ABSTRACT
Molluscum contagiosum virus (MCV), the only known extant human-adapted poxvirus, causes a long-duration infection characterized by skin lesions that typically display an absence of inflammation despite containing high titers of live virus. Despite this curious presentation, MCV is very poorly characterized in terms of host-pathogen interactions. The absence of inflammation around MCV lesions suggests the presence of potent inhibitors of human antiviral immunity and inflammation. However, only a small number of MCV immunomodulatory genes have been characterized in detail. It is likely that many more remain to be discovered, given the density of such sequences in other poxvirus genomes. NF-κB activation occurs in response to both virus-induced pattern recognition receptor (PRR) signaling and cellular activation by virus-induced proinflammatory cytokines like tumor necrosis factor and interleukin-1. Activated NF-κB drives cytokine and interferon gene expression, leading to inflammation and virus clearance. We report that MC005, which has no orthologs in other poxvirus genomes, is a novel inhibitor of PRR- and cytokine-stimulated NF-κB activation. MC005 inhibited NF-κB proximal to the IκB kinase (IKK) complex, and unbiased affinity purification revealed that MC005 interacts with the IKK subunit NEMO (NF-κB essential modulator). MC005 binding to NEMO prevents the conformational priming of the IKK complex that occurs when NEMO binds to ubiquitin chains during pathway activation. These data reveal a novel mechanism of poxvirus inhibition of human innate immunity, validate current dynamic models of NEMO-dependent IKK complex activation, and further clarify how the human-adapted poxvirus MCV can so effectively evade antiviral immunity and suppress inflammation to persist in human skin lesions.
IMPORTANCE Poxviruses adapt to specific hosts over time, evolving and tailoring elegantly precise inhibitors of the rate-limiting steps within the signaling pathways that control innate immunity and inflammation. These inhibitors reveal new features of the antiviral response, clarify existing models of signaling regulation while offering potent new tools for approaching therapeutic intervention in autoimmunity and inflammatory disease. Molluscum contagiosum virus (MCV) is the only known extant poxvirus specifically adapted to human infection and appears adept at evading normal human antiviral responses, yet it remains poorly characterized. We report the identification of MCV protein MC005 as an inhibitor of the pathways leading to the activation of NF-κB, an essential regulator of innate immunity. Further, identification of the mechanism of inhibition of NF-κB by MC005 confirms current models of the complex way in which NF-κB is regulated and greatly expands our understanding of how MCV so effectively evades human immunity.
KEYWORDS: NF-κB, immune evasion, innate immunity, molluscum contagiosum virus, poxvirus
INTRODUCTION
Host innate immune detection of virus infection employs pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) and cytosolic nucleic acid-sensing systems that stimulate signal transduction cascades leading to activation of the NF-κB (nuclear factor kappa light chain enhancer of activated B cells) and IRF (interferon [IFN] regulatory factor) transcription factor families. Such transcription factors induce type I IFNs and the proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin-1 (IL-1) (1). IFNs and cytokines then stimulate pathways that limit viral spread and direct antiviral acquired immunity. To overcome these host defense mechanisms, viruses evolve inhibitors that target the key rate-limiting steps in innate immune signaling. Thus, studying such inhibitors not only provides an understanding of viral pathogenesis but also reveals novel facets of the host innate signaling mechanisms that the inhibitors target, in addition to validating existing models describing the complexity of pathway activation. This provides avenues for therapeutic intervention in disorders defined by aberrant innate responses and inflammation. This is particularly true of poxviruses, which have evolved small inhibitory proteins by integrating host sequences into their genomes. These proteins then further evolve by refinement of their inhibitory activity through gradual mutation over long periods of virus-host evolution (2). Although the discovery and characterization of such poxvirus inhibitory proteins in nonhuman adapted poxviruses like vaccinia virus (VACV) have been highly informative in defining host-virus interactions and uncovering novel aspects of innate immunity (3), only a small number of these inhibitors have been described for the only known extant human-adapted poxvirus, molluscum contagiosum virus (MCV).
MCV is specifically adapted to human infection and has a genome predicted to encode 182 proteins, only 105 of which have orthologs in other orthopoxviruses (4). In contrast to non-human-adapted VACV, which causes local inflammation in human skin lesions, MCV can inhabit human dermal lesions over long periods of time with a minimal evident immune response and almost no inflammation, despite producing what are likely to be highly antigenic proteins. While this predicts that MCV has evolved unique, efficient inhibitors of human innate immunity, fewer than 10 MCV immunoregulators have been investigated in detail (5–7). Hence, we screened a library of open reading frames (ORFs) with no known function from the MCV genome for inhibition of human antiviral innate immune signaling networks that culminate in the activation of NF-κB, a critical proinflammatory transcription factor. This identified MC005, a protein with no orthologs in other poxvirus genomes, as a novel inhibitor of NF-κB activation. MC005 was previously shown to be expressed as an early gene product in MCV infection, consistent with a role in immunoregulation (8). Here we show that MC005 inhibited NF-κB activation stimulated by proinflammatory cytokines, PRR ligands, and DNA virus or RNA virus infection. Further functional analysis, including unbiased affinity purification to identify host targets of MC005, showed that MC005 binds the IκB kinase (IKK) complex through the regulatory subunit IKKγ/NEMO (NF-κB essential modulator). MC005 binds to NEMO in a specific region between the ubiquitin-binding UBAN (ubiquitin binding in ABIN and NEMO) domain and the region where NEMO binds the IKK subunits. As the current model of IKK activation suggests that ubiquitin binding causes a conformational change within the UBAN domain that conducts a change to the IKKs bound upstream, thereby priming them for activation, we propose that MC005 inhibits this conformational conduction of the priming signal, thus inhibiting IKK activation and thus downstream NF-κB activation. These discoveries not only validate the model of IKK regulation but also greatly extend our understanding of how MCV, the only known extant human-adapted poxvirus, so efficiently evades human immunity.
RESULTS
Identification of MC005 as a novel MCV inhibitor of PRR-driven NF-κB activation.
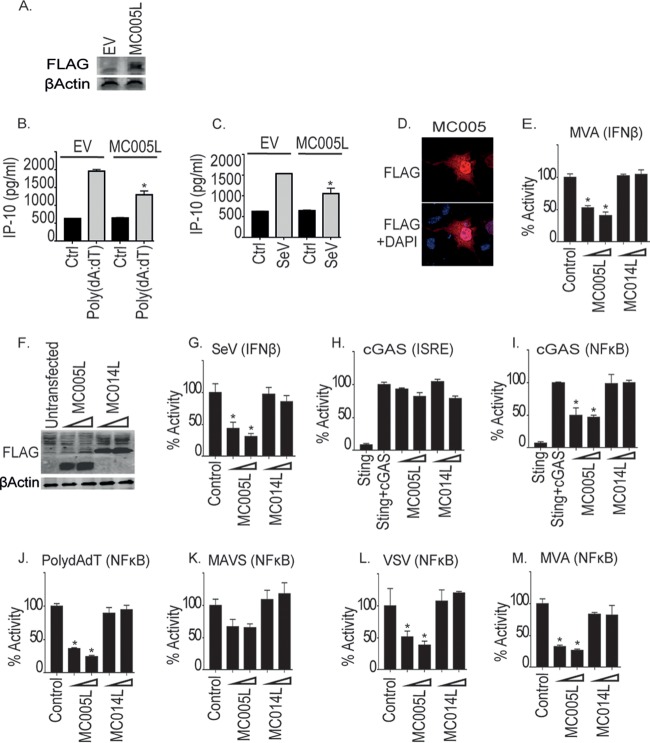
To identify novel MCV inhibitors of human innate antiviral sensing, we analyzed the MCV genome for ORFs predicted to encode small, soluble proteins of unknown function and tested the effect of the expression of a library of MCV ORFs on viral nucleic acid signaling, the first innate stimulus that occurs during viral infection and leads to the production of proinflammatory cytokines, chemokines, and type I IFNs (3). Initial screening identified MC005 as a potential inhibitor of nucleic acid signaling (data not shown). Because of the presence of a robust, well-characterized, endogenous nucleic acid sensing PRR system in the THP-1 human monocytic cell line, we generated stable MCV protein-expressing lines by lentivirus transduction and stable selection. Control empty-vector- and MC005-expressing stable cell lines were then stimulated by transfection with poly(dA-dT) double-stranded DNA prior to measurement of the production of IP-10 (CXCL10), an NF-κB- and IRF-dependent, PRR-inducible chemokine produced during poxvirus infection (9–11). Stable MC005 expression was detected (Fig. 1A), and this was correlated with a significant decrease in poly(dA-dT)-induced IP-10 that was observed in the presence of this viral protein (Fig. 1B). RNA virus-induced IP-10 production was also significantly inhibited by MC005 (Fig. 1C).
FIG 1.
Inhibition of cytosolic nucleic acid sensing pathway and virus-stimulated NF-κB activation by MC005. (A) THP-1 cells were transduced with lentiviruses containing the empty vector (EV) or MC005-FLAG-expressing constructs, and stable cells were generated. Extracts from cells were probed for expression of MC005-FLAG. Stable THP-1 cells were seeded at 1 × 106/ml and transfected with 1 μg/ml poly(dA-dT) (B) or infected with SeV (C) for 24 h. Supernatants were harvested from cells, and IP-10 production was assayed by ELISA. (D) Localization of MC005 HEK293T cells. Cells were transfected with 3 μg of the pCEP4-MC005-FLAG vector. Cells were fixed 24 h later and stained with DAPI (blue) or for MCV protein expression (red). Representative images are shown (n = 4). (E to L) HEK293T cells were seeded at 2 × 105/ml and transfected with the reporter genes indicated and 25 or 50 ng of the empty-vector (control [Ctrl]) or pCEP4 plasmid expressing MC005-FLAG or MC014-FLAG (indicated by a wedge). Cells were then infected with MVA virus (E, M), SeV (G), or VSV (L) for 16 h or transfected with 25 ng of cGAS and 25 ng of STING (H, I) or 1 μg/ml poly(dA-dT) (J). NF-κB or ISRE reporter activity was assayed 24 h later. Data are the mean ± the standard deviation of triplicate samples from a representative experiment (n = 3). *, P < 0.001 compared to the control.
To determine whether inhibition by MC005 was at the level of promoter induction, we next examined the effect of MC005 on the activity of the IFN-β promoter, which, like IP-10, is NF-κB and IRF dependent, by luciferase reporter gene assay. As an in vitro MCV infection model is not currently available, we examined the effect of MC005 expression on the activity of the IFN-β promoter induced by infection of human embryonic kidney 293T (HEK293T) cells with the distantly related poxvirus modified vaccinia virus Ankara (MVA). MC005 is a 9-kDa protein that, when expressed in HEK293T cells, is localized throughout the cells (Fig. 1D). Figure 1E shows a significant and dose-dependent inhibition of IFN-β promoter activation by MC005 but not by MC014, an irrelevant MCV protein that was expressed at levels similar to those of MC005 (Fig. 1F). A similar dose-dependent inhibition of IFN-β promoter activation by MC005 was seen after Sendai virus (SeV) infection of these cells (Fig. 1G).
As virus- and PRR-induced IFN-β and IP-10 induction occurs through both NF-κB and IRF family transcription factor activation, we next examined the effect of MC005 on the activation of these transcription factors. After entry into cells and uncoating, sensing of poxvirus during infection can occur through DNA genome sensing by the cyclic guanosine AMP synthase (cGAS) cytosolic DNA sensor, which then signals to transcription factors via STING (stimulator of IFN genes) (12, 13). Coexpression of cGAS and STING in HEK293T cells, which do not normally contain these proteins, reconstitutes cGAS-STING signaling (7, 14), leading to the activation of both NF-κB and IRF. Interestingly, while MC005 had no significant effect on cGAS-induced IRF activation (measured by IFN-sensitive response element [ISRE] promoter-driven luciferase expression), cGAS-induced NF-κB activation (measured by κB promoter-driven luciferase expression) was inhibited (Fig. 1H and I).
In HEK293T cells, RNA polymerase III transcribes AT-rich DNA such as poly(dA-dT) into an RNA ligand that activates the RIG-I/MAVS (mitochondrial antiviral signaling) sensing system, and this system has been implicated in the sensing of intracellular poxvirus DNA (15). Poly(dA-dT) stimulation of NF-κB activation was also inhibited by MC005, as was NF-κB activation with MAVS (Fig. 1J and K). We next examined the effect of MC005 on virus-induced NF-κB activation and found that MC005 also inhibited the activation of this transcription factor by infection with either the RNA virus vesicular stomatitis virus (VSV) or the poxvirus MVA (Fig. 1L and M).
TLR3 and TLR9 have been shown to play key roles in the sensing of poxvirus infection in vivo (16, 17), and both are characteristically upregulated in MCV-infected lesions despite a lack of inflammation or viral clearance in such lesions under most circumstances (18). We next examined the effect of MC005 expression on TLR9 and TLR3 signaling by using constitutively active CD4 fusions of these PRRs. Similar to the other PRR systems investigated, MC005 inhibited NF-κB activation, but not IRF activation, by these pathways (Fig. 2 A to D). Additionally, activation of the TLR3 pathway further downstream by expression of its adapter TRIF activated both NF-κB and IRFs, and as for the other stimuli, MC005 inhibited NF-κB, but not IRF, activity (Fig. 2E and F), suggesting that the point of inhibition lay further downstream. Together, these data suggested that MC005 targeted PRR-stimulated NF-κB, but not IRF, activity.
FIG 2.
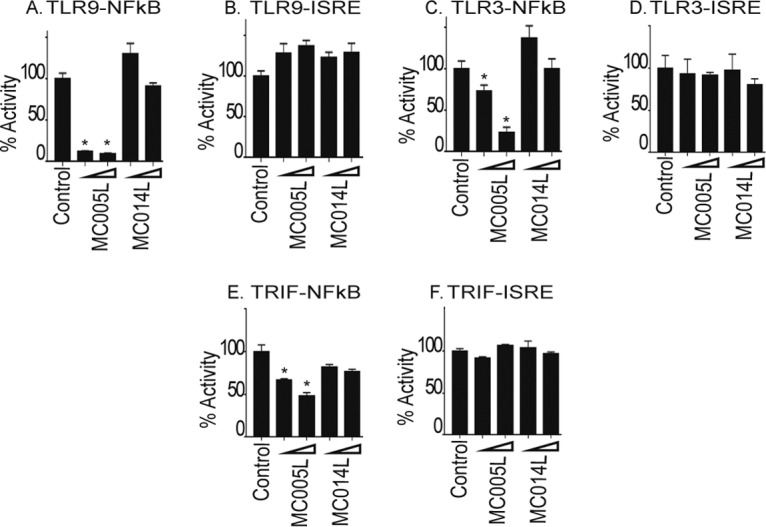
Inhibition of TLR-dependent NF-κB activation by MC005. The effect of MC005 on TLR9/3-stimulated NF-κB activation was examined. HEK293T cells were seeded at 2 × 105/ml and transfected with 80 ng of NF-κB or ISRE reporter gene, 40 ng of TK Renilla reporter gene, and 25 or 50 ng (indicated by a wedge) of empty-vector (control) or pCEP4 plasmid expressing the MCV ORFs indicated and CD4-TLR9 (A, B), CD4-TLR3 (C, D), or TRIF (E, F). Cells were harvested 24 h later and assayed for reporter gene activity. Data are percentages of the stimulation activity in control cells and are the mean ± the standard deviation of triplicate samples from a representative experiment (n = 3). *, P < 0.001 compared to the control.
MC005 inhibits NF-κB activation stimulated by the proinflammatory cytokines TNF and IL-1.
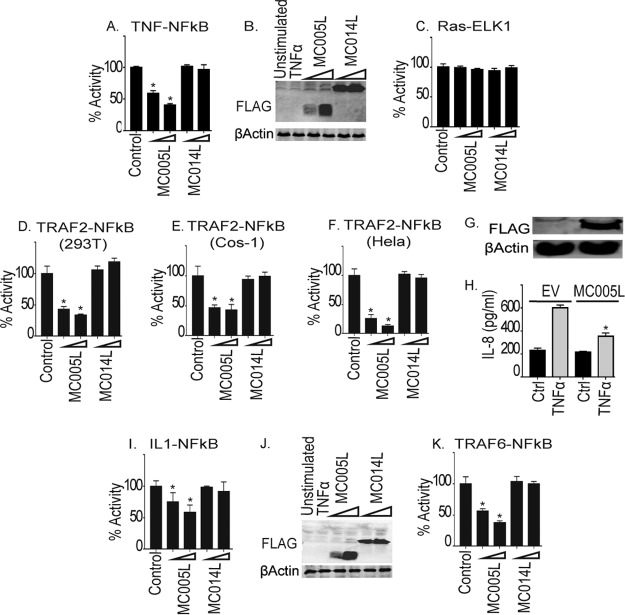
In addition to type I IFNs, primary virus sensing through PRRs leads to the upregulation and secretion of proinflammatory cytokines that directly drive inflammation in infected tissue to create the conditions necessary for an adaptive immune response to occur, leading to virus clearance. Such inflammation is absent for extended periods during MCV infection. Having demonstrated that MC005 inhibits NF-κB, but not IRF, signaling by PRRs, we next examined whether MC005 would also suppress proinflammatory cytokine-induced NF-κB activity. Similar to PRR-driven NF-κB activation, we observed that TNF-activated NF-κB was also inhibited by MC005 but not by MC014 at equivalent levels of expression (Fig. 3A and B). The specific effect of MC005 on NF-κB activation was further suggested by the lack of effect on an unrelated mitogenic signaling pathway activated by a constitutively active form of Ras, RasVHa (19) (Fig. 3C). There, RasVHa activation of Elk1 is monitored by using an Elk-1/GAL4 DNA-binding domain fusion that induces a GAL4 promoter luciferase system.
FIG 3.
MC005 inhibition of proinflammatory cytokine-stimulated NF-κB activation downstream of TRAFs. The effect of MC005 on TNF-stimulated NF-κB activation was examined. (A) HEK293T cells were seeded at 2 × 105/ml; transfected with 80 ng of NF-κB reporter gene, 40 ng of TK Renilla reporter gene, and 25 or 50 ng (indicated by a wedge) of empty-vector (control) or pCEP4 plasmid expressing the MCV ORFs indicated; stimulated with 50 ng/ml TNF-α for 6 h; harvested; and then assayed for NF-κB reporter gene activity. Data are percentages of the stimulation activity in control cells and are the mean ± the standard deviation of triplicate samples from a representative experiment (n = 4). (B) Extracts of samples from the experiment in panel A were probed for expression of FLAG-tagged viral proteins. (C) Elk1 activation by Ras was measured by reporter gene assay of cells transfected with empty-vector (control) or pCEP4 plasmid expressing MCV ORFs. (D to F) Same as for panel A, except that HEK293T (D), COS-1 (E), or HeLa (F) cells were transfected with 50 ng of TRAF2-FLAG instead of being subjected to TNF-α stimulation. (G, H) Stable cells containing the pCEP4 empty vector or pCEP4-MC005-FLAG were prepared, and expression of FLAG-tagged protein was probed (G). Stable cells were then seeded at 2 × 105/ml and stimulated with 50 ng/ml TNF-α for 24 h. IL-8 production was then assayed by ELISA (H). (I) Effect of MC005 on IL-1-stimulated NF-κB activation. Same as for panel A, except that cells were stimulated with 50 ng/ml IL-1β. (J) Extracts of samples from the experiment in panel I were probed for expression of FLAG-tagged viral proteins. (K) Same as for panel A, except that cells were transfected with 50 ng of plasmid expressing TRAF6. Data are percentages of the stimulation activity in control cells and are the mean ± the standard deviation of triplicate samples from a representative experiment (n = 3). *, P < 0.001 compared to the control.
We next examined the effect of MC005 on the activation of NF-κB by expression of TRAF2, a key regulatory component of the TNF receptor signaling pathway. MC005 inhibited NF-κB by TRAF2 in three different cell lines, HEK293T (Fig. 3D), Cos-1 (Fig. 3E), and HeLa (Fig. 3F), demonstrating that MC005 inhibition was independent of the cell type. We next established an MC005-expressing HEK293T stable cell line to investigate the effect of MC005 expression on TNF-α-driven cytokine production. Using empty-vector- and MC005-expressing, stable cells (Fig. 3G), we demonstrated that NF-κB inhibition by MC005 translated into suppression of secretion of IL-8, an important NF-κB-dependent chemokine produced during inflammation (Fig. 3H).
In addition to TNF-α, IL-1β plays a key role in local and systemic, virus-induced inflammation, and both production and activity of this cytokine are heavily targeted by poxviruses (3). As with other stimuli, MC005, but not MC014, inhibited the activation of NF-κB by IL-1 at similar levels of expression (Fig. 3I and J). MC005 also inhibited NF-κB activation by TRAF6, a key component of the IL-1 signaling pathway (Fig. 3K).
MC005 inhibits NF-κB activation at the level of the IKK complex.
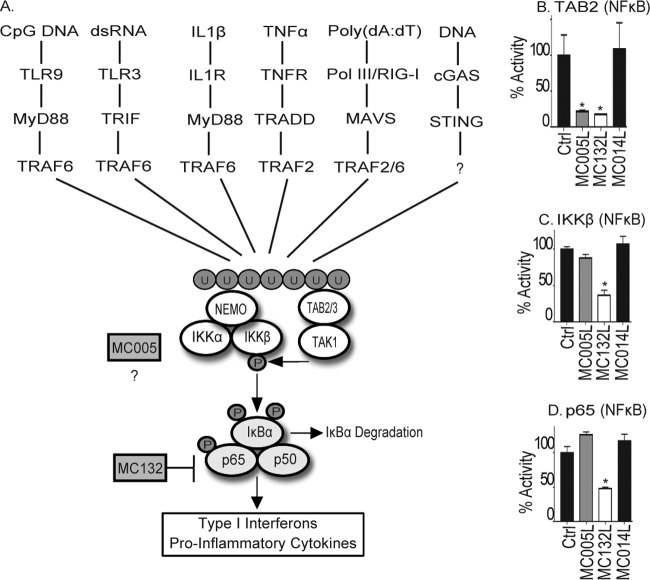
Having demonstrated thus far that MC005 can inhibit both PRR sensing and proinflammatory cytokine-stimulated activation of NF-κB, we suspected that this viral inhibitor was targeting a point in signaling common to both sets of pathways. PRR- and proinflammatory cytokine-stimulated NF-κB activation is catalyzed by the activation of RING domain-containing TRAF proteins like TRAF2 in the case of TNF and TRAF6 in the case of both IL-1 and PRR signaling (Fig. 4A), which generate long chains of ubiquitin at activation foci within cells. After the generation of these ubiquitin chains, two complexes bind to the chains in close proximity to each other, the TAB/TAK complex and the IKK complex. The IKK complex is composed of a regulatory ubiquitin-binding protein, NEMO (also known as IKKγ), that regulates the activity of the active kinase subunits IKKα and IKKβ. Ubiquitin binding by NEMO in the IKK complex causes a conformational change in NEMO, which primes IKKβ for phosphorylation and activation by TAK1 (20, 21). Once active, IKKβ, for example, then phosphorylates both the inhibitory protein IκBα (driving its proteasomal degradation) and the p65 subunit of NF-κB, releasing the transcription factor to translocate to the nucleus and induce antiviral and proinflammatory gene expression.
FIG 4.
MC005 inhibits PRR- and cytokine-stimulated NF-κB activation at a point proximal to the IKK complex. (A) Schematic showing multiple signal transduction pathways to NF-κB expected to be activated during a poxvirus infection, all of which are shown to be sensitive to MC005 inhibition (Fig. 1 to 3) and by MC132 targeting of p65. After the generation of ubiquitin chains, three key regulatory events occur that lead to canonical NF-κB activation. (i) The IKK complex (through NEMO) and the TAB/TAK complex (through the TABs) bind to ubiquitin chains. (ii) Ubiquitin binding of NEMO induces a conformational change that affects the entire IKK complex presenting the IKKs for autophosphorylation and phosphorylation by TAK1. (iii) Active phosphor-IKKβ dual phosphorylates both p65 and IκBα (triggering its degradation), leading to nuclear translocation of NF-κB and induction of antiviral and proinflammatory gene expression. (B to D) Comparison of MC005 with MC132 inhibition of signaling events surrounding IKK complex activation. HEK293T cells were seeded at 2 × 105/ml; transfected with 50 ng of the empty-vector (control [Ctrl]) or pCEP4 plasmid expressing the MCV ORFs indicated together with 10 ng of TAB2 (B), 10 ng of IKKβ (C), or 5 ng of p65 (D); harvested; and then assayed for NF-κB reporter gene activity 24 h later. Data are percentages of the stimulation activity in control cells and are the mean ± the standard deviation of triplicate samples from a representative experiment (n = 3). *, P < 0.001 compared to the control.
The data presented thus far suggested that MC005 targets NF-κB activation downstream of TRAF2/6. Thus, we next investigated the effect of MC005 expression on the signaling events proximal to NF-κB activation described above, in comparison to another MCV protein, MC132, which we recently showed targets p65 for degradation (7). Stimulating NF-κB activation by the TAB/TAK complex (by overexpressing TAB2), we observed that both MC005 and MC132 inhibited NF-κB activation (Fig. 4B). In contrast to this, stimulating NF-κB activation by IKKβ (where overexpression of the kinase simulates constitutive activation of the kinase without the normal regulation exerted on it by the IKK complex in the absence of an upstream signal) bypassed the inhibitory activity of MC005, but not that of MC132, which targets p65 directly (Fig. 4C). A similar result was observed when IKKα was expressed (data not shown). Consistent with MC005 inhibiting at the level of IKK complex activation, we found that activating the NF-κB-dependent reporter gene by overexpressing p65 similarly bypassed the inhibitory activity of MC005 but not that of MC132 (Fig. 4D). These data thus suggested that MC005 was targeting NF-κB activation at the level of the IKK complex.
MC005 interacts with NEMO.
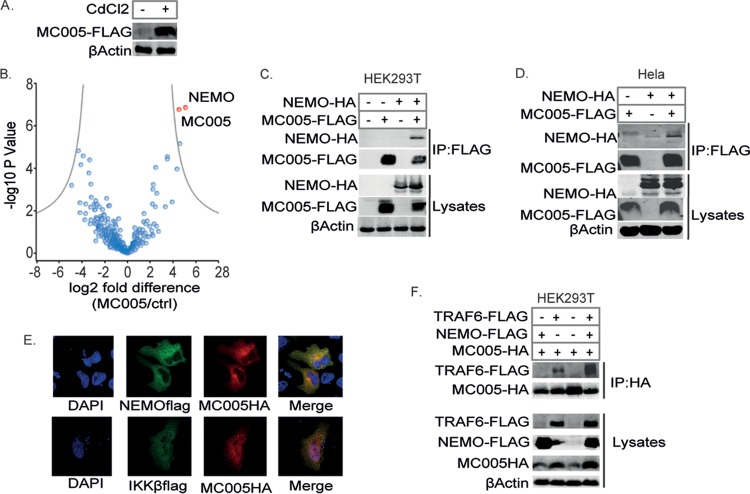
To gain insight into the mechanism by which MC005 was inhibiting at the level of the IKK complex, we investigated the cellular interactions of this viral protein by performing unbiased affinity purification combined with mass spectrometry (AP-MS). To do this, we generated a metallothionein promoter-inducible, MC005-expressing, stable HEK293T cell line in which high levels of MC005-FLAG could be induced with CdCl2 (Fig. 5A). Using this system, we performed an analysis of MC005 cellular interaction partners by AP-MS. This showed that NEMO was the most significantly enriched protein in the MC005 immunoprecipitates (Fig. 5B).
FIG 5.
MC005 interacts with NEMO. Analysis of cellular MC005-interacting proteins. (A) HEK293T cells stably transfected with pMEP4-MC005 and treated with (+) or without (−) 1 μM CdCl2 to induce expression. Lysates were probed for FLAG protein expression. (B) Volcano plot of MC005-interacting proteins identified by AP-MS from MC005-expressing HEK293T cells. (C to E) MC005 interacts with NEMO and the IKK complex. HEK293T (C) or HeLa (D) cells were seeded at 4 × 106/10-cm plate and transfected with 4 μg of expression vectors for NEMO-HA and MC005-FLAG as indicated. Twenty-four hours later, cells were lysed and immunoprecipitated (IP) with anti-FLAG antibody-coated beads and then immunoblotted with the antibodies indicated. Representative blots (n = 3) are shown. (E) HeLa cells seeded into 24-well dishes on coverslips were transfected with 0.5 μg of expression vectors for NEMO-FLAG, IKKβ-FLAG, and MC005-FLAG. Cells were fixed 24 h later and stained with DAPI (blue) or for HA (red) and FLAG (green). MC005 interacts with TRAF6 through NEMO association. (F) Same as for panel C, except that cells were transfected with NEMO-HA, MC005-HA, and TRAF6-FLAG. Representative blots (n = 3) are shown.
We confirmed this interaction by expressing NEMO and MC005 and demonstrating that these proteins coimmunoprecipitated in both HEK293T cells (Fig. 5C) and HeLa cells (Fig. 5D). We also demonstrated the colocalization of MC005 (predominantly localizing in the cytoplasm of HeLa cells) with both NEMO and IKKβ by confocal microscopy (Fig. 5E). These data suggested that MC005 inhibited NF-κB activation by binding NEMO within an intact IKK complex downstream of TRAF activation. To confirm this, we investigated the association of MC005 with TRAF6. While a low-level interaction with TRAF6 was evident, this interaction became significantly more pronounced when NEMO was overexpressed, suggesting that NEMO is the specific interaction target of MC005, bridging an association with activated TRAF6 (Fig. 5F).
A central 46-amino-acid motif is required for MC005 inhibition.
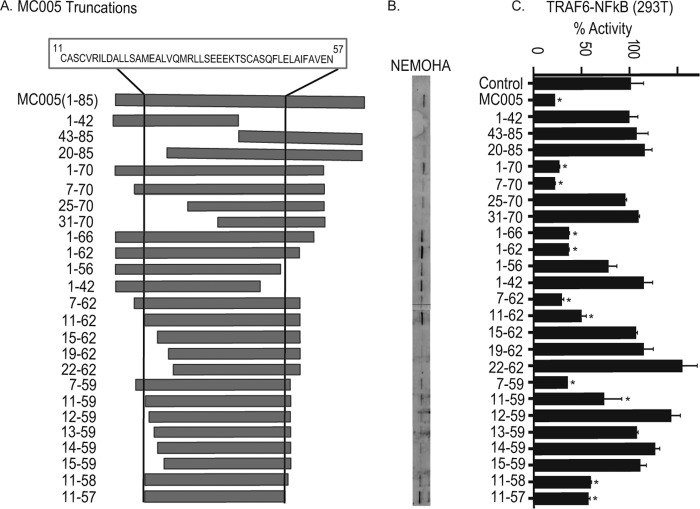
To determine the region of MC005 required for inhibition of NF-κB activation, we created a series of truncations of MC005 starting with the removal of large sections of the N and C termini and then narrowing down the region required for inhibition by progressive deletion of both the N and C termini. Through this systematic approach, we found that the first 10 amino acids of the N terminus and the last 29 amino acids of the C terminus were dispensable for inhibitory activity, since only MC005 fragments containing residues 11 to 57 gave more than 50% NF-κB inhibition (Fig. 6A and C). Interestingly, in most cases, the ability of an MC005 truncation to inhibit NF-κB correlated with its ability to bind NEMO (Fig. 6B and C), further confirming this protein as the specific target through which MC005 inhibits NF-κB activation.
FIG 6.
Inhibition of NF-κB by MC005 requires a central 46-amino-acid region. (A) Generation of MC005 truncations. Constructs expressing MC005 truncations were generated by truncation at the position in the protein indicated. The region and sequence required for activity are indicated at the top, and the region is marked in truncations by vertical lines. (B) Interaction of MC005 and truncations with NEMO correlates with inhibitory activity. Cells were seeded at 4 × 106/10-cm plate and transfected with 4 μg of expression vectors for NEMO-HA and MC005-FLAG as indicated. Twenty-four hours later, cells were lysed, immunoprecipitated with anti-FLAG antibody-coated beads, and then immunoblotted with the antibodies indicated. A representative blot is shown (n = 3). (C) Central 46-amino-acid region of MC005 required for NF-κB inhibition. HEK293T cells were seeded at 2 × 105/ml and transfected with 80 ng of NF-κB, 40 ng of TK Renilla reporter gene, 10 ng of TRAF6, and 50 ng of the empty vector (control) or pCEP4 plasmid expressing MC005 and truncations. Cells were harvested 24 h later and assayed for reporter gene activity. Data are percentages of the stimulation activity in control cells and are the mean ± the standard deviation of triplicate samples from a representative experiment (n = 3). *, P < 0.001 compared to the control.
MC005 prevents ubiquitin binding-dependent regulation of IKKβ activation by NEMO.
While purified NEMO is dimeric in solution (22), cross-linking experiments indicate that the endogenous IKK complex is composed of four NEMO molecules and two molecules each of IKKα and IKKβ, consistent with its apparent molecular mass of 700 to 900 kDa (23). This complex preexists prior to stimulation, and the intact, endogenous, oligomeric complex is necessary for canonical NF-κB activation (24). NEMO possesses a series of domains and regions that play key roles in complex assembly and in the regulation of IKK activation by bridging appropriate upstream signals to IKK activation. Thus, NEMO is a central regulatory cog in the control of the initiation of inflammation and a logical target for viral inhibition. Residues 1 to 80, which encompass the N terminus and half of the coiled coil 1 (CC1) domain, have been shown to be vital for NEMO dimerization (Fig. 7A) (20). Residues 40 to 120 are where NEMO interacts with the IKKs, while residues 249 to 339, a region consisting of the CC2 domain and a leucine zipper (LZ) motif, make up the UBAN domain, which binds TRAF-regulated ubiquitin chains (Fig. 7A). The UBAN domain also regulates the conformational change that occurs within NEMO that induces a “priming” of IKKα and IKKβ for subsequent autophosphorylation and phosphorylation by TAK1.
FIG 7.
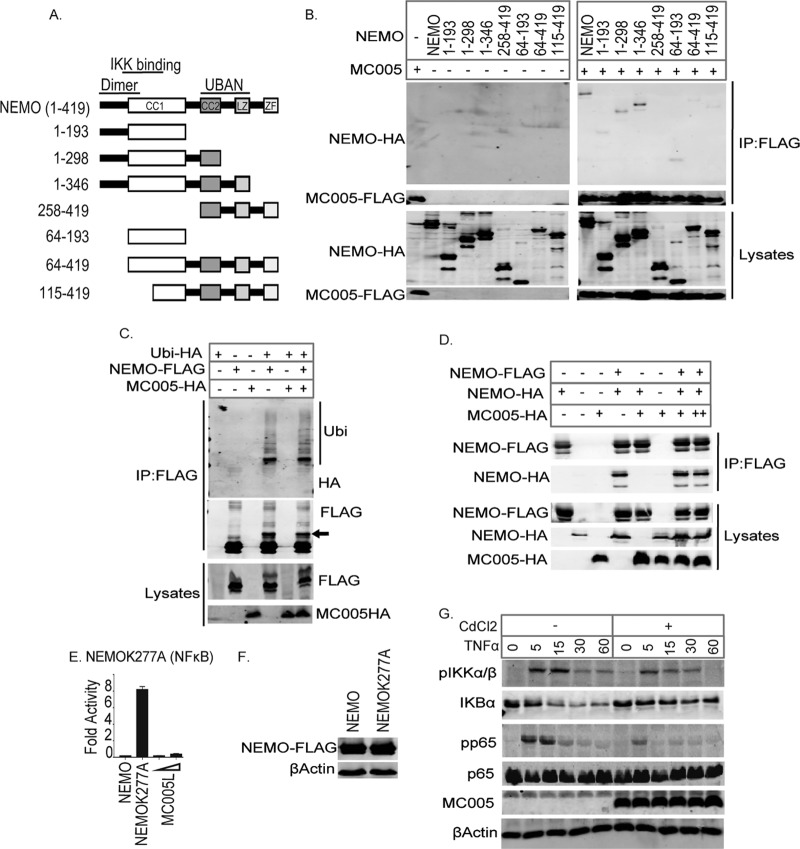
MC005 prevents ubiquitin-activated NEMO from positively regulating IKKβ. MC005 binds the second half of the NEMO CC1 domain. (A) Generation of NEMO truncations. The regions of note in NEMO are the dimer region (1 to 80), the IKK-binding region (40 to 120), and the UBAN domain (249 to 339). The CC, LZ, and zinc finger (ZF) regions are also indicated. Constructs expressing NEMO truncations were generated by truncation at the positions in the protein indicated. (B) Cells were seeded at 4 × 106/10-cm plate and transfected with 4 μg of expression vectors for NEMO-HA, NEMO truncation-HA, and MC005-FLAG as indicated. Twenty-four hours later, cells were lysed and immunoprecipitated (IP) with anti-FLAG antibody-coated beads and then immunoblotted with the antibodies indicated. A representative blot is shown (n = 3). MC005 does not disrupt NEMO ubiquitination or association with ubiquitinated proteins. (C) Cells were seeded at 4 × 106/10-cm plate and transfected with 4 μg of expression vectors for NEMO-FLAG, ubiquitin-HA (Ubi-HA), and MC005-HA as indicated. Twenty-four hours later, cells were lysed, immunoprecipitated with anti-FLAG antibody-coated beads, and then immunoblotted with the antibodies indicated. A representative blot is shown (n = 3). Arrow indicates ubiquitinated NEMO. MC005 does not prevent IKK complex assembly. (D) Cells were seeded at 4 × 106/10-cm plate and transfected with 4 μg of expression vectors for NEMO-HA, NEMO-FLAG, and MC005-HA as indicated. Twenty-four hours later, cells were lysed, immunoprecipitated with anti-FLAG antibody-coated beads, and then immunoblotted with the antibodies indicated. A representative blot is shown (n = 3). (E, F) Constitutively active K277A mutant NEMO simulating the conformation of ubiquitin-bound NEMO is inhibited by MC005. (E) HEK293T cells were seeded at 2 × 105/ml and transfected with 80 ng of NF-κB, 40 ng of TK Renilla reporter gene, 50 ng of NEMO or NEMOK277A, and 25 or 50 ng (indicated by a wedge) of empty-vector (control) or pCEP4 plasmid expressing MC005. Cells were harvested 24 h later and assayed for reporter gene activity. Data are percentages of the stimulation activity in control cells and are the mean ± the standard deviation of triplicate samples from a representative experiment (n = 3). (F) Extracts of samples from the experiment in panel E were probed for expression of FLAG-tagged MC005. MC005 interaction with the IKK complex inhibits IKKβ and p65 phosphorylation and IκB degradation. (G) HEK293T cells stably transfected with pMEP4 and pMEP4-MC005 were seeded at 6 × 105/well into six-well dishes and treated with (+) or without (−) 1 μM CdCl2 to induce MCV protein expression. Twenty-four hours later, cells were stimulated with 50 ng/ml TNF-α for the times indicated and cell lysates were immunoblotted with the antibodies indicated. Representative blots are shown (n = 3). *, P < 0.001 compared to the control.
To further characterize the mechanism by which MC005 was inhibiting the IKK complex by binding NEMO, we next investigated the region of NEMO to which MC005 was binding. To do this, we generated a series of truncations of NEMO corresponding to the known functional domains and regions within this protein (Fig. 7A). While all of the truncations of NEMO were expressed to approximately equivalent levels (Fig. 7B), MC005 was able to interact with all of these truncations except one truncation lacking a sequence N terminal of the CC2 domain (directly between the IKK-binding region and the UBAN domain) (Fig. 7B). Given that it effectively interacted with a truncation possessing the second half of the CC1 domain, this suggests that the viral protein is binding to a region between the second half of the CC2 domain and the beginning of the CC2 domain of NEMO.
We next examined the effect of MC005 on the regulatory events controlled by NEMO in inflammatory signaling and on IKK complex formation. We first investigated the effect of MC005 on the ubiquitination of NEMO and on its association with ubiquitin chains, which is critical for its regulation and activation of the IKK complex at signaling foci within the cell. While we observed that expression of ubiquitin-hemagglutinin (HA) induced both NEMO ubiquitination (indicated by a ubiquitin-HA-dependent shift in the mass of NEMO-FLAG) and the association of this protein with ubiquitinated proteins, MC005 had no effect on these processes and interactions (Fig. 7C). To investigate whether the association of MC005 was disrupting the formation or assembly of the multimeric IKK complex, we next examined the effect of MC005 on the ability of NEMO to oligomerize with itself. This demonstrated that the NEMO-NEMO interaction was not affected by the presence of the viral protein (Fig. 7D). Additionally, the close colocalization of MC005 with IKKβ (Fig. 5E) suggested that MC005 was not disrupting the association of the kinase subunits and thus was binding an intact IKK complex.
While heterologous expression of NEMO does not drive activation of the complex (as NEMO still requires ubiquitin chains from upstream activation events to mediate IKK activation), previous work demonstrated that a single mutation within the UBAN domain of murine NEMO simulates the conformational state that NEMO assumes after ubiquitin binding, which primes IKKβ for phosphorylation by TAK1 (25). We generated the human equivalent site mutation in NEMO (K277A) and demonstrated that while wild-type NEMO did not activate NF-κB, NEMOK277A potently activated this transcription factor even though mutant and wild-type NEMO were expressed to equivalent levels (Fig. 7E and F). Interestingly, MC005 inhibited constitutively active NEMOK277A, suggesting that MC005 binding to active NEMO prevents NEMO from conferring conformational priming to IKKβ. We next examined the direct consequences of MC005 binding to active NEMO by stimulating the metallothionein-inducible MC005 stable cells used previously (Fig. 5A) with TNF-α and observed that while TNF stimulated the IKK phosphorylation, IκB degradation, and p65 phosphorylation necessary for NF-κB activation, inducing the expression of MC005 inhibited all of these events (Fig. 7G).
The above data, taken in aggregate, suggest a model whereby MC005 binds NEMO and inhibits the activity of the conformational state that NEMO assumes on binding ubiquitin chains, which normally primes IKKβ for the phosphorylation events required for full catalytic activity (Fig. 8). Thus, MC005 binding to NEMO prevents IKKβ from phosphorylating IκBα and p65, thus inhibiting NF-κB activation and the induction of NF-κB-dependent gene expression. MC005 inhibition of active NEMO provides a rationale for inhibition of type I IFN, cytokine, and chemokine production after primary virus sensing and proinflammatory cytokine stimulation.
FIG 8.
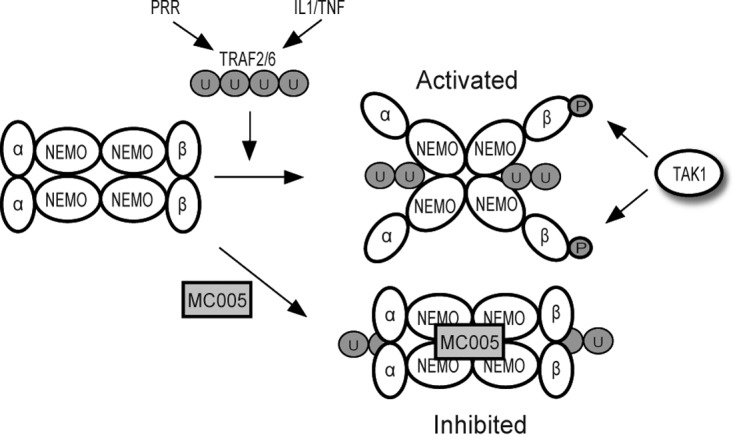
Model of MC005-mediated inhibition of human NF-κB signaling. PRR virus sensing and cytokine signaling pathways drive polyubiquitination events required for IKK complex activation. NEMO subunits bind TRAF-generated ubiquitin chains, which induces a conformational change within NEMO and a subsequent conformational change and rearrangement within the IKK complex leading to presentation of the IKKs (e.g., IKKβ) for phosphorylation by TAK1 and autophosphorylation. MC005 inhibits IKK complex activation by binding to active NEMO and preventing the conformational signal (induced by ubiquitin binding) from priming the IKKs for subsequent activation.
DISCUSSION
MCV is a common dermatotropic poxvirus that causes benign skin neoplasms in humans with a more serious presentation in immunocompromised individuals like HIV patients (6). Compared to other poxviruses like VACV, which causes local inflammation in human skin lesions (26), MCV causes less of an inflammatory response. One factor contributing to this may be the fact that MCV forms histologically distinct, walled-off lesions that are less accessible to the immune system. However, keratinocytes present in such lesions do have efficient viral sensing machinery, suggesting that MCV is better equipped than poxviruses like VACV to suppress human innate immunity, as a result of long-term coevolution and adaptation to human infection. NF-κB has a critical role in virus-sensing pathways and the initiation of both virus-induced inflammation and the adaptive response to viruses, and notwithstanding the ability of VACV to trigger local inflammation in skin lesions, VACV possesses numerous immunomodulatory proteins, including at least 10 inhibitors of NF-κB, with evidence of others yet to be identified (27). In contrast, prior to this study, fewer than 10 MCV immunomodulators had been reported, and only 3 of these affect NF-κB activation through partially characterized mechanisms (7, 28–30). We recently reported the discovery of an additional inhibitor, MC132, that inhibits NF-κB by targeting the p65 subunit for proteasomal degradation by recruiting the host cullin-5/elongin B/elongin C complex (7). Although MCV research has been hampered by the lack of an animal or cell line infection model, analysis of the function of isolated ORFs expressed in cell lines has previously revealed important insights into how MCV proteins suppress host immunity (6, 7).
Since the study of poxvirus immunoregulators that inhibit the host response has been instructive in defining host-pathogen interactions and discovering new aspects of innate immunity (3), MCV, having coevolved to specifically inhibit human immunity, is unparalleled as a model poxvirus for understanding human innate immunity. Thus, we screened isolated MCV ORFs for effects on known antiviral innate immune signaling pathways in human cells and here report the discovery of MC005 from MCV subtype I as an inhibitor of NF-κB activation. MC005 is a small, 89-amino-acid (9-kDa) protein encoded by MC005L, which is located at the left-hand terminus of the MCV genome; has no orthologues in any other known poxviruses; and exhibits no similarity to any other proteins.
TLRs and cytosolic nucleic acid detection PRRs are critically involved in initial viral sensing and type I IFN induction, while IL-1 and TNF production and subsequent signaling regulate virus-induced inflammation, which is required for mounting a full adaptive response leading to virus clearance (3). Compellingly, MC005, like MC132, inhibited both PRR and inflammatory cytokine signaling to NF-κB activation, as well as suppressing NF-κB activation and chemokine production following poxvirus or RNA virus infection.
Through systematic mapping of these pathways, we tracked the inhibitory effect of MC005 to the IKK complex and determined that it specifically interacted with NEMO, the regulatory subunit of the complex. As a focal point in IFN induction and inflammatory signaling, the subunits of the IKK complex are commonly targeted by viruses (31). The MCV protein MC159 has previously been shown to interact specifically with NEMO to inhibit NF-κB activation (28); however, the authors did not describe in detail how MC159 interacts with NEMO and inhibits IKK activation. Interestingly, although the cGAS-STING pathway induces NF-κB-dependent genes, the mechanism whereby STING activates NF-κB is unknown. Since MC005 inhibited cGAS-stimulated NF-κB activation, this suggests that NEMO does have a role in this pathway.
As our current understanding of the precise dynamic mechanism by which the IKK complex is activated is incomplete, we are in need of new tools to bolster existing models of activation of this critical regulatory crux in innate immunity and inflammation. What is clear is that ubiquitin chains (linked to upstream signaling proteins) serve as a “code” that is “read” and translated into a signal by ubiquitin-binding proteins like NEMO (which binds K63-linked and Met1-linked polyubiquitin chains), as well as TAB2/3 (which binds K63-linked polyubiquitin chains only), and several lines of evidence demonstrate that ubiquitin binding by these proteins is essential for activation of the IKK complex (20, 32, 33). This requirement for ubiquitin binding appears to be not solely for generating a scaffold for coclustering IKK and TAB/TAK complexes; rather, it also appears to be required for priming these complexes for cross- and autophosphorylation via conformational changes within complexes. A comparison of the crystal structure of the free and ubiquitin-bound forms of the NEMO UBAN domain revealed that ubiquitin binding induced a straightening of the CC2 domain (34), and those authors suggest that this may function as an allosteric means of inducing or conducting a conformational change in the IKK subunits of the multimeric complex to a form capable of autophosphorylation and phosphorylation by TAK1. Indeed, a single point mutation in the CC2 domain of murine NEMO (K270A) makes the IKK complex constitutively active without the requirement for ubiquitin binding by forcing NEMO into a conformation that simulates the ubiquitin-bound state (25). We observed that MC005 inhibited the constitutively active equivalent human point mutant form of NEMO (K277A).
We also demonstrated that MC005 binds to a point in NEMO directly between the regions that mediate IKK binding and ubiquitin binding. This binding does not impair NEMO dimerization in the multimeric IKK complex, nor does it prevent the binding of NEMO to ubiquitin. As the IKKs bind within the first half of the CC1 domain and the current model of IKK activation suggests that UBAN-ubiquitin binding causes a conformational straightening of the CC2 domain, which induces a conformational repositioning of the IKKs, priming them for phosphorylation by TAK1, we propose that MC005, by binding between these two regions, inhibits the conduction of this conformational signal within NEMO. Thus, the viral protein not only serves as a way of better understanding how MCV inhibits virus sensing and inflammation in human tissue but also serves as a useful tool for validating the evolutionarily conserved structural mechanism by which the IKK is regulated and activated. This better understanding might inform therapeutic strategies for inhibiting the IKK complex in inflammatory and autoimmune diseases.
Interestingly, the sequence of MC005 differs between MCV subtypes I and II. While subtypes I and IV are the most common in infection of immunocompetent individuals, subtype II is more common in HIV patients (35, 36), in whom MCV causes a more severe, disseminated disease, and is a good indicator of advancing immunosuppression (37). The subtype II MC005 variant is shorter than type I, including a 7-amino-acid C-terminal deletion and several conservative and nonconservative amino acid differences (residues 41, 44, 54, and 55) that lie within the region we have determined to be required for inhibitory activity (residues 11 to 57). It is possible that disrupted or altered activity of MC005 in MCV subtype II may affect the dynamics of infection in immunocompromised hosts, explaining this difference in its tropism and presentation.
Overall, our analysis of the MCV genome for ORFs that affect human signaling networks culminating in activation of NF-κB revealed MC005, an inhibitor of PRR- and cytokine-activated NF-κB unique to MCV among poxviruses, and delineated the precise mechanism of its action within the known dynamics of IKK complex regulation. Thus, four MCV proteins have now been shown to target NF-κB activation, namely, MC159, MC160, MC132, and MC005. Further screening of the MCV genome for novel inhibitors of human innate immunity will likely reveal additional inhibitors of innate immune signaling and novel details of the signaling pathways they antagonize and may present new strategies for selective inhibition of sensing and inflammatory events in disease.
MATERIALS AND METHODS
Cell culture and viruses.
HEK293T, HeLa, and COS-1 cells were maintained in Dulbecco's modified Eagle's medium containing 10% (vol/vol) fetal calf serum (FCS) and penicillin-streptomycin. THP1 cells were maintained in RPMI medium containing 10% (vol/vol) FCS and penicillin-streptomycin. Lentivirus-infected stable cells were maintained in or selected with 5 μg/ml puromycin, pCEP4- and pMEP4-carrying stable cells were selected with 300 μg/ml Hygrogold (InvivoGen), and expression of MC005 in pMEP4-carrying lines was induced with 1 μM cadmium chloride (Sigma). SeV (ECACC), VSV (a gift from John C. Bell, Ottawa Hospital Research Institute), and MVA virus (a gift from Ingo Drexler, Düsseldorf University) were all used at a multiplicity of infection of 10.
Plasmids and oligonucleotides.
MC005 was custom synthesized by GenScript and subcloned into the KpnI and NotI sites of the pCEP and cadmium chloride-inducible pMEP4 plasmids (Invitrogen) with a C-terminal FLAG (DYKDDDDK) or HA (YPYDVPDYA) tag. pCEP4-MC014-FLAG was described previously (7). The primer sets for cloning of full-length and truncated MC005 into pCEP4 corresponded to 18 bp of the sequence indicated with KpnI and HindIII overhanging restriction sites for insertion into the construct at those sites upstream of a FLAG tag. NEMO truncations were made with primers corresponding to 18-bp 5′ and 3′ regions indicated flanked by KpnI and HindIII overhanging restriction sites for insertion into pCEP4 at those sites upstream of a HA tag sequence. pdI-MC005, the retroviral expression construct, contained the direct 5′ and 3′ sequences of MC005L with SpeI and MluI overhangs, respectively, with a C-terminal FLAG tag. Plasmids expressing FLAG-IKKβ, FLAG-TAB2, FLAG-TRAF2, and FLAG-TRAF6 were from Tularik Inc. The sources of other expression plasmids were as follows: FLAG-TRIF, S. Akira (Osaka University, Osaka, Japan); Myc-MyD88, L. O'Neill (Trinity College, Dublin, Ireland); CD4-TLR9, A. Ozinsky (University of Washington, Seattle, WA); CD4-TLR3, R. Medzhitov (Yale University, New Haven, CT); FLAG-MAVS and cGAS, J. Chen (UT Southwestern Medical Center); RasVHa, D. Cantrell (University of Dundee, Dundee, United Kingdom). The human STING coding region was amplified by PCR from full-length I.M.A.G.E. cDNA clones (IRATp970D0274D and IRAVp968F0688D; imaGenes) and cloned into the vector pCMV-myc (Clontech). HA-ubiquitin was obtained from A. Mansell (Monash University, Melbourne, Australia). The NF-κB-luciferase reporter gene was from R. Hofmeister (Universitat Regensburg, Regensburg, Germany), ISRE-luciferase was from L. O'Neill, and the pFR-luciferase reporter and pFA2-Elk1 were from Agilent. Full-length NEMO-FLAG and NEMO-HA were from K. Fitzgerald (University of Massachusetts, Amherst, MA). K277A mutant NEMO was generated by site-directed mutagenesis of the region with primers FP (AGGAGGCCCTGGTGGCCGCACAGGAGGTGATCGATAAGCTG) and RP (CAGCTTATCGATCACCTCCTGTGCGGCCACCAG GGCCTCCT).
Antibodies.
The primary antibodies used for immunoblotting were anti-β-actin (AC-74), anti-FLAG (M2) (Sigma-Aldrich), anti-IκBα (R. Hay, University of Dundee, Dundee, United Kingdom), anti-phospho-p65 (Ser536, 93H1), anti-p65 (C-20; Santa Cruz), phospho-IKKα/β (Ser176/180) (16A6) rabbit monoclonal (CST), and anti-HA (Covance) antibodies. The secondary antibodies used for immunoblotting were IRDye 680LT anti-mouse, IRDye 800CW anti-rabbit, and IRDye 680LT anti-goat (LI-COR Biosciences) antibodies. The secondary antibodies used for confocal microscopy were Alexa Fluor 647-conjugated anti-mouse and Alexa Fluor 488-conjugated anti-rabbit antibodies (Invitrogen).
ELISA.
Cell culture supernatants were assayed for IL-8 and IP-10 proteins with an enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems) in accordance with the manufacturer's instructions.
Immunoblotting.
Cells were seeded at 5 × 105/well into six-well dishes and transfected with 3 μg of total DNA with GeneJuice (Novagen) the next day. Twenty-four hours later, cells were lysed in 200 μl of sample buffer (187.5 mM Tris [pH 6.8], 6% [wt/vol] SDS, 30% [vol/vol] glycerol, 0.3% [wt/vol] bromophenol blue, 150 mM dithiothreitol, and Benzonase), incubated on ice for 5 min, and then boiled for 5 min at 99°C. Twenty-microliter lysates were resolved by 10 to 20% SDS-PAGE, transferred to polyvinylidene difluoride membrane (Millipore), blocked for 1 h in 3% (wt/vol) bovine serum albumin (BSA) in phosphate-buffered saline (PBS), and probed overnight with a primary antibody (1:1,000 dilution in blocking solution). The next day, membranes were incubated with secondary antibodies (1:10,000 dilution in blocking solution) and blots were visualized with the Odyssey imaging system (LI-COR Biosciences).
Immunoprecipitation.
Cells were seeded at 4 × 106/10-cm dish and transfected with 8 μg of total plasmid the next day. Twenty-four hours later, the cells were washed with ice-cold PBS, scraped into lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 0.5% [vol/vol] NP-40, 30 mM NaF, 5 mM EDTA, 10% [vol/vol] glycerol, and 40 mM β-glycerophosphate containing the inhibitors 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, and 1% [vol/vol] aprotinin), and then left on ice for 45 min. Anti-FLAG antibody-coated beads (Sigma) were equilibrated with lysis buffer, and cleared lysates were mixed with beads and incubated for 2 h while rolling at 4°C. Beads were then washed three times with 1 ml of lysis buffer, and immunoprecipitated material was eluted with 3× FLAG peptide (Sigma) for 30 min while rolling at 4°C, after which it was separated and immunoblotted for the proteins indicated.
Confocal microscopy.
Cells were seeded onto glass coverslips at 3 × 105/ml in 24-well plates and stimulated the next day as indicated. Cells were fixed for 12 min in 4% (wt/vol) paraformaldehyde and permeabilized for 15 min with 0.5% (vol/vol) Triton X-100 in PBS. Coverslips were blocked for 1 h in 5% (wt/vol) BSA–0.05% (vol/vol) Tween 20 in PBS and stained overnight with primary antibodies (1:200 dilution in blocking solution). The following day, coverslips were incubated for 3 h with secondary antibodies (1:500 dilution in blocking solution) and mounted in Mowiol 4-88 (Calbiochem) containing 1 mg/ml 4′,6-diamidino-2-phenylindole (DAPI). Images were obtained with an Olympus FV1000 confocal microscope with a 360× oil immersion objective.
Reporter gene assays.
For reporter gene assays, cells were seeded into 96-well plates at 1 × 105/ml and transfected 16 h later with GeneJuice transfection reagent (Novagen) with 80 ng of luciferase reporter gene, 20 ng of pGL3-Renilla luciferase, and the indicated amounts of expression vectors and MCV ORFs and adjusted to 230 ng of total DNA with the empty vector pCMV-HA. Twenty-four hours after transfection, cells were stimulated with cytokines, infected with virus for 24 h or directly lysed in passive lysis buffer (Promega), and analyzed for luciferase activity. Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency. For the Ras-driven Elk1 reporter assay, 1 ng of the pFA-Elk1 expression vector and 80 ng of the pFR-luciferase reporter plasmid were employed, with a RasVHa expression vector for pathway activation.
Generation of stable cell lines using retroviral vectors.
Stable cell generation employed the pdlNotInPkMCSR expression construct (38) with cloning into the MluI and SpeI sites. Lentivirus was packaged with the Virapower packaging system (Invitrogen) as described in the manual. Stable cells were selected with 5 μg/ml puromycin.
Affinity purification and LC-MS/MS analysis.
Expression of FLAG-tagged MC005 in stably transfected HEK293T cells was induced with 1 μM CdCl2. After 24 h, cells were washed in 1× PBS, pelleted at 3,000 × g for 5 min at 4°C, and snap-frozen in liquid nitrogen. Cell pellets were thawed on ice and resuspended in 1 ml of ice-cold TAP lysis buffer (50 mM Tris-HCl [pH 7.5], 4.3% glycerol, 0.2% NP-40, 1.5 mM MgCl2, 100 mM NaCl) supplemented with EDTA-free cOmplete protease inhibitor cocktail (Roche) and 250 U of Benzonase (Sigma-Aldrich). After incubation on ice for 30 min, lysates were centrifuged at 12,000 × g for 5 min at 4°C. Cleared lysates were incubated with 40 μl of anti-FLAG M2 antibody affinity gel (Sigma-Aldrich) for 1 h on a rotating wheel at 4°C. After incubation, resin was washed in TAP lysis buffer (final two washes without NP-40) and resuspended in 40 μl of 6 M guanidinium-HCl in 100 mM Tris (pH 8.5) supplemented with 10 mM tris(2-carboxyethyl)phosphine (TCEP) and 40 mM chloroacetamide. After 30 min of incubation at room temperature (RT) in the dark, lysates were diluted 1:10 with digestion buffer (10% acetonitrile, 25 mM Tris [pH 8.5]) and incubated with 0.5 μg of EndoLysC (Wako Chemicals) and 0.5 μg of sequencing grade modified trypsin (Promega) overnight on a rotating wheel at RT. After digestion, peptides were acidified with trifluoroacetic acid, desalted on reversed-phase C18 StageTips, and eluted before liquid chromatography-tandem mass spectrometry (LC-MS/MS) with buffer B (80% acetonitrile, 0.5% acetic acid). Eluted peptides were analyzed on a nanoflow EASY-nLC system coupled to an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Peptide separation was achieved on a C18 reversed-phase column (ReproSil-Pur C18-AQ, 1.9 μm, 200 by 0.075 mm; Dr. Maisch GmbH) with a 120-min linear gradient of 2 to 60% acetonitrile in 0.1% formic acid. The LTQ-Orbitrap XL mass spectrometer was operated with a Top10 MS/MS spectrum acquisition method in the linear ion trap per MS full scan in the Orbitrap. Raw files were processed with MaxQuant (version 1.4.1.4) and searched with the built-in Andromeda search engine against a human protein database (UniProtKB release 2012_01) concatenated with a decoy of reversed sequences by using a label-free quantification (LFQ) algorithm as described previously (39). Carbamidomethylation was set as a fixed modification, while methionine oxidation and protein N-acetylation were included as variable modifications. The search was performed with an initial mass tolerance of 6 ppm for the precursor ion and 0.5 Da for the fragment ions. Search results were filtered in Perseus (version 1.4.1.8) with a false-discovery rate (FDR) of 0.01. Prior to statistical analysis, known contaminants and reverse hits were removed. Proteins identified with at least two unique peptides and a minimum of two quantitation events in at least one experimental group were considered for analysis. LFQ protein intensity values were log transformed, and missing values were filled in by imputation with random numbers drawn from a normal distribution. Significant interactors were determined with a two-sample t test with Welch correction after 250 permutations, the FDR threshold set to 0.01, and S0 empirically set to 1. Results were plotted with R (release version 2.15.3).
ACKNOWLEDGMENTS
This work was supported by Science Foundation Ireland grant 11/PI/1056 (to A.G.B.), the Max-Planck Free Floater program (to A.P.), the ERC (StG 311339-iViP to A.P.), and Marie Curie Intra-European Fellowship no. 332057 (to G.B. and A.G.B.). P.J.F. is partly supported by funding from the Imperial NIHR Biomedical Research Centre.
REFERENCES
- 1.Gürtler C, Bowie AG. 2013. Innate immune detection of microbial nucleic acids. Trends Microbiol 21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowie AG, Unterholzner L. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol 8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady G, Bowie AG. 2014. Innate immune activation of NFkappaB and its antagonism by poxviruses. Cytokine Growth Factor Rev 25:611–620. doi: 10.1016/j.cytogfr.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Senkevich TG, Koonin EV, Bugert JJ, Darai G, Moss B. 1997. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology 233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 5.Struzik J, Szulc-Dabrowska L, Niemialtowski M. 2014. Modulation of NF-κB transcription factor activation by molluscum contagiosum virus proteins. Postepy Hig Med Dosw (Online) 68:129–136. doi: 10.5604/17322693.1088053. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Anstey AV, Bugert JJ. 2013. Molluscum contagiosum virus infection. Lancet Infect Dis 13:877–888. doi: 10.1016/S1473-3099(13)70109-9. [DOI] [PubMed] [Google Scholar]
- 7.Brady G, Haas DA, Farrell PJ, Pichlmair A, Bowie AG. 2015. Poxvirus protein MC132 from molluscum contagiosum virus inhibits NF-B activation by targeting p65 for degradation. J Virol 89:8406–8415. doi: 10.1128/JVI.00799-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bugert JJ, Lohmuller C, Darai G. 1999. Characterization of early gene transcripts of molluscum contagiosum virus. Virology 257:119–129. doi: 10.1006/viro.1999.9649. [DOI] [PubMed] [Google Scholar]
- 9.Pascutti MF, Rodriguez AM, Falivene J, Giavedoni L, Drexler I, Gherardi MM. 2011. Interplay between modified vaccinia virus Ankara and dendritic cells: phenotypic and functional maturation of bystander dendritic cells. J Virol 85:5532–5545. doi: 10.1128/JVI.02267-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeruva S, Ramadori G, Raddatz D. 2008. NF-kappaB-dependent synergistic regulation of CXCL10 gene expression by IL-1beta and IFN-gamma in human intestinal epithelial cell lines. Int J Colorectal Dis 23:305–317. doi: 10.1007/s00384-007-0396-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. 2013. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai P, Wang W, Cao H, Avogadri F, Dai L, Drexler I, Joyce JA, Li XD, Chen Z, Merghoub T, Shuman S, Deng L. 2014. Modified vaccinia virus Ankara triggers type I IFN production in murine conventional dendritic cells via a cGAS/STING-mediated cytosolic DNA-sensing pathway. PLoS Pathog 10:e1003989. doi: 10.1371/journal.ppat.1003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. 2013. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valentine R, Smith GL. 2010. Inhibition of the RNA polymerase III-mediated dsDNA-sensing pathway of innate immunity by vaccinia virus protein E3. J Gen Virol 91:2221–2229. doi: 10.1099/vir.0.021998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchens M, Luker KE, Sottile P, Sonstein J, Lukacs NW, Nunez G, Curtis JL, Luker GD. 2008. TLR3 increases disease morbidity and mortality from vaccinia infection. J Immunol 180:483–491. doi: 10.4049/jimmunol.180.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuelsson C, Hausmann J, Lauterbach H, Schmidt M, Akira S, Wagner H, Chaplin P, Suter M, O'Keeffe M, Hochrein H. 2008. Survival of lethal poxvirus infection in mice depends on TLR9, and therapeutic vaccination provides protection. J Clin Invest 118:1776–1784. doi: 10.1172/JCI33940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ku JK, Kwon HJ, Kim MY, Kang H, Song PI, Armstrong CA, Ansel JC, Kim HO, Park YM. 2008. Expression of Toll-like receptors in verruca and molluscum contagiosum. J Korean Med Sci 23:307–314. doi: 10.3346/jkms.2008.23.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDermott EP, O'Neill LA. 2002. Ras participates in the activation of p38 MAPK by interleukin-1 by associating with IRAK, IRAK2, TRAF6, and TAK-1. J Biol Chem 277:7808–7815. doi: 10.1074/jbc.M108133200. [DOI] [PubMed] [Google Scholar]
- 20.Clark K, Nanda S, Cohen P. 2013. Molecular control of the NEMO family of ubiquitin-binding proteins. Nat Rev Mol Cell Biol 14:673–685. doi: 10.1038/nrm3644. [DOI] [PubMed] [Google Scholar]
- 21.Skaug B, Jiang X, Chen ZJ. 2009. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem 78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 22.Ivins FJ, Montgomery MG, Smith SJ, Morris-Davies AC, Taylor IA, Rittinger K. 2009. NEMO oligomerization and its ubiquitin-binding properties. Biochem J 421:243–251. doi: 10.1042/BJ20090427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tegethoff S, Behlke J, Scheidereit C. 2003. Tetrameric oligomerization of IkappaB kinase gamma (IKKgamma) is obligatory for IKK complex activity and NF-kappaB activation. Mol Cell Biol 23:2029–2041. doi: 10.1128/MCB.23.6.2029-2041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polley S, Huang DB, Hauenstein AV, Fusco AJ, Zhong X, Vu D, Schrofelbauer B, Kim Y, Hoffmann A, Verma IM, Ghosh G, Huxford T. 2013. A structural basis for IkappaB kinase 2 activation via oligomerization-dependent trans auto-phosphorylation. PLoS Biol 11:e1001581. doi: 10.1371/journal.pbio.1001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloor S, Ryzhakov G, Wagner S, Butler PJ, Smith DL, Krumbach R, Dikic I, Randow F. 2008. Signal processing by its coil zipper domain activates IKK gamma. Proc Natl Acad Sci U S A 105:1279–1284. doi: 10.1073/pnas.0706552105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wlodaver CG, Palumbo GJ, Waner JL. 2004. Laboratory-acquired vaccinia infection. J Clin Virol 29:167–170. doi: 10.1016/S1386-6532(03)00118-5. [DOI] [PubMed] [Google Scholar]
- 27.Sumner RP, Maluquer de Motes C, Veyer DL, Smith GL. 2014. Vaccinia virus inhibits NF-kappaB-dependent gene expression downstream of p65 translocation. J Virol 88:3092–3102. doi: 10.1128/JVI.02627-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Randall CM, Jokela JA, Shisler JL. 2012. The MC159 protein from the molluscum contagiosum poxvirus inhibits NF-kappaB activation by interacting with the IkappaB kinase complex. J Immunol 188:2371–2379. doi: 10.4049/jimmunol.1100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nichols DB, Shisler JL. 2006. The MC160 protein expressed by the dermatotropic poxvirus molluscum contagiosum virus prevents tumor necrosis factor alpha-induced NF-kappaB activation via inhibition of I kappa kinase complex formation. J Virol 80:578–586. doi: 10.1128/JVI.80.2.578-586.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murao LE, Shisler JL. 2005. The MCV MC159 protein inhibits late, but not early, events of TNF-alpha-induced NF-kappaB activation. Virology 340:255–264. doi: 10.1016/j.virol.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 31.Amaya M, Keck F, Bailey C, Narayanan A. 2014. The role of the IKK complex in viral infections. Pathog Dis 72:32–44. doi: 10.1111/2049-632X.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Husnjak K, Dikic I. 2012. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem 81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 33.Kulathu Y, Komander D. 2012. Atypical ubiquitylation—the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol 13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 34.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. 2009. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Thompson CH, de Zwart-Steffe RT, Donovan B. 1992. Clinical and molecular aspects of molluscum contagiosum infection in HIV-1 positive patients. Int J STD AIDS 3:101–106. doi: 10.1177/095646249200300205. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita H, Uemura T, Kawashima M. 1996. Molecular epidemiologic analysis of Japanese patients with molluscum contagiosum. Int J Dermatol 35:99–105. doi: 10.1111/j.1365-4362.1996.tb03270.x. [DOI] [PubMed] [Google Scholar]
- 37.Gur I. 2008. The epidemiology of molluscum contagiosum in HIV-seropositive patients: a unique entity or insignificant finding? Int J STD AIDS 19:503–506. doi: 10.1258/ijsa.2008.008186. [DOI] [PubMed] [Google Scholar]
- 38.Chinnakannan SK, Nanda SK, Baron MD. 2013. Morbillivirus V proteins exhibit multiple mechanisms to block type 1 and type 2 interferon signalling pathways. PLoS One 8:e57063. doi: 10.1371/journal.pone.0057063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. 2010. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol 189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]