ABSTRACT
The envelope glycoproteins (Envs) on the surfaces of HIV-1 particles are targeted by host antibodies. Primary HIV-1 isolates demonstrate different global sensitivities to antibody neutralization; tier-1 isolates are sensitive, whereas tier-2 isolates are more resistant. Single-site mutations in Env can convert tier-2 into tier-1-like viruses. We hypothesized that such global change in neutralization sensitivity results from weakening of intramolecular interactions that maintain Env integrity. Three strategies commonly applied to perturb protein structure were tested for their effects on global neutralization sensitivity: exposure to low temperature, Env-activating ligands, and a chaotropic agent. A large panel of diverse tier-2 isolates from clades B and C was analyzed. Incubation at 0°C, which globally weakens hydrophobic interactions, causes gradual and reversible exposure of the coreceptor-binding site. In the cold-induced state, Envs progress at isolate-specific rates to unstable forms that are sensitive to antibody neutralization and then gradually lose function. Agents that mimic the effects of CD4 (CD4Ms) also induce reversible structural changes to states that exhibit isolate-specific stabilities. The chaotropic agent urea (at low concentrations) does not affect the structure or function of native Env. However, urea efficiently perturbs metastable states induced by cold and CD4Ms and increases their sensitivity to antibody neutralization and their inactivation rates Therefore, chemical and physical agents can guide Env from the stable native state to perturbation-sensitive forms and modulate their stability to bestow tier-1-like properties on primary tier-2 strains. These concepts can be applied to enhance the potency of vaccine-elicited antibodies and microbicides at mucosal sites of HIV-1 transmission.
IMPORTANCE An effective vaccine to prevent transmission of HIV-1 is a primary goal of the scientific and health care communities. Vaccine-elicited antibodies target the viral envelope glycoproteins (Envs) and can potentially inhibit infection. However, the potency of such antibodies is generally low. Single-site mutations in Env can enhance the global sensitivity of HIV-1 to neutralization by antibodies. We found that such a hypersensitivity phenotype can also be induced by agents that destabilize protein structure. Exposure to 0°C or low concentrations of Env-activating ligands gradually guides Env to metastable forms that expose cryptic epitopes and that are highly sensitive to neutralization. Low concentrations of the chaotropic agent urea do not affect native Env but destabilize perturbed states induced by cold or CD4Ms and increase their neutralization. The concept of enhancing antibody sensitivity by chemical agents that affect the structural stability of proteins can be applied to increase the potency of topical microbicides and vaccine-elicited antibodies.
KEYWORDS: antibody neutralization, energy landscape, entry, envelope reactivity, envelope glycoproteins, HIV-1, microbicides
INTRODUCTION
The envelope glycoproteins (Envs) of human immunodeficiency virus type 1 (HIV-1) mediate entry of the viral core into the host cell cytoplasm (1, 2). On the surfaces of HIV-1 particles, Env is arranged as a trimer of dimers composed of three gp120 surface subunits and three gp41 transmembrane subunits. Membrane fusion is driven by the potential energy stored in the trimer, which is released in a stepwise manner during viral entry. Engagement of the CD4 receptor by gp120 causes a large favorable change in enthalpy that is partially offset by an unfavorable change in entropy (3–5). Restructuring of Env by CD4 allows engagement of the viral coreceptor, mainly CCR5 or CXCR4 (6, 7), causing additional release of energy (8). Subsequent steps in the entry pathway, which are less well defined, culminate with significant energy release during formation of the gp41 6-helix bundle (2). Therefore, retention of the potential energy contained in Env until both receptor and coreceptor are available is crucial for virus functionality. This energy is preserved by intra- and intersubunit interactions that maintain the structural integrity of the trimer. The strength of these interactions defines the energy barriers that prevent transitions to lower energy states, either functional or nonfunctional. Binding to different ligands can allow Env to overcome these barriers. For example, CD4 binding facilitates transitions to the CD4-activated state (4). Similarly, inhibitory ligands (e.g., antibodies) can bind to Env and cause restructuring, thus inducing transitions to nonfunctional forms (9–11).
Significant diversity exists in structural properties of Envs circulating in the population. Such differences are reflected in their patterns of recognition by antibodies (12–14). In addition to antigenic differences, Envs differ in their global sensitivity to neutralization by the antibodies. Laboratory-adapted strains are generally hypersensitive relative to primary patient isolates (15, 16). Primary isolates also exhibit different levels of global sensitivity. Based on their neutralization by sera from HIV-infected individuals, primary isolates are classified using a tiered system; tier-1 Envs are globally sensitive, whereas tier-2 or tier-3 Envs (more commonly encountered in patients) are more resistant (17). Single-site mutations in gp120 or gp41 can render resistant Envs globally sensitive to antibodies (18–25).
Our previous work has shown that exposure of Envs to 0°C, which is thought to destabilize protein structure by promoting hydration of hydrophobic residues (26, 27), can inactivate the infectivity of some HIV-1 isolates (23, 28). Strains that are cold sensitive are often also globally sensitive to antibody neutralization (23). These findings suggested that the same interactions that maintain the stability of Env also regulate global sensitivity to antibodies. We hypothesized that weakening of interactions that maintain the structural integrity of tier-2 Envs may increase their sensitivity to antibodies (i.e., render them tier-1-like). To test this hypothesis, we examined the structural and functional effects of treatments that perturb protein structure: (i) low temperature, (ii) a chaotropic chemical agent, and (iii) ligands that mimic the effects of the CD4 receptor. A large panel of structurally diverse full-length Envs from clades B and C was tested. For most isolates, exposure to 0°C induces gradual and reversible transitions to a state that exposes otherwise cryptic epitopes. Diverse isolates differ in their rates of transition to the cold-induced state and in their stabilities within this state, as manifested by their progressive increase in sensitivity to antibody neutralization and gradual loss of infectivity. Binding of small-molecule mimics of CD4 (CD4Ms) to Env induces rapid and reversible transition to an “open” conformation. Envs show different stabilities in the CD4M-induced state, manifested by different sensitivities to antibody neutralization, coreceptor activation, and (eventually) inactivation. The chaotropic agent urea (at low concentrations) does not affect the native Env trimer but efficiently destabilizes the cold- and CD4M-induced states and enhances their sensitivity to neutralization by monoclonal antibodies (MAbs) and sera from HIV-infected individuals. Therefore, primary tier-2 strains can be rendered tier-1-like, not only by mutations or Env-specific ligands, but also using a ligand-independent approach. Such treatments reduce the strength of interactions that stabilize Envs and guide them to nonnative forms that are prone to perturbations and globally sensitive to antibody neutralization.
RESULTS
Exposure to 0°C destabilizes the structure of tier-2 Envs by inducing gradual “opening” of the trimer.
The structure of Env differs among diverse HIV-1 isolates. Accordingly, Envs also differ in the types and strengths of interactions that maintain their integrity (23, 29). We investigated the diversity that exists among HIV-1 isolates in their structural robustness (i.e., propensity to undergo conformational changes upon exposure to protein-destabilizing treatments). For this purpose, we tested a panel of antigenically diverse full-length Envs, including 10 from clade B and 11 from clade C. All the Envs tested are classified as tier-2 isolates or exhibit tier-2-like patterns of sensitivity to patient sera and soluble CD4 (sCD4) (30–32). Envs are derived from transmitted/founder (T/F) viruses or from viruses isolated during the chronic phase of infection (Fig. 1A). The antigenic properties of the Envs (i.e., the integrity of specific epitopes) were measured using probes that engage well-defined targets, including MAbs and the CD4-Ig fusion protein. Binding of the probes to full-length cleaved Env trimers expressed on the surfaces of human osteosarcoma (HOS) cells was measured using a cell-based enzyme-linked immunosorbent assay (ELISA) system (33). The gp160 precursor is efficiently cleaved in HOS cells (34). Accordingly, these cells express on their surfaces primarily trimers containing the gp120 and gp41 subunits.
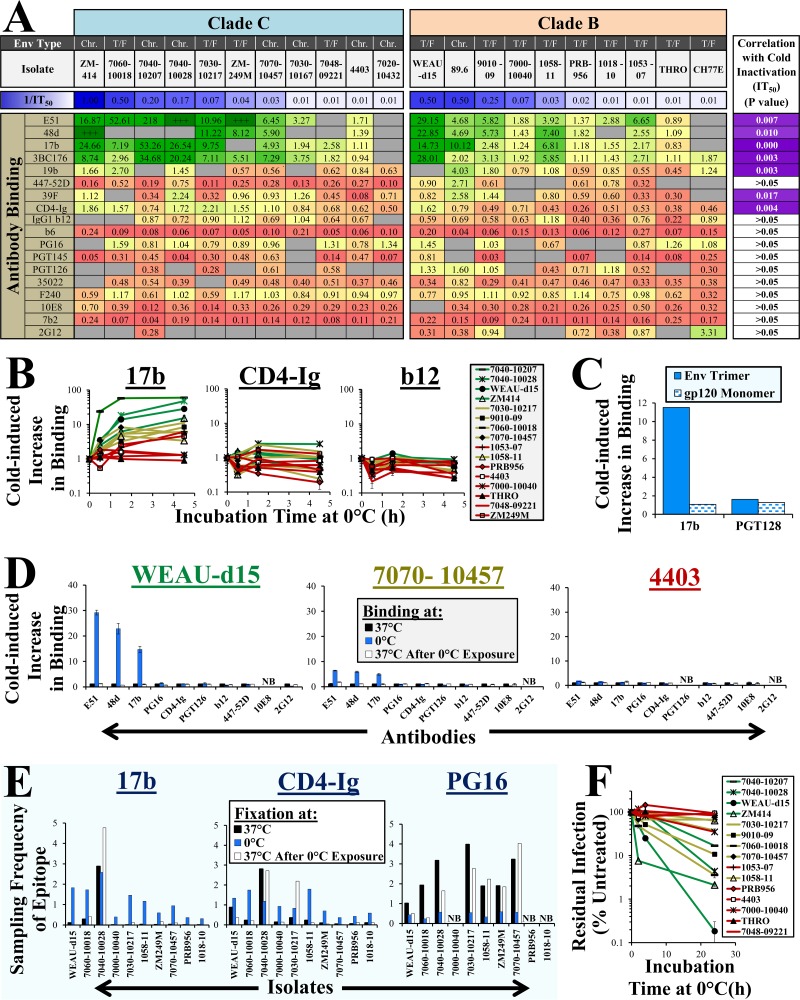
FIG 1.
Exposure of the membrane-bound Env trimer to 0°C induces gradual and reversible restructuring and inactivates virus infectivity. (A) Effect of cold on binding of MAbs. Full-length Envs of T/F and chronic-phase (Chr.) viruses were transiently expressed on HOS cells. Samples were incubated with the indicated MAbs or CD4-Ig at 0°C or 37°C for 45 min, and binding was measured by cell-based ELISA. The values represent ratios between binding at 0°C and 37°C (green, high; red, low). “+++” indicates high binding at 0°C but no binding at 37°C. Gray shading signifies no binding at either temperature. The sensitivity of each isolate to cold inactivation (1/IT50; h) is indicated and color coded (blue, sensitive; white, resistant). The right column indicates P values for a Spearman correlation between the IT50s of Envs and their cold-induced change in MAb binding. (B) Rates of cold-induced exposure of Env epitopes. Probe binding to Env-expressing HOS cells was measured after incubation at 0°C for 1.5 or 4.5 h. Binding is expressed relative to that of samples incubated at 37°C. (C) Effect of cold on binding of MAbs to soluble gp120 of strain 89.6. Samples were incubated for 6 h at 0°C with the MAbs, immunoprecipitated, and analyzed by Western blotting. The data represent the intensity of the gp120 band quantified by densitometry. (D) Cold-induced Env restructuring is reversible. HOS cells expressing the indicated Envs were incubated at 0°C for 2 h, reequilibrated to 37°C, and then incubated with probes. The Env labels are color coded by their sensitivity to cold-induced CoR-BS exposure. (E) Cold induces reversible changes in sampling frequencies of epitopes. HOS cells expressing the indicated Envs were incubated at 37°C or 0°C and fixed with glutaraldehyde at the same temperature. In addition, some samples were incubated at 0°C prior to fixation at 37°C. Glutaraldehyde activity was quenched, and binding of probes was measured for all the samples (fixed and nonfixed) at 37°C. The values represent ratios between binding of each probe to fixed and nonfixed samples. (F) Cold-induced inactivation of Env. Viruses were incubated for different times at 0°C and then added to cells at 37°C to quantify residual infectivity. The values represent measured luciferase activity expressed as a percentage of that measured for samples not treated with cold. P values were determined by a two-tailed test. The error bars indicate standard errors of the mean (SEM).
Exposure of some proteins to 0°C promotes the hydration of hydrophobic residues and can result in denaturation (26, 27). We examined whether exposure to 0°C disrupts the structure of Envs by measuring binding of different probes. For many Envs, incubation at 0°C increased the binding of MAbs 17b, 48d, and E51, which target otherwise cryptic epitopes that overlap the coreceptor-binding site (CoR-BS) of gp120 (Fig. 1A). Envs of diverse HIV-1 strains showed different degrees of cold-induced enhancement of CoR-BS MAb binding, ranging from no change to more than 50-fold increase. Binding of other CD4-induced MAbs, including 3BC176 and the V3 loop MAb 19b, was also increased at 0°C, whereas MAbs 447-52D and 39F demonstrated such trends primarily for clade B. Enhanced exposure of the CoR-BS at 0°C was also associated with increased binding of CD4-Ig. For other MAbs, cold generally exerted a uniform effect on the different Envs. No association was observed between the binding efficiency of probes at 25°C and their sensitivity to structural changes (data not shown).
We examined the progression of conformational changes at 0°C (Fig. 1B). Monitored over the course of 4.5 h, the Envs demonstrated different rates of increase in binding of MAb 17b; some reached maximal binding after 1.5 h at 0°C, whereas others continued to increase after that time. Relative to the significant changes in MAb 17b, binding of CD4-Ig and MAb b12 showed little change over time. In contrast to membrane-bound Env trimers, the structure of soluble gp120 monomers was unaffected after 6 h of incubation at 0°C (Fig. 1C).
We examined the reversibility of the cold-induced conformational changes. For this purpose, Env-expressing cells were incubated at 0°C for 2 h, followed by reequilibration to 37°C and measurement of MAb binding. We found that the conformational changes induced at 0°C were reversible. (Fig. 1D). To determine whether changes at 0°C were caused by the effects of cold on the antibody or Env, we used glutaraldehyde to fix the conformations of Env sampled at 0°C or 37°C and then examined the binding of the probes at a uniform temperature of 37°C (10, 22, 34). The frequency at which Env samples the binding-competent state for a given probe can be estimated by the ratio between the value for probe binding to fixed Env and the value for nonfixed Env (10, 22, 34). For most isolates, the sampling frequency of the 17b epitope was very low at 37°C and dramatically increased at 0°C (Fig. 1E). Return of the samples to 37°C after exposure to 0°C caused reversion of the sampling frequency to the low levels measured without cold exposure. Isolate 7040-10028 exhibited high sampling frequency of the 17b epitope at 37°C, which was maintained constant during and after cold treatment. Thus, the changes in MAb 17b binding and sampling frequency are generally reversible. Cold also increased the sampling frequency of the CD4-Ig epitope for most Envs (Fig. 1E). The sampling frequency of the trimer-dependent PG16 epitope was reduced at 0°C. However, the changes were not fully reversible for the cold-sensitive isolates. Such changes in PG16 were not observed in steady-state binding experiments at 0°C (Fig. 1A). Therefore, cold can induce an irreversible change to a conformational state that exhibits reduced sampling of the PG16 epitope. In this state, occupancy of conformations that can bind PG16 may be sufficient to allow engagement of this MAb under steady-state conditions.
We examined whether cold-induced restructuring was associated with changes in the fusion potential of Env. Viruses were incubated at 0°C for different times and then added to CD4+ CCR5+ cells to measure residual infectivity. The incubation time at 0°C required to reduce infectivity by 50% is defined as the half-life at 0°C (IT50) of each isolate. A wide range of sensitivities were observed, with IT50 values as low as 1 h (Fig. 1F). Strong correlations were found between the structural change induced at 0°C (i.e., the fold increase in exposure of CoR-BS epitopes) and the functional stability at 0°C (i.e., 1/IT50) (e.g., correlations in Fig. 1A, top row and right column).
We use HOS cells for antigenic profiling of Env trimers. These cells express primarily processed Env (i.e., gp120 and gp41 subunits) on their surfaces, whereas most other cell lines typically express uncleaved gp160 molecules (34). Viruses for our infectivity tests were produced in 293T cells. We compared the cold-induced structural changes of Envs expressed on HOS and 293T cells using 11 isolates and observed similar inducibility patterns (P = 0.016 in a Spearman correlation test) (data not shown). Furthermore, viruses produced in HOS and 293T cells show similar sensitivities to cold inactivation (IT50s, 24 and 18 h, respectively, for AD8). Therefore, Envs expressed on HOS and 293T cells show similar sensitivities to the structural and functional effects of cold.
In summary, exposure to 0°C disrupts interactions that maintain the structural integrity of membrane-bound trimers and induces gradual and mostly reversible conformational changes. Cold-induced restructuring is associated with gradual inactivation of Env function. Diverse tier-2 isolates show different propensities to undergo cold-induced restructuring and inactivation. These isolate-specific effects likely reflect the diversity in number and type of interactions that maintain Env in the native (unliganded) state.
Env sensitivity to cold inactivation is determined by the propensity to undergo structural changes and by the stability of the induced state.
The data presented in Fig. 1 suggested that conformational changes induced at 0°C may precede loss of function. For example, exposure of the 17b epitope was increased significantly for isolates 7040-10207 and 7040-10028 after 1 h at 0°C (Fig. 1A), whereas infectivity remained unaltered even after 4 h at 0°C (Fig. 1F). We performed a side-by-side analysis of the isolates to examine this apparent lag. Four-hour exposure to 0°C enhanced 17b binding to many Envs (Fig. 2A, bottom). These conformational changes corresponded well to inactivation at 24 h, but not at 4 h (Fig. 2A, top). In some cases, Envs exhibited significant changes in structure but no loss of function even after 24 h (e.g., isolate 7030-10217). These data suggest that some isolates are more stable in the cold-induced state.
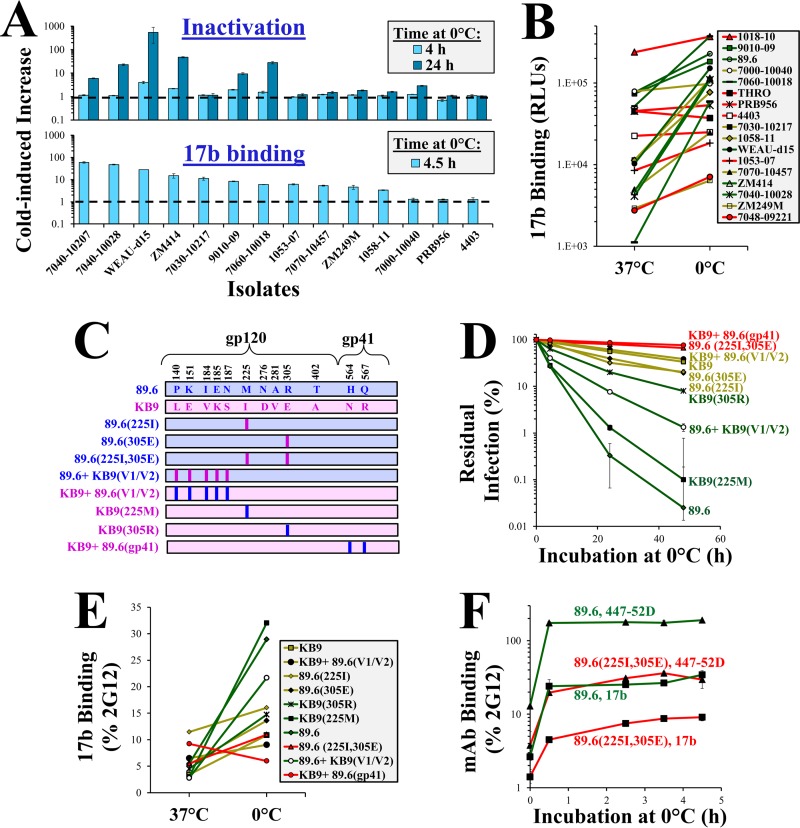
FIG 2.
Sensitivity to cold inactivation is determined by the propensity of Envs to undergo structural changes at 0°C and by the stability of the induced state. (A) Restructuring of Env precedes changes in function. The effects of incubation at 0°C on binding of MAb 17b (bottom) and virus infectivity (top) were compared for the indicated Envs. Binding and infectivity are expressed as the fold change in values relative to samples incubated at 37°C. (B) Effect of cold on MAb 17b binding to Envs. The sensitivity of each Env to inactivation at 0°C is represented by the color of the line (red, resistant; yellow, intermediate; green, sensitive). The data describe the absolute binding efficiency of MAb 17b (in relative light units [RLU]). (C) Variants of 89.6 Env tested in this study. Differences in sequence between 89.6 and KB9 Envs are shown. Vertical bars represent mutations introduced to revert sequence to the amino acid found in the similarly colored Env (89.6, blue; KB9, pink). (D) Cold-induced inactivation of virus containing variants of the 89.6 Env. (E) Relationship between binding of MAb 17b to Envs at 37°C and 0°C. To correct for differences in Env expression between variants, binding of MAb 17b was normalized to binding of MAb 2G12. The sensitivity of Envs to inactivation at 0°C is represented by the color of the lines (red, resistant; green, sensitive). (F) Rates of cold-induced exposure of CoR-BS. Probe binding to Env-expressing HOS cells incubated at 0°C for the indicated times is shown. Binding is expressed relative to that of samples incubated at 37°C.
We compared the degree of CoR-BS exposure in the native state (i.e., Env openness) with Env sensitivity to cold-induced structural changes. At 37°C, binding of 17b varied between Envs (Fig. 2B). Binding efficiency at 37°C was not associated with the effects of cold on binding or with Env sensitivity to cold inactivation (represented by the line color); Envs resistant to cold inactivation (red lines) displayed different degrees of openness at 37°C.
The different binding efficiencies of 17b to these genetically diverse Envs could result from different integrities of the epitope (rather than their exposure). We therefore performed the above-described tests in a set of closely matched variants of a single HIV-1 strain, which differ in their sensitivity to cold. The chimeric simian-human immunodeficiency virus (SHIV) strain 89.6 was created with a SIVmac239 backbone that was modified to include the Env of HIV-189.6. SHIV89.6 was nonpathogenic in rhesus macaques, but serial passage in this host gave rise to the pathogenic SHIVKB9, which induces rapid depletion of CD4+ T cells and AIDS-like disease (35, 36). Comparison of the sequences of 89.6 and KB9 Envs revealed only 12 amino acid differences in gp120 and gp41 (Fig. 2C), 2 of which were associated with the increased pathogenicity of KB9: (i) Arg to Glu at position 305 of the V3 loop and (ii) Met to Ile at position 225 of the gp120 inner domain (35, 37). The two changes also increase resistance of 89.6 Env to cold inactivation (38), suggesting a possible association between the cold sensitivity of Env and viral pathogenicity. We tested 89.6 variants that contained different combinations of 89.6 and KB9 sequences for the effects of cold. As expected, the variants differed in their sensitivity to cold inactivation (Fig. 2D). To determine if the Envs also differ in their sensitivity to cold-induced structural changes, we measured binding of MAb 17b. For increased accuracy, binding of this probe to the Envs was normalized for the expression level of each Env, which was quantified by the binding efficiency of MAb 2G12 (39). The epitope of MAb 2G12 is unaffected by changes at the positions that differentiate the 89.6 and KB9 Envs (Fig. 2C) (40). We found that at 37°C, binding of MAb 17b to the variants was relatively uniform (Fig. 2E). However, exposure to cold caused different structural changes in the variants. Similar to the panel of tier-2 isolates, the cold-induced increase in 17b binding correlated with Env sensitivity to inactivation at 0°C (P = 0.019; Spearman correlation test). A significant lag was observed between the occurrence of structural changes and inactivation at 0°C. For example, the cold-resistant 89.6(225I, 305E) Env achieved near-maximal exposure of the CoR-BS after incubation at 0°C for 3.5 h, whereas no loss of infectivity was observed at this time point (compare Fig. 2D and F). These findings support the notion that exposure to cold induces gradual rearrangement of the trimer; continued exposure to this structure-perturbing treatment causes progressive loss of Env function.
The metastable state induced by cold is sensitive to neutralization by antibodies.
The decreased functional stability of Envs in the cold-induced state suggested that such Envs could be more sensitive to the perturbing effects of antibodies. We therefore tested neutralization of a closely matched pair of 89.6 variants that differ in their propensities to assume this state and in their stabilities within it. The cold-sensitive wild-type 89.6 Env and the cold-resistant 89.6(225I, 305E) mutant were examined. Two different pools of serum (PS) from HIV-1 infected individuals and different MAbs were tested. Isolated exposure (for 2 h) of virus containing 89.6 Env to 0°C, PS(A), or PS(B) inhibited infection by 1.7-, 15-, or 9-fold, respectively (Fig. 3A). However, we observed that simultaneous exposure to 0°C and PS(A) or PS(B) inhibited infection by 91- or 40-fold, respectively. Synergy between the effects of cold (C) and inhibitor (I) on infection (SynI,C) is defined as follows:
| (1) |
where EC,I is the measured effect (reduction of infectivity) induced by simultaneous treatment of virus with cold and inhibitor, and EC and EI are the measured effects of treating virus with cold and antibody separately. Thus, coexposure of virus to these treatments for a brief (2-h) period induced moderate 3.7- and 2.6-fold synergistic effects (indicated by green circles in Fig. 3A). The neutralization potency of MAbs that target CD4-induced epitopes was also mildly enhanced by brief exposure to 0°C, including the V3 loop binding MAbs 447-52D and 19b and the CoR-BS MAbs 17b and 48d. For other inhibitors tested, neutralization potency was not enhanced by exposure to cold. This pattern is consistent with the epitopes that undergo changes at 0°C (Fig. 1A). Similar to the structural changes induced by cold, the state of increased sensitivity to neutralization was also reversible; exposure of virus to 0°C followed by reequilibration to 25°C before incubation with antibody resulted in a neutralization profile similar to that of virus not exposed to cold (Fig. 3B).
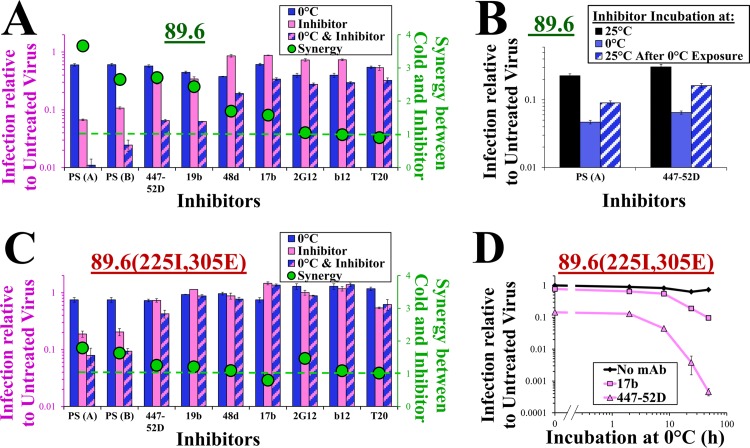
FIG 3.
The metastable states induced at 0°C show increased sensitivity to antibody neutralization. (A) Effect of brief exposure to cold on sensitivity of viruses containing 89.6 Env to neutralization. Viruses were incubated at 0°C or 37°C in the absence or presence of the indicated inhibitors (MAbs or polyclonal pooled sera from HIV-infected individuals) for 2 h and then added to the cells to measure residual infectivity. Infection is presented as the fold change relative to untreated virus. The green circles represent the calculated synergistic effect between cold and inhibitor (equation 1). The concentrations of all inhibitors used in neutralization assays are provided in Materials and Methods. (B) Reversibility of the effect of cold on HIV-1 neutralization by PS(A) and MAb 447-52D. Viruses were incubated for 1 h with antibody at 0°C, 25°C, or 25°C after 1-h exposure to 0°C. Samples were then added to CD4+ CCR5+ cells and further cultured at 37°C for 3 days to measure infectivity. (C) Effect of brief (2-h) exposure to 0°C on neutralization sensitivity of viruses containing 89.6(225I, 305E) Env, as shown in panel A. (D) Neutralization of virus containing the cold-resistant 89.6(225I, 305E) Env increases with incubation time at 0°C. Viruses were incubated with MAb 17b, MAb 447-52D, or no antibody at 0°C for the indicated times and then added to cells to measure residual infectivity.
We also tested the effects of cold on neutralization of the cold-resistant 89.6(225I, 305E) Env. At 37°C, virus containing 89.6(225I, 305E) Env was more resistant to neutralization by PS and MAbs than wild-type 89.6 Env (Fig. 3A and C) (23, 41). After exposure to 0°C for 2 h, the neutralization sensitivity of 89.6(225I, 305E) was not significantly increased (Fig. 3C). However, when the incubation time at 0°C was extended, we observed a dramatic increase in neutralization (e.g., sensitivity to MAb 447-52D was increased ∼230-fold when incubation at 0°C was extended to 48 h) (Fig. 3D).
Therefore, exposure of 89.6(225I, 305E) Env to cold induces exposure of the CoR-BS that progresses over ∼4 h (Fig. 2F). In this cold-induced conformation, viruses gradually increase in their sensitivity to antibodies; at 50 h, most viruses are still functional (IT50 = 84 h), but in a neutralization-sensitive form (90% are neutralized by MAb 17b, whereas no neutralization is observed without cold exposure). These results suggest that cold induces relatively rapid conformational changes, followed by gradual acquisition of antibody sensitivity and finally inactivation of function. These effects of cold are similar to the changes caused by CD4Ms (42–44). This likeness prompted us to compare the above-mentioned effects with the structural and functional changes induced by CD4Ms.
Stability of Envs in the CD4M-induced state determines their sensitivity to inhibition by these agents.
Ligands that mimic the effects of CD4 inhibit HIV-1 infection. Such agents include soluble forms of CD4 (44–48), peptides (49), and small molecules (33, 42). CD4Ms display a range of affinities, from the low-micromolar affinities of the first CD4Ms synthesized (42) to the low-nanomolar affinities of sCD4 and recent small-molecule agents (50). High-affinity CD4Ms or high concentrations of lower-affinity CD4Ms induce formation of the HR1 coiled coil (33). This activated state is unstable and gradually decays at isolate-specific rates. CD4Ms can also inactivate some HIV-1 isolates at lower concentrations that do not induce irreversible exposure of the HR1 coiled coil. We examined two CD4Ms, the high-affinity sCD4 and the lower-affinity synthetic small-molecule agent JRC-II-191 (molecular mass, 355 daltons), which is an analog of the compound NBD-556 (42, 51). The effects of sCD4 and JRC-II-191 on the structures and functions of seven Envs were tested. For all the Envs, sCD4 increased exposure of the CoR-BS and reduced virus infectivity by 2- to 281-fold (Fig. 4A). The lower-affinity JRC-II-191 (binding affinity of 0.76 μM to soluble gp120 of HIV-1 YU2 strain) also increased exposure of the CoR-BS for all the Envs (Fig. 4B). However, in several cases, the conformational change was not accompanied by loss of Env function, particularly at 4 μM. For example, JRC-II-191 increased binding of 17b to both 89.6 and 89.6(225I, 305E) by 4- and 11-fold, respectively. However, wild-type 89.6 was efficiently inactivated at 4 μM, whereas the infectivity of the mutant was unaffected at that concentration. These differences suggest that sensitivity to CD4Ms is also determined by stability within the induced state(s). This hypothesis was supported by the finding that CoR-BS exposure for the 1058-11 Env were similar at JRC-II-191 concentrations of 4 and 10 μM, but inactivation was ∼12-fold higher at 10 μM. Furthermore, several Envs showed similar structural changes induced by JRC-II-191 but different sensitivities to inactivation by the agent (marked by line color in Fig. 4C). The degree of initial Env “openness” did not correspond to CD4M-induced structural changes or functional inactivation (i.e., regardless of initial 17b binding, all the isolates underwent structural changes). Therefore, CD4Ms effectively induce structural changes to forms that exhibit isolate-specific stabilities; application of additional pressure (i.e., higher CD4M concentrations) induces inactivation of diverse Envs to different extents.
FIG 4.
The conformational state induced by CD4Ms demonstrates isolate-specific stabilities. (A and B) Effects of sCD4 and JRC-II-191 on structure and function of Env. HOS cells expressing the indicated Envs were incubated for 1 h at 37°C with MAb 17b in the presence of JRC-II-191 or sCD4 (Bottom). Changes in 17b binding relative to samples not containing CD4Ms (Top). In addition, viruses containing these Envs were incubated with cells in the absence or presence of sCD4 or JRC-II-191 for 6 h. The cells were then washed and further incubated to measure the residual infectivity of CD4M-treated virus relative to untreated virus. (C) Relationship between binding of MAb 17b to Envs in the absence and presence of JRC-II-191. The lines are color coded according to the sensitivity of each Env to inactivation by JRC-II-191 (red, resistant; green, sensitive). (D) Reversibility of structural changes induced by CD4Ms. HOS cells expressing 89.6 Env were incubated with MAb 17b and JRC-II-191 or sCD4. Alternatively, some samples were first incubated with the CD4M, which was then removed and MAb 17b was added. The data represent changes in binding of MAb 17b relative to samples incubated without CD4Ms. (E) Effect of JRC-II-191 on infection of CD4− CCR5+ cells. Viruses containing the indicated Envs were spinoculated onto cells at 10°C and then incubated for 2 h at 37°C in the absence or presence of JRC-II-191. The cells were then washed, and infectivity was measured 3 days later. The dashed lines represent infectivity measured in the same experiment by untreated virus bound to CD4+ CCR5+ cells. (F) Effect of JRC-II-191 on neutralization by PS and MAb 17b. Viruses containing the indicated Envs were added to cells in the presence of JRC-II-191 (4 μM), PS, or both for 6 h at 37°C. The cells were then washed and further incubated to measure residual infectivity. The blue triangles represent the calculated synergistic effects between JRC-II-191 and PS or MAb 17b. (G) Synergy between the effects of cold and JRC-II-191. Viruses were exposed to 0°C, JRC-II-191 (4 μM), or both for 2 h and then added to cells to measure residual infectivity. The values in the x axis represent the calculated products of the inhibitory effects of each treatment separately. The values in the y axis describe the measured effect on infectivity of virus exposed to both treatments simultaneously.
We tested the reversibility of the structural changes induced by the CD4Ms (Fig. 4D). At a low concentration, the effect of JRC-II-191 was fully reversible; removal of the agent before addition of 17b resulted in the return of Env to a “closed” conformation. At higher JRC-II-191 concentrations, structural changes were only partially reversible, suggesting that some Envs transitioned to a state that constitutively exposes the CoR-BS. The higher-affinity sCD4 showed partial reversibility at low concentrations and complete irreversibility at higher concentrations (Fig. 4D, bottom).
In addition to different sensitivities to inactivation by JRC-II-191, 89.6 and 89.6(225I, 305E) Envs differed in their abilities to utilize the agent to infect CD4− cells. Viruses were prebound to CD4− CCR5+ cells and then incubated with different concentrations of JRC-II-191. In the absence of the CD4M, no significant infection of the CD4− cells by either Env was observed (Fig. 4E). Addition of JRC-II-191 increased infection by both viruses in a dose-dependent manner to levels close to the infectivity of the CD4+ CCR5+ cells (indicated by dashed lines). Viruses containing 89.6 Env were responsive to significantly lower concentrations of the agent than 89.6(225I, 305E).
CD4Ms can increase HIV-1 sensitivity to patient plasma (52). We examined whether the 89.6 variants also differ in their sensitivity to JRC-II-191-mediated enhancement of neutralization. Wild-type 89.6 Env showed CD4M-mediated enhancement of neutralization by PS and MAb 17b (Fig. 4F). In contrast, variant 89.6(225I, 305E), which is more stable in the CD4M-induced state, showed little synergy between JRC-II-191 and PS or MAb 17b (Fig. 4F). Therefore, the two 89.6 variants show similar structural changes induced by CD4Ms. However, the wild-type Env is more readily rendered sensitive to antibody neutralization and responsive to coreceptor. In contrast, the structurally induced 89.6(225I, 305E) variant is more resistant to antibodies and coreceptor and requires higher concentrations of the CD4Ms to induce such a sensitive form.
The functional stability of the CD4M-induced state could be further reduced by cold treatment. Viruses containing different Envs were exposed to 0°C, JRC-II-191, or both treatments simultaneously, and infectivity was measured (Fig. 4G). For many Envs, JRC-II-191 and 0°C acted synergistically (i.e., inhibition by simultaneous treatment with JRC-II-191 and 0°C was greater than the calculated product of inhibition by treatment with JRC-II-191 or 0°C alone). For example, treatment of 89.6 Env with JRC-II-191 or 0°C alone resulted in 8.6- or 1.3-fold inhibition, whereas combined treatment reduced infectivity by 138-fold, representing a 12-fold synergistic effect. Diverse isolates demonstrated a range of synergies between these treatments, from ∼1-fold (i.e., no synergistic effect) to 160-fold.
In summary, at low concentrations or affinities, CD4Ms induce reversible transitions of Env to nonnative states. Different tier-2 isolates show comparable propensities to undergo restructuring by JRC-II-191 and expose the CoR-BS. However, the structurally induced Envs then show isolate-specific propensities for loss of fusion competence, sensitivity to antibody neutralization, and entry into CD4− cells. Increased concentrations of CD4Ms promote transitions from the (stable) structurally induced form to a perturbation-sensitive form. Such transitions can be enhanced by exposure to additional protein-destabilizing treatments (e.g., cold), implying that the observed moderate increases in antibody sensitivity could be further augmented.
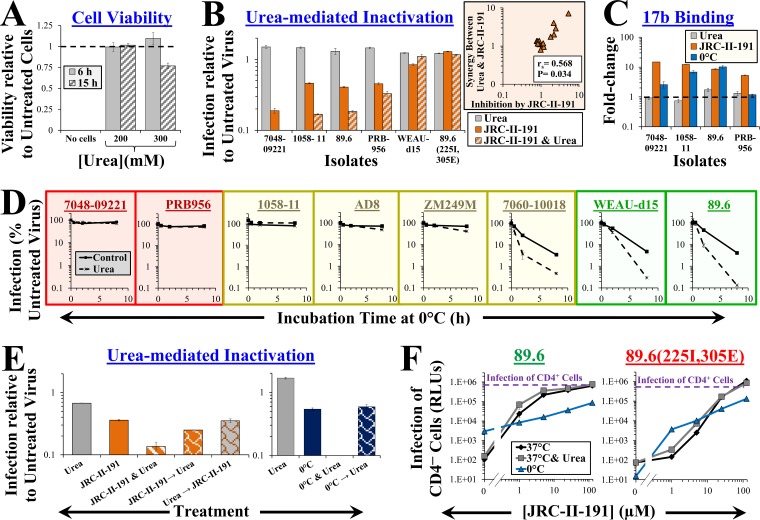
Exposure to low concentrations of the chaotropic agent urea enhances inactivation of the metastable states induced by CD4Ms and cold.
The above-mentioned data show that the neutralization sensitivity of tier-2 Envs can be increased by induction of metastable nonnative states, which appear to be dynamic and change over the course of time (for cold) or concentrations of the agent (for CD4Ms). We examined whether such effects can be achieved by chemical treatment. The chaotropic agent urea disrupts hydrogen bonds that stabilize protein structure (53, 54). At concentrations above 0.5 M, urea inactivates different HIV-1 isolates to similar extents (55). We examined whether at lower concentrations urea can induce destabilizing changes similar to those caused by cold. Exposure of cells to low concentrations of urea (≤0.3 M) for 6 h was not associated with toxicity (Fig. 5A). The infectivity of the different isolates was also not affected significantly at this concentration (Fig. 5B). However, urea enhanced the inactivating potency of JRC-II-191 by up to 7-fold. Urea-mediated enhancement of JRC-II-191 inactivation depends on the sensitivity of each isolate to this CD4M (Fig. 5B, inset). Interestingly, in contrast to the effects of cold or CD4Ms, urea does not increase exposure of the CoR-BS on native Envs (Fig. 5C).
FIG 5.
Low concentrations of urea perturb the metastable states induced by cold and CD4Ms but not the native state of Env. (A) Effect of urea on viability of Cf2Th CD4+ CCR5+ cells measured using an ATP-based assay. The data are presented as fractions of viability in samples incubated without urea. (B) Effects of urea and JRC-II-191 on infectivity. Viruses containing the indicated Envs were added to cells in the presence of urea, JRC-II-191 (4 μM), or both for 6 h at 37°C. The cells were then washed and further incubated to measure residual infectivity. The inset shows a correlation between the JRC-II-191-mediated inhibition of each isolate and the synergy it exhibited for the combined use of urea and JRC-II-191. (C) Effects of urea, JRC-II-191, and cold on CoR-BS exposure. The data represent the fold change in binding of MAb 17b relative to samples incubated at 37°C without urea or JRC-II-191. The change in 17b binding induced after 1 h at 0°C is shown for comparison. (D) Urea enhances inactivation of cold-induced Envs. Viruses containing the indicated Envs were incubated at 0°C in the absence or presence of urea for 0.5, 2, or 8 h and then equilibrated to 37°C. Samples were subsequently added to cells and incubated for 6 h. The cells were then washed, and infectivity was measured 3 days later. The graphs are color coded by Env sensitivity to cold-induced structural changes (red, resistant; yellow, intermediate; green, sensitive). (E) Reversibility of the effects of JRC-II-191 and cold on urea-mediated inactivation. Viruses containing 89.6 Env were exposed to 0°C or JRC-II-191 (4 μM) in the absence or presence of urea for 2 h. To test reversibility, the viruses were first exposed to 0°C or JRC-II-191 for 2 h, followed by removal of the treatment and incubation with urea for 2 h. As controls, some samples were treated with urea prior to addition of JRC-II-191. (F) Effect of urea or cold on JRC-II-191-induced infection of CD4− CCR5+ cells. Viruses were spinoculated onto CD4− CCR5+ cells at 10°C and then incubated for 2 h at 37°C or 0°C in the absence or presence of urea and JRC-II-191. The cells were then equilibrated to 37°C and washed, and infectivity was measured 3 days later. Data that describe infection at 37°C are also shown in Fig. 4E. The dashed lines represent infectivity measured in the same experiment by untreated virus bound to CD4+ CCR5+ cells. The error bars indicate SEM.
The fact that urea enhanced inactivation by JRC-II-191 but did not affect the structure or function of the native Env suggested that (at this concentration) it may destabilize only nonnative states. We examined whether urea also affects the state induced by cold. Viruses were incubated at 0°C for different time periods in the absence or presence of urea, and the changes in infectivity were measured. We observed that urea enhanced inactivation of cold-inducible Envs by up to 31-fold (Fig. 5D). Envs that showed gradual structural changes due to cold (e.g., ZM249M [Fig. 1B]) also showed urea-mediated enhancement of inactivation only after extended incubation times at 0°C. Consistent with the notion that urea (at these concentrations) affects only metastable states, removal of the destabilizing treatment (cold or CD4Ms) prior to urea exposure eliminated the inactivation-enhancing effects of the agent (Fig. 5E).
We examined the effects of Env destabilization (by urea or cold) on CD4M-mediated infection of CD4− cells. Exposure of cell-bound virus containing wild-type 89.6 Env to 0°C increased infection of CD4− cells (albeit to a limited extent). Treatment with JRC-II-191 at 0°C reduced infectivity relative to that at 37°C, likely due to simultaneous inactivation. In contrast, the cold-resistant Env 89.6(225I, 305E) showed similar levels of infection at 0°C and 37°C in the presence of JRC-II-191. Exposure to urea in the presence of JRC-II-191 enhanced infection of CD4− cells by ∼2-fold for the two Envs.
Taken together, these findings suggest that exposure to low, nontoxic concentrations of urea does not affect the structure or function of the native Env trimer. However, urea can enhance inactivation of metastable forms of Env induced by cold or CD4Ms.
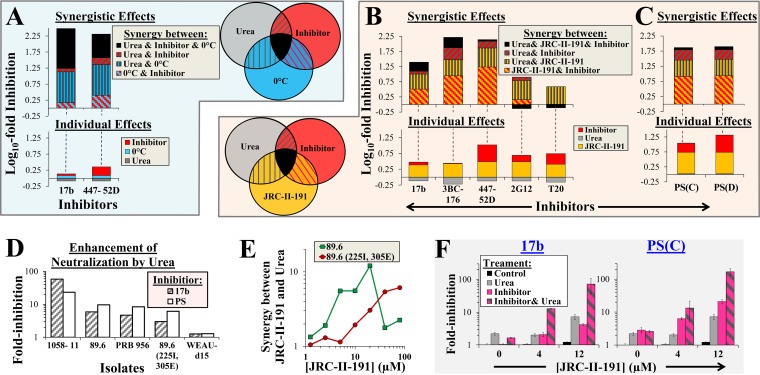
Env-destabilizing treatments act synergistically to enhance neutralization of tier-2 viruses by antibodies.
We examined whether urea can act synergistically with treatments that mobilize Env to metastable states to enhance antibody sensitivity. Mutations in the tier-2 Env 89.6 at positions 225 and 305 increased resistance to cold-induced structural changes (Fig. 2E) and antibody neutralization (i.e., rendered the Env tier-3-like) (Fig. 3C). Greater destabilization pressure (e.g., by extended exposure to 0°C) enhanced neutralization of this stable variant (Fig. 3D). We examined whether urea can perturb the metastable forms induced by cold to increase the sensitivity of this resistant virus to antibodies. The individual effects of 0°C, urea, and the CD4-induced MAbs 17b and 447-52D were measured. MAbs 17b and 447-52D alone showed minimal or no neutralization (Fig. 6A, bottom). Similarly, urea and 0°C alone did not significantly alter the infectivity of this stable Env. We then measured the reduction in infectivity induced by simultaneous exposure to all possible pairs of treatments (cold, urea, and inhibitors) and calculated the synergistic effects between them (Fig. 6A, Venn diagram, and equation 1). We also calculated the synergistic effect associated with exposure to the three treatments (SynU,I,C) based on the following expression:
| (2) |
where EU,I,C is the measured effect (fold reduction in infectivity) of simultaneous treatment by urea, inhibitor (e.g., antibody), and cold, and EU, EI, and EC are the measured effects of isolated treatment with urea, inhibitor, or cold. All values were log10 transformed for visualization of their additive effects in a stacked bar graph format. We found that the (log) sum contribution of synergistic effects to inhibition (Fig. 6A, top) was significantly greater than the individual effects of the treatments. For example, simultaneous exposure to 0°C, urea, and MAb 17b resulted in 341-fold inhibition relative to an expected inhibition of 1.12-fold based on the calculated product of their individual effects. Synergistic effects associated with urea accounted for up to ∼50% of the overall inhibition (166-fold for MAb 17b). Therefore, the combination of urea and cold-mediated destabilization can render a neutralization-resistant tier-3-like Env sensitive to these antibodies.
FIG 6.
Synergy between Env-destabilizing treatments enhances virus sensitivity to neutralization. (A) Enhancement of antibody neutralization by cold, urea, and their combination. Viruses containing 89.6(225I, 305E) Env were treated for 8 h with all possible combinations of cold, urea, or the indicated inhibitors (Venn diagram) and then added to cells to measure residual infectivity. The fold inactivation induced by each treatment and the calculated synergistic effects were determined as described in equation 1 and equation 2 and are expressed as their log10 values. The color coding of each section of the bars corresponds to that of the treatment combination shown in the Venn diagram. (B and C) Effects of urea, JRC-II-191, and their combination on HIV-1 neutralization. Viruses containing 89.6 Env were incubated with cells in the absence or presence of urea, JRC-II-191 (4 μM), and the indicated inhibitors for 6 h. The cells were then washed and further cultured to measure infectivity. Calculations were performed as described for panel A. (D) Effects of urea on antibody-mediated neutralization of different Envs. Viruses containing the indicated Envs were incubated with urea, JRC-II-191 (4 μM), and inhibitor (either MAb 17b or PS). The fold reduction in infection relative to samples similarly treated but without urea is shown (i.e., EU,I,J/EI,J). (E) Synergistic effects between urea and JRC-II-191. Synergy between treatments was calculated as described in equation 1. The data represent averages of at least two independent experiments. (F) Urea-mediated enhancement of antibody neutralization increases with the JRC-II-191 concentration. Viruses containing 89.6(225I, 305E) were added to cells in the presence of urea, MAb 17b, or PS and different concentrations of JRC-II-191 as described for panel B. The data represent fold changes in infectivity relative to untreated samples. The error bars indicate SEM.
We examined whether urea can also act synergistically with CD4Ms to enhance neutralization of tier-2 HIV-1 isolates. Inhibition of 89.6 by individual or combined treatment with JRC-II-191, urea, and different inhibitors (MAbs, T20, or PS) was measured. As expected, urea alone did not inhibit infection, whereas JRC-II-191 alone exerted a 3-fold inhibitory effect (Fig. 6B, bottom). Inhibition by the MAbs or T20 alone ranged between no effect and ∼3-fold inhibition. Synergy between JRC-II-191 and the CD4-induced antibodies accounted for ∼50% of the overall inhibitory effect, and the contribution of urea to synergy amounted to up to 50% of the total synergistic effect (representing ∼19-fold enhancement of inhibition). As expected, inhibition by T20 was not enhanced by urea, supporting the notion that this chemical agent does not efficiently promote transition to a fully activated state that contains a formed HR1 coiled coil. However, urea enhanced neutralization by both PS samples, accounting for ∼50% of the total synergistic effect (representing ∼10-fold enhancement of inhibition) (Fig. 6C).
We examined the effects of urea on neutralization of three additional tier-2 isolates and the tier-3-like mutant 89.6(225I, 305E) (Fig. 6D). Urea enhanced neutralization of isolate 1058-11 by MAb 17b and PS by 58- and 23-fold, respectively. In contrast, at this concentration of JRC-II-191 (4 μM), the enhancing effects of urea were relatively limited for isolates PRB956 and 89.6(225I, 305E) and completely absent for isolate WEAU-d15. The data presented in Fig. 3D and 4B, which show that higher concentrations of JRC-II-191 or extended cold incubation times can enhance inactivation even after maximal structural effects are achieved, suggested that increased CD4M “pressure” may further destabilize Env. We therefore examined whether higher concentrations of JRC-II-191 could augment the effects of urea. We found that for wild-type 89.6 Env, urea-mediated synergy peaked at 20 μM, whereas for isolate 89.6(225I, 305E), urea-associated synergy continued to increase at higher JRC-II-191 concentrations (Fig. 6E). Consequently, neutralization of this stable tier-3-like Env was enhanced 72- or 167-fold when exposed to JRC-II-191 (at 12 μM), urea, and MAb 17b or PS, respectively (Fig. 6F). Therefore, the antibody sensitivity of tier-2- or tier-3-like Envs can be enhanced by increasing induction pressure to assume a perturbation-sensitive form (by extended incubation at 0°C or higher concentrations of CD4Ms) in association with urea-mediated destabilization.
DISCUSSION
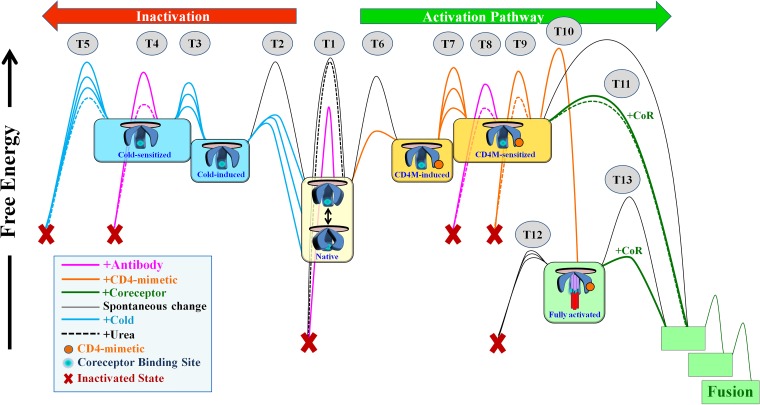
Architecture of the energy landscape of Env.
A large amount of potential energy is stored within the functional Env trimer and is harnessed to drive fusion between viral and cell membranes. This energy is retained by intramolecular interactions that maintain the integrity of the trimer and prevent changes to other conformations. The strength of these interactions defines the height of the energy barriers that keep Env in the native state (Fig. 7). Engagement of receptor or coreceptor disrupts specific interactions and allows Env to negotiate the barriers, facilitating transitions to defined intermediates along the activation pathway (4, 5, 8). Engagement of antibodies can also induce loss of energy by promoting changes to nonfunctional forms. Evolutionary selection of viruses with high infectivity and resistance to host antibodies dictates the requirement for conformational and functional stability of the native state. Disruption of barriers by mutations (22, 23, 56) or ligands (57) facilitates Env transfer between states (Fig. 7, numbered transitions [T]). The range of conformations that Envs can assume, the strengths of the interactions that maintain the integrity of each state, the conditions that facilitate transitions, and the reversibility of the changes define the architecture of the energy landscape of Env. We examined the effect of disrupting Env stability by three types of treatment: (i) cold, which globally weakens hydrophobic interactions; (ii) the chaotropic agent urea, which has local effects on interactions accessible to the chemical and the surrounding water shell (53, 54); and (iii) CD4Ms, which disrupt specific Env interactions. All three approaches modulate interactions that maintain trimer integrity (globally or locally), facilitating Env transitions to and between nonnative states. Such induced states are more sensitive to perturbations (e.g., antibody inactivation or coreceptor activation) and spontaneous loss of function.
FIG 7.
Model of the energy landscape of HIV-1 Env. Native (cleaved and unliganded) Env trimers on the surfaces of HIV-1 particles potentially sample different conformations; at equilibrium, the sampling frequency of each conformation is determined by its free energy. The structural integrity of native and nonnative states is maintained by intramolecular interactions that create energy barriers and prevent changes to other forms, functional or nonfunctional. The stability of the states is thus determined by the strength of such interactions (i.e., the height of the activation barriers) and the presence of agents that can allow Envs to negotiate them. In diverse HIV-1 strains, different interactions prevent transitions (T) between states, accounting for the diverse patterns of responsiveness to Env-perturbing agents (represented by similarly colored barriers connecting states). Engagement of CD4Ms and coreceptor facilitates transitions to states along the activation pathway (directed to the right in the schematic). Env can also change to nonfunctional forms. Incubation at 0°C or engagement of CD4Ms can induce changes to states that expose the CoR-BS. Increased exposure to these agents facilitates transitions to sensitized forms, which are more readily neutralized by antibodies and eventually inactivate in a spontaneous manner. Such changes occur at isolate-specific rates and are reversible; removal of the CD4Ms or cold allows Env to return to the closed functional native state. The chaotropic agent urea perturbs the unstable forms induced by cold or CD4Ms and enhances their sensitivity to antibodies and their rate of spontaneous inactivation (dashed lines). CD4M-sensitized Envs are responsive to coreceptor (i.e., they can infect CD4− cells); at the range of concentrations applied in the study, transition to the sensitized form is reversible. Higher concentrations of CD4Ms induce irreversible changes to a state characterized by formation of the HR1 coiled coil, which is unstable and rapidly decays (within minutes) to a nonfunctional form unless coreceptor is engaged (33).
Resistance of the robust native Env to structure-destabilizing treatments.
From the time of proteolytic cleavage in the producer cell to form the mature trimer, Env is maintained in the native state. Stability of the native state would presumably benefit HIV-1, since the time between virus budding and encounter with a CD4 receptor on a target cell (within the same host or during transmission) can extend over several hours. Therefore, the barriers should not be easily negotiated in a spontaneous manner. Properties of Envs circulating in the population are determined by their effects on virus fitness and resistance to antibodies and by constraints imposed during virus transmission (30–32, 58). The fact that native Env is unaffected by low concentrations of urea (both structurally and functionally [Fig. 7, T1]) may reflect the low-level (but extended) exposure of the virus to this denaturing chemical in semen (∼18 mM in human seminal plasma) (59). The urea-resistant nature of the native state may allow retention of Env functionality under these conditions. In contrast, cold affects both surface-exposed and cryptic elements. “Globally perturbing” pressure similar to that induced by cold is likely not applied on Env in vivo. Nevertheless, our data suggest that structural robustness (i.e., resistance to structural changes) and functional stability (resistance to neutralization) are likely associated with the strength of cold-sensitive interactions that maintain trimer integrity. Indeed, our studies in a rhesus macaque model of HIV-1 infection indicate that cold-sensitive viruses rapidly acquire resistance to cold exposure over the course of infection (38), likely due to association of the phenotype with antibody sensitivity (23).
Metastable, perturbation-sensitive states induced by cold and CD4Ms.
At 0°C, the Env trimer undergoes gradual rearrangement to a state characterized by increased exposure of the CoR-BS and decreased sampling of trimer-specific epitopes (Fig. 7, T2). Diverse tier-2 isolates have different propensities to undergo structural changes at 0°C, likely reflecting the strength (and/or number) of interactions that prevent such changes. If the pressure is relieved, many Envs can resume native structure and function. Accordingly, the cold-induced state is positioned at a higher energy level than the native state (Fig. 7). Such reversibility appears to be complete for the CoR-BS epitopes, although some residual changes (e.g., decreased sampling of trimer-dependent epitopes) may persist after return to 37°C. In the cold-induced open form, Env is functionally stable and resistant to antibody neutralization. Application of extended pressure (i.e., exposure time at 0°C) likely facilitates disruption of additional interactions and induces gradual change (T3) to a second (unstable) form that is sensitive to antibody neutralization (T4). The fact that changes in structure (Fig. 2F) precede the neutralization-sensitive form (Fig. 3D), which in turn precedes inactivation of function (Fig. 2D, and T5 in Fig. 7), suggests that Env assumes at least two phenotypically distinct forms upon exposure to cold: a functional and stable (“induced”) form followed by a functional but perturbation-sensitive (“sensitized”) form.
Similar to the effects of cold, exposure to low concentrations of CD4Ms induces a reversible transition (T6) to a conformational form that exhibits increased exposure of the CoR-BS. For some Envs, such changes are not accompanied by enhanced sensitivity to neutralization and spontaneous inactivation (Fig. 4B and E). Higher concentrations of CD4Ms promote transition (T7) to a form that exhibits enhanced neutralization sensitivity (T8), fusion with CD4− cells (T11), and spontaneous inactivation (T9) (Fig. 4B and E and 6E). Therefore, our findings also distinguish between (at least) two CD4M-induced forms; one functional (induced) and the other functional but unstable (sensitized). The propensity for transition between these states is isolate specific; 89.6 Env is more amenable to such changes, whereas 89.6(225I, 305E) is more resistant.
Both CD4M-induced forms are reversible; removal of the agent allows Env to resume native structure and function. In contrast, CD4M-induced transitions to the fully activated state (T10), which contains a formed HR1 coiled coil (60) and is committed to the activation pathway, are associated with significant restructuring of the trimer and are thus irreversible (2). The latter state is short-lived and decays within minutes (T12) to an inactive form unless CoR is engaged, allowing Env to proceed along the entry pathway (T13) (33). Formation of this activated intermediate requires concentrations of CD4Ms significantly higher than those applied in our study (a minimal JRC-II-191 concentration of ∼100 μM) (33). The fact that the low concentrations of CD4Ms applied in our study do not induce formation of the fully activated intermediate is also suggested by the reversibility of the observed effects (Fig. 4D and 5E) and by the absence of T20 inhibition enhancement (Fig. 6B).
The states induced at 0°C appear to be distinct from the CD4M-induced pathway. First, stabilities in the two states differ for many Envs. For example, Env WEAU-d15 is rapidly induced and inactivated by cold but is not inactivated in the open state induced by JRC-II-191 (Fig. 2A and 4B). Second, the states induced by cold and CD4Ms differ in their abilities to mediate CD4-independent infection; cold facilitates entry into CD4− cells inefficiently, even for 89.6 Env (∼1% of infection induced by CD4Ms) (Fig. 5F).
A recent study has suggested that different occupancy levels of the three Env protomers by CD4Ms define different functionally activated forms, as measured by the ability to mediate entry into CD4− cells (57). Here, we show that different treatments that destabilize Env structure can induce transitions to different Env forms that demonstrate distinct phenotypes. The four forms we describe may indeed correspond to different levels of Env protomer occupancy: the native state, structurally induced but stable (single occupancy), structurally induced and unstable (double occupancy), and irreversibly induced and fully activated (triple occupancy). Importantly, we show here that transitions among these forms can be controlled by agents that perturb the structure of proteins, allowing us to increase the sensitivity of the virus to neutralization.
Intrinsic reactivity of HIV-1 Envs.
Single-site mutations in gp120 or gp41 can increase the global sensitivity of Env to neutralization, often without changing the binding efficiencies of the antibodies (18–21, 61). Therefore, the responsiveness to the bound antibody also determines the outcome of the virus-antibody interaction (22). Such mutations likely facilitate Env transitions to forms that contain lower barriers for inactivation. Our results suggest that Env destabilization by chemical or temperature treatment acts in a similar manner, by inducing increased responsiveness to perturbations. For example, urea does not increase binding of MAb 17b in the absence or presence of cold or CD4Ms (Fig. 5C and data not shown) but enhances neutralization by the MAb in the presence of cold or CD4Ms by up to 166- and 58-fold, respectively (Fig. 6A and B). Therefore, global responsiveness to the bound antibody is increased, suggesting that a tier-1-like phenotype can also be induced by treatments that mobilize Env from the native state to perturbation-sensitive forms (compare the heights of the energy barriers that prevent antibody neutralization from the native state (T1) with those of similar barriers from the nonnative states (T4 and T8).
Hypersensitivity to antibodies is often associated with increased sensitivity to inactivation by cold and CD4Ms and increased responsiveness to coreceptor (manifested by infection of CD4− cells). This cluster of phenotypes collectively typifies HIV-1 Envs with high reactivity (23, 38, 56). Here, we observed that these reactivity-related phenotypes are more tightly associated in the context of closely matched Envs. For example, mutations at positions 225 and 305 of the tier-2 Env 89.6 increase the stability of native Env, as reflected by resistance to cold-induced structural changes (Fig. 2) and neutralization (Fig. 3A and C). This “hyperstabilized” Env also shows enhanced stability of the CD4M-induced state (manifested by resistance to urea-mediated inactivation) (Fig. 5B and 6F) and stability of the cold-induced state (manifested by absence of CD4− cell infection and urea-mediated inactivation) (Fig. 5F and 6E). Therefore, for the 89.6 panel of Envs, interactions that control sensitivity to antibodies, CD4Ms, cold, and urea overlap. For the diverse panel of isolates from clades B and C, the association between these phenotypes is less pronounced. For example, whereas sensitivities to structural changes by cold and CD4Ms are correlated (P = 0.04; Spearman correlation test), isolates can differ in their sensitivity to the two treatments. Isolate WEAU-d15 is sensitive to inactivation by cold but not by CD4Ms, whereas the opposite relationship is observed for isolate 7048-09221. Therefore, the sensitivities of Env interactions to each perturbing treatment (CD4Ms, cold, and antibody) overlap but still contain unique (isolate-specific) elements. The presence of a global perturbation sensitivity pattern is determined by this overlap. Accordingly, whereas the energy landscape in Fig. 7 is represented by a collection of states separated by isolate-specific barriers (implying that the barriers are independent of each other), a more accurate representation would acknowledge the potential association between them.
Enhancement of HIV-1 neutralization by facilitating Env transitions to perturbation-sensitive states and their destabilization.
Infection or vaccination often elicits low-potency antibodies that target relatively cryptic epitopes (22, 62, 63). Our data show that Env-destabilizing treatments can significantly increase neutralization by such antibodies. The fact that pooled sera from HIV-infected patients are enhanced suggests that antibody types normally elicited by infection can be rendered more potent. Neutralization of tier-3-like Envs [e.g., 89.6(225I, 305E)] can be significantly augmented in this manner by increasing Env transitions to perturbation-sensitive forms.
A wide range of inactivating agents have been tested as topical microbicides to prevent sexual transmission of HIV-1. Such agents include CCR5 inhibitors (64, 65), CD4Ms or other high-affinity Env-targeting ligands (66), and chemicals that directly inactivate virus (67). The efficacy of many inhibitors is hindered by the structural diversity of Envs circulating in the population. We describe a novel approach that is based on mobilization of Envs from the native state to metastable perturbation-sensitive forms. Thus, rather than direct inactivation of Env, this approach enhances the potency of other effectors (e.g., vaccine-elicited mucosal antibodies or topical microbicides). The conserved nature of many cryptic Env epitopes contributes to the efficacy of this strategy in inhibiting otherwise globally resistant (higher-tier) viruses. By broadening the range of chemical agents, we can likely further increase the potency of microbicides and antibodies elicited by vaccination.
MATERIALS AND METHODS
Antibodies and CD4-mimetic agents.
The MAbs indicated below were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. MAbs 447-52D and 39F, which target the V3 loop of Env, were contributed by Susan Zolla-Pazner and James Robinson, respectively (9, 68). James Robinson also provided MAbs 17b, 48d, and E51, which recognize gp120 epitopes that are induced by CD4 binding (44). MAb 10E8, which targets the membrane-proximal ectodomain region (MPER) of gp41 was contributed by Mark Connors (69). Hermann Katinger provided MAb 2G12, which targets a carbohydrate-dependent gp120 epitope (40), and MAb 2F5, which recognizes the MPER of gp41 (70). MAb IgG1 b12, which recognizes the CD4-binding site of gp120 (71, 72), was a kind gift from Dennis Burton. The International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Consortium kindly provided MAbs PG9 and PG16, which target trimer-dependent epitopes, and MAbs PGT121, PGT126, and PGT128 antibodies, which recognize glycan-dependent epitopes on gp120 (73–75). The nonneutralizing MAb F240 recognizes a nonhelical hydrophobic region on the gp41 ectodomain (76). MAb 7B2 recognizes the cluster I region of gp41 (77). MAb b6, which targets a CD4-BS epitope, was kindly provided by Peter Kwong (78). Plasmids encoding the heavy and light chains of MAb 3BC176 were kindly provided by Michel Nussenzweig; the MAb was purified as previously described (79). The CD4-Ig fusion protein is composed of the Fc region of human IgG1 linked to two copies of the first two N-terminal domains of the CD4 molecule. The CD4-Ig protein was produced and purified as previously described (60). Four-domain sCD4, which contains a 6-His tag, was purified using nickel-nitrilotriacetic acid (NTA) beads from the culture medium of a 293F producer cell line (80). The HIV-inhibitory peptide T20, which binds to the HR1 coiled coil of gp41, was provided by Trimeris/Roche through the NIH AIDS Reagent Program. The CD4-mimetic small-molecule inhibitor JRC-II-191 was synthesized by Joel Courter and Amos B. Smith III (42). Two separate pools of sera were generated, each from 16 different HIV-infected individuals, and heat inactivated at 55°C for 30 min before use.
Envelope glycoprotein constructs.
The Envs of HIV-1 isolate 89.6 and its variants were expressed from the pSVIIIenv plasmid under the control of the HIV-1 long terminal repeat (LTR) promoter (81). The construct was generated by replacing the KpnI(6347)-BamHI(8475) fragment of the above-mentioned plasmid with that of 89.6 (accession number U39362). Env amino acid numbering is based on positions in the HXBc2 strain (82). Mutations were introduced into the pSVIIIenv vector expressing the Envs by site-directed mutagenesis using the PfuUltra Hotstart PCR master mix (Stratagene) and the DpnI restriction enzyme. Complete sequencing of the entire Env gene was performed to verify that only the desired mutations were introduced.
A panel of Envs derived from transmitted/founder viruses or from viruses from chronically infected individuals were isolated as previously described (83, 84). The following clade C Envs were tested in this study (abbreviated isolate names and accession numbers are in parentheses): ZM414_200414_8 (ZM414, GU329416), 706010018_2E3 (7060-10018, FJ444047), 704010207_D11 (7040-10207, JQ777073), 704010028_F6 (7040-10028, JQ777039), 703010217_B6 (7030-10217, FJ443589), ZM249M-B10 (ZM249M, EU166862), 707010457.e10 (7070-10457, JQ779128), 703010167_w8_e15 (7030-10167, KC156213), 704809221_1B3 (7048-09221, FJ444116), 4403_A18 (4403, HM070677), and 702010432_w4_e16 (7020-10432, JQ779232). The clade B Envs tested in this study were WEAUd15.410.5017 (WEAU-d15, EU289202), 89.6 (89.6, U39362), 9010-09.A1.4924 (9010-09, EU575771), 700010040.C9.4520 (7000-10040, EU576418), 1058-11.B11.1550 (1058-11, AY331295), PRB956-04.B22.4267 (PRB956, EU576603), 1018-10.A5.1732 (1018-10, EU575091), 1053-07.B15.1648 (1053-07, EU575201), THRO.F4.2026 (THRO EU577077), and CH77E_flC7 (CH77E FJ496005). A list of all the Envs included in this study with accession numbers, antigenicity patterns at 25°C, and cold inducibility values can be found at http://www.haimlab.com/datasets. To improve the expression of these proteins, the entire Env gene and flanking regions (see below) were amplified from the original pCDNA3.1 expression vector and cloned into the pSVIIIenv vector using the InFusion system (Clontech). The primers used for amplifying the Envs of clade B viruses were Env5in and Env3in (85). For amplification and cloning of the Envs of clade C viruses, we used primers EnvA (5′-GGCTTAGGCATCTCCTATGGCAGGAAGAA-3′; nucleotides 5954 to 5982) and EnvN (5′-CTGCCAATCAGGGAAGTAGCCTTGTGT-3′; nucleotides 9145 to 9171).
Preparation of recombinant luciferase-expressing viruses.
Single-round, recombinant HIV-1 that expresses the luciferase gene was generated by transfection of human embryonic kidney (HEK) 293T cells (obtained from the American Type Culture Collection) using JetPrime transfection reagent (Polyplus). Briefly, cells were seeded in 6-well plates (8.5 × 105 cells per well) and transfected the next day with 0.4 μg of the HIV-1 packaging construct pCMVΔP1ΔenvpA, 1.2 μg of the firefly luciferase-expressing construct pHIvec2.luc, 0.4 μg of a plasmid expressing HIV-1 Env, and 0.2 μg of a plasmid expressing HIV-1 Rev. The next day, the transfection medium was changed to culture medium (Dulbecco's modified Eagle's medium [DMEM]-10% fetal bovine serum [FBS]). Virus-containing supernatants were collected on the following day, cleared of cell debris by low-speed centrifugation, and filtered through 0.45-μm filters. For virus samples that were subsequently attached to protein-binding plates, virus-containing preparations were first pelleted by ultracentrifugation at 100,000 × g for 2 h at 10°C and resuspended in phosphate-buffered saline (PBS). All the samples were then snap-frozen on dry ice immersed in ethanol for 15 min and stored at −80°C until use.
Infection by recombinant luciferase-expressing virus and antibody neutralization assays.
Canis familiaris thymus (Cf2Th) normal cells (obtained from the NIH AIDS Reagent Program) expressing CD4 and CCR5 (Cf2Th CD4+ CCR5+) or only CCR5 (Cf2Th CD4− CCR5+) were used as target cells for measuring infection. Approximately 5 h before infection, the target cells were detached from culture plates using PBS containing 7.5 mM EDTA and seeded in 96- or 384-well luminometer-compatible plates (at a density of 4.5 × 103 or 1.8 × 103 cells per well, respectively). For neutralization assays, virus preparations were incubated on ice or at 37°C in the absence or presence of the inhibitor for 2 h. Unless otherwise indicated, all neutralization assays were conducted using the following concentrations of inhibitors: PS(A) and PS(B), 1:50 dilution; PS(C) and PS(D), 1:200 dilution; 447-52d and 19b, 2 μg/ml; 10E8 and VRC01, 5 μg/ml; 17b, 3BC176, and 48d, 8 μg/ml; 2G12 and b12, 0.5 μg/ml; T20, 150 nM. Samples were then added to CD4+ CCR5+ cells, which were incubated for 3 days to allow infection. In experiments that examined the effect of urea on infection, cells were incubated with inhibitor (in the absence or presence of urea) for 6 h at 37°C. The cells were then washed and further cultured for 3 days. To measure infection, the medium was removed, and the cells were lysed with passive lysis buffer (Promega) and subjected to three freeze-thaw cycles. To measure luciferase activity, 100 μl of luciferin buffer (15 mM MgSO4, 15 mM KPO4 [pH 7.8], 1 mM ATP, and 1 mM dithiothreitol) and 50 μl of 1 mM d-luciferin potassium salt (Syd Laboratories, MA) were added to each sample in 96-well plates (30 μl and 15 μl for samples in 384-well plates). Luminescence was recorded using a Synergy H1 microplate reader (BioTek Instruments). The synergistic effects between two- and three-treatment combinations were calculated as described by equation 1 and equation 2, respectively.
Virus sensitivity to cold inactivation.
Recombinant viruses were generated as described above, diluted, and divided into aliquots (one sample for each prospective time point). All the samples were then snap-frozen on dry ice immersed in ethanol for 15 min and stored at −80°C. This freezing method significantly reduces virus inactivation during freezing/thawing. At different time points, the samples were thawed by incubation in a 37°C water bath for 2 min and then placed on ice. The last sample thawed was not incubated on ice. All the virus samples were then placed in a 37°C water bath for 2 min and added to Cf2Th CD4+ CCR5+ cells. Three days later, the cells were lysed and assayed for luciferase activity as described above.
Activation of cell-bound virus.
To measure activation of HIV-1 infection by cold and urea, recombinant viruses were suspended in DMEM-10% FBS supplemented with 20 mM HEPES, pH 7.3. The viruses were then spinoculated onto confluent monolayers of CD4+ CCR5+ or CD4− CCR5+ cells (cultured in 96-well plates) at 10°C for 2 h at 2,000 rpm. To test the effects of cold, the samples were then placed on ice or at 25°C for 2 or 4 h and then reequilibrated to 37°C. To test the effect of urea, postspinoculation, samples were incubated for 2 h at 37°C in the absence or presence of urea (250 mM). The culture medium was then replaced by DMEM-10% FBS equilibrated to 37°C, and the samples were further cultured for 3 days for infectivity using the luciferase activity.
Reversibility of the JRC-II-191-induced activated state.
To measure the reversibility of urea-enhanced inactivation of the JRC-II-191-induced state, recombinant viruses suspended in PBS were bound to 96-well protein-binding plates (PerkinElmer) by spinoculation for 2 h at 10°C. The plates were then blocked with DMEM-10% FBS supplemented with 20 mM HEPES (pH 7.3) for 2 h. Plate-bound viruses were incubated in the absence or presence of JRC-II-191 for 2 h, followed by removal of JRC-II-191 and incubation with 225 mM urea or 30 mM NaCl as a control for an additional 2 h. As controls, some viruses were first incubated with 225 mM urea or 30 mM NaCl for 2 h, followed by removal of the agent and addition of JRC-II-191 for 2 h. Samples were then washed with DMEM-10% FBS, and Cf2Th CD4+ CCR5+ cells (1.8 × 103 per well) were added and cultured for 3 days before luciferase activity was measured.
Cell-based ELISA to measure binding of probes to cell surface-expressed Envs.
Binding of antibodies and CD4-Ig to HIV-1 Env trimers expressed on HOS cells was measured using a modified protocol of the cell-based ELISA system described previously (22, 85). Briefly, HOS cells were seeded in 96-well plates (1.2 × 104 cells per well) and transfected after 6 h with 60 ng of plasmid expressing the Env, 6 ng of Rev-expressing plasmid, and 12 ng of Tat-expressing plasmid per well using 0.18 μl per well of JetPrime (Polyplus Inc.) transfection reagent. For experiments performed in 384-well plates, each well contained 4.5 × 103 cells, which were transfected with 26 ng of Env-expressing plasmid and 5.5 ng of a Tat-expressing plasmid using 0.08 μl JetPrime reagent. In all the experiments, a negative-control plasmid that contained a stop mutation in amino acid position 46 of Env (according to standard HXBc2 numbering [82]) was used to determine background binding to the cells. Three days after transfection, the cells were washed twice with blocking buffer (20 mg/ml bovine serum albumin [BSA], 1.8 mM CaCl2, 1 mM MgCl2, 25 mM Tris, pH 7.5, and 140 mM NaCl) and incubated with the probes in blocking buffer for 45 min. Unless otherwise indicated, all MAbs were added at 0.5 μg/ml, whereas CD4-Ig was added at 2 μg/ml. In some cases, antibodies were incubated with Env-expressing cells at 0°C or at 37°C. For this purpose, the cells were first equilibrated to 0°C by incubation on ice for 15 min, followed by addition of ice-cold blocking buffer containing the MAbs for 45 min. All the samples were then washed twice with blocking buffer equilibrated to the same temperature as that containing the primary antibody and 6 times with blocking buffer equilibrated to room temperature. Samples were then incubated with a horseradish peroxidase (HRP)-conjugated goat anti-human IgG polyclonal antibody preparation for 45 min. The cells were subsequently washed 6 times with blocking buffer and 6 times with washing buffer (140 mM NaCl, 1.8 mM CaCl2, 1 mM MgCl2, and 20 mM Tris, pH 7.5). HRP enzyme activity was determined after the addition of 35 μl per well of a 1:1 mixture of SuperSignal West Pico chemiluminescent peroxide and luminol enhancer solutions (Thermo Scientific) supplemented with 150 mM NaCl. Samples in 384-well plates were incubated with 25 μl of the reagent mixture. Light emission was measured with a Synergy H1 microplate reader.
Glutaraldehyde fixation of cell surface Env.
The sampling frequencies of different Env epitopes were measured during or after exposure of the Env-expressing cells to 0°C. For this purpose, HOS cells were transfected with Env constructs and 3 days later were fixed using glutaraldehyde, as previously described (22, 34). Briefly, the cells were first equilibrated to the desired temperature, washed 3 times with fixation buffer (140 mM NaCl, 1.8 mM CaCl2, 1 mM MgCl2, and 10 mM HEPES, pH 7.4), and then incubated for 15 min with fixation buffer (adjusted to the desired temperature) containing 5 mM glutaraldehyde. As controls, some samples were incubated with fixation buffer with no glutaraldehyde added. Glutaraldehyde activity was halted by addition of 50 mM glycine in fixation buffer, which was added to all samples. The cells were then washed twice with fixation buffer containing 25 mM glycine, twice with fixation buffer containing 12.5 mM glycine, and then twice with washing buffer. Subsequently, fixed and nonfixed samples were examined for binding of antibodies using the cell-based ELISA method described above.
Effect of cold on antibody binding to soluble gp120.
HEK293T cells were seeded in 6-well plates (1 × 106 cells/well) and transfected the next day with plasmids that expressed full-length Env and Tat (1.2 and 0.3 μg/well, respectively) using Lipofectamine LTX transfection reagent (Invitrogen). On the following day, the culture medium was replaced by Pro293sCDM serum-free medium (Lonza) supplemented with 10 μg/ml penicillin-streptomycin solution and 2 mM glutamine. The cells were incubated at 37°C for 2 days before the medium was harvested and cleared of cells and debris by centrifugation (2,000 × g for 5 min at room temperature), followed by filtration (0.45-μm-pore-size filter). Supernatants containing shed gp120 were then incubated at 25°C or 0°C for 7 h in the presence of MAb 17b or PGT128 (2 μg/ml). Protein A-Sepharose beads were then added in a total volume of 500 μl and further incubated for 2 h at 25°C to precipitate the antibody-bound gp120. The beads were then washed once with Pro293sCDM medium, three times with NP-40 buffer, and once with Tris-NaCl buffer (0.15 M NaCl, 10 mM Tris, pH 7.5). Subsequently, the beads were boiled in sample buffer containing 4 mM β-mercaptoethanol for 10 min. The precipitated proteins were separated by SDS-PAGE and blotted onto a polyvinylidene difluoride (PVDF) membrane. Western blots were probed with pooled sera from HIV-infected individuals and detected using HRP-conjugated goat anti-human IgG polyclonal antibody. Band intensity was quantified by densitometry using ImageLab 5.2.1 software (Bio-Rad).
Effect of urea on cell viability.
Cf2Th CD4+ CCR5+ cells cultured in 96-well plates (1.8 × 103 cells per well) were incubated at 37°C in DMEM-10% FBS supplemented with different concentrations of urea or with 30 mM NaCl as a control. After 6 or 15 h, the cells were washed twice with DMEM-10% FBS and further cultured for 3 days. The culture medium was then removed, and 100 μl of CellTiter-Glo luminescent viability assay reagent (Promega) was added to each well. Samples were then incubated for 10 min, and luminescence was detected using a Synergy H1 luminometer, which reflects the ATP content of the samples as a measure of cell culture viability.
ACKNOWLEDGMENTS
We are grateful to Beatrice Hahn for providing Envs of transmitted/founder and chronic-phase viruses from clades B and C.
The work was partly supported by a UIRF ICE Fund Award to H.H. J.J. was supported by an NIH Graduate Student Training Program Fellowship in Virology (NIH T32 AI007533-17). O.D. was supported in part by a fellowship from the Sloan Foundation. J.S. was supported by NIH grants AI24755, GM56550, and AI100645 and by a gift from the late William F. McCarty-Cooper.
We have no conflicting financial interests.
REFERENCES
- 1.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 2.Blumenthal R, Durell S, Viard M. 2012. HIV entry and envelope glycoprotein-mediated fusion. J Biol Chem 287:40841–40849. doi: 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci U S A 97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schon A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, Smith AB III, Sodroski J, Freire E. 2006. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry 45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ray N, Doms RW. 2006. HIV-1 coreceptors and their inhibitors. Curr Top Microbiol Immunol 303:97–120. [DOI] [PubMed] [Google Scholar]
- 7.Berger EA, Murphy PM, Farber JM. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol 17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 8.Brower ET, Schon A, Klein JC, Freire E. 2009. Binding thermodynamics of the N-terminal peptide of the CCR5 coreceptor to HIV-1 envelope glycoprotein gp120. Biochemistry 48:779–785. doi: 10.1021/bi8021476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 10.Yuan W, Bazick J, Sodroski J. 2006. Characterization of the multiple conformational states of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker. J Virol 80:6725–6737. doi: 10.1128/JVI.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guttman M, Cupo A, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. 2015. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun 6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rambaut A, Posada D, Crandall KA, Holmes EC. 2004. The causes and consequences of HIV evolution. Nat Rev Genet 5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 13.Barouch DH. 2008. Challenges in the development of an HIV-1 vaccine. Nature 455:613–619. doi: 10.1038/nature07352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoxie JA. 2010. Toward an antibody-based HIV-1 vaccine. Annu Rev Med 61:135–152. doi: 10.1146/annurev.med.60.042507.164323. [DOI] [PubMed] [Google Scholar]
- 15.Moore JP, Cao Y, Qing L, Sattentau QJ, Pyati J, Koduri R, Robinson J, Barbas CF III, Burton DR, Ho DD. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J Virol 69:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrin T, Loh TP, Vennari JC, Schuitemaker H, Nunberg JH. 1995. Adaptation to persistent growth in the H9 cell line renders a primary isolate of human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J Virol 69:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Rourke SM, Schweighardt B, Scott WG, Wrin T, Fonseca DP, Sinangil F, Berman PW. 2009. Novel ring structure in the gp41 trimer of human immunodeficiency virus type 1 that modulates sensitivity and resistance to broadly neutralizing antibodies. J Virol 83:7728–7738. doi: 10.1128/JVI.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Rourke SM, Schweighardt B, Phung P, Fonseca DP, Terry K, Wrin T, Sinangil F, Berman PW. 2010. Mutation at a single position in the V2 domain of the HIV-1 envelope protein confers neutralization sensitivity to a highly neutralization-resistant virus. J Virol 84:11200–11209. doi: 10.1128/JVI.00790-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang PF, Bouma P, Park EJ, Margolick JB, Robinson JE, Zolla-Pazner S, Flora MN, Quinnan GV Jr. 2002. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J Virol 76:644–655. doi: 10.1128/JVI.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park EJ, Gorny MK, Zolla-Pazner S, Quinnan GV Jr. 2000. A global neutralization resistance phenotype of human immunodeficiency virus type 1 is determined by distinct mechanisms mediating enhanced infectivity and conformational change of the envelope complex. J Virol 74:4183–4191. doi: 10.1128/JVI.74.9.4183-4191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haim H, Salas I, McGee K, Eichelberger N, Winter E, Pacheco B, Sodroski J. 2013. Modeling virus- and antibody-specific factors to predict human immunodeficiency virus neutralization efficiency. Cell Host Microbe 14:547–558. doi: 10.1016/j.chom.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB III, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog 7:e1002101. doi: 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding S, Tolbert WD, Prevost J, Pacheco B, Coutu M, Debbeche O, Xiang SH, Pazgier M, Finzi A. 2016. A highly conserved gp120 inner domain residue modulates Env conformation and trimer stability. J Virol 90:8395–8409. doi: 10.1128/JVI.01068-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradley T, Trama A, Tumba N, Gray E, Lu X, Madani N, Jahanbakhsh F, Eaton A, Xia SM, Parks R, Lloyd KE, Sutherland LL, Scearce RM, Bowman CM, Barnett S, Abdool-Karim SS, Boyd SD, Melillo B, Smith AB III, Sodroski J, Kepler TB, Alam SM, Gao F, Bonsignori M, Liao HX, Moody MA, Montefiori D, Santra S, Morris L, Haynes BF. 2016. Amino acid changes in the HIV-1 gp41 membrane proximal region control virus neutralization sensitivity. EBioMedicine 12:196–207. doi: 10.1016/j.ebiom.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunugi S, Tanaka N. 2002. Cold denaturation of proteins under high pressure. Biochim Biophys Acta 1595:329–344. doi: 10.1016/S0167-4838(01)00354-5. [DOI] [PubMed] [Google Scholar]
- 27.Privalov PL. 1990. Cold denaturation of proteins. Crit Rev Biochem Mol Biol 25:281–305. [DOI] [PubMed] [Google Scholar]
- 28.Kassa A, Finzi A, Pancera M, Courter JR, Smith AB III, Sodroski J. 2009. Identification of a human immunodeficiency virus type 1 envelope glycoprotein variant resistant to cold inactivation. J Virol 83:4476–4488. doi: 10.1128/JVI.02110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agrawal N, Leaman DP, Rowcliffe E, Kinkead H, Nohria R, Akagi J, Bauer K, Du SX, Whalen RG, Burton DR, Zwick MB. 2011. Functional stability of unliganded envelope glycoprotein spikes among isolates of human immunodeficiency virus type 1 (HIV-1). PLoS One 6:e21339. doi: 10.1371/journal.pone.0021339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker ZF, Iyer SS, Wilen CB, Parrish NF, Chikere KC, Lee FH, Didigu CA, Berro R, Klasse PJ, Lee B, Moore JP, Shaw GM, Hahn BH, Doms RW. 2013. Transmitted/founder and chronic HIV-1 envelope proteins are distinguished by differential utilization of CCR5. J Virol 87:2401–2411. doi: 10.1128/JVI.02964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilen CB, Parrish NF, Pfaff JM, Decker JM, Henning EA, Haim H, Petersen JE, Wojcechowskyj JA, Sodroski J, Haynes BF, Montefiori DC, Tilton JC, Shaw GM, Hahn BH, Doms RW. 2011. Phenotypic and immunologic comparison of clade B transmitted/founder and chronic HIV-1 envelope glycoproteins. J Virol 85:8514–8527. doi: 10.1128/JVI.00736-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haim H, Si Z, Madani N, Wang L, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, Gabuzda D, Smith AB III, Sodroski J. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog 5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haim H, Salas I, Sodroski J. 2013. Proteolytic processing of the human immunodeficiency virus envelope glycoprotein precursor decreases conformational flexibility. J Virol 87:1884–1889. doi: 10.1128/JVI.02765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cayabyab M, Karlsson GB, Etemad-Moghadam BA, Hofmann W, Steenbeke T, Halloran M, Fanton JW, Axthelm MK, Letvin NL, Sodroski JG. 1999. Changes in human immunodeficiency virus type 1 envelope glycoproteins responsible for the pathogenicity of a multiply passaged simian-human immunodeficiency virus (SHIV-HXBc2). J Virol 73:976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cayabyab M, Rohne D, Pollakis G, Mische C, Messele T, Abebe A, Etemad-Moghadam B, Yang P, Henson S, Axthelm M, Goudsmit J, Letvin NL, Sodroski J. 2004. Rapid CD4+ T-lymphocyte depletion in rhesus monkeys infected with a simian-human immunodeficiency virus expressing the envelope glycoproteins of a primary dual-tropic Ethiopian clade C HIV type 1 isolate. AIDS Res Hum Retroviruses 20:27–40. doi: 10.1089/088922204322749477. [DOI] [PubMed] [Google Scholar]
- 37.Etemad-Moghadam B, Rhone D, Steenbeke T, Sun Y, Manola J, Gelman R, Fanton JW, Racz P, Tenner-Racz K, Axthelm MK, Letvin NL, Sodroski J. 2001. Membrane-fusing capacity of the human immunodeficiency virus envelope proteins determines the efficiency of CD+ T-cell depletion in macaques infected by a simian-human immunodeficiency virus. J Virol 75:5646–5655. doi: 10.1128/JVI.75.12.5646-5655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGee K, Haim H, Korioth-Schmitz B, Espy N, Javanbakht H, Letvin N, Sodroski J. 2014. The selection of low envelope glycoprotein reactivity to soluble CD4 and cold during simian-human immunodeficiency virus infection of rhesus macaques. J Virol 88:21–40. doi: 10.1128/JVI.01558-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu JC, Chen F, Xu HR, Huang YF, Lu NQ. 2007. Standardization and quality control for determination of fructose in seminal plasma. J Androl 28:207–213. doi: 10.2164/jandrol.106.001552. [DOI] [PubMed] [Google Scholar]
- 40.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol 70:1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Etemad-Moghadam B, Sun Y, Nicholson EK, Karlsson GB, Schenten D, Sodroski J. 1999. Determinants of neutralization resistance in the envelope glycoproteins of a simian-human immunodeficiency virus passaged in vivo. J Virol 73:8873–8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madani N, Schon A, Princiotto AM, Lalonde JM, Courter JR, Soeta T, Ng D, Wang L, Brower ET, Xiang SH, Kwon YD, Huang CC, Wyatt R, Kwong PD, Freire E, Smith AB III, Sodroski J. 2008. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure 16:1689–1701. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiang SH, Doka N, Choudhary RK, Sodroski J, Robinson JE. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res Hum Retroviruses 18:1207–1217. doi: 10.1089/08892220260387959. [DOI] [PubMed] [Google Scholar]
- 44.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daar ES, Li XL, Moudgil T, Ho DD. 1990. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc Natl Acad Sci U S A 87:6574–6578. doi: 10.1073/pnas.87.17.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deen KC, McDougal JS, Inacker R, Folena-Wasserman G, Arthos J, Rosenberg J, Maddon PJ, Axel R, Sweet RW. 1988. A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature 331:82–84. doi: 10.1038/331082a0. [DOI] [PubMed] [Google Scholar]
- 47.Traunecker A, Luke W, Karjalainen K. 1988. Soluble CD4 molecules neutralize human immunodeficiency virus type 1. Nature 331:84–86. doi: 10.1038/331084a0. [DOI] [PubMed] [Google Scholar]
- 48.Wu X, Sambor A, Nason MC, Yang ZY, Wu L, Zolla-Pazner S, Nabel GJ, Mascola JR. 2008. Soluble CD4 broadens neutralization of V3-directed monoclonal antibodies and guinea pig vaccine sera against HIV-1 subtype B and C reference viruses. Virology 380:285–295. doi: 10.1016/j.virol.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin L, Stricher F, Misse D, Sironi F, Pugniere M, Barthe P, Prado-Gotor R, Freulon I, Magne X, Roumestand C, Menez A, Lusso P, Veas F, Vita C. 2003. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat Biotechnol 21:71–76. [DOI] [PubMed] [Google Scholar]
- 50.Melillo B, Liang S, Park J, Schon A, Courter JR, LaLonde JM, Wendler DJ, Princiotto AM, Seaman MS, Freire E, Sodroski J, Madani N, Hendrickson WA, Smith AB III. 2016. Small-molecule CD4-mimics: structure-based optimization of HIV-1 entry inhibition. ACS Med Chem Lett 7:330–334. doi: 10.1021/acsmedchemlett.5b00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Q, Ma L, Jiang S, Lu H, Liu S, He Y, Strick N, Neamati N, Debnath AK. 2005. Identification of N-phenyl-N′-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology 339:213–225. doi: 10.1016/j.virol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Madani N, Princiotto AM, Schon A, LaLonde J, Feng Y, Freire E, Park J, Courter JR, Jones DM, Robinson J, Liao HX, Moody MA, Permar S, Haynes B, Smith AB III, Wyatt R, Sodroski J. 2014. CD4-mimetic small molecules sensitize human immunodeficiency virus to vaccine-elicited antibodies. J Virol 88:6542–6555. doi: 10.1128/JVI.00540-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennion BJ, Daggett V. 2003. The molecular basis for the chemical denaturation of proteins by urea. Proc Natl Acad Sci U S A 100:5142–5147. doi: 10.1073/pnas.0930122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hua L, Zhou R, Thirumalai D, Berne BJ. 2008. Urea denaturation by stronger dispersion interactions with proteins than water implies a 2-stage unfolding. Proc Natl Acad Sci U S A 105:16928–16933. doi: 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leaman DP, Zwick MB. 2013. Increased functional stability and homogeneity of viral envelope spikes through directed evolution. PLoS Pathog 9:e1003184. doi: 10.1371/journal.ppat.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herschhorn A, Ma X, Gu C, Ventura JD, Castillo-Menendez L, Melillo B, Terry DS, Smith AB III, Blanchard SC, Munro JB, Mothes W, Finzi A, Sodroski J. 2016. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. mBio 7:e01598. doi: 10.1128/mBio.01598-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Madani N, Princiotto AM, Zhao C, Jahanbakhshsefidi F, Mertens M, Herschhorn A, Melillo B, Smith AB III, Sodroski J. 2017. Activation and inactivation of primary human immunodeficiency virus (HIV-1) envelope glycoprotein trimers by CD4-mimetic compounds. J Virol 91:e01880-16. doi 10.1128/JVI.01880-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP Jr, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A 110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Owen DH, Katz DF. 2005. A review of the physical and chemical properties of human semen and the formulation of a semen simulant. J Androl 26:459–469. doi: 10.2164/jandrol.04104. [DOI] [PubMed] [Google Scholar]
- 60.Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S, Chaiken I, Freire E, Smith AB III, Sodroski JG. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci U S A 101:5036–5041. doi: 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park EJ, Vujcic LK, Anand R, Theodore TS, Quinnan GV Jr. 1998. Mutations in both gp120 and gp41 are responsible for the broad neutralization resistance of variant human immunodeficiency virus type 1 MN to antibodies directed at V3 and non-V3 epitopes. J Virol 72:7099–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, Kwong PD, Nabel GJ, Mascola JR, Wyatt RT. 2009. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog 5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neff CP, Kurisu T, Ndolo T, Fox K, Akkina R. 2011. A topical microbicide gel formulation of CCR5 antagonist maraviroc prevents HIV-1 vaginal transmission in humanized RAG-hu mice. PLoS One 6:e20209. doi: 10.1371/journal.pone.0020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Veazey RS, Ketas TJ, Dufour J, Moroney-Rasmussen T, Green LC, Klasse PJ, Moore JP. 2010. Protection of rhesus macaques from vaginal infection by vaginally delivered maraviroc, an inhibitor of HIV-1 entry via the CCR5 co-receptor. J Infect Dis 202:739–744. doi: 10.1086/655661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gibson RM, Arts EJ. 2012. Past, present, and future of entry inhibitors as HIV microbicides. Curr HIV Res 10:19–26. doi: 10.2174/157016212799304616. [DOI] [PubMed] [Google Scholar]
- 67.Friend DR, Kiser PF. 2013. Assessment of topical microbicides to prevent HIV-1 transmission: concepts, testing, lessons learned. Antiviral Res 99:391–400. doi: 10.1016/j.antiviral.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 68.Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol 66:7538–7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol 67:6642–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, Lamacchia M, Garratty E, Stiehm ER, Bryson YJ, Cao YZ, Moore JP, Ho DD, Barbas CF III. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 72.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cavacini LA, Emes CL, Wisnewski AV, Power J, Lewis G, Montefiori D, Posner MR. 1998. Functional and molecular characterization of human monoclonal antibody reactive with the immunodominant region of HIV type 1 glycoprotein 41. AIDS Res Hum Retroviruses 14:1271–1280. doi: 10.1089/aid.1998.14.1271. [DOI] [PubMed] [Google Scholar]
- 77.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol 74:627–643. doi: 10.1128/JVI.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mouquet H, Klein F, Scheid JF, Warncke M, Pietzsch J, Oliveira TY, Velinzon K, Seaman MS, Nussenzweig MC. 2011. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PLoS One 6:e24078. doi: 10.1371/journal.pone.0024078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang X, Tomov V, Kurteva S, Wang L, Ren X, Gorny MK, Zolla-Pazner S, Sodroski J. 2004. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J Virol 78:12975–12986. doi: 10.1128/JVI.78.23.12975-12986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol 69:4413–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Korber B, Foley B, Kuiken C, Pillai S, Sodroski J. 1998. Numbering positions in HIV relative to HXBc2, p iii-102–iii-103. Los Alamos National Laboratory, Los Alamos, NM. [Google Scholar]
- 83.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol 82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abrahams MR, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping LH, Athreya GS, Treurnicht FK, Keele BF, Wood N, Salazar-Gonzalez JF, Bhattacharya T, Chu H, Hoffman I, Galvin S, Mapanje C, Kazembe P, Thebus R, Fiscus S, Hide W, Cohen MS, Karim SA, Haynes BF, Shaw GM, Hahn BH, Korber BT, Swanstrom R, Williamson C, CAPRISA Acute Infection Study Team, Center for HIV-AIDS Vaccine Immunology Consortium. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol 83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.DeLeon O, Hodis H, O'Malley Y, Johnson J, Salimi H, Zhai Y, Winter E, Remec C, Eichelberger N, Van Cleave B, Puliadi R, Harrington RD, Stapleton JT, Haim H. 2017. Accurate predictions of population-level changes in sequence and structural properties of HIV-1 Env using a volatility-controlled diffusion model. PLoS Biol 15:e2001549. doi: 10.1371/journal.pbio.2001549. [DOI] [PMC free article] [PubMed] [Google Scholar]