ABSTRACT
Arenaviruses are enveloped negative-strand RNA viruses that cause significant human disease. These viruses encode only four proteins to accomplish the viral life cycle, so each arenavirus protein likely plays unappreciated accessory roles during infection. Here we used immunoprecipitation and mass spectrometry to identify human proteins that interact with the nucleoproteins (NPs) of the Old World arenavirus lymphocytic choriomeningitis virus (LCMV) and the New World arenavirus Junín virus (JUNV) strain Candid #1. Bioinformatic analysis of the identified protein partners of NP revealed that host translation appears to be a key biological process engaged during infection. In particular, NP associates with the double-stranded RNA (dsRNA)-activated protein kinase (PKR), a well-characterized antiviral protein that inhibits cap-dependent protein translation initiation via phosphorylation of eIF2α. JUNV infection leads to increased expression of PKR as well as its redistribution to viral replication and transcription factories. Further, phosphorylation of PKR, which is a prerequisite for its ability to phosphorylate eIF2α, is readily induced by JUNV. However, JUNV prevents this pool of activated PKR from phosphorylating eIF2α, even following exposure to the synthetic dsRNA poly(I·C), a potent PKR agonist. This blockade of PKR function is highly specific, as LCMV is unable to similarly inhibit eIF2α phosphorylation. JUNV's ability to antagonize the antiviral activity of PKR appears to be complete, as silencing of PKR expression has no impact on viral propagation. In summary, we provide a detailed map of the host machinery engaged by arenavirus NPs and identify an antiviral pathway that is subverted by JUNV.
IMPORTANCE Arenaviruses are important human pathogens for which FDA-approved vaccines do not exist and effective antiviral therapeutics are needed. Design of antiviral treatment options and elucidation of the mechanistic basis of disease pathogenesis will depend on an increased basic understanding of these viruses and, in particular, their interactions with the host cell machinery. Identifying host proteins critical for the viral life cycle and/or pathogenesis represents a useful strategy to uncover new drug targets. This study reveals, for the first time, the extensive human protein interactome of arenavirus nucleoproteins and uncovers a potent antiviral host protein that is neutralized during Junín virus infection. In so doing, it shows further insight into the interplay between the virus and the host innate immune response and provides an important data set for the field.
KEYWORDS: Junín, PKR, arenavirus, eIF2alpha, host-pathogen interactions, interactome, lymphocytic choriomeningitis virus, nucleoprotein, protein-protein interactions, proteomics
INTRODUCTION
The arenaviruses are a family of enveloped RNA viruses that can cause severe human disease. In particular, several family members, including Lassa virus (LASV) and Junín virus (JUNV), cause hemorrhagic fever syndromes (1). Additionally, lymphocytic choriomeningitis virus (LCMV), which has a global distribution, is an underappreciated human pathogen that represents a significant teratogenic threat to developing fetuses and is a danger to immunosuppressed populations (1, 2). There are currently no FDA-approved vaccines to prevent arenavirus infection, and effective antiviral treatments are limited (3, 4). Several of the arenaviruses require biosafety level 4 (BSL-4) containment and are designated select agents and potential bioterrorism threats (5). This highlights the critical need for new prevention and treatment options for these dangerous viruses, and the successful development of a new antiviral therapeutic will depend on an improved understanding of the basic biology of the arenavirus life cycle.
Arenaviruses have a single-stranded, bisegmented, negative-sense RNA genome. Each genomic RNA segment contains 2 open reading frames arranged in an ambisense fashion (6). The virus encodes only 4 proteins: the nucleoprotein (NP) and glycoprotein (GPC) on the S genomic segment and the RNA-dependent RNA polymerase (L) and viral matrix protein (Z) on the L genomic segment (6). These four viral proteins are sufficient to achieve all of the steps of the viral life cycle. Nevertheless, it is likely that each is highly multifunctional and relies upon interactions with multiple host proteins to facilitate key steps of the viral life cycle and/or to subvert an effective host immune response.
The canonical role of arenavirus NPs is to encapsidate the viral genomic RNA and aid the viral polymerase in the process of genome replication (6). NP also acts to prevent the induction of type I interferon (IFN) via its ability to degrade double-stranded RNA (dsRNA) substrates that may activate cytoplasmic RIG-I-like receptors (7, 8). Work by other groups has already demonstrated the ability of arenavirus NPs to engage in protein-protein interactions with host cellular proteins. However, only a few host factors engaged by NP are known, and the functional importance of these interactions necessitates further investigation. Among the Old World arenaviruses, it was demonstrated that ALIX/AIP1, an endosomal sorting complex required for transport (ESCRT)-associated protein, associates with NP and is necessary for the efficient recruitment of NP into Z-induced virus-like particles (9). LCMV NP has been shown to interact with IKKε, which contributes to its repression of type I IFN induction (10). Keratin 1, also an interacting partner of LCMV NP, is important for the virus's ability to spread from cell to cell (11). The NP of the New World arenavirus JUNV was shown to associate with eukaryotic translation initiation factor 4A (eIF4A) and eIF4GI, and these interactions were important for translation of viral mRNAs (12). Lastly, JUNV NP was shown to interact with hnRNP A/B proteins, which are required for viral propagation (13). While a small number of cellular proteins have been shown to interact and/or colocalize with arenavirus NPs, a large-scale mapping of the host cellular interactome of NP is necessary to fully appreciate the multifunctional role that this viral protein plays during the viral life cycle.
Besides our appreciation of NP's role in interfering with type I IFN induction and its interaction with a few cellular proteins, little is known about the additional accessory role(s) that NP may play in infected cells and how these may be affected by its interaction with host proteins. To address this gap in our knowledge, we mapped the cellular protein interactome of the arenavirus NP by using LCMV, an Old World arenavirus member, and the New World JUNV strain Candid #1 (14). Our studies reveal that arenavirus NPs interact with an extensive array of cellular proteins. In particular, host protein translation appears to be a major cellular function targeted by arenavirus NPs. We identified that dsRNA-activated protein kinase (PKR), an important antiviral effector, interacts with both the JUNV and LCMV NPs. We show that despite its becoming highly activated during JUNV infection, PKR is unable to carry out its canonical antiviral function, which is to phosphorylate eIF2α and trigger a global shutdown of cap-dependent translation. LCMV similarly activates PKR, but unlike JUNV, it appears to be unable to fully suppress PKR's kinase activity, as transient eIF2α phosphorylation occurs during LCMV infection. Nevertheless, the replication of both viruses was unaffected by PKR silencing, suggesting that arenaviruses have developed strategies to neutralize this critical arm of the host innate immune response.
RESULTS
Identification of human proteins that associate with arenavirus nucleoproteins.
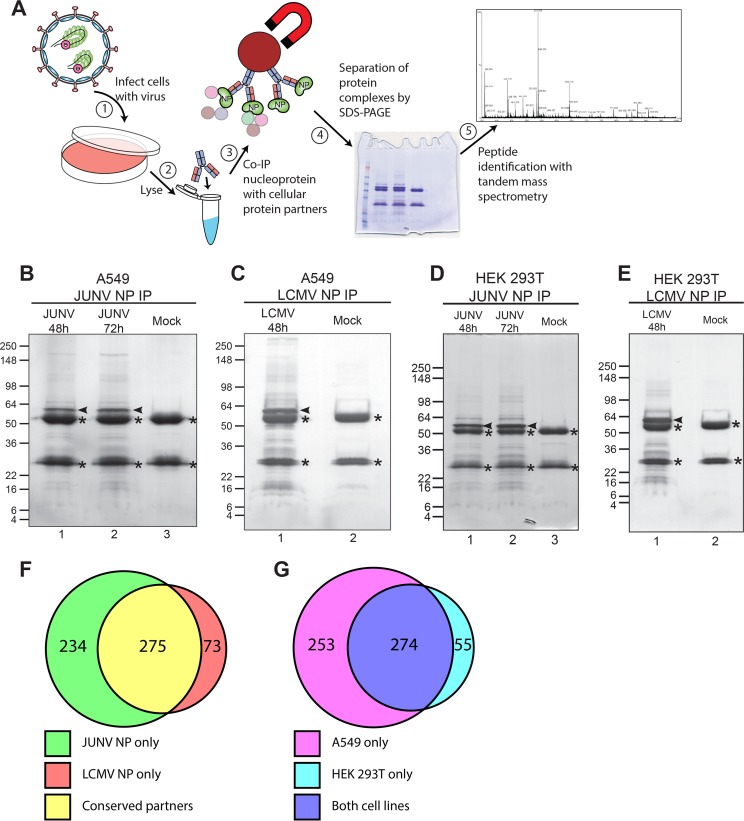
Viruses hijack cellular factors to complete necessary steps of their life cycle and/or to evade the host immune response. Thus, discovering these host factors has the potential to identify drug targets or to help illuminate cellular pathways targeted by the virus. To identify the cellular protein partners of arenavirus NPs, we employed a strategy in which human cells (A549 or HEK 293T) were first infected with either the Old World arenavirus LCMV strain Armstrong 53b or the New World arenavirus JUNV strain Candid #1 (here referred to as JUNV) (Fig. 1A). Following establishment of acute infection, each NP and its associated cellular protein partners were immunoprecipitated from cell lysates by use of an NP-specific monoclonal antibody. The immune complexes were separated by SDS-PAGE and stained with Coomassie blue to allow visualization of the captured NP (bait) as well as its interacting host proteins (prey) (Fig. 1B to E). The stained SDS-PAGE gels were cut into sections, and the proteins contained in each section were digested with trypsin and extracted for subsequent liquid chromatography-tandem mass spectrometry (LC-MS/MS). Identified proteins were considered valid interactors in a given run when (i) 2 or more unique tryptic peptides were detected from a particular protein in the infected sample but not the mock control or (ii) a minimum of 5-fold more total tryptic peptides were detected from a particular protein in the infected sample than from the mock control. Using these criteria, we identified 509 human proteins that associated with JUNV NP and 348 that associated with LCMV NP in at least one experiment (Fig. 1F; see Table S1 in the supplemental material). Of these proteins, 275 had a conserved interaction with JUNV NP and LCMV NP (Fig. 1F). Additionally, there was a high degree of reproducibility in comparing the interacting host proteins between the 2 human cell lines, HEK 293T and A549, as 83% of interacting proteins identified in HEK 293T cells were also observed in A549 cells (Fig. 1G). Thus, this data set represents the first large-scale map of the arenavirus NP-human protein interactome.
FIG 1.
Identification of human proteins that associate with arenavirus nucleoproteins. (A) Overview of experimental approach to identify human cellular protein partners of arenavirus nucleoproteins in the context of infected cells. A549 or HEK 293T cells were infected with either JUNV Candid #1 or LCMV Armstrong 53b or left uninfected (mock). Cells were lysed, and NP was immunoprecipitated with monoclonal antibodies. The immunoprecipitated fractions were separated by SDS-PAGE, and lanes were cut into multiple slices. Proteins were digested with trypsin, peptides were extracted from each slice, and proteins were identified by tandem mass spectrometry. (B to E) Representative Coomassie blue-stained gels of immunoprecipitated NP from JUNV- or mock-infected A549 cells (B), LCMV- or mock-infected A549 cells (C), JUNV- or mock-infected HEK 293T cells (D), or LCMV- or mock-infected HEK 293T cells (E). Immunoprecipitated NP is indicated by an arrowhead in each gel. Background bands composed of IgG heavy and light chains are denoted with asterisks. (F) Venn diagram representing the numbers of host proteins interacting with JUNV NP, LCMV NP, or both. (G) Venn diagram representing the numbers of host protein partners (of either JUNV or LCMV NP) identified in A549 cells, HEK 293T cells, or both. The Coomassie blue-stained gels shown in panels B to E are representative of 2 independent experiments.
Cellular processes targeted by arenavirus NPs.
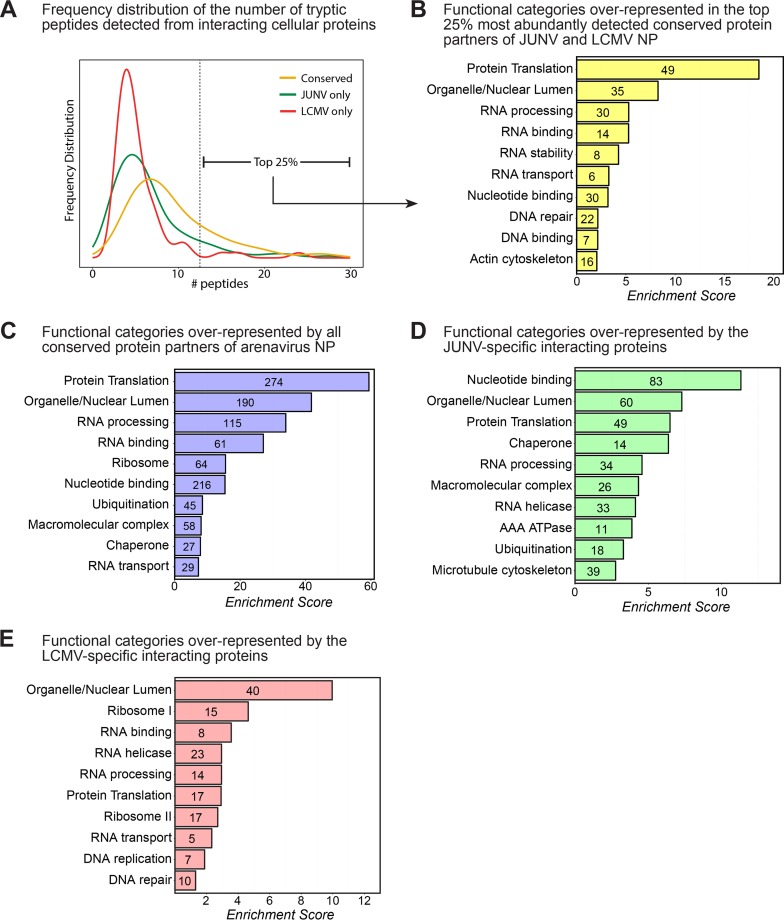
We were especially interested in the 275 proteins that exhibited a conserved interaction with both JUNV NP and LCMV NP, as these may represent fundamental and highly conserved aspects of arenavirus biology. Indeed, conserved hits for both viruses, on average, were identified by more spectral counts than those for JUNV NP or LCMV NP alone (Fig. 2A). Because the strongest interactors were likely those identified by multiple tryptic peptides, we examined in greater detail the “strong interactors,” which were those in the top 25% of all interacting host proteins by spectral counts (Fig. 2A and Table 1). To better characterize this subset of proteins, we used the NIH DAVID functional annotation and gene enrichment tool (15, 16) to identify functional groups of proteins that may be overrepresented in our data set. Among the annotation clusters identified within this group, protein translation was the top process engaged by the arenavirus NPs (Fig. 2B), suggesting that this may be a key cellular process regulated by these viruses during infection. Two important host protein partners involved in translation were the eukaryotic translation initiation factor 2 alpha subunit (eIF2α), a translation initiation factor critical for the cap-dependent assembly of the 43S ribosomal preinitiation complex (17), and the double-stranded RNA-activated protein kinase (PKR), which can lead to arrest of cellular translation in response to viral infection via phosphorylation of eIF2α (17) (Table 1; Table S1). In addition, NIH DAVID analysis of the subset of proteins interacting with only JUNV NP or LCMV NP revealed the functional processes of nucleotide binding and/or the organellar/nuclear lumen being engaged by the respective NPs (Fig. 2D and E). The functional categories enriched in this “strong interactor” subset were very similar to the categories represented by the data set as a whole (Fig. 2B and C).
FIG 2.
Bioinformatic analysis of host protein partners of arenavirus NPs. (A) Frequency distribution histogram showing the number of individually identified peptides per protein for host proteins interacting with JUNV NP only, LCMV NP only, or both viral NPs (conserved). The vertical dashed line indicates the threshold above which the 25% most abundant conserved interacting proteins were detected. (B) Bioinformatic NIH DAVID functional annotation clustering analysis showing the most highly enriched biological function categories represented in the list of the 25% most abundantly detected and conserved interacting proteins (n = 69) displayed in Table 1. (C) Bioinformatic NIH DAVID functional annotation clustering analysis showing the most highly enriched biological function categories represented in the entire list of interacting proteins (n = 582). (D) NIH DAVID analysis of the subset of proteins displaying specific interactions with only the JUNV NP (n = 234). (E) NIH DAVID analysis of the subset of proteins displaying specific interactions with only the LCMV NP (n = 73). For panels A to E, interacting human proteins that were detected in at least 1 of the 2 independent experiments for JUNV in either A549 or HEK 293T cells and/or 1 of the 2 independent experiments for LCMV in either A549 or HEK 293T cells were used for bioinformatic analysis.
TABLE 1.
Most abundantly detected (top 25%) conserved protein partners of JUNV Candid #1 and LCMV Armstrong 53b NPsa
| Gene symbol | Gene description | IPI ID | Peptide spectral count |
|
|---|---|---|---|---|
| JUNV | LCMV | |||
| AIFM1 | Programmed cell death 8 (apoptosis-inducing factor) | IPI00000690 | 89 | 3 |
| ASPH | Aspartate beta-hydroxylase | IPI00294834 | 23 | 8 |
| CCT6A | Chaperonin containing TCP1, subunit 6A (zeta 1) | IPI00027626 | 43 | 3 |
| CCT7 | Chaperonin containing TCP1, subunit 7 (eta) | IPI00018465 | 40 | 4 |
| CKAP4 | Cytoskeleton-associated protein 4 | IPI00141318 | 22 | 9 |
| CKAP5 | Cytoskeleton-associated protein 5 | IPI00028275 | 13 | 12 |
| CLTC | Clathrin, heavy polypeptide (Hc) | IPI00024067 | 19 | 18 |
| DDX3X | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | IPI00215637 | 18 | 18 |
| DDX5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | IPI00017617 | 12 | 24 |
| DHX9 | DEAH box helicase 9 | IPI00844578 | 32 | 27 |
| EEF1A2 | Eukaryotic translation elongation factor 1 alpha 2 | IPI00014424 | 16 | 8 |
| FLNA | Filamin A, alpha (actin binding protein 280) | IPI00302592 | 16 | 77 |
| FLNB | Filamin B, beta (actin binding protein 278) | IPI00289334 | 13 | 21 |
| HADHA | Hydroxyacyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase/enoyl-coenzyme A hydratase (trifunctional protein), alpha subunit | IPI00031522 | 22 | 15 |
| HIST2H2BE | Histone 2, H2be | IPI00003935 | 8 | 39 |
| HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | IPI00215965 | 13 | 16 |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoprotein A2/B1 | IPI00396378 | 17 | 12 |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoprotein A2/B1 | IPI00386854 | 3 | 27 |
| HNRNPK | Heterogeneous nuclear ribonucleoprotein K | IPI00216049 | 24 | 15 |
| HNRNPM | Heterogeneous nuclear ribonucleoprotein M | IPI00171903 | 25 | 32 |
| HNRNPU | Heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) | IPI00479217 | 15 | 12 |
| HSPA8 | Heat shock 70-kDa protein 8 | IPI00003865 | 31 | 18 |
| IGF2BP1 | Insulin-like growth factor 2 mRNA binding protein 1 | IPI00008557 | 20 | 19 |
| ILF2 | Interleukin enhancer binding factor 2, 45 kDa | IPI00005198 | 12 | 19 |
| IQGAP1 | IQ motif-containing GTPase activating protein 1 | IPI00009342 | 14 | 20 |
| KIAA1618 | Chromosome 17 open reading frame 27 | IPI00642126 | 76 | 3 |
| LMNA | Lamin A/C | IPI00021405 | 31 | 105 |
| MAP1B | Microtubule-associated protein 1B | IPI00008868 | 14 | 10 |
| MATR3 | Matrin 3 | IPI00017297 | 12 | 20 |
| MOV10 | Mov10, Moloney leukemia virus 10, homolog (mouse) | IPI00444452 | 17 | 16 |
| MYBBP1A | MYB binding protein (P160) 1a | IPI00005024 | 28 | 6 |
| MYO1B | Myosin IB | IPI00376344 | 10 | 30 |
| MYO1C | Myosin IC | IPI00010418 | 27 | 55 |
| MYO6 | Myosin VI | IPI00008455 | 9 | 18 |
| NCL | Nucleolin | IPI00183526 | 22 | 17 |
| NCL | Nucleolin | IPI00444262 | 22 | 13 |
| NPM1 | Anaplastic lymphoma kinase (Ki-1) | IPI00220740 | 14 | 26 |
| PABPC1 | Poly(A) binding protein, cytoplasmic 1 | IPI00008524 | 29 | 39 |
| PARP1 | Poly(ADP-ribose) polymerase family, member 1 | IPI00449049 | 24 | 30 |
| PLEC1 | Plectin 1, intermediate filament binding protein, 500 kDa | IPI00014898 | 55 | 307 |
| PKR | Double-stranded RNA-activated protein kinase (eukaryotic translation initiation factor 2 alpha kinase 2) | IPI00019463 | 18 | 7 |
| PRKDC | Protein kinase, DNA activated, catalytic polypeptide | IPI00296337 | 60 | 46 |
| RPL10A | Ribosomal protein L10a | IPI00412579 | 15 | 12 |
| RPL12 | Ribosomal protein L12 | IPI00024933 | 15 | 11 |
| RPL13P12 | Ribosomal protein L13 pseudogene 12 | IPI00397611 | 13 | 13 |
| RPL23 | Ribosomal protein L23 | IPI00010153 | 14 | 19 |
| RPL23A | Ribosomal protein L23a | IPI00021266 | 12 | 15 |
| RPL3 | Ribosomal protein L3 | IPI00550021 | 19 | 14 |
| RPL4 | Ribosomal protein L4 | IPI00003918 | 20 | 9 |
| RPL5 | Ribosomal protein L5 | IPI00000494 | 22 | 17 |
| RPL6 | Ribosomal protein L6 | IPI00329389 | 25 | 28 |
| RPL7 | Ribosomal protein L7 | IPI00030179 | 14 | 18 |
| RPL7A | Ribosomal protein L7a | IPI00299573 | 16 | 17 |
| RPLP0 | Ribosomal protein, large, P0 | IPI00008530 | 39 | 31 |
| RPLP2 | Ribosomal protein, large, P2 | IPI00008529 | 22 | 15 |
| RPS18 | Ribosomal protein S18 | IPI00013296 | 12 | 14 |
| RPS19 | Ribosomal protein S19 | IPI00215780 | 9 | 21 |
| RPS3 | Ribosomal protein S3 | IPI00011253 | 14 | 31 |
| RPS3A | Ribosomal protein S3A | IPI00419880 | 11 | 20 |
| RPS4X | Ribosomal protein S4, X-linked | IPI00217030 | 13 | 22 |
| RRBP1 | Ribosome binding protein 1 homolog, 180 kDa (dog) | IPI00215743 | 40 | 6 |
| SFPQ | Splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) | IPI00010740 | 17 | 16 |
| SLC25A5 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | IPI00007188 | 27 | 27 |
| TMPO | Thymopoietin | IPI00030131 | 9 | 43 |
| TMPO | Thymopoietin | IPI00216230 | 4 | 34 |
| TUBB2C | Tubulin, beta 2C | IPI00007752 | 14 | 8 |
| XRCC5 | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining, Ku autoantigen, 80 kDa) | IPI00220834 | 14 | 13 |
| XRCC6 | X-ray repair complementing defective repair in Chinese hamster cells 6 (Ku autoantigen, 70 kDa) | IPI00644712 | 15 | 18 |
The most abundant interactors were defined as those having the largest average numbers of spectral counts (total peptides) from the JUNV (n = 8 independent experiments) or LCMV (n = 4 independent experiments) mass spectrometry experiments.
Biochemical validation of interactions between cellular proteins and arenavirus NPs.
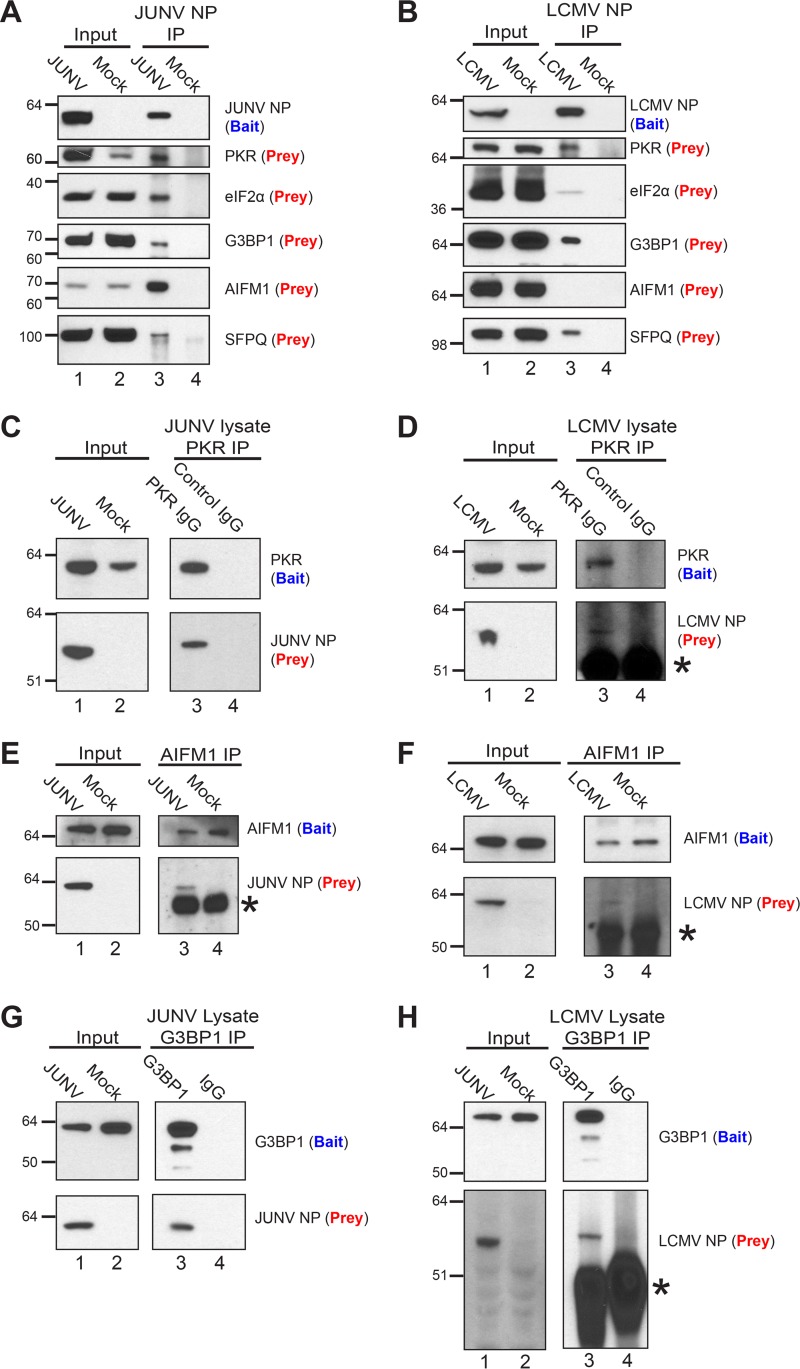
We next chose a subset of the identified host proteins and attempted to further validate their association with a particular arenavirus NP via immunoprecipitation and Western blotting. This subset of proteins included PKR, eIF2α, and Ras GTPase-activating protein-binding protein 1 (G3BP1) (proteins involved in translation); splicing factor, proline- and glutamine-rich (SFPQ) (part of several top clusters identified by the DAVID analysis, including RNA processing, binding, and stability); and apoptosis-inducing factor 1, mitochondrial (AIFM1) (the most abundantly detected interacting partner of JUNV NP; identified as being part of the organellar/nuclear lumen cluster by the DAVID analysis). Each protein, with the exception of AIFM1, coimmunoprecipitated as prey with NP (bait) from cells infected with JUNV or LCMV (Fig. 3A, lane 3, and B, lane 3). AIFM1 interacted specifically with JUNV NP but not with LCMV NP, while eIF2α interacted weakly with LCMV NP compared to its interaction with JUNV NP. Reciprocally, PKR, G3BP1, and AIFM1, when serving as bait for immunoprecipitation from JUNV- or LCMV-infected lysates, were each able to coprecipitate both viral NPs, but JUNV NP interacted more strongly with both AIFM1 and PKR than LCMV NP did (Fig. 3C to H). Finally, we addressed whether this subset of host proteins could associate with an arenavirus NP in the absence of other viral proteins or the full viral genome. G3BP1, eIF2α, AIFM1, and SFPQ interacted with the Lassa virus, LCMV, and JUNV NPs (expressed via plasmids in HEK 293T cells), while PKR associated only with JUNV NP (Fig. 4A). Notably, the strength of interaction between each host protein and a particular NP was variable (Fig. 4A). Additionally, immunoprecipitation of PKR and AIFM1 from transfected HEK 293T cells was able to coprecipitate each of the arenaviral NPs tested (Fig. 4B and C).
FIG 3.
Biochemical validation of interactions between cellular proteins and arenavirus NPs in infected cells. (A) JUNV NP (bait) was immunoprecipitated from lysates of A549 cells infected (JUNV) or not (mock) with JUNV, and associated cellular proteins (prey) were detected by Western blotting. (B) LCMV NP (bait) was immunoprecipitated from lysates of A549 cells infected (LCMV) or not (mock) with LCMV Armstrong 53b, and associated cellular proteins (prey) were detected by Western blotting. (C and D) A549 cells were infected with JUNV (C) or LCMV (D) or left uninfected (mock). At 72 hpi (JUNV) or 48 hpi (LCMV), immunoprecipitations from infected lysates were performed using either a monoclonal antibody to the C terminus of PKR (bait) or a species-matched control IgG, and the associated JUNV NP (C) or LCMV NP (D) prey was detected by Western blotting. (E and F) AIFM1 (bait) was immunoprecipitated from lysates of A549 cells infected or not (mock) with JUNV (E) or LCMV Armstrong 53b (F) by use of an AIFM1-specific polyclonal antibody, and AIFM1 (bait) and the associated NP (prey) were detected by Western blotting. (G and H) G3BP1 (bait) was immunoprecipitated from lysates of A549 cells infected with JUNV (72 hpi) (G) or LCMV Armstrong 53b (48 hpi) (H) by use of either a polyclonal antibody to G3BP1 (bait) or a control rabbit IgG, and G3BP1 (bait) and the associated NP (prey) were detected by Western blotting. Protein bands composed of the IgG heavy chain in panels D, E, F, and H are denoted with asterisks. Data are representative of 2 (A and B), 4 (C), 5 (D), or 1 (E to H) independent experiment.
FIG 4.
Biochemical validation of interactions between arenavirus NPs expressed from plasmids and endogenous host proteins. (A) HEK 293T cells were cotransfected with a plasmid encoding the respective arenavirus NP with a C-terminal HA epitope tag, the TEV cleavage site, and a biotin acceptor peptide, along with a second plasmid that encodes BirA, a bacterial biotin ligase, to ensure biotinylation of the viral NPs. As a control, cells were cotransfected with the BirA plasmid and an empty vector (p0). Biotinylated NPs and associated host proteins were affinity purified from cell lysates (input) by use of magnetic streptavidin beads, and captured proteins were detected by Western blotting. (B and C) HEK 293T cells were transfected with a plasmid encoding the respective arenavirus NP with a C-terminal HA epitope tag, the TEV cleavage site, and a biotin acceptor peptide or, as a control, an empty vector (p0). (B) PKR (bait) was immunoprecipitated with a monoclonal antibody to the C terminus of PKR or with an irrelevant rabbit IgG. PKR (bait) and the associated viral NP (prey) were detected by Western blotting. (C) AIFM1 (bait) was immunoprecipitated with a polyclonal antibody. AIFM1 (bait) and the associated viral NP (prey) were detected by Western blotting. Data are representative of 2 (A), 3 (B), or 2 (C) independent experiments.
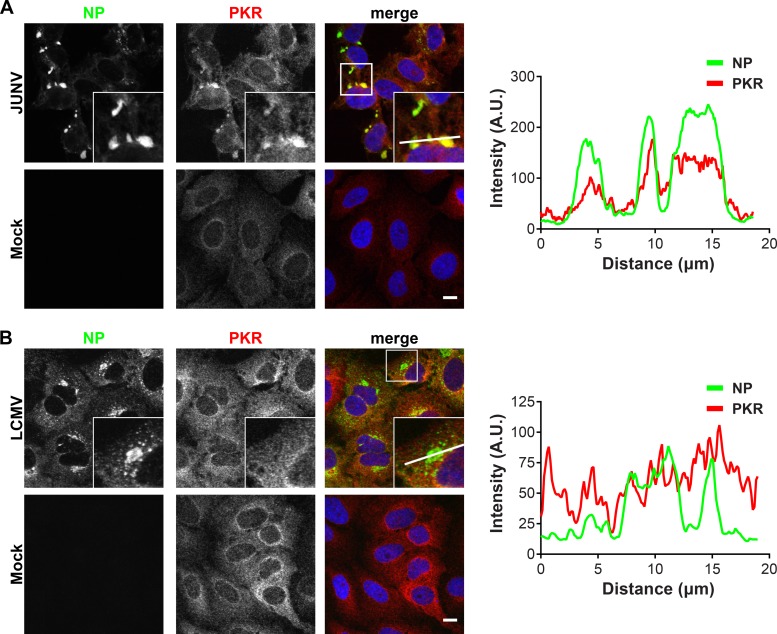
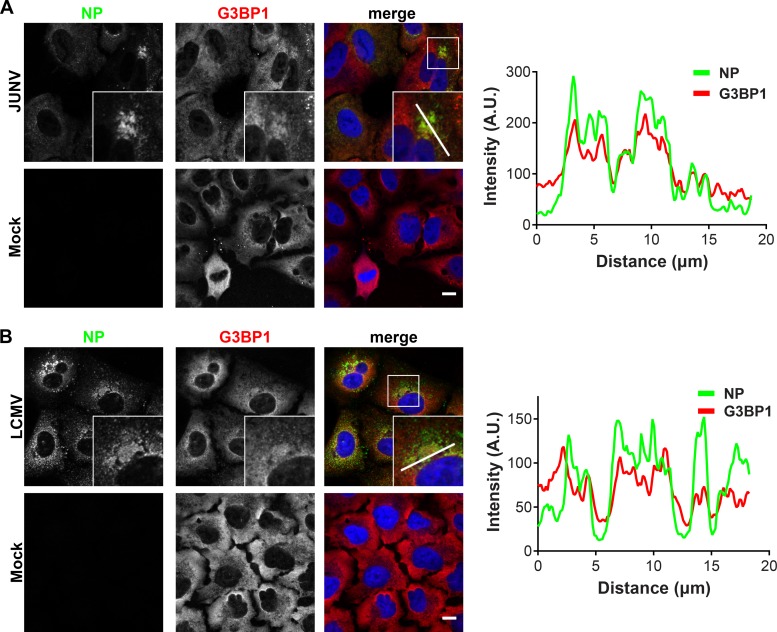
PKR and G3BP1 colocalize with JUNV NP.
We next used fluorescence microscopy to determine whether PKR and G3BP1 were recruited to the replication and transcription factories of JUNV or LCMV. In JUNV-infected cells at 72 h postinfection (hpi), the majority of the cells were infected, and NP formed large, perinuclear puncta in most cells (Fig. 5A). In LCMV-infected cells at 48 hpi, most cells were infected and displayed a variable pattern of cytoplasmic NP staining. LCMV NP concentrated in puncta that were either small and scattered throughout the cytoplasm or large and located near the nucleus (Fig. 5B). In the setting of JUNV infection, the total PKR signal was increased compared to that in mock cells (Fig. 5A). While PKR staining was cytoplasmic in both infected and uninfected cells, JUNV infection resulted in a relocalization and concentration of PKR in JUNV NP-containing puncta (Fig. 5A, fluorescence intensity profiles). This was specific to JUNV, as PKR protein levels were not upregulated in LCMV-infected cells and PKR remained diffusely localized, without specifically colocalizing with LCMV NP (Fig. 5B, fluorescence intensity profiles). This finding is consistent with biochemical evidence suggesting that PKR has a stronger association with JUNV NP than with LCMV NP (Fig. 3C and D and 4A).
FIG 5.
PKR colocalizes with JUNV NP but not with LCMV NP. A549 cells were infected (JUNV) or not (mock) with JUNV and fixed at 72 hpi (n = 5) (A) or infected (LCMV) or not (mock) with LCMV Armstrong 53b and fixed at 48 hpi (n = 4) (B). Cells were stained for NP (green) and PKR (red) and visualized by confocal immunofluorescence microscopy. The insets show higher-magnification views of the boxed areas. Fluorescence plot profiles are included to show NP and PKR signal intensities along the white line indicated in the inset of the merge panel for either JUNV- or LCMV-infected cells. Bars = 10 μm. A.U., arbitrary units.
G3BP1 generally displayed a diffuse cytoplasmic staining pattern in uninfected cells (Fig. 6), although some minority of uninfected cells spontaneously displayed distinct cytoplasmic puncta of G3BP1 staining in presumed stress granule (SG) structures (Fig. 6A). Similar to the results for PKR, a portion of the cytoplasmic G3BP1 concentrated in JUNV NP puncta (Fig. 6A, fluorescence intensity profiles). In LCMV-infected cells, G3BP1 maintained a predominantly diffuse cytoplasmic staining pattern, with slight enrichment at NP puncta (Fig. 6B, fluorescence intensity profiles).
FIG 6.
G3BP1 colocalizes with JUNV and LCMV NPs. A549 cells were infected (JUNV) or not (mock) with JUNV and fixed at 72 hpi (n = 1) (A) or infected (LCMV) or not (mock) with LCMV Armstrong 53b and fixed at 48 hpi (n = 1) (B). Cells were stained for NP (green) and G3BP1 (red) and visualized by confocal immunofluorescence microscopy. The insets show higher-magnification views of the boxed areas. Fluorescence plot profiles are included to show NP and G3BP1 signal intensities along the white line indicated in the inset of the merge panel for either JUNV- or LCMV-infected cells. Bars = 10 μm.
Collectively, these experiments further validate the biological interaction of PKR and G3BP1 with JUNV NP and define these host proteins as components/markers of presumptive viral replication and transcription factories. Further, they show the specificity of the NP-PKR interaction for JUNV.
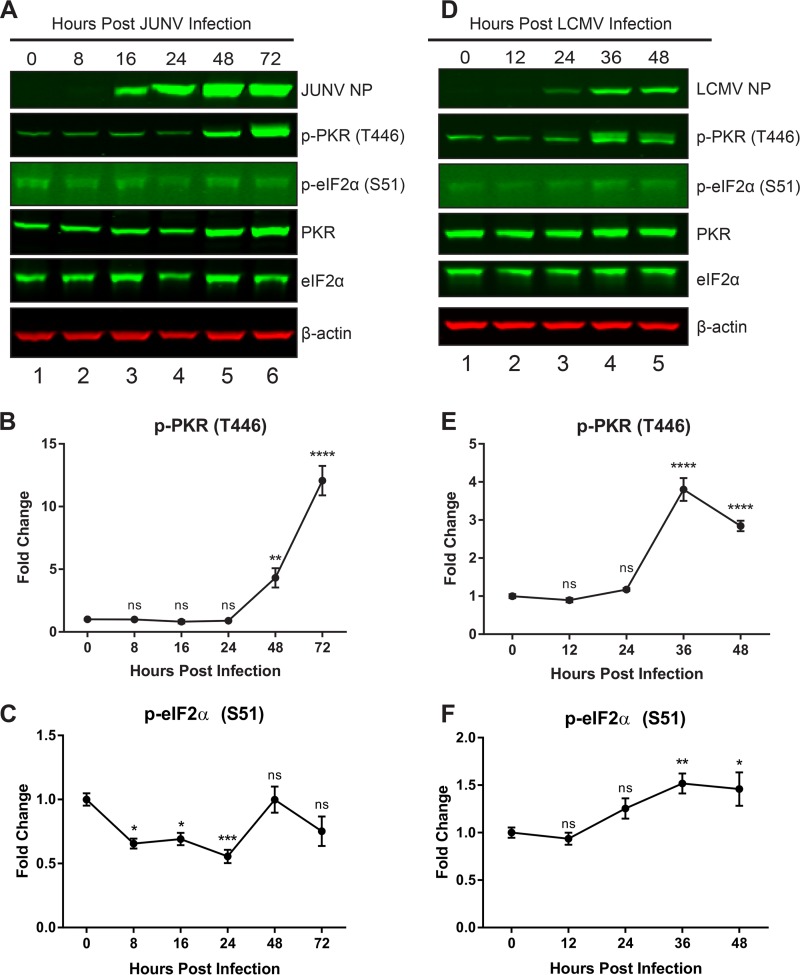
PKR is activated following JUNV infection but does not phosphorylate eIF2α.
Because of the importance of PKR as an antiviral innate immune mediator (17) and the specificity of the interaction with the JUNV NP (Fig. 3C and D, 4A, and 5), we next sought to functionally characterize the importance of this interaction in the context of both New World and Old World arenavirus infections. Over a time course of acute infection, cellular protein lysates were probed for phospho-PKR (T446), which is phosphorylated at a site in the activation loop and is a marker of activated PKR. We also probed for phospho-eIF2α (S51), the target of PKR's kinase activity and a marker of global translational shutdown. Both expression and phosphorylation of PKR were strongly induced late in infection with JUNV, yet there was no concomitant increase in the phosphorylation of PKR's target, eIF2α, at these time points (Fig. 7A to C). In LCMV-infected cells, overall PKR levels did not increase, but there was an induction of phosphorylation of PKR at 36 and 48 hpi. However, in contrast to the results with JUNV, there was a small yet significant increase in the phosphorylation of eIF2α at these time points (Fig. 7D to F). This suggested an ability of JUNV to prevent activated PKR from phosphorylating its target (eIF2α).
FIG 7.
PKR is activated following JUNV infection but cannot phosphorylate eIF2α. A549 cells were infected with JUNV (A to C) or LCMV (D to F). Infected cell lysates were collected during a time course of acute infection. (A and D) Viral NPs, phosphorylated PKR (T446), total PKR, phosphorylated eIF2α (S51), total eIF2α, and β-actin were visualized by Western blotting. Phosphorylated PKR (B and E) and phosphorylated eIF2α (C and F) were quantified and compared using one-way ANOVA. Data are presented as mean fold changes ± standard errors of the means (SEM) for 2 independent experiments featuring 3 technical replicates each. ns, not significant (P > 0.05); *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
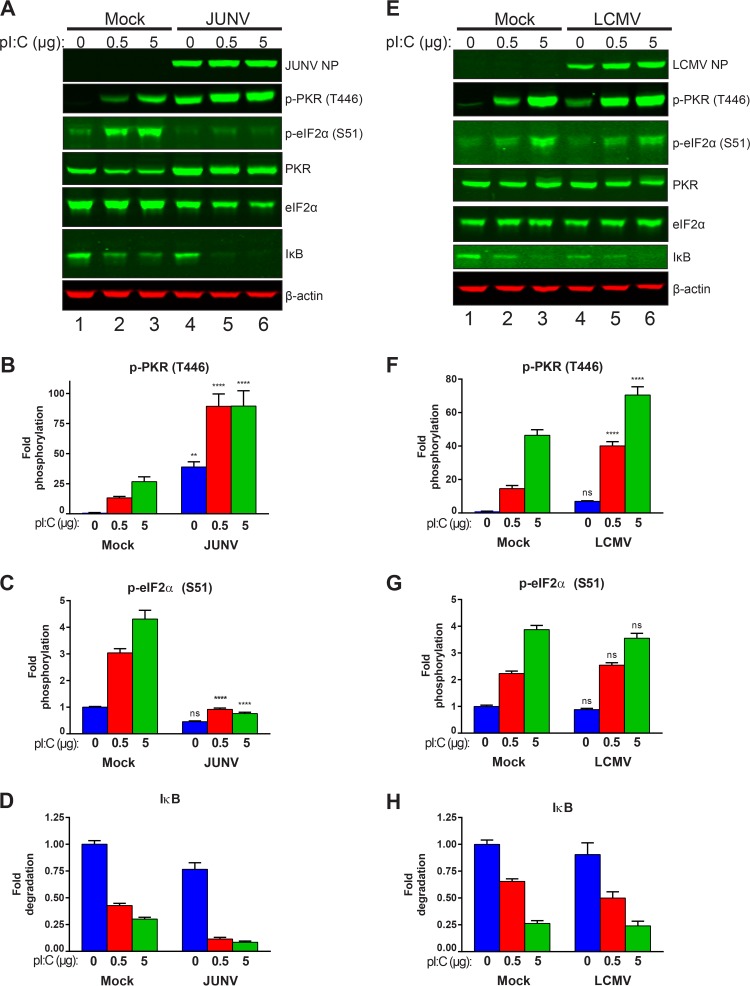
JUNV infection blocks poly(I·C)-induced phosphorylation of eIF2α.
The observation that PKR was highly activated (phosphorylated) in JUNV-infected cells yet the activated PKR failed to phosphorylate eIF2α suggested that the virus was evading this innate surveillance pathway. To determine if we could overcome the ability of the virus to block active PKR from phosphorylating eIF2α, we transfected cells with poly(I·C), a synthetic dsRNA analog that strongly activates PKR (17). Comparing mock-transfected cells that had been treated with Lipofectamine but no poly(I·C) (vehicle control) to those transfected with increasing quantities of poly(I·C) showed that phosphorylation of PKR was effectively induced by increasing concentrations of poly(I·C) (Fig. 8A, B, E, and F). In JUNV-infected cells, there were already high levels of phospho-PKR (Fig. 8A and B). However, transfection with poly(I·C) further increased the induction of PKR phosphorylation (Fig. 8A and B). In mock-infected cells, the increased PKR phosphorylation following poly(I·C) transfection led to increased phosphorylation of eIF2α (Fig. 8A, C, E, and G). However, there was no increase in the phosphorylation of eIF2α in JUNV-infected cells (Fig. 8A and C). In LCMV-infected cells, phosphorylation of PKR could be induced further following poly(I·C) transfection (Fig. 8E and F). However, this active PKR was able to phosphorylate eIF2α, and there was no difference in phospho-eIF2α levels between mock-infected and LCMV-infected cells following poly(I·C) transfection (Fig. 8E and G). Thus, JUNV appears to specifically block activated PKR from phosphorylating its downstream target, eIF2α.
FIG 8.
JUNV infection blocks poly(I·C)-induced phosphorylation of eIF2α. A549 cells infected with either JUNV (A to D) or LCMV (E to H) or left uninfected (mock) were transfected with the indicated quantities of poly(I·C) to induce PKR activation. (A and E) Viral NPs, phosphorylated PKR (T446), total PKR, phosphorylated eIF2α (S51), total eIF2α, IκB, and β-actin were visualized by Western blotting. Phosphorylated PKR (B and F), phosphorylated eIF2α (C and G), and IκB (D and H) were quantified and compared using two-way ANOVA. Data are presented as mean fold changes ± SEM for 2 independent experiments featuring 3 technical replicates each. ns, not significant (P > 0.05); *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
JUNV selectively blocks PKR's functionality.
Though activated PKR's canonical target is eIF2α, PKR can also activate alternate signaling pathways. Among these alternate pathways is the activation of NF-κB signaling (18), through an incompletely defined pathway. While the exact mechanism is unclear, it is known that IκB is degraded following PKR activation and that this allows the nuclear translocation of the NF-κB transcription factor and the subsequent expression of NF-κB-responsive genes (18). To determine whether this alternate PKR signaling pathway remained functional in cells expressing arenavirus NPs, we also probed cellular protein lysates for IκB to see if it was degraded following poly(I·C) transfection, as would be expected. We saw that whether cells were uninfected or infected with JUNV or LCMV, they were able to efficiently degrade IκB following transfection with poly(I·C), suggesting that this alternate PKR signaling function remained intact (Fig. 8A, D, E, and H). An alternate possibility is that the observed IκB degradation was the result of signaling downstream of other innate immune sensors capable of responding to dsRNA, such as Toll-like receptor 3 (TLR3), RIG-I, or MDA5 (19).
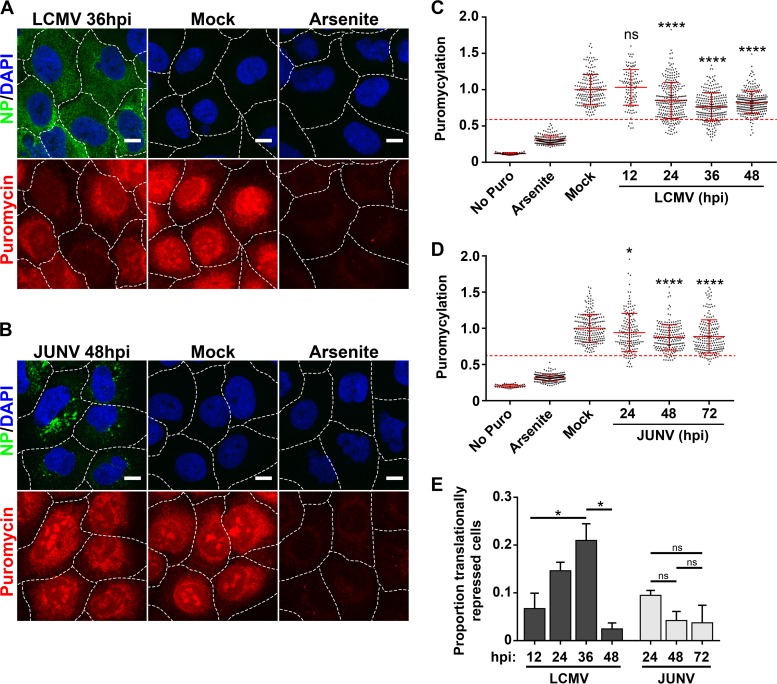
Translational profile of cells infected with JUNV or LCMV.
We wanted to examine whether the eIF2α phosphorylation observed in LCMV- but not JUNV-infected cells (Fig. 7 and 8) was functionally correlated with repression of global translation in these cells. To assess rates of active translation, we utilized a newly described assay to examine protein translation levels in individual cells infected with either LCMV or JUNV (20, 21). At various time points following infection, cells were pulse labeled with puromycin, which is covalently incorporated into growing peptide chains. Nascent peptides were detected with a puromycin-specific monoclonal antibody and were visualized by confocal microscopy. Mock-infected cells were labeled with puromycin to determine normal translation rates, whereas mock-infected cells that were pretreated with sodium arsenite were labeled to determine protein production levels in a repressed translational state (Fig. 9A to D). While cells infected with either LCMV or JUNV exhibited reduced rates of translation compared to those of uninfected cells, the biological importance of this subtle decrease is unclear, as the majority of cells maintained translation rates that were in the “normal range” observed for mock-infected cells (Fig. 9A to D). However, at intermediate time points following infection with LCMV (24 to 36 hpi), there was a significant increase in cells exhibiting translation rates that were highly reduced compared to those of mock-infected cells (Fig. 9A, C, and E). A similar pattern was not observed for JUNV-infected cells (Fig. 9B, D, and E). These data further support the observations shown in Fig. 7 and 8 indicating that LCMV is more sensitive than JUNV to PKR-dependent phosphorylation of eIF2α. Importantly, these experiments demonstrate that, in most instances, JUNV and LCMV are capable of infecting host cells without inducing a potent global shutdown of translation, despite highly activating PKR.
FIG 9.
Translational profiles of cells infected with JUNV or LCMV. (A and B) Cells were either infected with LCMV (n = 2) (A) or JUNV (n = 2) (B), mock infected, or treated with 500 μM sodium arsenite, and rates of translation were assessed by labeling newly synthesized peptides with puromycin. (C and D) Puromycin levels were quantitated for individual cells receiving no puromycin, mock-infected cells labeled with puromycin, mock-infected cells treated with sodium arsenite prior to puromycin labeling, and cells infected with LCMV (C) or JUNV (D) at different time points following infection. The level for each individual cell was normalized to the mean level of puromycylation for mock-infected cells. The puromycin labeling of individual cells is represented by single dots, the mean puromycin labeling is represented by a solid red horizontal line for each condition, and red error bars represent standard deviations (SD) and were compared between the mock group and each time point by one-way ANOVA. A red dashed line representing the threshold for translational repression was set as the mean level of puromycylation in mock-infected cells labeled with puromycin minus (1.96 × SD). (C) n values were as follows: no Puro, n = 44 cells; arsenite, n = 194 cells; mock, n = 191 cells; LCMV 12 h, n = 113 cells; LCMV 24 h, n = 276 cells; LCMV 36 h, n = 260 cells; and LCMV 48 h, n = 269 cells. (D) n values were as follows: no Puro, n = 40 cells; arsenite, n = 205 cells; mock, n = 215 cells; JUNV 24 h, n = 161 cells; JUNV 48 h, n = 198 cells; and JUNV 72 h, n = 205 cells. (E) The proportions of individual cells that fell below the translational repression threshold under each condition were compared by one-way ANOVA. Though all possible comparisons between LCMV-infected cultures at different times postinfection were made for panel E, for clarity, only significant differences are shown. In panels A and B, the borders between adjacent cells are represented by dashed white lines. Bars = 10 μm. ns, not significant (P > 0.05); *, P ≤ 0.05; ****, P ≤ 0.0001.
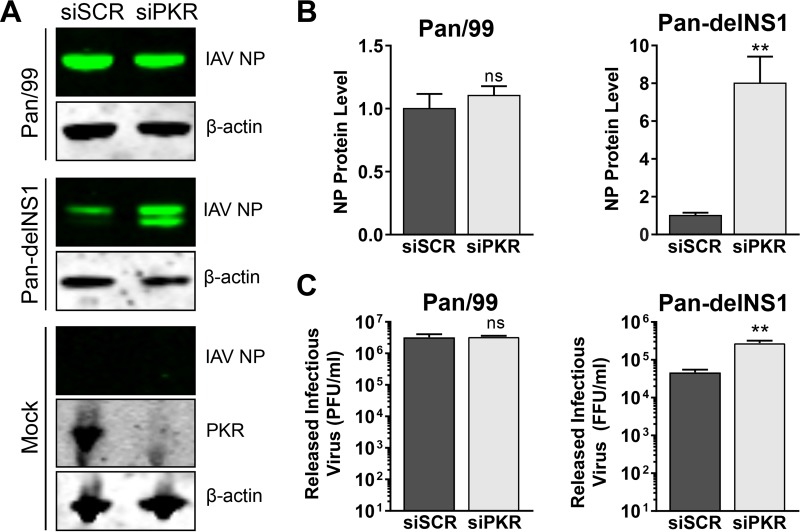
Replication of IAV lacking expression of the NS1 protein is rescued by siRNA-mediated knockdown of PKR.
We wanted to examine whether arenavirus growth would be affected by the knockdown of PKR by small interfering RNA (siRNA). In order to validate our siRNA knockdowns, we infected siRNA-treated cells with an influenza A virus (IAV; strain Pan-delNS1) lacking expression of the nonstructural protein NS1, a well-documented PKR antagonist (22, 23), to demonstrate that we were able to effectively knock down PKR expression to a level that could functionally rescue the replication of this highly PKR-sensitive virus. Protein levels of PKR were greatly reduced in cells transfected with a PKR-specific siRNA compared to those in cells transfected with a scrambled, nontargeting control siRNA (Fig. 10A). As expected, the level of viral nucleoprotein in cells and the quantity of infectious virus released by cells infected with the wild-type parental strain Pan/99 were unaffected by PKR knockdown (Fig. 10A to C). However, when cells were infected with the mutant virus Pan-delNS1, the viral nucleoprotein was expressed at higher levels (∼8-fold) and significantly more (∼6-fold) infectious virus was released from cells in which PKR had been knocked down following siRNA transfection (Fig. 10A to C). Together these results show that our knockdown of PKR by siRNA was sufficient to rescue a PKR-sensitive virus.
FIG 10.
Loss of functional PKR enhances growth of mutant delNS1 but not wild-type (WT) influenza A virus. A549 cells were transfected with a nontargeting scrambled siRNA (siSCR) or with a PKR-specific siRNA. Three days following siRNA transfection, cells were infected with either WT influenza A virus (IAV) Pan/99 or a mutant IAV that does not express NS1 (Pan-delNS1) or mock infected, and supernatants and cellular protein lysates were collected at 16 hpi. Protein levels of IAV nucleoprotein (NP) were detected by Western blotting (A) and were quantified, normalized to β-actin levels, and normalized to the mean NP level in siSCR-transfected cells (B). In panel A, PKR expression in mock-infected cells was visualized by Western blotting to confirm knockdown. (C) Quantities of infectious virus in supernatants were determined by plaque assay (for IAV Pan/99) or focus assay (for IAV Pan-delNS1). For panels B and C, the effect of PKR knockdown on viral NP levels as well as on the amounts of infectious virus released was determined using a two-tailed, unpaired t test. Data are presented as mean PFU per milliliter ± SEM or mean focus-forming units (FFU) per milliliter ± SEM for 2 independent experiments featuring 2 technical replicates each. ns, not significant (P > 0.05); **, P ≤ 0.01.
JUNV's antagonism of PKR's antiviral activity is complete.
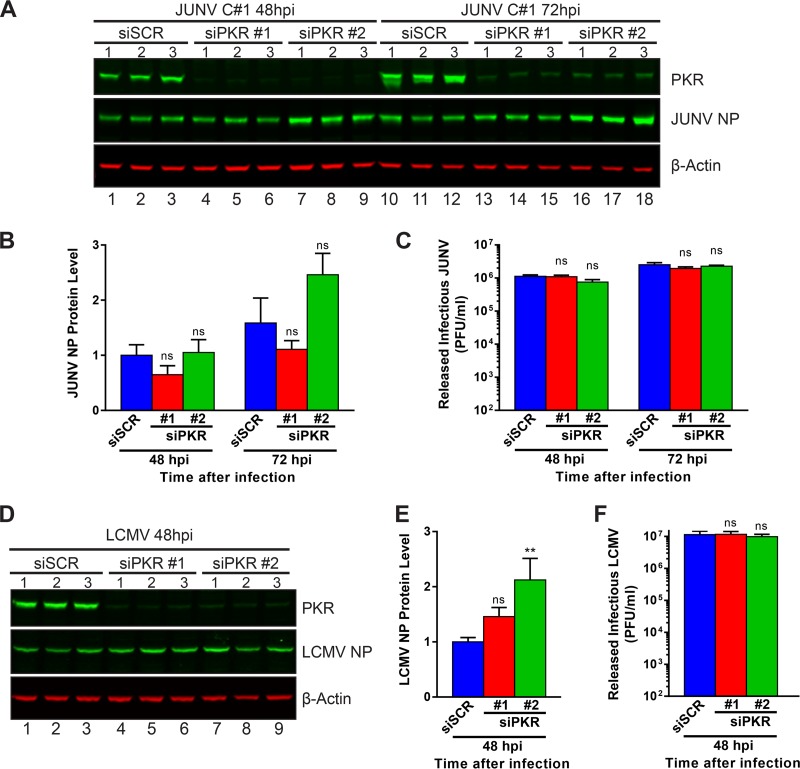
We next wished to test whether PKR, which appears to be unable to phosphorylate its downstream substrate, eIF2α, during JUNV infection (Fig. 7A and C), could still exert an antiviral effect on the propagation of JUNV. Additionally, we were curious to see if knockdown of PKR could improve the replication of LCMV by preventing the activation of eIF2α (Fig. 7 and 8) and the transient translation repression (Fig. 9) observed during LCMV infection. For these purposes, we measured the levels of NP produced in infected cells (Fig. 11A, B, D, and E) and the levels of infectious virus released (Fig. 11C and F) for cells treated with siRNAs to silence PKR expression. Despite nearly complete knockdown of PKR with either of two distinct PKR-specific siRNAs (Fig. 11A and D), release of infectious JUNV and LCMV was not affected compared to that from cells treated with a scrambled, nonspecific siRNA (Fig. 11C and F). Levels of JUNV NP were similarly unaffected by the knockdown of PKR (Fig. 11B). LCMV NP levels were slightly increased when PKR was knocked down with one of the two PKR-specific siRNAs (Fig. 11E). This result suggests that JUNV is capable of completely neutralizing the antiviral actions of PKR and, further, that PKR is not fundamentally required for the efficient propagation of JUNV. On the other hand, while eIF2α is activated and translation is repressed during LCMV infection, knockdown of PKR had no effect on the release of infectious LCMV and a modest effect on production of viral NP, which we believe to represent the transience of PKR's restriction on LCMV infection.
FIG 11.
Loss of functional PKR does not affect arenavirus propagation. A549 cells were transfected with a nontargeting scrambled siRNA (siSCR) or one of two PKR-specific siRNAs (siPKR #1 and siPKR #2). Three days after siRNA transfection, cells were infected with JUNV (A to C) or LCMV (D to F), and supernatants and cellular protein lysates were collected at 48 hpi (JUNV and LCMV) and 72 hpi (JUNV only). (A and D) Protein levels of PKR and viral NP were visualized by Western blotting. (B and E) NP protein levels were quantified, normalized to β-actin levels, and normalized to the mean NP level in siSCR-transfected cells at 48 hpi and then compared by one-way ANOVA. (C and F) Quantities of infectious virus in supernatants were determined by plaque assay and were compared by one-way ANOVA. Data are presented as mean PFU per milliliter ± SEM for 2 independent experiments featuring 3 technical replicates each for panels A to C (JUNV) or for 3 independent experiments featuring 3 technical replicates each for panels D to F (LCMV). ns, not significant (P > 0.05); **, P ≤ 0.01.
DISCUSSION
The small size of the arenavirus proteome suggests that each viral protein is highly multifunctional. One strategy to identify additional accessory roles for these proteins is to identify the host machinery that they engage during infection. In this study, we conducted a large-scale mapping of the human cellular interactome of Old World and New World arenavirus NPs. While the identified interactome was complex, a large percentage of the cellular protein partners were shared among Old World and New World NPs (Fig. 1F). Bioinformatic analysis revealed a significant enrichment in proteins involved in protein translation, including the antiviral protein PKR (Fig. 2 and Table 1). Our studies demonstrate that JUNV, despite activating PKR during infection (Fig. 7A and B), selectively and potently inhibits PKR's ability to phosphorylate its downstream substrate, eIF2α (Fig. 7A and C and 8A and C), and to subsequently interfere with viral propagation (Fig. 11C).
PKR is one of four cellular kinases able to phosphorylate eIF2α and thus to globally shut down cap-dependent translation in response to viral infection (24). This type I IFN-stimulated protein consists of two domains: an N-terminal domain that contains two double-stranded RNA binding motifs and a C-terminal kinase domain. In nonstressed cells, monomeric PKR is expressed at low basal levels. Upon exposure to a dsRNA ligand, the N-terminal domain binds the dsRNA, PKR dimerizes, and each PKR subunit trans-autophosphorylates at T446 and T451. Phosphorylation of these residues leads to full activation of PKR, which is then able to phosphorylate eIF2α at S51, leading to cap-dependent translational shutdown (Fig. 12) (17, 25).
FIG 12.
JUNV infection blocks PKR's antiviral activity. Inactive PKR exists in an unphosphorylated, monomeric form. Upon binding of a dsRNA ligand with its N-terminal dsRNA binding domains, PKR dimerizes and undergoes an autophosphorylation event (at T446 and T451). This activated form of PKR phosphorylates eIF2α at serine 51, which leads to a global cap-dependent translational shutdown. The results of this study show that PKR is able to become phosphorylated in cells infected with JUNV but is deficient in its ability to phosphorylate its target, eIF2α. We hypothesize that the viral NP may be responsible for blocking this step, as it was shown to interact with both PKR and eIF2α. Other functions of active PKR, such as the activation of NF-κB (as assayed by IκB degradation), remain intact.
A key finding of our proteomic studies was that both the JUNV and LCMV NPs interact with PKR. However, our follow-up biochemical validation (Fig. 3 and 4) and immunofluorescence microscopy (Fig. 5) studies suggest that this interaction may be more biologically relevant for JUNV NP than for LCMV NP. We demonstrated that PKR is strongly activated in cells infected with JUNV or LCMV (Fig. 7), and even more so in infected cells that are transfected with the PKR agonist poly(I·C) (Fig. 8). Nevertheless, this active PKR is able to phosphorylate eIF2α only in LCMV-infected cells, as eIF2α phosphorylation by active PKR is strongly blunted in JUNV-infected cells (Fig. 7 and 8). This effect was recapitulated in experiments measuring rates of translation in infected cells (Fig. 9), with the majority of cells infected with JUNV or LCMV maintaining normal rates of translation throughout the course of infection. However, at 36 hpi, a significant proportion of LCMV-infected cells enter a state of translation repression (Fig. 9E), which happens to coincide with the spike in observed eIF2α phosphorylation observed by Western blotting (Fig. 7D and F). Interestingly, this effect of translation repression appears to be transient, as the proportion of LCMV-infected cells with normal levels of translation recovers by 48 hpi (Fig. 9A, C, and E). The finding that siRNA silencing of PKR, while sufficient to rescue the PKR-sensitive virus IAV Pan-delNS1 (Fig. 10), has no effect on the JUNV life cycle (Fig. 11A to C) and minimal to no effect on that of LCMV (Fig. 11D to F) suggests that these viruses are both resistant to PKR's antiviral activity. The observation that LCMV remains resistant to PKR is surprising in light of the previous findings of this paper and may reflect the transience of PKR's ability to control LCMV infection and/or the fact that LCMV possesses complementary mechanisms to inhibit PKR activity. A final possibility is that PKR may play a proviral role in the context of arenavirus infection as has previously been described for hepatitis C virus (26, 27), though we consider this possibility unlikely, because PKR knockdown did not reduce the fitness of JUNV or LCMV (Fig. 11).
Because protein translation is an absolutely critical process for the production of new viral components, many viruses, including those that rely on cap-dependent translation, have evolved mechanisms to interfere with the canonical antiviral role of PKR (28). For example, the dsDNA poxvirus vaccinia virus expresses the E3L protein, which prevents PKR from binding to dsRNA (29, 30). The paramyxovirus measles virus, a negative-strand RNA virus, antagonizes PKR activation via its nonstructural protein C, which negatively regulates the production of viral dsRNA intermediates during infection (31). In A549 cells, it was shown that the paramyxovirus respiratory syncytial virus (RSV) antagonizes PKR activity via its N protein, which binds PKR and decreases its ability to bind eIF2α, thus shielding eIF2α from PKR's kinase activity (32). Another report confirmed that RSV infection leads to the activation of PKR but that it also leads to significant phosphorylation of eIF2α in HEp-2 cells (33). The discrepancy between these two studies may be due to cell type-specific expression of protein phosphatase 2, the enzyme responsible for dephosphorylating eIF2α. Nonstructural protein 1 (NS1) of the orthomyxovirus influenza A virus directly interacts with PKR and appears to interfere with its ability to bind dsRNA and become activated (34, 35). A common theme from these examples is that PKR antagonism represents a common strategy employed by diverse viruses, but the means by which they accomplish this feat are similarly various. For the viruses cited herein, the ability of JUNV to prevent eIF2α phosphorylation is most similar to that previously described for RSV, as in both cases PKR becomes activated but is unable to phosphorylate eIF2α (Fig. 7, 8, and 12) (32).
The exact mechanism used by JUNV to antagonize PKR's ability to phosphorylate eIF2α remains unclear. Our studies suggest that NP itself, as well as its ability to interact with the cellular factors PKR and eIF2α, may be critical. Indeed, in a related study, Linero et al. demonstrated that acute JUNV infection or expression of JUNV NP alone is sufficient to prevent eIF2α phosphorylation and the subsequent formation of cellular stress granules (SGs) following exposure to the oxidative stressor sodium arsenite (36). Phosphorylation of eIF2α is a key event in driving the nucleation of SGs, which contain stalled cellular mRNAs and their associated translation initiation factors and are associated with inhibition of cap-dependent translation in response to various cellular stresses, including virus infection (37). Sodium arsenite is thought to induce SG formation via activation of PKR and/or heme-regulated inhibitor (38, 39), each of which ultimately phosphorylates eIF2α. Therefore, if sodium arsenite drives eIF2α phosphorylation via PKR, NP may similarly block PKR's ability to phosphorylate eIF2α in response to both oxidative stress (sodium arsenite) and dsRNA intermediates [poly(I·C)].
JUNV has also been shown to block SG formation in response to challenge with the endoplasmic reticulum (ER) stressors dithiothreitol (DTT) and thapsigargin, both of which lead to the phosphorylation of eIF2α via activation of protein kinase-like endoplasmic reticulum kinase (PERK), not PKR (40, 41). Paradoxically, despite failing to drive SG formation in JUNV-infected cells, both DTT and thapsigargin potently induced the phosphorylation of eIF2α in the same cells (36). We identified G3BP1, another cellular protein critical for the nucleation of stress granules (42), as an interacting partner of JUNV and LCMV NPs (Fig. 3A, B, G, and H and 4A; see Table S1 in the supplemental material). Further, a related study demonstrated that G3BP1 colocalizes with the arenavirus Tacaribe virus NP during infection (43). It is possible that JUNV NP can prevent SG nucleation despite the presence of phosphorylated eIF2α by interfering with key SG proteins, such as G3BP1. Indeed, several other SG proteins were identified as partners of arenavirus NPs in our study, including eIF3 components, G3BP2, PABC, and several small ribosomal subunit proteins (Tables S1 and S2) (44, 45).
In addition to the antagonism of PKR's antiviral function reported here, arenaviruses have been shown to interfere with other innate immune pathways responsible for antiviral defense, including the RIG-I/MAVS pathway that drives type I IFN production. In particular, arenavirus NPs have been shown to associate with IKKε and to block its ability to phosphorylate/activate IRF3, which is required for type I IFN production (10). LCMV NP has also been shown to block NF-κB activation (46). Arenavirus NPs further antagonize the type I IFN response via 3′-to-5′ exonuclease activity, which limits the availability of viral dsRNA replicative intermediates for recognition by the RIG-I/MAVS pathway (7, 8). Notably, our results suggest that NP cannot clear all of these dsRNA intermediates, as we presume that these drive the activation of PKR during infection. Intriguingly, arenaviruses appear to be highly selective in their antagonism of these innate immune pathways. For example, Pythoud et al. demonstrated that despite blocking RIG-I/MAVS-dependent type I IFN production, LCMV leaves MAVS-dependent apoptosis fully functional (47). Likewise, we report here that JUNV can block activated PKR from phosphorylating eIF2α (Fig. 7A and C and 8A and C) yet leaves a phosphorylated form of PKR that is capable of inducing the degradation of IκB in response to poly(I·C) (Fig. 8A and D), which is a key step upstream of NF-κB activation (17, 18). We recommend caution in the interpretation of this finding, though, as other innate immune sensors are also able to activate NF-κB in response to stimulation with dsRNA ligands (19). These examples of selective antagonism suggest that arenaviruses have coevolved with their reservoir rodents so that arenavirus-infected cells within the host, while unable to respond to the infecting arenavirus, can still mount an effective antimicrobial response against other environmental pathogens to ensure the fitness and survival of the host.
The cellular protein partners of the arenavirus NPs identified in this study may provide clues to advance our understanding of how these viruses hijack the host cell machinery to facilitate the viral life cycle. Importantly, each protein partner represents a candidate target for future antiviral screening. For example, several host proteins that interact with NP and are required for viral propagation were validated in our study, including hnRNPA1 and hnRNPA2B1 (13), as well as eIF4A and eIF4G, which are components of the eIF4F translation initiation complex (12, 43) (Table 1). Also consistent with these previous studies, eIF4E, the cap-binding component of the eIF4F complex, was not detected in our work, supporting the hypothesis that JUNV and other New World arenavirus NPs can replace the cap-binding function of eIF4E to facilitate translation of viral mRNAs (12). Finally, 8 additional host proteins identified in our screen—COPA, DDX60L, EIF3A, EIF3G, FAU, FBL, HNRNPK, and NOP56 (Table S1)—were previously shown to be required for LCMV and vesicular stomatitis virus (VSV) replication (48). Our results suggest that these proteins may be critical for viral propagation due to their interaction with the LCMV NP. These examples highlight the feasibility and utility of using a proteomics-based approach to identify antiviral targets.
In summary, we have provided the first detailed map of the human proteins engaged by arenavirus NPs and also highlighted host processes that are likely important for arenavirus propagation and/or pathogenesis. This information will serve as a useful resource to guide future studies investigating the importance of arenavirus-host interactions and defining additional functions of NP. By showing that JUNV can selectively block the canonical antiviral role of PKR, we have advanced our understanding of the seemingly multifaceted strategy employed by arenaviruses to curtail an effective innate immune response. Important areas to address in the future will include dissecting the specific mechanism used by JUNV to inhibit PKR's antiviral activity as well as fully interrogating the importance of the remaining cellular interacting proteins identified in the proteomic screen.
MATERIALS AND METHODS
Cells and viruses.
HEK 293T/17 cells (CRL-11268) and A549 cells (CCL-185) were procured from the American Type Culture Collection (ATCC, Manassas, VA). Vero E6 cells were generously given by J. L. Whitton (The Scripps Research Institute, La Jolla, CA). HEK 293T/17 cells were cultured in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, 1% MEM nonessential amino acid solution, 1% HEPES buffer solution, and 1% GlutaMAX, all purchased from Thermo Fisher Scientific (Waltham, MA). A549 cells were cultured in DMEM-F12 (Thermo Fisher) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Vero E6 cells were maintained in DMEM containing 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% HEPES buffer solution. JUNV strain Candid #1 was generously provided by R. Tesh (The University of Texas Medical Branch at Galveston) and M. J. Buchmeier (University of California, Irvine). LCMV Armstrong 53b was obtained from J. L. Whitton. Titers of LCMV and JUNV were determined by standard plaque assay on Vero E6 cells. Recombinant influenza A/Panama/2007/99 (Pan/99) wild-type virus and recombinant influenza A Panama/2007/99 ΔNS1 (Pan-delNS1) virus were generously provided by T. Wolff (Robert Koch Institut, Berlin, Germany) (49). Cells were infected with IAV at a multiplicity of infection (MOI) of 1 after being washed with serum-free medium. Infected cells were cultured in serum-free DMEM containing 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)–trypsin (Sigma-Aldrich) and 0.14% bovine serum albumin (BSA). Titers of IAV Pan/99 were determined by standard plaque assay on MDCK cells, using a low-viscosity overlay (0.5× DMEM, 0.5 μg/ml TPCK-trypsin, 0.07% BSA, 1.2% Avicel [FMC Biopolymer]) (50). Titers of IAV Pan-delNS1 were determined by an immunological focus assay on MDCK cells, essentially as described previously (51), using a low-viscosity overlay and an IAV NP monoclonal antibody (ab20343; Abcam) (1:5,000).
Immunoprecipitations and affinity purifications.
Cells were scraped into the medium following infection or transfection, pelleted, washed with cold phosphate-buffered saline (PBS), and gently lysed on ice in 25 mM Tris-HCl, pH 7.6, containing 1% Triton X-100 (Sigma-Aldrich, St. Louis, MO), 0.5% Nonidet P-40 IGEPAL CA-630 (MP Biomedicals, Solon, OH), 140 mM NaCl, 1 mM calcium chloride (Sigma-Aldrich), and a Complete Mini EDTA-free protease inhibitor cocktail tablet (Roche Applied Science). Cell lysates were clarified of insoluble material by centrifugation at 10,000 rpm at 4°C. To preclear proteins nonspecifically interacting with magnetic protein G beads (Dynabeads protein G beads; Thermo Fisher), beads were added to the lysate and incubated on a rotating platform for 15 min at 4°C. An appropriate antibody was added to cell lysates and incubated for 2 h at 4°C. Magnetic protein G beads were added to lysates and incubated for 1 h at 4°C. The beads were washed 4 times with ice-cold lysis buffer to remove nonspecific proteins. Bound protein was stripped from the beads into 1× Laemmli buffer, 5% β-mercaptoethanol by heating at 100°C for 5 min. Immunoprecipitated proteins were separated by SDS-PAGE as described below. Antibodies used for immunoprecipitations were as follows: antibodies to JUNV NP (KA03-AA01; BEI Resources), LCMV NP (1.1.3 and 24A; generously provided by M. J. Buchmeier) (52), AIF (sc-9416; Santa Cruz), G3BP1 (A302-033A; Bethyl), and PKR (Y117; Abcam).
Affinity purification of biotinylated viral NPs was performed as follows. Viral NPs were cloned into a modified pCAGGS plasmid and carried a C-terminal hemagglutinin (HA) epitope tag, the tobacco etch virus (TEV) protease cleavage site, and a biotin acceptor peptide (BAP). Viral NPs were biotinylated when cotransfected with a plasmid expressing BirA, a bacterial biotin ligase. As a control to validate the specificity of the protein-protein interaction, cells were transfected with an empty vector along with the BirA plasmid. Cells were lysed as described above. Biotinylated proteins were captured by incubating cleared lysate with magnetic streptavidin beads (Dynabeads MyOne streptavidin T1; Thermo Fisher) at 4°C for 2.5 h. Beads were washed 4 times with cold lysis buffer, and interacting proteins were stripped from beads into 1× Laemmli buffer containing 5% β-mercaptoethanol by heating at 100°C for 5 min.
Mass spectrometry.
To define the cellular protein interactome of JUNV NP and LCMV NP, A549 or HEK 293T cells were infected with either JUNV Candid #1 or LCMV Armstrong 53b. Following infection, the viral NPs were immunoprecipitated (as described above) along with associated cellular proteins. After denaturation, the immune complexes were separated by SDS-PAGE on Novex 4 to 20% Tris-glycine polyacrylamide gels. Gels were stained with Coomassie blue (0.02% Brilliant Blue R [Sigma-Aldrich] in 32% methanol and 22% acetic acid) at room temperature overnight. Gels were destained for 6 to 8 h in a solution containing 30% methanol and 10% acetic acid. Each lane was cut out of the gel in 13 (JUNV NP) or 15 (LCMV NP) slices (cut maps are available upon request). In-gel trypsin digestion of proteins was performed with sequencing-grade modified trypsin (6 ng/μl) (Promega, Madison, WI) in 50 mM ammonium bicarbonate overnight at 37°C, as previously reported (53). Peptides were extracted from the digested gel slices by use of 50% acetonitrile (MeCN) and 2.5% formic acid (FA) and dried using a vacuum centrifuge. Dried peptides were resuspended in 2.5% MeCN and 2.5% FA and loaded onto a 12-cm reverse-phase Magic C18 microcapillary column (5 μm by 200 Å; Michrom Bioresources, Inc., Auburn, CA) by utilizing a MicroAS autosampler (Thermo Scientific, Pittsburgh, PA). Peptides were eluted with a 5 to 35% MeCN (0.15% FA) gradient by use of a Surveyor Pump Plus high-pressure liquid chromatograph (HPLC) (Thermo Scientific) over 40 min, after a 15-min isocratic loading with 2.5% MeCN and 0.15% FA. An LTQ-XL linear ion trap mass spectrometer (Thermo Scientific) was used to acquire mass spectra of eluted peptides over the entire run, using 10 MS/MS scans following each survey scan. Raw data were searched against the human IPI forward and reverse concatenated databases by using SEQUEST software, allowing a 2-Da mass tolerance for peptide matches. Cysteine residues were required to have a static increase of 71.0 Da for acrylamide adduction. A 16.0-Da differential modification on methionine residues was permitted. Host proteins were accepted as legitimate NP protein partners if they were identified by 2 or more unique tryptic peptides in samples infected with either JUNV or LCMV but not in the corresponding uninfected control. Alternatively, proteins were included if there was a 5-fold larger quantity of total tryptic peptides that were detected for a particular human protein from a virus-infected sample than for the sample from the corresponding uninfected control. Using these filters, the false discovery rate for peptides was <1%.
Bioinformatic analysis of cellular protein partners was performed with the NIH DAVID functional annotation tool (version 6.7; https://david.ncifcrf.gov/) (15, 16).
SDS-PAGE and Western blotting.
Novex 4 to 20% Tris-glycine polyacrylamide gels or NuPAGE Novex 4 to 12% Bis-Tris Midi protein gels (Thermo Scientific) were used to separate protein lysates by SDS-PAGE. MOPS (morpholinepropanesulfonic acid) SDS running buffer (Thermo Scientific) was used with the NuPage Bis-Tris polyacrylamide gels. Protein was transferred from gels to nitrocellulose membranes by use of an iBlot gel transfer device and iBlot Transfer Stack nitrocellulose membranes (Thermo Scientific). Membranes were blocked with 5% milk in PBS for 1 h at room temperature. Primary antibodies and secondary antibodies were diluted in PBS containing 5% milk, 3% FBS, and 0.05% Nonidet P-40 IGEPAL CA-630. Blots were incubated in diluted primary antibody overnight at 4°C. Primary antibodies used for Western blotting were as follows: PKR (sc-707; Santa Cruz) (1:1,000), IκBα (9242l Cell Signaling) (1:1,000), rabbit anti-actin (A2066; Sigma-Aldrich) (1:5,000), mouse anti-actin (A5441; Sigma-Aldrich) (1:5,000), SFPQ (NB100-61044; Novus) (1:2,500), eIF2α (sc-11386; Santa Cruz) (1:1,000), p-eIF2α S51 (3398; Cell Signaling) (1:1,000), G3BP1 (A302-033A; Bethyl) (1:2,500), AIF (sc-9416; Santa Cruz) (1:1,000), p-PKR T446 (E120; Abcam) (1:1,000), anti-JUNV NP (NA05-AG12; BEI Resources) (1:200), anti-LCMV NP (2165; obtained from M. J. Buchmeier) (1:10,000), and IAV NP (ab20343; Abcam) (1:1,000) antibodies. Unbound primary antibody was washed from blots by 3 consecutive washes in Western wash solution (PBS with 0.5% Nonidet P-40 IGEPAL CA-630). Blots were incubated with diluted secondary antibody for 2 h at room temperature. The secondary antibodies used were as follows: goat anti-mouse (H+L) (71045-3; Novagen) (1:10,000), goat anti-mouse (light chain only) (AP200P; Millipore) (1:10,000), goat anti-rabbit (H+L) (111-035-045; Jackson) (1:10,000), mouse anti-rabbit (light chain only) (211-032-171) (1:10,000), and rabbit anti-goat peroxidase (401515; Calbiochem) (1:10,000). Finally, blots were washed 3 more times with Western wash solution before being developed with chemiluminescence substrate (either Pierce ECL Western blotting substrate or SuperSignal West Pico or Femto chemiluminescence substrate [Thermo Scientific]). Alternatively, blots were probed with fluorescently labeled secondary antibodies, namely, IRDye 800CW-labeled goat anti-rabbit IgG (H+L) (926-32211; Li-Cor) (1:20,000), IRDye 800CW-labeled goat anti-mouse IgG (H+L) (926-32210; Li-Cor) (1:20,000), IRDye 680RD-labeled goat anti-mouse IgG (H+L) (926-68070; Li-Cor) (1:20,000), and IRDye 680RD-labeled goat anti-rabbit IgG (H+L) (926-68071; Li-Cor) (1:20,000), and visualized with an Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE).
Plasmids and transfection.
To validate the interaction of identified cellular proteins, the arenaviral NPs were subcloned into a modified pCAGGS expression vector as previously described (54, 55). This vector expresses an NP fusion protein containing 3 C-terminal elements: an HA epitope tag (YPYDVPDYA), the TEV protease cleavage site (ENLYFQG), and a 23-amino-acid BAP (MASSLRQILDSQKMEWRSNAGGS). The BAP sequence can be biotinylated when cells are cotransfected with a plasmid that encodes the bacterial biotin ligase BirA, and the biotinylated NP can be affinity purified as described previously (54, 55). The NP sequences subcloned into the pCAGGS expression vector were as follows (for each NP, an NCBI gene identifier number and a protein locus number are listed): LASV strain Josiah (NC_004296 and NP_694869), LCMV Armstrong 53b (DQ408671 and ABD73126), and JUNV strain Candid #1 (HQ126699 and AEB32437). Transfection of HEK 293T cells was done using polyethylenimine (PEI) (Polysciences, Inc., Warrington, PA) (5 μg of PEI [Thermo Fisher] per 1 μg of DNA).
Confocal microscopy.
Localization of the viral NP and cellular proteins in JUNV- or LCMV-infected A549 cells was visualized by confocal microscopy. A549 cells were seeded onto no. 1.5 12-mm glass coverslips (Thermo Scientific). The day after seeding, cells were infected or not (mock) with JUNV at an MOI of 0.1 or with LCMV at an MOI of 0.01. Cells were fixed 48 or 72 h after infection with 4% paraformaldehyde (PFA; Electron Microscopy Sciences) in 1× PBS. Cells were permeabilized in PBS with 0.1% Triton X-100 and 1% BSA and then blocked in PBS containing 3% BSA for 30 min at room temperature.
Cells were incubated with a primary antibody diluted in 1% BSA in 1× PBS at room temperature for 1 h. The NA05-AG12 (mouse) antibody to detect JUNV NP was diluted 1:100, the 1.1.3 (mouse) antibody for LCMV NP was diluted 1:500, the anti-PKR antibody (rabbit monoclonal Y117; Abcam) was diluted 1:100, and anti-G3BP1 (A302-033A; Bethyl) was diluted 1:100. Coverslips were washed 4 times in 1× PBS at room temperature. Coverslips were incubated with secondary antibody diluted in 1% BSA in 1× PBS for 30 min at room temperature. Secondary antibodies were Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) (A-11029; Thermo Scientific) (1:800) and Alexa Fluor 647-conjugated goat anti-rabbit IgG (H+L) (A-21245; Thermo Scientific) (1:800). Coverslips were washed 3 times in PBS, stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich), washed a final time in PBS, and mounted onto glass slides by use of ProLong Gold antifade reagent (Thermo Fisher). Confocal microscopy was performed with a Zeiss LSM 510 laser scanning confocal microscope. Images were acquired with a 63× objective lens with a numerical aperture of 1.4. Images were acquired at 1.0 Airy unit for the Alexa Fluor 647 dye. Pinhole diameters for the DAPI and Alexa Fluor 488 channels were set accordingly.
Puromycylation of nascent polypeptides.
To label newly synthesized peptides in infected cells growing on glass coverslips, cells were incubated for 5 min at 37°C in puromycylation medium (DMEM-F12, 10% FBS, 1% penicillin-streptomycin supplemented with 91 μM puromycin [Sigma-Aldrich] and 208 μM emetine [Sigma-Aldrich]) as previously described (21). As a negative control, some cells were pretreated with complete medium containing 500 μM sodium arsenite (Sigma-Aldrich) for 15 min at 37°C before labeling with puromycylation medium. Following the labeling reaction, cells were briefly washed with cold Dulbecco's PBS (DPBS; with calcium and magnesium) (Thermo Fisher). Cells were washed with cold permeabilization buffer (50 mM Tris-HCl, 5 mM MgCl2, 25 mM KCl, 0.015% digitonin [Sigma-Aldrich]) to remove free puromycin before being fixed in 3% PFA in 1× PBS for 15 min at room temperature. Cells were prepared for confocal microscopy as described above. LCMV NP was labeled with the 1.1.3 antibody (at 6.8 μg/ml), and JUNV NP was labeled with the NA05 antibody (at 10 μg/ml), with both antibodies directly conjugated to Alexa Fluor 488 (Thermo Fisher). Puromycin was detected with the monoclonal antibody 12D10 (at 1 μg/ml) directly conjugated to Alexa Fluor 647 (Sigma-Aldrich). Puromycin levels in individual cells were quantitated by use of a customized image analysis pipeline in CellProfiler (Broad Institute) (56).
Poly(I·C) transfections.
For a single well in a six-well plate, 0 μg, 0.5 μg, or 5 μg of poly(I·C) (Sigma-Aldrich) was added to 125 μl Opti-MEM medium (Thermo Fisher) and mixed well. Three microliters of Lipofectamine 2000 (Thermo Fisher) was added to a tube containing 125 μl Opti-MEM and mixed gently. The solution containing poly(I·C) was added to the solution containing Lipofectamine 2000, mixed gently, and incubated at room temperature for 10 min. The medium was aspirated from the wells and replaced with 2 ml of fresh warm complete A549 medium. The poly(I·C)-Lipofectamine transfection mix was added dropwise to the wells. The transfected cells were incubated at 37°C for 6 h, at which time cells were lysed and analyzed by Western blotting.
siRNA transfection.
siRNAs were reverse transfected into A549 cells in a 12-well plate format as follows. To 100 μl of Opti-MEM, 2 μl of Lipofectamine RNAi Max (Thermo Fisher) was added and mixed gently. To another 100 μl of Opti-MEM, 12 pmol of siRNA was added and mixed. The siRNA-containing solution was added to the Lipofectamine-containing solution and allowed to incubate at room temperature for 5 min. Two hundred microliters of this mixture was added to an empty well of a 12-well plate. Next, 40,000 A549 cells in 1 ml of complete A549 medium were added to the Opti-MEM/siRNA-containing well and incubated at 37°C. At 2 days posttransfection, the medium was replaced with fresh prewarmed complete A549 medium. siRNA-transfected A549 cells were infected at 72 h post-siRNA transfection. The following Silencer Select siRNAs were used for knockdown experiments: Silencer Select negative control 1 siRNA siSCR (Thermo Fisher), siPKR-1 (s11185; Thermo Fisher) (Fig. 10 and 11), and siPKR-2 (s229501; Thermo Fisher) (Fig. 11).
Statistics.
Statistical analyses were performed in GraphPad software. Two-way analysis of variance (ANOVA) with post hoc Tukey's multiple-comparison test was used for Fig. 8. One-way ANOVA with Dunnett's multiple-comparison test was used for Fig. 7, 9C, 9D, and 11. One-way ANOVA with Tukey's multiple-comparison test was used for Fig. 9E. Unpaired two-tailed Student's t test was used for Fig. 10.
Supplementary Material
ACKNOWLEDGMENTS
We thank the UVM Immunobiology Group for insightful discussions. We are grateful to Michael Buchmeier, J. Lindsay Whitton, Bob Tesh, and Thorsten Wolff for providing critical reagents as described in Materials and Methods. We acknowledge Jamie Kelly and Sarah Girome for their help with experiments. We thank the UVM Neuroscience Imaging and Physiology Core and the UVM Microscopy Imaging Center for microscopy and imaging support. Confocal microscopy was performed on a Zeiss 510 Meta laser scanning confocal microscope supported by NIH award 1S10RR019246 from the National Center for Research Resources.
We gratefully acknowledge NIH grants T32 HL076122-10 (B.R.K.), T32 AI055402 (C.M.Z.), R21 AI088059 (J.B.), and P20RR021905 (Immunobiology and Infectious Disease COBRE) (J.B.). Additionally, the mass spectrometry analysis was supported by the Vermont Genetics Network through NIH grant 8P20GM103449 from the INBRE program and the NIGMS (B.A.B. and M.E.W.).
Funding Statement
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00763-17.
REFERENCES
- 1.Buchmeier MJ, de la Torre JC, Peters CJ. 2007. Arenaviridae: the viruses and their replication, p 1791–1827. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Fischer SA, Graham MB, Kuehnert MJ, Kotton CN, Srinivasan A, Marty FM, Comer JA, Guarner J, Paddock CD, DeMeo DL, Shieh WJ, Erickson BR, Bandy U, DeMaria A Jr, Davis JP, Delmonico FL, Pavlin B, Likos A, Vincent MJ, Sealy TK, Goldsmith CS, Jernigan DB, Rollin PE, Packard MM, Patel M, Rowland C, Helfand RF, Nichol ST, Fishman JA, Ksiazek T, Zaki SR, LCMV in Transplant Recipients Investigation Team. 2006. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N Engl J Med 354:2235–2249. doi: 10.1056/NEJMoa053240. [DOI] [PubMed] [Google Scholar]
- 3.Enria DA, Briggiler AM, Sanchez Z. 2008. Treatment of Argentine hemorrhagic fever. Antiviral Res 78:132–139. doi: 10.1016/j.antiviral.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCormick JB, King IJ, Webb PA, Scribner CL, Craven RB, Johnson KM, Elliott LH, Belmont-Williams R. 1986. Lassa fever. Effective therapy with ribavirin. N Engl J Med 314:20–26. [DOI] [PubMed] [Google Scholar]
- 5.Botten J, King B, Klaus J, Ziegler C. 2013. Pathogenic Old World arenaviruses, p 233–259. In Singh SK, Ruzek D (ed), Viral hemorrhagic fevers. CRC Press, Boca Raton, FL. [Google Scholar]
- 6.Buchmeier MJ, Bowen MD, Peters CJ. 2001. Arenaviridae: the viruses and their replication, p 1635–1668. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 7.Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, Ly H, Liang Y, Dong C. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468:779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. 2011. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci U S A 108:2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shtanko O, Watanabe S, Jasenosky LD, Watanabe T, Kawaoka Y. 2011. ALIX/AIP1 is required for NP incorporation into Mopeia virus Z-induced virus-like particles. J Virol 85:3631–3641. doi: 10.1128/JVI.01984-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pythoud C, Rodrigo WW, Pasqual G, Rothenberger S, Martinez-Sobrido L, de la Torre JC, Kunz S. 2012. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKKepsilon. J Virol 86:7728–7738. doi: 10.1128/JVI.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labudova M, Tomaskova J, Skultety L, Pastorek J, Pastorekova S. 2009. The nucleoprotein of lymphocytic choriomeningitis virus facilitates spread of persistent infection through stabilization of the keratin network. J Virol 83:7842–7849. doi: 10.1128/JVI.00309-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linero F, Welnowska E, Carrasco L, Scolaro L. 2013. Participation of eIF4F complex in Junin virus infection: blockage of eIF4E does not impair virus replication. Cell Microbiol 15:1766–1782. doi: 10.1111/cmi.12149. [DOI] [PubMed] [Google Scholar]
- 13.Maeto CA, Knott ME, Linero FN, Ellenberg PC, Scolaro LA, Castilla V. 2011. Differential effect of acute and persistent Junin virus infections on the nucleo-cytoplasmic trafficking and expression of heterogeneous nuclear ribonucleoproteins type A and B. J Gen Virol 92:2181–2190. doi: 10.1099/vir.0.030163-0. [DOI] [PubMed] [Google Scholar]
- 14.Goni SE, Iserte JA, Ambrosio AM, Romanowski V, Ghiringhelli PD, Lozano ME. 2006. Genomic features of attenuated Junin virus vaccine strain candidate. Virus Genes 32:37–41. doi: 10.1007/s11262-005-5843-2. [DOI] [PubMed] [Google Scholar]
- 15.Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Garcia MA, Meurs EF, Esteban M. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Zamanian-Daryoush M, Mogensen TH, DiDonato JA, Williams BR. 2000. NF-kappaB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-kappaB-inducing kinase and IkappaB kinase. Mol Cell Biol 20:1278–1290. doi: 10.1128/MCB.20.4.1278-1290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai T, Akira S. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci 1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 21.David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, Pierre P, Bennink JR, Yewdell JW. 2012. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol 197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergmann M, Garcia-Sastre A, Carnero E, Pehamberger H, Wolff K, Palese P, Muster T. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J Virol 74:6203–6206. doi: 10.1128/JVI.74.13.6203-6206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatada E, Saito S, Fukuda R. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J Virol 73:2425–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donnelly N, Gorman AM, Gupta S, Samali A. 2013. The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci 70:3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, Romano PR, Nagamura-Inoue T, Tian B, Devert TE, Mathews MB, Ozato K, Hinnebusch AG. 2001. Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J Biol Chem 276:24946–24958. doi: 10.1074/jbc.M102108200. [DOI] [PubMed] [Google Scholar]
- 26.Garaigorta U, Chisari FV. 2009. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe 6:513–522. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnaud N, Dabo S, Maillard P, Budkowska A, Kalliampakou KI, Mavromara P, Garcin D, Hugon J, Gatignol A, Akazawa D, Wakita T, Meurs EF. 2010. Hepatitis C virus controls interferon production through PKR activation. PLoS One 5:e10575. doi: 10.1371/journal.pone.0010575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reineke LC, Lloyd RE. 2013. Diversion of stress granules and P-bodies during viral infection. Virology 436:255–267. doi: 10.1016/j.virol.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang HW, Watson JC, Jacobs BL. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci U S A 89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langland JO, Jacobs BL. 2002. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299:133–141. doi: 10.1006/viro.2002.1479. [DOI] [PubMed] [Google Scholar]
- 31.Toth AM, Devaux P, Cattaneo R, Samuel CE. 2009. Protein kinase PKR mediates the apoptosis induction and growth restriction phenotypes of C protein-deficient measles virus. J Virol 83:961–968. doi: 10.1128/JVI.01669-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groskreutz DJ, Babor EC, Monick MM, Varga SM, Hunninghake GW. 2010. Respiratory syncytial virus limits alpha subunit of eukaryotic translation initiation factor 2 (eIF alpha) phosphorylation to maintain translation and viral replication. J Biol Chem 285:24023–24031. doi: 10.1074/jbc.M109.077321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindquist ME, Mainou BA, Dermody TS, Crowe JE Jr. 2011. Activation of protein kinase R is required for induction of stress granules by respiratory syncytial virus but dispensable for viral replication. Virology 413:103–110. doi: 10.1016/j.virol.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Min JY, Krug RM, Sen GC. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y, Wambach M, Katze MG, Krug RM. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 36.Linero FN, Thomas MG, Boccaccio GL, Scolaro LA. 2011. Junin virus infection impairs stress-granule formation in Vero cells treated with arsenite via inhibition of eIF2alpha phosphorylation. J Gen Virol 92:2889–2899. doi: 10.1099/vir.0.033407-0. [DOI] [PubMed] [Google Scholar]
- 37.Thomas MG, Loschi M, Desbats MA, Boccaccio GL. 2011. RNA granules: the good, the bad and the ugly. Cell Signal 23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel CV, Handy I, Goldsmith T, Patel RC. 2000. PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J Biol Chem 275:37993–37998. doi: 10.1074/jbc.M004762200. [DOI] [PubMed] [Google Scholar]
- 39.McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 280:16925–16933. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- 40.Harding HP, Zhang Y, Ron D. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 41.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 5:897–904. doi: 10.1016/S1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 42.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Baird NL, York J, Nunberg JH. 2012. Arenavirus infection induces discrete cytosolic structures for RNA replication. J Virol 86:11301–11310. doi: 10.1128/JVI.01635-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem Sci 33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 45.Buchan JR, Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodrigo WW, Ortiz-Riano E, Pythoud C, Kunz S, de la Torre JC, Martinez-Sobrido L. 2012. Arenavirus nucleoproteins prevent activation of nuclear factor kappa B. J Virol 86:8185–8197. doi: 10.1128/JVI.07240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pythoud C, Rothenberger S, Martinez-Sobrido L, de la Torre JC, Kunz S. 2015. Lymphocytic choriomeningitis virus differentially affects the virus-induced type I interferon response and mitochondrial apoptosis mediated by RIG-I/MAVS. J Virol 89:6240–6250. doi: 10.1128/JVI.00610-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panda D, Das A, Dinh PX, Subramaniam S, Nayak D, Barrows NJ, Pearson JL, Thompson J, Kelly DL, Ladunga I, Pattnaik AK. 2011. RNAi screening reveals requirement for host cell secretory pathway in infection by diverse families of negative-strand RNA viruses. Proc Natl Acad Sci U S A 108:19036–19041. doi: 10.1073/pnas.1113643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matthaei M, Budt M, Wolff T. 2013. Highly pathogenic H5N1 influenza A virus strains provoke heterogeneous IFN-alpha/beta responses that distinctively affect viral propagation in human cells. PLoS One 8:e56659. doi: 10.1371/journal.pone.0056659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matrosovich M, Matrosovich T, Garten W, Klenk HD. 2006. New low-viscosity overlay medium for viral plaque assays. Virol J 3:63. doi: 10.1186/1743-422X-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Battegay M, Cooper S, Althage A, Banziger J, Hengartner H, Zinkernagel RM. 1991. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24- or 96-well plates. J Virol Methods 33:191–198. doi: 10.1016/0166-0934(91)90018-U. [DOI] [PubMed] [Google Scholar]
- 52.Buchmeier MJ, Lewicki HA, Tomori O, Oldstone MB. 1981. Monoclonal antibodies to lymphocytic choriomeningitis and Pichinde viruses: generation, characterization, and cross-reactivity with other arenaviruses. Virology 113:73–85. doi: 10.1016/0042-6822(81)90137-9. [DOI] [PubMed] [Google Scholar]
- 53.Ballif BA, Cao Z, Schwartz D, Carraway KL III, Gygi SP. 2006. Identification of 14-3-3epsilon substrates from embryonic murine brain. J Proteome Res 5:2372–2379. doi: 10.1021/pr060206k. [DOI] [PubMed] [Google Scholar]
- 54.Cornillez-Ty CT, Liao L, Yates JR III, Kuhn P, Buchmeier MJ. 2009. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J Virol 83:10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klaus JP, Eisenhauer P, Russo J, Mason AB, Do D, King B, Taatjes D, Cornillez-Ty C, Boyson JE, Thali M, Zheng C, Liao L, Yates JR III, Zhang B, Ballif BA, Botten JW. 2013. The intracellular cargo receptor ERGIC-53 is required for the production of infectious arenavirus, coronavirus, and filovirus particles. Cell Host Microbe 14:522–534. doi: 10.1016/j.chom.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. 2011. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.