ABSTRACT
Interactions between hepatitis C virus (HCV) and lipoproteins in humans play an important role in the efficient establishment of chronic infection. Apolipoprotein E (ApoE) on the HCV envelope mediates virus attachment to host cells as well as immune evasion. This interaction is thought to occur in hepatocytes, as ApoE plays dual functions in HCV assembly and maturation as well as cell attachment. In the present study, we found that secreted ApoE (sApoE) can also bind to viral particles via its C-terminal domain after HCV is released from the cell. Furthermore, the binding affinity of interactions between the sApoE N terminus and cell surface receptors affected HCV infectivity in a dose-dependent manner. The extracellular binding of sApoE to HCV is dependent on HCV envelope proteins, and recombinant HCV envelope proteins are also able to bind to sApoE. These results suggest that extracellular interactions between HCV and sApoE may potentially complicate vaccine development and studies of viral pathogenesis.
IMPORTANCE End-stage liver disease caused by chronic HCV infection remains a clinical challenge, and there is an urgent need for a prophylactic method of controlling HCV infection. Because host immunity against HCV is poorly understood, additional investigations of host-virus interactions in the context of HCV are important. HCV is primarily transmitted through blood, which is rich in lipoproteins. Therefore, it is of interest to further determine how HCV interacts with lipoproteins in human blood. In this study, we found that secreted ApoE (sApoE), an exchangeable component found in lipoproteins, participates in extracellular interactions with HCV virions. More significantly, different variants of sApoE differentially affect HCV infection efficiency in a dose-dependent manner. These findings provide greater insight into HCV infection and host immunity and could help propel the development of new strategies for preventing HCV infection.
KEYWORDS: CRISPR-Cas9, hepatitis C virus, infectivity, secreted apolipoprotein E, envelope protein
INTRODUCTION
Approximately 170 million people are chronically infected with hepatitis C virus (HCV) worldwide (1). Most HCV infections become chronic in humans and can progress to hepatic fibrosis, cirrhosis, or even hepatocellular carcinoma (2). This process is normally accompanied by high morbidity due to extrahepatic manifestations, including mixed cryoglobulinemia, diabetes mellitus, and atherosclerosis (3). In most developing countries, the current standard treatment for chronic hepatitis C is a combination of pegylated interferon with ribavirin (4), although unfortunately, the effects are unsatisfactory. Recently, highly efficacious direct-acting antiviral drugs (DAAs) have been approved; however, their clinical benefits may be limited by their lack of improvement of HCV-associated end-stage liver disease, their high cost, the emergence of drug-resistant HCV variants, and the potential for HCV reinfection in previously cured patients. Therefore, HCV might not be eliminated in the short term (5), and as a consequence, a prophylactic vaccine is highly desirable. Because the development of efficient HCV vaccines has proven difficult, further understanding of the molecular mechanisms underlying HCV infection is necessary.
HCV is a single-stranded, positive-sense RNA virus of the Flaviviridae family (6). The HCV genome is ∼9.6 kb in length and encodes a long polyprotein (of more than 3,000 amino acids [aa]) that is proteolytically processed to generate 10 mature viral proteins. Viral structural proteins reside in the N-terminal third of the polyprotein and include core or capsid protein (C) and the envelope glycoproteins E1 and E2. p7 (a viroporin) and nonstructural proteins are encoded in the remaining C-terminal two-thirds of the polyprotein; these proteins play a variety of roles in virus assembly and RNA replication (7–9). HCV virions consist of a nucleocapsid containing the viral genome enveloped by an endoplasmic reticulum (ER)-derived lipid bilayer in which E1 and E2 are assembled as heterodimers (10, 11).
Highly efficient establishment of chronic HCV infection relies on not only the effective inhibition of the host's innate immunity through the activities of viral proteins (12–17) but also the chimeric formation of lipoviral particles (LVPs) by HCV virions and blood lipoproteins, which allow HCV to efficiently spread through blood vessels and effectively escape from host humoral immunity (18, 19). These relationships imply that lipoprotein components could be involved in the mechanism of HCV escape from humoral immunity.
Apolipoprotein E (ApoE) is abundant in plasma (20 to 50 μg/ml), where it functions as an exchangeable surface ligand for several classes of lipoproteins to facilitate receptor recognition and lipid transport regulation, and it is also involved in immune regulation and nerve tissue regeneration (20, 21). ApoE is polymorphic, with three common alleles (ApoE2 [Cys112 and Cys158], ApoE3 [Cys112 and Arg158], and ApoE4 [Arg112 and Arg158]) and tens of rare alleles (22). The ApoE isoforms are classified based on their relative charge. Different mutations causing the same migration pattern after isoelectric focusing define the different isoform subtypes. Although these allelic forms differ by only one or two amino acids, the differences often alter the structure and function of the protein. According to crystallography studies, a hinge region connects the N- and C-terminal regions of ApoE (23, 24). The N-terminal region (residues 1 to 167) forms an antiparallel four-helix bundle that contains a receptor-binding site (residues 136 to 150) (25). The C-terminal domain (residues 206 to 299) contains three α-helices that form a large exposed hydrophobic surface and that interact with residues in the N-terminal helix bundle domain through hydrogen bonds and salt bridges (26).
In an in vitro hepatocyte culture model, the role of ApoE in the HCV replication cycle was elucidated in several previous studies. For example, small interfering RNA (siRNA)-induced downregulation of cellular ApoE (cApoE) expression led to decreased HCV yield and infectivity (27–29). Moreover, interactions of cApoE with the viral protein NS5A or the viral envelope proteins have also been detected (30–34). As a consequence, cApoE is thought to be involved in HCV virion assembly and maturation (34, 35). Furthermore, immunoelectron microscopy has indicated the presence of unequal numbers of ApoE molecules on the surfaces of HCV LVPs, and it was found that HCV LVP attachment to cells is mediated through the binding of ApoE to cell surface heparin sulfate (36–39).
It has been shown that during the spread of dengue virus through blood vessels, the virus can associate with host antibodies to alter infectivity in vivo. The antibody-dependent enhancement (ADE) formed by dengue virus is an important factor to consider when studying the pathogenic mechanism of the virus and when developing vaccines (40, 41). Notably, both dengue virus and HCV belong to the Flaviviridae family. Secreted ApoE (sApoE) is abundant in blood and can dissociate from the lipoprotein surface (42, 43). Dissociated free sApoE is structurally labile and can rapidly bind to other lipoproteins, become recruited to the plasma membrane, or associate with other complexes. Therefore, we sought to understand whether sApoE molecules on lipoproteins in blood could associate with HCV to affect viral infectivity and whether these interactions are involved in the immune escape mechanism of the virus.
RESULTS
Binding of secreted ApoE to HCV particles.
The liver is the major organ of ApoE synthesis, and several studies have reported that HCV can associate with cApoE before being secreted from infected hepatocytes during the HCV LVP assembly and release process. However, there are high concentrations of sApoE protein present in blood, and the ApoE molecules found on HCV LVPs are likely obtained from extracellular exchange of sApoE, although there is currently no clear understanding of the details defining these extracellular interactions.
To distinguish cApoE carried by HCV virions assembled in the Huh7.5 cell line, we used sApoE protein tagged with anti-hemagglutinin (anti-HA) secreted by heterologous cells and its deletion mutant in this study (Fig. 1A). It has been shown that using a flexible linker (SGGRGG) between the HA tag and the C terminus of ApoE does not affect the secretion or normal biological function of the protein (44). ApoE expression was not detected in the cell culture medium or cell lysates of 293T cells, which eliminates the concern of interference due to endogenous ApoE. sApoE or sApoE-HA was secreted into the cell culture medium following transient transfection of pcDNA3-ApoE3 or pCDNA3-ApoE-HA into 293T cells. Density gradient centrifugation analysis confirmed that sApoE-HA was in a lipoprotein-associated state, equivalent to sApoE (Fig. 1B), and could therefore be used as a mimic of sApoE in blood.
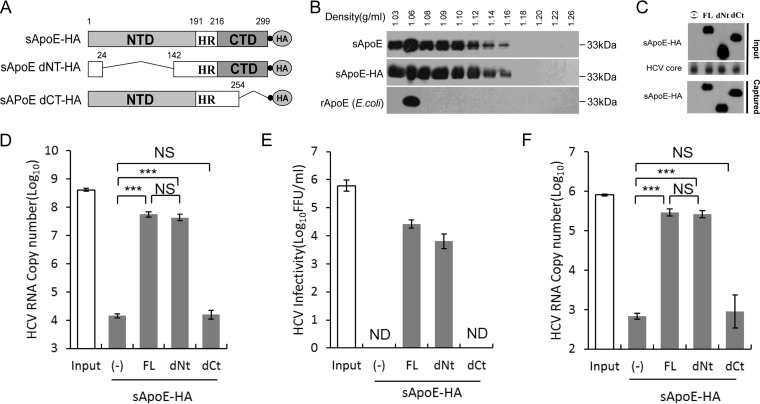
FIG 1.
Secreted ApoE interacts with extracellular HCV particles in a manner dependent on the C-terminal domain of ApoE. (A) Schematic representation of HA-tagged, full-length, N- or C-terminal domain deletion mutants of sApoE. (B) Isopycnic gradient centrifugation analysis showing the distribution of exogenously expressed sApoE or sApoE-HA in 293T cell culture medium as well as recombinant ApoE (rApoE) protein expressed by E. coli and run through a density gradient as a control. ApoE protein was detected by Western blotting with WUE-4 anti-ApoE antibody. (C) sApoE-HA and concentrated HCV core proteins in cell culture media were detected by Western blotting and are shown at the top. Proteins are specified on the left. The mixtures were separately captured by HA-specific immunoprecipitation, and the presence of sApoE-HA in the immunocomplexes was verified by Western blotting using HA antibody (bottom). (D) HCV genome copies in the immunocomplexes were quantified using qRT-PCR. (E) Determination of the infectivity of immunocaptured HCV particles. Equivalent amounts of HCVcc particles and full-length sApoE-HA or deletion mutants were mixed and subjected to HA affinity capture. Captured particles were eluted with an HA peptide. Infectivity titers in the input and the corresponding eluent were determined by FF-IHC. (F) PHH derived-HCV genomes in the immunocomplexes were pulled down using anti-HA agarose and quantified using qRT-PCR. The data in panels D to F represent the mean values and standard deviations from three independent experiments. **, P < 0.01; ***, P < 0.001 (unpaired Student's t test). NS, not significant; ND, not detected.
We mixed sApoE-HA secreted from 293T cells with HCV derived from a cell culture system (HCVcc) and captured the HA antibody. Compared with 293T cell supernatant, which did not express sApoE (Fig. 1C), beads coupled with the HA antibody could efficiently pull down the HCV genome when associated with sApoE-HA protein (Fig. 1D). To confirm that sApoE-HA could associate with infectious HCV virions, we then used the HA peptide to competitively elute the immunoprecipitate associated with the HA antibody. Infectious HCV virus was detected in the eluent containing sApoE-HA (Fig. 1E), indicating that extracellularly secreted sApoE-HA protein can associate with infectious HCVcc.
According to crystallography studies, there are two independent domains in the ApoE protein: the N-terminal domain (NTD) and the C-terminal domain (CTD). We therefore designed a pulldown assay using the HA antibody to identify which ApoE domain interacts with HCV by using secreted sApoE-HA mutated so that either the NTD or CTD was removed (dNT or dCT sApoE-HA, respectively) (Fig. 1A). Our results showed that the ApoE protein with no NTD interacted with HCV in a manner similar to that of full-length ApoE protein (Fig. 1D and E), whereas the ApoE protein with no CTD could not associate with HCV virions. These results suggest that the interaction of sApoE-HA with extracellular HCVcc virions depends on the CTD of ApoE.
It has been reported that Huh7-derived cells, which are deficient in very-low-density lipoprotein (VLDL) secretion, do not possess typical lipoprotein production machinery. Therefore, to reflect the natural behavior of authentic viruses, we also detected whether HCV particles produced by primary human hepatocytes (PHHs) could bind to sApoE. To accomplish this, PHH-derived HCV particles were separately mixed with sApoE-HA mutants (FL-HA, dNt-HA, and dCt-HA) secreted by 293T cells and subjected to pulldown using anti-HA agarose. HCV viral genome copies were then measured. The results (Fig. 1F) indicated that sApoE can also bind with PHH-derived HCV particles and that this process is also dependent on the CTD of ApoE.
ApoE polymorphisms influence HCV infectivity in a dose-dependent manner.
ApoE exhibits genetic polymorphisms among the population, and these variants differ in their receptor binding affinities. Clinical studies have shown that HCV susceptibilities vary among populations with different ApoE genotypes. Therefore, we hypothesized that differences in sApoE variants may directly affect HCV infectivity.
To investigate whether different sApoE variants differentially affect HCV infectivity, we examined four ApoE natural variants in humans, termed ApoE1 (Harrisburg type) (45), 2, 3 and 4 (Table 1). We obtained cell culture medium from 293T cells individually containing each of the 4 sApoE variants, as well as a blank control (Fig. 2A). Equal amounts of HCVcc were separately mixed with the cell supernatants, and the mixtures were then used to infect Huh7.5 hepatocytes (Fig. 2B). For accurate measurement of infectivity, HCV viral titers were determined using focus forming assays based on immunohistochemistry (FF-IHC).
TABLE 1.
List of ApoE variants used in this study
| Isoform | Amino acid at position (1–299): |
Charge relative to ApoE3 | Variant to which the isoform corresponds (dsSNPa identifier) | |||
|---|---|---|---|---|---|---|
| 112 | 139 | 146 | 158 | |||
| ApoE1 | Cys | Ser | Glu | Arg | −2 | rs121918394 |
| ApoE2 | Cys | Ser | Lys | Cys | −1 | rs7412 |
| ApoE3b | Cys | Ser | Lys | Arg | 0 | |
| ApoE4 | Arg | Ser | Lys | Arg | +1 | rs429358 |
| ApoE4Ac | Cys | Arg | Lys | Arg | +1 | |
dbSNP URL: https://www.ncbi.nlm.nih.gov/SNP/.
Represents the prototype ApoE (UniProt accession number P02649 and GenBank accession number K00396).
Represents an artificial ApoE mutant.
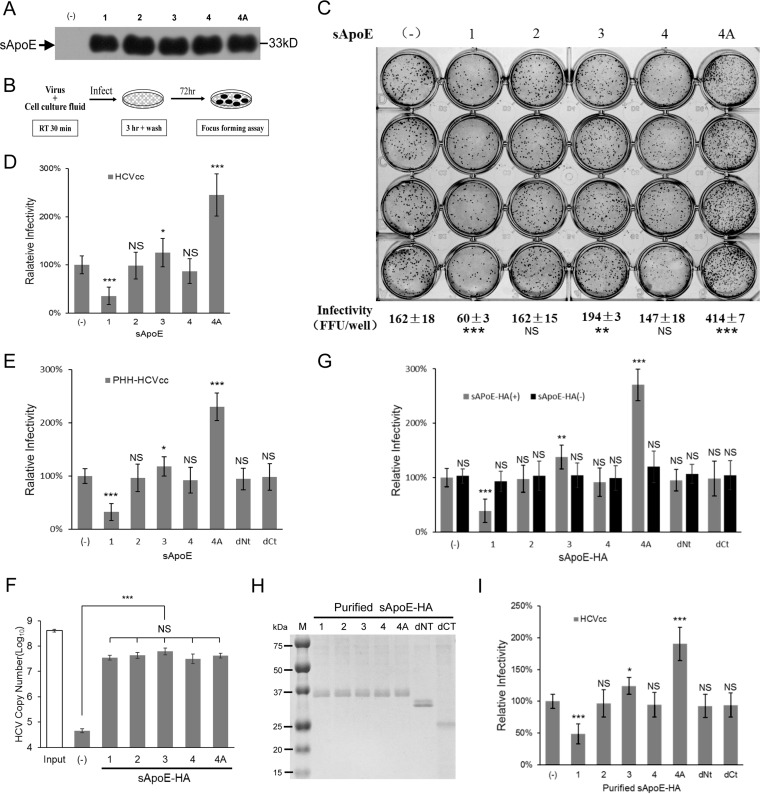
FIG 2.
Infectivity of HCV is influenced by sApoE isoform. (A) Forty-eight hours after transient transfection of plasmids, the blank sample and five sApoE variants were secreted by 293T cells and detected in cell culture media by Western blotting. (B) Schematic diagram of the experimental procedure. (C to E) Equivalent amounts of HCVcc derived from Huh7.5 (C and D) or PHHs (E) were mixed with the same amounts of cell culture media including different sApoE variants and deletion mutants used to infect Huh7.5 cells in a 24-well cell culture plate. The infectivity of HCV was determined by FF-IHC in quadruplicate. (F) The ability of different sApoE isoforms and mutants to interact with HCV. Equivalent amounts of HCVcc particles and different full-length sApoE isoforms and mutants were mixed and subjected to HA affinity capture. HCV genome copies in the immunocomplexes were quantified using qRT-PCR. (G) The same amounts of cell culture media including different sApoE-HA variants or anti-HA agarose as used to remove sApoE-HA were mixed with equivalent amounts of HCVcc and used to infect Huh7.5 cells. The infectivity of HCV was determined. (H) Different sApoE-HA variants immunoprecipitated by anti-HA agarose were eluted with EF solution and assayed by SDS-PAGE utilizing Coomassie blue staining. (I) The effects of purified ApoE variants and deletion mutants on HCV infectivity were detected. The HCV infectivity data are shown as the mean ± SDs pooled from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired Student's t test). NS, not significant.
The supernatant containing sApoE1 exhibited lower receptor affinity than did the 293T culture supernatant without sApoE and led to a significant decrease in HCVcc infectivity (Fig. 2C and D). Among the three most common ApoE variants (ApoE2, -3, and -4) in the population, the sApoE3 supernatant led to a slight increase in HCVcc infectivity, whereas the sApoE4 supernatant decreased HCVcc infectivity.
It has been shown that increasing the positive charge on the ApoE receptor interaction area (aa 136 to 150) can enhance the affinity of ApoE in binding to heparan sulfate proteoglycan (HSPG) on the hepatocyte cell surface. We therefore replaced the serine at amino acid position 139 in ApoE3 with arginine to artificially construct an ApoE4A mutant with an additional positive charge and therefore higher receptor affinity for HSPG (Table 1). The sApoE4A mutant secreted by 293T cells was mixed with an equal amount of HCVcc, and viral infectivity was measured (Fig. 2A and B). As expected, the results indicated that the sApoE4A mutant significantly increased the infectivity of HCVcc in vitro (Fig. 2C); similar results were obtained upon repeating the experiment (Fig. 2D).
To measure the effects on infectivity caused by different ApoE variants using PHH-derived HCV, the same method as that described above was applied. The results indicated that different ApoE variants altered the infectivity of HCV particles derived from PHHs (Fig. 2E). To further ensure that these alterations were specifically caused by sApoE variants in the supernatant, we expressed sApoE1, -2, -4, and -4A as well as sApoE3-HA, all carrying the HA tag. We found that the sApoE1, -2, -3, -4, and -4A were all able to associate with HCV particles (Fig. 2F). Furthermore, the infectivity results (Fig. 2G) showed that the presence of sApoE caused fluctuations in HCV infectivity compared to results from experiments using a cellular supernatant control containing no ApoE. Likewise, when sApoE-HA proteins were removed from the cell supernatant using anti-HA agarose, HCV infectivity was no longer altered. After this, we allowed HCV particles to bind to purified ApoE-HA protein (Fig. 2H), and the resultant effects on HCV infectivity were again observed (Fig. 2I). The results indicated that the presence of sApoE in the supernatant was the key factor altering HCV infectivity.
Because the detection of high-concentration HCVcc titers requires continuous serial dilution, the concentration of sApoE in solution is also diluted during these measurements. Therefore, in order to examine the dose dependency of the relationship between HCV infectivity and sApoE concentration in the supernatant, we used HCV reporter virus, which has a wider linear range that allows detection with no serial dilution (Fig. 3A). We used sApoE1, -3, and -4A proteins secreted by heterologous cells and the sApoE deletion mutant in this study (Fig. 3B). As sApoE1 has the lowest receptor affinity, amounts of reporter virus equivalent to those in the experiments described above were used, with exposure to increasing concentrations of sApoE1. The results showed a dose-dependent relationship, with increasing sApoE1 concentrations resulting in continuously decreasing HCV infectivity. When the sApoE1 concentration approached the level found in human blood (20 μg/ml), HCV infectivity was decreased by 90% (Fig. 3C). Moreover, as the concentration of sApoE3 in the mixture gradually increased, the HCV infectivity first increased and then decreased (Fig. 3D). In contrast, as the concentration of sApoE4A in the mixture gradually increased, the infectivity of HCV quadrupled (Fig. 3E). These results suggest that both the concentration and the receptor affinity of sApoE in the host alter the infectivity of HCV.
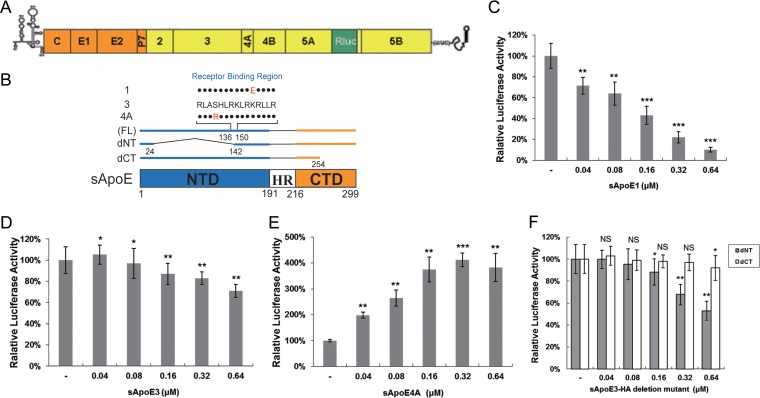
FIG 3.
Infectivity of reporter HCV is influenced by sApoE in a dose-dependent manner. (A) Genomic organization of the JFH1-derived luciferase reporter construct (HCV-Rluc). (B) Schematic representation of full-length (FL; 1, 3, and 4A) and N- or C-terminal domain deletion mutants of sApoE. (C to F) Equivalent amounts of reporter HCV were incubated with increasing concentrations of sApoE1 (C), sApoE3 (D), sApoE4A (E), or sApoE3 deletion mutants (F) at room temperature for 30 min and subsequently used to infect Huh7.5 cells in 24-well culture plates. At 5 h h.p.i., the mixture of HCV-Rluc and sApoE was removed, and the cells were washed twice with PBS and then incubated with DMEM containing 10% FBS. After 24 h, the cells were lysed for luciferase assays. The quantitative results derived from the data were converted to percentages of the control in the bar graph. The data are shown as the mean ± SDs (error bars) pooled from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired Student's t test). NS, not significant.
Theoretically, sApoE alters viral infectivity not only by associating with HCV but also by competing with HCV for receptor binding. It has been shown that ApoE has two functional domains: the NTD, which can competitively associate with receptors, and the CTD, which can associate with HCV particles. Notably, each of these domains can separately inhibit HCV infectivity. Under the experimental conditions used in this study, the ApoE construct with no NTD exhibited stronger inhibition of HCV infectivity (Fig. 3F) than the ApoE construct with no CTD, indicating that the effect of CTD of this protein on HCV infectivity was more notable than that of NTD competitively associated with the HCV receptor.
Secreted ApoE binds HCV independent of cellular ApoE.
It has been shown that ApoE can be expressed in liver cells and secreted extracellularly. ApoE is also able to form homodimers. Previous reports have indicated that HCVcc can associate with intracellular cApoE. Therefore, we further investigated whether the interaction between sApoE and HCVcc depends on the virus associating with cApoE intracellularly.
To test this dependence, it was necessary to examine the behavior of HCVcc particles that had not been exposed to cApoE. Using siRNA to knock down host cellular ApoE expression in Huh7-HCV cells would not be sufficient for this purpose, as siRNA cannot completely eliminate ApoE expression.
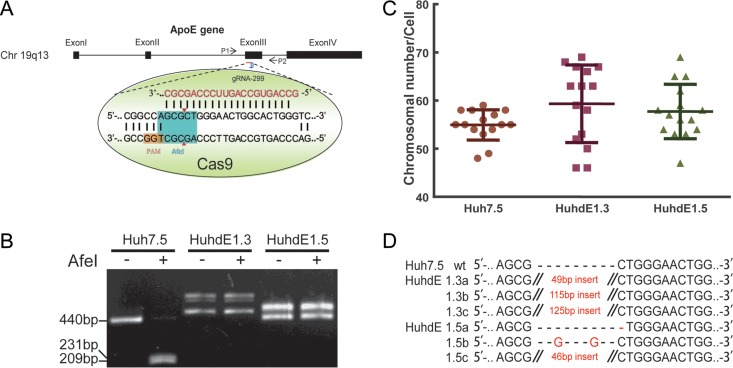
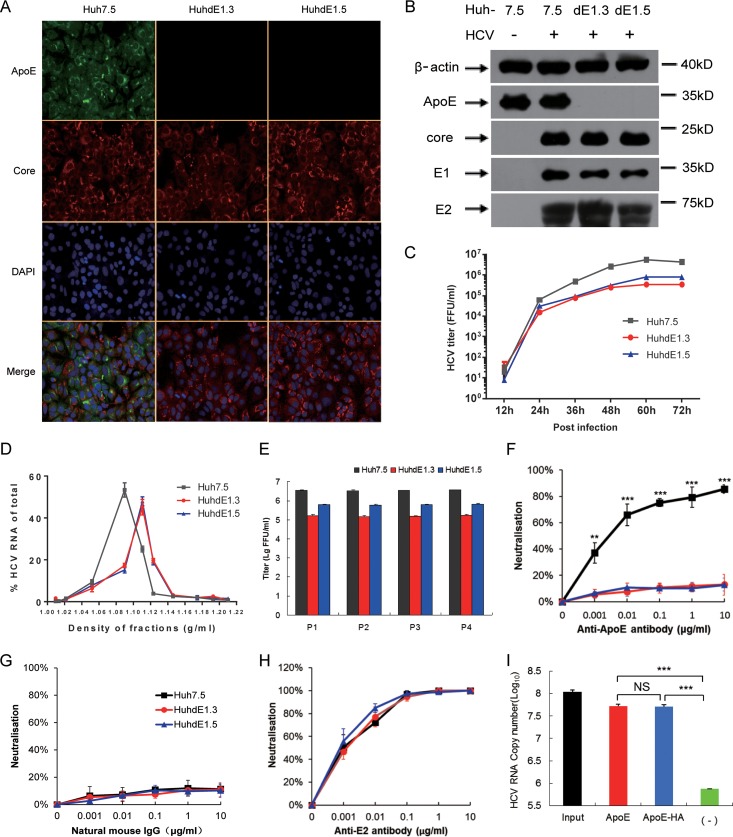
To overcome this limitation, we utilized the recently developed genome editing technology known as CRISPR-Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein). This system can induce site-specific cleavage of the cellular genome and preferentially introduce an indel mutation while the genome is repaired via error-prone nonhomologous end joining (NHEJ). For detection convenience, we chose the AfeI restriction site on the third exon of the ApoE gene as our target (Fig. 4A). The CRISPR-Cas9 system was transfected into Huh7.5 cells for transient expression. After two rounds of single-cell clone selection and amplification, ApoE protein expression was undetectable in 2 cell clones, which were named HuhdE1.3 and HuhdE1.5. To confirm the absence of ApoE, we extracted the genomes of the cells and subjected them to analysis via PCR and AfeI enzyme digestion. Only 1 PCR product was amplified from wild-type Huh7.5 cells, and the PCR product could be digested with AfeI. In contrast, multiple PCR products were amplified from both the HuhdE1.3 and HuhdE1.5 cells, and AfeI digestion was no longer possible (Fig. 4B). These results suggested that no wild-type ApoE gene was present in either of the modified cell lines. Additionally, karyotype analysis showed that the wild-type and derivative Huh7 cells were hypotriploid (Fig. 4C). Next-generation sequencing analysis showed 3 ApoE mutants in both the HuhdE1.3 and HuhdE1.5 cells (Fig. 4D), and the wild-type ApoE sequence was not detected. Amino acid sequence analysis revealed that the open reading frame of the ApoE gene was destroyed in both the HuhdE1.3 and HuhdE1.5 cells and that the cells no longer encoded or produced full-length ApoE protein (<100 aa). Thus, we successfully constructed an ApoE gene knockout hepatocyte cell line using the CRISPR-Cas9 system.
FIG 4.
Establishement of ApoE knockout human hepatic cell lines. (A) Schematic showing the CRISPR-Cas9 system and the target site for gRNA 299 in the ApoE gene locus used in this study. Protospacer adjacent motif (PAM) sequences are in orange. Fragments amplified by p1 with p2 and the AfeI restriction site (in aqua blue) were used in a restriction fragment length polymorphism gel assay. (B) Restriction fragment length polymorphism gel analysis. The CRISPR-Cas9 system was transiently expressed in Huh7.5 cells. After two rounds of subcloning and testing for protein expression, two monoclonal ApoE knockout cell lines were isolated and termed HuhdE1.3 and HuhdE1.5. The short lines indicate fragments generated by AfeI digestion. (C) Karyotype analysis of the Huh7.5 and ApoE knockout cell lines, Huh7dE1.3 and Huh7dE1.5. (D) Sequences of indel mutations identified in the ApoE gene using next-generation sequencing for each cell line. Red dashes, deletions; red bases, insertions.
We next used HCVcc to infect Huh7.5, HuhdE1.3, and HuhdE1.5 cells, and all three cell lines were then infected with HCV (Fig. 5A). HCV protein expression was not inhibited by ApoE knockout in any of the cell lines (Fig. 5B), but viral infectivity was reduced (Fig. 5C), and the density distribution of virus particles over an iodixanol density gradient also shifted toward a higher density (Fig. 5D). HCV was able to be passaged both in Huh7.5 cells and in the ApoE gene knockout HuhdE1.3 and HuhdE1.5 cells (Fig. 5E). To further confirm the absence of ApoE on the HCV particles, the particles were subjected to a neutralization assay using ApoE antibody. In contrast to the HCV produced in Huh7.5 cells, the viral particles produced in the ApoE gene knockout cell lines could not be neutralized by the ApoE antibody (Fig. 5F and G), whereas viral particles from all three cell lines could be neutralized using an E2 antibody (Fig. 5H). After confirming that no cApoE was present on the HCV particles, HCV produced in HuhdE1.5 cells was mixed with sApoE-HA or sApoE for a pulldown assay using beads coupled with ApoE antibody, and viral genome copy number was quantitatively analyzed. Compared with the control group, both sApoE-HA and sApoE could associate with HCVcc lacking cApoE (Fig. 5I). These results indicate that the interaction of sApoE with HCV virions occurs independently of the presence of cApoE during intracellular virus assembly. The results also illustrate that the presence of the HA tag at the C-terminal domain of sApoE did not affect the ability of HCV virions to interact with ApoE.
FIG 5.
sApoE bound to HCVcc produced from ApoE knockout cell lines. (A) Immunofluorescence analysis of wild-type and ApoE knockout Huh7.5 cell lines infected with HCVcc. Cell lines (top) were infected by HCVcc, and 48 h later, immunofluorescence analysis was performed using ApoE (green) and HCV core (red) antibodies and 4′,6-diamidino-2-phenylindole (DAPI; blue). Three images were merged in the bottom row. (B) Expression of cellular ApoE and viral proteins analyzed by Western blotting. (C) Huh7.5 and ApoE knockout cell lines were infected with HCVcc (multiplicity of infection [MOI] = 0.01). The supernatants were harvested to quantify virus production by FF-IHC assay. (D) Biophysical properties of HCV particles produced from Huh7.5 and ApoE knockout cell lines. Huh7.5, HuhdE1.3, and HuhdE1.5 cells were infected with HCVcc. After 60 h, the supernatants were concentrated and fractionated by density gradient centrifugation. Twelve fractions were harvested and used for the determination of density and HCV RNA concentration. (E) HCVcc was continuously passaged separately in the Huh7.5, HuhdE1.3, and HuhdE1.5 cell lines. The Huh7.5 and ApoE knockout cell lines were infected with HCVcc (MOI = 0.01). The supernatants were harvested to quantify virus production 60 h postinfection. Virus titers were determined using focus forming assays at the indicated number of passages (P1 to P4) and are shown as the means ± SDs (error bars) pooled from three independent wells. (F to H) HCV produced from the Huh7.5 and ApoE knockout cell lines, HuhdE1.3 and HuhdE1.5, were neutralized by anti-ApoE or anti-E2 antibodies. The natural mouse IgG was used as a control. (I) HCVcc produced by the ApoE knockout cell line HuhdE1.5 was mixed with sApoE or HA-tagged sApoE (sApoE-HA) secreted by 293T cells in cell culture media. The negative control was blank 293T cell culture supernatant. The mixtures were separately captured by ApoE-specific immunoprecipitation. HCV genome copies in the immunocomplexes were quantified using qRT-PCR. The data represent the means ± SDs from three independent experiments. **, P < 0.01; ***, P < 0.001 (unpaired Student's t test). NS, not significant.
Binding of sApoE to HCV depends on envelope proteins.
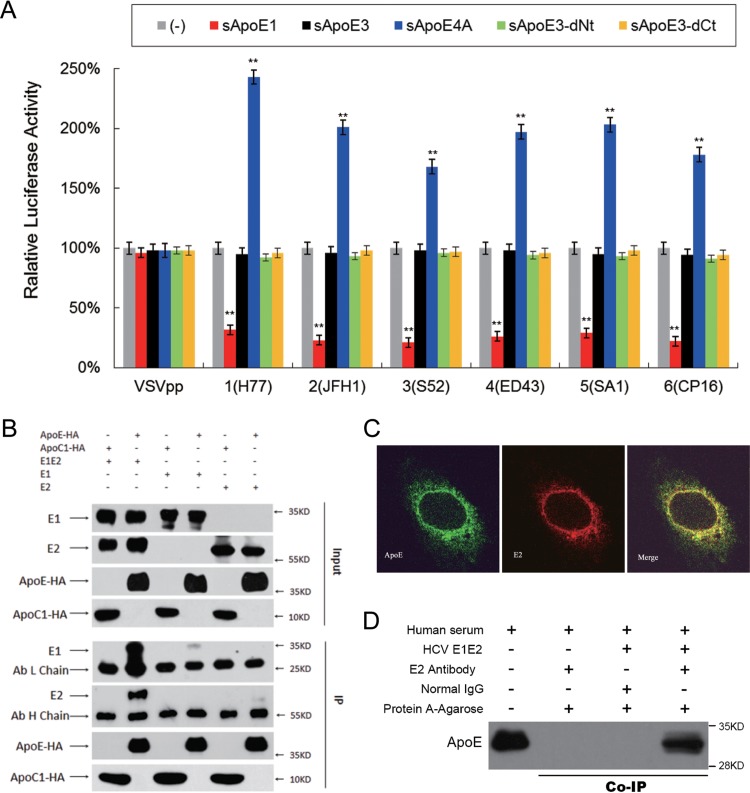
Heterodimers of the E1 and E2 viral envelope glycoproteins are present on the surfaces of HCV particles. Because the extracellular interaction of sApoE with HCVcc occurs independently of the intracellular interaction of the virus with cApoE during virus assembly, we hypothesized that the interaction is mediated through the HCV envelope proteins. To exclude the effects of other HCV viral proteins, we used HCV envelope proteins from six HCV genotypes to create lentiviral vectors capable of producing HCV pseudoparticles (HCVpps). The six HCVpps were separately mixed with sApoE1, sApoE3, sApoE4A, and ApoE3 deletion mutants in the cell supernatant, and infectivity assays were performed. A vesicular stomatitis virus envelope protein pseudotyped particle (VSVpp) was used as a control, and the results indicated that the presence of different sApoE variants had no differential effects on the infectivity of this pseudovirus (Fig. 6A). However, the infectivity of the HCVpps was significantly affected, with ApoE1 causing inhibition of infectivity and sApoE4A increasing it. As the only difference in the assembly processes of the VSVpp and HCVpps was the presence of the HCV envelope proteins, these results suggest that the effects of sApoE on the infectivity of HCV are mediated through these envelope proteins.
FIG 6.
Interactions between recombinant HCV envelope proteins and sApoE. (A) The infectivity of pseudoviral particles influenced by sApoE variants depends on HCV envelope proteins. The HCV or VSV envelope protein pseudotyped lentiviral particles containing a luciferase reporter gene were produced and incubated with cell culture supernatant containing different sApoE variants or mutants secreted from 293T cells at room temperature for 30 min and subsequently used to infect Huh7.5 cells in a 24-well culture plate. At 5 h.p.i., the mixture was removed, and the cells were washed twice with PBS and then incubated with DMEM containing 10% FBS. After 24 h, the cells were lysed for luciferase assays. The quantitative results derived from the data were converted to percentages of the control in the bar graph, and the data are shown as the means ± SDs (error bars) pooled from three independent experiments. **, P < 0.01 (unpaired Student's t test). (B) Interactions between sApoE and HCV envelope proteins were analyzed by coimmunoprecipitation. 293T cells stably expressing the HCV envelope protein E1E2, E1, or E2 were lysed and separately incubated with cell culture supernatant containing sApoE3-HA or sApoC-HA secreted by 293T cells. These mixtures were then captured by HA-specific immunoprecipitation. The proteins (input) or immunocomplexes (IP) in the mixture were analyzed by Western blotting. Proteins are specified on the left, and molecular mass standards are labeled on the right. (C) Colocalization of HCV envelope proteins and cApoE in hepatocytes. Recombinant HCV envelope proteins were expressed in Huh7.5 cells. cApoE (green) and HCV E2 (red) proteins were analyzed by immunofluorescence with specific antibodies and observed via confocal microscopy. (D) Co-IP of recombinant HCV envelope proteins with sApoE in human serum. Lysates from naive 293T cells or cells stably expressing HCV envelope proteins (E1E2) were incubated with human serum and protein G-conjugated agarose beads (Invitrogen) bound with normal mouse IgG or an E2-specific MAb. Upon extensive washing, precipitated proteins were separated by 10% SDS-PAGE and then transferred onto a PVDF membrane. ApoE protein was subsequently detected by Western blotting using an ApoE-specific MAb.
To assess what interactions form between sApoE and the HCV envelope proteins, we constructed cell lines stably expressing E1E2 dimers, E1 monomers, or E2 monomers. Cell lysates were mixed with sApoE-containing supernatant and detected via coimmunoprecipitation (Co-IP). An obvious interaction was observed between sApoE3 and the E1E2 dimer, whereas only weak interactions were observed for E1 and E2 monomers (Fig. 6B). The interaction of ApoE with fused envelope proteins was further evaluating using a two-hybrid system with truncated envelope proteins missing the transmembrane region. The results showed no detectable interactions (unpublished data), indicating that sApoE efficiently interacts with HCV envelope proteins only when they are in their native spatial conformation, with a preference for the E1E2 dimer.
As the HCV viral envelope proteins in their native conformation induce the production of neutralizing antibodies in vivo, they are therefore the main candidate antigens for vaccine development. However, it must be considered that antigen immunogenicity is influenced by numerous factors. In the present study, we showed that recombinant HCV envelope proteins can bind to ApoE protein in both cells and blood. To demonstrate this, we first examined the distribution of cell-expressed recombinant E1E2 envelope proteins, and the results indicated that the HCV envelope proteins can colocalize with cApoE in cells (Fig. 6C). Then, we mixed E1E2 protein expressed by 293T cells with normal human serum and found that the HCV envelope proteins coimmunoprecipitated with sApoE (Fig. 6D). These results imply that HCV envelope proteins interact with both cApoE and sApoE proteins in both cells and blood.
DISCUSSION
Typically, host-virus interactions begin with viral attachment to the cellular membrane. However, a study reported in 1964 showed that blood components can enhance the infectivity of many viral species, including Japanese encephalitis virus and West Nile virus (46), through a phenomenon termed ADE. The correlation of ADE with virus pathogenicity was later clarified in a dengue virus study (47) which reported that when a virus enters the host through blood, the interaction of the virus with blood components is a key determining factor for pathogenicity. It has been shown that HCV interacts with cApoE in hepatocytes and is secreted into blood for transmission, and abundant exchangeable sApoE on the surfaces of lipoproteins in blood can dissociate and bind to other biological complexes. In the present study, the three major ApoE isoforms and two ApoE variants, ApoE1 and ApoE4A, which have a significantly decreased or increased affinity for HSPG on hepatocytes than ApoE3, were employed. We reported that HCV also binds to extracellular sApoE and found that both sApoE protein concentration and variant type affect virus infectivity. For example, sApoE4 caused a reduction in HCV infectivity, a finding that is consistent with clinical data indicating that the ApoE4 genotype is associated with a degree of resistance against HCV infection (48, 49). During the preparation of the manuscript, another similar result was also reported (50).
Interactions between HCV and lipoprotein components in blood can affect not only virus infectivity but also viral immunogenicity. A recent study showed that binding of ApoE to HCV virions mediates viral evasion from neutralizing antibodies (51). Although the binding of HCV with ApoE intracellularly or extracellularly may have only a slight influence on the virus life cycle, the interaction of either pseudo-viral particles or recombinant E1E2 protein with extracellular sApoE could have a significant role in vaccine development. These findings imply that antigens containing HCV envelope proteins could bind to host sApoE and that this interaction might lead to decreased antigen immunogenicity. Therefore, disrupting these interactions might be a novel strategy to enhance the immune response for vaccine antigen design.
The neutralizing antibody level in serum is a key indicator when evaluating humoral immunity. HCV can form LVPs with lipoproteins to attach to HSPG and very low-density lipoprotein receptors on hepatocyte membranes. At low neutralizing antibody levels (resulting from a lack of high dilutions), serum lipoprotein could competitively interact with receptors to interfere with results. In the present study, we further confirmed that sApoE, as a component of lipoprotein, can form extracellular interactions with HCV virions. Furthermore, different ApoE variants and protein concentrations differentially affect HCV infectivity.
Human ApoE functions in lipid metabolism, immune regulation, and neural development, and as a consequence, interactions between HCV and cApoE mainly occur within hepatocytes and thus cause pathogenic effects through the liver. However, these interactions could also affect the systemic function of sApoE and thereby cause indirect effects on other organs. It has been shown that chronic HCV patients have a high incidence of extrahepatic manifestations, including atherosclerosis, diabetes mellitus, and other abnormal lipid metabolism-related disorders. The phenomenon of HCV forming extracellular interactions with carrier lipoproteins provides a new avenue for clinical studies on HCV-related extrahepatic syndrome and might impact clinical safety evaluations of HCV envelope protein-based vaccines.
Because no crystal structures of the E1E2 protein complex exist, we can only hypothesize that deletion of E2 or the expression of fusion proteins directly disturbed the ability of the viral proteins to interact with ApoE (unpublished data), and no interaction domain on the envelope proteins could be defined. Although Lee et al. (34) showed that the interaction of ApoE with HCV E2 protein was not affected when using detergent to remove lipids, indicating that a direct interaction exists between the two proteins, more accurate studies utilizing amino acid mutation are required to further confirm the residues that mediate the interaction. Lee et al. (34) also indicated that the interaction of envelope proteins with cApoE depends on the protein's transmembrane domain. However, the presence of sApoE on the viral surface is not involved in intracellular HCV LVP assembly. Therefore, the HCV E2 transmembrane domain may not act directly on ApoE but rather help maintain the native conformation of the E2 protein to facilitate its association with ApoE. E1 and E2 assist each other in correct folding, and as a consequence, when E1 or E2 protein was expressed alone, the binding affinity toward ApoE markedly decreased (Fig. 6B). Moreover, a recent study also demonstrated that the HVR1 of the HCV E2 protein modulates the conformation and/or epitope exposure of virus particle-associated ApoE (51), which also indicates that the structural conformation of E2 could affect its affinity toward binding with ApoE. Notably, although the association of sApoE with viral particles relies on interactions with the viral envelope proteins, prior immunoelectron microscopy studies have revealed various amounts of ApoE proteins on individual HCV virion surfaces (36, 39). On the one hand, this indicates that a single HCV particle could interact with several ApoE proteins, which supports our results showing that the interaction of sApoE with HCV is dose dependent. On the other hand, this finding implies that the interaction efficiency of different HCV LVPs with sApoE varies. In the present study, we observed that sApoE more markedly affected the infectivity of HCVccs than HCVpps, although the reason behind this difference remains unclear.
Several earlier studies have indicated that ApoE can associate with HCV intracellularly and participate in the maturation of HCV viral particles. In our research, we further proved that HCV can form extracellular associations with secreted sApoE. Thus, the presence of ApoE on HCV viral particles might result not only from hepatic cells but also from blood constituents. Therefore, future analyses of HCV infection should consider that extracellular ApoE may not only competitively associate with HCV receptors but also directly interact with HCV particles. Moreover, ApoE could also competitively associate with several blood constituents. Therefore, when assessing the effects of ApoE protein on HCV infectivity, factors including cApoE production within hepatic cells, the secretion of ApoE from hepatocytes, the concentration of sApoE in blood circulation, HCV concentration, and the concentration of several blood constituents, including VLDL and LDL, must all be considered with regard to HCV infectivity in vivo. Further probing of the extracellular interactions that exist between HCV and sApoE could provide new insights into the aspects of the HCV life cycle that affect viral infectivity.
MATERIALS AND METHODS
Cells and virus.
The Huh7.5 cell line was provided by Charles Rice (Rockefeller University). 293T cells were obtained from the Kunming Cell Bank of Type Culture Collection, Chinese Academy of Sciences. All cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, nonessential amino acids, 100 U/ml of penicillin, and 100 μg/ml of streptomycin (HyClone) at 37°C in 5% CO2. Stable cell lines induced to express HCV envelope proteins were maintained by the addition of 6 μg/ml of puromycin (Sigma-Aldrich). Primary human hepatocytes (PHHs) (lot number FCL; BioreclamationIVT) were plated at 1.5 × 105/cm2 in 6-well collagen I-coated plates with InVitroGRO CP medium (BioreclamationIVT). Four hours after plating, nonadherent cells were removed, and the PHHs were maintained in InVitroGRO HI medium (BioreclamationIVT). Infectious HCVcc was produced by transfection of Huh7.5 cells with in vitro-transcribed RNA derived from JFH1A or JFH1A-Rluc.
Plasmid constructs.
Plasmids were constructed according to standard methods. All plasmid vectors were cloned and propagated in Escherichia coli DH5α, DH10B, or Stbl3 grown at 30°C. The DH10B strain was used for vectors containing HCV genomes, and the Stbl3 strain was used for constructs containing duplicated sequences. Constructs were verified by restriction enzyme digestion and sequencing. Descriptions of the cloning strategies are provided below.
The p94-JFH1A plasmid, which contains the T7 promoter sequence fused to the JFH1 complete genome sequence with adaptive mutations, the hepatitis delta virus genomic ribozyme, and the T7 terminator sequence, was described previously (52). To introduce an MluI monoclonal restriction site in the NS5A C-terminal coding sequence, the PCR primers JFH1-7453-RsrIIF/JFH1-7525-MluR and JFH1-7525-MluF/JFH1-7796-BsrGIR (see Table 2 for primer sequences) were used to amplify two products from the p94-JFH1A template that were combined in a second round of PCR, cleaved by RrsII and BsrGI, and then ligated back into p94-JFH1A digested by SpeI and BsrGI with the SpeI-RrsII fragment from p94-JFH1A to create p94-JFH1A-M420. The p94-JFH1A-Rluc vector containing the HCV reporter genome was created by amplifying the Renilla luciferase-coding region from pRL-SV40 (Promega) using primers NS5A-Rluc-HF and NS5A-Rluc-HR and then inserted into MluI-linearized p94-JFH1A-M420 using the In-Fusion HD enzyme (TaKaRa). The amino acid substitutions T197S, S199G, S200L, and M202H were introduced into E1 by overlapping PCR amplification using synthetic oligonucleotide primers 2a-78F, 2a-1371R, 2a-E1A4F, and 2a-E1A4R. The PCR DNA fragments were digested with the restriction enzymes AgeI and BsiWI and cloned into JFH1A cDNA vectors, which were similarly cut by both AgeI and BsiWI, as previously described (53). The E1 protein containing these substitutions can be recognized by the E1-specific monoclonal antibody (MAb) A4.
TABLE 2.
Primers used to generate the viral constructs
| Primer name | Sequence (5′–3′)a |
|---|---|
| JFH1-7453-RsrIIF | ATCCGGCGGTCCGACGT |
| JFH1-7525-MluR | AGGCTCACGCGTGAGGGGGGGCATAGAGGAGGC |
| JFH1-7525-MluF | CTCACGCGTGAGCCTGGAGATCCGGACCTGGAGTC |
| JFH1-7796-BsrGIR | TCTTTGATGTTGTACAGTACACCTTG |
| NS5A-Rluc-HF | GCCCCCCCTCACGCGTATGGCTTCCAAGGTGTACG |
| NS5A-Rluc-HR | CTCCAGGCTCACGCGTCTGCTCGTTCTTCAGCACGCGC |
| 2a-78F | CTAGCCATGGCGTTAGTATGAGTGT |
| 2a-1731R | ATGACCTCGGGGACGCG |
| 2a-E1A4F | CAGGTGAAGAATTCCAGTGGCCTCTACCATGTGACCAATGACTGC |
| 2a-E1A4R | GCAGTCATTGGTCACATGGTAGAGGCCACTGGAATTCTTCACCTG |
| ApoE3-KasIF | GAGGGCGCCGAGCGCGG |
| ApoE-SG-HAR | GTATGGGTATCCTCCTCTTCCTCCTGAGTGATTGTCGCTGGGCACA |
| ApoE dCT-SG-HAR | GTATGGGTATCCTCCTCTTCCTCCTGAGGCCTGCAGGCGTATCTG |
| HA-SacI-XbaIR | GGTCTAGAGCTCAAGCGTAATCTGGAACATCGTATGGGTATCCTCCTCTTCCTCCTG |
| Hbactin-KpnIF | AGTGGTACCATGGATGATGATATCGCCGCG |
| Hbactin-XbaIR | AGTTCTAGACTAGAAGCATTTGCGGTGGACG |
| ApoE129-g20F | CACCGCCAGTGCCAGTTCCCAGCGC |
| ApoE129-g20R | AAACGCGCTGGGAACTGGCACTGGC |
| H77-741-BglIIF | AATAGATCTGCCACCATGGGGTACATACCGCTCGTCGGCG |
| H77-2579-XbaIR | GAATCTAGATCACGCCTCCGCTTGGGATATGAGtaac |
| JFH1-740- BglIIF | AATAGATCTGCCACCATGGGGTACATCCCCGTCGTAGGCG |
| JFH1-2590-XbaIR | GAATCTAGATCATGCTTCGGCCTGGCCCAACAAGATG |
| S52-739-BglIIF | AATAGATCTGCCACCATGGGGTACATCCCGCTCGTCGGCG |
| S52-2592-XbaIR | GAATCTAGATCATGCTTCTGCTTGTGATACCATCAGCATC |
| ED43-740-BglIIF | AATAGATCTGCCACCATGGGATACATCCCGCTCGTAGGCG |
| ED43-2575-XbaIR | GAATCTAGATCACGCCTCAACTTGACTTACCATAAACATCATCC |
| SA1-739-BglIIF | AATAGATCTGCCACCATGGGGTACATCCCGCTCGTAGGCG |
| SA1-2577-XbaIR | GAATCTAGATCATGCTTCTGCCTGACAAACTAGAAGC |
| CP16-740-BglIIF | AATAGATCTGCCACCATGGGGTACATCCCAGTCCTAGGAG |
| CP16-2584-XbaIR | GAATCTAGATCAGGCCTCCGCTTGGCCCACCAGGAAC |
| H77-171-NheF | TTGGCTAGCATGGGTTGCTCTTTCTCTATCTTCCTTCTGG |
| H77-383-BglIIR | TTGAGATCTTACGCGTCGACGCCGGCAAATAG |
| H77-384-NheF | TTGGCTAGCATGGAAACCCACGTCACCGGGGGAAATG |
| H77-746-BglIIR | TTGAGATCTTACGCCTCCGCTTGGGATATGAG |
The restriction sites used to facilitate cloning are underlined.
The pcDNA3.1-derived vectors pcDNA3-ApoE1, -2, -3, -4, and -4A, pcDNA3-ApoE dNT, and pcDNA3-ApoE dCT expressing different full-length ApoE variants and deletion mutants were described previously (32). An HA tag was fused to the C terminus of ApoE (Fig. 1A) using overlap extension PCR. For ApoE-HA and ApoE dNT-HA, PCR products were amplified from pcDNA3-ApoE3 using the primer ApoE3-KasIF paired with ApoE-SG-HAR. For ApoE dCT-HA, PCR products were amplified from pcDNA3-ApoE dCT using primer ApoE3-KasIF paired with ApoE dCT-SG-HAR. These PCR products were gel purified and extended using ApoE3-KasIF and HA-SacI-XbaIR. The resulting products were cloned into pcDNA-ApoE and cleaved using the restriction enzymes KasI and XbaI to create pcDNA3-ApoE-HA, pcDNA3-ApoE dNT-HA, and pcDNA3-ApoE dCT-HA.
The human β-actin gene was amplified from Huh7 cell total RNA using primers Hbactin-KpnIF and Hbactin-XbaIR with a GoldScript one-step reverse transcription-PCR (RT-PCR) kit (Life Technologies, Guangzhou, China) according to the manufacturer's instructions. The RT-PCR product was inserted into a pcDNA3.1 vector between the KpnI and XbaI restriction sites to create the pcDNA3.1-HBactin plasmid.
To produce CRISPR-Cas9 bicistronic expression vectors, pCW299 with gRNA 299 and oligonucleotides ApoE129-g20F and ApoE129-g20F were annealed at 95°C for 5 min and then cooled to room temperature, followed by ligation to plasmid pX330-U6-Chimeric_BB-CBh-hSpCas9 (a gift from Feng Zhang; Addgene plasmid 42230), which was linearized using the BbsI restriction enzyme.
pUC57-H77C, pUC57-S52, pUC57-ED43, and pUC57-SA1, containing the genomes of HCV genotypes 1a (H77C, GenBank accession no. AF011751 [54]), 3a (S52, GenBank accession no. GU814264 [55]), 4a (ED43, GenBank accession no. GU814266 [55]), and 5a (SA1, GenBank accession no. KJ925146 [56]), respectively, and flanked with the EcoRI-T7 promoter and an XbaI restriction site, were synthesized by Sangon Biotech Co. Ltd. (Shanghai, China). Some restriction enzyme sites were deleted via silent mutation. pBR-CP16 contains the genome of HCV genotype 6n, which is prevalent among intravenous drug users in the Yunnan province of southwestern China (GenBank accession no. KY014622). Using the plasmids containing various HCV genotypes as templates, PCR was used to amplify DNA fragments encoding the last 58 residues of the HCV core and all of the E1 and E2 proteins using the primers H77-741-BglIIF/H77-2579-XbaIR, JFH1-740-BglIIF/JFH1-2590-XbaIR, S52-739-BglIIF/S52-2592-XbaIR, ED43-740-BglIIF/ED43-2575-XbaIR, SA1-739-BglIIF/SA1-2577-XbaIR, and CP16-740-BglIIF/CP16-2584-XbaIR. The DNA fragments were amplified separately and inserted into a pEGFPN2 vector (Clontech) using BglII and XbaI to create plasmids pN-Env HCV1 to pN-Env HCV6.
PCR with primers H77-171-NheF/H77-383-BglIIR, H77-384-NheF/H77-746-BglIIR, and H77-171-NheF/H77-746-BglIIR was used to amplify E1 (polyprotein residues 171 to 383), E2 (polyprotein residues 384 to 746), and E1 plus E2 (polyprotein residues 171 to 746) from the pN-Env HCV1 template; the products were cleaved by NheI and BglII, ligated with pCW57.1 (a gift from David Root; Addgene plasmid 41393), and digested by NheI and BamHI to create the lentiviral plasmids pCW-E1, pCW-E2, and pCW-E1E2.
pEGFP-puro was a gift from Michael McVoy (Addgene plasmid 45561). pMD2.G and psPAX2 were obtained from Didier Trono (Addgene plasmids 12259 and 122560). pNL4-3.Luc.R−.E− was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH.
Antibodies.
A hybridoma cell line producing the ApoE-specific monoclonal antibody WUE-4 was obtained from the American Type Culture Collection (CRL-2247). A human ApoE enzyme-linked immunosorbent assay (ELISA) kit was obtained from Mabtech (number 3212-1A-20). Goat anti-human ApoE polyclonal antibody was purchased from Calbiochem (number 178479). HCV core (number 1868), E1 (number 1879), and E2 (number 1876) monoclonal antibodies were purchased from Virostat. Anti-β-actin (AC15) and HA monoclonal antibodies (HA-7), an anti-HA immunoprecipitation kit, an HA peptide, and a horseradish peroxidase-conjugated goat anti-mouse IgG were purchased from Sigma-Aldrich. Alexa Fluor 488-labeled donkey anti-goat IgG and Alexa Fluor 594-labeled donkey anti-mouse IgG were purchased from Thermo Fisher Scientific. The mouse natural IgG protein (ab198772) was purchased from Abcam. Mouse anti-E1 MAb A4 (11) was kindly provided by Jean Dubuisson (Institut de Biologie de Lille, France). The mouse anti-E2 MAb AP33 (57) was kindly provided by Arvind H. Patel (Institute of Virology, Glasgow, Scotland).
HCV RNA synthesis and transfection of cultured cells.
Plasmids carrying full-length HCV genomes were linearized with XbaI and extracted with phenol and chloroform, precipitated with ethanol, and dissolved in RNase-free water. RNA was transcribed from 3 μg of purified template using a T7 RiboMAX expression large-scale RNA production system (Promega, Beijing, China). Complete removal of the template DNA was ensured by an additional treatment with RNase-free DNase and purification over Qiagen RNA isolation columns. The quality of the in vitro-transcribed RNA was examined by agarose gel electrophoresis. RNA transcripts were introduced into cells by transfection using a TransIT-mRNA transfection kit (Mirus). For transfection, cells plated in 10-cm cell culture dishes were grown overnight to approximately 80% confluence. The culture medium was replaced with fresh complete growth medium 30 min before transfection. Transfection complexes, prepared by adding 10 μg of HCV RNA, 20 μl of mRNA boost reagent, and 20 μl of transfection reagent to 500 μl of Opti-MEM in sterile tubes, were incubated for 3 min at room temperature and added dropwise to each plate.
Isopycnic gradient centrifugation analysis.
Media containing sApoE or HCV were layered on top of 9 to 30% OptiPrep (Axis-Shield, Norway) gradients prepared in phosphate-buffered saline (PBS). The gradients were centrifuged at 40,000 rpm (197,000 × g) for 6 h at 4°C in an SW-41 rotor. Eleven fractions (1 ml each) were collected from the bottom of the tube. The density of the fractions was determined using a refractometer (Abbemat 350; Antton Paar).
Western blot analysis.
Protein concentrations were determined using protein assay reagent (Thermo Scientific, Beijing, China). Each sample was analyzed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Separated proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane. After blocking with 5% skim milk, immunoblotting analysis for ApoE, β-actin, HA tag, HCV core, E1, and E2 proteins was performed using primary monoclonal antibodies specific to each protein, a secondary goat anti-mouse antibody, and an enhanced chemiluminescence kit (Thermo Scientific).
DNA transfection and ApoE knockout cell line selection.
DNA transfection was performed using a lipofection method according to the manufacturer's instructions. For sApoE production, cell culture dishes were coated with 0.1 mg/ml of poly-l-lysine. The day before transfection, 3.2 × 106 293T cells in 8 ml of cell culture medium without antibiotics were seeded onto a 10-cm dish to reach 80% confluence by the time of transfection. A volume of 15 μg of ApoE expression plasmid was diluted to 1 ml in Opti-MEM medium (Life Technologies). In addition, 37.5 μl of Lipofectamine 2000 (Life Technologies) was added to 1 ml of Opti-MEM medium, and the DNA and Lipofectamine 2000 were gently mixed, incubated at room temperature for 20 min, and added to the cells in a dropwise manner. Eight hours after transfection, the cells were transferred to DMEM complete medium, and the supernatants were collected after 48 h. For ApoE gene knockout cell line selection, 4 × 105 Huh7.5 cells in 6-well plates were cotransfected with plasmids pEGFP-puro and pCW299 in a 3:1 ratio. Then 2 μg of DNA was mixed with 5 μl of DMRIE-C reagent (Life Technologies) in Opti-MEM and transferred to the Huh7.5 cells. At 24 h posttransfection, 10 μg/ml of puromycin was added and selected for 2 days. Then the cells were split into 10-cm cell culture dishes at various cell densities in growth medium. Well-isolated cell colonies were selected and amplified. sApoE protein expression was detected by ELISA. A cell line not expressing sApoE was subjected to two rounds of subcloning by limiting dilution, and the ApoE knockout cell lines were confirmed by restriction fragment length polymorphism (RFLP) assays or Sanger sequencing of the mutated ApoE gene.
HCVcc particle pulldown by HA-specific immunocapture.
HCVcc (500 μl) and an equivalent volume of cell culture supernatant containing ApoE-HA were preincubated at 4°C overnight. The mixture was subjected to HA tag-specific immunocapture using 40 μl of HA affinity gel (Sigma-Aldrich). After washing with 800 μl of PBS, the captured proteins were eluted by boiling in Laemmli sample buffer and subjected to SDS-PAGE and Western blot analysis. Captured HCV RNAs were extracted using TRIzol A+ and quantified by quantitative RT-PCR (qRT-PCR). Immunocaptured HCV particles were eluted with 5 volumes of PBS containing 100 μg/ml of HA peptide (Sigma-Aldrich). Infectivity titers were determined by immunohistochemical assays.
RNA extraction and quantification by qRT-PCR.
HCV RNA was isolated using TRIzol LS (Life Technologies) according to the manufacturer's protocols. RNA levels were adjusted to internal control RNA input (2 ng of β-actin RNA), which was added in excess prior to RNA isolation. The absolute levels of HCV RNA were quantified. Oligonucleotide primers w168 (5′-CTA GCC ATG GCG TTA GTA TGA GTG T-3′) and w169 (5′-CGG CAA TTC CGG TGT ACT CA-3′) and probe w170 (5′-FAM-TCC CGG GAG AGC CAT AGT GGT CTG CG-BHQ1-3′), which are complementary to the HCV 5′ untranslated region (UTR), were used. For β-actin mRNA, the primers used were w159 (5′-TGC CGA CAG GAT GCA GAA G-3′) and w160 (5′-CTC AGG AGG AGC AAT GAT CTT GAT-3′), and the TaqMan probe was 5′-Joe-ATC ACT GCC CTG GCA CCC AGC A-BHQ1-3′. Reactions were run using an ABI 7500 real-time PCR system (Life Technologies) as follows: 5 min at 42°C, 10 s at 95°C, and 40 cycles at 95°C for 5 s and 60°C for 35 s. Fluorescence detection was performed using the preset channels Fam, Joe, and Rox. RNA quantities were derived by comparison with RNA concentration standards run in parallel. The RNA standards were in vitro T7 transcripts of the JFH1 genome and β-actin RNA, which were DNase treated, purified on an RNeasy minicolumn (Qiagen, Shanghai, China), and quantified via spectrophotometry.
FF-IHC.
For the focus-forming assay based on immunohistochemistry (FF-IHC), HCV in culture supernatant was 10-fold serially diluted, and 250 μl of diluted HCV was used per well to infect Huh7.5 cells in 24-well plates. After 6 h of incubation at 37°C, the cells were washed twice with PBS, and 250 μl of fresh complete medium containing 1% methylcellulose was added. At 72 h postinfection (h.p.i.), the cells were fixed with 10% neutralized formalin. Focus-forming units were determined by immunohistochemistry using a core-specific monoclonal antibody, peroxidase-conjugated secondary anti-mouse antibody, and 3,3-diaminobenzidine substrate (Vector Laboratories, Burlingame, CA). Viral titer was determined based on the average number of core-positive focus-forming units in triplicate or quadruplet assays.
Quantification of ApoE.
Human ApoE levels in culture medium or normal human serum were quantified using a human apolipoprotein E enzyme-linked immunosorbent assay kit (Mabtech AB, Sweden) according to the manufacturer's protocol; the recombinant ApoE3 standard protein was also provided by the manufacturer.
ApoE protein purification.
A total of 500 μl of anti-HA affinity matrix beads was washed 3 times with 10 mM PBS (pH 7.4) and mixed with 10 to 15 ml of 293T cell supernatant with or without HA-tagged ApoE protein for 2 h. After centrifugation at 3,000 × g for 5 min, the supernatant was collected, and the beads were washed 3 times with PBS; this was followed by the addition of 500 μl of EF (1 mg/ml HA peptide in PBS) to elute the ApoE-HA protein.
HCV neutralization assays.
Antibody inhibition assays were performed using Huh7.5 cells, and virus infectivity was determined by FF-IHC. Briefly, Huh-7.5 cells were plated at a density of 1 × 104 per well in a 96-well plate, and ∼50 FFU of virus was preincubated at 37°C for 1 h with the appropriate inhibitory or control antibody prior to infecting cells. At 3 h postinfection, the inoculum was replaced with fresh medium containing 1% methylcellulose and incubated for 48 h. The infectivity was determined as FF-IHC following immunostaining of the cells for viral core proteins as described above.
Lentiviral pseudoparticles.
To generate pseudoparticles for the delivery of inducible HCV envelope protein, 1 × 107 Lenti-X 293T cells (Clontech) in a T175 tissue culture flask were cotransfected with plasmids encoding (i) pCW-E1, pCW-E2, or pCW-E1E2 provirus, (ii) psPAX2, and (iii) pMD2.G in a ratio of 4:3:2. To generate HCV pseudoparticles, 4 × 106 Lenti-X 293T cells (Clontech) in a 10-cm cell culture dish were cotransfected with a pNL4-3.Luc.R−.E− proviruses vector containing the firefly luciferase reporter gene and plasmids pN-HCV1 to -6, encoding the Env proteins of various HCV genotypes, in a ratio of 3:1. A total of 12 μg of DNA was transfected using Lipofectamine 2000 reagent. Transfections were carried out for 16 h, followed by a medium change to DMEM containing 10% FBS. The culture medium was collected at 48 h and 72 h, pooled, filtered through a 0.45-mm filter, ultracentrifuged, and stored at −80°C.
Infectivity measurement via luciferase assay.
Huh7.5 cells were seeded at a density of 2 × 105 per well in 12-well plates and allowed to adhere overnight. A volume of 100 μl of HCV-Rlu reporter virus was incubated with 200 μl of DMEM complete medium containing increasing concentrations of secreted ApoE at room temperature for 1 h, after which the mixture was added to the Huh7.5 cells. At 5 h postinfection, the mixture was removed, and the infected cells were washed twice with PBS and then incubated with DMEM complete medium. The supernatants were removed after a 24-h culture. The cells were then washed with PBS and lysed in 100 μl of lysis buffer, and 10 μl of lysate was added to 100 μl of assay buffer (Promega) in a 96-well assay plate. A microplate reader (Synergy 4, BioTek, Shanghai, China) was then used to perform a 10-s measurement. All luciferase assays were performed at least in triplicate.
Coimmunoprecipitation.
293T cells overexpressing HCV H77C E1E2, E1, and E2 were cultured in T225 cell culture flasks for 16 h to become adherent, and doxycycline was added at a final concentration of 1 μg/ml. After 48 h, the cells were lysed using nondenaturing lysis buffer (20 mM Tris-HCl [pH 8.0], 137 mM NaCl, 2 mM EDTA, 1% Triton X-100, and protease inhibitor cocktail tablets). After 1 h of incubation on ice, cell debris was removed by 10 min of centrifugation at 3,000 × g. To detect interactions between sApoE in the cell culture medium and HCV envelope proteins, 10 μl of anti-HA agarose beads (Sigma-Aldrich) and 250 μl of sApoE-HA secreted by 293T cells transfected with pcDNA3.1-ApoE-HA using Lipofectamine 2000 were added to 250-μl aliquots of cell lysates containing E1, E2, or E1E1. ApoC1-HA secreted by 293T cells was used as a negative control. To detect interactions between E1E2 and sApoE in serum, 250 μl of E1E2 protein in cell lysis buffer was mixed with 250 μl of normal human serum and added to protein A agarose bound to anti-E2 antibody (Thermo Scientific). Normal mouse IgG was used as a negative control. The mixtures were incubated overnight by continuously inverting the tubes at 4°C, and the beads were washed three times by resuspension in 20 mM cold PBS followed by centrifugation at 3,000 × g. The samples were resuspended with 50 μl of 1× SDS-PAGE loading buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 0.1% bromophenol blue [BPB], 10% glycerol, and 1% β-mercaptoethanol), and the samples were incubated for 5 min at 95°C, followed by Western blotting.
Statistical analysis.
Data are expressed as the means and standard deviations (SDs). Statistical analyses were performed using Student's t test. A P value of <0.01 was considered highly significant, and a value of <0.001 was considered extremely significant.
Accession number(s).
Complete sequences for pcDNA3.1-derived vectors pcDNA3-ApoE1, -2, -3, -4, and -4A are available from GenBank under accession numbers KY924484 to KY924488.
ACKNOWLEDGMENTS
We thank Charles Rice for kindly providing the Huh-7.5 cell line; Feng Zhang, David Root, Michael McVoy, and Didier Trono for plasmids; and Arvind Patel and Jean Dubuisson for antibodies.
W.C. and S.D. designed the study; Z.L., Y.L., Y.B., H.Z., Y.Y., and W.C. performed the experiments; Q.L., W.C., and S.D. analyzed the data; and W.C. wrote the manuscript.
We have no conflicts of interest to declare.
This work was supported by the Natural Science Foundation of Yunnan Province (2013FA053 and 2015BC010), Fundamental Research Funds for the Central Universities (2015PT310001 and 2016ZX310183-5), and the CAMS Initiative for Innovative Medicine (2016-I2M-1-013 and 2016-I2M-1-019).
REFERENCES
- 1.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. 2013. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 2.Chen SL, Morgan TR. 2006. The natural history of hepatitis C virus (HCV) infection. Int J Med Sci 3:47–52. doi: 10.7150/ijms.3.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Negro F. 2014. Facts and fictions of HCV and comorbidities: steatosis, diabetes mellitus, and cardiovascular diseases. J Hepatol 61:S69–S78. doi: 10.1016/j.jhep.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 4.McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med 339:1485–1492. [DOI] [PubMed] [Google Scholar]
- 5.Baumert TF, Fauvelle C, Chen DY, Lauer GM. 2014. A prophylactic hepatitis C virus vaccine: a distant peak still worth climbing. J Hepatol 61:S34–S44. doi: 10.1016/j.jhep.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Lemon SM, Walker C, Alter MJ, Yi M. 2007. Hepatitis C virus, p 1253–1304. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 7.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol 68:5045–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol 67:1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci U S A 88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones DM, McLauchlan J. 2010. Hepatitis C virus: assembly and release of virus particles. J Biol Chem 285:22733–22739. doi: 10.1074/jbc.R110.133017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol 68:6147–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding Q, Cao X, Lu J, Huang B, Liu YJ, Kato N, Shu HB, Zhong J. 2013. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J Hepatol 59:52–58. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 13.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M Jr, Lemon SM. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A 102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 15.Nitta S, Sakamoto N, Nakagawa M, Kakinuma S, Mishima K, Kusano-Kitazume A, Kiyohashi K, Murakawa M, Nishimura-Sakurai Y, Azuma S, Tasaka-Fujita M, Asahina Y, Yoneyama M, Fujita T, Watanabe M. 2013. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology 57:46–58. doi: 10.1002/hep.26017. [DOI] [PubMed] [Google Scholar]
- 16.Otsuka M, Kato N, Moriyama M, Taniguchi H, Wang Y, Dharel N, Kawabe T, Omata M. 2005. Interaction between the HCV NS3 protein and the host TBK1 protein leads to inhibition of cellular antiviral responses. Hepatology 41:1004–1012. doi: 10.1002/hep.20666. [DOI] [PubMed] [Google Scholar]
- 17.Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 18.Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M Jr, Ye J. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci U S A 104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto M, Aizaki H, Fukasawa M, Teraoka T, Miyamura T, Wakita T, Suzuki T. 2011. Structural requirements of virion-associated cholesterol for infectivity, buoyant density and apolipoprotein association of hepatitis C virus. J Gen Virol 92:2082–2087. doi: 10.1099/vir.0.032391-0. [DOI] [PubMed] [Google Scholar]
- 20.Weisgraber KH. 1994. Apolipoprotein E: structure-function relationships. Adv Protein Chem 45:249–302. doi: 10.1016/S0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 21.Mahley RW, Rall SC Jr. 2000. Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet 1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 22.de Knijff P, van den Maagdenberg AM, Frants RR, Havekes LM. 1994. Genetic heterogeneity of apolipoprotein E and its influence on plasma lipid and lipoprotein levels. Hum Mutat 4:178–194. doi: 10.1002/humu.1380040303. [DOI] [PubMed] [Google Scholar]
- 23.Aggerbeck LP, Wetterau JR, Weisgraber KH, Wu CS, Lindgren FT. 1988. Human apolipoprotein E3 in aqueous solution. II. Properties of the amino- and carboxyl-terminal domains. J Biol Chem 263:6249–6258. [PubMed] [Google Scholar]
- 24.Wetterau JR, Aggerbeck LP, Rall SC Jr, Weisgraber KH. 1988. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J Biol Chem 263:6240–6248. [PubMed] [Google Scholar]
- 25.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. 1991. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science 252:1817–1822. [DOI] [PubMed] [Google Scholar]
- 26.Westerlund JA, Weisgraber KH. 1993. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J Biol Chem 268:15745–15750. [PubMed] [Google Scholar]
- 27.Chang KS, Jiang J, Cai Z, Luo G. 2007. Human apolipoprotein E is required for infectivity and production of hepatitis C virus in cell culture. J Virol 81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itzhaki RF, Irving WL, Wozniak MA. 2003. Apolipoprotein E and hepatitis C virus. Hepatology 38:1060. doi: 10.1002/hep.1840380437. [DOI] [PubMed] [Google Scholar]
- 29.Jiang J, Luo G. 2009. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J Virol 83:12680–12691. doi: 10.1128/JVI.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans MJ, Rice CM, Goff SP. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci U S A 101:13038–13043. doi: 10.1073/pnas.0405152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benga WJ, Krieger SE, Dimitrova M, Zeisel MB, Parnot M, Lupberger J, Hildt E, Luo G, McLauchlan J, Baumert TF, Schuster C. 2010. Apolipoprotein E interacts with hepatitis C virus nonstructural protein 5A and determines assembly of infectious particles. Hepatology 51:43–53. doi: 10.1002/hep.23278. [DOI] [PubMed] [Google Scholar]
- 32.Cun W, Jiang J, Luo G. 2010. The C-terminal alpha-helix domain of apolipoprotein E is required for interaction with nonstructural protein 5A and assembly of hepatitis C virus. J Virol 84:11532–11541. doi: 10.1128/JVI.01021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazumdar B, Banerjee A, Meyer K, Ray R. 2011. Hepatitis C virus E1 envelope glycoprotein interacts with apolipoproteins in facilitating entry into hepatocytes. Hepatology 54:1149–1156. doi: 10.1002/hep.24523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JY, Acosta EG, Stoeck IK, Long G, Hiet MS, Mueller B, Fackler OT, Kallis S, Bartenschlager R. 2014. Apolipoprotein E likely contributes to a maturation step of infectious hepatitis C virus particles and interacts with viral envelope glycoproteins. J Virol 88:12422–12437. doi: 10.1128/JVI.01660-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hishiki T, Shimizu Y, Tobita R, Sugiyama K, Ogawa K, Funami K, Ohsaki Y, Fujimoto T, Takaku H, Wakita T, Baumert TF, Miyanari Y, Shimotohno K. 2010. Infectivity of hepatitis C virus is influenced by association with apolipoprotein E isoforms. J Virol 84:12048–12057. doi: 10.1128/JVI.01063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merz A, Long G, Hiet MS, Brugger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. 2011. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem 286:3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang J, Cun W, Wu X, Shi Q, Tang H, Luo G. 2012. Hepatitis C virus attachment mediated by apolipoprotein E binding to cell surface heparan sulfate. J Virol 86:7256–7267. doi: 10.1128/JVI.07222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S, McCormick KD, Zhao W, Zhao T, Fan D, Wang T. 2012. Human apolipoprotein E peptides inhibit hepatitis C virus entry by blocking virus binding. Hepatology 56:484–491. doi: 10.1002/hep.25665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Catanese MT, Uryu K, Kopp M, Edwards TJ, Andrus L, Rice WJ, Silvestry M, Kuhn RJ, Rice CM. 2013. Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci U S A 110:9505–9510. doi: 10.1073/pnas.1307527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardosa MJ. 1987. Dengue virus isolation by antibody-dependent enhancement of infectivity in macrophages. Lancet i:193–194. [DOI] [PubMed] [Google Scholar]
- 41.Halstead SB. 1988. Pathogenesis of dengue: challenges to molecular biology. Science 239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 42.Sundaram M, Yao Z. 2012. Intrahepatic role of exchangeable apolipoproteins in lipoprotein assembly and secretion. Arterioscler Thromb Vasc Biol 32:1073–1078. doi: 10.1161/ATVBAHA.111.241455. [DOI] [PubMed] [Google Scholar]
- 43.Saito H, Lund-Katz S, Phillips MC. 2004. Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins. Prog Lipid Res 43:350–380. doi: 10.1016/j.plipres.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Long G, Hiet MS, Windisch MP, Lee JY, Lohmann V, Bartenschlager R. 2011. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology 141:1057–1066. doi: 10.1053/j.gastro.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Mann WA, Gregg RE, Sprecher DL, Brewer HB Jr. 1989. Apolipoprotein E-1Harrisburg: a new variant of apolipoprotein E dominantly associated with type III hyperlipoproteinemia. Biochim Biophys Acta 1005:239–244. doi: 10.1016/0005-2760(89)90043-X. [DOI] [PubMed] [Google Scholar]
- 46.Hawkes RA. 1964. Enhancement of the infectivity of arboviruses by specific antisera produced in domestic fowls. Aust J Exp Biol Med Sci 42:465–482. doi: 10.1038/icb.1964.44. [DOI] [PubMed] [Google Scholar]
- 47.Halstead SB. 2014. Dengue antibody-dependent enhancement: knowns and unknowns. Microbiol Spectr 2(6). doi: 10.1128/microbiolspec.AID-0022-2014. [DOI] [PubMed] [Google Scholar]
- 48.Wozniak MA, Itzhaki RF, Faragher EB, James MW, Ryder SD, Irving WL. 2002. Apolipoprotein E-epsilon 4 protects against severe liver disease caused by hepatitis C virus. Hepatology 36:456–463. doi: 10.1053/jhep.2002.34745. [DOI] [PubMed] [Google Scholar]
- 49.Toniutto P, Fabris C, Fumo E, Apollonio L, Caldato M, Mariuzzi L, Avellini C, Minisini R, Pirisi M. 2004. Carriage of the apolipoprotein E-epsilon4 allele and histologic outcome of recurrent hepatitis C after antiviral treatment. Am J Clin Pathol 122:428–433. doi: 10.1309/YLEXQA2R6R2N95JU. [DOI] [PubMed] [Google Scholar]
- 50.Yang Z, Wang X, Chi X, Zhao F, Guo J, Ma P, Zhong J, Niu J, Pan X, Long G. 2016. Neglected but important role of apolipoprotein E exchange in hepatitis C virus infection. J Virol 90:9632–9643. doi: 10.1128/JVI.01353-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fauvelle C, Felmlee DJ, Crouchet E, Lee J, Heydmann L, Lefevre M, Magri A, Hiet MS, Fofana I, Habersetzer F, Foung SK, Milne R, Patel AH, Vercauteren K, Meuleman P, Zeisel MB, Bartenschlager R, Schuster C, Baumert TF. 2016. Apolipoprotein E mediates evasion from hepatitis C virus neutralizing antibodies. Gastroenterology 150:206–217 e204. doi: 10.1053/j.gastro.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 52.Xu J, Zhang X, Wang X, Wu W, Wu Z, Zhang Z, Xiang H, Yao Y, Cun W, Dong S. 2013. Screening of highly permissive cell subclone for hepatitis C virus culture with reporter gene on viral genome. Zhong Hua Wei Sheng Wu Xue He Mian Yi Xue Za Zhi 33:161–167. (In Chinese.) [Google Scholar]
- 53.Goueslain L, Alsaleh K, Horellou P, Roingeard P, Descamps V, Duverlie G, Ciczora Y, Wychowski C, Dubuisson J, Rouille Y. 2010. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol 84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yanagi M, Purcell RH, Emerson SU, Bukh J. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci U S A 94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gottwein JM, Scheel TK, Callendret B, Li YP, Eccleston HB, Engle RE, Govindarajan S, Satterfield W, Purcell RH, Walker CM, Bukh J. 2010. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J Virol 84:5277–5293. doi: 10.1128/JVI.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wose Kinge CN, Espiritu C, Prabdial-Sing N, Sithebe NP, Saeed M, Rice CM. 2014. Hepatitis C virus genotype 5a subgenomic replicons for evaluation of direct-acting antiviral agents. Antimicrob Agents Chemother 58:5386–5394. doi: 10.1128/AAC.03534-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clayton RF, Owsianka A, Aitken J, Graham S, Bhella D, Patel AH. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J Virol 76:7672–7682. doi: 10.1128/JVI.76.15.7672-7682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]