Abstract
The vertebrate inner ear houses highly specialized sensory organs, tuned to detect and encode sound, head motion and gravity. Gene expression programs under the control of transcription factors orchestrate the formation and specialization of the non-sensory inner ear labyrinth and its sensory constituents. More recently, epigenetic factors and non-coding RNAs emerged as an additional layer of gene regulation, both in inner ear development and disease. In this review, we provide an overview on how epigenetic modifications and non-coding RNAs, in particular microRNAs (miRNAs), influence gene expression and summarize recent discoveries that highlight their critical role in the proper formation of the inner ear labyrinth and its sensory organs. In contrast to non-mammalian vertebrates, adult mammals lack the ability to regenerate inner ear mechano-sensory hair cells. Finally, we discuss recent insights into how epigenetic factors and miRNAs may facilitate, or in the case of mammals, restrict sensory hair cell regeneration.
Keywords: microRNA, DNA methylation, histone modification, inner ear development, hair cell regeneration, deafness
1. Introduction
The development of the inner ear is a dynamic process, eventually leading to a complex tissue enabling hearing and balance, represented by the auditory and vestibular systems, respectively. In the auditory system, the sensory epithelium embedded within the cochlea contains the organ of Corti responsible for hearing. The vestibular system contains five organs, including the three semicircular canals with cristae sensory epithelia that detect angular acceleration by fluid motion and the saccule and the utricle, which contain the macula sensory epithelium that detects linear acceleration due to gravity.
The inner ear sensory apparatus and its innervating sensory neurons originate from the otic placode, a thickening of surface ectoderm adjacent to the hindbrain. Otic induction and its regionalization occur in a series of steps that requires the reiterative interplay of inductive signals and the action of transcription factors [1]. Once formed, the otic vesicle grows in size and undergoes extensive morphological changes, forming the auditory and vestibular apparatus and the endolymphatic duct. The first cell lineage specified within the developing otic placode is the neuronal lineage. As early as the otic cup stage, neuroblasts begin to delaminate from a broad anteriorly located neurogenic domain and differentiate into sensory neurons of the auditory-vestibular ganglion. The auditory and vestibular hair cells, sensory receptors of the inner ear, and their surrounding supporting cells are derived from their respective vestibular and auditory pro-sensory patches within the developing otocyst [2].
Non-mammalian vertebrates such as birds, fish and amphibians have the capacity to regenerate hair cells in response to injury. In fish and amphibians, hair cells are added to the inner ear and the lateral line sensory organs throughout the lifetime of the animal as part of ongoing growth [3]. The newly generated hair cells originate from neighboring non-sensory supporting cells. Such ongoing hair cell production is dramatically up-regulated in response to injury [4, 5]. In the avian inner ear, a steady turnover of hair cells is observed within vestibular sensory organs but not within the auditory sensory organ [6, 7]. However, upon damage, auditory hair cells are readily regenerated from supporting cells through both mitotic [8, 9] and non-mitotic mechanisms [10]. The mammalian inner ear has a very limited ability to replace lost hair cells. Once matured, the auditory sensory epithelium lacks the ability to regenerate lost hair cells [11] and the vestibular sensory epithelia mount only a limited regenerative response to trauma [12, 13] (see chapter by Burns and Stone in this issue).
During early development, transcriptional pathways have been elucidated that govern the differentiation of the otocyst towards sensory or non-sensory regions [reviewed in [14]. Temporal and spatial triggers of otic development and maturation have been characterized, including the molecular controls that guide otic induction, patterning, morphogenesis and neurosensory development [15] (see chapter by Whitfield and Alsina in this issue). Knowledge about transcriptional pathways has laid the groundwork for establishing early and late developmental pathways of the inner ear. Moreover, pathogenic mutations in several transcription factors are associated with deafness [16, 17].
In this review we will discuss a new layer of gene regulation involving mechanisms of noncoding RNAs, chromatin remodeling, histone modifications, DNA methylation and research progress in these areas in the auditory and vestibular systems. It has become clear in recent years that the epigenetic layer of regulation plays an essential role in the development and maintenance of the inner ear, and has potential in regenerative capacities, with a great deal yet to be explored.
2. Gene regulation by epigenetic pathways and non-coding RNAs
2.1 Epigenetic gene regulation
Epigenetic regulation, above the DNA sequence, is controlled by a combination of dynamic nucleosome occupancy, post-translational modifications (PMTs) of histones, DNA methylation, and nuclear regulatory non-coding RNAs (Fig. 1) [18–20]. This level of regulation plays an essential role in development from a single totipotent oocyte through defined subsets of multipotent progenitor cells, eventually yielding the entire variety of cell types comprising the highly complex adult organism. Global epigenomic maps are essential to our understanding of gene regulation, or misregulation, as well as critical for controlling cell fate commitment and maintenance [21–23].
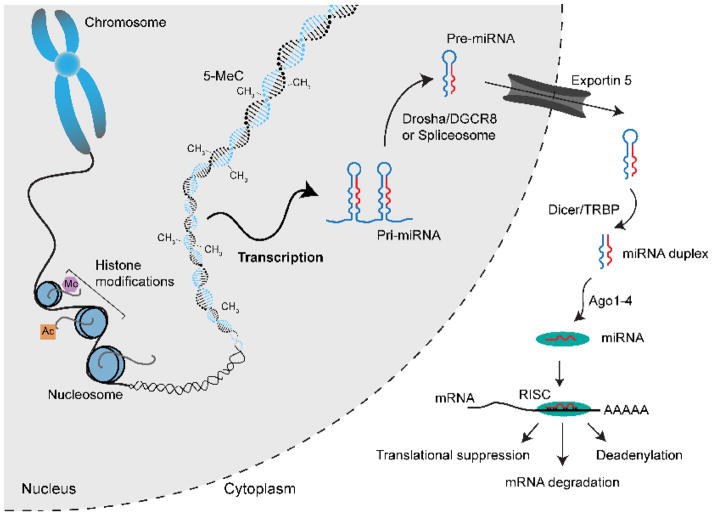
Fig. 1.
Schematic diagram of miRNA biogenesis and epigenetic mechanisms in the mammalian cell. Common histone PMTs such as histone methylation (me) and acetylation (ac) are indicated. DNA methylation is shown on the DNA double helix, as 5-MeC and CH3. Gene transcription produces a pri-miRNA that is processed by Drosha to form a pre-miRNA. Exportin-5 facilitates its export to the cytoplasm, where Dicer cleaves the pre-miRNA into short double-stranded RNA. The RISC complex is activated and its catalytic component argonaute (Ago) leads to degradation of mRNA, translational suppression and deadenylation.
Chromatin allows for DNA to be tightly packed within eukaryotic nuclei. Nucleosomes constitute the fundamental subunit of chromatin. Nucleosome cores are comprised of 145–147 base pairs (bps) of DNA, wrapped around an octamer of two copies of histone proteins H2A, H2B, H3 and H4 [24].
Open chromatin structure is an identifier of regulatory elements. Areas of chromatin with lower nucleosome occupancy have markedly increased accessibility to the nuclease DNaseI and are referred to as DNase Hypersensitive sites (DHS). Using a DNaseI hypersensitivity assay coupled with high-throughput sequencing, DNaseI-seq, DHS sites were found to be associated with most of the defined cis-regulatory elements such as promoters, insulators, silencers, enhancers and locus control region [25].
The ENCODE (The Encyclopedia Of DNA Elements) Project analysis clearly demonstrates the overlap between transcription factor binding sites (TFBS) and a less dense nucleosome structure. Furthermore, DHS and transcription factor binding appear in a developmental stage or cell lineage dependent context [26, 27]. Genome-wide DHS maps will reveal high resolution, ~150–200 bp, open regulatory segments that allow more accurate predictions of TFBS.
Histone modifications are present as enrichment patterns at promoters and enhancers. The core histones comprising the nucleosome undergo various modifications at numerous positions across their N-terminal tails that project out of the mostly globular octamer. Common PTMs of histone proteins include acetylation, methylation, phosphorylation and ubiquitination. The modification pattern of histones is highly dynamic and is regulated by the opposing action of enzyme families. For instance, histone acetyltransferases catalyze the transfer of an acetyl group to the ε-amino group of lysine side chains, whereas histone deacetylases (HDACs) remove the acetyl group [28].
The overall chromatin architecture is directly linked to the composition of histone PMTs at a certain genomic location [29]. Different chemical modifications imparted on the histones have different effects over the degree of association between the negatively charged DNA and the nucleosome. They can tighten the DNA loop around each nucleosome and/or compress nucleosomes together, or it can loosen each nucleosome’s hold upon the DNA and separate nucleosomes from each other. In addition, histone modifications serve as docking sites for regulatory proteins and chromatin remodelers that may alter the placement or position of nucleosomes in an ATP-dependent manner [30]. As a result, histone PTM compositions can be analyzed for its effect on gene expression resulting from its underlying influence on chromatin architecture.
Histone PTMs allow exploration of the basic epigenetic landscape dynamics. For example, histone 3 lysine 4 tri-methylation (H3K4me3) is positively correlated to the transcription start sites of actively transcribed genes [31]. Histone 3 lysine 27 tri-methylation (H3K27me3) is a repressive modification that spans over inactive genes; however, it is typically restricted to repressed key lineage developmental genes, not all inactive genes. Co-localization of H3K4me3 and H3K27me3 is known to mark genes found in a poised expression state, mainly in pluripotent stem cells or multipotent progenitor cells. This bivalent state is likely enriched in cell populations earlier in development to confer the ability to respond to signaling cues and readily activate gene expression (by loss of H3K27me3) or long-term repression (by loss of H3K4me3) as cells commit to their terminal fates [32, 33]. ChIP-Seq combines chromatin immunoprecipitation (ChIP) and high-throughput sequencing [34] and is used to establish genome-wide histone modification profiles [35].
DNA methylation is a key regulator of promoter and enhancer activity. DNA methylation, carried out by DNA methyltransferases (DNMTs) involves the covalent addition of a methyl group at the 5′ carbon of the cytosine ring, resulting in 5-methylcytosine [36]. In general, DNA methylation of promoters and enhancers negatively correlates to their activity and the expression of genes [37] through the disruption of transcription factor binding. For example, there is a gain of DNA methylation in pluripotency-related enhancers and gene promoters as differentiation progresses, and differential methylation in lineage specific enhancers between various lineages of blood and skin stem cells [38]. Altered patterns of promoter and enhancer DNA methylation are frequent in tumorigenic and aging cells. Connexin 26 is the most prevalent gene associated with profound congenital deafness [39]. Aberrant methylation in its promoter region has been found in breast [40] and lung cancer [41]. Aging rats have hypermethylation of the connexin 26 promotor region associated with a reduction in connexin 26 expression [42]. Recent advances and manipulations of next-generation sequencing platforms have presented a great advance in mapping the cell’s methylome in a genome-wide fashion, in particular MethylCseq [43]. This method provides genome-wide, nucleotide resolution of DNA methylation.
2.2 Gene regulation by non-coding RNAs
The central dogma of molecular biology that genetic information begins with DNA and ends with protein was challenged with the discovery of the wide range of non-protein coding RNA transcription and RNA-mediated genetic and epigenetic events in higher organisms [44]. The vast majority of the genome, approximately 97%, is non-coding. While less is known about all the regulatory and other elements of this larger portion, a significant role is attributed to RNA. Non-coding genome contains multiple forms of RNAs, including long non-coding (lncRNA) [45], piwi-interacting RNAs (piRNAs), circular RNAs (circRNAs), small nucleolar RNAs (sno-RNAs) and microRNAs (miRNAs). Small or short non-coding RNA (ncRNA), which are under 200 nucleotides (nt) in length, include miRNAs (Fig. 1). The primary miRNA (pri-miRNA) transcripts are processed in the nucleus by a microprocessor complex that includes Drosha, an RNase III-like enzyme, and DGCR8 [46, 47]. A precursor miRNA (pre-miRNA) hairpin product 70 nt long is generated, which is exported by the receptor Exportin 5 to the cytoplasm. There, a second RNase III enzyme, Dicer, converts the RNA into mature miRNA duplexes. These duplexes are separated to produce a mature 22-nt miRNA, which is incorporated into the RNA-induced silencing complex (RISC). Finally, the complex targets mRNAs in order to repress them by degradation or translational repression.
In mammals, a single miRNA can bind any of hundreds of mRNA targets by complementation of the seed region of the miRNA to the target’s 3′ untranslated region (UTR) binding site. This leads to a reduction in gene expression by translational suppression and mRNA destabilization [48]. The second major class of ncRNA, greater than 200 nt, is lncRNA [45]. They can either be cis-acting and function in local gene silencing (e.g., Xist RNA, a key regulator of X chromosome inactivation [49]); or trans-acting and target multiple genes throughout the genome, such as lncRNA-p21 [45]. lncRNAs mechanisms of action are likely unique and can be difficult to identify, as they do not have structural characteristics and their biogenesis and processing may differ.
In the inner ear field, the history of research in non-coding RNAs goes back to 2005, when the triad miRNA-96, 182 and 183 were detected in zebrafish hair cells of sensory epithelia by in situ hybridization [50]. Microarray analysis of mouse inner ears revealed expression of miRNAs and in particular, miRNA-96, 182 and 183 in mammalian hair cells [51]. Since then profiling using microarray and next generation sequencing platforms uncovered hundreds of miRNAs in the auditory and vestibular portions of the inner ear, including 74 differentially expressed miRNAs [52, 53]. MiRNAs specific to age-related hearing loss were identified in a microarray screen, and differential regulation of specific miRNAs led to the suggestion that pro-apoptotic miRNAs and those promoting proliferation and differentiation are both involved in age-related degeneration of the organ of Corti [54].
Much less is known about lncRNAs in the developing and mature inner ear. The Meg3 RNA has been proposed to function as a lncRNA and based on expression studies, to play a role in pattern specification, differentiation and maintenance of inner ear cells [55]. Rubie is a lncRNA positioned upstream of the Bmp4 gene, and its disruption by an intronic endogenous retrovirus is predicted to be the cause of the Epistatic circler (Ecl) mouse vestibular phenotype [56].
3. Role of epigenetic factors and miRNAs in inner ear development
3.1 Epigenetics and otic induction, patterning and morphogenesis
Cranial placodes, including the otic placode, arise from a common precursor field, the preplacodal region, adjacent to the neural plate. Mutual repression between the homeodomain transcription factors OTX2 and GBX2 segregate placode progenitors of different fate, with GBX2 being required for otic placode specification [57]. A recent study identified DNMT3A, an enzyme required for de-novo DNA methylation as an essential upstream regulator of otic GBX2 expression. De-novo DNA methylation, which creates new patterns of DNA methylation, occurs predominantly during early development and gametogenesis [58]. In the early chick embryo, Dnmt3a is expressed in the pre-placodal region and later in the otic placode itself. Knockdown of Dnmt3a selectively reduces the expression of early otic marker and specifier genes Gbx2 and Pax2, followed by a loss of late otic marker genes and a severe reduction in otic vesicle size [59]. How DNMT3A regulates Gbx2 expression has yet to be established. DNA methylation of cis-regulatory elements undergoes dynamic changes during development and cell differentiation [60, 61]. The authors propose a model by which DNMT3A activates Gbx2 gene expression by selectively methylating a repressor binding site in the Gbx2 promotor.
Following otic placode formation, the otic placode invaginates to form the otic cup, which pinches closed dorsally to form the otic vesicle (otocyst) [62]. A study by Uribe and colleagues revealed a critical role for histone demethylase KDM4B (alternative name JMJD2B) in otic invagination [63]. KDM4B is a histone demethylase that preferentially catalyzes demethylation of tri and di-methylated lysine residue 9 on histone 3 (H3K9me2/3). H3K9me3 and H3K9me2 are considered repressive epigenetic marks and are correlated with transcriptional silencing of a gene locus [64]. In the developing chick embryo KDM4B is expressed in pre-otic ectoderm and later becomes restricted to the border of the otocyst. In the absence of KDM4B, cytoskeletal rearrangements and cell polarization, required for otic invagination, fail to occur. The authors provide compelling evidence that the observed defects are due to a failure of Dlx3 induction. Dlx3, a member of the vertebrate Distal-less related (Dlx) homeodomain genes, is initially expressed throughout the otic placode and during otic invagination becomes restricted to the rim of the invaginating otic pit [65]. The authors show that KDM4B directly binds to the Dlx3 regulatory region and is required for demethylation of H3K9me3 at the promoter of Dlx3. Knock down of Dlx3 causes similar otic invagination defects, and forced expression of Dlx3 is sufficient to rescue otic defects due to the loss of KDM4B [63].
Otic morphogenesis requires a highly coordinated and complex pattern of growth. The histone deacetylase HDAC1 has recently emerged as an important regulator of cell proliferation and cell survival. In mice, targeted deletion of Hdac1 dramatically impairs proliferation and is associated with elevated levels of the cyclin-dependent kinase (CDK) inhibitor expression [66]. In zebrafish, HDAC1 is highly expressed in the developing otocyst and posterior lateral line primordium. Hdac1 knockdown results in a smaller otic vesicle, fused otoliths, malformed or absent semicircular canals, and fewer sensory hair cells [67]. These defects can at least in part be attributed to defects in otic cell proliferation and cell survival, which in Hdac1 morphants is severely reduced. In addition, knockdown of Hdac1 reduced otic expression of fibroblast growth factor (FGF) ligands FGF3 and 8 [67]. FGF3 and FGF8 play a critical role in otic patterning and hair cell fate specification in zebrafish [68, 69], and disrupted FGF signaling likely contributes to the observed inner ear defects in Hdac1 morphants. Whether HDAC1 plays a similar role in murine inner ear morphogenesis has yet to be established.
Haploinsufficiency in Chd7, a gene that encodes for a chromatin modifier protein, causes CHARGE Syndrome, a multiple congenital anomaly disorder, which among other effects is characterized by inner ear defects [70]. CHD7 is a member of the chromodomain helicase DNA-binding (CHD) family of ATP-dependent chromatin remodeling enzymes. ATP-dependent chromatin remodeling complexes facilitated by ATP hydrolysis shift, exchange or remove adjacent nucleosomes, enabling binding of transcription factors to DNA (reviewed in [71]). In both humans and mice, Chd7 haplo-insufficiency results in severe semicircular canal malformation, and accompanying vestibular defects [72–75]. These defects are in part due to an abnormal pattern of cell proliferation. In addition, analysis of heterozygous Chd7 mutant and conditional Chd7 knockout mice revealed that CHD7 regulates the expression of vestibular regulatory genes in a gene dosage dependent manner [76]. Interestingly, inhibition of retinoic acid (RA) signaling can rescue semicircular canal defects in heterozygous Chd7 mutant mice, suggesting that hyperactive RA signaling contributes to inner ear malformations in Chd7 mutants [77]. The exact mechanism through which CHD7 regulates vestibular–specific genes has yet to be determined. CHD7 occupancy is cell-type specific, and most CHD7 sites show features of gene enhancer elements. It has been proposed that CHD7 provides access to transcriptional activators and co-factors by locally remodeling chromatin structure [78, 79]. In neuronal stem cells CHD7 directly interacts with HMG-box transcription factor SOX2 and cooperatively regulates the expression of key developmental regulators including the Notch ligand Jagged1 (JAG1) [80]. JAG1 is an essential regulator of inner ear neuro-sensory development and is required for semicircular canal formation [81]. Jag1 expression is severely reduced in E10.5 heterozygous Chd7 mutant otocysts, suggesting that Jag1 deficiency may in part contribute to the vestibular defects observed in Cdh7 mutants [80]. Future studies are warranted to address whether SOX2 and CHD7 directly interact in otic progenitor cells and cooperate in regulating otic gene expression.
3.2 MiRNAs in otic patterning and morphogenesis
Most of our understanding of how miRNAs might control inner ear development stem from studies of Dicer1 mutant animals. Dicer1 function is essential for the processing of pre-miRNAs into functional mature miRNAs. In addition, Dicer1 is required for processing of double stranded RNA into small-interfering RNAs (siRNAs) [82].
In zebrafish, Dicer1 mutant larvae have a severely malformed inner ear and lack otoconia [83]. Similarly, in mice, early otic deletion of the Dicer1 gene (Pax2-Cre Dicer1flox/flox or Foxg1Cre/+ Dicer1flox/flox) results in gross inner ear malformations. The overall size of the inner ear is reduced, cochlear outgrowth is severely stunted and the development of semicircular canals is arrested at the canal plate stage [84, 85]. FGF10 dependent signaling plays a critical role in otic morphogenesis. Loss of FGF ligand FGF10 leads to severe inner ear malformations including partial failure of semicircular canal formation and cochlear outgrowth defects [86, 87]. Fgf10 expression is severely diminished in Dicer1 mutants, suggesting that partial loss FGF10 function contributes to Dicer1 mutant phenotype [84]. The contribution of individual miRNAs to inner ear morphogenesis has yet to be examined.
A potential key regulator of inner ear morphogenesis is miR-200b, which is selectively expressed in cochlear and vestibular epithelial cells [88]. MiR-200b belongs to the miR-200 family, which is comprised of miR- 200a, miR- 200b, miR- 200c, miR- 141 and miR- 429 [89]. Epithelial-to-mesenchymal transition (EMT) and its reversal (MET) are critical for tissue remodeling during development (reviewed in [90]). Recent studies revealed that EMT and MET are controlled by a double negative feedback loop between members of miR-200 family and the transcription factors ZEB1 and ZEB2. miR-200 family members enforce an epithelial fate and oppose EMT by transcriptionally silencing Zeb1 and Zeb2 genes [91, 92]. The pro-mesenchymal fate transcription factors ZEB1 and ZEB2 in turn repress the transcription of epithelial genes, including miR-200 genes [93]. In the developing inner ear, epithelial remodeling is required for the formation of semicircular canals [94]. Twirler mice, which have a noncoding point mutation in the first intron of the Zeb1 gene, have severe vestibular and auditory defects [95]. Transcriptome analysis revealed that in Twirler mice, ZEB1 target genes are de-repressed, suggesting that ZEB1 function is compromised in Twirler mice and that the resulting disruption of epithelial and mesenchymal cell identities underlies the inner ear malformations observed in the Twirler mice [88]. It has yet to be determined whether and to what extend miR-200b expression is de-regulated in Twirler mice.
3.3 Epigenetic factors in neurogenesis and hair cell formation
Central to otic neurogenesis are the basic helix-loop-helix (bHLH) transcription factors Neurogenin1 (NEUROG1) and NEUROD1 [96]. NEUROG1 is required for neuronal fate specification [97, 98], whereas NEUROD1, which functions downstream of NEUROG1 is required for the progression of the otic neurogenic program [99].
A recent study found that subunits of SWI/SNF-like chromatin remodeling complex cooperate with EYA1/SIX1 in the transcriptional activation of Neurog1 and Neurod1 in the developing inner ear [100]. Haplo-insufficiency for EYA1 leads to branchio-oto-renal syndrome, which is characterized by combinations of craniofacial defects and hearing loss with or without kidney anomalies [101, 102]. Animal models revealed that the transcriptional cofactor EYA1 and the homeobox transcription factor SIX1 are required for formation of the cochlea and vestibulocochlear ganglion [103, 104]. EYA1 and SIX1 are members of Eyes absent (Eya) and Sine oculis (So/Six) gene families. EYA proteins have a potent transactivation domain but lack a DNA binding domain and require the presence of transcription factors to act as potent transcriptional activator (reviewed in [105]). Employing gain and loss-of-function strategies, the authors uncovered that EYA1/SIX1 co-activator complex controls otic neural induction and neural differentiation by positively regulating the expression of Neurog1 and Neurod1. Interestingly, the neural inductive activity of EYA1/SIX1 depended on the presence and enzymatic function of core components of SWI/SNF like BAF complex. Mammalian SWI/SNFlike BAF complexes are ATP-dependent chromatin-remodeling complexes that are made up of a core ATPase subunit BRG1 (official name SMARCA4) or BRM (SMARCA2), and BRG1 Associated Factors (BAFs) (reviewed in [106]). The authors found that EYA1 and SIX1 directly interact with SWI/SNF subunits BRG1 and BAF170, and BRG1 ATPase activity is required for normal otic neurogenesis as well as ectopic neurogenesis induced by Eya1/Six1 overexpression, indicating a central role for SWI/SNF-like chromatin remodeling complex in otic neurogenesis [100]. A parallel pathway of Neurog1 regulation involves the ATP-dependent chromatin-remodeler CHD7. In Chd7-deficient otic tissue the cochlear vestibule ganglion is missing or severely reduced. TBX1, a T-box transcription factor selectively expressed in the non-neurogenic domain of the developing otocyst, is known to limit the extent of the neurogenic domain within the otic vesicle [107]. Analysis of Chd7-deficient otic tissue revealed that the expression domain of Tbx1 is expanded into the neurosensory domain, and Neurog1 expression is initially induced but fails to be maintained [75]. Future studies are needed to clarify whether CHD7 occupies Tbx1 and Neurog1 cis regulatory sequences.
The bHLH transcription factor ATOH1 plays a central role in mechano-sensory hair cell development. ATOH1 is both necessary and sufficient for hair cell formation and differentiation [108–110]. ATOH1 expressing hair cells and their surrounding supporting cells derive from sensory progenitor cells commonly referred to as pro-sensory cells. Pro-sensory cells and later differentiating supporting cells are inhibited from inducing Atoh1 expression through a lateral inhibition mechanism mediated by the Notch signaling pathway [111–113]. A recent study by Stojanova and colleagues examined whether epigenetic mechanisms control Atoh1 expression in the developing murine cochlea. Developmentally regulated genes often exhibit both repressive (H3K27me3) and active (H3K4me3) epigenetic marks within their promoter and enhancer sequences, referred to as bivalent state [33]. The authors found that these bivalent epigenetic marks are frequent within the Atoh1 locus of pro-sensory cells and perinatal supporting cells, indicating that Atoh1 is poised for expression in these cell types. In differentiating hair cells, these bivalent marks were infrequent and H3K9 acetylation, a mark, associated with actively transcribed genes, was prevalent [114]. Inhibition of histone acetyltransferase activity reduced H3K9 acetylation at the Atoh1 locus and prevented Atoh1 induction, suggesting a central role for histone acetyltransferases in Atoh1 induction. In turn, shut down of Atoh1 expression in maturating hair cells required H3K9 deacetylation and was accompanied by an increased frequency of H3K9 tri-methylation (H3K9me3) [115], a mark associated with gene silencing [116].
3.4 MiRNAs in neurogenesis and hair cell formation
Early otic Dicer1 conditional knockouts (Foxg1Cre/+ Dicer1flox/flox or Pax2-Cre Dicer1flox/flox) have, in addition to defects in otic morphology, severe neuro-sensory defects. In these mutants, both vestibular and auditory sensory epithelia are severely reduced in size, innervating neurons are largely missing and the organization and apical specialization of remaining hair cells is severely disrupted [84, 117] [84, 85]. Innervating sensory neurons depend on trophic support from hair cells and supporting cells. Defects in neuronal survival may be a consequence of the stunted sensory development in early otic Dicer1 conditional knockouts; alternatively it may be due to defects in neuronal differentiation as previously reported for Dicer1 mutant cortical neurons [118]. Early otic Dicer1 conditional knockouts were not viable so hearing could not be tested. The targeted removal of functional mature miRNAs from hair cells using Pou4f3-Cre mice crossed with Dicer1flox/flox led to a mouse that survived until approximately one month of age, enabling hearing to be tested [52]. These mice were profoundly deaf, as evaluated by auditory brainstem response. The Dicer1 mutant hair cells began degenerating soon after birth, with surviving cells having an abnormal morphology, suggesting a critical role for miRNAs in hair cell differentiation/ maturation.
Re-examination of the cochlear phenotype of early otic Dicer1 mouse mutants by Huyghe and colleagues revealed previously undetected defects in pro-sensory cell proliferation and hair cell fate specification. Dicer1 deficient pro-sensory cells were delayed in their cell cycle withdrawal and showed a bias to differentiate as hair cells. In addition, defects in planar cell polarity (PCP) as well as defects in formation or maintenance of kinocilia were noted. These defects are likely due to de-regulation of Wnt signaling. Among the genes de-repressed in the absence of Dicer1 were secreted frizzled-related protein (Sfrp)4 and Sfrp5, which encode for known modulators of both canonical and non-canonical Wnt signaling [119]. Exposure of cochlear tissue to SFRP4 and SFRP5 protein caused cell fate specification and PCP defects that largely resembled the defects observed in Dicer1 mutants. Sfrp4 and Sfrp5 expression in the developing cochlea is likely repressed by miR-124. The authors found that miR-124 is selectively expressed in the differentiating auditory sensory epithelium, miR-124 negatively regulates Sfrp4 and Sfrp5 expression, and similar to Sfrp4/5 overexpression, miR-124 inhibition biases pro-sensory cells towards a hair cell fate [117].
The failure of Dicer1 deficient auditory pro-sensory cells to exit the cell cycle at the appropriate time may, at least in part, be due to the loss of let-7 miRNA function. A recent study by Golden and colleagues revealed that a double negative feedback loop between let-7 miRNAs and the RNA binding protein (RBP) LIN28B times and coordinates cell cycle exit and differentiation of pro-sensory cells in the murine cochlea. Let-7 (lethal-7), originally identified as a regulator of developmental timing (heterochrony) in C. elegans [120], was among the first miRNAs to be discovered [121, 122]. In human and mice, members of the let-7 family (let-7a, b, c, d, e, f, g, i, mir-98) function as tumor suppressors and are highly associated with a terminal differentiated state. Let-7 miRNAs target and transcriptionally silence pro-growth genes including Lin28a and Lin28b. In turn, LIN28 RBPs bind let-7 primary and precursor transcript and interfere with their processing by Drosha and Dicer1 (reviewed in [123]). In the developing inner ear, Lin28b is selectively expressed in pro-sensory cells, whereas let-7 miRNAs are expressed in terminally differentiated cells including hair cells and supporting cells [124]. Using inducible LIN28B and let-7g transgenic mouse lines the authors found that higher than normal levels of let-7 miRNAs resulted in premature pro-sensory cell cycle exit, whereas lower than normal let-7 levels, due to LIN28B overexpression delayed pro-sensory cell cycle exit [124]. LIN28B overexpression led to an increase in cochlear N-Myc (Mycn) and cyclin D1 (Ccnd1) expression. N-Myc and cyclin D1 are known let-7 target genes [125, 126] and loss of either gene has been shown to disrupt cell proliferation in the developing inner ear [127, 128], suggesting that the effects of LIN28B and let-7 on proliferation are at least in part due to deregulation of N-Myc and cyclin D1 expression. Over-expression of let-7 miRNAs accelerates development and induces precocious differentiation in many tissues including the retina [129]. However, even though lower than normal let-7 levels due to LIN28B overexpression delayed auditory hair cell differentiation, let-7g overexpression failed to induce precocious differentiation [124]. These findings suggest that in the developing cochlea, let-7 mainly functions as an inhibitor of progenitor-specific gene expression.
Sensory cells including hair cells express three homologous miRNAs, miR-96, miR-182, and miR-183 [51]. These evolutionary conserved miRNAs are expressed from a single genetic locus and are commonly referred to as miR-183 cluster or family [130, 131]. Overexpression of miR-183 family members in sensory progenitors biases them towards a hair cell fate. In zebrafish overexpression of miR-96 or miR-182 results in ectopic sensory patches and increases the number of hair cells within the inner ear vestibular sensory organs, whereas knockdown of miR-96, -182, and/or-183 generates fewer hair cells [132]. A similar bias towards a hair cell fate was observed when miR-183 family members were overexpressed in the developing chicken auditory organ. However, in contrast to zebrafish, overexpression of miR-183 cluster members failed to induce ectopic sensory patches [133]. Mutations in the seed region of miR-96 are associated with hearing loss in humans and mice [134, 135]. The mouse mutant diminuendo (Dmdo) has a single base change in the seed region of the miR-96 gene [135]. In homozygous and heterozygous Dmdo mice, hair cells form relatively normal but have misshapen stereocilia and their biophysical maturation and specialization is arrested [135, 136]. At one month of age, Dmdo homozygous mice have virtually no hair cells and Dmdo heterozygous mice show severe outer hair cell loss [135]. The observed defects could be due loss of miR-96 dependent target regulation and/or gain of novel targets. Qualitative similar hair cell defects are observed in hair cell-specific Dicer1 mutants, suggesting the former to be the case.
4. Non-coding RNAs and epigenetics in human sensory disease
4.1 miRNAs in deafness
Mutations in miRNAs in humans have been found in three families [134, 137]. Of the miR-183, miR-182, and miR-96 triad that is specifically expressed in inner and outer hair cells, only miR-96 is associated with human disease. While attempts have been made to discover if there is a larger number of miRNAs contributing directly to human deafness, this is unlikely to be the case [138].
4.2 Epigenetic factors associated with deafness
Mutations in epigenetic factors recently emerged as potential causes for deafness in humans. Germline mutations in histone methyltransferases have been identified in human syndromes associated with sensorineural hearing loss. WHSC1 (Wolf-Hirschhorn Syndrome candidate 1, also known as NSD2 and MMSET) is a histone methyltransferase that preferentially catalyzes H3K36 methylation, an epigenetic modification highly associated with transcriptional activation [139]. Haploinsufficiency in Whsc1 is linked to Wolf-Hirschhorn syndrome (WHS), a complex genetic disorder that causes severe growth retardation, craniofacial defects, seizures, and in some cases hearing impairment [140]. Whsc1 mutant mice are developmentally delayed and manifest similar craniofacial defects and external ear and eye anomalies seen in humans with WHS [141, 142]. The authors found that cochlear hair cells form in Whsc1 heterozygous and homozygous mutants, but their shape, arrangement, morphological specialization and innervation are disrupted, suggesting that WHSC1 dysfunction, might contribute to sensorineural hearing loss in individuals with WHS [142]. How WHSC1 controls cochlear hair cell arrangement, specialization and innervation are currently unknown, but components of the Wnt/ PCP machinery are among the likely targets of WHSC1 mediated regulation.
5. Role of epigenetic factors and miRNAs in hair cell regeneration
5.1 Supporting cell proliferation in birds and fish
Histone modifying enzymes and miRNAs recently emerged as critical regulators of supporting cell proliferation in fish and birds.
In the mature vestibular epithelia of birds, supporting cells divide at a low rate and continually and spontaneously replace lost hair cells. Treatment of hair cell-damaged chicken utricle with broad spectrum HDAC inhibitors or class I selective HDAC inhibitors resulted in a decrease in supporting cell proliferation, but did not affect the differentiation of replacement hair cells [143]. In developing zebrafish, class I HDACs, HDAC1 and HDAC3 are selectively expressed in lateral line primordium and neuromast [144, 145]. Knockdown of HDAC3 or broad inhibition of HDAC activity severely impaired cell proliferation within the lateral line primordium [144] [146]. Similarly, in hair cell-damaged zebrafish larvae, inhibition of HDAC activity with broad spectrum HDAC inhibitors reduced supporting cell proliferation and subsequent hair cell regeneration. In mice, CDK inhibitors p27/Kip1 (CDKN1B) and p21/Cip1 (CDKN1A) are required for maintaining auditory supporting cells and hair cells in a postmitotic state [147–149]. The authors found that in the hair cell-damaged zebrafish larvae HDAC inhibition increased p21/Cip1 and p27/Kip1 expression, suggesting that HDACs facilitate supporting cell proliferation through restricting p27/Kip1 and p21/Cip1 expression [145].
Lysine-specific demethylase 1 (LSD1, official name KDM1A) is a FAD-dependent enzyme that catalyzes the demethylation of mono- and di-methylated lysine 4 and 9 on histone 3 (H3K4me1/2 and H3K9me1/2) repressing and activating transcription, respectively (reviewed in [150]). Two recent studies revealed an important role for LSD1 in regulating sensory progenitor cell proliferation. In the developing zebrafish larvae, pharmacological inhibition of LSD1 using trans-2-phenylcyclopropylamine (2-PCPA) disrupted cell proliferation and induced apoptosis in neuromast cells, causing a reduction in the numbers of sensory hair cells and supporting cells produced [67]. Similarly, in the hair cell-damaged zebrafish, 2-PCPA treatment significantly decreased supporting cell proliferation and subsequent production of hair cells. Based on gene expression data the authors propose that LSD1 promotes cell proliferation by inhibiting the expression of p21/Cip1 and p27/kip1 as well as by promoting the expression Wnt/β-cateninsignaling components [151].
Supporting cells within the auditory sensory epithelia (basilar papilla) of birds are quiescent, but re-enter the cell cycle within hours following hair cell damage [152]. Enrichment analysis of gene sets differentially expressed in the quiescent and proliferating basilar papilla uncovered miR-181a as a potential upstream regulator of supporting cell proliferation. The authors found that in the intact chicken basilar papilla miR-181a overexpression was sufficient to stimulate supporting cell proliferation and the production of new hair cells [153]. In a follow up study the authors found that knock-down of endogenous miR-181a in basilar papilla inhibited the regenerative proliferation of supporting cells in response to hair cell damage [154]. miR-181a has been shown to interfere with p27/Kip1 transcription and translation in tumor cell lines [155, 156], suggesting that miR-181a might trigger cell cycle re-entry of supporting cells in a p27/Kip1 dependent manner. Whether miR-181a is capable of stimulating supporting cell proliferation in mammalian species is currently unknown.
5.2 Targets for regenerative strategies in mammals
Prior to the onset of hearing, mammalian supporting cells have a latent ability to spontaneously regenerate hair cells. Isolated auditory supporting cells placed in culture re-enter the cell cycle and generate hair cells through both mitotic and non-mitotic mechanisms [157, 158]. In response to hair cell damage, auditory supporting cells spontaneously regenerate hair cells in vivo [159, 160]. The limited regenerative response of immature auditory supporting cells can be further enhanced by either inhibiting the Notch signaling pathway [161, 162], or by over-activating Wnt/ β-catenin signaling [163–165].
The polycomb group protein BMI1 is required to sustain Wnt/β-catenin signaling in neonatal supporting cells. BMI1 is part of the multi-protein polycomb repressive complex 1 (PRC1), which is needed for ubiquitination of histone H2A lysine 119. This epigenetic mark is associated with gene repression (reviewed in [166]). In cochlear explants, obtained from neonatal Bmi1 mutant mice, supporting cells failed to re-enter the cell cycle in response to hair cell damage and BMI1 deficiency reduced the sphere-forming ability of unfractionated cochlear epithelial cells and Wnt-responsive supporting cells. The authors found that in the Bmi1-deficient cochlear tissue, Wnt antagonist DKK1 was increased and Wnt/β-catenin signaling was attenuated. Over-activation of Wnt signaling using GSK-3β inhibitor partially reversed the proliferation deficit of Bmi1 mutant supporting cells, suggesting that BMI1 facilitates supporting cell proliferation by sustaining high levels of canonical Wnt signaling [167].
DNA methylation may restrict hair cell lineage potential. DNMT1 activity is required to maintain patterns of DNA methylation in daughter cells after DNA replication [168]. The nucleotide analog 5-azacytidine (5-aza) incorporates into DNA and interferes with DNMT function, leading to a decrease in global DNA methylation [169]. A recent study used a prosensory-like cell line (MUCS), initially derived from adult mouse utricle, to determine the role of DNA methylation in hair cell fate induction and differentiation. MUCS cells have mesenchymal characteristics and the authors found that treatment of MUCS cells with 5-aza increased expression of epithelial genes and hair cell genes on both transcript and protein levels, which facilitated the formation of hair cell-like cells [170].
The newt, a urodele amphibian, possesses a remarkable ability for tissue and cell regeneration independently of its age. Regeneration in adult newts is mediated primarily by dedifferentiation and re-differentiation (trans-differentiation) of terminally differentiated cells [171]. Gene expression profiling of newt inner ear and lens tissue revealed that the early phase of lens and hair cell regeneration is accompanied by a sharp drop in let-7 miRNA expression, suggesting that high let-7 miRNA levels may be refractory to trans-differentiation of terminal differentiated cells [172]. A recent study indicates that indeed, high let-7 miRNA levels impair hair cell regeneration in the early postnatal murine cochlea. In early postnatal cochlear explants, pharmacological inhibition of Notch signaling allows for the production of new hair cells through supporting cell-to-hair cell conversion (trans-differentiation) [173]. Using a similar experimental approach, the authors found that let-7g overexpression significantly decreased the production of new hair cells in response to Notch inhibition, whereas overexpression of let-7 antagonist LIN28B enhanced hair cell production in response to Notch inhibition [124]. Expression of mature let-7 miRNAs rise within the auditory sensory epithelium during postnatal maturation, and it is intriguing to speculate that such rise in let-7 miRNA expression may be functionally linked to the decline in regenerative capacity of supporting cells.
Another miRNA recently implicated in auditory hair cell regeneration is miR-210. miR-210, an evolutionary conserved miRNA, is critical for the cells response to hypoxia, with roles in mitochondrial metabolism, angiogenesis, DNA repair, and cell survival [174]. miR-210 was identified in a screen for miRNAs that are differentially expressed during hair cell differentiation using the inner ear cell line (UB/OC-1). MiR-210 knockdown in UB/OC-1 cells induces premature hair cell marker expression, suggesting that miR-210 functions in maintenance of a progenitor state. Overexpression of miR-210 in early postnatal cochlear explants resulted in spontaneous formation of new hair cells, which based on Cre-loxP mediated lineage tracing, originated from supporting cells. A subset of in silico predicted miR-210 targets were experimentally validated. However, none of the validated miR-210 targets have yet been associated with sensory cell development [175]. Futures studies are needed to resolved how miR-210 facilitates supporting cell-mediated hair cell regeneration.
6. Conclusions
The past decade has seen a remarkable increase in our understanding of non-coding RNAs and chromatin modifying enzymes that has changed how we view gene expression and regulation in every organism and tissue studied. Advances in genomic technologies have enabled a comprehensive look at the key mechanisms of epigenetics. Investigations of the auditory and vestibular systems have lagged behind partly due to the lack of a robust cell culture system for the inner ear, the prior need for a large number of cells for genomic experiments, and the requirement for a model organisms rather than direct examination of the human inner ear. Despite these hindrances, we predict that the next decade will lead to dramatic discoveries, following the progress made to date and summarized in this review. Most promising, epigenomic dynamics uncovered during development or hair cell regeneration may help identify key regulators, which will in turn aid in prioritizing genes for future studies.
Acknowledgments
We would like to thank Kathy Ushakov for creating the figure. This work was supported by the National Institutes of Health/NIDCD R01DC011835 (KBA), R01 DC011571 (AD), the I-CORE Gene Regulation in Complex Human Disease Center No. 41/11 (KBA), the United States-Israel Binational Science Foundation (BSF) 2013027 (KBA) and the David M. Rubinstein Fund for Hearing Research (AD).
Abbreviations
- bps
base pairs
- DHS
DNase Hypersensitive sites
- TFBS
transcription factor binding sites
- PTMs
post-translational modifications
- HDACs
histone deacetylases
- H3K4me3
histone 3 lysine 4 tri-methylation
- H3K27me3
histone 3 lysine 27 tri-methylation
- ChIP
chromatin immunoprecipitation
- DNMTs
DNA methyltransferases
- lncRNA
long non-coding RNA
- piRNA
piwi-interacting RNA
- circRNA
circular RNA
- sno-RNA
small nucleolar RNA
- miRNA
microRNAs
- pri
primary
- pre
precursor
- ncRNA
non-coding RNA
- nt
nucleotides
- RISC
RNA-induced silencing complex
- UTR
untranslated region
- CDK
cyclin-dependent kinase
- FGF
fibroblast growth factor
- CHD
chromodomain helicase DNA-binding
- RA
retinoic acid
- siRNAs
small-interfering RNAs
- EMT
epithelial-to-mesenchymal transition
- MET
mesenchymal-to-epithelial transition
- bHLH
basic helix-loop-helix
- H3K9me3
H3K9 trimethylation
- PCP
planar cell polarity
- Sfrp
secreted frizzled-related protein
- RBP
RNA binding protein
- WHS
Wolf-Hirschhorn syndrome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Singh S, Groves AK. The molecular basis of craniofacial placode development. Wiley Interdiscip Rev Dev Biol. 2016;5:363–376. doi: 10.1002/wdev.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fekete DM, Wu DK. Revisiting cell fate specification in the inner ear. Curr Opin Neurobiol. 2002;12:35–42. doi: 10.1016/s0959-4388(02)00287-8. [DOI] [PubMed] [Google Scholar]
- 3.Corwin JT. Postembryonic production and aging in inner ear hair cells in sharks. J Comp Neurol. 1981;201:541–553. doi: 10.1002/cne.902010406. [DOI] [PubMed] [Google Scholar]
- 4.Balak KJ, Corwin JT, Jones JE. Regenerated hair cells can originate from supporting cell progeny: evidence from phototoxicity and laser ablation experiments in the lateral line system. J Neurosci. 1990;10:2502–2512. doi: 10.1523/JNEUROSCI.10-08-02502.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones JE, Corwin JT. Regeneration of sensory cells after laser ablation in the lateral line system: hair cell lineage and macrophage behavior revealed by time-lapse video microscopy. J Neurosci. 1996;16:649–662. doi: 10.1523/JNEUROSCI.16-02-00649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jorgensen JM, Mathiesen C. The avian inner ear. Continuous production of hair cells in vestibular sensory organs, but not in the auditory papilla. Naturwissenschaften. 1988;75:319–320. doi: 10.1007/BF00367330. [DOI] [PubMed] [Google Scholar]
- 7.Roberson DF, Weisleder P, Bohrer PS, Rubel EW. Ongoing production of sensory cells in the vestibular epithelium of the chick. Hear Res. 1992;57:166–174. doi: 10.1016/0378-5955(92)90149-h. [DOI] [PubMed] [Google Scholar]
- 8.Corwin JT, Cotanche DA. Regeneration of sensory hair cells after acoustic trauma. Science. 1988;240:1772–1774. doi: 10.1126/science.3381100. [DOI] [PubMed] [Google Scholar]
- 9.Ryals BM, Rubel EW. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science. 1988;240:1774–1776. doi: 10.1126/science.3381101. [DOI] [PubMed] [Google Scholar]
- 10.Adler HJ, Raphael Y. New hair cells arise from supporting cell conversion in the acoustically damaged chick inner ear. Neurosci Lett. 1996;205:17–20. doi: 10.1016/0304-3940(96)12367-3. [DOI] [PubMed] [Google Scholar]
- 11.Roberson DW, Rubel EW. Cell division in the gerbil cochlea after acoustic trauma. Am J Otol. 1994;15:28–34. [PubMed] [Google Scholar]
- 12.Kawamoto K, Izumikawa M, Beyer LA, Atkin GM, Raphael Y. Spontaneous hair cell regeneration in the mouse utricle following gentamicin ototoxicity. Hear Res. 2009;247:17–26. doi: 10.1016/j.heares.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin V, Golub JS, Nguyen TB, Hume CR, Oesterle EC, Stone JS. Inhibition of Notch activity promotes nonmitotic regeneration of hair cells in the adult mouse utricles. J Neurosci. 2011;31:15329–15339. doi: 10.1523/JNEUROSCI.2057-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7:837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 15.Wu DK, Kelley MW. Molecular mechanisms of inner ear development. Cold Spring Harb Perspect Biol. 2012;4:a008409. doi: 10.1101/cshperspect.a008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vahava O, Morell R, Lynch ED, Weiss S, Kagan ME, Ahituv N, Morrow JE, Lee MK, Skvorak AB, Morton CC, Blumenfeld A, Frydman M, Friedman TB, King MC, Avraham KB. Mutation in transcription factor POU4F3 associated with inherited progressive hearing loss in humans. Science. 1998;279:1950–1954. doi: 10.1126/science.279.5358.1950. [DOI] [PubMed] [Google Scholar]
- 17.de Kok YJ, van der Maarel SM, Bitner-Glindzicz M, Huber I, Monaco AP, Malcolm S, Pembrey ME, Ropers HH, Cremers FP. Association between X-linked mixed deafness and mutations in the POU domain gene POU3F4. Science. 1995;267:685–688. doi: 10.1126/science.7839145. [DOI] [PubMed] [Google Scholar]
- 18.Neph S, Stergachis A, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos J. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson M, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Shu W, Chen H, Bo X, Wang S. Genome-wide analysis of the relationships between DNaseI HS, histone modifications and gene expression reveals distinct modes of chromatin domains. Nucleic Acids Res. 2011;39:7428–7443. doi: 10.1093/nar/gkr443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hawkins RD, Hon GC, Yang C, Antosiewicz-Bourget JE, Lee LK, Ngo QM, Klugman S, Ching KA, Edsall LE, Ye Z, Kuan S, Yu P, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Ren B. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res. 2011;21:1393–1409. doi: 10.1038/cr.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 25.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Consortium EP. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barski A, Cuddapah S, Cui K, Roh T-Y, Schones D, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 32.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O’Meara S, Teague J, Butler A, Hinton J, Latimer C, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Cole J, Forbes S, Jia M, Jones D, Kok CY, Leroy C, Lin ML, McBride DJ, Maddison M, Maquire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, Pleasance E, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Collins VP, Ichimura K, Law S, Wong J, Yuen ST, Leung SY, Tonon G, DePinho RA, Tai YT, Anderson KC, Kahnoski RJ, Massie A, Khoo SK, Teh BT, Stratton MR, Futreal PA. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 34.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, Chen Y, DeSalvo G, Epstein C, Fisher-Aylor KI, Euskirchen G, Gerstein M, Gertz J, Hartemink AJ, Hoffman MM, Iyer VR, Jung YL, Karmakar S, Kellis M, Kharchenko PV, Li Q, Liu T, Liu XS, Ma L, Milosavljevic A, Myers RM, Park PJ, Pazin MJ, Perry MD, Raha D, Reddy TE, Rozowsky J, Shoresh N, Sidow A, Slattery M, Stamatoyannopoulos JA, Tolstorukov MY, White KP, Xi S, Farnham PJ, Lieb JD, Wold BJ, Snyder M. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol. 2009;5:e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie W, Schultz M, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker J, Tian S, Hawkins R, Leung D, Yang H, Wang T, Lee A, Swanson S, Zhang J, Zhu Y, Kim A, Nery J, Urich M, Kuan S, Yen C-a, Klugman S, Yu P, Suknuntha K, Propson N, Chen H, Edsall L, Wagner U, Li Y, Ye Z, Kulkarni A, Xuan Z, Chung W-Y, Chi N, Antosiewicz-Bourget J, Slukvin I, Stewart R, Zhang M, Wang W, Thomson J, Ecker J, Ren B. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Estivill X, Fortina P, Surrey S, Rabionet R, Melchionda S, D’Agruma L, Mansfield E, Rappaport E, Govea N, Mila M, Zelante L, Gasparini P. Connexin-26 mutations in sporadic and inherited sensorineural deafness. Lancet. 1998;351:394–398. doi: 10.1016/S0140-6736(97)11124-2. [DOI] [PubMed] [Google Scholar]
- 40.Tan LW, Bianco T, Dobrovic A. Variable promoter region CpG island methylation of the putative tumor suppressor gene Connexin 26 in breast cancer. Carcinogenesis. 2002;23:231–236. doi: 10.1093/carcin/23.2.231. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, Huhn D, Knosel T, Pacyna-Gengelbach M, Deutschmann N, Petersen I. Downregulation of connexin 26 in human lung cancer is related to promoter methylation. Int J Cancer. 2005;113:14–21. doi: 10.1002/ijc.20498. [DOI] [PubMed] [Google Scholar]
- 42.Wu X, Wang Y, Sun Y, Chen S, Zhang S, Shen L, Huang X, Lin X, Kong W. Reduced expression of Connexin26 and its DNA promoter hypermethylation in the inner ear of mimetic aging rats induced by d-galactose. Biochem Biophys Res Commun. 2014;452:340–346. doi: 10.1016/j.bbrc.2014.08.063. [DOI] [PubMed] [Google Scholar]
- 43.Lister R, Ecker JR. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- 45.Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 47.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nesterova TB, Johnston CM, Appanah R, Newall AE, Godwin J, Alexiou M, Brockdorff N. Skewing X chromosome choice by modulating sense transcription across the Xist locus. Genes Dev. 2003;17:2177–2190. doi: 10.1101/gad.271203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 51.Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 52.Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, Biesemeier DJ, Shomron N, Fekete DM, Hornstein E, Avraham KB. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci USA. 2009;106:7915–7920. doi: 10.1073/pnas.0812446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rudnicki A, Isakov O, Ushakov K, Shivatzki S, Weiss I, Friedman LM, Shomron N, Avraham KB. Next-generation sequencing of small RNAs from inner ear sensory epithelium identifies microRNAs and defines regulatory pathways. BMC Genomics. 2014;15:484. doi: 10.1186/1471-2164-15-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Q, Liu H, McGee J, Walsh EJ, Soukup GA, He DZ. Identifying microRNAs involved in degeneration of the organ of Corti during age-related hearing loss. PLoS One. 2013;8:e62786. doi: 10.1371/journal.pone.0062786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manji SS, Sorensen BS, Klockars T, Lam T, Hutchison W, Dahl HH. Molecular characterization and expression of maternally expressed gene 3 (Meg3/Gtl2) RNA in the mouse inner ear. J Neurosci Res. 2006;83:181–190. doi: 10.1002/jnr.20721. [DOI] [PubMed] [Google Scholar]
- 56.Roberts KA, Abraira VE, Tucker AF, Goodrich LV, Andrews NC. Mutation of Rubie, a novel long non-coding RNA located upstream of Bmp4, causes vestibular malformation in mice. PLoS One. 2012;7:e29495. doi: 10.1371/journal.pone.0029495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steventon B, Mayor R, Streit A. Mutual repression between Gbx2 and Otx2 in sensory placodes reveals a general mechanism for ectodermal patterning. Dev Biol. 2012;367:55–65. doi: 10.1016/j.ydbio.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 59.Roellig D, Bronner ME. The epigenetic modifier DNMT3A is necessary for proper otic placode formation. Dev Biol. 2016;411:294–300. doi: 10.1016/j.ydbio.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schubeler D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 61.Wiench M, John S, Baek S, Johnson TA, Sung MH, Escobar T, Simmons CA, Pearce KH, Biddie SC, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, Hager GL. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 2011;30:3028–3039. doi: 10.1038/emboj.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meier S. Development of the embryonic chick otic placode. II. Electron microscopic analysis. Anat Rec. 1978;191:459–477. doi: 10.1002/ar.1091910406. [DOI] [PubMed] [Google Scholar]
- 63.Uribe RA, Buzzi AL, Bronner ME, Strobl-Mazzulla PH. Histone demethylase KDM4B regulates otic vesicle invagination via epigenetic control of Dlx3 expression. J Cell Biol. 2015;211:815–827. doi: 10.1083/jcb.201503071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Curr Opin Genet Dev. 2008;18:159–168. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 65.Brown ST, Wang J, Groves AK. Dlx gene expression during chick inner ear development. J Comp Neurol. 2005;483:48–65. doi: 10.1002/cne.20418. [DOI] [PubMed] [Google Scholar]
- 66.Lagger G, O’Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He Y, Yu H, Sun S, Wang Y, Liu I, Chen Z, Li H. Trans-2-phenylcyclopropylamine regulates zebrafish lateral line neuromast development mediated by depression of LSD1 activity. Int J Dev Biol. 2013;57:365–373. doi: 10.1387/ijdb.120227hl. [DOI] [PubMed] [Google Scholar]
- 68.Phillips BT, Bolding K, Riley BB. Zebrafish fgf3 and fgf8 encode redundant functions required for otic placode induction. Dev Biol. 2001;235:351–365. doi: 10.1006/dbio.2001.0297. [DOI] [PubMed] [Google Scholar]
- 69.Millimaki BB, Sweet EM, Dhason MS, Riley BB. Zebrafish atoh1 genes: classic proneural activity in the inner ear and regulation by Fgf and Notch. Development. 2007;134:295–305. doi: 10.1242/dev.02734. [DOI] [PubMed] [Google Scholar]
- 70.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, Brunner HG, Veltman JA, van Kessel AG. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 71.Basson MA, van Ravenswaaij-Arts C. Functional insights into chromatin remodelling from studies on CHARGE syndrome. Trends Genet. 2015;31:600–611. doi: 10.1016/j.tig.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adams ME, Hurd EA, Beyer LA, Swiderski DL, Raphael Y, Martin DM. Defects in vestibular sensory epithelia and innervation in mice with loss of Chd7 function: implications for human CHARGE syndrome. J Comp Neurol. 2007;504:519–532. doi: 10.1002/cne.21460. [DOI] [PubMed] [Google Scholar]
- 73.Bosman EA, Penn AC, Ambrose JC, Kettleborough R, Stemple DL, Steel KP. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum Mol Genet. 2005;14:3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- 74.Hurd EA, Adams ME, Layman WS, Swiderski DL, Beyer LA, Halsey KE, Benson JM, Gong TW, Dolan DF, Raphael Y, Martin DM. Mature middle and inner ears express Chd7 and exhibit distinctive pathologies in a mouse model of CHARGE syndrome. Hear Res. 2011;282:184–195. doi: 10.1016/j.heares.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hurd EA, Poucher HK, Cheng K, Raphael Y, Martin DM. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development. 2010;137:3139–3150. doi: 10.1242/dev.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hurd EA, Micucci JA, Reamer EN, Martin DM. Delayed fusion and altered gene expression contribute to semicircular canal defects in Chd7 deficient mice. Mech Dev. 2012;129:308–323. doi: 10.1016/j.mod.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Micucci JA, Layman WS, Hurd EA, Sperry ED, Frank SF, Durham MA, Swiderski DL, Skidmore JM, Scacheri PC, Raphael Y, Martin DM. CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum Mol Genet. 2014;23:434–448. doi: 10.1093/hmg/ddt435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schnetz MP, Bartels CF, Shastri K, Balasubramanian D, Zentner GE, Balaji R, Zhang X, Song L, Wang Z, Laframboise T, Crawford GE, Scacheri PC. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG, Adams DJ, Flicek P, Crawford GE, Laframboise T, Tesar P, Wei CL, Scacheri PC. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6:e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Engelen E, Akinci U, Bryne JC, Hou J, Gontan C, Moen M, Szumska D, Kockx C, van Ijcken W, Dekkers DH, Demmers J, Rijkers EJ, Bhattacharya S, Philipsen S, Pevny LH, Grosveld FG, Rottier RJ, Lenhard B, Poot RA. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43:607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- 81.Kiernan AE, Ahituv N, Fuchs H, Balling R, Avraham KB, Steel KP, Hrabe de Angelis M. The Notch ligand Jagged1 is required for inner ear sensory development. Proc Natl Acad Sci USA. 2001;98:3873–3878. doi: 10.1073/pnas.071496998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 83.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 84.Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009;328:328–341. doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kersigo J, D’Angelo A, Gray BD, Soukup GA, Fritzsch B. The role of sensory organs and the forebrain for the development of the craniofacial shape as revealed by Foxg1-cre-mediated microRNA loss. Genesis. 2011;49:326–341. doi: 10.1002/dvg.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, Fritzsch B. Expression and function of FGF10 in mammalian inner ear development. Dev Dyn. 2003;227:203–215. doi: 10.1002/dvdy.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Urness LD, Wang X, Shibata S, Ohyama T, Mansour SL. Fgf10 is required for specification of non-sensory regions of the cochlear epithelium. Dev Biol. 2015;400:59–71. doi: 10.1016/j.ydbio.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hertzano R, Elkon R, Kurima K, Morrisson A, Chan SL, Sallin M, Biedlingmaier A, Darling DS, Griffith AJ, Eisenman DJ, Strome SE. Cell type-specific transcriptome analysis reveals a major role for Zeb1 and miR-200b in mouse inner ear morphogenesis. PLoS Genet. 2011;7:e1002309. doi: 10.1371/journal.pgen.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz HR, Schier AF, Dulac C. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lamouille S, Subramanyam D, Blelloch R, Derynck R. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell Biol. 2013;25:200–207. doi: 10.1016/j.ceb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 93.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martin P, Swanson GJ. Descriptive and experimental analysis of the epithelial remodellings that control semicircular canal formation in the developing mouse inner ear. Dev Biol. 1993;159:549–558. doi: 10.1006/dbio.1993.1263. [DOI] [PubMed] [Google Scholar]
- 95.Kurima K, Hertzano R, Gavrilova O, Monahan K, Shpargel KB, Nadaraja G, Kawashima Y, Lee KY, Ito T, Higashi Y, Eisenman DJ, Strome SE, Griffith AJ. A noncoding point mutation of Zeb1 causes multiple developmental malformations and obesity in Twirler mice. PLoS Genet. 2011;7:e1002307. doi: 10.1371/journal.pgen.1002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang T, Kersigo J, Jahan I, Pan N, Fritzsch B. The molecular basis of making spiral ganglion neurons and connecting them to hair cells of the organ of Corti. Hear Res. 2011;278:21–33. doi: 10.1016/j.heares.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 98.Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J Assoc Res Otolaryngol. 2000;1:129–143. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu M, Pereira FA, Price SD, Chu MJ, Shope C, Himes D, Eatock RA, Brownell WE, Lysakowski A, Tsai MJ. Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev. 2000;14:2839–2854. doi: 10.1101/gad.840500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ahmed M, Xu J, Xu PX. EYA1 and SIX1 drive the neuronal developmental program in cooperation with the SWI/SNF chromatin-remodeling complex and SOX2 in the mammalian inner ear. Development. 2012;139:1965–1977. doi: 10.1242/dev.071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- 102.Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RM, Jr, Brophy PD, Berkman J, Gattas M, Hyland V, Ruf EM, Schwartz C, Chang EH, Smith RJ, Stratakis CA, Weil D, Petit C, Hildebrandt F. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci U S A. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- 104.Zheng W, Huang L, Wei ZB, Silvius D, Tang B, Xu PX. The role of Six1 in mammalian auditory system development. Development. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tadjuidje E, Hegde RS. The Eyes Absent proteins in development and disease. Cell Mol Life Sci. 2013;70:1897–1913. doi: 10.1007/s00018-012-1144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Raft S, Nowotschin S, Liao J, Morrow BE. Suppression of neural fate and control of inner ear morphogenesis by Tbx1. Development. 2004;131:1801–1812. doi: 10.1242/dev.01067. [DOI] [PubMed] [Google Scholar]
- 108.Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 109.Cai T, Seymour ML, Zhang H, Pereira FA, Groves AK. Conditional deletion of Atoh1 reveals distinct critical periods for survival and function of hair cells in the organ of Corti. J Neurosci. 2013;33:10110–10122. doi: 10.1523/JNEUROSCI.5606-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3:580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- 111.Lanford PJ, Lan Y, Jiang R, Lindsell C, Weinmaster G, Gridley T, Kelley MW. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21:289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- 112.Kiernan AE, Cordes R, Kopan R, Gossler A, Gridley T. The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development. 2005;132:4353–4362. doi: 10.1242/dev.02002. [DOI] [PubMed] [Google Scholar]
- 113.Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- 114.Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012;13:424. doi: 10.1186/1471-2164-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stojanova ZP, Kwan T, Segil N. Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea. Development. 2015;142:3529–3536. doi: 10.1242/dev.126763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 117.Huyghe A, Van den Ackerveken P, Sacheli R, Prevot PP, Thelen N, Renauld J, Thiry M, Delacroix L, Nguyen L, Malgrange B. MicroRNA-124 regulates cell specification in the cochlea through modulation of Sfrp4/5. Cell Rep. 2015;13:31–42. doi: 10.1016/j.celrep.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 118.De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cruciat CM, Niehrs C. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb Perspect Biol. 2013;5:a015081. doi: 10.1101/cshperspect.a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 121.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 122.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 123.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 124.Golden EJ, Benito-Gonzalez A, Doetzlhofer A. The RNA-binding protein LIN28B regulates developmental timing in the mammalian cochlea. Proc Natl Acad Sci U S A. 2015;112:E3864–3873. doi: 10.1073/pnas.1501077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Buechner J, Tomte E, Haug BH, Henriksen JR, Lokke C, Flaegstad T, Einvik C. Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br J Cancer. 2011;105:296–303. doi: 10.1038/bjc.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 127.Dominguez-Frutos E, Lopez-Hernandez I, Vendrell V, Neves J, Gallozzi M, Gutsche K, Quintana L, Sharpe J, Knoepfler PS, Eisenman RN, Trumpp A, Giraldez F, Schimmang T. N-myc controls proliferation, morphogenesis, and patterning of the inner ear. J Neurosci. 2011;31:7178–7189. doi: 10.1523/JNEUROSCI.0785-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Laine H, Sulg M, Kirjavainen A, Pirvola U. Cell cycle regulation in the inner ear sensory epithelia: role of cyclin D1 and cyclin-dependent kinase inhibitors. Dev Biol. 2010;337:134–146. doi: 10.1016/j.ydbio.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 129.La Torre A, Georgi S, Reh TA. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc Natl Acad Sci U S A. 2013;110:E2362–2370. doi: 10.1073/pnas.1301837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 131.Dambal S, Shah M, Mihelich B, Nonn L. The microRNA-183 cluster: the family that plays together stays together. Nucleic Acids Res. 2015;43:7173–7188. doi: 10.1093/nar/gkv703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li H, Kloosterman W, Fekete DM. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J Neurosci. 2010;30:3254–3263. doi: 10.1523/JNEUROSCI.4948-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang KD, Stoller ML, Fekete DM. Expression and misexpression of the miR-183 family in the developing hearing organ of the chicken. PLoS One. 2015;10:e0132796. doi: 10.1371/journal.pone.0132796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MA. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 135.Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, Dalmay T, Moreno-Pelayo MA, Enright AJ, Steel KP. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kuhn S, Johnson SL, Furness DN, Chen J, Ingham N, Hilton JM, Steffes G, Lewis MA, Zampini V, Hackney CM, Masetto S, Holley MC, Steel KP, Marcotti W. miR-96 regulates the progression of differentiation in mammalian cochlear inner and outer hair cells. Proc Natl Acad Sci U S A. 2011;108:2355–2360. doi: 10.1073/pnas.1016646108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Solda G, Robusto M, Primignani P, Castorina P, Benzoni E, Cesarani A, Ambrosetti U, Asselta R, Duga S. A novel mutation within the MIR96 gene causes non-syndromic inherited hearing loss in an Italian family by altering pre-miRNA processing. Hum Mol Genet. 2012;21:577–585. doi: 10.1093/hmg/ddr493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hildebrand MS, Witmer PD, Xu S, Newton SS, Kahrizi K, Najmabadi H, Valle D, Smith RJ. miRNA mutations are not a common cause of deafness. Am J Med Genet A. 2010;152A:646–652. doi: 10.1002/ajmg.a.33299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, Xi Y, Park BH, Shi X, Garcia BA, Li W, Gozani O. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44:609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stec I, Wright TJ, van Ommen GJ, de Boer PA, van Haeringen A, Moorman AF, Altherr MR, den Dunnen JT. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- 141.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 142.Ahmed M, Ura K, Streit A. Auditory hair cell defects as potential cause for sensorineural deafness in Wolf-Hirschhorn syndrome. Dis Model Mech. 2015;8:1027–1035. doi: 10.1242/dmm.019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Slattery EL, Speck JD, Warchol ME. Epigenetic influences on sensory regeneration: histone deacetylases regulate supporting cell proliferation in the avian utricle. J Assoc Res Otolaryngol. 2009;10:341–353. doi: 10.1007/s10162-009-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.He Y, Wang Z, Sun S, Tang D, Li W, Chai R, Li H. HDAC3 Is Required for Posterior Lateral Line Development in Zebrafish. Mol Neurobiol. 2015 doi: 10.1007/s12035-015-9433-6. [DOI] [PubMed] [Google Scholar]
- 145.He Y, Tang D, Li W, Chai R, Li H. Histone deacetylase 1 is required for the development of the zebrafish inner ear. Sci Rep. 2016;6:16535. doi: 10.1038/srep16535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.He Y, Wu J, Mei H, Yu H, Sun S, Shou J, Li H. Histone deacetylase activity is required for embryonic posterior lateral line development. Cell Prolif. 2014;47:91–104. doi: 10.1111/cpr.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen P, Segil N. p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development. 1999;126:1581–1590. doi: 10.1242/dev.126.8.1581. [DOI] [PubMed] [Google Scholar]
- 148.Lowenheim H, Furness DN, Kil J, Zinn C, Gultig K, Fero ML, Frost D, Gummer AW, Roberts JM, Rubel EW, Hackney CM, Zenner HP. Gene disruption of p27(Kip1) allows cell proliferation in the postnatal and adult organ of corti. Proc Natl Acad Sci U S A. 1999;96:4084–4088. doi: 10.1073/pnas.96.7.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Laine H, Doetzlhofer A, Mantela J, Ylikoski J, Laiho M, Roussel MF, Segil N, Pirvola U. p19(Ink4d) and p21(Cip1) collaborate to maintain the postmitotic state of auditory hair cells, their codeletion leading to DNA damage and p53-mediated apoptosis. J Neurosci. 2007;27:1434–1444. doi: 10.1523/JNEUROSCI.4956-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rudolph T, Beuch S, Reuter G. Lysine-specific histone demethylase LSD1 and the dynamic control of chromatin. Biol Chem. 2013;394:1019–1028. doi: 10.1515/hsz-2013-0119. [DOI] [PubMed] [Google Scholar]
- 151.He Y, Tang D, Cai C, Chai R, Li H. LSD1 is required for hair cell regeneration in zebrafish. Mol Neurobiol. 2016;53:2421–2434. doi: 10.1007/s12035-015-9206-2. [DOI] [PubMed] [Google Scholar]
- 152.Warchol ME, Corwin JT. Regenerative proliferation in organ cultures of the avian cochlea: identification of the initial progenitors and determination of the latency of the proliferative response. J Neurosci. 1996;16:5466–5477. doi: 10.1523/JNEUROSCI.16-17-05466.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Frucht CS, Uduman M, Duke JL, Kleinstein SH, Santos-Sacchi J, Navaratnam DS. Gene expression analysis of forskolin treated basilar papillae identifies microRNA181a as a mediator of proliferation. PLoS One. 2010;5:e11502. doi: 10.1371/journal.pone.0011502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Frucht CS, Santos-Sacchi J, Navaratnam DS. MicroRNA181a plays a key role in hair cell regeneration in the avian auditory epithelium. Neurosci Lett. 2011;493:44–48. doi: 10.1016/j.neulet.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang X, Gocek E, Liu CG, Studzinski GP. MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle. 2009;8:736–741. doi: 10.4161/cc.8.5.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cuesta R, Martinez-Sanchez A, Gebauer F. miR-181a regulates cap-dependent translation of p27(kip1) mRNA in myeloid cells. Mol Cell Biol. 2009;29:2841–2851. doi: 10.1128/MCB.01971-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.White PM, Doetzlhofer A, Lee YS, Groves AK, Segil N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature. 2006;441:984–987. doi: 10.1038/nature04849. [DOI] [PubMed] [Google Scholar]
- 158.Sinkkonen ST, Chai R, Jan TA, Hartman BH, Laske RD, Gahlen F, Sinkkonen W, Cheng AG, Oshima K, Heller S. Intrinsic regenerative potential of murine cochlear supporting cells. Sci Rep. 2011;1:26. doi: 10.1038/srep00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bramhall NF, Shi F, Arnold K, Hochedlinger K, Edge AS. Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Reports. 2014;2:311–322. doi: 10.1016/j.stemcr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cox BC, Chai R, Lenoir A, Liu Z, Zhang L, Nguyen DH, Chalasani K, Steigelman KA, Fang J, Rubel EW, Cheng AG, Zuo J. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development. 2014;141:816–829. doi: 10.1242/dev.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Korrapati S, Roux I, Glowatzki E, Doetzlhofer A. Notch signaling limits supporting cell plasticity in the hair cell-damaged early postnatal murine cochlea. PLoS One. 2013;8:e73276. doi: 10.1371/journal.pone.0073276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mizutari K, Fujioka M, Hosoya M, Bramhall N, Okano HJ, Okano H, Edge AS. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron. 2013;77:58–69. doi: 10.1016/j.neuron.2012.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chai R, Kuo B, Wang T, Liaw EJ, Xia A, Jan TA, Liu Z, Taketo MM, Oghalai JS, Nusse R, Zuo J, Cheng AG. Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc Natl Acad Sci U S A. 2012;109:8167–8172. doi: 10.1073/pnas.1202774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shi F, Kempfle JS, Edge AS. Wnt-responsive Lgr5-expressing stem cells are hair cell progenitors in the cochlea. J Neurosci. 2012;32:9639–9648. doi: 10.1523/JNEUROSCI.1064-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shi F, Hu L, Edge AS. Generation of hair cells in neonatal mice by beta-catenin overexpression in Lgr5-positive cochlear progenitors. Proc Natl Acad Sci U S A. 2013;110:13851–13856. doi: 10.1073/pnas.1219952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gil J, O’Loghlen A. PRC1 complex diversity: where is it taking us? Trends Cell Biol. 2014;24:632–641. doi: 10.1016/j.tcb.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 167.Lu X, Sun S, Qi J, Li W, Liu L, Zhang Y, Chen Y, Zhang S, Wang L, Miao D, Chai R, Li H. Bmi1 Regulates the Proliferation of Cochlear Supporting Cells Via the Canonical Wnt Signaling Pathway. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-9686-8. [DOI] [PubMed] [Google Scholar]
- 168.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 170.Zhou Y, Hu Z. Epigenetic DNA demethylation causes inner ear stem cell differentiation into hair cell-like cells. Front Cell Neurosci. 2016;10:185. doi: 10.3389/fncel.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sandoval-Guzman T, Wang H, Khattak S, Schuez M, Roensch K, Nacu E, Tazaki A, Joven A, Tanaka EM, Simon A. Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell. 2014;14:174–187. doi: 10.1016/j.stem.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 172.Tsonis PA, Call MK, Grogg MW, Sartor MA, Taylor RR, Forge A, Fyffe R, Goldenberg R, Cowper-Sal-lari R, Tomlinson CR. MicroRNAs and regeneration: Let-7 members as potential regulators of dedifferentiation in lens and inner ear hair cell regeneration of the adult newt. Biochem Biophys Res Commun. 2007;362:940–945. doi: 10.1016/j.bbrc.2007.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell. 2009;16:58–69. doi: 10.1016/j.devcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle. 2010;9:1072–1083. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Riccardi S, Bergling S, Sigoillot F, Beibel M, Werner A, Leighton-Davies J, Knehr J, Bouwmeester T, Parker CN, Roma G, Kinzel B. MiR-210 promotes sensory hair cell formation in the organ of corti. BMC Genomics. 2016;17:309. doi: 10.1186/s12864-016-2620-7. [DOI] [PMC free article] [PubMed] [Google Scholar]