ABSTRACT
Rotavirus infection is one of the most common causes of diarrheal illness in humans. In neonatal mice, rhesus rotavirus (RRV) can induce biliary atresia (BA), a disease resulting in inflammatory obstruction of the extrahepatic biliary tract and intrahepatic bile ducts. We previously showed that the amino acid arginine (R) within the sequence SRL (amino acids 445 to 447) in the RRV VP4 protein is required for viral binding and entry into biliary epithelial cells. To determine if this single amino acid (R) influences the pathogenicity of the virus, we generated a recombinant virus with a single amino acid mutation at this site through a reverse genetics system. We demonstrated that the RRV mutant (RRVVP4-R446G) produced less symptomatology and replicated to lower titers both in vivo and in vitro than those seen with wild-type RRV, with reduced binding in cholangiocytes. Our results demonstrate that a single amino acid change in the RRV VP4 gene influences cholangiocyte tropism and reduces pathogenicity in mice.
IMPORTANCE Rotavirus is the leading cause of diarrhea in humans. Rhesus rotavirus (RRV) can also lead to biliary atresia (a neonatal human disease) in mice. We developed a reverse genetics system to create a mutant of RRV (RRVVP4-R446G) with a single amino acid change in the VP4 protein compared to that of wild-type RRV. In vitro, the mutant virus had reduced binding and infectivity in cholangiocytes. In vivo, it produced fewer symptoms and lower mortality in neonatal mice, resulting in an attenuated form of biliary atresia.
KEYWORDS: RRV, biliary atresia, cholangiocyte, reverse genetics
INTRODUCTION
Biliary atresia (BA) is a unique disease of infancy leading to inflammatory obstruction of the extrahepatic biliary tract. It is the most common cause of pathological jaundice in the pediatric population, and it may progress to hepatic cirrhosis and end-stage liver disease, requiring liver transplantation (1, 2). The etiology of BA remains unknown. It has been proposed that a viral agent may be responsible. Several viruses, including rotavirus group C (3), reovirus type 3 (4), Epstein-Barr virus (5), cytomegalovirus (6), and human papillomavirus (7), have been found in explanted livers of infants with BA. In the mouse model of BA, rhesus rotavirus (RRV) infection of newborn BALB/c mice results in an inflammatory cholangiopathy and a pathological phenotype similar to those in human BA (8, 9). This manifests as bilirubinuria, jaundice, acholic stools, growth retardation, and, ultimately, death (9). The morphological and histological changes in the biliary tree parallel those found in infants (8), and susceptibility is limited to the neonatal period (10).
Rotaviruses are distributed throughout the world, with a wide range of host susceptibilities. In humans, these viruses induce diarrhea and are estimated to cause 215,000 deaths in children under 5 years of age every year (11). Rotaviruses are 65- to 75-nm icosahedral nonenveloped particles with a triple-layered protein capsid containing 11 double-stranded RNA (dsRNA) gene segments encoding either one or two proteins each, with conserved 5′ and 3′ noncoding regions. The outer capsid is composed of a triple-layered protein consisting of VP4 and VP7 (12). VP4 is the spike protein that binds to cellular receptors and is proteolytically activated by trypsin-like proteases (13, 14).
The pathogenesis of rotavirus infection is dependent on its attachment and entry into the cell. The mechanism by which rotavirus attaches to a host cell is a multistep process involving both sialic acid-labeled glycoproteins and specific cellular surface proteins that serve as viral “receptors” (15–17). Rotavirus receptors include the integrins α2β1, α4β1, αVβ3, and αXβ2 (16, 18–23), along with heat shock cognate protein 70 (Hsc70) (16).
Reverse genetics systems for the double-stranded RNA viruses, including reovirus and bluetongue virus, have been established and used to study mechanistic roles of various proteins (24–27). Until recently, reverse genetics has been difficult to achieve with rotavirus. Several studies reported a reverse genetics system in which overexpression of recombinant vaccinia virus-mediated T7 RNA polymerase facilitated the transcription of the plasmid-cloned rotavirus genes, supported by antibody-, small interfering RNA (siRNA)-, or temperature-sensitive mutation-based selection systems allowing for the generation of a recombinant rotavirus (28, 29).
We previously showed that in BA, the cellular target of RRV is the biliary epithelial cell (cholangiocyte) (30). Our previous studies showed that the VP4 gene of RRV is the major determinant in modulating disease pathogenesis (31, 32). We recently found that the VP4 gene plays a role in primary binding to cells (cholangiocytes and MA104 cells) prior to entry and that treating cells with the VP4-derived peptide TRTRVSRLY (amino acids 440 to 449 of VP4) can inhibit virus binding and subsequent replication (33). In that study, we identified the specific amino acid sequence SRL within TRTRVSRLY and determined that it binds to Hsc70, driving cell entry. For the current report, we used a reverse genetics system to generate an RRV VP4 mutant (RRVVP4-R446G) with one amino acid change from wild-type (WT) RRV (arginine [R] at position 446 to glycine [G]). The mutated virus resulted in reduced cholangiocyte binding and infectivity. When it was injected into newborn mice, the virus resulted in an attenuation of the BA phenotype, with significantly reduced symptoms and mortality.
RESULTS
Sequence comparison of various strains of rotavirus used in the murine model of BA.
Several rotavirus strains were used to induce the murine model of BA, as shown in Table 1. We previously showed that the VP4 gene of RRV is the major determinant in the murine model of BA (31, 32). The SPPIDER program (34) was utilized to predict potential protein-protein interactions and to analyze the VP4 sequences of various rotavirus strains. The program predicted that amino acid 446 (arginine) of VP4 plays an important role. When several different rotavirus sequences were compared, 3 strains of rotavirus (RRV, SA-11, and GRV) had an arginine (R) at position 446. All these strains were capable of inducing the murine model of BA (31). Computer-generated analysis using published crystalline structures illustrated that the amino acid sequence SRL was exposed on the surface of the VP4 protein, allowing for binding (Fig. 1).
TABLE 1.
VP4 protein sequence comparison for different rotavirus strains
| Rotavirus strain | Amino acid at VP4 protein positiona: |
Induction of murine BAb | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 440 | 441 | 442 | 443 | 444 | 445 | 446 | 447 | 448 | ||
| RRV | T | R | T | R | V | S | R | L | Y | Yes |
| TUCH | V | R | T | R | I | S | G | L | Y | No |
| Ro1845 | M | R | T | R | V | S | G | L | Y | No |
| SA-11 | T | R | T | R | L | D | R | L | Y | Yes |
| NCDV | L | R | T | R | V | S | G | L | Y | No |
| Wa | L | R | T | R | T | V | N | L | Y | No |
| EDIM | T | R | T | R | V | S | G | L | Y | No |
| CU-1 | M | R | T | R | V | S | G | L | Y | No |
| WC-3 | T | R | T | R | V | S | K | L | Y | No |
| VA-70 | M | R | T | R | I | V | N | L | Y | No |
| EB | T | R | T | R | V | S | G | L | Y | No |
| EMcN | T | R | T | R | V | S | G | L | Y | No |
| ST-3 | T | R | T | R | V | S | K | L | Y | No |
| GRV | T | R | T | R | V | S | R | L | Y | Yes |
| KU | L | R | T | R | T | V | N | L | Y | No |
According to analysis of rotavirus VP4 amino acid sequences obtained from the NCBI database. The arginine residue at position 446 is indicated in bold.
Bile duct obstruction was determined by histology.
FIG 1.
RRV VP4 structure. (A) Cryo-electron microscopy (cryoEM)-resolved rotavirus particle structure (PDB code 4V7Q) showing the location of the SRL motif on the surface of the outer capsid protein VP4. The SRL-containing motif (residues 444 to 450; VSRLYGL) is shown in red on the surface of the isolated chain BX of structure 4V7Q (shown using yellow surface and backbone rendering). (B) One-dimensional profile of the same chain, with red braids, green arrows, and blue segments representing helices, beta strands, and loops, respectively, and shaded boxes representing the solvent accessibility of individual amino acid residues (with black corresponding to fully buried and gray to partially exposed residues). The location of the SRL motif (residues 445 to 447; highlighted in yellow and circled) within the BX chain of structure 4V7Q is indicated, with mapping of all protein interaction interfaces shown in magenta. As can be seen, the SRL motif is located in a generally accessible region. (Adapted from reference 33 with permission of the publisher.)
Binding of the SRL region to Hsc70 is dependent on arginine (R) and is not a function of charge.
We recently demonstrated that RRV utilizes the SRL region of its VP4 protein to bind to extracellular Hsc70 (33). Through blocking assays, we demonstrated that the peptide VSRLY is capable of blocking RRV's ability to bind to cholangiocytes. To determine if this binding is specific to arginine or is a function of amino acid charge, we treated cholangiocytes with a synthetic peptide containing a mutation of arginine to lysine (K), the VSKLY peptide, which has a charge similar to that of arginine, followed by infection with RRV. The VSRLY peptide blocked RRV binding similarly to what has been published before, while the VSKLY peptide had no effect (Fig. 2). This illustrated that the binding of SRL to Hsc70 is not an effect of amino acid charge.
FIG 2.
Blocking of viral binding by use of synthetic peptides. Cholangiocytes were pretreated with 1 mM synthetic peptides VSRLY, VSGLY, and VSKLY, followed by infection with WT RRV. The VSRLY peptide was able to inhibit binding of WT RRV, while VSGLY and VSKLY had no effect. *, P < 0.05.
Construction of an RRV VP4-encoding plasmid and generation of RRV VP4 mRNA.
RNA was isolated by a standard procedure from virus grown in MA104 cells. The VP4-encoding genome segment of RRV was amplified as described in Materials and Methods and cloned into the pBacT7 vector, yielding the plasmid pBacT7-VP4(RRV) (Fig. 3A). The purified plasmid was transfected into BHK21-T7 cells, which express T7 RNA polymerase. Thus, transcription of the resulting vector, pBacT7-VP4(RRV), allowed production of a 2,362-nucleotide (nt) positive-strand RNA with the correct 5′ and 3′ ends, corresponding to the ends of RRV VP4 mRNA (Fig. 3B).
FIG 3.
Schematic representation of transcription plasmids carrying the full-length RRV VP4 gene. (A) Plasmid pBacT7-VP4(RRV) contains the authentic full-length VP4 gene cDNA of RRV, flanked by the T7 RNA polymerase promoter at the 5′ end and the HDV ribozyme at the 3′ end, followed by the T7 RNA polymerase terminator. Manipulation of the VP4 gene by means of silent mutations (positions are indicated by arrows below the sequences) was carried out in pBacT7-VP4(RRV): the mutant plasmids, pBacT7-VP4(RRVXbaI) and pBacT7-VP4(RRVR446G), contain a mutation creating a unique XbaI site and a mutation encoding an amino acid change from R to G, respectively. Numbers indicate the nucleotide positions in the RRV VP4 gene sequence. (B) Sequences at the 5′ and 3′ termini of the RRV VP4 gene in the transcription vectors. PT7, ribozyme, TT7, and UTR denote the T7 RNA polymerase promoter, HDV ribozyme, T7 RNA polymerase terminator, and untranslated region, respectively. ORF, open reading frame.
Rescue of rotavirus containing recombinant gene 4.
BHK21-T7 cells were transfected with the pBacT7-VP4(RRV) plasmid. Twenty-four hours after transfection, the cells were infected with the KU strain of rotavirus (helper virus) and incubated at 37°C to allow the helper virus to replicate. The virus was subsequently passaged in MA104 cells as described in Materials and Methods. After subsequent passages, higher viral titers were observed in the pBacT7-VP4(RRV)-transfected samples, suggesting the presence of a recombinant virus, since RRV grew better than KU (data not shown). The samples were collected at each passage level for detection of RRV VP4 by sequence-specific reverse transcription-PCR (RT-PCR). The plasmid-transfected 5th-passage supernatants confirmed the presence of recombinant gene 4. To verify the presence of recombinant virus in the pBacT7-VP4(RRV)-transfected samples, a single viral isolate was obtained by plaque purification, grown, and then concentrated by pelleting using ultracentrifugation. After rescue of the reassortant virus, KU with RRV VP4, we backcrossed it to another RRV reassortant containing TUCH VP4 [RT(VP4); a reassortant with 10 genes from RRV, except for the VP4 gene, which was from strain TUCH] by conventional reassortment techniques, as previously described (32), to ultimately obtain an RRV with recombinant VP4.
Analysis of recombinant viruses.
All viruses were pelleted by ultracentrifugation and used for dsRNA extraction and analysis by PAGE. The migration patterns of KU VP4 and RRV VP4 differ and can be distinguished as shown in Fig. 4A. The migration profiles of dsRNAs of different viruses are shown in the figure, including those for the RRV used for VP4 gene cloning (lane 1), KU with RRV VP4 (lane 2), and KU used as a helper virus (lane 3). The in vitro-generated VP4 gene dsRNA (lane 2) migrated to the same position as that of the corresponding dsRNA of WT RRV (lane 1), with a mobility slower than that of the VP4 gene segment of the KU helper virus (lane 3). We amplified the VP4 fragments by RT-PCR and sequenced them to confirm that the VP4 gene of the rescued virus was derived from RRV (data not shown). Therefore, an infectious rotavirus transfectant containing the cDNA-derived VP4 gene segment of RRV, named the KUVP4(RRV) virus, could be recovered by cDNA transfection followed by infection with helper virus (KU).
FIG 4.
Rescue of viruses containing the cDNA-derived VP4 gene. (A) PAGE analysis of dsRNAs extracted from the rescued VP4 gene transfectants. Lane 1, dsRNAs from WT RRV; lane 2, dsRNAs from KU helper virus; lane 3, dsRNAs from KU-VP4(RRV). (B) PAGE analysis of dsRNAs extracted from single-gene reassortants. Lane 1, dsRNAs from WT RRV; lane 2, dsRNAs from strain TUCH; lane 3, dsRNAs from KU helper virus; lane 4, dsRNAs from KU-VP4(RRVR446G); lane 5, dsRNAs from RRVVP4-R446G.
Introduction of site-specific mutations into the genome of an infectious rotavirus.
RRV infection of cells is dependent on cell surface attachment, uncoating, replication, and release of viral progeny. We previously showed through in vitro peptide binding assays that the synthetic peptide TRTRVSRLY (amino acid positions 440 to 446), derived from the RRV VP4 protein, is able to block the virus's ability to bind and replicate in a dose-dependent manner. The SRL portion of this peptide binds to Hsc70 (33). As previously described, the arginine in SRL is present in all tested strains of rotavirus which can induce BA. In this study, we attempted to mutate the arginine (R) to glycine (G), which is present in some non-BA-causing strains.
We introduced 2 mutations into the coding region of the VP4 gene in plasmid pBacT7-VP4(RRV) through PCR-based site-directed mutagenesis. The 2 mutations in the cDNA comprised an A-to-T change at position 1029, creating a unique XbaI site as a gene marker, and an A-to-G change at position 1345, giving an R-to-G amino acid change at position 446 in the VP4 protein. Transfection of each mutagenized plasmid into BHK21-T7 cells, followed by superinfection with the helper virus KU, resulted in the rescue and isolation of transfectant virus clones, named KU-VP4(RRVXbaI) and KU-VP4(RRVR446G). As described above, RRV VP4 was detected after the 5th passage by PCR and after the 9th passage by gel electrophoresis (Fig. 4B). KU-VP4(RRVXbaI) and KU-VP4(RRVR446G) were subsequently backcrossed to RT(VP4) to obtain the desired RRV VP4 mutations on an RRV background, and the resultant strains were named RRVVP4-XbaI and RRVVP4-R446G, respectively. After plaque purification, each virus clone was amplified and used for subsequent characterization experiments. The viruses were extracted, and the resulting dsRNAs were subjected to PAGE and RT-PCR of the entire VP4 sequences to confirm the mutations.
RT-PCR and restriction enzyme digestion analysis revealed that the 919-bp PCR product of the VP4 gene digested with XbaI resulted in products of 361 and 558 bp, as shown in Fig. 5A. In contrast, the corresponding DNA fragments from WT RRV VP4 transfectant viruses could not be cut with XbaI (Fig. 5B). Direct sequence analysis of the PCR products also revealed the correct presence of these mutations within the genomes of the rescued viruses (Fig. 6). Therefore, successful rescue of infectious rotaviruses containing site-specific mutations in their genomes was achieved via transfection of cloned cDNA followed by helper virus infection in a T7 RNA polymerase-driven system.
FIG 5.
Site-specific mutations introduced into the genomes of the rescued VP4 gene transfectants. The VP4 gene of each virus was amplified by RT-PCR, using the primers shown in Table 2, to yield a 686-bp product. (A) The VP4 gene was amplified by use of RRV VP4-specific primers (lanes 1 to 3) and rotavirus VP4 degenerate primers (lanes 4 to 6). (B) The amplified fragments were digested with XbaI, followed by separation in a 2% agarose gel. Uncut WT VP4 (RRV) and VP4XbaI (RRV) were run in a 2% agarose gel (lanes 1 and 2), along with WT VP4 (RRV) and VP4XbaI (RRV) after digestion with XbaI (lanes 3 and 4).
FIG 6.
Confirmation of site-specific mutations introduced into the genomes of the rescued VP4 gene transfectants. The full-length VP4 genes were directly sequenced, which confirmed the site-specific mutations introduced within the genomes of the infectious VP4 gene transfectants.
Physical properties of the mutated virus.
In order to study the role of arginine (R) in the SRL region of RRV VP4, we replaced this amino acid with a glycine (G) (present in non-BA-inducing strains [Table 1]) in the VP4 protein (RRVVP4-R446G) and performed binding and infectivity assays in cholangiocytes. Wild-type RRV and the recombinant virus RRVVP4-XbaI bound to cholangiocytes at rates significantly higher than that of the mutated virus (12.0% ± 0.4% and 7.7% ± 0.1%, respectively) (Fig. 7A). Following binding, plates were incubated at 37°C and assayed for infectivity at 24 and 48 h. At 24 h, there was no significant difference between RRVVP4-R446G and WT RRV; however, at 48 h postinfection, the recombinant virus titer was significantly reduced compared to that of the wild type (1.6 × 107 ± 0.2 × 107 versus 5.6 × 107 ± 0.2 × 107 focus-forming units [FFU]/ml, respectively) (Fig. 7B).
FIG 7.
Viral binding and replication in murine cholangiocytes. (A) Percentages of RRV, RRVVP4-XbaI, and RRVVP4-R446G binding to murine cholangiocytes at 4°C. The quantity of bound virus was measured using FFA. (B) Quantification of infectious rotavirus strains in murine cholangiocytes at 24 and 48 h postinfection, measured using FFA. For both sets of experiments, values (n = 3) are expressed as mean FFU per milliliter, with standard errors; each assay was repeated three times. (C) Cholangiocytes were pretreated with 1 mM synthetic peptides TRTRVSRLY, DGEA, and GHRP, followed by infection with RRV or RRVVP4-R446G. The DGEA peptide was able to inhibit binding of both strains, while TRTRVSRLY was able to block only WT RRV binding. *, P < 0.05.
To determine if the mutant strain RRVVP4-R446G was still able to bind to Hsc70, we performed binding blocking assays employing synthetic peptides. As previously published, RRV's binding was reduced when cholangiocytes were treated with 1 mM TRTRVSRLY and DGEA peptides, which block binding to Hsc70 and integrin α2β1, respectively. RRVVP4-R446G binding was similarly inhibit by the DGEA peptide; however, the TRTRVSRLY peptide was no longer able to inhibit binding of the mutated virus. GHRP is a nonspecific peptide that was used as a negative control, and it had no effect on the binding of either strain (Fig. 7C).
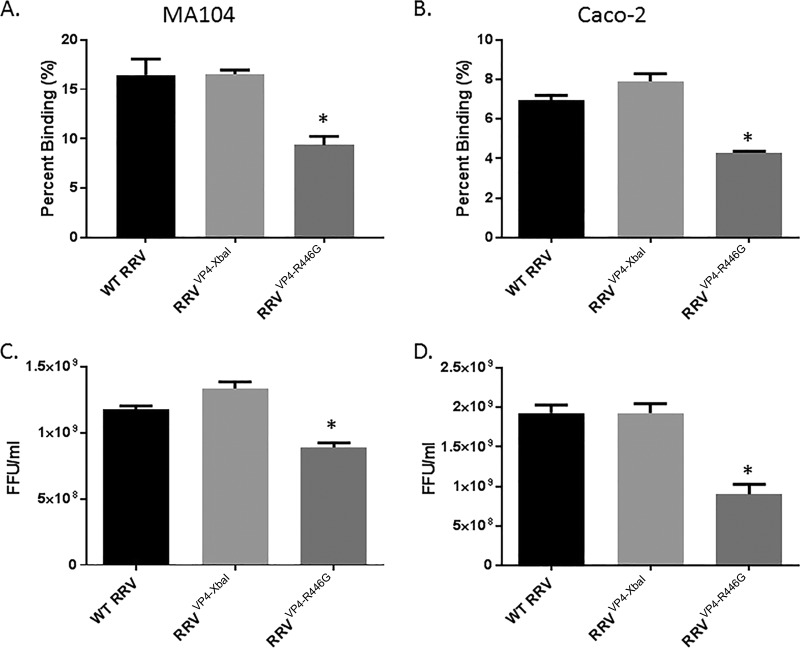
To establish if the reduced properties of RRVVP4-R446G were unique to cholangiocyte infection, we performed binding and infectivity assays with MA104 and Caco-2 cells. The binding of RRVVP4-R446G in both MA104 and Caco-2 cells was significantly lower than that of WT RRV (Fig. 8A and B). Correspondingly, the infectivity of RRVVP4-R446G in both cell lines was also reduced at 48 h compared to that of WT RRV (Fig. 8C and D).
FIG 8.
Viral binding and replication in MA104 and Caco-2 cells. Percentages of RRV, RRVVP4-XbaI, and RRVVP4-R446G binding to MA104 cells (A) and Caco-2 cells (B) at 4°C are shown. For both cell types, RRVVP4-R446G bound at a significantly lower percentage than that for RRV. Quantification of infectious rotavirus strains in MA104 cells (C) and Caco-2 cells (D) at 48 h postinfection revealed a similar pattern. For both sets of experiments (n = 3), values are expressed as mean FFU per milliliter, with standard errors; the assay was repeated three times.
Mutant virus leads to altered BA symptoms in infected mice.
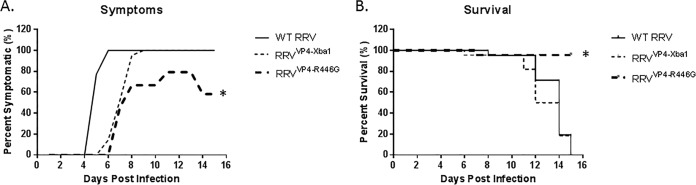
Pups were inoculated with the virus within 24 h of birth, and symptoms were observed for 16 days. Mice inoculated with RRVVP4-XbaI developed symptoms of biliary obstruction, such as acholic stool, bilirubinuria, and jaundice with weight loss, similar to those of WT RRV-injected mice. In contrast, when the mutant virus (RRVVP4-R446G) was injected, mice developed significantly fewer symptoms (Fig. 9A). The development of symptoms in mutant mice was delayed 2 days, and by day 12, only 80% of mice were symptomatic, which was reduced to 50% by day 21. WT RRV- and RRVVP4-XbaI-injected mice had 90 to 100% mortality by day 15, while RRVVP4-R446G-injected mice had a mortality rate of 10% (Fig. 9B).
FIG 9.
In vivo effect of mutant virus in the mouse model of biliary atresia. (A) Symptoms were recorded for different groups of mice for 16 days. One hundred percent of the mice in the RRV (n = 21)- and RRVVP4-XbaI (n = 22)-treated groups showed symptoms, but only 80% of those in the RRVVP4-R446G-injected group (n = 23) showed symptoms, and this gradually decreased to 40% later. (B) Survival was noted to be significantly improved (85%) for mice injected with RRVVP4-R446G compared to that for mice injected with RRV (0%) or RRVVP4-XbaI (10%). *, P < 0.05.
The bile duct viral titer for RRVVP4-XbaI-infected mice at 1 day postinjection was similar to that for RRV-infected mice (8.1 × 102 ± 0.7 × 102 versus 9.1 × 102 ± 0.8 × 102 FFU/ml/mg). RRVVP4-R446G-injected mice, however, had a viral titer of 4.8 × 102 ± 1.0 ×102 FFU/ml/mg, which is significantly lower than those of WT RRV- and RRVVP4-XbaI-injected mice (Fig. 10A). This significant difference was also witnessed at 7 days postinfection, with RRVVP4-R446G-injected mice having a titer in the bile ducts of 3.3 × 104 ± 1.1 × 104 FFU/ml/mg, versus 1.2 × 105 ± 0.5 × 105 and 1.1 × 105 ± 0.3 × 105 FFU/ml/mg for WT RRV- and RRVVP4-XbaI-injected mice, respectively. In addition, at this time point there was also a significantly smaller amount of RRVVP4-R446G than that of WT RRV or RRVVP4-XbaI in the intestine (Fig. 10B).
FIG 10.
Virus titers in organs following RRVVP4-R446G injection. (A) Viral titers from extrahepatic bile ducts harvested at 1 and 7 days postinfection were both significantly lower for mice injected with RRVVP4-R446G than for those injected with RRV or RRVVP4-XbaI (n = 6 per group). (B) Similarly, RRVVP4-R446G-injected mice expressed significantly lower viral titers in the intestine at 7 days postinfection than those for RRV-injected mice. *, P < 0.05.
Histological findings of biliary injury and cholangiogram.
Histological evaluation of livers at 12 days postinjection for the wild-type RRV and RRVVP4-XbaI infection groups showed more accumulation of inflammatory cells, with bile duct proliferation and bile plugs. In contrast, RRVVP4-R446G-infected mouse livers showed mild inflammation (Fig. 11A and B). Cholangiograms performed on mice inoculated with wild-type RRV and RRVVP4-XbaI showed a complete bile duct obstruction in which the dye could not pass beyond a certain point. The RRVVP4-R446G-infected mice, however, had patent bile ducts, as demonstrated by a continuous flow of dye from the gallbladder to the duodenum (Fig. 11C and D).
FIG 11.
Histologic evaluation of the liver and cholangiograms performed 12 days after infection with RRV and RRVVP4-R446G. (A) Hematoxylin and eosin staining of the liver after infection with RRV shows severe inflammation with an accumulation of inflammatory cells. (B) In contrast, mice injected with RRVVP4-R446G had mild inflammation. Magnification, ×10. (C and D) Cholangiograms demonstrating injection of methylene blue dye into the gallbladders of mice at 12 to 14 days postinoculation. The cholangiograms show the presence of biliary obstruction with RRV and the absence of biliary obstruction with RRVVP4-R446G. There were 7 mice in the RRV group and 8 in the RRVVP4-R446G group.
Mutant virus-injected mice have reduced NK cell activation.
It was previously shown that there is an increased number of activated NK cells in the livers of RRV-infected mice at the time of bile duct obstruction (7 days postinjection), implicating these cells as effector cells in the pathogenesis of experimental BA (31, 35). Among neonatal mice inoculated with WT RRV, RRVVP4-R446G, or saline, WT RRV-injected mice had a larger amount of activated NK cells (75.1% ± 2.7%), while the mutant virus RRVVP4-R446G-injected mice had a significantly smaller amount of NK cell activation (59.8% ± 2.8%) (P < 0.05) that was similar to that of saline-injected mice (55.5% ± 1.1%) (Fig. 12A and B).
FIG 12.
NK cell activation. (A and B) Flow cytometry of lymphocyte populations harvested from the livers of pups at 7 days postinfection illustrated a significant decrease in the number of activated NK cells present in the RRVVP4-R446G-injected mice compared to that for RRV-injected mice but a similar number compared to that for saline-injected mice. Red quadrants show the percentages of CD69+ cells in the CD49b+ population (2 livers per sample and 3 samples per strain). *, P < 0.05 (n = 3).
DISCUSSION
Biliary atresia (BA) is a devastating disease of newborns that occurs within the first months of life. The disease is characterized by liver inflammation, fibrosis and obstruction of the bile duct, and, without treatment, death. Although the etiology of BA is still unclear, human and mouse data suggest a perinatal viral infection as the possible cause of its pathogenesis (36). Several viruses, including rotavirus, may play a role in the activation of both inflammatory and autoimmune pathways (37). In the murine model of BA, RRV targets the biliary epithelium, leading to activation of a host immune response leading to liver inflammation and extrahepatic biliary obstruction (30, 38).
We previously showed that the VP4 gene of RRV plays an integral role in the induction of the murine model of BA by infection of cholangiocytes (32) and immune cell activation (31). Subsequent studies demonstrated that the peptide TRTRVSRLY within the VP4 protein was able to block RRV's ability to bind to and infect cholangiocytes (33), and when this peptide was injected into mice in vivo, it could attenuate the disease phenotype. Furthermore, we determined that the 3-amino-acid sequence SRL within this region, most importantly the amino acid arginine (R), plays a critical role. Through computer analysis, we identified this arginine to play an important role in protein-protein interaction. Using several strains in the mouse model, we identified that the strains capable of inducing BA in the mouse model all have an arginine in the same place in the VP4 protein (position 446). Several human rotavirus strains containing the TRTRVSRLY sequence in their VP4 protein have also been isolated (39, 40), suggesting a relevance to human BA.
Rotavirus belongs to the family Reoviridae and consists of 11 double-stranded RNA segments. Although the functions of most of its proteins are known, for the development of a safe vaccine and to study the effects of different mutations on virus replication and disease pathogenesis, there is always a need for development of a reverse genetics system. In 2006, Komoto et al. (28) first described a reverse genetics system for rotavirus, using a helper virus and antibody selection method. Since then, other approaches have been reported for rotavirus reverse genetics (29, 41, 42), all requiring the presence of a helper virus, until recently, when one study generated a recombinant virus using all 11 cloned genes (43). In the present study, we utilized a reverse genetics system as previously described (28, 29), with some modifications, using a helper virus but simplifying the mechanism by eliminating the need for a vaccinia virus, antibodies, and siRNA to achieve a recombinant rotavirus strain.
Rotavirus mRNA possesses a 5′ cap structure but lacks a 3′ poly(A) tail (44, 45); both ends, however, are required for genome replication and translation (46–48). Utilizing BHK21-T7 cells, which express high levels of T7 RNA polymerase, we used the 5′ (with the T7 polymerase promoter) and 3′ (with the hepatitis delta virus [HDV] ribozyme) ends of RRV VP4 in the vector so that, during transcription, the authentic viral mRNA would be obtained with the 5′ and 3′ signal sequences to be incorporated into the helper virus core to rescue the virus with a cloned VP4 gene.
Using the helper virus KU and serial passages, we attempted to rescue a transfectant virus containing the RRV cDNA-derived VP4 gene segment in the background of the helper virus KU to obtain KU-VP4(RRV). We took advantage of the better growth characteristics of rotavirus with the RRV VP4 gene than those of KU to generate the recombinant virus. The KU-VP4(RRV) construct was subsequently backcrossed with RT(VP4) (RRV with TUCH VP4) by using conventional methods and MA104 cells, yielding RRVrVP4 (RRV with recombinant RRV VP4). Using this method, we rescued three transfectant viruses: one with no mutations, derived from pBacT7-VP4(RRV); one with a silent mutation to create an XbaI restriction enzyme site in the VP4 gene as a gene marker, derived from plasmid pBacT7-VP4(RRVXbaI); and a third virus, derived from plasmid pBacT7-VP4(RRVR446G). We verified the presence of these mutations in the rescued mutant viruses, RRVVP4-XbaI and RRVVP4-R446G, through RT-PCR–restriction enzyme digestion analysis and sequence analysis of the virion dsRNAs. The mutation used to create the XbaI site did not result in any amino acid change in the VP4 protein and thus was not expected to change the biological properties of RRVVP4-XbaI compared to those of virus with the authentic RRV-derived VP4 gene. The RRVVP4-R446G mutation resulted in an amino acid substitution of glycine for arginine at position 446 of the VP4 protein.
All the viruses were generated by reverse genetics, and therefore their sequences were precisely defined and identical to each other, with the exception of the single nucleotide change A to T at position 1029 (no amino acid change) to create RRVVP4-XbaI and the nucleotide change A to G at position 1345 (amino acid change of R to G) to create RRVVP4-R446G. This procedure allowed us to eliminate other extrinsic variables and to precisely assess the effect of the single amino acid change. We previously showed that arginine at position 446 in RRV VP4 plays an important role in protein-protein interaction. However, this is the first time that a single engineered amino acid substitution in this region has been shown to directly confer an attenuated phenotype in an animal model.
To investigate the role of arginine at amino acid 446 in vivo, we generated the mutant virus RRVVP4-R446G, in which the arginine is replaced with a glycine (glycine is present at the same position in other strains which do not cause BA), through reverse genetics. When it was injected into newborn pups, the mutant virus resulted in reduced symptoms and mortality and also resulted in lower viral titers in the biliary system. In vitro, the replication yield of the mutant virus RRVVP4-R446G was similar to that of the WT at 24 h but was significantly reduced compared to that of the WT at 48 h. Similarly, the replication rate of RRVVP4-R446G was reduced in MA104 and Caco-2 cells. The basis for the reduction in replication is unclear; it may be a function of binding and entry but also may be a factor of how a virus traffics intracellularly following entry and how this, in turn, triggers the host immune response. Our previous data support this possibility, as we found that binding of arginine (R) to Hsc70 was required (33), which in turn governed translocation of RRV inside the cell. Alternatively, the mutant virus may not infect immune cells (dendritic cells), which also governs how the host immune system responds and has been shown to be a crucial step required for the development of BA (49). We have begun studies focused on differentiating between these alternative mechanisms.
This is the first report of a single reverse-engineered amino acid substitution in the VP4 protein of a rotavirus strain that conferred an attenuated phenotype in a mouse model. That a single nucleotide change in VP4 can lead to such a dramatically attenuated phenotype implies that the VP4 protein performs a critical function in virulence that may not always be readily identifiable in cell culture. In a previous study, Feng et al. (50) showed that rotavirus species-specific replication in the biliary tract is regulated by both VP4, which mediates viral entry into target cells, and NSP1, which suppresses the host interferon response. In our study, both the wild-type and mutant viruses had the same NSP1 protein, indicating that the changes we observed were because of a function of VP4.
In summary, we developed a T7-driven reverse genetics system for RRV that enabled the rescue of wild-type virus and the generation of VP4 mutants. This reverse genetics system may become a useful tool for evaluating the molecular determinants of virulence. It will allow us to insert sequences into noncoding regions and to determine the growth properties of such recombinant viruses as well as to examine RNA synthesis directly. Chimeric viruses carrying various VP4 proteins are currently being generated and tested under appropriate containment in vitro and in vivo in both cell lines and the murine model of BA. Using this system, we will investigate the hypothesis that the pathogenicity of human BA is regulated by VP4.
MATERIALS AND METHODS
Ethics statement.
All animal research was performed in accordance with regulations and protocols approved by the Institutional Animal Care and Use Committee at Cincinnati Children's Hospital Medical Center (protocol number IACUC2013-0039), which adheres to the NIH OLAW regulation (animal assurance number A3108) and the Animal Welfare Act (certification number 31-8-001).
Cells and viruses.
Monkey kidney cells (MA104 cells) (BioWhittaker, Walkersville, MD) were grown in Dulbecco's modified Eagle's medium (DMEM) (Cellgro) supplemented with 10% fetal bovine serum (FBS), 0.01% penicillin-streptomycin (Gibco/BRL, Gaithersburg, MD), 0.01% l-glutamine (Gibco/BRL), and 0.005% amphotericin B (Cellgro). BHK21-T7 cells (baby hamster kidney cell line expressing T7 polymerase; obtained from Ursula Buchholz, NIH, Bethesda, MD) were maintained in DMEM supplemented with nonessential amino acids, l-glutamine, penicillin-streptomycin, amphotericin B, and 10% fetal calf serum. The mouse cholangiocyte cell line (mCL), generously provided by the laboratory of James Boyer (Yale Liver Care Center, Hartford, CT), was cultured as previously described (51). All media were purchased from Gibco/BRL, and cells were grown at 37°C with 5% CO2. The viruses used included RRV, a simian strain of genotype G3P[3] (kindly provided by H. Greenberg, Stanford University, Palo Alto, CA), and strain KU (G1P1A[8]), a human strain (generously provided by Koki Taniguchi, Fujita Health University, Toyoake, Aichi, Japan).
Structural modeling of VP4 protein.
The mapping of interaction sites and relevant functional motifs as well as the structure visualization was performed using the Polyview Web server (http://polyview.cchmc.org) and resolved structures of capsid proteins (Protein Data Bank [PDB] code 4V7Q for rotavirus) (52, 53).
Peptides.
Synthetic peptides VSKLY, VSGLY, VSRLY, TRTRVSRLY, and DGEA, corresponding to different regions of the VP4 protein of RRV, were synthesized and purified by high-performance liquid chromatography (HPLC) and kept lyophilized (GenScript, Piscataway, NJ). An unrelated peptide (GHRP) of the same size and random-sequence peptides with identical amino acid compositions were synthesized for use as controls. Peptides were resuspended to 1 mM in DMEM before use in replication assays.
Generation of VP4-encoding plasmid.
RNA was isolated from virus grown in MA104 cells. The full-length RRV VP4 (accession no. AY033150) coding segment was amplified by reverse transcription-PCR (RT-PCR) using primers 1 and 2 (Table 2) and a Qiagen Long Range 2-Step RT-PCR kit (Qiagen, Hilden, Germany). The amplified product was cloned into the vector pBacT7-S1T3D (a gift from Terence Dermody, Vanderbilt University), which contained T7 RNA polymerase promoter, hepatitis delta virus ribozyme, and T7 RNA polymerase terminator sequences. The pBacT7-S1T3D vector was digested first with KpnI and then blunt end digested with the Klenow fragment, followed by digestion with the SacII enzyme. The amplified RRV VP4 plasmid was digested with SmaI and SacII. Both the vectors and the PCR product were gel purified and ligated using standard protocols (54). After transformation and screening, the constructs were confirmed by digestion and by Sanger DNA sequencing using an ABI 3730 DNA analyzer (Applied Biosystems). The positive plasmids, named pBacT7-VP4(RRV), were purified using a Promega plasmid kit (Promega Inc., Madison, WI) prior to transfection.
TABLE 2.
Primers used for this study
| Primer no. | Nucleotide sequence (5′–3′)a | Comment |
|---|---|---|
| 1 | CAGCATCCCGGGTAATACGACTCACTATAGGCTATAAAATGGCTTCGCTCAT | SmaI site |
| 2 | ATCGACCGCGGTCACATCCTCTGGAAATTGCTTAC | SacII site |
| 3 | ATCGTTACCAACGGATTTTGTAATATCTAGATATGAAGTAATTAAAGAGAATTC | |
| 4 | GAATTCTCTTTAATTACTTCATATCTAGATATTACAAAATCCGTTGGTAACGAT | |
| 5 | CACCAGAACTAGAGTTAGTGGATTGTATGGGTTACCTGC | R-to-G change |
| 6 | GCAGGTAACCCATACAATCCACTAACTCTAGTTCTGGTG | |
| 7 | CATATACCGTTGACTTATCTGATG | RRV VP4 |
| 8 | TAATGGTCCGTATTGTGAATAGC | |
| 9 | GGGGTCCTGGTGAAACTAATG | Rotavirus VP4 |
| 10 | AAGCGCCAATGGAACAATG | |
| 11 | ATACGGACCATTACAATCTACTCCA | RRV462 |
| 12 | ATTCTTTTCCATTATTTGGGTTAGC | RRV1381 |
The restriction enzyme site for XbaI is underlined.
Introduction of genetic markers and mutation in the RRV VP4 gene.
Manipulation of the VP4 gene was carried out in plasmid pBacT7-VP4(RRV). The creation of an XbaI site was performed by changing the nucleotide at position 1029 from A to T, keeping the amino acid sequence unchanged. The amino acid change from R (arginine) to G (glycine) at position 446 was carried out with a nucleotide change at position 1345 (A to G). Both changes were carried out using a QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer's instructions. Briefly, pBacT7-VP4(RRV) was amplified by use of mutated nucleotide primers 3 and 4 for creation of the XbaI site and primers 5 and 6 to change the amino acid at position 446 from R to G in RRV VP4. After digestion with DpnI to eliminate the wild-type (WT) pBacT7-VP4(RRV) used as a template, the PCR products were used to transform XL-10 competent cells. The generated plasmids, pBacT7-VP4(RRVXbaI) (WT plasmid with XbaI site) and pBacT7-VP4(RRVR446G) (WT plasmid encoding R-to-G substitution at position 446), were confirmed by sequencing.
Reverse genetics system.
BHK21-T7 cells were seeded at a concentration of 5 × 105 cells/ml in 6-well plates. After overnight culture at a confluence of 70%, 2.5 μg of pBacT7-VP4(RRV) plasmid (or mutant VP4 plasmid) was transfected into BHK21-T7 cells by use of 4 μl of TransIT LT-1 (Mirus, Madison, WI)/μg of DNA according to the manufacturer's instructions. Twenty-four hours after transfection, the transfected cells were washed twice with serum-free medium and then infected with KU as the helper virus at a multiplicity of infection (MOI) of 10. Twenty-four hours after the helper virus infection, the BHK21-T7 cells were harvested, freeze-thawed three times, and centrifuged at low speed to remove cell debris. To rescue recombinant viruses, supernatants containing the viral progenies produced in BHK21-T7 cells were propagated in MA104 cells. At the first passage, confluent monolayers of MA104 cells in 25-cm2 flasks were inoculated with the supernatant corresponding to one well of BHK21-T7 cells. For further serial passage in MA104 cell culture, 1/5 of the undiluted MA104 cell lysate was used to infect MA104 cells in 25-cm2 flasks for 48 h, and at each passage, aliquots were kept frozen at −80°C for further analysis. Recombinant KU viruses carrying the RRV VP4s [KU-VP4(RRV)] were rescued from the 10th passage in MA104 cells by two plaque-to-plaque cloning steps in MA104 cells, as previously described (32). KU-VP4(RRV) was backcrossed to RT(VP4) (an RRV reassortant containing TUCH VP4) to obtain RRVrVP4 (RRV with recombinant VP4). Therefore, the viruses obtained from the plasmids pBacT7-VP4(RRV), pBacT7-VP4(RRVXbaI), and pBacT7-VP4(RRVR446G) were named RRVrVP4, RRVVP4-XbaI, and RRVVP4-R446G, respectively.
RT-PCR and sequencing.
Total RNA was isolated from virus supernatants after pelleting through ultracentrifugation, using an RNA isolation kit (Zymo Research, Irvine, CA). RT-PCR was performed using a SuperScript III One-Step RT-PCR kit (Invitrogen, Carlsbad, CA) and primers 7 and 8 (for detection of RRV VP4), primers 9 and 10 (for detection of all rotavirus VP4s), or primers 11 and 12 (for amplification of the region containing XbaI). For sequence analysis of virus populations, RT-PCR products were purified, and sequencing was carried out using an ABI Prism BigDye v3.1 Terminator sequencing kit and a model 3730 DNA analyzer (Applied Biosystems, Foster City, CA). Sequence files were analyzed using Finch TV (Geospiza, PerkinElmer, WA, USA).
Electrophoretic analysis of dsRNA.
Viral RNA was extracted using phenol-chloroform, and the dsRNA segments were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by silver staining as described previously (32). The migration patterns of the reassortants' dsRNA genes were compared with those from the parental strains, and gene assignments were made based on the migration of RRV gene segments. Progeny with single-gene substitutions were identified and reamplified. The parental origins of the single-gene reassortants were reconfirmed by gel electrophoresis and sequencing after amplification.
Measurement of RRV replication in cholangiocytes.
Cholangiocytes were seeded in 24-well plates in DMEM as previously described (55) and then incubated at 37°C until 100% confluence. Plates were washed with Earle's balanced salt solution (EBSS) and infected with RRV at various MOIs at 37°C for 1 h. The cultures were washed and incubated with serum-free DMEM at 37°C for 24 and 48 h, and viral yields were assessed by a focus-forming assay (FFA).
Viral binding and attachment assay in cholangiocytes.
Virus binding to cholangiocytes and MA104 and Caco-2 cells has been described previously (51). In brief, cholangiocytes were grown to confluence in 24-well plates, and attachment assays were performed in triplicate. At the time of assay, the cells, medium, and inoculating virus were cooled to 4°C. Cells were washed, treated with an inoculum of RRV at an MOI of 0.5, and incubated for 1 h at 4°C. After 1 h, the inoculum was removed, and the cells were washed twice to remove any unbound virus. All washes and residual inoculum were combined to account for all unbound virus. The cells were then frozen and later underwent 2 freeze-thaw cycles. Any virus found within the final cell fraction reflected bound virus. The quantities of bound and unbound virus were determined by FFA. The amount of bound virus was expressed as a percentage of the total amount of virus used to inoculate the cells.
Quantification of infectious rotavirus.
Cell suspensions were analyzed for the presence of infectious RRV by fluorescent FFA as previously described (56). Ninety-six-well plates were seeded with MA104 cells and grown for 4 days. Once they were confluent, the cells were infected with serially diluted virus isolates (1:10) and centrifuged at 2,000 rpm for 1.5 h. The cells were washed with DMEM and incubated at 37°C for 14 to 16 h with DMEM containing 4 μg/ml of trypsin. The medium was aspirated, and cells were fixed with cold 80% acetone for 15 min at 20°C. Following a wash with phosphate-buffered saline (PBS), a guinea pig anti-rotavirus immunoglobulin G (IgG) (1:1,000) (Jackson Immunoresearch, USA) primary antibody was added and incubated for 30 min. Wells were washed with PBS, and a fluorescein isothiocyanate (FITC)-tagged goat anti-guinea pig IgG (1:500) (MP Biomedicals, Solon, OH) secondary antibody was added and incubated for 30 min at 37°C. Wells were washed twice and allowed to dry completely. Fluorescent cells were counted using a UV microscope (10× objective), and the amount of infectious virus was reported in focus-forming units (FFU) per milliliter. For titers in tissue, the amount of live virus per sample was normalized to the weight of each sample at the time of harvest, resulting in the number of FFU per milliliter per milligram of tissue.
Mouse model of biliary atresia.
The experimental model of BA was induced in BALB/c mice (Envigo Labs, Indianapolis, IN). Newborn pups within 24 to 48 h of life were injected intraperitoneally (i.p.) with RRV, RRVVP4-XbaI, or RRVVP4-R446G at 1.5 × 106 FFU/pup or with saline as a control. Pups were monitored daily for 21 days for clinical signs of hepatobiliary injury (i.e., acholic stools, bilirubinuria, and jaundice of non-fur-covered skin) and for survival. Subsets of mice were sacrificed at 7 and 10 days postinfection, and their extrahepatic biliary tracts and livers were harvested. One set of tissue was analyzed for the presence of infectious virus by a focus-forming assay. Another set of tissues was processed for hematoxylin and eosin (H&E) staining.
Flow cytometry.
Livers were harvested from pups 7 days after injection and pooled at three per sample in RPMI medium plus 2% FBS. Samples were processed as described before (31) to purify the mononuclear cells. After red blood cell lysis, the cells were washed two times and suspended in 1× PBS plus 1% FBS. About 1 × 106 cells were treated with Fc block (BD Biosciences, San Jose, CA) in a 96-well v-bottomed plate and then incubated at 4°C for 30 min. A panel of anti-mouse CD49b (an NK cell marker) and anti-mouse CD69 (a marker of activation) (eBioscience, San Diego, CA) antibodies (1:100 dilution) was added and incubated for 30 min at 4°C. Cells were washed three times, filtered, and fixed with 4% paraformaldehyde. Samples were then read on an Accuri C6 flow cytometer (BD, Franklin Lakes, NJ) and analyzed with FlowJo, version 7.6.5 (FlowJo).
Histopathology and cholangiograms.
Livers and extrahepatic bile ducts were harvested from neonatal mice 10 days after injection with RRV, RRVVP4-XbaI, or RRVVP4-R446G, using a dissecting microscope. Liver samples and the extrahepatic bile ducts were fixed with formalin, embedded in paraffin, sectioned with a microtome (5-μm thickness), and stained with H&E for microscopic analysis.
Cholangiograms were performed at 10 days postinfection. Pups were sacrificed and the abdominal cavity opened, and the liver was flipped to expose the extrahepatic bile duct. The gallbladder was held with a pair of forceps, and methylene blue dye was injected through a 30-gauge needle. Passage of dye through the duct to the duodenum confirmed ductal patency. Pictures were taken through an inverted dissecting microscope.
Statistical analysis.
Analysis of noncontinuous variables was performed with the chi-square and Fisher exact tests. Results for continuous variables were expressed as means ± standard errors of the means (SEM) and were analyzed by analysis of variance (ANOVA) with post hoc testing, where appropriate. P values of <0.05 were considered significant.
ACKNOWLEDGMENTS
Sequence and structure analyses were performed with the help of the Cincinnati Children's Hospital Medical Center Protein Informatics Core and the University of Cincinnati Bioinformatics Core. We thank Terence Dermody for providing the pBacT7-S1T3D plasmid.
This study was funded by National Institutes of Health (NIH) grant R01 DK-091566.
REFERENCES
- 1.Balistreri WF, Grand R, Hoofnagle JH, Suchy FJ, Ryckman FC, Perlmutter DH, Sokol RJ. 1996. Biliary atresia: current concepts and research directions. Summary of a symposium. Hepatology 23:1682–1692. [DOI] [PubMed] [Google Scholar]
- 2.Bessho K, Bezerra JA. 2011. Biliary atresia: will blocking inflammation tame the disease? Annu Rev Med 62:171–185. doi: 10.1146/annurev-med-042909-093734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riepenhoff-Talty M, Gouvea V, Evans MJ, Svensson L, Hoffenberg E, Sokol RJ, Uhnoo I, Greenberg SJ, Schakel K, Zhaori G, Fitzgerald J, Chong S, el-Yousef M, Nemeth A, Brown M, Piccoli D, Hyams J, Ruffin D, Rossi T. 1996. Detection of group C rotavirus in infants with extrahepatic biliary atresia. J Infect Dis 174:8–15. doi: 10.1093/infdis/174.1.8. [DOI] [PubMed] [Google Scholar]
- 4.Glaser JH, Balistreri WF, Morecki R. 1984. Role of reovirus type 3 in persistent infantile cholestasis. J Pediatr 105:912–915. doi: 10.1016/S0022-3476(84)80076-1. [DOI] [PubMed] [Google Scholar]
- 5.Fjaer RB, Bruu AL, Nordbo SA. 2005. Extrahepatic bile duct atresia and viral involvement. Pediatr Transplant 9:68–73. doi: 10.1111/j.1399-3046.2005.00257.x. [DOI] [PubMed] [Google Scholar]
- 6.Domiati-Saad R, Dawson DB, Margraf LR, Finegold MJ, Weinberg AG, Rogers BB. 2000. Cytomegalovirus and human herpesvirus 6, but not human papillomavirus, are present in neonatal giant cell hepatitis and extrahepatic biliary atresia. Pediatr Dev Pathol 3:367–373. doi: 10.1007/s100240010045. [DOI] [PubMed] [Google Scholar]
- 7.Drut R, Drut RM, Gomez MA, Cueto Rua E, Lojo MM. 1998. Presence of human papillomavirus in extrahepatic biliary atresia. J Pediatr Gastroenterol Nutr 27:530–535. doi: 10.1097/00005176-199811000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Petersen C, Grasshoff S, Luciano L. 1998. Diverse morphology of biliary atresia in an animal model. J Hepatol 28:603–607. doi: 10.1016/S0168-8278(98)80283-3. [DOI] [PubMed] [Google Scholar]
- 9.Riepenhoff-Talty M, Schaekel K, Clark HF, Mueller W, Uhnoo I, Rossi T, Fisher J, Ogra PL. 1993. Group A rotaviruses produce extrahepatic biliary obstruction in orally inoculated newborn mice. Pediatr Res 33:394–399. [DOI] [PubMed] [Google Scholar]
- 10.Czech-Schmidt G, Verhagen W, Szavay P, Leonhardt J, Petersen C. 2001. Immunological gap in the infectious animal model for biliary atresia. J Surg Res 101:62–67. doi: 10.1006/jsre.2001.6234. [DOI] [PubMed] [Google Scholar]
- 11.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD. 2016. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin Infect Dis 62:S96–S105. doi: 10.1093/cid/civ1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Baker ML, Jiang W, Estes MK, Prasad BV. 2009. Rotavirus architecture at subnanometer resolution. J Virol 83:1754–1766. doi: 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dormitzer PR, Sun ZY, Wagner G, Harrison SC. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J 21:885–897. doi: 10.1093/emboj/21.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert JM, Greenberg HB. 1998. Cleavage of rhesus rotavirus VP4 after arginine 247 is essential for rotavirus-like particle-induced fusion from without. J Virol 72:5323–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez S, Arias CF. 2004. Multistep entry of rotavirus into cells: a Versaillesque dance. Trends Microbiol 12:271–278. doi: 10.1016/j.tim.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Arias CF, Isa P, Guerrero CA, Mendez E, Zarate S, Lopez T, Espinosa R, Romero P, Lopez S. 2002. Molecular biology of rotavirus cell entry. Arch Med Res 33:356–361. doi: 10.1016/S0188-4409(02)00374-0. [DOI] [PubMed] [Google Scholar]
- 17.Arias CF, Silva-Ayala D, Lopez S. 2015. Rotavirus entry: a deep journey into the cell with several exits. J Virol 89:890–893. doi: 10.1128/JVI.01787-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coulson BS, Londrigan SL, Lee DJ. 1997. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc Natl Acad Sci U S A 94:5389–5394. doi: 10.1073/pnas.94.10.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham KL, Halasz P, Tan Y, Hewish MJ, Takada Y, Mackow ER, Robinson MK, Coulson BS. 2003. Integrin-using rotaviruses bind alpha2beta1 integrin alpha2 I domain via VP4 DGE sequence and recognize alphaXbeta2 and alphaVbeta3 by using VP7 during cell entry. J Virol 77:9969–9978. doi: 10.1128/JVI.77.18.9969-9978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerrero CA, Mendez E, Zarate S, Isa P, Lopez S, Arias CF. 2000. Integrin alpha(v)beta(3) mediates rotavirus cell entry. Proc Natl Acad Sci U S A 97:14644–14649. doi: 10.1073/pnas.250299897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hewish MJ, Takada Y, Coulson BS. 2000. Integrins alpha2beta1 and alpha4beta1 can mediate SA11 rotavirus attachment and entry into cells. J Virol 74:228–236. doi: 10.1128/JVI.74.1.228-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Londrigan SL, Graham KL, Takada Y, Halasz P, Coulson BS. 2003. Monkey rotavirus binding to alpha2beta1 integrin requires the alpha2 I domain and is facilitated by the homologous beta1 subunit. J Virol 77:9486–9501. doi: 10.1128/JVI.77.17.9486-9501.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarate S, Espinosa R, Romero P, Guerrero CA, Arias CF, Lopez S. 2000. Integrin alpha2beta1 mediates the cell attachment of the rotavirus neuraminidase-resistant variant nar3. Virology 278:50–54. doi: 10.1006/viro.2000.0660. [DOI] [PubMed] [Google Scholar]
- 24.Boyce M, Celma CC, Roy P. 2008. Development of reverse genetics systems for bluetongue virus: recovery of infectious virus from synthetic RNA transcripts. J Virol 82:8339–8348. doi: 10.1128/JVI.00808-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Gennip RG, van de Water SG, Potgieter CA, Wright IM, Veldman D, van Rijn PA. 2012. Rescue of recent virulent and avirulent field strains of bluetongue virus by reverse genetics. PLoS One 7:e30540. doi: 10.1371/journal.pone.0030540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi T, Ooms LS, Ikizler M, Chappell JD, Dermody TS. 2010. An improved reverse genetics system for mammalian orthoreoviruses. Virology 398:194–200. doi: 10.1016/j.virol.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komoto S, Kawagishi T, Kobayashi T, Ikizler M, Iskarpatyoti J, Dermody TS, Taniguchi K. 2014. A plasmid-based reverse genetics system for mammalian orthoreoviruses driven by a plasmid-encoded T7 RNA polymerase. J Virol Methods 196:36–39. doi: 10.1016/j.jviromet.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komoto S, Sasaki J, Taniguchi K. 2006. Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc Natl Acad Sci U S A 103:4646–4651. doi: 10.1073/pnas.0509385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trask SD, Taraporewala ZF, Boehme KW, Dermody TS, Patton JT. 2010. Dual selection mechanisms drive efficient single-gene reverse genetics for rotavirus. Proc Natl Acad Sci U S A 107:18652–18657. doi: 10.1073/pnas.1011948107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen SR, Jafri M, Donnelly B, McNeal M, Witte D, Bezerra J, Ward R, Tiao GM. 2007. Effect of rotavirus strain on the murine model of biliary atresia. J Virol 81:1671–1679. doi: 10.1128/JVI.02094-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walther A, Mohanty SK, Donnelly B, Coots A, Lages CS, Lobeck I, Dupree P, Meller J, McNeal M, Sestak K, Tiao G. 2015. Rhesus rotavirus VP4 sequence-specific activation of mononuclear cells is associated with cholangiopathy in murine biliary atresia. Am J Physiol Gastrointest Liver Physiol 309:G466–G474. doi: 10.1152/ajpgi.00079.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, Donnelly B, Bondoc A, Mohanty SK, McNeal M, Ward R, Sestak K, Zheng S, Tiao G. 2011. The rhesus rotavirus gene encoding VP4 is a major determinant in the pathogenesis of biliary atresia in newborn mice. J Virol 85:9069–9077. doi: 10.1128/JVI.02436-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohanty SK, Donnelly B, Lobeck I, Walther A, Dupree P, Coots A, Meller J, McNeal M, Sestak K, Tiao G. 2017. The SRL peptide of rhesus rotavirus VP4 protein governs cholangiocyte infection and the murine model of biliary atresia. Hepatology 65:1278–1292. doi: 10.1002/hep.28947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porollo A, Meller J. 2007. Prediction-based fingerprints of protein-protein interactions. Proteins 66:630–645. doi: 10.1002/prot.21248. [DOI] [PubMed] [Google Scholar]
- 35.Shivakumar P, Sabla GE, Whitington P, Chougnet CA, Bezerra JA. 2009. Neonatal NK cells target the mouse duct epithelium via Nkg2d and drive tissue-specific injury in experimental biliary atresia. J Clin Invest 119:2281–2290. doi: 10.1172/JCI38879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bezerra JA. 2005. Potential etiologies of biliary atresia. Pediatr Transplant 9:646–651. doi: 10.1111/j.1399-3046.2005.00350.x. [DOI] [PubMed] [Google Scholar]
- 37.Mack CL. 2007. The pathogenesis of biliary atresia: evidence for a virus-induced autoimmune disease. Semin Liver Dis 27:233–242. doi: 10.1055/s-2007-985068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shivakumar P, Campbell KM, Sabla GE, Miethke A, Tiao G, McNeal MM, Ward RL, Bezerra JA. 2004. Obstruction of extrahepatic bile ducts by lymphocytes is regulated by IFN-gamma in experimental biliary atresia. J Clin Invest 114:322–329. doi: 10.1172/JCI200421153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khamrin P, Maneekarn N, Peerakome S, Yagyu F, Okitsu S, Ushijima H. 2006. Molecular characterization of a rare G3P[3] human rotavirus reassortant strain reveals evidence for multiple human-animal interspecies transmissions. J Med Virol 78:986–994. doi: 10.1002/jmv.20651. [DOI] [PubMed] [Google Scholar]
- 40.Li K, Lin XD, Huang KY, Zhang B, Shi M, Guo WP, Wang MR, Wang W, Xing JG, Li MH, Hong WS, Holmes EC, Zhang YZ. 2016. Identification of novel and diverse rotaviruses in rodents and insectivores, and evidence of cross-species transmission into humans. Virology 494:168–177. doi: 10.1016/j.virol.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johne R, Reetz J, Kaufer BB, Trojnar E. 2015. Generation of an avian-mammalian rotavirus reassortant by using a helper virus-dependent reverse genetics system. J Virol 90:1439–1443. doi: 10.1128/JVI.02730-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Troupin C, Dehee A, Schnuriger A, Vende P, Poncet D, Garbarg-Chenon A. 2010. Rearranged genomic RNA segments offer a new approach to the reverse genetics of rotaviruses. J Virol 84:6711–6719. doi: 10.1128/JVI.00547-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanai Y, Komoto S, Kawagishi T, Nouda R, Nagasawa N, Onishi M, Matsuura Y, Taniguchi K, Kobayashi T. 2017. Entirely plasmid-based reverse genetics system for rotaviruses. Proc Natl Acad Sci U S A 114:2349–2354. doi: 10.1073/pnas.1618424114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCrae MA, McCorquodale JG. 1983. Molecular biology of rotaviruses. V. Terminal structure of viral RNA species. Virology 126:204–212. [DOI] [PubMed] [Google Scholar]
- 45.Imai M, Akatani K, Ikegami N, Furuichi Y. 1983. Capped and conserved terminal structures in human rotavirus genome double-stranded RNA segments. J Virol 47:125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patton JT, Wentz M, Xiaobo J, Ramig RF. 1996. cis-Acting signals that promote genome replication in rotavirus mRNA. J Virol 70:3961–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wentz MJ, Patton JT, Ramig RF. 1996. The 3′-terminal consensus sequence of rotavirus mRNA is the minimal promoter of negative-strand RNA synthesis. J Virol 70:7833–7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chizhikov V, Patton JT. 2000. A four-nucleotide translation enhancer in the 3′-terminal consensus sequence of the nonpolyadenylated mRNAs of rotavirus. RNA 6:814–825. doi: 10.1017/S1355838200992264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saxena V, Shivakumar P, Sabla G, Mourya R, Chougnet C, Bezerra JA. 2011. Dendritic cells regulate natural killer cell activation and epithelial injury in experimental biliary atresia. Sci Transl Med 3:102ra194. doi: 10.1126/scitranslmed.3002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng N, Sen A, Wolf M, Vo P, Hoshino Y, Greenberg HB. 2011. Roles of VP4 and NSP1 in determining the distinctive replication capacities of simian rotavirus RRV and bovine rotavirus UK in the mouse biliary tract. J Virol 85:2686–2694. doi: 10.1128/JVI.02408-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jafri M, Donnelly B, Allen S, Bondoc A, McNeal M, Rennert PD, Weinreb PH, Ward R, Tiao G. 2008. Cholangiocyte expression of alpha2beta1-integrin confers susceptibility to rotavirus-induced experimental biliary atresia. Am J Physiol Gastrointest Liver Physiol 295:G16–G26. doi: 10.1152/ajpgi.00442.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. 2011. Atomic model of an infectious rotavirus particle. EMBO J 30:408–416. doi: 10.1038/emboj.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Porollo A, Meller J. 2007. Versatile annotation and publication quality visualization of protein complexes using POLYVIEW-3D. BMC Bioinformatics 8:316. doi: 10.1186/1471-2105-8-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 55.Jafri M, Donnelly B, McNeal M, Ward R, Tiao G. 2007. MAPK signaling contributes to rotaviral-induced cholangiocyte injury and viral replication. Surgery 142:192–201. doi: 10.1016/j.surg.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 56.Mohanty SK, Donnelly B, Bondoc A, Jafri M, Walther A, Coots A, McNeal M, Witte D, Tiao GM. 2013. Rotavirus replication in the cholangiocyte mediates the temporal dependence of murine biliary atresia. PLoS One 8:e69069. doi: 10.1371/journal.pone.0069069. [DOI] [PMC free article] [PubMed] [Google Scholar]