Abstract
Purpose of review
Here, we discuss a recently developed experimental strategy for discovering small molecules with potential to prevent and treat skeletal muscle atrophy.
Recent findings
Muscle atrophy involves and requires widespread changes in skeletal muscle gene expression, which generate complex but measurable patterns of positive and negative changes in skeletal muscle mRNA levels (a.k.a. mRNA expression signatures of muscle atrophy). Many bioactive small molecules generate their own characteristic mRNA expression signatures, and by identifying small molecules whose signatures approximate mirror images of muscle atrophy signatures, one may identify small molecules with potential to prevent and/or reverse muscle atrophy. Unlike a conventional drug discovery approach, this strategy does not rely on a predefined molecular target but rather exploits the complexity of muscle atrophy to identify small molecules that counter the entire spectrum of pathological changes in atrophic muscle. We discuss how this strategy has been used to identify two natural compounds, ursolic acid and tomatidine, that reduce muscle atrophy and improve skeletal muscle function.
Summary
Discovery strategies based on mRNA expression signatures can elucidate new approaches for preserving and restoring muscle mass and function.
Keywords: connectivity map, expression signatures, skeletal muscle atrophy, tomatidine, ursolic acid
INTRODUCTION
Maintaining adequate skeletal muscle mass and strength is critical for quality of life and healthy ageing. However, muscle mass and strength slowly decline after the age of 30 and rapidly decline with malnutrition, muscle disuse and essentially any serious illness or injury. This process of decline, called skeletal muscle atrophy, causes weakness and fatigability, impairs whole body metabolism, delays recovery from acute illness and injury, increases morbidity and mortality from chronic disease, and is a major cause of falls, extended hospital stays and loss of independent living.
Despite the broad impact of skeletal muscle atrophy, we lack reliably effective ways to prevent and treat it. Although a number of pharmaceutical and nutritional approaches for muscle atrophy are being actively investigated, none are yet proven to be both well tolerated and effective in humans, and each appears to have its own advantages and disadvantages [1▪,2]. It seems likely that preventing and treating muscle atrophy on a population-wide basis will require a repertoire of modalities that can be used alone or in combination depending on the clinical circumstances, similar to other complex chronic diseases such as hypertension, type 2 diabetes and dyslipidemia.
Most of the therapeutic approaches for muscle atrophy currently under investigation are conceptually derived from serendipitous discoveries of interventions that increase muscle mass. Discovery of entirely new approaches for muscle atrophy has been hampered by the fact that muscle atrophy is a highly complex molecular process that remains poorly understood. This concept is well illustrated by genome-wide investigations of skeletal muscle mRNA expression. Such studies have revealed that muscle atrophy involves complex pathological patterns, or signatures, of skeletal muscle mRNA expression that change with time, vary with the cause of muscle atrophy and involve hundreds of exquisitely coordinated positive and negative changes in levels of specific skeletal muscle mRNAs (some recent examples include [3–5]).
Approximately 25 of the specific mRNAs that are induced or repressed in atrophying muscle have been investigated in detail and found to encode important regulators of muscle mass, key evidence that muscle atrophy requires alterations in muscle mRNA expression (some recent examples and older references may be found in [5–14]). However, the downstream mechanisms by which those proteins regulate muscle mass are not entirely clear, and the upstream signalling network that regulates muscle mRNA expression is labyrinthine and just beginning to be characterized. Furthermore, the vast majority of skeletal muscle mRNAs whose levels are altered during muscle atrophy remain completely unstudied. Thus, although important advances have been made, it has become increasingly clear that we are not yet close to having a detailed understanding of how muscle atrophy occurs at the molecular level. This creates additional challenges for conventional pharmaceutical development strategies, which begin with a single well defined molecular target and are frequently unsuccessful for a variety of reasons, even when a highly rational molecular target is in hand.
Although genome-wide data have emphasized how far we are from a comprehensive molecular understanding of muscle atrophy, they also offer new and potentially powerful opportunities for therapeutic discovery. Here, we will discuss how we have used genome-wide data to discover two small molecules that are potentially applicable to the prevention and treatment of muscle atrophy.
DISCOVERY OF URSOLIC ACID AS A SMALL MOLECULE INHIBITOR OF MUSCLE ATROPHY
As discussed above, muscle atrophy involves and requires many positive and negative changes in muscle mRNA levels. This overall pattern of changes represents a molecular signature, a.k.a. an mRNA expression signature of muscle atrophy. Because many bioactive small molecules generate their own characteristic mRNA expression signatures, we hypothesized that we might identify candidate small molecule inhibitors of muscle atrophy by identifying small molecules whose mRNA expression signatures negatively correlate to mRNA expression signatures of muscle atrophy [4].
The first step towards testing this hypothesis was to determine mRNA expression signatures of muscle atrophy. To this end, we obtained skeletal muscle samples from humans and mice under basal conditions and during two conditions that cause muscle atrophy (fasting and spinal cord injury). We then analysed mRNA from the skeletal muscle samples with genome-wide mRNA expression arrays. Because genome-wide methods have high false-positive rates, we reasoned that the discovery process would ultimately be more efficient and productive if each small molecule signature was compared with not one but two muscle atrophy signatures. In addition, because the individual mRNAs that are induced or repressed as a muscle undergoes atrophy may be species-specific and may vary with the cause of atrophy, we reasoned that our chance of discovering broad spectrum inhibitors of muscle atrophy would be increased if the two muscle atrophy signatures were filtered in a way that captured mRNAs that were conserved across two species and two atrophy stimuli. Finally, because our ultimate goal was to find small molecules that would be applicable to human muscle atrophy, both muscle atrophy signatures contained data from humans. Guided by these concepts, we used the genome-wide array data to build two distinct mRNA expression signatures of muscle atrophy. The first signature (muscle atrophy signature 1) consists of evolutionarily conserved mRNAs that are induced or repressed by fasting in both human and mouse skeletal muscle [4]. The second signature (muscle atrophy signature 2) consists of mRNAs that are induced or repressed by two very different types of muscle atrophy stimuli (fasting and spinal cord injury) in human skeletal muscle [4].
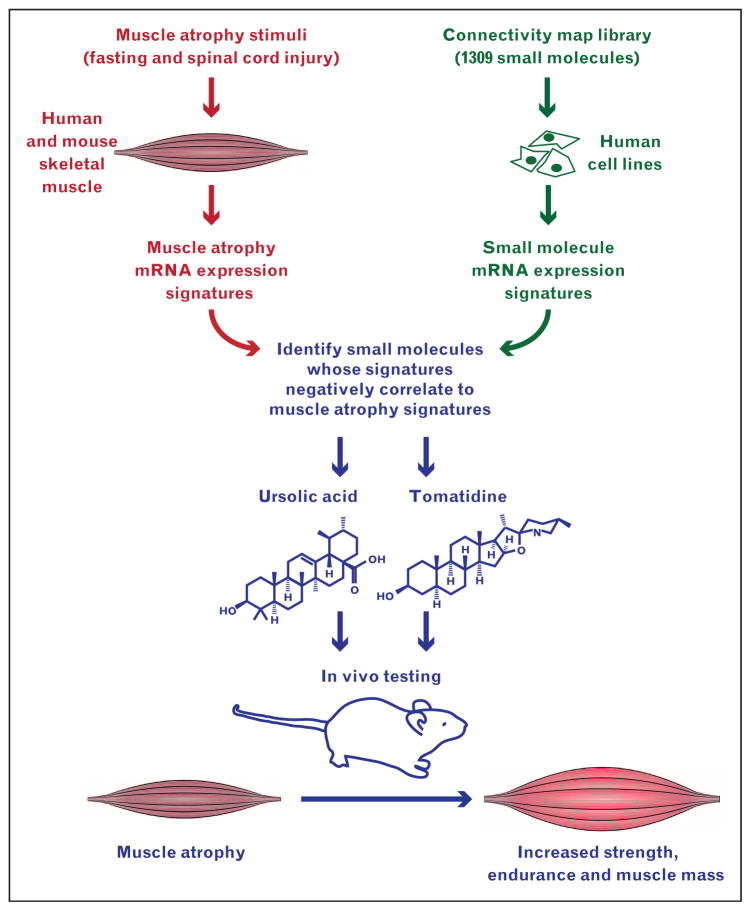
Next, to identify small molecules whose mRNA expression signatures negatively correlate to mRNA expression signatures of muscle atrophy, we used the Connectivity Map (a discovery resource developed by Lamb et al. [15]) to compare muscle atrophy signatures 1 and 2 with the mRNA expression signatures of 1309 small molecules in several human cell lines (Fig. 1). We focused on small molecules with favourable safety characteristics and became particularly interested in ursolic acid, whose signature in PC3 cells negatively correlated to muscle atrophy signatures 1 and 2 [4]. Ursolic acid (Fig. 1) is a naturally occurring pentacyclic triterpene acid present in several edible herbs and fruits, including apples, which on average contain about 50 mg ursolic acid [16]. Ursolic acid is known to possess a variety of anticancer properties [17] and is widely regarded as having a favourable safety profile, with an oral LD50 more than 8000 mg/kg in mice [18].
FIGURE 1.
Schematic illustration of the strategy used to discover ursolic acid and tomatidine as small molecule inhibitors of muscle atrophy. We compared two mRNA expression signatures of skeletal muscle atrophy with mRNA expression signatures of 1309 small molecules in the Connectivity Map, searching for negative correlations. We found that the mRNA expression signatures of ursolic acid and tomatidine negatively correlated to both muscle atrophy signatures, and then used mouse models to discover that ursolic acid and tomatidine reduce muscle atrophy and increase strength, endurance and muscle mass.
The effects of ursolic acid on skeletal muscle were not known. However, the finding that ursolic acid’s signature negatively correlated to signatures of muscle atrophy suggested that ursolic acid might inhibit muscle atrophy. Using mouse models, we found that ursolic acid reduces muscle atrophy induced by two distinct causes: lack of nutrients (fasting) and lack of muscle innervation (denervation) [4]. Importantly, ursolic acid’s positive effects on muscle mass were accompanied by increased strength [4] and exercise capacity [19]. Furthermore, we found that ursolic acid increases muscle specific force (i.e. strength per unit muscle mass), indicating a greater effect on strength than muscle mass [4].
In many ways, muscle atrophy is the opposite of muscle hypertrophy. Thus, a small molecule whose mRNA expression signature is opposite to signatures of muscle atrophy could be predicted to not only reduce muscle atrophy but also promote muscle hypertrophy. Consistent with this prediction, we found that ursolic acid stimulates muscle hypertrophy in healthy mice lacking muscle atrophy [4].
The way that ursolic acid was identified, coupled with its effects in skeletal muscle, implied that ursolic acid would alter muscle mRNA expression in a manner that reduces atrophy and promotes hypertrophy. Indeed, a genome-wide investigation of muscle mRNA expression showed that ursolic acid generates numerous positive and negative changes in muscle mRNA levels, including some changes that are known to reduce muscle atrophy and promote muscle hypertrophy (e.g. induction of insulin-like growth factor I (IGF-I) and spermine oxidase mRNAs, and repression of MuRF1 mRNA) [4].
At this point, the way(s) in which ursolic acid alters muscle gene expression remain(s) unclear. However, we do know that ursolic acid acts directly on muscle cells, and its effects in muscle are mediated in part by Akt, a protein kinase known to reduce muscle atrophy and promote muscle hypertrophy [20]. This is supported by our findings that ursolic acid increases skeletal muscle Akt activity in vivo [4,19], and direct addition of ursolic acid to cultured skeletal myotubes rapidly stimulates Akt and one of its key downstream effectors, mTOR complex 1 (mTORC1) [4]. In cultured myotubes, ursolic acid mediated stimulation of Akt/mTORC1 signalling requires growth factors (insulin or IGF-I) and is associated with enhanced activation of the insulin and IGF-I receptors [4].
Interestingly, ursolic acid’s effects in skeletal muscle are accompanied by reductions in adiposity, fasting blood glucose and plasma cholesterol and triglycerides [4]. This led us to test ursolic acid’s effects in diet-induced obese mice, where we found that ursolic acid reduces obesity, glucose intolerance and nonalcoholic fatty liver disease (NAFLD) [19]. Importantly, ursolic acid does not alter spontaneous activity or decrease food intake, but rather reduces obesity by increasing energy expenditure [19]. This can be at least partially explained by ursolic acid’s capacity to increase skeletal muscle Akt activity, which is sufficient to increase resting energy expenditure and provide protection against obesity, glucose intolerance and NAFLD (see references within [19]). In addition, we found that ursolic acid also increases the amount of brown fat [19]. Like skeletal muscle, brown fat has a high rate of energy expenditure and provides protection against obesity [21], suggesting that ursolic acid may increase resting energy expenditure and reduce obesity by increasing both muscle and brown fat.
Some of these findings have been confirmed and extended by other groups. For example, Figueiredo and Nader [22] found that ursolic acid increases the amount of total cellular protein in serum-treated but not serum-starved myotubes. Since myotubes are postmitotic, this protein accretion reflects myotube hypertrophy. In a recent in-vivo study, Ogasawara et al. [23] found that a single dose of ursolic acid acutely increases Akt activity in rat skeletal muscle and sustains the activation of mTORC1 after resistance exercise, suggesting that ursolic acid may facilitate the anabolic response to physical therapy. In addition, several other groups have used mouse or rat models to demonstrate that ursolic acid reduces adiposity, blood glucose and plasma lipids, and prevents and/or reverses obesity and obesity-related insulin resistance, dyslipidemia and NAFLD [24–26].
Most recently, Bang et al. [27▪] reported the results of a randomized, placebo-controlled study of orally administered ursolic acid in humans. Sixteen healthy male individuals (average age 29) with a more than 3-year history of resistance exercise training were randomized to receive either placebo or ursolic acid (450 mg/day) for 8 weeks, while continuing resistance exercise training [27▪]. This regimen of ursolic acid significantly increased muscle strength by 6–12% (depending on the muscle group) and significantly decreased fat mass by 26% [27▪], indicating that short-term treatment with a moderate dose of ursolic acid increases strength and reduces fat in healthy humans.
DISCOVERY OF TOMATIDINE AS A SMALL MOLECULE INHIBITOR OF MUSCLE ATROPHY
Ursolic acid served as a proof-of-concept for the discovery strategy that was used to identify it. Thus, we recently used the same general strategy to search for other small molecules whose mRNA expression signatures negatively correlate to muscle atrophy signatures 1 and 2.
We found that both muscle atrophy signatures negatively correlated to mRNA expression signatures of tomatidine [28▪], a steroidal alkaloid that is structurally dissimilar to ursolic acid (Fig. 1).
In nature, tomatidine exists as the aglycone of alpha-tomatine, an abundant glycoalkaloid in tomato plants that mediates plant defense [29]. The level of alpha-tomatine in tomatoes is typically highest in immature, green tomatoes and declines as tomatoes ripen [29,30]. When consumed by animals, alpha-tomatine is hydrolyzed in the gut to tomatidine, which is absorbed [30]. Similar to ursolic acid, tomatidine is thought to have a favourable safety profile [29,30], and it has some beneficial effects in preclinical models including reduced plasma cholesterol and prevention of atherosclerosis [31].
Tomatidine’s effects on skeletal muscle were unknown. Using mouse models, we found that tomatidine shares many of the same effects as ursolic acid, including reduced muscle atrophy during fasting and muscle disuse; enhanced recovery from muscle atrophy; increased strength, exercise capacity and muscle specific force; inhibition of the loss of specific force during atrophy; muscle hypertrophy; and reduced adiposity [28▪]. Also like ursolic acid, tomatidine increases the size of all muscle fibre types but does not alter the relative amounts of muscle fibre types; and tomatidine acts directly on muscle cells, stimulating hypertrophy of cultured C2C12 myotubes and primary human myotubes with an EC50 of approximately 200 nmol/l, and a maximal effect at 1 μmol/l [28▪]. Tomatidine and ursolic acid have similar efficacies in vivo and in vitro, but tomatidine is nearly 10-fold more potent than ursolic acid.
Tomatidine’s hypertrophic effect depends at least in part on mTORC1, which is activated by tomatidine in both mouse muscle and cultured myotubes, leading to altered muscle mRNA expression, increased protein synthesis and mitochondrial biogenesis, and ultimately cellular hypertrophy [28▪]. Interestingly, tomatidine differs from ursolic acid in that it does not appear to activate Akt in mouse skeletal muscle or cultured myotubes [28▪], suggesting that ursolic acid and tomatidine utilize different proximal signalling pathways to activate mTORC1.
CONCLUSION
In our view, three areas stand out as being especially important related areas for future research.
The general strategy used to identify ursolic acid and tomatidine (Fig. 1) could potentially be used to discover additional small molecules that improve muscle function and reduce muscle atrophy.
Additional mechanistic studies are needed to better understand how ursolic acid and tomatidine improve muscle function and reduce muscle atrophy. Such studies may provide key insights into the pathogenesis of muscle atrophy and potential links between muscle atrophy, obesity and type 2 diabetes.
We believe that it is very important to continue efforts aimed at translating ursolic acid and tomatidine to humans. For example, ursolic acid and tomatidine could potentially be developed as nutritional ingredients to help preserve muscle mass and function during ageing, muscle disuse and illness. They also represent attractive lead compounds for pharmaceuticals targeting various forms of muscle atrophy, as well as obesity, type 2 diabetes, dyslipidemia and NAFLD. Nutritional and pharmaceutical approaches based on ursolic acid and tomatidine could potentially be used in conjunction with other existing and emerging nutritional, pharmaceutical and physical approaches to muscle atrophy.
KEY POINTS.
The molecular pathogenesis of muscle atrophy is highly complex and involves widespread changes in muscle mRNA expression, which are not well understood but can be fully ascertained through the use of methods such as genome-wide mRNA expression arrays.
The overall pattern of mRNA expression during muscle atrophy represents a molecular signature that can be used to discover small molecules that have opposite effects on cellular gene expression and thus potential to inhibit muscle atrophy.
mRNA expression signatures of muscle atrophy have been used to discover two naturally occurring bioactive lipids (ursolic acid and tomatidine) that alter muscle gene expression in a manner that improves muscle function and reduces muscle atrophy.
Further development of ursolic acid, tomatidine and structural analogues may lead to new nutritional and pharmacologic approaches for muscle atrophy.
Acknowledgments
We thank Drs. John Talley, Vitor Lira, Peter Snyder and Daryl Granner for critical review of the manuscript.
Financial support and sponsorship
The authors’ work is supported by the National Institutes of Health (grants 5R01AR059115-05, F31AG04603801, 1R43AG044898-01 and 1R41AG047684-01), the Department of Veterans Affairs (grants 5I01BX 000976-05 and 1I01RX001477-01), the Fraternal Order of Eagles Diabetes Research Center at the University of Iowa and Emmyon, Inc.
Footnotes
Conflicts of interest
The authors are inventors on patent applications related to ursolic acid and tomatidine, which have been filed by the University of Iowa Research Foundation and licensed to Emmyon, Inc. C.M.A. and S.M.E. hold equity in Emmyon, Inc. C.M.A. is a cofounder and officer of Emmyon, Inc. S.M.E. is an employee of Emmyon, Inc.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
- 1▪.Sepulveda PV, Bush ED, Baar K. The pharmacology of manipulating lean body mass. Clin Exp Pharmacol Physiol. 2015;42:1–13. doi: 10.1111/1440-1681.12320. Recent review discussing some of the approaches to muscle atrophy that are currently being investigated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brotto M, Abreu EL. Sarcopenia: pharmacology of today and tomorrow. J Pharmacol Exp Ther. 2012;343:540–546. doi: 10.1124/jpet.112.191759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llano-Diez M, Gustafson AM, Olsson C, et al. Muscle wasting and the temporal gene expression pattern in a novel rat intensive care unit model. BMC Genomics. 2011;12:602. doi: 10.1186/1471-2164-12-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunkel SD, Suneja M, Ebert SM, et al. mRNA expression signatures of human skeletal muscle atrophy identify a natural compound that increases muscle mass. Cell Metab. 2011;13:627–638. doi: 10.1016/j.cmet.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebert SM, Dyle MC, Kunkel SD, et al. Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. J Biol Chem. 2012;287:27290–27301. doi: 10.1074/jbc.M112.374777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebert SM, Monteys AM, Fox DK, et al. The transcription factor ATF4 promotes skeletal myofiber atrophy during fasting. Mol Endocrinol. 2010;24:790–799. doi: 10.1210/me.2009-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mittal A, Bhatnagar S, Kumar A, et al. The TWEAK-Fn14 system is a critical regulator of denervation-induced skeletal muscle atrophy in mice. J Cell Biol. 2010;188:833–849. doi: 10.1083/jcb.200909117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Vermulst M, Wang YE, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moresi V, Williams AH, Meadows E, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelleher AR, Kimball SR, Dennis MD, et al. The mTORC1 signaling repressors REDD1/2 are rapidly induced and activation of p70S6K1 by leucine is defective in skeletal muscle of an immobilized rat hindlimb. Am J Physiol Endocrinol Metab. 2013;304:E229–E236. doi: 10.1152/ajpendo.00409.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bongers KS, Fox DK, Ebert SM, et al. Skeletal muscle denervation causes skeletal muscle atrophy through a pathway that involves both Gadd45a and HDAC4. Am J Physiol Endocrinol Metab. 2013;305:E907–E915. doi: 10.1152/ajpendo.00380.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee D, Goldberg AL. SIRT1 protein, by blocking the activities of transcription factors FoxO1 and FoxO3, inhibits muscle atrophy and promotes muscle growth. J Biol Chem. 2013;288:30515–30526. doi: 10.1074/jbc.M113.489716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox DK, Ebert SM, Bongers KS, et al. p53 and ATF4 mediate distinct and additive pathways to skeletal muscle atrophy during limb immobilization. Am J Physiol Endocrinol Metab. 2014;307:E245–E261. doi: 10.1152/ajpendo.00010.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bongers KS, Fox DK, Kunkel SD, et al. Spermine oxidase maintains basal skeletal muscle gene expression and fiber size, and is strongly repressed by conditions that cause skeletal muscle atrophy. Am J Physiol Endocrinol Metab. 2015;308:E144–E158. doi: 10.1152/ajpendo.00472.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 16.Jager S, Trojan H, Kopp T, et al. Pentacyclic triterpene distribution in various plants: rich sources for a new group of multipotent plant extracts. Molecules. 2009;14:2016–2031. doi: 10.3390/molecules14062016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shanmugam MK, Dai X, Kumar AP, et al. Ursolic acid in cancer prevention and treatment: molecular targets, pharmacokinetics and clinical studies. Biochem Pharmacol. 2013;85:1579–1587. doi: 10.1016/j.bcp.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Lee AW, Chen TL, Shih CM, et al. Ursolic acid induces allograft inflammatory factor-1 expression via a nitric oxide-related mechanism and increases neovascularization. J Agric Food Chem. 2010;58:12941–12949. doi: 10.1021/jf103265x. [DOI] [PubMed] [Google Scholar]
- 19.Kunkel SD, Elmore CJ, Bongers KS, et al. Ursolic acid increases skeletal muscle and brown fat and decreases diet-induced obesity, glucose intolerance and fatty liver disease. PLoS One. 2012;7:e39332. doi: 10.1371/journal.pone.0039332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiaffino S, Mammucari C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skelet Muscle. 2011;1:4. doi: 10.1186/2044-5040-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell Metab. 2010;11:257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Figueiredo VC, Nader GA. Ursolic acid directly promotes protein accretion in myotubes but does not affect myoblast proliferation. Cell Biochem Funct. 2012;30:432–437. doi: 10.1002/cbf.2821. [DOI] [PubMed] [Google Scholar]
- 23.Ogasawara R, Sato K, Higashida K, et al. Ursolic acid stimulates mTORC1 signaling after resistance exercise in rat skeletal muscle. Am J Physiol Endocrinol Metab. 2013;305:E760–E765. doi: 10.1152/ajpendo.00302.2013. [DOI] [PubMed] [Google Scholar]
- 24.Rao VS, de Melo CL, Queiroz MG, et al. Ursolic acid, a pentacyclic triterpene from Sambucus australis, prevents abdominal adiposity in mice fed a high-fat diet. J Med Food. 2011;14:1375–1382. doi: 10.1089/jmf.2010.0267. [DOI] [PubMed] [Google Scholar]
- 25.Li S, Liao X, Meng F, et al. Therapeutic role of ursolic acid on ameliorating hepatic steatosis and improving metabolic disorders in high-fat diet-induced nonalcoholic fatty liver disease rats. PLoS One. 2014;9:e86724. doi: 10.1371/journal.pone.0086724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundaresan A, Harini R, Pugalendi KV. Ursolic acid and rosiglitazone combination alleviates metabolic syndrome in high fat diet fed C57BL/6J mice. Gen Physiol Biophys. 2012;31:323–333. doi: 10.4149/gpb_2012_037. [DOI] [PubMed] [Google Scholar]
- 27▪.Bang HS, Seo DY, Chung YM, et al. Ursolic Acid-induced elevation of serum irisin augments muscle strength during resistance training in men. Korean J Physiol Pharmacol. 2014;18:441–446. doi: 10.4196/kjpp.2014.18.5.441. Recent randomized, placebo-controlled study showing that ursolic acid increases muscle strength in humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28▪.Dyle MC, Ebert SM, Cook DP, et al. Systems-based discovery of tomatidine as a natural small molecule inhibitor of skeletal muscle atrophy. J Biol Chem. 2014;289:14913–14924. doi: 10.1074/jbc.M114.556241. Recent study showing how mRNA expression signatures of muscle atrophy were used to discover tomatidine as a small molecule that reduces muscle atrophy and improves muscle function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman M. Anticarcinogenic, cardioprotective, and other health benefits of tomato compounds lycopene, alpha-tomatine, and tomatidine in pure form and in fresh and processed tomatoes. J Agric Food Chem. 2013;61:9534–9550. doi: 10.1021/jf402654e. [DOI] [PubMed] [Google Scholar]
- 30.Koh E, Kaffka S, Mitchell AE. A long-term comparison of the influence of organic and conventional crop management practices on the content of the glycoalkaloid alpha-tomatine in tomatoes. J Sci Food Agric. 2013;93:1537–1542. doi: 10.1002/jsfa.5951. [DOI] [PubMed] [Google Scholar]
- 31.Fujiwara Y, Kiyota N, Tsurushima K, et al. Tomatidine, a tomato sapogenol, ameliorates hyperlipidemia and atherosclerosis in apoE-deficient mice by inhibiting acyl-CoA:cholesterol acyl-transferase (ACAT) J Agric Food Chem. 2012;60:2472–2479. doi: 10.1021/jf204197r. [DOI] [PubMed] [Google Scholar]