). To denature the proteins 1× Laemmli buffer was added and the samples were incubated at 95°C for 5 min. The protein lysates were separated with a 12% SDS-PAGE gel and transferred onto a polyvinylidene difluoride (PVDF) membrane (Merck Millipore). Detection of the primary antibodies was performed using fluorescence-labeled secondary antibodies (Li-Cor).
For immunofluorescence analysis HeLa cells were seeded on coverslips and transfected with 1 μg of the Mx1 expression plasmid per well of a six-well plate. Twenty-four hours posttransfection, cells were fixed with 3% paraformaldehyde (Roth) for 20 min and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) for 10 min. Then cells were incubated with 5% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 1 h at room temperature. Detection of Mx proteins was performed in 1% BSA in PBS for 1 h at room temperature using the Mx-specific M143 antibody, which recognizes the highly conserved G domain in Mx proteins of different vertebrate species (35), and a secondary donkey anti-mouse IgG (Alexa Fluor 488; Invitrogen). Nuclear genomic DNA was stained with TO-PRO-3 iodide (Invitrogen). Cells were analyzed by confocal fluorescence microscopy.
Orthomyxovirus minireplicon systems.
To reconstitute the polymerase of FLUAV H5N1 (A/Vietnam/1203/04) (58) or THOV (strain SiAr126) (74), 293T cells were cultured in 12-well plates for 24 h and then cotransfected with 10 ng of expression plasmids encoding the polymerase subunits PB2, PB1, and PA (for FLUAV and THOV) and 100 ng (FLUAV) or 25 ng (THOV) of NP plasmids as described previously (41, 44). In addition, 50 ng of the artificial viral minigenomes encoding firefly luciferase in negative-sense orientation flanked by viral noncoding regions (Pol-I FF-Luc) and 10 ng of a plasmid coding for a Renilla luciferase under the control of the constitutive SV40 promoter (SV40-RL) were added. To determine the effect of Mx1 on the activity of the viral polymerase, 300-ng quantities of the Mx1 expression constructs were cotransfected. At 24 h posttransfection the firefly and Renilla luciferase activities were measured (dual-luciferase reporter kit; Promega). Firefly luciferase activity was normalized to Renilla luciferase activity, and the empty vector control was set to 100%. Western blot analyses were performed with specific antibodies directed against FLAG (Sigma-Aldrich) or Mx-specific M143 (35), FLUAV-NP (Serotec), THOV-NP (44), and β-actin (Sigma-Aldrich).
Infection experiments with FLUAV (H7N7).
To analyze the effect of bat Mx1 on the replication of FLUAV, 293T cells seeded in 6-well plates were transfected with 800 ng of FLAG-tagged Mx1 encoding plasmids 16 h prior to infection with an MOI of 0.001 of recombinant SC35M (H7N7) encoding NP of A/Thailand/01/04 (Kan-1, H5N1) (42). At 48 h postinfection, the supernatants were harvested and viral titers were measured using plaque assay on MDCK cells. Western blot analyses on the transfected cell lysates were performed with antibodies directed against FLAG (Sigma-Aldrich), FLUAV-NP (Thermo Fisher Scientific), and β-actin (Sigma-Aldrich).
H17N10 VLP system.
H17N10 virus-like particles (VLPs) were produced in the presences of Mx1. 293T donor cells were transfected (12-well format) with expression plasmids coding for PB2 (50 ng), PB1 (50 ng), PA (10 ng), and NP (100 ng) of H17N10. In addition, expression plasmids encoding the viral minigenome Pol-I FF-Luc (50 ng) as well as HA (100 ng), neuraminidase (NA; 100 ng), M1 (125 ng), M2 (20 ng), and nuclear export protein (NEP) (25 ng) of SC35M (H7N7) were transfected as previously described (45), together with 300-ng quantities of the different Mx1 constructs. As a negative control, the HA plasmid was omitted. At 48 h posttransfection, the supernatants were harvested and the firefly luciferase activity in the lysates of the VLP-producing cells was measured. Residual plasmids in the supernatants were digested via DNase treatment for 1 h at 37°C. To detect the VLPs released from the transfected 293T donor cells, MDCK II indicator cells (in a 24-well format) were infected with 50 μl per well of the VLP-containing supernatants of the producer cells. Because the viral polymerase enclosed in the VLPs is not strong enough to robustly express the luciferase gene from the minigenome, the indicator cells were additionally infected with an SC35M helper virus (H7N7) at an MOI of 1. At 24 h postinfection, the MDCK cells were lysed, the firefly luciferase activity was measured, and the empty vector control was set to 100%. Western blot analyses of the VLP producer and indicator cell lysates were performed with antibodies directed against FLAG (Sigma-Aldrich), FLUAV NP (Thermo Fisher Scientific), and β-actin (Sigma-Aldrich).
VSVΔG replicon system.
The VSV*ΔG(FLuc) replicon in which the viral G gene is replaced by firefly luciferase cDNA was produced as previously described (46). 293T cells were cultured in 12-well plates and transfected with 300 ng of Mx1 or cotransfected with 300 ng of Mx1 and 300 ng of VSV-G expression plasmids. At 24 h posttransfection, the cells were infected with VSV*ΔG(FLuc) using an MOI of 1 (VLP infection). Cells were lysed after 24 h, and firefly luciferase activity was determined. The supernatants of the transfected cells were harvested and were used to infect 293T cells in 12-well plates (VLP titration). At 24 h postinfection, firefly luciferase activity was measured in the cell lysates. The luciferase activity detected in the cells that received VLPs of the empty vector control-transfected producer cells was set to 100%. Western blot analyses were performed with lysates of the transfected producer cells using antibodies directed against FLAG (Sigma-Aldrich) and β-actin (Sigma-Aldrich).
EBOV VLP systems.
EBOV VLPs were produced as previously described (47) by transfection of 293T cells in 90-mm dishes with plasmids coding for NP (900 ng), VP35 (900 ng), VP30 (500 ng), L (7,000 ng), T7 (1,800 ng) and an artificial viral tetracistronic minigenome, p4cis-vRNA-RenLuc (1,800 ng), which encodes Renilla luciferase, VP40, GP1,2, and VP24. As a negative control, L was omitted. At 24 h after transfection, the medium was replaced with DMEM with 5% FCS. At 72 h posttransfection, the VLP-containing supernatant was harvested and clarified at 500 × g for 10 min.
For infection experiments, 293T cells were cultured in 12-well plates for 24 h and cotransfected with 50 ng of NP, 375 ng of L, 50 ng of VP35, 30 ng of VP30, 90 ng of TIM-1 (adhesion factor of EBOV), and 300 ng of Mx1 expression plasmids. At 24 h posttransfection, the transfected cells were infected with 50 μl of the VLP containing supernatant per well. Control cells were treated with comparable amounts of a VLP preparation which was produced in the absence of L. At 24 h postinfection, the activity of the Renilla luciferase encoded by the viral minigenome was determined (luciferase reporter kit; Promega). The empty vector control was set to 100%. Western blot analysis of the infected cell lysates was performed with specific antibodies directed against FLAG (Sigma-Aldrich) and β-actin (Sigma-Aldrich).
RVFV polymerase assay.
To determine the effect of Mx1 on RVFV polymerase activity, 293T cells were transfected with expression plasmids encoding Mx1 (250 ng), RVFV L, M, and N (250 ng each), a minigenome construct coding for the full-length RVFV S segment with the NSs ORF replaced by Renilla luciferase (250 ng), firefly luciferase under the control of the constitutively active SV40 promoter (50 ng), and a dominant negative PKR construct (125 ng) as described previously (48). As a control, the RVFV L plasmid was omitted and the Mx1 plasmid was replaced by an empty vector. At 4 h posttransfection, the medium was exchanged and the cells further incubated for 48 h at 37°C. Cells were lysed and the luciferase activities were determined. The activity of Renilla luciferase was normalized to firefly luciferase, and empty vector control with RVFV L was set to 100%. Expression of FLAG-Mx1, RVFV-N (antibody; a gift from Alejandro Brun), and tubulin was monitored by Western blotting.
Statistical analyses.
Statistics were determined with the GraphPad Prism 5 software. Each experiment was done in technical duplicates and performed three times. The mean values of the technical duplicates were used for the statistical analysis. A one-sided Student t test comparing Mx1 with its corresponding antivirally inactive mutant was performed. Mean values and standard errors of the means (SEM) are reported. For viral growth analysis, the geometric mean of 4 independent experiments was calculated and statistics were performed on log-transformed values.
Data collection, alignments, and evolutionary analysis.
Mx1 sequences from the following were obtained from GenBank and incorporated in the evolutionary analysis: Myotis lucifugus (XM_006104436), Myotis davidii (XM_006754325), Myotis brandtii (XM_005885691), Pteropus alecto (NM_001290174), and Eptesicus fuscus (XM_008145691). We downloaded transcriptome sequencing data of Artibeus jamaicensis (75), a Jamaican fruit bat of the family of Phyllostomidae, from EMBL-EBI (SRR539297; HiSeq 2000; 100-bp paired-end reads) and built a de novo transcriptome assembly utilizing three different assembly tools: Velvet (v1.2.10) (76) with Oases (v0.2.08) (77), SOAPDenovo-Trans (v1.0.3) (78), and Trinity (v20131110) (79) with default parameters and multiple k-mer values. The resulting contigs of each assembly tool were merged and clustered by sequence identity using CD-HIT-EST (-c 0.95, v4.6) (80), resulting in a final assembly comprising 462,445 contigs. The assembly was further searched by BLASTN using the Mx1 cDNAs of closely related bat species (C. perspicillata and S. lilium). We identified full-length open reading frames of homologous Mx1 genes in the de novo transcriptome assembly of A. jamaicensis and incorporated the best-matching sequence in our analyses. Mx1 homologous sequences of other mammals were downloaded from Ensembl, including those of Homo sapiens (ENSG00000157601), Pan troglodytes (ENSPTRG00000013927), Canis lupus (ENSCAFG00000010172), Equus caballus (ENSECAG00000011776), Bos taurus (ENSBTAG00000030913), Mus musculus (ENSMUSG00000000386 and ENSMUSG00000023341), Rattus norvegicus (ENSRNOG00000001959 and ENSRNOG00000001963), Sus scrofa (ENSSSCG00000012077), Felis catus (ENSFCAG00000008068), and Ovis aries (ENSOARG00000010283). For M. musculus and R. norvegicus, both Mx genes were used, because of the known homology to human Mx1.
In-frame multiple-sequence alignments for all 13 bat Mx1 cDNA sequences and additional mammalian Mx1 genes were conducted using TranslatorX (v1.1) (81) and the aligner Muscle (v3.8.31) (82). All alignments were automatically adjusted, and gaps/stop codons were removed prior to subsequent analyses. To determine the evolutionary context of bat and other mammalian Mx1 genes, we constructed maximum likelihood trees using RAxML (v8.0.25) (83) using the GTRGAMMA substitution model and 1,000 bootstrap replicates.
Recombination events and positive selection analysis.
We used the GARD (51) tool to detect possible recombination events in the bat Mx1 alignment. First, an automatic model selection was applied to suggest the best-fitting nucleotide substitution model for the alignment. Then, the in-frame alignment and corresponding model were conducted to the GARD algorithm to estimate recombination breakpoints using the general discrete model of site-to-site rate variation and 3 rate classes. Furthermore, we performed an RDP (v4.80) analysis (52) to validate the GARD results.
We performed maximum likelihood tests on nested “site” models implemented in CODEML, part of the PAML software suite (49), and the HyPhy suite (84) to detect significantly positive selected sites under various models in the full bat alignment and corresponding fragments, previously identified with GARD. Alignment-specific input trees were calculated with RAxML. The coding sequence alignments were fit to paired site models that disallow (M1a and M7) or allow (M2a and M8) positive selection (ω > 1). A likelihood ratio test between paired models was performed to derive P values for each alignment. If a significant difference (P < 0.01) between model M7 versus M8 was detected, a Bayes empirical Bayes (BEB) analysis was performed to identify codons in the alignment with ω value of >1 and to calculate their impact on the positive selection (significant if posterior probability is ≥0.95). We run CODEML multiple times with different starting ω values (0, 0.5, 1, 1.5, and 2.0) and under various codon frequency models (F3x4, F1x4, and F61).
Nevertheless, one must be aware of the possibility that lower sampling of Mx1 sequences could lead to reduced power to detect positively selected sites (85). Therefore, we extended the novel bat Mx1 sequences presented in this study by additional sequences from public databases to achieve a final set of 13 Mx1 cDNAs out of three bat families. These were used for positive selection analysis. Likelihood ratio tests, using 2 degrees of freedom, between M7 (null model; positive selection not allowed) and M8 (positive selection model) showed a significant (P < 0.05) rejection of the null model for the full Mx1 bat alignment (Fig. 6A). We therefore conclude that positive selection can be adequately detected in this 13-species data set, although the need for more sequenced bat genes is obvious.
All of the above-described procedures were generalized and implemented in a web-based pipeline called PoSeiDon (M. Hölzer and M. Marz, submitted for publication) freely available at http://www.rna.uni-jena.de/en/poseidon.
Accession number(s).
GenBank accession numbers for Mx1 cDNA sequences that were generated in the present study are as follows: C. perspicillata Mx1, KR362561; E. helvum Mx1, KR362562; H. monstrosus Mx1, KR362563; M. daubentonii Mx1, KR362564; P. pipistrellus Mx1, KR362565; R. aegyptiacus Mx1, KR362566; and S. lilium Mx1, KR362567.
Supplementary Material
ACKNOWLEDGMENTS
We thank Martin Schwemmle and Mindagaus Juozapaitis for providing us with RNA from Sturnira lilium and the expression plasmids encoding the bat influenza A-like viral proteins.
This work was supported by funding from the Deutsche Forschungsgemeinschaft, priority program-SSP1596/2, to Georg Kochs (KO1579/9-2), Friedemann Weber (WE2616/7-2), Manja Marz (MA5082/7-1), and Marcel A. Müller (MU3564/1-1) and by the Intramural Research Program of the National Institutes of Health, NIAID, to Thomas Hoenen.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00361-17.
REFERENCES
- 1.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. 2006. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev 19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang LF, Walker PJ, Poon LL. 2011. Mass extinctions, biodiversity and mitochondrial function: are bats ‘special’ as reservoirs for emerging viruses? Curr Opin Virol 1:649–657. doi: 10.1016/j.coviro.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wynne JW, Wang LF. 2013. Bats and viruses: friend or foe? PLoS Pathog 9:e1003651. doi: 10.1371/journal.ppat.1003651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binger T, Annan A, Drexler JF, Muller MA, Kallies R, Adankwah E, Wollny R, Kopp A, Heidemann H, Dei D, Agya-Yao FC, Junglen S, Feldt T, Kurth A, Oppong S, Adu-Sarkodie Y, Drosten C. 2015. A novel rhabdovirus isolated from the straw-colored fruit bat Eidolon helvum, with signs of antibodies in swine and humans. J Virol 89:4588–4597. doi: 10.1128/JVI.02932-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arguin PM, Murray-Lillibridge K, Miranda ME, Smith JS, Calaor AB, Rupprecht CE. 2002. Serologic evidence of Lyssavirus infections among bats, the Philippines. Emerg Infect Dis 8:258–262. doi: 10.3201/eid0803.010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field H, McCall B, Barrett J. 1999. Australian bat lyssavirus infection in a captive juvenile black flying fox. Emerg Infect Dis 5:438–440. doi: 10.3201/eid0503.990316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Delicat A, Paweska JT, Gonzalez JP, Swanepoel R. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 8.Pourrut X, Souris M, Towner JS, Rollin PE, Nichol ST, Gonzalez JP, Leroy E. 2009. Large serological survey showing cocirculation of Ebola and Marburg viruses in Gabonese bat populations, and a high seroprevalence of both viruses in Rousettus aegyptiacus. BMC Infect Dis 9:159. doi: 10.1186/1471-2334-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New World bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss S, Witkowski PT, Auste B, Nowak K, Weber N, Fahr J, Mombouli JV, Wolfe ND, Drexler JF, Drosten C, Klempa B, Leendertz FH, Kruger DH. 2012. Hantavirus in bat, Sierra Leone. Emerg Infect Dis 18:159–161. doi: 10.3201/eid1801.111026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo WP, Lin XD, Wang W, Tian JH, Cong ML, Zhang HL, Wang MR, Zhou RH, Wang JB, Li MH, Xu J, Holmes EC, Zhang YZ. 2013. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog 9:e1003159. doi: 10.1371/journal.ppat.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller MA, Devignot S, Lattwein E, Corman VM, Maganga GD, Gloza-Rausch F, Binger T, Vallo P, Emmerich P, Cottontail VM, Tschapka M, Oppong S, Drexler JF, Weber F, Leroy EM, Drosten C. 2016. Evidence for widespread infection of African bats with Crimean-Congo hemorrhagic fever-like viruses. Sci Rep 6:26637. doi: 10.1038/srep26637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almeida MF, Martorelli LF, Aires CC, Sallum PC, Durigon EL, Massad E. 2005. Experimental rabies infection in haematophagous bats Desmodus rotundus. Epidemiol Infect 133:523–527. doi: 10.1017/S0950268804003656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandl JN, Ahmed R, Barreiro LB, Daszak P, Epstein JH, Virgin HW, Feinberg MB. 2015. Reservoir host immune responses to emerging zoonotic viruses. Cell 160:20–35. doi: 10.1016/j.cell.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker ML, Schountz T, Wang LF. 2013. Antiviral immune responses of bats: a review. Zoonoses Public Health 60:104–116. doi: 10.1111/j.1863-2378.2012.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biesold SE, Ritz D, Gloza-Rausch F, Wollny R, Drexler JF, Corman VM, Kalko EK, Oppong S, Drosten C, Muller MA. 2011. Type I interferon reaction to viral infection in interferon-competent, immortalized cell lines from the African fruit bat Eidolon helvum. PLoS One 6:e28131. doi: 10.1371/journal.pone.0028131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowled C, Baker M, Tachedjian M, Zhou P, Bulach D, Wang LF. 2011. Molecular characterisation of Toll-like receptors in the black flying fox Pteropus alecto. Dev Comp Immunol 35:7–18. doi: 10.1016/j.dci.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowled C, Baker ML, Zhou P, Tachedjian M, Wang LF. 2012. Molecular characterisation of RIG-I-like helicases in the black flying fox, Pteropus alecto. Dev Comp Immunol 36:657–664. doi: 10.1016/j.dci.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papenfuss AT, Baker ML, Feng ZP, Tachedjian M, Crameri G, Cowled C, Ng J, Janardhana V, Field HE, Wang LF. 2012. The immune gene repertoire of an important viral reservoir, the Australian black flying fox. BMC Genomics 13:261. doi: 10.1186/1471-2164-13-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou P, Cowled C, Wang LF, Baker ML. 2013. Bat Mx1 and Oas1, but not Pkr are highly induced by bat interferon and viral infection. Dev Comp Immunol 40:240–247. doi: 10.1016/j.dci.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Zhang G, Cheng D, Ren H, Qian M, Du B. 2015. Molecular characterization of RIG-I, STAT-1 and IFN-beta in the horseshoe bat. Gene 561:115–123. doi: 10.1016/j.gene.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P, Tachedjian M, Wynne JW, Boyd V, Cui J, Smith I, Cowled C, Ng JH, Mok L, Michalski WP, Mendenhall IH, Tachedjian G, Wang LF, Baker ML. 2016. Contraction of the type I IFN locus and unusual constitutive expression of IFN-alpha in bats. Proc Natl Acad Sci U S A 113:2696–2701. doi: 10.1073/pnas.1518240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holzinger D, Jorns C, Stertz S, Boisson-Dupuis S, Thimme R, Weidmann M, Casanova JL, Haller O, Kochs G. 2007. Induction of MxA gene expression by influenza A virus requires type I or type III interferon signaling. J Virol 81:7776–7785. doi: 10.1128/JVI.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, Staeheli P. 2008. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog 4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haller O, Staeheli P, Schwemmle M, Kochs G. 2015. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol 23:154–163. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Gao S, von der Malsburg A, Dick A, Faelber K, Schroder GF, Haller O, Kochs G, Daumke O. 2011. Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity 35:514–525. doi: 10.1016/j.immuni.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Verhelst J, Parthoens E, Schepens B, Fiers W, Saelens X. 2012. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J Virol 86:13445–13455. doi: 10.1128/JVI.01682-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kochs G, Haller O. 1999. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc Natl Acad Sci U S A 96:2082–2086. doi: 10.1073/pnas.96.5.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kochs G, Janzen C, Hohenberg H, Haller O. 2002. Antivirally active MxA protein sequesters La Crosse virus nucleocapsid protein into perinuclear complexes. Proc Natl Acad Sci U S A 99:3153–3158. doi: 10.1073/pnas.052430399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwemmle M, Weining KC, Richter MF, Schumacher B, Staeheli P. 1995. Vesicular stomatitis virus transcription inhibited by purified MxA protein. Virology 206:545–554. doi: 10.1016/S0042-6822(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 32.Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan A, Campbell EM, Brandariz-Nunez A, Diaz-Griffero F. 2014. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11:68. doi: 10.1186/s12977-014-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mänz B, Dornfeld D, Gotz V, Zell R, Zimmermann P, Haller O, Kochs G, Schwemmle M. 2013. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog 9:e1003279. doi: 10.1371/journal.ppat.1003279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Busnadiego I, Kane M, Rihn SJ, Preugschas HF, Hughes J, Blanco-Melo D, Strouvelle VP, Zang TM, Willett BJ, Boutell C, Bieniasz PD, Wilson SJ. 2014. Host and viral determinants of Mx2 antiretroviral activity. J Virol 88:7738–7752. doi: 10.1128/JVI.00214-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flohr F, Schneider-Schaulies S, Haller O, Kochs G. 1999. The central interactive region of human MxA GTPase is involved in GTPase activation and interaction with viral target structures. FEBS Lett 463:24–28. [DOI] [PubMed] [Google Scholar]
- 36.Bouloy M, Janzen C, Vialat P, Khun H, Pavlovic J, Huerre M, Haller O. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J Virol 75:1371–1377. doi: 10.1128/JVI.75.3.1371-1377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stertz S, Reichelt M, Krijnse-Locker J, Mackenzie J, Simpson JC, Haller O, Kochs G. 2006. Interferon-induced, antiviral human MxA protein localizes to a distinct subcompartment of the smooth endoplasmic reticulum. J Interferon Cytokine Res 26:650–660. doi: 10.1089/jir.2006.26.650. [DOI] [PubMed] [Google Scholar]
- 38.Pavlovic J, Zurcher T, Haller O, Staeheli P. 1990. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J Virol 64:3370–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavlovic J, Haller O, Staeheli P. 1992. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J Virol 66:2564–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dick A, Graf L, Olal D, von der Malsburg A, Gao S, Kochs G, Daumke O. 2015. Role of nucleotide binding and GTPase domain dimerization in dynamin-like myxovirus resistance protein A for GTPase activation and antiviral activity. J Biol Chem 290:12779–12792. doi: 10.1074/jbc.M115.650325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmermann P, Manz B, Haller O, Schwemmle M, Kochs G. 2011. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J Virol 85:8133–8140. doi: 10.1128/JVI.00712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Götz V, Magar L, Dornfeld D, Giese S, Pohlmann A, Hoper D, Kong BW, Jans DA, Beer M, Haller O, Schwemmle M. 2016. Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci Rep 6:23138. doi: 10.1038/srep23138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frese M, Kochs G, Meier-Dieter U, Siebler J, Haller O. 1995. Human MxA protein inhibits tick-borne Thogoto virus but not Dhori virus. J Virol 69:3904–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell PS, Patzina C, Emerman M, Haller O, Malik HS, Kochs G. 2012. Evolution-guided identification of antiviral specificity determinants in the broadly acting interferon-induced innate immunity factor MxA. Cell Host Microbe 12:598–604. doi: 10.1016/j.chom.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Juozapaitis M, Aguiar Moreira E, Mena I, Giese S, Riegger D, Pohlmann A, Hoper D, Zimmer G, Beer M, Garcia-Sastre A, Schwemmle M. 2014. An infectious bat-derived chimeric influenza virus harbouring the entry machinery of an influenza A virus. Nat Commun 5:4448. doi: 10.1038/ncomms5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berger Rentsch M, Zimmer G. 2011. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS One 6:e25858. doi: 10.1371/journal.pone.0025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watt A, Moukambi F, Banadyga L, Groseth A, Callison J, Herwig A, Ebihara H, Feldmann H, Hoenen T. 2014. A novel life cycle modeling system for Ebola virus shows a genome length-dependent role of VP24 in virus infectivity. J Virol 88:10511–10524. doi: 10.1128/JVI.01272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Habjan M, Penski N, Wagner V, Spiegel M, Overby AK, Kochs G, Huiskonen JT, Weber F. 2009. Efficient production of Rift Valley fever virus-like particles: the antiviral protein MxA can inhibit primary transcription of bunyaviruses. Virology 385:400–408. doi: 10.1016/j.virol.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 49.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 50.Mitchell PS, Young JM, Emerman M, Malik HS. 2015. Evolutionary analyses suggest a function of MxB immunity proteins beyond lentivirus restriction. PLoS Pathog 11:e1005304. doi: 10.1371/journal.ppat.1005304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol 23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 52.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kishino H, Hasegawa M. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol 29:170–179. [DOI] [PubMed] [Google Scholar]
- 54.Luis AD, Hayman DT, O'Shea TJ, Cryan PM, Gilbert AT, Pulliam JR, Mills JN, Timonin ME, Willis CK, Cunningham AA, Fooks AR, Rupprecht CE, Wood JL, Webb CT. 2013. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc Biol Sci 280:20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones KE, Purvis A, MacLarnon A, Bininda-Emonds OR, Simmons NB. 2002. A phylogenetic supertree of the bats (Mammalia: Chiroptera). Biol Rev Camb Philos Soc 77:223–259. doi: 10.1017/S1464793101005899. [DOI] [PubMed] [Google Scholar]
- 56.Simmons NB. 2005. Evolution. An Eocene big bang for bats. Science 307:527–528. [DOI] [PubMed] [Google Scholar]
- 57.Mitchell PS, Emerman M, Malik HS. 2013. An evolutionary perspective on the broad antiviral specificity of MxA. Curr Opin Microbiol 16:493–499. doi: 10.1016/j.mib.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dittmann J, Stertz S, Grimm D, Steel J, Garcia-Sastre A, Haller O, Kochs G. 2008. Influenza A virus strains differ in sensitivity to the antiviral action of Mx-GTPase. J Virol 82:3624–3631. doi: 10.1128/JVI.01753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arthur DR. 1956. The Ixodes ticks of Chiroptera (Ixodoidea, Ixodidae) J Parasitol 42:180–196. [PubMed] [Google Scholar]
- 60.Klompen JSH, Black WC, Keirans JE, Oliver JH Jr. 1996. Evolution of ticks. Annu Rev Entomol 41:141–161. [DOI] [PubMed] [Google Scholar]
- 61.Hefti HP, Frese M, Landis H, Di Paolo C, Aguzzi A, Haller O, Pavlovic J. 1999. Human MxA protein protects mice lacking a functional alpha/beta interferon system against La Crosse virus and other lethal viral infections. J Virol 73:6984–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haller O, Frese M, Rost D, Nuttall PA, Kochs G. 1995. Tick-borne Thogoto virus infection in mice is inhibited by the orthomyxovirus resistance gene product Mx1. J Virol 69:2596–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayman DT, Emmerich P, Yu M, Wang LF, Suu-Ire R, Fooks AR, Cunningham AA, Wood JL. 2010. Long-term survival of an urban fruit bat seropositive for Ebola and Lagos bat viruses. PLoS One 5:e11978. doi: 10.1371/journal.pone.0011978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frese M, Kochs G, Feldmann H, Hertkorn C, Haller O. 1996. Inhibition of bunyaviruses, phleboviruses, and hantaviruses by human MxA protein. J Virol 70:915–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andersson I, Bladh L, Mousavi-Jazi M, Magnusson KE, Lundkvist A, Haller O, Mirazimi A. 2004. Human MxA protein inhibits the replication of Crimean-Congo hemorrhagic fever virus. J Virol 78:4323–4329. doi: 10.1128/JVI.78.8.4323-4329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reichelt M, Stertz S, Krijnse-Locker J, Haller O, Kochs G. 2004. Missorting of LaCrosse virus nucleocapsid protein by the interferon-induced MxA GTPase involves smooth ER membranes. Traffic 5:772–784. doi: 10.1111/j.1600-0854.2004.00219.x. [DOI] [PubMed] [Google Scholar]
- 67.Nigg PE, Pavlovic J. 2015. Oligomerization and GTP-binding requirements of MxA for viral target recognition and antiviral activity against influenza A virus. J Biol Chem 290:29893–29906. doi: 10.1074/jbc.M115.681494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao H, Killip MJ, Staeheli P, Randall RE, Jackson D. 2013. The human interferon-induced MxA protein inhibits early stages of influenza A virus infection by retaining the incoming viral genome in the cytoplasm. J Virol 87:13053–13058. doi: 10.1128/JVI.02220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alcaide M, Rico C, Ruiz S, Soriguer R, Munoz J, Figuerola J. 2009. Disentangling vector-borne transmission networks: a universal DNA barcoding method to identify vertebrate hosts from arthropod bloodmeals. PLoS One 4:e7092. doi: 10.1371/journal.pone.0007092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kopp A, Gillespie TR, Hobelsberger D, Estrada A, Harper JM, Miller RA, Eckerle I, Muller MA, Podsiadlowski L, Leendertz FH, Drosten C, Junglen S. 2013. Provenance and geographic spread of St. Louis encephalitis virus. mBio 4:e00322-13. doi: 10.1128/mBio.00322-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kühl A, Hoffmann M, Muller MA, Munster VJ, Gnirss K, Kiene M, Tsegaye TS, Behrens G, Herrler G, Feldmann H, Drosten C, Pohlmann S. 2011. Comparative analysis of Ebola virus glycoprotein interactions with human and bat cells. J Infect Dis 204(Suppl 3):S840–S849. doi: 10.1093/infdis/jir306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Horisberger MA, de Staritzky K. 1987. A recombinant human interferon-alpha B/D hybrid with a broad host-range. J Gen Virol 68(Part 3):945–948. [DOI] [PubMed] [Google Scholar]
- 73.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. [DOI] [PubMed] [Google Scholar]
- 74.Albanese M, Bruno-Smiraglia C, Di di Cuonzo G, Lavagnino A, Srihongse S. 1972. Isolation of Thogoto virus from Rhipicephalus bursa ticks in western Sicily. Acta Virol 16:267. [PubMed] [Google Scholar]
- 75.Shaw TI, Srivastava A, Chou WC, Liu L, Hawkinson A, Glenn TC, Adams R, Schountz T. 2012. Transcriptome sequencing and annotation for the Jamaican fruit bat (Artibeus jamaicensis). PLoS One 7:e48472. doi: 10.1371/journal.pone.0048472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schulz MH, Zerbino DR, Vingron M, Birney E. 2012. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S, Zhou X, Lam TW, Li Y, Xu X, Wong GK, Wang J. 2014. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30:1660–1666. doi: 10.1093/bioinformatics/btu077. [DOI] [PubMed] [Google Scholar]
- 79.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 81.Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pond SL, Frost SD, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 85.McBee RM, Rozmiarek SA, Meyerson NR, Rowley PA, Sawyer SL. 2015. The effect of species representation on the detection of positive selection in primate gene data sets. Mol Biol Evol 32:1091–1096. doi: 10.1093/molbev/msu399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.