Abstract
We demonstrate that closure tables are an effective data structure for developing database-driven applications that query biomedical ontologies and that require cross-querying between multiple ontologies. A closure table stores all available paths within a tree, even those without a direct parent-child relationship; additionally, a node can have multiple ancestors which gives the foundation for supporting linkages between controlled ontologies. We augment the meta-data structure of the ICD9 and ICD10 ontologies included in i2b2, an open source query tool for identifying patient cohorts, to utilize a closure table. We describe our experiences in incorporating existing mappings between ontologies to enable clinical and health researchers to identify patient populations using the ontology that best matches their preference and expertise.
I. Introduction
Biomedical ontologies, such as those available through the Unified Medical Language System (UMLS) [1] or through BioPortal [2], are driving an increasingly larger proportion of modern biomedical applications for both research and clinical operation tasks and the need for efficient and effective data structures and modeling techniques is apparent [3]. The i2b2 (Informatics for Integrating Biology and the Bedside) query tool is a successful example of an open-source project that relies directly upon publicly available ontologies [4].
The goal of i2b2 is to assist clinical and health informatics researchers in identifying patient cohorts for clinical research or trials [4]. Users drag-and-drop concepts from an ontology cell that visualizes the concepts as a tree and form Boolean queries of inclusion and exclusion criteria that pinpoint a necessary population for the researcher’s study. i2b2 is open-source and is made available world wide for adoption. i2b2 is reported to be used by over half of all institutions receiving a Clinical and Translational Science Award (CTSA), over 60 academic medical centers, and over 10 international medical centers [5]. We have demonstrated that i2b2 can bootstrap rural health analytics and learning networks [6] and quickly give infrastructure to institutions that otherwise lack the resources to support clinical and health informatics research [7].
In this paper we demonstrate that closure tables are an effective data structure for developing database-driven applications and share our experience with improving the open-source i2b2 project by providing extensions that support closure table adoption. Using ICD9 and ICD10 as an example [8], we will show that closure tables enable cross-querying of ontologies within i2b2 in order for users to interact with the ontology that bests matches their preference and expertise, allowing clinical research to be expedited. We compare our solution to the native path-to-root structure found within i2b2 and show that closure tables offer a solution that is more manageable and optimized for relational databases.
II. Closure Tables
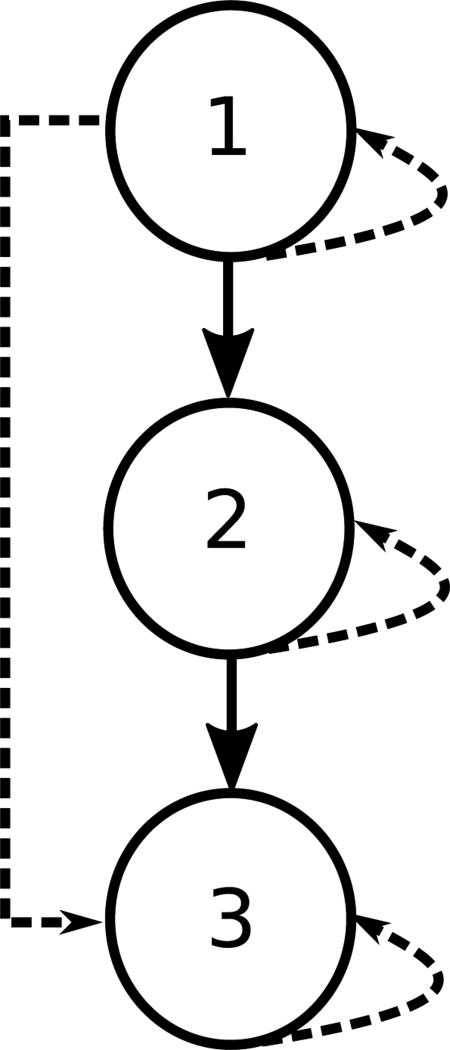
A closure table is a data structure that represents a tree as a table of ancestor and descendant pairs; specifically, it stores both direct and indirect ancestor-descendant relationships [9]. For the small graph in Figure 2, direct relationships between nodes 1 and 2 and between nodes 2 and 3 exist; the indirect relationship between node 1 and node 3 is also recorded in the table. Each node has a self-referential link to itself as well. This data is summarized in Table 1.
Fig. 2.
This basic example of a closure table shows three nodes; direct ancestor and descendant relations are represented by solid lines and indirect descendants are represented by dashed lines.
TABLE I.
A closure table corresponding to the example in Figure 2.
| Ancestor | Descendant |
|---|---|
|
| |
| 1 | 1 |
| 1 | 2 |
| 1 | 3 |
| 2 | 2 |
| 2 | 3 |
| 3 | 3 |
Additionally, the flexibility of closure tables allow a node to have more than one ancestor. For example, if a new node was introduced in the example illustrated in Figure 2, it could be the ancestor of node 2, even if node 2 already has node 1 as an ancestor. We will use this flexibility to insert linkages between ontologies later.
Mapping ontologies into relational databases is an open area of research [10] where a cohesive partnership between relational databases and semantic web technologies, such as OWL, must manifest in order for database-driven applications to leverage semantic knowledge [11]. For example, curation and interchange of ontologies through standards such as OWL have a long record of success [11], but the proliferation of database-driven web-applications must also integrate semantic knowledge natively. In the next section, we discuss how to natively support ontologies as closure tables in i2b2.
III. Integrating Closure Tables with i2b2
We recommended the adoption of closure tables as future work in an unrelated contribution to the i2b2 project [12] which was followed up and independently shown to perform well for SNOMED-CT, a commonly-used polyhierarchy with millions of components [13]. The types of ontologies integrated into i2b2’s ontology cell varies, ranging from simpler ICD9 and ICD10 light-weight ontologies to fuller, more expansive ontologies such as the Gene Ontology (GO) [14].
We provide an i2b2 closure table toolkit [15] to assist system administrators in adopting closure tables for the ontology cell of i2b2. This requires an additional table to hold the ancestor and descendant pairs that we call metatree; within this table, we have added some additional columns to simplify future expressions, such as including the text label of the concept corresponding to the ancestor and descendant. We also track the depth of the tree at this particular relationship and the length of the path between the ancestor and descendant. Knowing depth assists in drawing the actual meta-data because one can query for any given node with any given offset. For example, if an ontology has a large number of concepts, an interactive visualization of the concept tree may wish to load smaller sub-trees dynamically within its web interface to avoid loading delays. The table also contains a column that represents the paths as arrays for quick expansion.
By default, the meta-data in i2b2 is constructed dynamically by specifying a concept table and a concept column; the closure table is enabled simply by replacing the default concept table with a join between the i2b2 meta-data tables and the newly-constructed metatree table of ancestor and descendant pairs. The keys of the metatree table are unique pairs of integers that represent ancestors and descendants. We add an additional column to the default meta-data table to hold the meta-id from the metatree; our metatree holds foreign key relationships between the meta-data in i2b2 and the ancestor and descendant pairs. The result of having unique integer keys is that joins and look-ups are easily optimized; the foreign key relationships ensure referential integrity between the metatree and i2b2 meta-data.
IV. Augmenting Ontologies
Recall that i2b2 enables researchers to develop queries via dragging-and-dropping from an ontology cell that illustrates the concepts available as a tree; concepts can include facets of patients and visits, such as demographics, diagnoses, lab results, and so on. As an example, we focus on the ICD9 and ICD10 [8] components of our implementation of i2b2.
A. Issues with Retrospective Analysis
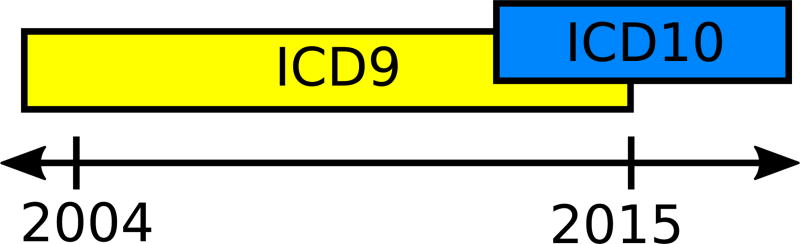
In 2015, a mandated switch from the ICD9 standard to ICD10 complicated the ability of researchers to query diagnostic codes in i2b2. As seen in Figure 3, patient visits within our data that were coded between 2004 and 2013 have only ICD9 codes for diagnoses, and patient visits that were coded between 2013 and 2015 have either ICD9 or ICD10 codes. After October 2015, patient visits in our data are only coded with ICD10. This switch in standards complicates a clinical researcher’s ability to query for patient populations with i2b2. In our experience, many clinical researchers have little knowledge or training with medical coding, while others have become familiar with common groups of codes over time from conducting and coordinating studies and have not had time to adjust to the new standards.
Fig. 3.
In our implementation of i2b2, patient data is tagged with ICD9 concepts for a 10 year period and later tagged with ICD10 concepts for nearly a three year period; a small period of time contains concepts from either ontology.
Without familiarity with ICD10 and without our proposed cross-querying solution, it is difficult for i2b2 users to create drag-and-drop queries to target patients in the most recent years of data. Furthermore, ICD10 contains particularly specific codes yet researchers tend to categorize patients into abstract buckets that more closely resemble higher-level, non-specific codes. These higher-level codes in ICD10 are not suitable for billing due to regulations that require specificity to the fifth digit; consequently, these concept codes do not appear naturally in our data sources despite being available in the ontology. In the next section, we describe how closure tables can enable cross-querying of ontologies in order for researchers to search patients using standards that best fit their preference and expertise.
B. Cross-Querying Ontologies
General Equivalence Mappings (GEMs) between the ICD9 and ICD10 standards were developed and made publicly available for assisting in analysis of data generated before and after the transition to ICD10 [16]. These equivalence mappings pose known challenges in data analysis because there is rarely an exact match between new and old concepts [16]. Simply adding the GEMs as additional synonyms of concepts within i2b2 is insufficient due to i2b2 being based largely upon aggregation of concepts and the fidelity of ancestor and descendant pairs require the ability to roll up and aggregate as needed. For example, querying for ICD9:250 would also query for ICD9:250.00, ICD9:250.01, and so on. Additionally, the native path-to-root meta-data method that i2b2 uses is known to be slow for overly large ontologies [13]; adding synonyms per every concept (and child concept for aggregation) is not feasible without radically increasing the size of the meta-data table.
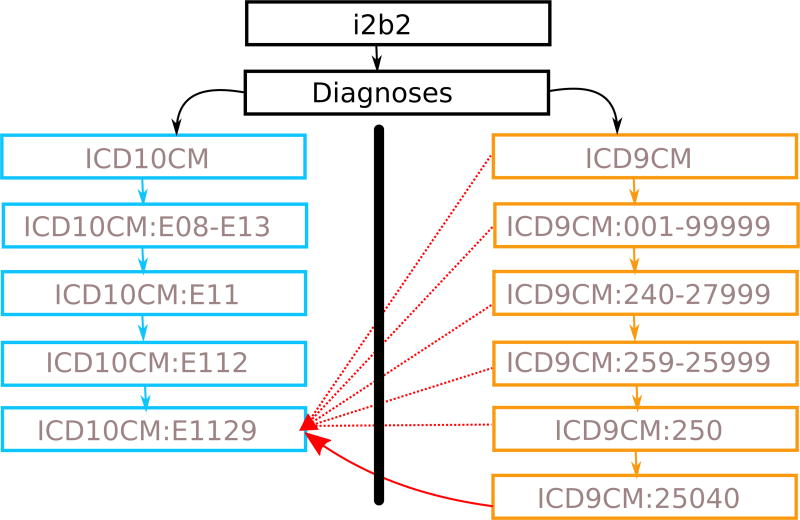
Because i2b2 generates aggregate counts of patient populations, specificity is not typically a priority and over-specifying diagnostic criteria can limit results unnecessarily. Without the closure table, if the GEMs were simply used as additional facts, aggregation potentially misses concepts. We use diabetes as an example in Figure 5 and illustrate an abbreviated closure table for a small subsection of ICD9 (orange boxes) and ICD10 (blue boxes) standards. Due to space considerations, we omit self-referential and indirect links within each ontology and draw only direct or derived links from the GEMs. The GEMs map ICD10CM:E1129 onto ICD9CM:25040. Using the closure table, i2b2 queries for ICD9CM:250 would consider all descendants of ICD9CM:250 and return matches for ICD10CM:E1129. Suppose instead that we had an ICD9 concept mapped to ICD10CM:E112, only adding ICD10CM:E112 as a synonym in the default i2b2 meta-data would not cover its relationship with ICDCM:E1129 unless it was also added as a synonym. The closure table enables us to bridge the gap between ICD9 and ICD10 by simply adding one link between the corresponding codes and also allows us to avoid enumerating synonym or child relationships.
Fig. 5.
We give an abbreviated closure table for the ICD9/ICD10 concepts corresponding to diabetes; ICD9 codes (orange boxes) and ICD10 codes (blue boxes) appear as two children of the diagnoses node, which are not connected by definition. The solid red arrow is taken directly from the GEMs and bridges ICD9 and ICD10 at a leaf node; the dotted red arrows are indirect links from the closure table structure. At this point, the ICD10CM:E1129 concept is considered part of the ICD9CM tree as a direct descendant of ICD9CM:25040, an indirect descendant of ICD9:250, and so on. The reverse holds true when an ICD9 concept is treated as a descendant of its ICD10 equivalent.
The potential downside to closure tables is that they create long tables as a result of storing indirect relations; however, this is not a true concern because ancestor and descendant pairs are unique integers that can be easily indexed for fast retrieval. Once the structure is created, bridging and manipulating sub-trees is as simple as inserting or modifying a single link. Additionally, scale is not of concern because the metatree is dwarfed by the actual patient data being described by the included ontologies.
The benefits of closure tables extend beyond ICD9 and ICD10. The Agency for Healthcare Research and Quality (AHRQ) released the Clinical Classifications Software (CCS) as a way of clustering patient diagnoses [17] and mappings exist for both ICD9 and ICD10; we leave implementing this as future work, but propose that a closure tree could unify CCS groups to simplify patient selection without redundant copies of concepts from the related ontologies.
V. Conclusion
In terms of making ontology management easier for database-driven applications, we wish to explore the impact that closure tables could have in federated configurations of i2b2, such as those constructed with the Shared Health Research Information Network (SHRINE) extension of i2b2 [18]. In particular, we wish to explore how closure tables can help overcome the shortcomings of rural networks [7] and assist in boostrapping i2b2 for rural health analytics and learning networks [6]. We have shown that closure tables are an effective and efficient data structure for developing database-driven applications that leverage ontologies and that require cross-querying between multiple ontologies. Although designed with i2b2 in mind, our closure table toolkit [15] can assist developers in generating closure tables for any database-driven application that needs to integrate standardized ontologies.
Fig. 1.
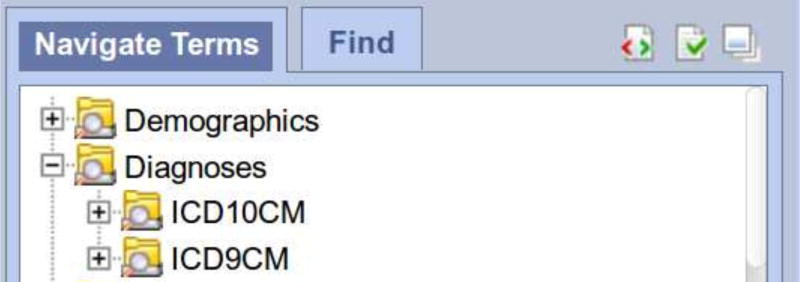
We arrange our meta-data for i2b2 to enable users to construct queries using either ICD9 or ICD10 diagnosis codes.
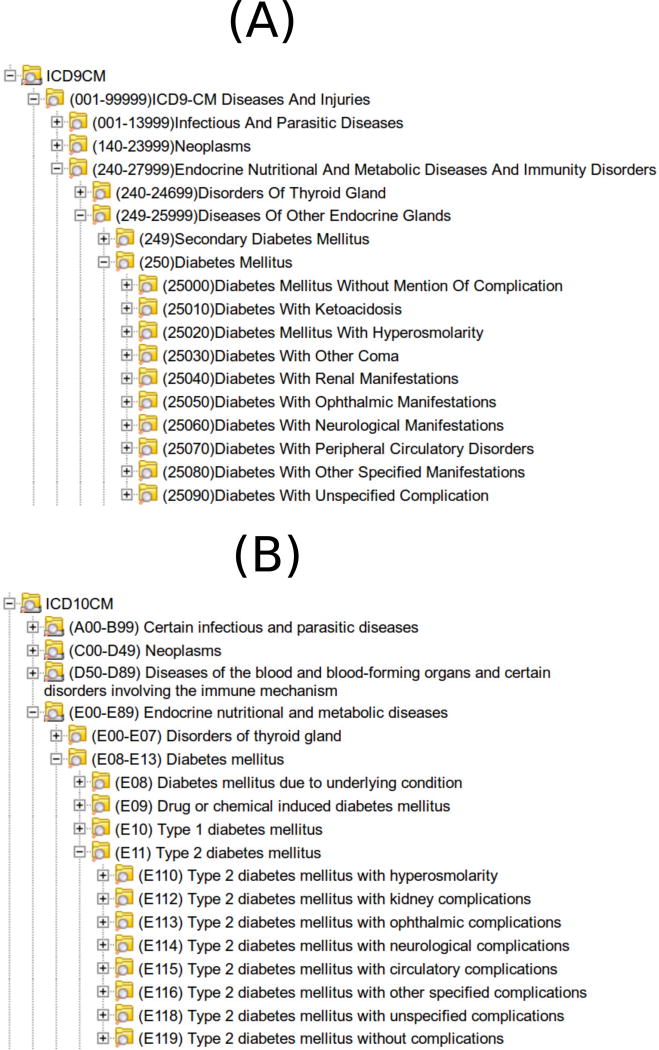
Fig. 4.
From within i2b2, users select from either the ICD9 ontology (A above) or the ICD10 ontology (B above) to identify patients with a history of a particular concept, such as diabetes.
Acknowledgments
The project described was supported by the NIH National Center for Advancing Translational Sciences through grant numbers UL1TR001998 and UL1TR000117. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Contributor Information
Daniel R. Harris, Center for Clinical and Translational Sciences, University of Kentucky, Lexington, Kentucky 40506 Institute for Pharmaceutical Outcomes and Policy, College of Pharmarcy, University of Kentucky, Lexington, Kentucky 40506.
Darren W. Henderson, Center for Clinical and Translational Sciences, University of Kentucky, Lexington, Kentucky 40506
Jeffery C. Talbert, Center for Clinical and Translational Sciences, University of Kentucky, Lexington, Kentucky 40506 Institute for Pharmaceutical Outcomes and Policy, College of Pharmarcy, University of Kentucky, Lexington, Kentucky 40506.
References
- 1.Bodenreider O. The unified medical language system (umls): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, Musen MA. Bioportal: enhanced functionality via new web services from the national center for biomedical ontology to access and use ontologies in software applications. Nucleic acids research. 2011;39(suppl 2):W541–W545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saripalle RK. International Conference on Smart Health. Springer; 2015. Need for a specialized metamodel for biomedical and health informatics domain; pp. 99–104. [Google Scholar]
- 4.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, Kohane I. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) Journal of the American Medical Informatics Association. 2010;17(2):124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohane IS, Churchill SE, Murphy SN. A translational engine at the national scale: informatics for integrating biology and the bedside. Journal of the American Medical Informatics Association. 2012;19(2):181–185. doi: 10.1136/amiajnl-2011-000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris DR, Baus AD, Harper TJ, Jarrett TD, Pollard CR, Talbert JC. Engineering in Medicine and Biology Society (EMBC), 2016 IEEE 38th Annual International Conference of the. IEEE; 2016. Using i2b2 to bootstrap rural health analytics and learning networks; pp. 2533–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris DR, Harper TJ, Henderson DW, Henry KW, Talbert JC. 2016 IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI) IEEE; 2016. Informatics-based challenges of building collaborative healthcare research and analysis networks from rural community health centers; pp. 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.International statistical classification of diseases and related health problems. Vol. 1. World Health Organization; 2004. [PubMed] [Google Scholar]
- 9.Karwin B. SQL antipatterns: avoiding the pitfalls of database programming. Pragmatic Bookshelf; 2010. [Google Scholar]
- 10.Konstantinou N, Spanos D, Mitrou N. Ontology and database mapping: a survey of current implementations and future directions. Journal of Web Engineering. 2008;7(1):001–024. [Google Scholar]
- 11.Bechhofer S. Encyclopedia of Database Systems. Springer; 2009. Owl: Web ontology language; pp. 2008–2009. [Google Scholar]
- 12.Harris DR, Henderson DW, Kavuluru R, Stromberg AJ, Johnson TR. Using common table expressions to build a scalable boolean query generator for clinical data warehouses. IEEE journal of biomedical and health informatics. 2014;18(5):1607–1613. doi: 10.1109/JBHI.2013.2292591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell JR, Campbell WS, Hickman H, Pedersen J, Mc-Clay J. AMIA Annual Symposium Proceedings. Vol. 2015. American Medical Informatics Association; 2015. Employing complex polyhierarchical ontologies and promoting interoperability of i2b2 data systems; p. 359. [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nature genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.i2b2-cttk. 2016 [Online]. Available: https://bitbucket.org/_harris/i2b2-cttk.
- 16.Butler RR. Icd-10 general equivalence mappings: Bridging the translation gap from icd-9. Journal of AHIMA. 2007;78(9):84–86. [PubMed] [Google Scholar]
- 17.Elixhauser A, Steiner C, Palmer L. Clinical classifications software (ccs). us agency for healthcare research and quality; 2012. 2013 [Google Scholar]
- 18.Weber GM, Murphy SN, McMurry AJ, MacFadden D, Nigrin DJ, Churchill S, Kohane IS. The shared health research information network (shrine): a prototype federated query tool for clinical data repositories. Journal of the American Medical Informatics Association. 2009;16(5):624–630. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]