ABSTRACT
Crimean-Congo hemorrhagic fever virus (CCHFV) is a tick-borne Nairovirus of the Bunyaviridae family, causing severe illness with high mortality rates in humans. Here, we demonstrate that CCHFV nucleocapsid protein (CCHFV-NP) augments mRNA translation. CCHFV-NP binds to the viral mRNA 5′ untranslated region (UTR) with high affinity. It facilitates the translation of reporter mRNA both in vivo and in vitro with the assistance of the viral mRNA 5′ UTR. CCHFV-NP equally favors the translation of both capped and uncapped mRNAs, demonstrating the independence of this translation strategy on the 5′ cap. Unlike the canonical host translation machinery, inhibition of eIF4F complex, an amalgam of three initiation factors, eIF4A, eIF4G, and eIF4E, by the chemical inhibitor 4E1RCat did not impact the CCHFV-NP-mediated translation mechanism. However, the proteolytic degradation of eIF4G alone by the human rhinovirus 2A protease abrogated this translation strategy. Our results demonstrate that eIF4F complex formation is not required but eIF4G plays a critical role in this translation mechanism. Our results suggest that CCHFV has adopted a unique translation mechanism to facilitate the translation of viral mRNAs in the host cell cytoplasm where cellular transcripts are competing for the same translation apparatus.
IMPORTANCE Crimean-Congo hemorrhagic fever, a highly contagious viral disease endemic to more than 30 countries, has limited treatment options. Our results demonstrate that NP favors the translation of a reporter mRNA harboring the viral mRNA 5′ UTR. It is highly likely that CCHFV uses an NP-mediated translation strategy for the rapid synthesis of viral proteins during the course of infection. Shutdown of this translation mechanism might selectively impact viral protein synthesis, suggesting that an NP-mediated translation strategy is a target for therapeutic intervention against this viral disease.
KEYWORDS: bunyavirus, negative-strand RNA virus, nucleocapsid protein, translation
INTRODUCTION
Crimean-Congo hemorrhagic fever virus (CCHFV) is a tick-borne Nairovirus within the Bunyaviridae family that causes severe hemorrhagic fever with a mortality rate of 30% in more than 30 countries worldwide (1–4). There is no vaccine against CCHFV, and therapeutic interventions are limited. This virus is transmitted to humans by either tick bites or direct contact with blood or tissue samples from the infected hosts (5, 6). The recent frequent outbreaks of CCHFV in Mediterranean countries are likely associated with the broad habitat and population size of the CCHFV tick vector (7). The increased tick vector population could be associated with climate change (7). The CCHFV genome is composed of three negative-sense RNA segments (S, M, and L), which encode nucleocapsid protein (NP), glycoprotein precursor (GPC), and RNA-dependent RNA polymerase (RdRp), respectively (8). Although NP is mainly considered to be an integral component of the viral capsid, recent studies on Lassa fever virus (LASV) (Arenaviridae) and Sin Nombre hantavirus (SNV) (Bunyaviridae) have revealed the diverse roles of NP in the virus replication cycle (9–13). Recently, the X-ray crystallographic structure at 2.3-Å resolution has demonstrated a unique metal-based endonuclease activity in CCHFV-NP (14–16). The interaction between NP and cellular heat shock protein HSP70 has been reported to play a role in CCHFV replication (17). Recently, cryoelectron microscopy reconstruction studies revealed a ring-shaped architecture of CCHFV-NP-RNA oligomer, suggesting the interaction between head and stalk domains of CCHFV-NP results in NP multimerization while encapsidating the viral genome.
The rapid synthesis of viral proteins and replication of viral genome in the host cell is critical for the rapid spread of new virions to the neighboring cells during the course of infection. Thus, the efficiency of viral protein synthesis may impact the viral load in infected hosts and, hence, the outcome of the viral disease. Viruses have evolved numerous strategies to boost the translation of their mRNAs by avoiding competition from the host cell transcripts for the same translation machinery (18). We have previously reported that hantaviruses, from another genus in the Bunyaviridae family, have adopted a unique translation mechanism mediated by hantavirus nucleocapsid protein (N protein) that likely facilitates the translation of viral mRNA in the host cell during the course of infection (12). Hantavirus N protein specifically binds to the mRNA 5′ cap and ribosomal protein S19, a structural component of the 40S ribosomal subunit (19). N protein engages the 40S ribosomal subunit at the mRNA 5′ cap independent of eIF4F cap binding complex (19). The eIF4F complex engages the 43S preinitiation ribosome complex at the mRNA 5′ cap during the canonical host translation strategy. We demonstrated that the hantavirus translation strategy preferentially facilitates the translation of an mRNA harboring the viral mRNA 5′ untranslated region (UTR) (20).
To elucidate whether nucleocapsid protein from other genera of the Bunyaviridae family have a similar function, here we demonstrate that CCHFV-NP facilitates mRNA translation in conjunction with the viral mRNA 5′ UTR. Unlike hantavirus N protein, the CCHFV-NP requires eIF4G to initiate translation. Our results show that an CCHFV-NP-mediated translation mechanism is conceptually similar to but mechanistically different from the hantavirus N protein-mediated translation strategy. The studies reported here suggest that CCHFV-NP lures the host translation machinery for the preferential translation of viral mRNA in virus-infected cells where cellular transcripts are competing for the same translation machinery.
RESULTS
CCHFV-NP facilitates mRNA translation with the assistance of viral mRNA 5′ UTR.
Similar to hantaviruses, CCHFV initiates transcription by a cap-snatching mechanism by which viral RdRp cleaves the host cell mRNA 6 to 16 nucleotides downstream of the 5′ cap and uses the resulting capped oligoribonucleotide as a primer to initiate transcription (21). The capped RNA primer remains attached to the viral mRNA 5′ terminus, which further increases the UTR length. Viral S-segment transcripts contain a truncated 3′ UTR and lack the 3′ poly(A) tail. To our knowledge, these characteristic features are unknown for M and L segment-derived mRNAs.
To demonstrate the role of NP in translation, we generated a few green fluorescent protein (GFP) reporter constructs expressing mRNAs that structurally mimic the transcripts expressed during CCHFV infection in cells (Fig. 1A). HeLa cells in 12-well plates were cotransfected with a fixed concentration (0.1 μg) of either pCCHFG1 plasmid or pCDGFP plasmid, along with increasing input concentrations of pCCHF-NP plasmid, expressing CCHFV-NP (strain 10200) (NCBI accession number U88410.1). The GFP mRNA expressed from the pCCHFG1 plasmid is partially a structural mimic of CCHFV S-segment mRNA (strain 10200). It contains a 70-nucleotide-long 5′ UTR composed of a 5′-terminal nonviral 15 nucleotides representing the cap-snatched primer, followed by a 55-nucleotide-long 5′ UTR of CCHFV S-segment mRNA. The mRNA lacks the 3′ UTR. In comparison, the GFP mRNA expressed from the pCDGFP plasmid contains a nonviral 71-nucleotide-long 5′ UTR and 268-nucleotide-long 3′ UTR with a poly(A) tail (Fig. 1A). Cells were harvested 24 h posttransfection, and GFP expression was measured by flow cytometry (Fig. 1B). Increased input concentrations of pCCHF-NP plasmid during transfection selectively increased the GFP signal in cells cotransfected with pCCHFG1 plasmid (Fig. 1B). Although the increased input concentrations of pCCHF-NP plasmid resulted in a corresponding increase in NP expression in cells (Fig. 1B1), it is possible that cotransfection of pCCHF-NP plasmid differentially impacted the transfection efficiencies of two reporter plasmids, resulting in differential reporter expression. Alternatively, it is equally probable that NP expression differentially impacted the stability of two reporter mRNAs which differentially influenced GFP expression in HeLa cells. To rule out these possibilities, we quantified the GFP mRNA levels in transfected cells by real-time PCR. As shown in Fig. 1C, the GFP mRNA levels remained constant in transfected cells, suggesting that NP selectively facilitated the translation of reporter mRNA expressed from pCCHFG1 plasmid.
FIG 1.
CCHFV-NP augments mRNA translation in vivo. (A) mRNA expressed from plasmids pCCHF G1, pCCHF G2, and pCCHF G3 contain the same 15-nucleotide-long nonviral sequence upstream of the 5′ UTR, shown under dotted lines. This sequence is different in mRNA expressed from the pCDGFP plasmid. The exact sequence of the viral 5′ UTR, nonviral UTR, and scrambled 5′ UTR is shown in panel L. HDV represents the hepatitis delta virus ribozyme sequence. Arrows show the transcription initiation site. (B) HeLa cells in 12-well plates were cotransfected with a fixed concentration (0.1 μg) of either pCCHF G1 plasmid (filled bars) or pCDGFP plasmid (open bars) with increasing input concentrations of pCCHF-NP plasmid (0.0 to 0.4 μg). The total plasmid in each well was balanced to 1.0 μg using empty-vector pcDNA 3.1(+). Cells were harvested 24 h posttransfection, and total GFP fluorescence signal from each well was quantified by flow cytometry, as previously reported (36). The GFP readout from three triplicates of an experiment was averaged. Three such averaged readouts from three independent experiments were further averaged and normalized relative to the control lacking transfection with pCCHF-NP plasmid (first bar from the left). Error bars represent the standard deviations calculated from three independent experiments. (B1) Western blot (WB) analysis showing the expression levels of CCHFV NP and β actin in cell lysates from panel B. CCHFV-NP was detected using a monoclonal antibody against (mAb) the C-terminal His tag. (C) Total RNA was purified from cells cotransfected with plasmids as mentioned for panel B. GFP mRNA was quantitatively examined by real-time PCR analysis (relative quantification method) using power Sybr green PCR master mix from Applied Biosystems as previously reported (35). (D and E) The experiment performed is similar to the experiment performed for panel B, except the pCCHF G1 plasmid was replaced with pCCHF G2 (D) or pCCHF G3 (E). (D1 and E1) Western blot analysis showing the expression levels of CCHFV NP and β actin in cell lysates from panels D and E, respectively. (F) SDS-PAGE analysis of GST-NP fusion protein (left) and GST (right), purified from the Rosetta (DE3) PLacI E. coli strain. Crude lysate (CL), flowthrough (FL), and eluted protein (EL) are shown. Gels were truncated to fit the space. (G) Diagrammatic representation of 5′-capped reporter mRNAs, synthesized by in vitro T7 transcription reaction. The 3′ end of FLuc control mRNA was posttranscriptionally poly(A) tailed as previously reported (35). The exact sequences of viral, nonviral, and scrambled 5′ UTRs are shown in panel L. (H, I, J, and K) One microgram each of purified CCHF FL1 (H), CCHF FL2 (I), CCHF FL3 (J), and Fluc control (K) mRNAs was translated in 50 μl rabbit reticulocyte lysates (RRL) in the absence or presence of either 50 nM or 100 nM purified GST-NP (filled bars) or GST (open bars). Each reaction was carried out in triplicate and repeated three times. Luciferase signal in each sample was normalized relative to the control lacking the GST or GST-NP fusion protein. Error bars represent the standard deviations from three independent experiments. (L) Sequence of viral, nonviral, and scrambled 5′ UTRs used in the above-described panels. The viral mutant 5′ UTR sequence was used for Fig. 6. (M) Five percent denaturing polyacrylamide gels showing radiolabeled transcripts, as mentioned in the text.
In negative-strand RNA viruses, the 3′ termini of mRNA molecules are usually truncated. The 3′ terminus of CCHFV S-segment cRNA (strain 10200) is 167 nucleotides in length. We deleted the 3′-terminal 20 nucleotides of the 3′ UTR and incorporated the remaining 147 nucleotides downstream of the GFP stop codon, preceding the HDV sequence in pCCHFG2 plasmid (Fig. 1A). HeLa cells in 12-well plates again were cotransfected with the new construct (pCCHFG2) to monitor the impact of CCHFV-NP upon the translation of GFP reporter harboring the viral mRNA 3′ UTR sequence (Fig. 1D). Again, increased NP expression (Fig. 1D1) selectively facilitated the translation of GFP reporter mRNA expressed from pCCHFG2 plasmid (Fig. 1D), suggesting that the 3′ UTR does not play a role in NP-mediated translation of GFP mRNA. To further confirm that NP, with the assistance of the viral mRNA 5′ UTR, alone can facilitate the translation of the downstream open reading frame (ORF), we generated pCCHFG3 plasmid expressing GFP mRNA, similar to pCCHFG2 plasmid, except the sequence of the viral mRNA 5′ UTR was randomized (Fig. 1A). HeLa cells in 12-well plates again were cotransfected with pCCHFG3 plasmid to determine whether randomization of viral mRNA 5′ UTR sequence affects the GFP signal in cells expressing CCHFV-NP (Fig. 1E). It is evident that increased NP expression (Fig. 1E1) did not support the translation of reporter mRNA harboring the randomized viral mRNA 5′ UTR (Fig. 1E). This suggests that the viral mRNA 5′ UTR is required for the preferential translation of GFP reporter mRNA by an NP-mediated translation strategy. It must be noted that both HEK and HeLa cells used in this study are permissive to CCHFV infection (22).
We next asked whether purified CCHFV-NP can facilitate the translation of reporter mRNAs in rabbit reticulocyte lysates, a commonly used in vitro translation system. CCHFV-NP was cloned in the pGX4T3 backbone and expressed as an N-terminally glutathione S-transferase (GST)-tagged fusion protein (Fig. 1F). A similar strategy was used for cloning, expression, and purification of GST tag in Escherichia coli (Fig. 1F). Using T7 RNA polymerase, we synthesized capped luciferase mRNAs containing either the 5′ UTR of CCHFV S-segment mRNA without the 3′ UTR (CCHF FL1), both the 5′ UTR and truncated 3′ UTR of S-segment mRNA (CCHF FL2), or randomized 5′ UTR and truncated 3′ UTR of S-segment mRNA (CCHF FL3) (Fig. 1G). These transcripts also contained 15 nonviral nucleotides at the 5′ terminus between the m7G cap and viral UTR, representing the cap-snatched sequence (Fig. 1G). It must be noted that the UTRs present in CCHF FL1, CCHF FL2, and CCHF FL3 transcripts were also present in transcripts expressed in cells from plasmids pCCHF G1, pCCHF G2, and pCCHF G3, respectively (Fig. 1A). We also synthesized firefly luciferase (FLuc) control RNA containing both 5′ and 3′ nonviral UTRs. These transcripts were individually translated in rabbit reticulocyte lysate in the absence or the presence of either purified GST or GST-NP fusion protein to further examine the role of NP in mRNA translation, as previously reported (12). Interestingly, the addition of purified GST-NP in the translation mix selectively facilitated the translation of luciferase mRNA containing the 5′ UTR of CCHFV S-segment mRNA (Fig. 1H to K). It is evident that GST-NP equally facilitated the translation of both CCHF FL1 and CCHF FL2 mRNAs (Fig. 1, compare panels H and I), confirming that the viral mRNA 5′ UTR alone is sufficient for NP-mediated translation. As expected, mutation of the viral mRNA 5′ UTR abrogated the NP-mediated translation of reporter luciferase mRNA (Fig. 1J). Similarly, NP did not influence the translation of FLuc control mRNA lacking both viral mRNA 5′ and 3′ UTRs (Fig. 1K). To ensure that exogenously added GST-NP did not affect the stability of luciferase mRNAs during translation, all four luciferase transcripts (Fig. 1G) were radioactively labeled with [32P]GTP during synthesis. The radiolabeled mRNAs were incubated with the translation mix containing purified GST-NP. The reaction mixtures were separated on denaturing polyacrylamide gels. We did not notice any change in the integrity of radiolabeled mRNAs (Fig. 1M), confirming that purified GST-NP did not affect the mRNA stability in the translation mix. Taken together, these results suggest that CCHFV-NP, similar to hantavirus N protein, facilitates the translation of reporter mRNA containing the viral mRNA 5′ UTR.
Identification of correct termini of CCHFV mRNA.
To generate the reporter transcripts correctly mimicking the structure of CCHFV mRNA, the 5′ and 3′ termini of CCHFV mRNA needed to be identified. The CCHFV S-segment genomic RNA contains 5′ and 3′ UTR sequences of 167 and 55 nucleotides in length. The sequences at 5′ and 3′ termini are complementary and likely undergo base pairing, and they form the panhandle structure that has been proposed to serve as a promoter for the Bunyaviridae RdRp (23) (Fig. 2A). CCHFV RdRp uses a short capped RNA primer to initiate transcription, possibly by the prime-and-realign mechanism, similar to hantaviruses (Fig. 2A). However, similar to other genera of the Bunyaviridae family, the Nairovirus transcription terminated before the 5′ UTR of vRNA was completely transcribed, generating the viral mRNA with a truncated 3′ UTR (21) (Fig. 2A). We used 5′ and 3′ rapid amplification of cDNA ends (RACE) to identify the exact sequence of the cap-snatched primer and the exact terminus of the 3′ UTR of CCHFV S-segment mRNA, as discussed in Materials and Methods. These experiments revealed that CCHFV snatched 9- to 16-nucleotide-long capped RNA primers from host cell transcripts. The primers contained a C residue at the 3′ terminus. The RdRp terminated transcription at the G residue located at the 126th position from the 5′ terminus of S-segment vRNA (Fig. 2A and B). Due to the prior termination, the 3′ UTR of S-segment mRNA was truncated by 126 nucleotides (Fig. 2A).
FIG 2.
Identification of viral mRNA termini. (A) A hypothetical panhandle structure formed by the base pairing of complementary nucleotides at the 5′ and 3′ termini of the viral genome. Shown is the CCHFV S-segment vRNA having 5′ and 3′ UTRs of 167 and 55 nucleotides in length, respectively (black dotted line). The ORF sequence is shown by a blue line. RdRp uses a capped RNA fragment (green) as a primer to initiate transcription. The primer is extended by RdRp, and the transcription is terminated before the entire 5′ terminus of the vRNA is transcribed, generating a viral mRNA with a 3′ UTR truncated by 126 nucleotides. (B) The 5′ and 3′ RACE experiments were performed as described in Materials and Methods. The 5′ RACE experiment shows the sequence of capped RNA primers (lowercase letters) used by the RdRp to initiate transcription. The 3′ RACE experiment shows the terminal sequence of the viral mRNA 3′ UTR (uppercase letters).
Further confirmation of CCHFV-NP-mediated translation mechanism using luciferase reporter with flanking S-segment UTRs.
The reporter mRNAs used in Fig. 1 contained a 148-nucleotide-long 3′ UTR of S-segment mRNA. However, the 3′ RACE experiment (Fig. 2B) revealed that the 3′ UTR of CCHFV S-segment mRNA is truncated by 126 nucleotides and is only 41 nucleotides in length. We generated additional constructs expressing luciferase reporter mRNA harboring the 55- and 41-nucleotide-long 5′ and 3′ UTR sequences of CCHFV S-segment mRNA. In addition, the 14-nucleotide-long cap-snatched sequence, verified by 5′ RACE (Fig. 2B), was incorporated upstream of the 5′ UTR to correctly mimic the cap-snatched sequence of viral mRNA. The impact of CCHFV-NP upon the translation of these newly designed transcripts was examined in cell culture. Briefly, cells were cotransfected with increased input concentrations of pCCHFV-NP plasmid expressing CCHFV-NP along with a fixed concentration of either pcDLuc1, pCDLuc2, pCDLuc3, pCDLuc4, or pGL3 control plasmid (Fig. 3). The transcripts expressed by these constructs are shown in Fig. 3. Cells were lysed 36 h posttransfection, and resulting cell lysates were examined for luciferase and NP expression. An examination by Western blot analysis revealed that increased input concentrations of pCCHFV-NP plasmid in cotransfection experiments yielded increased endogenous levels of NP in cells (Fig. 3A1 and B1). It is evident from Fig. 3A that luciferase mRNA, having a viral mRNA 5′ UTR and a nonviral 3′ UTR along with a poly(A) tail expressed from pCDLuc1 plasmid, was not favored for translation by CCHFV-NP. Similar results were obtained in cells cotransfected with control PGL3 plasmid. However, the deletion of the 3′ nonviral UTR along with a poly(A) tail by the engineered HDV ribozyme sequence selectively favored the translation of the resulting mRNA, expressed from pCDLuc2 plasmid, by the CCHFV-NP (Fig. 3A). This observation suggests that either the 3′ nonviral UTR or the 3′ poly(A) tail prevented the preferential translation of the reporter transcript by the CCHFV-NP-mediated translation strategy by an unknown mechanism.
FIG 3.
Further confirmation of NP-mediated translation strategy using firefly luciferase reporter. The plasmids pCDLuc1, pCDLuc2, pCDLuc3, and pCDLuc4 were constructed as mentioned in Materials and Methods. The mRNA molecules expressed from these constructs under the CMV promoter are shown. The viral and scrambled 5′ UTR sequences present in these transcripts are shown in panel L of Fig. 1. The 14-nucleotide nonviral sequence identified by the 5′ RACE shown in Fig. 2 (CUUCGUUCUCAGAAC) was incorporated upstream of the 5′ UTR. This sequence represents the cap-snatched primer. The truncated 3′ UTR sequence of the CCHFV S-segment mRNA, identified by 3′ RACE (Fig. 2), was incorporated preceding the HDV sequence. (A) HeLa cells in 12-well plates were cotransfected with a fixed amount (100 ng) of pGL3 (blue), pCDLuc1 (red), or pCDLuc2 (green), along with increasing input concentrations of pCCHFV-NP. Cells were lysed 36 h posttransfection, and lysates were examined for luciferase expression, as mentioned in Materials and Methods. Luciferase signal was normalized to the mock control. Experiments were repeated three times for the calculation of standard deviations, which are shown by error bars. (B) HeLa cells were cotransfected with pCDLuc3 or pCDLuc4 along with pCCHFV-NP, followed by lysis and examination of luciferase activity, as shown in panel A. (A1 and B1) Western blot analysis showing the expression levels of CCHFNV NP and β actin in cell lysates from panels A and B, respectively.
The luciferase reporter mRNA expressed from pCDLuc3 plasmid structurally mimics the CCHFV S-segment mRNA (Fig. 3B). It has both 5′ and 3′ viral mRNA UTR sequences of the correct size and 14-nucleotide-long nonviral sequence upstream of the 5′ UTR mimicking the cap-snatched sequence. This transcript was preferentially translated in cells by the NP-mediated translation strategy (Fig. 3B). However, the randomization of 5′ UTR sequence (pCDLuc4 plasmid) abolished the preferential translation of the reporter mRNA. Taken together, the results from Fig. 3A and B clearly demonstrate that the 5′ UTR of CCHFV S-segment mRNA is sufficient for the preferential translation of the downstream open reading frame by the NP-mediated translation strategy.
A mixture of cells receiving NP expression plasmid, reporter plasmid, or both were analyzed in the experiments depicted in Fig. 1 and 3. It will be more accurate to specifically examine only those cells receiving both the NP expression construct and the reporter plasmid of interest to precisely delineate the impact of NP upon the translation of the desired reporter mRNA. We generated another NP expression construct (pNP-IRES-mCherry) (Fig. 4F) expressing NP by cap-dependent translation and mCherry reporter from the internal ribosomal entry site (IRES). Thus, all mCherry-positive cells contain the CCHFV-NP. HeLa cells were cotransfected with the reporter construct of interest (pCCHFG2 or pCCHFG3) along with either pNP-IRES-mCherry or pmCherry control plasmid. As shown in Fig. 4F, pCCHFG2 expresses the GFP reporter mRNA containing 5′ and 3′ viral UTR devoid of 3′ poly(A) tail. In comparison, pCCHFG3 plasmid expresses the same mRNA except the 5′ UTR sequence was randomized. The cotransfected cells were examined by flow cytometry, and GFP signal in cells positive for both GFP and mCherry is quantified and plotted in Fig. 4E and K. It is evident that NP selectively favored the translation of GFP reporter mRNA expressed from pCCHFG2 plasmid in cells expressing both the GFP reporter and NP (Fig. 4E). Interestingly, the fold change in GFP signal due to NP expression was similar to that seen in Fig. 1 using a mixture of cells. Again, the randomization of 5′ viral UTR sequence abolished the preferential translation of the reporter mRNA by the NP-mediated translation strategy (Fig. 4K). These studies clearly demonstrate that NP, with the assistance of viral mRNA 5′ UTR, favors the translation of the downstream ORF.
FIG 4.
Examination by flow cytometry to further confirm the NP-mediated translation of reporter mRNA. HeLa cells in 24-well plates were transfected with pCCHFG2 plasmid (B), cotransfected with pCCHFG2 and pmCherry plasmids (C), or cotransfected with pCCHFG2 and pNP-IRES-mCherry plasmids (D). (A) Mock-transfected cells. Cells were examined by flow cytometry 24 h posttransfection. Mean GFP signal from GFP-positive cells in quadrant III (B) and both GFP- and RFP-positive cells from quadrant IV (C and D) was calculated. The mean GFP fluorescence values from panels A, B, C, and D were normalized relative to values for panel C and are plotted in panel E. (F) mRNA molecules expressed by plasmids pCCHFG2, pmCherry, pNP-IRES-mCherry, and pCCHFG3. See Materials and Methods for details about the construction of these plasmids. Cells in panels G, H, I, and J were transfected and examined similarly to cells shown in panels A, B, C, and D, except the pCCHFG2 plasmid was replaced by the pCCHFG3 plasmid. (K) The quantified GFP signal was analyzed and plotted as described for panel E.
CCHFV-NP-mediated translation mechanism is cap independent.
Since both cellular and viral transcripts contain a 5′ m7G cap, we next wanted to determine whether CCHFV-NP preferentially facilitates the translation of capped viral mRNAs. Using an in vitro T7 transcription system, we synthesized the luciferase mRNA flanked by the 5′ and 3′ UTR sequences of CCHFV S-segment mRNA. The mRNA was the same in sequence as the mRNA produced by the pCDLuc3 plasmid shown in Fig. 3B. The mRNA was either uncapped or capped during synthesis. Both uncapped and capped mRNA were translated in rabbit reticulocyte lysates in the absence or presence of purified CCHFV-NP. The lysates were examined for luciferase expression to determine the impact of CCHFV-NP upon the translation of luciferase reporter mRNA. It is evident from Fig. 5 that CCHFV-NP equally facilitated the translation of both capped and uncapped transcripts, demonstrating that the mRNA 5′ cap does not play a role in the CCHFV-NP-mediated translation strategy. This observation is consistent with the previously reported finding that CCHFV-NP does not bind to the mRNA 5′ cap (14–16).
FIG 5.
Translation in rabbit reticulocyte lysate. The luciferase reporter mRNA containing the CCHFV S-segment mRNA 5′ UTR and truncated 3′ UTR, along with an additional 14 nonviral nucleotides upstream of the 5′ UTR, was synthesized by T7 transcription reaction. The mRNA was either uncapped or capped during synthesis. One microgram of the reporter mRNA was translated in rabbit reticulocyte lysates in the absence or presence of purified CCHFV-NP (100 ng or 300 ng), as mentioned in Materials and Methods. The luciferase signal was quantified and normalized to the RNA control.
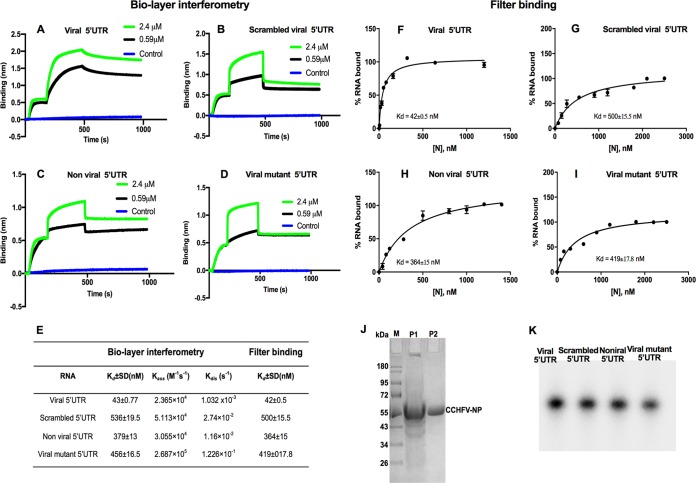
CCHFV-NP binds to viral mRNA 5′ UTR with high affinity.
Since CCHFV-NP facilitated the translation of reporter mRNAs harboring the viral mRNA 5′ UTR, we asked whether binding of NP to the viral mRNA 5′ UTR is required to facilitate the translation of the downstream open reading frame. Both the CCHFV S-segment mRNA 5′ UTR and its scrambled counterpart that abrogates the NP-mediated translation strategy were synthesized by the in vitro T7 transcription and biotinylated during synthesis, as previously reported (24). As shown in Fig. 1L, the scrambled 5′ UTR has the same nucleotide composition as the viral mRNA 5′ UTR, but the primary sequences of the two UTRs are different. We used biolayer interferometry to quantitatively examine the binding of CCHFV-NP with the UTR sequences, as described in Materials and Methods. As shown in Fig. 6, scrambling the UTR sequence affects both the forward and reverse kinetics for binding to CCHFV-NP. CCHFV-NP binds to the viral UTR with 12-fold stronger affinity than scrambled UTR. The tight binding of CCHFV-NP with the viral UTR is consistent with favorable translation of reporter mRNA harboring the viral UTR. In comparison, the randomization of viral UTR sequence abrogated the translation of reporter mRNAs by the NP-mediated translation strategy (Fig. 1, 3, and 4) and also the binding of CCHFV-NP with the scrambled UTR (6B). Taken together, these results suggest that high-affinity binding of CCHFV-NP to the viral mRNA 5′ UTR favors the translation of the downstream open reading frame of the reporter mRNA.
FIG 6.
Binding analysis of purified CCHFV-NP with RNA molecules using biolayer interferometry (BLI). The CCHFV S-segment mRNA 5′ UTR (A), scrambled 5′ UTR (B), nonviral 5′ UTR (C), and viral mutant 5′ UTR (D) were synthesized by T7 transcription and biotinylated during synthesis, as described in Materials and Methods. The biotinylated RNA was immobilized on a streptavidin biosensor. The purified N-terminally His-tagged CCHFV-NP was passed over the immobilized RNA molecule at the indicated concentrations. (E) The data points shown in panels A to D were fit to a 1:1 binding model for the calculation of association (Kass) and dissociation (Kdis) rate constants. The binding affinity (Kd = Kdis/Kass) was calculated from the kinetic data. The standard deviations (SD) were calculated from three independent experiments. The experiments shown in panels A, B, C, and D were verified by filter binding assay shown in panels F, G, H, and I, respectively. The dissociation constants (Kd) reported in panels F to I were calculated as mentioned in Materials and Methods. (J) A 12% SDS-PAGE gel showing purified His-tagged CCHFV-NP (lane P2). The molecular mass marker and a partially purified NP fraction are shown in lanes M and P1, respectively. (K) A 12% acrylamide urea gel showing four radiolabeled UTR molecules that were used in RNA-protein binding assays.
To further characterize the RNA binding of CCHFV-NP, we synthesized the nonviral 5′ UTR (Fig. 1L), whose sequence of nonviral origin did not support the translation of reporter mRNA, with the assistance of CCHFV-NP. In addition, we incorporated mutations that changed both the nucleotide composition and the sequence of the viral mRNA 5′ UTR. The resulting mutant (viral mutant 5′ UTR; Fig. 1L) was also synthesized by T7 transcription. The UTRs were biotinylated during synthesis, and their interaction with CCHFV-NP was studied using biolayer interferometry. It is evident from Fig. 6C and D that, similar to the scrambled UTR, NP bound to both of these UTRs with weaker affinity.
To further confirm the results shown in Fig. 6A to D, we studied the interaction of CCHFV-NP with the radiolabeled UTR sequences by filter binding assay, as mentioned in Materials and Methods. To ensure the integrity of UTR preparations used in these experiments, the purified radiolabeled UTR sequences, synthesized by T7 transcription, were separated on 12% acrylamide urea gel. As shown in Fig. 6K, the UTR sequences were intact without any noticeable degradation. The filter binding analysis demonstrated that CCHFV-NP bound to the viral mRNA 5′ UTR with higher affinity than scrambled UTR, nonviral UTR, or the viral mutant 5′ UTR (Fig. 6F to I). The results from filter binding analysis are consistent with the results from biolayer interferometry (Fig. 6E). These observations collectively demonstrate that high-affinity binding of CCHFV-NP with the viral mRNA 5′ UTR likely is required for the preferential translation of the downstream open reading frame in the reporter mRNA. The mFold analysis revealed that the viral 5′ UTR has a strong hairpin secondary structure. The remaining RNAs used in binding analysis shown in Fig. 6 have weak secondary structures that likely do not exist at room temperature. These observations suggest that CCHFV-NP prefers structured RNAs for high-affinity binding.
CCHFV-NP does not require eIF4F complex to facilitate mRNA translation.
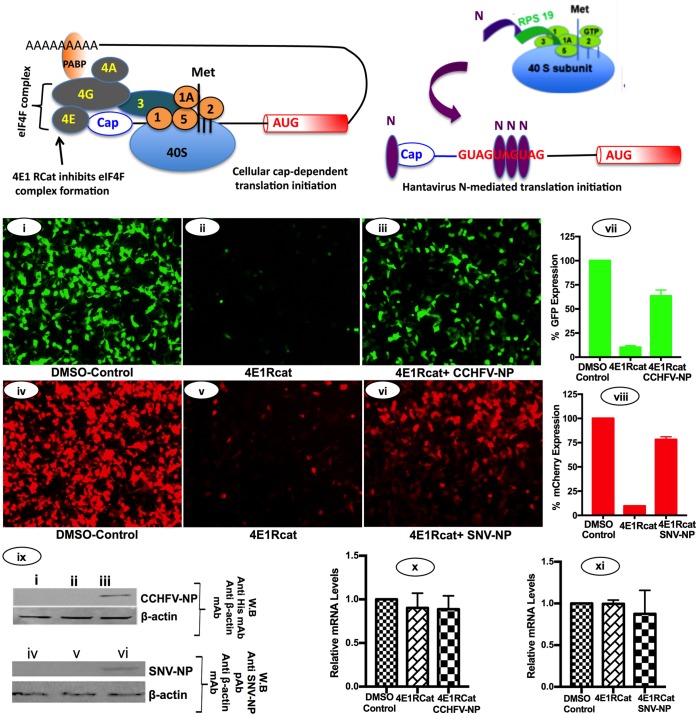
The canonical host translation machinery requires the cap binding eIF4F complex, composed of eIF4E, eIF4G, and eIF4A, to initiate translation on capped mRNAs. The eIF4F complex recruits 43S preinitiation ribosome complex by direct interaction between eIF4G and eIF3, a component of the 43S preinitiation ribosome complex (Fig. 7). We previously reported that hantavirus N protein facilitates mRNA translation without the requirement of eIF4F complex. N protein binds to the ribosomal protein S19 (RPS19), a structural component of the 40S ribosomal subunit (19). In addition, monomeric and trimeric N protein molecules bind to the mRNA 5′ cap and highly conserved triplet repeat sequence (UAGUAGUAG) of the viral mRNA 5′ UTR, respectively (20). N protein molecules individually associated with the 43S preinitiation complex and the mRNA 5′ cap likely undergo protein-protein interaction and load the ribosome on the mRNA 5′ cap without the assistance of eIF4F complex. Thus, N protein facilitates the translation of any capped mRNA. However, the mRNA molecules harboring the viral mRNA 5′ UTR are preferentially translated due to high-affinity binding of trimeric N protein molecules with the triplet repeat sequence of the viral UTR (Fig. 7). N protein-associated ribosomes are internally loaded on the viral mRNA 5′ UTR irrespective of the 5′ cap and thus are preferentially translated by the N protein-mediated translation strategy, as shown in the model (Fig. 7).
FIG 7.
Translation shutdown using 4E1RCat. A model showing the canonical translation machinery of the host cell. Some initiation factors are omitted for simplicity. The inhibitor 4E1RCat inhibits the interaction between eIF4G and eIF4E. The hantavirus N-mediated translation mechanism does not require eIF4F complex, as mentioned in the text. Binding of hantavirus N protein (N) to the ribosomal protein RSP19 (green), mRNA 5′ cap (blue), and the triplet repeat sequence at the 5′ UTR of hantaviral mRNA (red) is shown. HEK293T cells seeded in 24-well plate were transfected with pTriEx1.1 empty vector (i, ii, iv, and v), pCCHFV-NP (iii), or pTriEx-SNVN (vi). Twenty hours posttransfection, cells were again transfected with either pCCHFG2 plasmid (i to iii) or pTriEX-mCherry plasmid, expressing mCherry reporter mRNA flanked with 5′ and 3′ UTR sequences of Sin Nombre hantavirus S-segment mRNA, as previously reported (25). Six hours after the second transfection, medium was replaced with fresh medium containing either DMSO (1%) or 4E1RCat (30 μM). Cells were washed with PBS for 14 to 16 h after the treatment with 4E1RCat, and reporter expression was visualized under the fluorescence microscope and analyzed by fluorescence-activated cell sorting (FACS). The percentage of reporter expression was calculated from the FACS data and plotted in panels vii and viii. Error bars represent the standard deviations from three independent experiments. (ix) Western blot analysis of CCHFV-NP, SNVN-N protein, and β-actin in samples obtained from panels i to vii. CCHFV-NP was detected using anti-His tag monoclonal antibody, and SNV-N protein was detected using anti-N protein polyclonal antibody. (x and xi) Reporter mRNA levels in cells from panels i, ii, and iii (x) and iv, v, and vi (xi). The reporter mRNA levels were quantified by real-time PCR (relative quantification method).
We next wanted to determine whether CCHFV-NP uses a similar strategy to engage ribosomes on the viral mRNA 5′ UTR. HEK293T cells were cotransfected with pCCHFG2 plasmid expressing GFP reporter mRNA harboring the CCHFV S-segment mRNA 5′ and 3′ UTR sequences along with pCCHFV-NP plasmid expressing CCHFV-NP (Fig. 7i to iii). Note that this GFP reporter mRNA is preferentially translated by CCHFV-NP, as shown in Fig. 1 and 4. In a control experiment, HEK293T cells were cotransfected with pTriEx-mCherry plasmid expressing mCherry reporter mRNA harboring the 5′ and 3′ UTR sequences of Sin Nombre hantavirus S-segment mRNA along with pTriEx-SNVN plasmid expressing the SNV N protein. The reporter mRNA from pTriEx-mCherry plasmid is also preferentially translated by the SNV N protein (25). Twenty-six hours posttransfection cells were treated with 4E1RCat, a known chemical inhibitor of eIF4F complex. 4E1RCat binds to eIF4E, disrupts the interactions between eIF4E and eIF4G, and prevents the formation of eIF4F complex on the mRNA 5′ cap (26). As a result, the cap-dependent translation machinery of the host cell is shut down. The microscopic examination of cells 24 h after 4E1RCat treatment revealed that translational shutdown inhibited the translation of GFP and mCherry reporter mRNAs in cells (Fig. 7ii and v). The treatment of 4E1RCat failed to inhibit the translation of mCherry reporter mRNA in SNV N protein-expressing cells (Fig. 7vi and viii), further confirming the previously reported observations that N protein does not require eIF4F complex to facilitate mRNA translation. Interestingly, 4E1RCat also failed to inhibit the translation of GFP reporter mRNA in CCHFV-NP-expressing cells (Fig. 7iii and vii), suggesting that similar to hantaviruses, CCHFV-NP does not require eIF4F complex to facilitate the translation of GFP reporter mRNA. We did not observe a noticeable change in the reporter mRNA levels, examined by real-time PCR analysis (Fig. 7x and xi), demonstrating that changes in reporter expression occurred at the translational level. Since the translation of both SNV N protein and CCHFV-NP is dependent upon the canonical translation machinery, their translation would also be shut down by treatment with 4E1RCat inhibitor. To ensure that endogenous levels of both of these proteins generated in a time period of 26 h prior to 4E1RCat treatment likely supported the translation of reporter mRNAs, we carried out Western blot analysis to examine the endogenous levels of both SNV-N protein and CCHFV-NP in cells. Figure 7ix clearly demonstrates that both of these proteins existed in cells 24 h after 4E1RCat treatment.
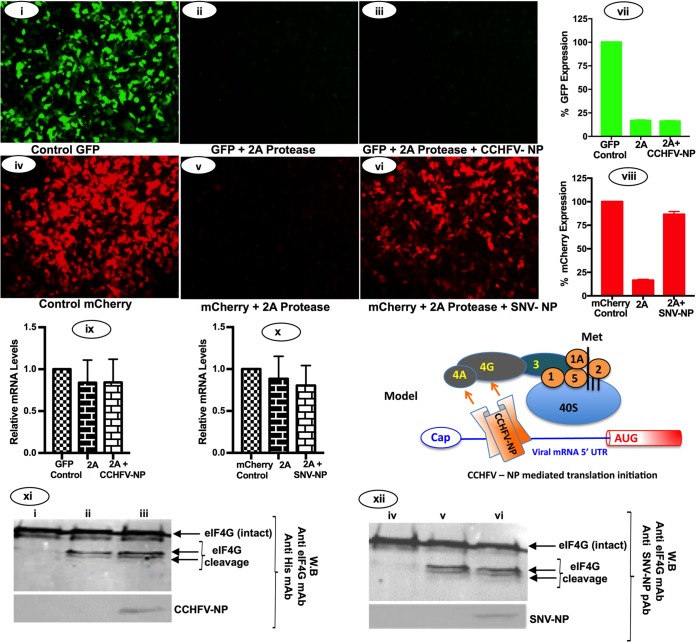
CCHFV-NP requires eIF4G to facilitate mRNA translation.
Although the treatment of 4E1RCat prevents the formation of eIF4F complex at the mRNA 5′ cap, the structural components of eIF4F complex (eIF4E, eIF4G, and eIF4A) physically exist in 4E1RCat-treated cells. We next used human rhinovirus-16 2A protease to inhibit the formation of eIF4F complex and monitor the impact of host translation shutdown in the CCHFV-NP-mediated translation strategy. The 2A protease specifically cleaves eIF4G and inhibits the cap-dependent canonical host translation machinery (27–29). It plays important roles in virus replication (30–32). HEK293T cells were cotransfected with GFP and mCherry reporter constructs, along with plasmids expressing either CCHFV-NP or hantavirus N protein (Fig. 8). The experiment was performed exactly as described for Fig. 7, except the formation of eIF4F complex was inhibited by the enzymatic degradation of eIF4G using human rhinovirus-16 2A protease and not 4E1RCat (Fig. 8). As shown in Fig. 8xi and xii, the expression of human rhinovirus-16 2A protease cleaved the eIF4G in cells. Consistent with our previous observations (12), the translation shutdown by the enzymatic degradation of eIF4G was rescued by hantavirus N protein expression in cotransfected cells (compare Fig. 8v to vi). However, it is evident from Fig. 8 that unlike hantavirus N protein, the translation shutdown of GFP reporter mRNA due to the expression of 2A protease was not rescued by CCHFV-NP (compare Fig. 8ii to 8iii, as well as 8vii to iii), demonstrating that CCHFV-NP requires the intact eIF4G to facilitate mRNA translation by an unknown mechanism. To ensure that changes in the GFP and mCherry signal observed in Fig. 8 were not due to alterations in the endogenous reporter mRNA levels, we quantified the GFP (Fig. 8ix) and mCherry (Fig. 8x) mRNA levels by real-time PCR. The lack of noticeable change in the reporter mRNA levels demonstrates that changes observed in reporter expression occurred at the translational level. Although NP expression plasmids were transfected in cells 24 h before the second transfection with 2A protease construct was performed, we carried out Western blot analysis to ensure that 2A protease expression did not dramatically impact the endogenous NP levels in cells. We observed that both CCHFV-NP and SNV N protein existed in cells 24 h after the second transfection with a plasmid expressing the 2A protease. Taken together, the results from Fig. 8 clearly demonstrate that although the formation of eIF4F complex is not required for the CCHFV-NP-mediated translation strategy, the structural integrity of eIF4G plays a critical role in this translation mechanism.
FIG 8.
Translation shutdown using human rhinovirus 2A protease. HEK293T cells seeded in 24-well plates were transfected with empty vector (i, ii, iv, and v), pCCHFV-NP (iii), or pTriEx-SNVN (vi). Twenty-four hours posttransfection, cells were again transfected with either pCCHFG2 (i) or pCCHFG2 along with pF/HRV-16 2A expressing human rhinovirus 16 2A protease (ii and iii), pTriEX-mCherry (iv), or pTriEX-mCherry along with pF/HRV-16 2A (v and vi). Reporter gene expression was observed under the microscope 24 h after the second transfection. (vii and viii) GFP (vii) and mCherry (viii) signals were quantified by FACS and plotted. (ix and x) GFP mRNA levels in cells from panels i, ii, and iii (ix) and mCherry mRNA levels in cells from panels iv, v, and vi (x) were quantified by real-time PCR. Cell lysates from panels i to vi were examined by Western blot analysis for the expression of eIF4G and NP. (xi and xii) The corresponding Western blots from panels i to iii (xi) and iv to vi (xii). It is evident from panels xi and xii that expression of 2A protease resulted in the cleavage of eIF4G. The cleavage products are shown by an arrow. A hypothetical model, showing that NP bound to the viral mRNA 5′ UTR may also interact with either eIF4A or eIF4G to engage the ribosome on the viral mRNA 5′ UTR, is shown on the right side of panel x.
DISCUSSION
The rapid elevation of viral load in infected hosts is linked at the molecular level to the rapid multiplication of viral genome and efficient synthesis of viral proteins in the host cell. It is well understood that viruses have evolved selfish strategies to favor the translation of viral mRNAs in the host cell cytoplasm, where cellular transcripts are competing for the same translation machinery. For example, suspension of cap-dependent translation initiation by picornaviruses and degradation of cellular mRNAs by herpes simplex virus are well-known viral mechanisms that eliminate the competition of host cell transcripts for the same translation apparatus (33). Similarly, the nsp1 protein of severe acute respiratory syndrome coronavirus suppresses host gene expression by promoting both the degradation of host cell mRNA and inactivating the translational activity of the 40S ribosomal subunit (18). However, it is interesting to understand how negative-strand RNA viruses such as CCHFV or hantaviruses, whose infection does not induce a translation shutoff or cytopathic effects to the host cell (6), manage the efficient synthesis of viral proteins during the course of infection. Our results suggest that these viruses use their nucleocapsid proteins to preferentially engage the host translation machinery at the viral mRNA 5′ UTR for efficient synthesis of viral proteins.
To this end, our results demonstrate that reporter GFP mRNAs harboring the viral mRNA 5′ UTR, irrespective of 3′ poly(A) tail, were efficiently translated in cells coexpressing the CCHFV-NP (Fig. 1A to E). A similar moderate increase in the translation of GFP reporter transcripts by purified CCHFV-NP was also observed in the rabbit reticulocyte lysates. The fusion of truncated S-segment 3′ UTR (147 nucleotides) downstream of the GFP reporter did not impact the translation of the reporter mRNA, demonstrating that 3′ UTR sequence does not play a role in the NP-mediated translation strategy. Interestingly, the preferential translation of GFP reporter mRNA was abrogated when the 5′ UTR sequence was scrambled, confirming that the 5′ UTR sequence was required for the NP-mediated translation strategy (Fig. 1). To generate a luciferase reporter mRNA structurally mimicking the viral S-segment mRNA, 5′ and 3′ RACE were used to map the exact sequence of the 5′ and 3′ UTR of the CCHFV S-segment mRNA (Fig. 2). The RACE experiment revealed that CCHFV RdRp uses 9- to 16-nucleotide-long capped mRNA primers to initiate transcription. Interestingly, due to the truncation of 146 nucleotides, the 3′ UTR of CCHFV S-segment mRNA was 41 nucleotides in length. The mechanism for transcription termination at the 146th G residue from the 5′ terminus of vRNA template remains unclear (Fig. 2).
Identification of correct 5′ and 3′ termini of CCHFV S-segment mRNA (Fig. 2) leads to the design of a construct expressing luciferase reporter mRNA structurally mimicking the CCHFV S-segment mRNA (Fig. 3). This reporter mRNA contained all structural features of S-segment mRNA, except the NP ORF was replaced by the luciferase ORF. The reporter transcript was preferentially translated in cells coexpressing CCHFV-NP from a transfected plasmid. Again, unlike the case for the 3′ UTR, the mutations in the 5′ UTR impacted the preferential translation of the reporter transcript. An examination of selected cells coexpressing both CCHFV-NP and reporter mRNA harboring the viral mRNA 5′ UTR further confirmed that NP, with the assistance of the viral 5′ UTR, favors the translation of downstream ORF (Fig. 4). Interestingly, despite having the viral mRNA 5′ UTR, a luciferase reporter mRNA having a nonviral 3′ UTR and a poly(A) tail was not favored for translation by the CCHFV-NP (Fig. 3A). Poly(A)-tailed mRNAs are circularized during translation by direct contact between eIF4G and poly(A) binding protein (PABP) (34). However, the lack of 3′ poly(A) tail in CCHFV mRNA likely prevents such circularization. It is possible that CCHFV-NP does not favor the translation of circularized mRNAs to avoid competition from the host cell transcripts for the CCHFV-NP-mediated translation strategy. As a result, CCHFV-NP might selectively boost the translation of viral transcripts in virus-infected cells.
The observation that CCHFV-NP equally favors the translation of both capped and uncapped mRNAs is consistent with previous reports that CCHFV-NP does not bind to the mRNA 5′ cap (16). However, the high-affinity binding between CCHFV-NP and the viral mRNA 5′ UTR (Fig. 6) is required for the NP-mediated translation strategy. This is supported by the observation that abrogation of NP-UTR interaction by the randomization of viral UTR sequence impacted the NP-mediated translation strategy (Fig. 1 to 3).
Shutdown of host cap-dependent translation machinery by 4E1RCat, a known inhibitor of eIF4F complex formation (26), had no impact on NP-mediated translation strategy (Fig. 7), confirming that the NP-mediated translation strategy is cap independent and does not require eIF4F cap binding complex to initiate translation. Interestingly, the inhibition of eIF4F complex formation by the enzymatic degradation of eIF4G using human rhinovirus-16 2A protease (27–29) specifically impacted the NP-mediated translation strategy (Fig. 8). Consistent with our previous reports (12), the enzymatic degradation of eIF4G had no impact on the hantavirus N protein-mediated translation mechanism. This observation clearly demonstrates that CCHFV-NP requires eIF4G for translation. Our model for the hantavirus N protein-mediated translation mechanism (Fig. 7) depicts that monomeric and trimeric N protein molecules individually associated with ribosomal protein S19 (RPS19) and viral mRNA 5′ UTR, respectively, undergo protein-protein interaction and internally load the N protein-associated ribosomes onto the viral mRNA 5′ UTR (Fig. 7). However, it remains unclear how CCHFV-NP engages ribosomes on the viral mRNA 5′ UTR. Based on our observations in Fig. 7 and 8, we propose a hypothetical model that NP simultaneously binds to both the viral mRNA 5′ UTR and eIF4G or eIF4A to engage ribosome internally on the viral mRNA 5′ UTR (Fig. 8).
MATERIALS AND METHODS
Cells and transfection.
HeLa and HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and penicillin-streptomycin (100 μg/ml) in a CO2 incubator. All plasmid DNA transfections were performed using TurboFect transfection reagent (Thermo Fisher Scientific) according to the manufacturer's instructions.
Plasmid construction.
Straightforward cloning techniques were used to generate the constructs used in this study. Briefly, PCR was used to fuse the DNA sequences encoding the 5′ UTR of CCHFV S-segment mRNA and hepatitis delta virus ribozyme (HDV) upstream of the AUG codon and downstream of the 3′ terminus of the GFP ORF, respectively. The resulting PCR product was cloned 15 nucleotides downstream of the transcription initiation site in the pCDNA 3.1(+) backbone between SacI and XhoI restriction sites to generate the pCCHFG1 construct used for Fig. 1. The same strategy was used for the construction of pCCHF G2 and pCCHF G3 constructs, with both having the truncated 3′ UTR (147 nucleotides) of CCHFV S-segment mRNA. For the construction of pCDGFP plasmid, the GFP ORF was cloned between NheI and XhoI restriction sites in the pCDNA 3.1(+) backbone. The pCCHF-NP plasmid was generated by subcloning the NP ORF in the pTriEX1.1 backbone between PacI and XhoI restriction sites. The cloning strategy used for the construction of pCCHFG1 plasmid was also used for the construction of plasmids pCDLuc1, pCDLuc2, pCDLuc3, and pCDLuc4, except the GFP ORF was replaced by the firefly luciferase (FLuc) ORF. The pGL3 plasmid was from Promega. The plasmid pmCherry has an IRES sequence upstream of the mCherry ORF and is expressed under the cytomegalovirus (CMV) promoter. This plasmid was a gift from Jainming Que (Kansas University Medical Center). The plasmid pNP-IRES-mCherry was constructed by the incorporation of CCHFV-NP ORF upstream of the IRES sequence in the pmCherry plasmid. The mRNA expressed from the pNP-IRES-mCherry plasmid under the CMV promoter is translated by both cap-dependent and IRES-mediated translation strategies, generating CCHFV-NP and mCherry, respectively. The plasmids pTriEx-SNV and pTriEx-mCherry were constructed as previously reported (25). The plasmid pF/HRV-16 2A was generated as previously reported (12).
Cloning, expression, and purification of CCHFV-NP.
The plasmid expressing CCHFV-NP from strain 10200 was a gift from Stuart T. Nichol (CDC, Atlanta, GA). The NP ORF was PCR amplified and cloned between NdeI and XhoI restriction sites in the pET30a backbone. The resulting plasmid (pET-CCHFNP) expressing CCHFV-NP as an N-terminally His-tagged fusion protein was transformed into Escherichia coli Rosetta (DE3) cells (Novagen). The bacterial cultures were induced by 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when the optical density at 600 nm reached 0.6. The culture was grown for an additional 20 h at 16°C. Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM dithiothreitol [DTT], 1% Triton X-100, 5 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate, 0.1 mM phenyl methyl sulfonyl fluoride [PMSF]). The cleared cell lysate was loaded onto a nickel-nitrilotriacetic acid column (Gold Bio) preequilibrated with the lysis buffer. After two washes with lysis buffer, the column was washed once with wash buffer (50 mM Tris, 500 mM NaCl, 0.05% Triton X-100, and 30 mM imidazole). The bound NP was eluted with the elution buffer (50 mM Tris, 500 mM NaCl, 0.05% Triton X-100, and 250 mM imidazole). The eluted protein was quantified and stored in 100-μl aliquots at −80°C.
For the expression of GST-tagged CCHFV-NP in bacteria, the CCHFV-NP gene was PCR amplified and inserted between BamHI and XhoI restriction sites in the pGX4T3 vector backbone. The resulting pGST-NP plasmid expressing NP with N-terminally fused GST tag was transformed into bacteria. The expression and purification of GST-tagged NP was carried out as previously reported (12). The same strategy was used for the expression of GST tag.
5′ RACE.
5′ UTR sequence of the CCHFV S segment was determined by 5′ RACE, performed using a 5′ RACE kit (catalog no. 03353621001; Roche Applied Science) by following the manufacturer's instructions. Total RNA was purified from the inactivated CCHFV-infected Vero E6 cell lysate, acquired from the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA). 5′ RACE was performed as previously reported (35).
3′ RACE.
3′ RACE was performed using a 3′ RNA/DNA anchoring method. First, total RNA was extracted as in 5′ RACE. A single-strand DNA linker then was polyadenylated and phosphorylated at the 5′ terminus using a 5′ DNA adenylation kit (NEB) and ligated to the 3′ end of the extracted RNA using T4 RNA ligase 2, truncated (NEB). The linked RNA was converted to cDNA using an anchoring primer complementary to the DNA linker sequence. The cDNA then was used to generate a PCR product with the forward primer complementary to the CCHF sequence and the anchoring primer as the reverse primer. The resulting PCR product was then used as a template for a second PCR using a CCHF-specific forward primer further downstream of the first forward primer and the same anchoring reverse primer. The resulting PCR product then was cloned using a TA cloning kit, and the plasmid was sequenced to read the 3′ UTR of the CCHFV S-segment mRNA.
Luciferase assays.
HeLa cells were cotransfected with a luciferase reporter construct of interest along with pCCHFV-NP plasmid, as mentioned in Fig. 3. Cells from each well were lysed with 200 μl of lysis buffer 36 h posttransfection. The resulting lysates were centrifuged at 12,000 × g for 2 min at 4°C, and supernatant was assayed for luciferase activity using a luciferase reagent (Promega) by following the manufacturer's protocol.
Flow cytometry.
Flow cytometry was used for quantitative examination of GFP expression in cells coexpressing the GFP reporter transcript of interest and CCHFV-NP. Briefly, HeLa cells seeded in 24-well plates were transfected with pTriEx1.1 empty vector (mock), pCCHFG2, or pCCHFG3, along with either the pmCherry or pNP-IRES-mChery construct. Cells were harvested 48 h posttransfection and analyzed on a flow cytometer (BD Accuri C6 flow cytometer; BD Biosciences, USA). Briefly, 50,000 events for each sample were examined for GFP and mCherry expression. The mean GFP fluorescence value from cells positive for both mCherry and GFP expression was examined to delineate the impact of CCHFV-NP upon the translation of the GFP reporter transcript of interest. Mean GFP fluorescence signal from each sample was normalized to the respective control for the calculation of fold change in GFP expression. The experiments were repeated independently three times for the calculation of standard deviations.
Biolayer interferometry.
Biolayer interferometry was carried out in a FortéBio BLItz instrument to monitor the interaction of CCHFV-NP with the CCHFV S-segment mRNA 5′ UTR, scrambled viral 5′ UTR, nonviral 5′ UTR, and viral mutant 5′ UTR. Briefly, the UTR sequences were synthesized in vitro by T7 RNA polymerase. The RNA was biotinylated during synthesis by the addition of biotinylated CTP (04739205001; Roche) to the reaction mixture. The biotinylated RNA was purified using TRIzol reagent (ThermoFisher). The RNA was immobilized on a high-precision streptavidin biosensor (18-5117; ForteBio). All reactions were carried out in buffer containing 20 mM Tris-HCl (pH 7.4), 300 mM NaCl, 20 mM KCl, and 1 mM DTT at room temperature. The reaction cycles were comprised of 30 s of initial baseline, 60 s of UTR binding onto the probe, 30 s of baseline, 300 s of association of CCHFV-NP at different concentrations (0.59 μM and 2.4 μM), and then a 500-s dissociation phase. The association and dissociation phases of each reaction cycle were analyzed by fitting the data into a 1:1 binding model using BLItz software. The CCHFV-NP used in these experiments was bacterially expressed and purified using an N-terminal His tag.
Filter binding analysis.
The UTR molecules of interest were synthesized by T7 transcription and labeled with [α32-P]GTP during synthesis, as previously reported (24). The dissociation constant (Kd) was obtained by GraphPad Prism 7.0 software using nonlinear regression analysis based on a single-site binding model.
In vitro translation.
The capped and uncapped luciferase mRNA molecules were synthesized by in vitro T7 transcription, and the translation of the resulting mRNA was carried out in rabbit reticulocyte lysates in the presence or absence of purified CCHFV-NP, as previously reported (12).
Real-time PCR.
Cells were lysed and total RNA was purified by TRIzol reagent (Invitrogen). The reporter mRNA levels (GFP and mCherry) were quantified by real-time PCR using a relative quantification method, as previously reported (25).
ACKNOWLEDGMENTS
We acknowledge Stuart Nichole and Eric Bergeron from the CDC for providing the CCHFV-NP expression construct.
Figures 1 and 2 and Fig. S1 in the supplemental material were provided by E.C., and all remaining figures were provided by S.J. E.C. designed and generated most of the plasmids used in this work, but the plasmid pNP-IRES-mCherry was generated by S.S.G.
This work was supported by an internal grant from the College of Veterinary Medicine, Western University of Health Sciences.
REFERENCES
- 1.Elliott RM, Bouloy M, Calisher CH, Goldbach R, Moyer JT, Nichol ST, Pettersson R, Plyusnin A, Schmaljohn CS. 2000. Family Bunyaviridae, p 599–621. In van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon S, Maniloff J, Mayo MA, McGeogch D, Pringle CR, Wickner RB (ed), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, San Diego, CA. [Google Scholar]
- 2.Hoogstraal H. 1979. The epidermology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe and Africa. J Med Entomol 15:307–417. doi: 10.1093/jmedent/15.4.307. [DOI] [PubMed] [Google Scholar]
- 3.Swanepoel R. 1994. Crimean-Congo hemorrhagic fever, p 723–729. In Coetzer JAW, Thomson GR, Tustin RC (ed), Infectious diseases of livestock, with reference to South Africa. Oxford University Press, Cape Town, South Africa. [Google Scholar]
- 4.Swanepoel R, Shepherd AJ, Leman PA, Shepherd SP, McGillivray GM, Erasmus MJ, Searle LA, Gill DE. 1987. Epidemiologic and clinical features of Crimean-Congo hemorrhagic fever in southern Africa. Am J Trop Med Hyg 36:120–132. doi: 10.4269/ajtmh.1987.36.120. [DOI] [PubMed] [Google Scholar]
- 5.Mourya DT, Yadav PD, Shete AM, Gurav YK, Raut CG, Jadi RS, Pawar SD, Nichol ST, Mishra AC. 2012. Detection, isolation and confirmation of Crimean-Congo hemorrhagic fever virus in human, ticks and animals in Ahmadabad, India, 2010-2011. PLoS Negl Trop Dis 6:e1653. doi: 10.1371/journal.pntd.0001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitehouse CA. 2004. Crimean-Congo hemorrhagic fever. Antiviral Res 64:145–160. doi: 10.1016/j.antiviral.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Estrada-Pena A, Ruiz-Fons F, Acevedo P, Gortazar C, de la Fuente J. 2013. Factors driving the circulation and possible expansion of Crimean-Congo haemorrhagic fever virus in the western Palearctic. J Appl Microbiol 114:278–286. doi: 10.1111/jam.12039. [DOI] [PubMed] [Google Scholar]
- 8.Schmaljohn CS, Hooper JW. 2001. Bunyaviridae: the viruses and their replication, p 1581–1602. In Knipe DM, Howley PM (ed), Fields virology, 4th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 9.Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. 2011. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci U S A 108:2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hastie KM, Liu T, Li S, King LB, Ngo N, Zandonatti MA, Woods VL Jr, de la Torre JC, Saphire EO. 2011. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc Natl Acad Sci U S A 108:19365–19370. doi: 10.1073/pnas.1108515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mir MA, Duran WA, Hjelle BL, Ye C, Panganiban AT. 2008. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc Natl Acad Sci U S A 105:19294–19299. doi: 10.1073/pnas.0807211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mir MA, Panganiban AT. 2008. A protein that replaces the entire cellular eIF4F complex. EMBO J 27:3129–3139. doi: 10.1038/emboj.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, Ly H, Liang Y, Dong C. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468:779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter SD, Surtees R, Walter CT, Ariza A, Bergeron E, Nichol ST, Hiscox JA, Edwards TA, Barr JN. 2012. Structure, function, and evolution of the Crimean-Congo hemorrhagic fever virus nucleocapsid protein. J Virol 86:10914–10923. doi: 10.1128/JVI.01555-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YDS, Karlberg H, Devignot S, Weber F, Hao Q, Tan YJ, Mirazimi A, Kotaka M. 2012. Structure of Crimean-Congo hemorrhagic fever virus nucleoprotein: superhelical homo-oligomers and the role of caspase-3 cleavage. J Virol 86:12294. doi: 10.1128/JVI.01627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y, Wang W, Ji W, Deng M, Sun Y, Zhou H, Yang C, Deng F, Wang H, Hu Z, Lou Z, Rao Z. 2012. Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc Natl Acad Sci U S A 109:5046–5051. doi: 10.1073/pnas.1200808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surtees R, Dowall SD, Shaw A, Armstrong S, Hewson R, Carroll MW, Mankouri J, Edwards TA, Hiscox JA, Barr JN. 2016. Heat shock protein 70 family members interact with Crimean-Congo hemorrhagic fever virus and Hazara virus nucleocapsid proteins and perform a functional role in the Nairovirus replication cycle. J Virol 90:9305–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol 16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haque A, Mir MA. 2010. Interaction of hantavirus nucleocapsid protein with ribosomal protein S19. J Virol 84:12450–12453. doi: 10.1128/JVI.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mir MA, Panganiban AT. 2010. The triplet repeats of the Sin Nombre hantavirus 5′ untranslated region are sufficient in cis for nucleocapsid-mediated translation initiation. J Virol 84:8937–8944. doi: 10.1128/JVI.02720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin H, Elliott RM. 1993. Non-viral sequences at the 5′ ends of Ugbe nairovirus S mRNAs. J Gen Virol 74:2293–2297. doi: 10.1099/0022-1317-74-10-2293. [DOI] [PubMed] [Google Scholar]
- 22.Flick R, Flick K, Feldmann H, Elgh F. 2003. Reverse genetics for Crimean-Congo hemorrhagic fever virus. J Virol 77:5997–6006. doi: 10.1128/JVI.77.10.5997-6006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flick R, Elgh F, Pettersson RF. 2002. Mutational analysis of the Uukuniemi virus (Bunyaviridae family) promoter reveals two elements of functional importance. J Virol 76:10849–10860. doi: 10.1128/JVI.76.21.10849-10860.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganaie SS, Haque A, Cheng E, Bonny TS, Salim NN, Mir MA. 2014. Ribosomal protein S19 binding domain provides insights into hantavirus nucleocapsid protein-mediated translation initiation mechanism. Biochem J 464:109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salim NN, Ganaie SS, Roy A, Jeeva S, Mir MA. 2016. Targeting a novel RNA-protein interaction for therapeutic intervention of Hantavirus disease. J Biol Chem 291:24702–24714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cencic R, Hall DR, Robert F, Du Y, Min J, Li L, Qui M, Lewis I, Kurtkaya S, Dingledine R, Fu H, Kozakov D, Vajda S, Pelletier J. 2011. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc Natl Acad Sci U S A 108:1046–1051. doi: 10.1073/pnas.1011477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem 257:14806–14810. [PubMed] [Google Scholar]
- 28.Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. 1996. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J Virol 70:8444–8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liebig HD, Ziegler E, Yan R, Hartmuth K, Klump H, Kowalski H, Blaas D, Sommergruber W, Frasel L, Lamphear B, Rhoads R, Keuchler E, Skern T. 1993. Purification of two picornaviral 2A proteinases: interaction with eIF-4 gamma and influence on in vitro translation. Biochemistry 32:7581–7588. doi: 10.1021/bi00080a033. [DOI] [PubMed] [Google Scholar]
- 30.Park N, Skern T, Gustin KE. 2010. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. J Biol Chem 285:28796–28805. doi: 10.1074/jbc.M110.143404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker E, Jensen L, Croft S, Wei K, Fulcher AJ, Jans DA, Ghildyal R. 2016. Rhinovirus 16 2A protease affects nuclear localization of 3CD during infection. J Virol 90:11032–11042. doi: 10.1128/JVI.00974-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park N, Schweers NJ, Gustin KE. 2015. Selective removal of FG repeat domains from the nuclear pore complex by enterovirus 2A(pro). J Virol 89:11069–11079. doi: 10.1128/JVI.00956-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gale M Jr, Tan SL, Katze MG. 2000. Translational control of viral gene expression in eukaryotes. Microbiol Mol Biol Rev 64:239–280. doi: 10.1128/MMBR.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wakiyama M, Imataka H, Sonenberg N. 2000. Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr Biol 10:1147–1150. doi: 10.1016/S0960-9822(00)00701-6. [DOI] [PubMed] [Google Scholar]
- 35.Cheng E, Mir MA. 2012. Signatures of host mRNA 5′ terminus for efficient hantavirus cap snatching. J Virol 86:10173–10185. doi: 10.1128/JVI.05560-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hussein IT, Cheng E, Ganaie SS, Werle MJ, Sheema S, Haque A, Mir MA. 2012. Autophagic clearance of Sin Nombre hantavirus glycoprotein Gn promotes virus replication in cells. J Virol 86:7520–7529. doi: 10.1128/JVI.07204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]